Abstract
We have defined the optimal binding sites for Stat5a and Stat5b homodimers and found that they share similar core TTC(T/C)N(G/A)GAA interferon gamma-activated sequence (GAS) motifs. Stat5a tetramers can bind to tandemly linked GAS motifs, but the binding site selection revealed that tetrameric binding also can be seen with a wide range of nonconsensus motifs, which in many cases did not allow Stat5a binding as a dimer. This indicates a greater degree of flexibility in the DNA sequences that allow binding of Stat5a tetramers than dimers. Indeed, in an oligonucleotide that could bind both dimers and tetramers, it was possible to design mutants that affected dimer binding without affecting tetramer binding. A spacing of 6 bp between the GAS sites was most frequently selected, demonstrating that this distance is favorable for Stat5a tetramer binding. These data provide insights into tetramer formation by Stat5a and indicate that the repertoire of potential binding sites for this transcription factor is broader than expected.
Signal transducers and activators of transcription (STAT proteins) are latent transcription factors located in the cytosol of resting cells (8). Following stimulation with cytokines or growth factors, these proteins are rapidly tyrosine phosphorylated, allowing them to dimerize and translocate to the nucleus, where they bind to target genes. To date, seven mammalian STAT proteins have been identified. While Stat2 is activated by alpha/beta interferon, Stat4 by interleukin-12 (IL-12), and Stat6 by IL-4–IL-13, Stat1, Stat3, Stat5a, and Stat5b are activated by a wider range of stimuli (8, 13, 17).
Human Stat5a and Stat5b proteins are 91% identical at the amino acid level and are encoded by two closely linked genes located on chromosome 17q11.2 (12, 19, 31). Stat5 proteins can be activated by multiple cytokines and growth factors including prolactin, growth hormone, IL-2, IL-7, IL-9, IL-15, IL-3, granulocyte-macrophage colony-stimulating factor, IL-5, erythropoietin, and thrombopoietin (reviewed in reference 17). Stat5a−/−, Stat5b−/−, and Stat5a−/− Stat5b−/− mice have been generated. Whereas Stat5a−/− mice have defective mammary gland development and lactogenesis due to defective prolactin signaling (21), Stat5b−/− mice exhibit a loss of sexually dimorphic growth due to defective growth hormone signaling (34). The analysis of Stat5a−/− Stat5b−/− mice has shown more severe defects associated with prolactin and growth hormone (32), as well as defects in fetal erythropoiesis (31a). In the immune system, both Stat5a−/− and Stat5b−/− mice exhibit decreased IL-2-induced IL-2Rα expression (14, 27); however, high doses of IL-2 can overcome defective IL-2-induced proliferation in Stat5a−/− mice but not in Stat5b−/− mice. Moreover, Stat5b−/− mice have a greater defect in their natural killer (NK) cell function (14). Finally, mice lacking expression of both Stat5a and Stat5b have a greater immunological defect than seen in either alone, both in terms of T-cell proliferation and in the development of NK cells (25). Thus, Stat5a and Stat5b appear to have both overlapping and distinctive functions.
Most STATs bind to GAS (IFN-γ-activated sequence) motifs, with a consensus TTCNmGAA, with m = 4 for Stat6 and m = 3 for the optimal binding of other STATs (11, 30, 39). However, differences in the fine DNA binding specificities for the various STAT proteins were shown to depend on the sequence of nucleotides in the GAS element as well as of those immediately adjacent to it (reviewed in reference 9). An added complexity related to STAT binding properties can occur based on the ability of STATs to bind to DNA as higher-order complexes occurring through N-terminal interactions between adjacent dimers (15, 23, 36, 37, 39). Tetramer formation can stabilize the binding of STAT dimers to two tandem low-affinity sites by decreasing the off-rate of the complex (15, 36, 37). Moreover, such oligomerization of STAT proteins has been suggested to contribute to their DNA binding specificity (39).
Since different STATs activate nonidentical sets of genes, one mechanism contributing to this selectivity is their differential DNA binding specificities. Therefore, we determined the optimal binding sites for Stat5a and Stat5b to evaluate if any binding differences were found for these highly homologous proteins. In so doing, we have established the consensus sequences for optimal binding of Stat5a and Stat5b homodimers and found that they are very similar. We have also identified sequences that efficiently bound Stat5a tetramers, providing new insights into the DNA sequence requirements for Stat5a dimer versus tetramer formation.
MATERIALS AND METHODS
Expression and purification of recombinant Stat5 proteins.
The production and purification of Stat5a, Stat5b, and Stat5aW37A in baculovirus-infected insect cells were previously described (15). The Stat5aY694F mutant was generated from the Stat5a-encoding transfer vector by using the MORPH Site-Specific Plasmid DNA Mutagenesis Kit (5′-3′, Inc.). Immunoblotting was performed with a pan-Stat5 antibody (Transduction Labs), Stat5a- or Stat5b-specific antisera (19), or the PY20 antiphosphotyrosine monoclonal antibody mAb (Santa Cruz Biotechnology). The purity of protein preparations was evaluated by silver staining (SilverXpress; Novex).
DNA binding site selection.
Optimal Stat5a and Stat5b binding sites were selected according to the method of Pollock and Treisman (29) except that the pool of double-stranded oligonucleotides (R76) had the following sequence: 5′-CAGGTCAGTATAGCGGATCCTGTCGN26GAGGCCACTCGAGTGCA ACTGCAGC-3′ (top strand; BamHI and XhoI sites are underlined; N26 corresponds to nucleotides where all four bases were randomized). Approximately 300 ng of purified recombinant Stat5a, Stat5b, and Stat5aY694F were used for binding reactions. Antisera specific for human Stat5a or Stat5b (19) were used to immunoprecipitate DNA-protein complexes. A binding site selection was previously performed for Stat6 (30).
Electrophoretic mobility shift assays (EMSAs).
In some cases, probes were prepared by PCR labeling of oligonucleotides containing flanking sequences (using forward primer 5′-TAGTGGATCCTGTGG-3′ and reverse primer 5′-CCCTCGAGTGGCCTC-3′); certain mutant oligonucleotides (e.g., 947M4) contained changes in the 3′ flank, and in these cases the reverse primer was 5′-CCCTCGAGTGGCCGA-3′. Each flanking primer used for PCR amplification inadvertently contained a single nucleotide change from the R76 sequence. Two picograms of each relevant oligonucleotide were amplified in PCR buffer containing 10 mM dGTP, dATP, and dTTP, 80 μM dCTP, 80 ng of each primer, and Taq polymerase and labeled with 0.5 μl of [α-32P]dCTP, in a 20-μl total reaction volume for five cycles of 45 s at 94°C, 45 s at 40°C with the RAMP set to 5.00, and 45 s at 72°C. This was followed by 25 cycles of 30 s at 94°C, 30 s at 40°C, and 30 s at 72°C. PCR products were run on 8% polyacrylamide gels, and the bands were excised and purified. In other cases, oligonucleotides corresponding to both strands were synthesized with XmaI and BglII ends, annealed, end labeled with [α-32P]dCTP by using Klenow fill-in (New England Biolabs), and gel purified. Purified Stat5 proteins (150 ng unless otherwise stated) were incubated on ice with 1.34 μg of poly(dI-dC) in binding buffer (20 mM Tris-HCl, pH 7.5; 20 mM HEPES, pH 7.9; 400 mM KCl; 1 mM EDTA; 1 mM dithiothreitol, 1 mM AEBSF; 1 mM Na3VO4; 10 mM NaF; and 1 μg of bovine serum albumin per ml) for 15 min before addition of 40,000 cpm (corresponding to 0.2 to 0.6 ng) of probe. Reaction mixtures (20 μl) were incubated on ice for another 15 min and then resolved on 6% nondenaturing polyacrylamide gels (59:1 acrylamide-bisacrylamide).
Methylation interference assays.
Oligonucleotides were cloned in pGL3 basic (Promega Corp.) between the XmaI and BglII sites. Plasmids were linearized at either end with XmaI or BglII and end labeled with [α-32P]dGTP and [α-32P]dCTP (both >3,000 Ci/mmol; Amersham) by a fill-in reaction with Klenow enzyme. Probes were released by a second digestion with XmaI or BglII, as appropriate, and purified on nondenaturing polyacrylamide gels. Then, 2 × 106 cpm of probe and 0.6 to 3 μg of Stat5a protein or an equivalent amount of Stat5aW37A were used in each binding reaction. Methylation interference assays were performed as described earlier (2).
Molecular modeling of Stat5.
The best alignment between the amino acid sequences of human Stat5a, Stat5b, and the other five known STAT proteins was obtained by using the Needleman-Wunsch algorithm (28) implemented in Look version 3 (Molecular Application Group, Palo Alto, Calif.). Importantly, Stat1 and Stat5 showed marked amino acid sequence conservation, with 37% identity in a sequence alignment and the preservation of a number of residues throughout all of the STAT protein sequences examined. We therefore built molecular homology models of Stat5a and Stat5b based on the high-resolution (2.9 Å) structure of Stat1/DNA complex (7; see also http://www.rockefeller.edu/kuriyan/). An alignment analysis revealed several regions that markedly differ between Stat5a/Stat5b and Stat1, such as the segment between β1 and β2 and the β4/α5/β5 region (see Fig. 1B). However, homology modeling of both Stat5a and Stat5b could be performed with Segment Match Modeling (SegMod; also implemented in Look version 3), which uses both the backbone and side chains of fragments to model a complete system in one step. The average of 10 independent models was minimized by using the molecular mechanics program ENCAD (18) until convergence was reached, as judged by root mean square of the energy gradient (average derivative of <0.1 kcal/mol/Å). The quality of the model was assessed with PROCHECK (16) by using comparison values typical for a 2.0-Å resolution X-ray structure. After the homology modeling, DNA-Stat5a and DNA-Stat5b complexes were assembled. The DNA atomic coordinates corresponding to the M67 GAS element (5′-TTCCCGTAA-3′), which was cocrystallized with Stat1, were substituted by the ones for the FcγRI GAS element (5′-TTCCCAGAA-3′), since the latter motif binds Stat5 well, whereas the former does not (data not shown and reference 3). Double-helical B-form DNA corresponding to the M67 and FcγRI sequences were built by using the Biopolymer Builder implemented in Sybyl 6.4 (TRIPOS Associates, St. Louis, Mo.).
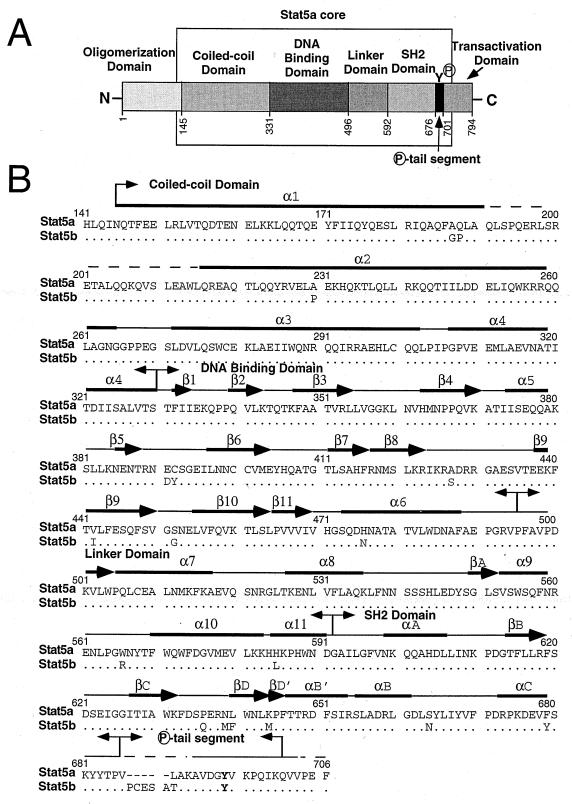
FIG. 1.
Domain structure and sequence alignment of Stat5 proteins. (A) Schematic representation of Stat5 proteins: the domain boundaries of human Stat5a are shown. The Stat5a core region (residues 141 to 706) used to construct the computer model shown in Fig. 6 is indicated by the open box. (B) Sequence alignment of the core regions of human Stat5a and Stat5b. Dots indicate identical nucleotides; hyphens are gaps introduced to optimize alignment. Amino acids in the DNA binding domain which differ between Stat5a and Stat5b are indicated. The loop which intercalates into the major groove of DNA is in the “DNA binding domain” box. Tyr694 of Stat5a and Tyr699 of Stat5b are in boldface. Arrows indicate β strands, and solid bars indicate α-helices, as predicted from the molecular modeling of Stat5. Panels A and B were adapted from Fig. 1A and B in reference 7, with the permission of Cell and J. Kuriyan.
For building models of two Stat5a dimers bound to DNA, we used B-form double-helical DNAs corresponding to oligonucleotide 928 (5′-GTTTCGTGGAATCGTGGCACTATGAACCA-3′, which has a spacing of six positions between the GAS motifs [underlined]) or to a modified version of this oligonucleotide in which the spacing was increased to 11, 5′-GTTTCGTGGAATCGTagctaGGCACTATGAACCA-3′ (lowercase nucleotides are those inserted in the 928 sequence). The protein-DNA complex was modeled by using manual docking procedures (Dock module in Sybyl) and minimized until convergence was achieved, as judged by the root mean square of the energy gradient (average derivative of <0.1 kcal/mol/Å) by using the AMBER force field (38), implemented in Macromodel 6.0 (24).
RESULTS
Stat5a and Stat5b have similar optimal DNA binding preferences.
Human Stat5a and Stat5b differ by six amino acids in their DNA-binding domains (shown in Fig. 1B), which suggested that they might differ in their DNA binding specificities. We therefore used a DNA binding site selection method (29) to investigate this possibility. Human Stat5a and Stat5b proteins were expressed by using a baculovirus expression system and purified from insect cells; the purity of these proteins was evaluated by silver staining (Fig. 2A; for lanes 1 and 2, two concentrations of protein were loaded). Western blotting with pan-Stat5-, Stat5a-, and Stat5b-specific antisera confirmed the identities of the proteins (Fig. 2B to D, lanes 1 and 2). These proteins were constitutively phosphorylated on tyrosine, as shown by Western blotting with PY20 (Fig. 2E, lanes 1 and 2). Stat5a mutants that cannot form tetramers (W37A) or that are not tyrosine phosphorylated and therefore cannot dimerize or bind DNA (Y694F) were also prepared; these proteins were expressed at a lower purity and concentration (Fig. 2A, lanes 3 and 4 versus lane 1), so the amount of these preparations added in experiments was increased to adjust for the lower purity of Stat5aW37A and Stat5aY694F proteins. As expected, Stat5aY694F was nonreactive on PY20 blots (Fig. 2E, lane 3), indicating that this tyrosine residue was phosphorylated in the wild-type Stat5a protein.
FIG. 2.
Characterization of purified Stat5 proteins by Western blotting. Baculoviruses encoding Stat5a, Stat5b, Stat5aY694F, or Stat5aW37A were used to infect insect cells. Recombinant proteins were purified, analyzed on sodium dodecyl sulfate-polyacrylamide gels, and either silver stained (A) or immunoblotted by using antibodies to Stat5a and Stat5b (Stat5; B), anti-Stat5a (C), anti-Stat5b (D), or phosphotyrosine (PY20; E).
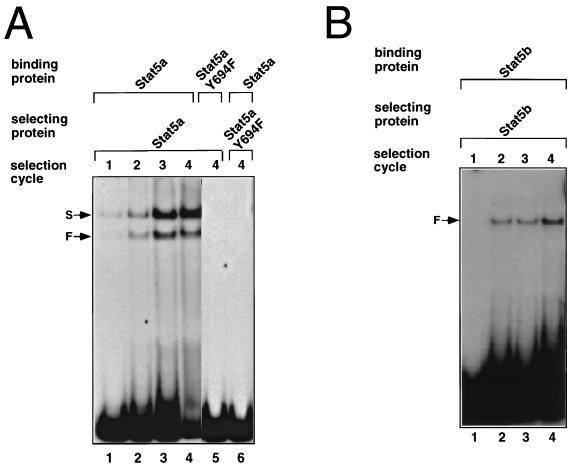
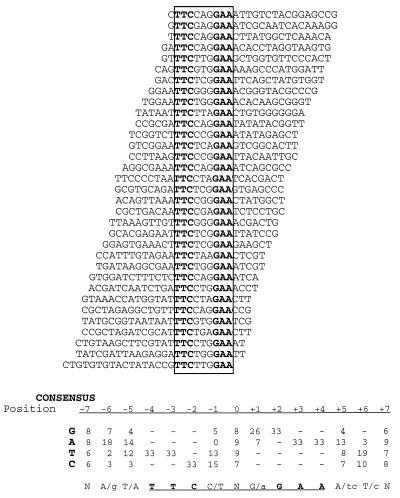
Optimal Stat5a and Stat5b binding sites were selected from a pool of double-stranded oligonucleotides, as described in Materials and Methods. After four cycles of selection, the selected oligonucleotide pools were PCR amplified and used as probes in binding reactions with the selecting protein (Fig. 3). Stat5a formed faster- and slower-migrating complexes with the oligonucleotide pools selected in each cycle (Fig. 3A, lanes 1 to 4). As expected, neither complex formed with Stat5aY694F (lane 5), which cannot form dimers or bind DNA and, correspondingly, no Stat5a DNA binding activity was detected when the binding site selection was performed with Stat5aY694F protein (lane 6). The faster-migrating complex comigrated with Stat5a dimers, while the slower one comigrated with tetramers (see below). After the fourth round of selection, the faster complex (lane 4) was excised, PCR amplified, cloned, and sequenced. By aligning the sequences of 33 independent clones (Fig. 4), we established a consensus motif for the binding of Stat5a homodimers: (A/g)(T/A)TTC(C/T)N(G/a)GAA(A/tc)(T/c) (the GAS element is underlined; Fig. 4 shows each of the selected sequences and the derived consensus motif for Stat5a binding). All of the sequences displayed in Fig. 4 are in the same orientation with respect to the flanking sequences. The position of the selected GAS motif within the 26 random nucleotides in R76 therefore appeared to be random, suggesting that the constant flanking sequences did not influence the sites that were selected.
FIG. 3.
Binding site selection for Stat5a. (A) DNA selected at sequential cycles by Stat5a were used as probes in EMSAs with Stat5a (lanes 1 to 4). The faster complex (F) comigrated with Stat5a dimers, and the slower complex (S) migrated with Stat5a tetramers. Stat5aY694F did not bind to DNA selected by the wild-type protein (lane 5) and did not select for Stat5a binding sites (lane 6). (B) Same as for panel A, except that Stat5b was used.
FIG. 4.
Alignment of 33 Stat5a dimer-selected DNAs. Sequences and nucleotide frequency in 33 binding sites selected by Stat5a homodimers after four cycles of selection (lane 4 of Fig. 3A). Nine of these DNAs were tested with Stat5a in EMSAs, and each bound Stat5a dimers. Not included in the figure are 10 sequences that lacked canonical GAS motifs, since the five of these that were tested in EMSAs were all proven to be falsely selected sequences that could not bind dimeric Stat5a. All sequences are shown in the same orientation relative to the flanking sequences (top strand). The conserved consensus GAS motif is boxed; its central nucleotide was assigned position zero. The consensus Stat5a binding motif was derived from the nucleotide frequencies in the binding sites in the figure. At each position, the more-favored nucleotides are shown in upper case letters, while less-favored nucleotides are in lower case.
Interestingly, Stat5b formed only one complex with the selected oligonucleotides (Fig. 3B, lanes 2 to 4) that comigrated with the faster one formed by Stat5a (dimer). Alignment of 45 independent oligonucleotide sequences selected by Stat5b (Fig. 5) in the fourth cycle of selection (Fig. 3B, lane 4) yielded a consensus for Stat5b dimers of: (A/tg)(T/A)TTC(C/T)(T/cag)(G/a)GAA(T/A)(T/ca) (the GAS element is underlined; Fig. 5 shows each of the selected sequences and the derived consensus motif for Stat5b binding). Thus, both Stat5a and Stat5b homodimers selected similar consensus GAS motifs. Consistent with the recognition by Stat5a and Stat5b of similar GAS motifs, each protein could bind to the oligonucleotide pools selected by the other (data not shown). Interestingly, formation of the faster mobility complex by Stat5b was favored, while the opposite was true for Stat5a. EMSAs performed with GAS elements from different genes (whey acidic protein, β-casein, oncostatin M, serine protease inhibitor 2.1, IL-2 response element from murine and human IL-2Rα genes, FcγRI, M67 SIE, and cyclin D2) did not reveal any selectivity for binding of Stat5a versus Stat5b (data not shown). Taken together, our results indicate that homodimers of Stat5a and Stat5b have similar DNA binding specificities.
FIG. 5.
Alignment of 45 Stat5b dimer-selected DNAs. Sequences and nucleotide frequency in 45 binding sites selected by Stat5b homodimers after four cycles of selection (lane 4 of Fig. 3B). Ten of these DNAs were tested in EMSAs and shown to bind Stat5b dimers. Not included are 27 sequences that lacked consensus GAS motifs, since the 12 of these that were tested in EMSAs were found to be falsely selected sequences that could not bind dimeric Stat5b. All sequences are shown in the same orientation relative to the flanking sequences (top strand). The conserved consensus Stat5b binding motif was derived from the nucleotide frequencies in the binding sites in the figure. At each position, the more-favored nucleotides are shown in upper case letters, while less-favored nucleotides are in lower case.
The amino acids that differ in Stat5a and Stat5b are predicted not to contact DNA.
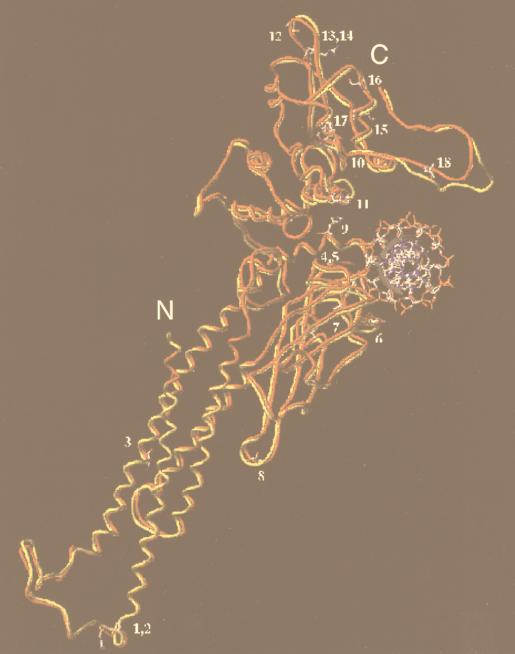
To investigate the positions of the amino acid differences between Stat5a and Stat5b, we performed molecular modeling studies based on the coordinates of the crystal structure of the Stat1 core bound to DNA (7). As expected based on the structures for Stat1 (7) and Stat3β (4) bound to DNA, Stat5 proteins are predicted to bind DNA as dimers in which the two monomers contact each other only through the phosphotyrosine-SH2 domain interaction and are related by a twofold axis of symmetry which passes through the center of the DNA. Analogous to the situation for Stat1, the major site of protein-DNA interaction appears to be the loop formed by amino acids 470 to 474, which intercalates in the major groove of DNA. Interestingly, the amino acid residues in this loop are identical between Stat5a and Stat5b. Moreover, superimposition of the core models for Stat5a and Stat5b (Fig. 6) predicted that all 18 amino acid differences (see Fig. 1B) do not contact the DNA. This is consistent with our experimental data indicating that the DNA binding specificities of Stat5a and Stat5b are very similar.
FIG. 6.
The 18 amino acid differences between the Stat5a and Stat5b cores appear to be remote from DNA binding surface. Superimposed structures of human Stat5a (red) and Stat5b (yellow) core monomers were obtained by homology modeling. The 18 amino acids which differ between Stat5a and Stat5b (see Fig. 1B) are indicated as follows: 1, A187G; 2, Q188P; 3, A230P; 4, E391D; 5, C392Y; 6, A427S; 7, V442I; 8, S452G; 9, H476N; 10, W566R; 11, H585L; 12, P636Q; 13, N639M; 14, L640F; 15, K644M; 16, S664N; 17, F679Y; and 18, L687T. The first letter and number represent the residue in Stat5a, while the second letter represents the corresponding residue in Stat5b. Five residues, PCESA, are present in Stat5b (amino acids 687 to 691) but not Stat5a.
Stat5a forms tetramers with oligonucleotides selected in the slower-migrating complex.
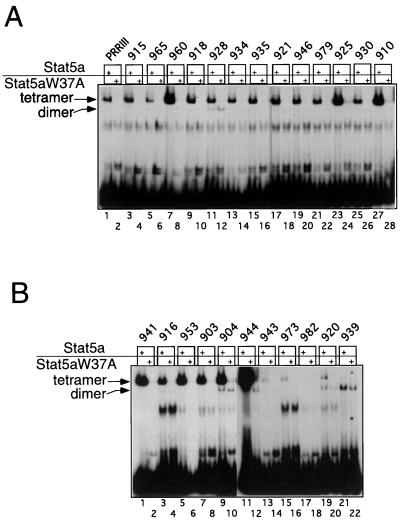
Since Stat5 proteins can cooperatively bind to tandem GAS motifs as tetramers (15, 23), we speculated that the slower-migrating complex formed by Stat5a with the selected oligonucleotides (Fig. 3A) was a tetramer. We therefore cloned the DNAs that were bound to Stat5a in the slower-migrating complex after four cycles of selection (Fig. 3A, lane 4), sequenced the inserts, and tested 59 independent clones for their abilities to bind Stat5a in EMSAs. The sequences of the 50 oligonucleotides that bound Stat5a tetramers are shown in Fig. 7; the other nine selected DNAs either did not bind Stat5a (e.g., oligonucleotide 982; Fig. 8B, lane 17), or bound only as dimers (e.g., oligonucleotide 939, lane 21) and therefore were not included in Fig. 7.
FIG. 7.
Alignment of 50 Stat5a tetramer-selected DNAs. Sequences of 50 binding sites selected by Stat5a tetramers after four cycles of selection (lane 4 of Fig. 3A). All of the sequences shown bound Stat5a tetramers in EMSAs. Not shown are nine sequences that were selected but which were unable to bind Stat5 tetramers. Shown are sequences containing a consensus GAS motif (top 18 sequences) or a nonconsensus GAS motif (middle 25 sequences). As noted in the text, the bottom seven sequences bound Stat5a tetramers relatively poorly. The nucleotides in italics correspond to sequences from the nonrandom flanking sequences of R76; these are included as the GAG in the 3′ flank was shown in Fig. 11 to be important for tetrameric Stat5a binding to oligonucleotide 947. Note that this GAG was often 3 bp downstream of a TTC, thus forming nonconsensus GAS motifs. On the right are sequence identifiers. Those containing sequences with GASc motifs are in boldface. An “r” before a sequence identifier indicates that the bottom rather than top strand is shown to align the GASc or GASn shown in the open box 5′ to the more-divergent sequence shown in the shaded boxes. Underlined numbers refer to sequences that were evaluated by EMSA in Fig. 8. Superscripted “○” and “∗” symbols refer to sequences with spacings of 5 and 7 bp between the boxed regions, respectively. The ability of the tetramer-selected sequences to bind Stat5a as a dimer is indicated.
FIG. 8.
Representative EMSAs from oligonucleotides selected in the binding site selection with Stat5a corresponding to the slower-mobility complex. The panels show binding with Stat5a (150 ng) or an equivalent amount of mutant Stat5aW37A protein. The sequences for the oligonucleotides used are shown in Fig. 7 except for 982 and 939 (5′-TCTTCGTGGAAGCAGCGTGGCAGGTA-3′ and 5′-TTCCTGGAAATGGATATTAGTACCCC-3′, respectively). In each oligonucleotide, the consensus GAS motif is underlined with the TTC and GAA shown in boldface; a 9-bp segment downstream which could represent a second divergent GAS motif is also underlined. In each case, the similarity of these putative GAS motifs is remote, as indicated by the nucleotides in boldface. Accordingly, neither of these sequences bound Stat5a tetramers (panel B, lanes 17 and 21). The sequence of the PRRIII oligonucleotide is 5′-TCTTCTAGGAAGTACCAAACATTTCTGATAATA-3′. All of the oligonucleotides also included 15 constant nucleotides both 5′ and 3′ to facilitate labeling by PCR (see Materials and Methods).
Remarkably, only 18 of the 50 tetrameric binding sites contained a consensus TTCN3GAA motif (GASc), while all of the others contained a GAS motif with a single nucleotide change from the TTCN3GAA consensus (GASn). In addition to the GASc or GASn motif present in all 50 binding sites (Fig. 7, open boxes, on the left), the most striking feature, found in 33 of the sequences, was the presence of either a TTC or GAA half-GAS motif at a distance of ≥5 bp (e.g., oligonucleotides 978, 972, and 915) away from the GAS motif. This spacing is presumably the minimal distance for stable tetramer formation (reference 36 and see also Discussion). Notably, the majority of the TTC half-sites were located at a 6-bp distance downstream from the GASc or GASn (e.g., 972, 924, r926, and r957), while most of the GAA half-sites were located 12 bp apart from the GASc or GASn (e.g., oligonucleotides r944, 915, r916, and 941). Such a spacing is consistent with the GAA being 6 bp downstream of the TTC in consensus TTCN3GAA GAS motifs. These observations led us to speculate that a second, more-divergent GAS motif spaced 6 bp from a more-conserved one (GASc or GASn) was responsible for the binding of one of the two dimers of Stat5a in the tetrameric complex. These putative divergent GAS motifs are shown in shaded boxes in Fig. 7. For two of the binding sites (oligonucleotides 936 and 978) such an element was located at an inter-GAS spacing of 5 bp, while for seven sites (oligonucleotides 934, 918, 921, 960, 909, 905, and 920) a spacing of 7 bp was seen. Interestingly, there was greater divergence in such accessory sites when the sequence contained a GASc rather than a GASn motif (compare the nucleotides in the shaded boxes for the upper 18 sequences to the middle 25 sequences). Many of the binding sites which did not contain a half-GAS motif had four of six of the key nucleotides in GAS motifs (e.g., oligonucleotides 930, r935, and 923; the conserved nucleotides are in boldface in the shaded boxes). However, certain sequences were more divergent and correspondingly these were typically weak tetramer binding sites (see below). Overall, the nucleotide preferences of the Stat5a tetramer were much less stringent than those found for the Stat5a dimer (Fig. 4), particularly related to the TTC and GAA sequence of the GAS elements (positions ±2, ±3, and ±4).
Representative EMSAs were performed with selected DNAs and wild-type Stat5a or the tetramerization-defective W37A mutant of Stat5a (15) (Fig. 8A and 5B). As expected, the slower-migrating complexes were formed only with wild-type Stat5a, whereas the faster complexes were detected with both wild-type and W37A mutant Stat5a proteins, indicating that the slower complexes required N-terminal interactions for their formation. Consistent with this conclusion, the slower complexes formed by the selected oligonucleotides comigrated with the complex formed by PRRIII (lane 1), which contains two tandemly linked GAS motifs and is known to bind Stat5a tetramers (15). Most of the selected sites (top 43 sequences in Fig. 7) bound Stat5a with an apparent affinity similar to that seen for PRRIII (e.g., oligonucleotides 915, 965, and 918; Fig. 8A, lanes 3, 5, and 9 versus lane 1) or higher than that seen with PRRIII (e.g., oligonucleotides 960, 925, and 910; Fig. 8A, lanes 7, 23, and 27 versus lane 1). A few of the selected sites (the bottom seven sequences in Fig. 7) bound Stat5a tetramers weakly, even at a concentration of 150 ng (e.g., oligonucleotides 973 and 920; Fig. 8B, lanes 15 and 19), and no binding was detected with these oligonucleotides when only 20 ng of Stat5a was used. In contrast, all 9 of the 43 higher-affinity oligonucleotides tested (903, 916, 918, 928, 934, 941, 943, 944, and 953) could bind Stat5a tetramers at this concentration of Stat5a.
Nucleotide requirements for Stat5a tetramer binding.
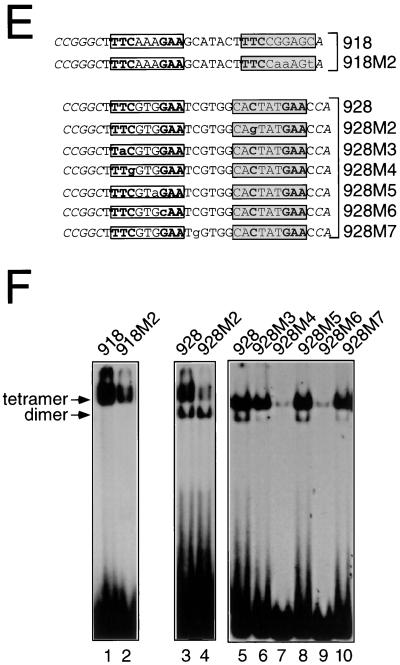
We next sought to determine the nucleotide requirements for formation of tetramers. To directly test the role of the half-GAS sites in the binding of Stat5a tetramers, we mutated them in the context of oligonucleotides 918, 928, and 934 and evaluated these mutants (Fig. 9A, M1 mutants) for binding to Stat5a in EMSAs (Fig. 9B). The half-GAS sites were necessary for the binding of Stat5a tetramers to each of these oligonucleotides (lanes 5 versus 3, 9 versus 7, and 13 versus 11). Nevertheless, mutation of the half-GAS site in oligonucleotide 928 did not affect its ability to bind Stat5a dimers, indicating that its consensus GAS was sufficient to allow the formation of dimers (lanes 9 and 10) but not of tetramers. Correspondingly, Stat5aW37A dimers bound to either the wild-type or the mutant 928 probes (lanes 8 and 10). This was not the case for oligonucleotides 918 and 934, which could bind Stat5a only as tetramers (lanes 6 and 14). The fact that tetramer formation was favored for oligonucleotides 918, 928, and 934 (Fig. 9B, lanes 3, 7, and 11) indicated cooperative binding.
FIG. 9.
Obligatory role of half-GAS sites present in binding sites selected in the slower-migrating complex that corresponds to Stat5a tetramers. (A) Schematic representation of the probes used in the EMSAs shown in panel B. The 26 nucleotides selected by the Stat5a slower complex in the clones analyzed (918, 928, and 934) are in plain text, while the artificially introduced flanking nucleotides are in italics. The nucleotide changes introduced in their mutated versions (918M1, 928M1, and 934M1) are in lowercase letters. The consensus GAS elements are in the open box, while the nonconsensus GAS elements are in the shadowed boxes. Consensus half-sites are in boldface. (B) EMSAs were performed with Stat5a (150 ng) or with an equivalent amount of Stat5aW37A, and the probes shown in panel A, which were labeled by Klenow fill-in.
To further elucidate the sequence requirements for tetramer formation, we performed methylation interference assays on oligonucleotides 918 and 928 (Fig. 10). These studies revealed the importance of the Gs in the “GAA” of the GASc in oligonucleotide 918 (open boxes, positions ± 2; Fig. 10A and D) for binding of Stat5a tetramers. Moreover, strong binding interference was also observed for the G in the half-GAS site (bottom strand, position −2), as well as for two other Gs in the top strand (positions 0 and +1) and another G in the bottom strand (position +4). Thus, the half-GAS site and these other contacts together may functionally form a “full” nonconsensus binding site (shadowed box).
FIG. 10.
Methylation interference analysis of dimeric or tetrameric Stat5a. (A) Oligonucleotide 918 was labeled either on the sense (TOP) or antisense (bottom [BOT]) strand, methylated with dimethyl sulfate, and incubated with Stat5a. Shown are piperidine-mediated cleavages of free probe (F) or of probe bound (B) to a Stat5a tetramer. (B and C), Comparison of the nucleotide contacts between tetrameric or dimeric Stat5a and oligonucleotide 928. The same analysis as in panel A was performed with oligonucleotide 928, either free (F) or bound (B) to a Stat5a tetramer (B) or to a Stat5aW37A dimer (C). Nucleotides which interfered with binding of tetramers but not of dimers are indicated with an arrow. Filled circles indicate strong interference, while asterisks indicate hypermethylation. The artificially introduced flanking nucleotides are in italics. The consensus GAS element is in an open box, while the nonconsensus GAS element is in a shadowed box. (D) Summary of methylation interference analyses shown in panels A, B, and C. (E) Wild-type and mutant forms of oligonucleotides 918 and 928. (F) The oligonucleotides in panel E were labeled by Klenow fill-in and used in EMSAs with 150 ng of Stat5a.
To compare the nucleotides critical for binding Stat5a tetramers versus dimers, we used oligonucleotide 928, which bound Stat5a mainly as a tetramer and Stat5aW37A only as a dimer (Fig. 9B, lanes 7 and 8). The tetrameric (Fig. 10B) and dimeric (Fig. 10C) complexes contacted not only the Gs in the GAA of the consensus GAS (open boxes; positions ± 2 in Fig. 10D) but also the neighboring G (position +1 in the consensus GAS), a finding in agreement with the preference for this nucleotide observed in the binding site selection analysis (Fig. 4). However, only the tetramer contacted the G at position +2 on the top strand of the adjacent half-GAS site and the G at position −2 (relative to this half-GAS site) on the bottom strand. Thus, the half-GAS site and at least one other nucleotide were important for the formation of Stat5a tetramers but not dimers.
We next mutated these nucleotide contacts in the non-consensus GAS motif in oligonucleotides 918 and 928 (see Fig. 10E) and evaluated their Stat5a binding activity in EMSAs (Fig. 10F). Mutation of the two Gs at positions 0 and +1 in the top strand and the one at position +4 in the bottom strand of oligonucleotide 918 (mutant 918M2; Fig. 10E) diminished Stat5a tetramer binding (Fig. 10F, lane 2 versus 1). Analogously, in oligonucleotide 928, mutation of the G at position −2 in the bottom strand of the nonconsensus GAS motif (mutant 928M2) diminished tetramer formation without altering dimer formation (lane 4 versus lane 3). This is consistent with the importance of this G for binding Stat5a tetramers but not dimers (Fig. 10D). Thus, the half-GAS site and the neighboring divergent nucleotides appear to form a nonconsensus GAS motif that is essential for binding Stat5a tetramers. The analysis of additional mutants of oligonucleotide 928 (oligonucleotides 928 M3 through M7) confirmed the essential role of the C in the TTC and the G in the GAA of the consensus GAS motif for both tetramer and dimer binding (mutants M4 and M6, Fig. 10F, lanes 7 and 9 versus lane 5), in accord with the observed strong interference of methylation of these residues (Fig. 10B and D). In contrast, mutation of GTG to GTA in the central part of the consensus GAS motif (M5, lane 8 versus lane 5) had no appreciable effect on the binding of either tetramers or dimers. Importantly, mutant M3 showed that mutation of the TTC to TAC did not affect tetramer binding but substantially diminished dimer binding (lane 6 versus lane 5). Finally, the M7 mutant confirmed the important role, as suggested by the methylation interference analysis, of a nucleotide downstream of the GASc for dimer but not tetramer formation (lane 10 versus lane 5). Thus, we have demonstrated the greater tolerance for deviation from consensus GAS motifs for tetramer binding, a result consistent with an overall larger number of contact points with DNA than exists for dimers, and have identified mutants that can selectively affect either tetramer binding (928M1; Fig. 9B, lane 10 versus lane 9) or dimer binding (928 M3 and M7 Fig. 10F, lanes 6 and 10 versus lane 5).
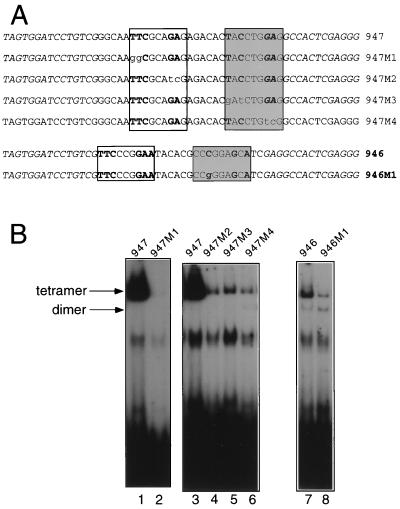
As mentioned above, certain binding sites did not contain a half-GAS TTC or GAA sequence but nevertheless efficiently bound Stat5a tetramers. To clarify the basis of binding to these sequences, we introduced mutations in two representative oligonucleotides (946 and 947, Fig. 11A) to understand which nucleotides were critical for Stat5a tetramer binding. When the putative highly divergent GAS motif (shaded box in Fig. 11A) in oligonucleotide 946 was altered from CCCGGAGCA to CCgGGAGCA (Fig. 11A), binding activity of Stat5a tetramers (but not dimers) was decreased (Fig. 11B, lane 8 versus lane 7). This confirms that a nucleotide located within a region that would not normally be recognized as contributing to Stat5a binding was important for anchoring the tetrameric complex. We also analyzed oligonucleotide 947 in which both GAS motifs are nonconsensus motifs and where the downstream GASn includes the GAG from the constant R76 flanking sequence. Interestingly, each of the four putative half-GAS sites were important for binding of Stat5a tetramers, as shown by the strong reduction in binding shown by mutants 947 M1 to M4 (Fig. 11A; Fig. 11B, lane 2 versus lane 1 and lanes 4 to 6 versus lane 3). This also confirms that the GAG in the constant flanking sequence was often involved in Stat5a tetramer binding when separated by three nucleotides from a TTC or TAC, as occurred in a number of the selected oligonucleotides (Fig. 7; e.g., oligonucleotides 917, 906, and 932). This is in contrast to the random positioning of a single GAS motif within the random 26 nucleotides in the Stat5a dimer-selected sequences (Fig. 4).
FIG. 11.
Critical role of nucleotides which do not form half-GAS sites for binding of Stat5a tetramers. (A) Sequences of wild-type and mutant forms of oligonucleotides 946 and 947. (B) EMSAs performed with the oligonucleotides in panel A were labeled by PCR (see Materials and Methods) and 150 ng of Stat5a.
A spacing of 6 bp between GAS sites is optimal for tetramer formation.
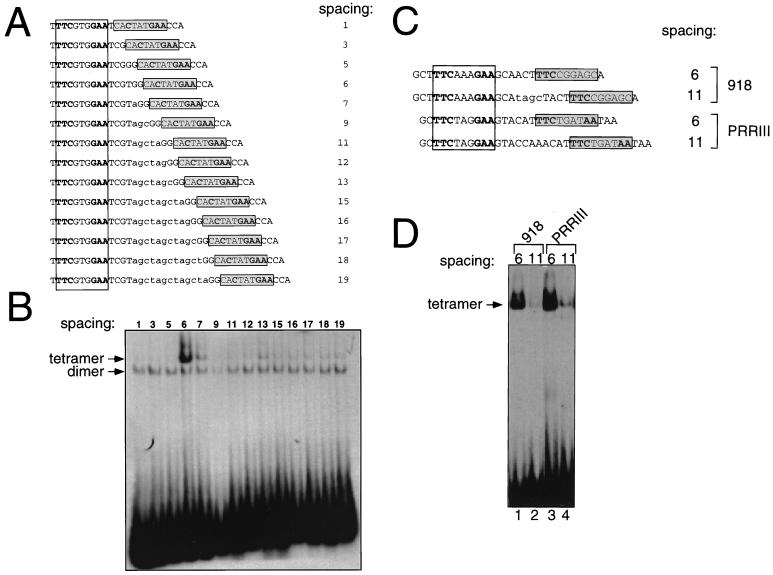
As noted above, many oligonucleotides selected in the slower migrating complex of Stat5a contained a GAS (consensus and/or nonconsensus) motif separated by 6 or 7 bp from an often more divergent motif (Fig. 7). Since the length of the random sequence of the oligonucleotide pool used in the binding site selection was 26 bp, the maximal distance between two 9-bp GAS elements that we could detect is 8 bp (actually, a slightly greater spacing could have been seen because of the use of the GAG in the 3′ flanking sequence as an alternate to a GAA half-site in some sequences such as oligonucleotide 947). To overcome this bias, we synthesized a series of mutants of oligonucleotide 928 in which the number of base pairs between the GAS motifs ranged between 1 and 19 (Fig. 12A) and used them as probes in EMSAs with Stat5a. For a spacing of greater than 6 bp, additional nucleotides were inserted within the six inter-GAS motif nucleotides. Based on methylation interference, there was no evidence that the six inter-GAS residues contributed to tetramer binding (Fig. 10B), although this possibility is not excluded since methylation interference assays only can evaluate the importance of G and A residues. As shown in Fig. 12B and consistent with the binding site selection analysis, distances less than 6 bp did not allow efficient tetramer binding (lanes 1 to 3), while a spacing of 6 bp was optimal (lane 4). Spacing greater than 6 bp showed weaker but visible tetramer formation (lanes 5 to 14). As expected, the variation in the inter-GAS spacing had no effect on dimer binding (lanes 1 to 14). Interestingly, for two oligonucleotides tested (918 and 934), a spacing of 7 bp was as efficient as a spacing of 6 bp (data not shown). We extended this analysis by comparing a spacing of 6 versus 11 bp for oligonucleotide 918 and for the naturally occurring tandem GAS elements in PRRIII (ref. 15; Fig. 12C). In both cases, a spacing of 6 bp was much more efficient in allowing Stat5a tetramer formation than was a spacing of 11 bp (Fig. 12D, lanes 1 and 3 versus lanes 2 and 4). Thus, a spacing of 6 bp (and in some cases also of 7 bp, depending on the nucleotide context) between GAS motifs allowed maximal binding of Stat5a tetramers in vitro. Although the binding site selection identified a few sequences with an inter-GAS spacing of 5 bp, it appears that tetramers are more favored at a spacing of at least 6 bp, possibly due to steric hindrance at a spacing of less than 5 bp. This is consistent with the observation that Stat1 tetramer binding was unstable at a spacing of 5 bp (36). It is possible that optimal binding is seen at a spacing of 6 bp rather than at larger distances due to additional stabilizing interactions that do not occur with a larger spacing (see Discussion). Nevertheless, Stat5 tetramers can form at greater distances. For example, Stat5 tetramers can physiologically interact with an element such as PRRIII (spacing of 11; see reference 15).
FIG. 12.
A spacing of 6 bp between GAS sites is optimal for Stat5a tetramer formation. (A) Schematic representation of the probes used in EMSAs shown in panel B. This series of oligonucleotides was based on oligonucleotide 928 (see Fig. 9A for the artificially introduced flanking nucleotides), in which the consensus (open box) and nonconsensus (shadowed box) GAS elements were 6 bp apart. This spacing was varied either by removing intermediate base pairs (spacing of 1, 3, or 5 bp) or by adding them (lowercase letters, spacing of ≥7 bp). (B) EMSAs were performed with 20 ng of Stat5a and the probes shown in panel A. (C) Schematic representation of the probes used in EMSAs shown in panel D; see Fig. 9A for the artificially introduced flanking nucleotides. The distance between the consensus (open box) and nonconsensus (shadowed box) GAS elements present in oligonucleotide 918 (7 bp) was either reduced to 6 bp (lane 1) or increased to 11 bp (lane 2). The naturally occurring inter-GAS distance of 11 bp in PRRIII (lane 4) was reduced to 6 bp (lane 3). Stat5a was used at 150 ng in EMSAs shown in lanes 1 and 2 and 20 ng for lanes 3 and 4. Probes were labeled by Klenow fill-in.
DISCUSSION
Based on the phenotype of Stat5a−/−, Stat5b−/−, and Stat5a−/− Stat5b−/− mice, Stat5a and Stat5b appear to mediate both overlapping and distinct functions. For this reason and because of the amino acid differences in the Stat5a and Stat5b DNA binding domains, we had anticipated that we might identify differences in the preferred binding sites for homodimers of these two proteins. However, we found similar optimal binding sites for human Stat5a and Stat5b homodimers (Fig. 4 and 5). Thus, our data do not provide a basis for the different roles of these STATs based on differences in DNA binding specificity of Stat5a versus Stat5b homodimers. The similarity in the optimal binding sites for Stat5a and Stat5b is consistent with the prediction that the amino acids that differ in these proteins do not contact DNA, as indicated by our modeling studies of the Stat5a versus Stat5b cores (Fig. 6). However, since the motifs that we have defined represent optimal binding sites, it is possible that lower-affinity sites that are selective for Stat5a versus Stat5b binding might exist. The presence of a glycine at residue 433 in murine Stat5b versus a glutamic acid in Stat5a has been reported to confer distinct DNA binding specificities (6). However, all published human Stat5a and Stat5b proteins have a Glu at this position (12, 19, 31), suggesting that the Gly versus Glu results are not relevant to potential binding differences between human Stat5a and Stat5b. Furthermore, Gly433 in murine Stat5b (20) was Glu433 in the two other murine Stat5b sequences (1, 26). It is possible, however, that Stat5a and Stat5b could exert different actions based on potential differences (i) in their expression levels in different cellular lineages, (ii) in the efficiency of their respective recruitment and activation in response to different stimuli, (iii) in their abilities to interact with other transcription factors and/or coactivators, or (iv) in their abilities to form tetramers. Indeed, the most striking difference between the Stat5a and Stat5b selections was the absence of a tetrameric complex for Stat5b. This may reflect that Stat5b bound DNA less efficiently than Stat5a (Fig. 3B versus 3A) even though both proteins showed comparable levels of tyrosine phosphorylation (Fig. 2E, lane 2 versus 1), or that Stat5b is intrinsically less efficient at forming tetramers, as suggested by Verdier et al. (35). Consistent with this possibility, we reproducibly have observed less Stat5b than Stat5a tetramer DNA binding activity in nuclear extracts prepared by using a 293T cell-based IL-2-induced Stat5 binding reconstitution system (reference 15 and data not shown).
We also established the sequence requirements for the binding of Stat5a tetramers to DNA. We found that there are consensus GAS motifs that do not bind Stat5a except when tandemly linked to a TTC or GAA half-GAS site (Fig. 9). TTC or GAA half-GAS sites appear to be able to function as part of nonconsensus “full” GAS motifs, since other nucleotides are also important to anchor the tetrameric complex to DNA (e.g., oligonucleotides 918 and 928). This is consistent with the degree of divergence of nonconsensus GAS motifs that were selected (Fig. 7). Indeed, mutational analysis of oligonucleotide 928, which can bind Stat5a dimers and tetramers, revealed that it was possible to introduce specific mutations that virtually abolished the binding of Stat5a as dimers without affecting its binding as tetramers (Fig. 10). This diminished nucleotide stringency is presumably allowed by the increased cooperativity of binding fostered by tetramerization. Conversely, we have previously shown that improving the affinity of imperfect GAS motifs can compensate for defective tetramerization of the W37A mutant of Stat5a (15). Thus, the overall stability of the tetrameric complex is due to DNA-protein as well as to protein-protein interactions.
It was striking that none of the selected sequences contained two consensus GAS motifs even though we have confirmed that sequences containing two consensus GAS motifs can efficiently bind Stat5 tetramers (reference 15 and data not shown). We hypothesize that the fact that such sequences were not identified reflects the large range of tandem imperfect GAS-like sequences that can bind, in accord with considerable flexibility in the binding motifs for tetramers, and that such sequences would be identified if a sufficiently large number of tetrameric binding sites were sequenced.
For the binding of Stat5a tetramers, an inter-GAS spacing of less than 5 bp was not favored and a spacing of 6 bp appeared optimal. Presumably, this reflects steric interference that occurs at short distances. Although the random oligonucleotide used in the binding site selection could allow one to observe a larger spacing (up to 8 bp within the random core and up to 11 bp if one allows for the utilization of the GAG in the 3′ flank as an acceptable 3′ end of a GAS motif), spacing above 7 bp was not seen. The only exception to this may be oligonucleotide r944 which has a TTC at a spacing of 15 bp but which also has a TTTN3GAA motif at the typical spacing of 6 (Fig. 7).
To further understand Stat5a binding as a tetramer, we used our model of the Stat5a core dimer bound to DNA (based on the Stat1 structure; see Methods) to build a model which could accommodate the independent binding of two core dimers of Stat5a to oligonucleotide 928 in which the GAS motifs are 6 bp apart. In this model, the two Stat5a dimer cores were positioned at a distance so that they could potentially form contacts. If proven to be correct, this predicted protein-protein interaction could further stabilize the tetrameric complex, providing a molecular basis for the efficient tetramerization observed at a spacing of approximately 6 bp and potentially decreasing the stringency for nucleotide requirements in the DNA. In contrast, at a spacing of 11 bp the distance between the two adjacent Stat5a core dimers was predicted to be large enough so that these additional interactions could not occur, lowering the predicted stability of the complex.
The relevance of the optimal binding of Stat5 to GAS elements 6 or 7 bp apart observed in vitro for Stat5-driven transcription has not been fully elucidated. A single copy of oligonucleotide 918 or 928 linked to a minimal cytomegalovirus promoter could drive transcription of the luciferase reporter gene in a Stat5-dependent manner in response to IL-2 in a 293T cell reconstitution system (data not shown), showing that a Stat5 tetramer bound to these elements is transcriptionally active. We also compared the activity of a 928 reporter oligonucleotide (spacing of 6 bp) to one with a spacing of 11 bp. Although transfection of a Stat5 expression vector resulted in significantly higher activity of the former construct (spacing of 6 bp) than the latter construct (spacing of 11 bp), the basal activity of the former construct was also higher, so that the fold induction at the two different spacings was similar. We hypothesize that the higher basal level at a spacing of 6 bp may result from the presence of low levels of endogenous activated Stat5 in 293T cells; thus, the similar fold induction but higher absolute level of the construct with a spacing of 6 bp might have been predicted. It is interesting that transcription of the serine protease inhibitor 2.1 (Spi2.1) gene has been shown to depend on the cooperative binding of Stat5 to a consensus and a nonconsensus GAS elements 6 bp apart (5; see Fig. 13 for the sequence). Nonconsensus GAS motifs 6 or 7 bp apart from a consensus GAS are present in other Stat5-responsive genes (Fig. 13). Whether Stat5 binds to these sequences as tetramers and whether this is important for the transcriptional regulation of these genes awaits direct testing. Interestingly, formation of a tetrameric Stat5 complex is essential for the IL-2-inducible activation of PRRIII (15), which has a natural spacing of 11 bp between its GAS motifs. In PRRIII, the sequences flanking the GAS motifs bind important accessory factors that are critical for maximal transcriptional activation. This is in accord with transcriptional activation of eukaryotic genes involving the assembly of multiprotein complexes which, at least in some cases, requires a highly specific three-dimensional architecture (33). In this regard, in the context of the murine IL-2Rα gene, while increasing the naturally occurring inter-GAS spacing from 11 to 16 bp strongly reduced the IL-2-driven transcription, increasing it to 21 bp fully maintained its transcriptional activity (23). These results might be explained by the very different three-dimensional structures formed by Stat5 tetramers that are bound to GAS elements spaced so that the two dimers are on opposite sides of the DNA (inter-GAS spacing of 6 bp; the same relative orientation as at a 16-bp spacing) versus the structure in which Stat5 dimers are bound on the same side of the DNA (inter-GAS spacing of 11 bp; same as at a 21-bp spacing).
FIG. 13.
Putative tetramer binding sites in Stat5 responsive gene. An “r” before the sequence name indicates that the bottom rather than top strand is shown to align the GASc shown in the open box 5′ to the more divergent sequence shown in the shaded boxes, analogous to the sequences in Fig. 7. The numbers in parentheses are the references from which the sequences were derived; for porcine β-casein, a GenBank accession number is shown.
Interestingly, the consensus GAS motifs selected by Stat5a and Stat5b dimers are relatively similar to those reported for Stat1, Stat3, and Stat4 (11, 39), although in each of these cases the core GAS motif was TTCCSGGAA, while it was TTCYNRGAA for both Stat5a and Stat5b. Because these analyses were performed in different labs, it is unclear whether these differences correspond to distinct binding specificities or simply reflect differences in the stringency of selection. However, all of these differ from the optimal site selected by Stat6 (TTCNTNGGAA [reference 30 and data not shown]), which has a spacing of four nucleotides rather than three between the two half-GAS motifs (30). Optimal sites for tetrameric binding of Stat1, Stat3, Stat4, and Stat6 have not been reported and therefore a comparison of tetrameric STAT binding is not yet possible. However, presumably differences in affinity and specificity for different STATs may help to determine the cytokines which can activate the transcription of specific target genes.
In conclusion, our data indicate that the repertoire of potential binding sites for Stat5a is broader than expected, a finding likely to be relevant to other STATs. It is possible that different spacing of GAS motifs may also influence the degree of STAT-mediated DNA bending, which could also influence the potency of STAT-induced transcriptional activation of target genes. The existence of naturally occurring suboptimal GAS sites and variable spacing between them may represent strategies that allow greater specificity by requiring cooperative binding of STAT oligomers. In turn, this could also influence the ability of STAT proteins to interact with other components of the transcription machinery.
ACKNOWLEDGMENTS
We thank Enrico Balducci for valuable discussions and support, David Margulies, Jian-Xin Lin, and Anne Puel for critical comments on the manuscript and Maria Berg for technical assistance.
REFERENCES
- 1.Azam M, Erdjument-Bromage H, Kreider B L, Xia M, Quelle F, Basu R, Saris C, Tempst P, Ihle J N, Schindler C. Interleukin-3 signals through multiple isoforms of Stat5. EMBO J. 1995;14:1402–1411. doi: 10.1002/j.1460-2075.1995.tb07126.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baldwin A S J. Methylation interference assay for analysis of DNA-protein interactions. In: Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. Vol. 2. New York, N.Y: Greene Publishing Associates/Wiley-Interscience; 1987. pp. 12.3.1–12.3.4. [DOI] [PubMed] [Google Scholar]
- 3.Beadling C, Ng J, Babbage J W, Cantrell D A. Interleukin-2 activation of STAT5 requires the convergent action of tyrosine kinases and a serine/threonine kinase pathway distinct from the Raf1/ERK2 MAP kinase pathway. EMBO J. 1996;15:101–112. [PMC free article] [PubMed] [Google Scholar]
- 4.Becker S, Groner B, Muller C W. Three-dimensional structure of the Stat3β homodimer bound to DNA. Nature. 1998;394:145–151. doi: 10.1038/28101. [DOI] [PubMed] [Google Scholar]
- 5.Bergad P L, Shih H-M, Towle H C, Schwarzenberg S J, Berry S A. Growth hormone induction of hepatic serine protease inhibitor 2.1 transcription is mediated by a Stat5-related factor binding synergistically to two γ-activated sites. J Biol Chem. 1995;270:24903–24910. doi: 10.1074/jbc.270.42.24903. [DOI] [PubMed] [Google Scholar]
- 6.Boucheron C, Dumon S, Santos S C R, Moriggl R, Hennighausen L, Gisselbrecht S, Gouilleux F. A single amino acid in the DNA binding regions of STAT5A and STAT5B confers distinct DNA binding specificities. J Biol Chem. 1998;273:33936–33941. doi: 10.1074/jbc.273.51.33936. [DOI] [PubMed] [Google Scholar]
- 7.Chen X, Vinkemeier U, Zhao Y, Jeruzalmi D, Darnell J E, Jr, Kuriyan J. Crystal structure of a tyrosine phosphorylated STAT-1 dimer bound to DNA. Cell. 1998;93:827–839. doi: 10.1016/s0092-8674(00)81443-9. [DOI] [PubMed] [Google Scholar]
- 8.Darnell J E., Jr STATs and gene regulation. Science. 1997;277:1630–1635. doi: 10.1126/science.277.5332.1630. [DOI] [PubMed] [Google Scholar]
- 9.Decker T, Kovarik P, Meinke A. GAS elements: A few nucleotides with a major impact on cytokine-induced gene expression. J Interferon Cytokine Res. 1997;17:121–134. doi: 10.1089/jir.1997.17.121. [DOI] [PubMed] [Google Scholar]
- 10.Grillot D A M, González-García M, Ekhterae D, Duan L, Inohara N, Otha S, Seldin M F, Núñez G. Genomic organization, promoter region analysis, and chromosome localization of the mouse bcl-x gene. J Immunol. 1997;158:4750–4757. [PubMed] [Google Scholar]
- 11.Horvath C M, Wen Z, Darnell J E., Jr A STAT protein domain that determines DNA sequence recognition suggests a novel DNA-binding domain. Genes Dev. 1995;9:984–994. doi: 10.1101/gad.9.8.984. [DOI] [PubMed] [Google Scholar]
- 12.Hou J, Schindler U, Henzel W J, Wong S C, McKnight Identification and purification of human Stat proteins activated in response to interleukin-2. Immunity. 1995;2:321–329. doi: 10.1016/1074-7613(95)90140-x. [DOI] [PubMed] [Google Scholar]
- 13.Ihle J N. STATs: signal transducers and activators of transcription. Cell. 1996;84:331–334. doi: 10.1016/s0092-8674(00)81277-5. [DOI] [PubMed] [Google Scholar]
- 14.Imada K, Bloom E T, Nakajima H, Horvath-Arcidiacono J A, Udy G B, Davey H, Leonard W J. Stat5b is essential for natural killer cell-mediated proliferation and cytolytic activity. J Exp Med. 1998;188:2067–2074. doi: 10.1084/jem.188.11.2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.John S, Vinkemeier U, Soldaini E, Darnell J E, Jr, Leonard W J. The significance of tetramerization in promoter recruitment by Stat5. Mol Cell Biol. 1999;19:1910–1918. doi: 10.1128/mcb.19.3.1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Laskowski R A, Macarthur M W, Moss D S, Thornton J M. PROCHECK—a program to check the stereochemical quality of protein structures. J Appl Crystallogr. 1993;26:283–291. [Google Scholar]
- 17.Leonard W J, O'Shea J J. Jaks and STATs: Biological implications. Annu Rev Immunol. 1998;16:293–322. doi: 10.1146/annurev.immunol.16.1.293. [DOI] [PubMed] [Google Scholar]
- 18.Levitt M, Hirshberg M, Sharon R, Daggett V. Potential-energy function and parameters for simulations of the molecular-dynamics of proteins and nucleic-acids in solution. Comput Phys Commun. 1995;91:215–231. [Google Scholar]
- 19.Lin J-X, Mietz J, Modi W S, John S, Leonard W J. Cloning of human Stat5B. Reconstitution of interleukin-2-induced Stat5A and Stat5B DNA binding activity in COS-7 cells. J Biol Chem. 1996;271:10738–10744. [PubMed] [Google Scholar]
- 20.Liu X, Robinson G W, Gouilleux F, Groner B, Hennighausen L. Cloning and expression of Stat5 and an additional homologue (Stat5b) involved in prolactin signal transduction in mouse mammary tissue. Proc Natl Acad Sci USA. 1995;92:8831–8835. doi: 10.1073/pnas.92.19.8831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu X, Robinson G W, Wagner K U, Garrett L, Wynshaw-Boris A, Hennighausen L. Stat5a is mandatory for adult mammary gland development and lactogenesis. Genes Dev. 1997;9:2266–2278. doi: 10.1101/gad.11.2.179. [DOI] [PubMed] [Google Scholar]
- 22.Meeker T C, Loeb J, Ayres M, Sellers W. The human Pim-1 gene is selectively transcribed in different hemato-lymphoid cell lines in spite of a G+C-rich housekeeping promoter. Mol Cell Biol. 1990;10:1680–1688. doi: 10.1128/mcb.10.4.1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meyer W K-H, Reichenbach P, Schindler U, Soldaini E, Nabholz M. Interaction of STAT5 dimers on two low affinity binding sites mediates interleukin 2 (IL-2) stimulation of IL-2 receptor α gene transcription. J Biol Chem. 1997;272:31821–31828. doi: 10.1074/jbc.272.50.31821. [DOI] [PubMed] [Google Scholar]
- 24.Mohamadi F, Richards N G J, Guida W C, Liskamp R, Lipton M, Caufield C, Chang G, Hendrickson T, Still W C. MacroModel-An integrated software system for modeling organic and bioorganic molecules using molecular mechanics. J Comput Chem. 1990;11:440–450. [Google Scholar]
- 25.Moriggl R, Topham D J, Teglund S, Sexl V, McKay C, Wang D, Hoffmeyer A, van Deursen J, Sangster M Y, Bunting K D, Grosveld G C, Ihle J N. Stat5 is required for IL-2-induced cell cycle progression of peripheral T cells. Immunity. 1999;10:249–259. doi: 10.1016/s1074-7613(00)80025-4. [DOI] [PubMed] [Google Scholar]
- 26.Mui A L, Wakao H, O'Farrell A M, Harada N, Miyajima A. Interleukin-3, granulocyte-macrophage colony stimulating factor and interleukin-5 transduce signals through two STAT5 homologues. EMBO J. 1995;14:1166–1175. doi: 10.1002/j.1460-2075.1995.tb07100.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nakajima H, Liu X W, Wynshaw-Boris A, Rosenthal L A, Imada K, Finbloom D S, Hennighausen L, Leonard W J. An indirect effect of Stat5a in IL-2-induced proliferation: a critical role for Stat5a in IL-2-mediated IL-2 receptor α chain induction. Immunity. 1997;7:691–701. doi: 10.1016/s1074-7613(00)80389-1. [DOI] [PubMed] [Google Scholar]
- 28.Needleman S B, Wunsch C D. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J Mol Biol. 1970;48:443–453. doi: 10.1016/0022-2836(70)90057-4. [DOI] [PubMed] [Google Scholar]
- 29.Pollock R, Treisman R. A sensitive method for the determination of protein-DNA binding specificities. Nucleic Acids Res. 1990;18:6197–6204. doi: 10.1093/nar/18.21.6197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schindler U, Wu P, Rothe M, Brasseur M, McKnight S L. Components of a Stat recognition code: evidence for two layers of molecular selectivity. Immunity. 1995;2:689–697. doi: 10.1016/1074-7613(95)90013-6. [DOI] [PubMed] [Google Scholar]
- 31.Silva C M, Lu H. Characterization and cloning of STAT5 from IM-9 cells and its activation by growth hormone. Mol Endocrinol. 1996;10:508–518. doi: 10.1210/mend.10.5.8732682. [DOI] [PubMed] [Google Scholar]
- 31a.Socolovsky M, Fallon A E, Wang S, Brugnara C, Lodish H F. Fetal anemia and apoptosis of red cell progenitors in Stat5a−/−5b−/− mice: a direct role for Stat5 in Bcl-X(L) induction. Cell. 1999;98:181–191. doi: 10.1016/s0092-8674(00)81013-2. [DOI] [PubMed] [Google Scholar]
- 32.Teglund S, McKay C, Schuetz E, van Dursen J M, Stravopodis D, Wang D, Brown M, Bodner S, Grosveld G, Ihle J N. Stat5a and Stat5b proteins have essential and nonessential, or redundant, roles in cytokine responses. Cell. 1998;93:841–850. doi: 10.1016/s0092-8674(00)81444-0. [DOI] [PubMed] [Google Scholar]
- 33.Tijan R, Maniatis T. Transcriptional activation: a complex puzzle with few easy pieces. Cell. 1994;77:5–8. doi: 10.1016/0092-8674(94)90227-5. [DOI] [PubMed] [Google Scholar]
- 34.Udy G B, Towers R P, Snell R G, Wilkins R J, Park S-H, Ram P A, Waxman D J, Davey H W. Requirement of STAT5b for sexual dimorphism of body growth rates and liver gene expression. Proc Natl Acad Sci USA. 1997;94:7239–7244. doi: 10.1073/pnas.94.14.7239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Verdier F, Rabionet R, Gouilleux F, Beisenherz-Huss C, Varlet P, Muller O, Mayeux P, Lacombe C, Gisselbrecht S, Chretien S. A sequence of the CIS promoter interacts preferentially with two associated STAT5A dimers: a distinct biochemical difference between STAT5A and STAT5B. Mol Cell Biol. 1998;18:5852–5860. doi: 10.1128/mcb.18.10.5852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vinkemeier U, Cohen S L, Moarefi I, Chait B T, Kuriyan J, Darnell J E., Jr DNA binding of in vitro activated Stat1a, Stat1b and truncated Stat1: interaction between NH2-terminal domains stabilizes binding of two dimers to tandem DNA sites. EMBO J. 1996;15:5616–5626. [PMC free article] [PubMed] [Google Scholar]
- 37.Vinkemeier U, Moarefi I, Darnell J E, Jr, Kuriyan J. Structure of the amino-terminal protein interaction domain of Stat4. Science. 1998;279:1048–1052. doi: 10.1126/science.279.5353.1048. [DOI] [PubMed] [Google Scholar]
- 38.Weiner S J, Kollman P A, Nguyen D T, Case D A. An all-atom force field for stimulation of protein and nucleic acids. J Comput Chem. 1986;7:230–252. doi: 10.1002/jcc.540070216. [DOI] [PubMed] [Google Scholar]
- 39.Xu X, Sun Y-L, Hoey T. Cooperative DNA binding and sequence-selective recognition conferred by the STAT amino-terminal domain. Science. 1996;273:794–797. doi: 10.1126/science.273.5276.794. [DOI] [PubMed] [Google Scholar]
- 40.Ye S-K, Maki K, Kitamura T, Sunaga S, Akashi K, Domen J, Weissman I L, Honjo T, Ikuta K. Induction of germline transcription in the TCRγ locus by Stat5: implications for accessibility control by the IL-7 receptor. Immunity. 1999;11:213–223. doi: 10.1016/s1074-7613(00)80096-5. [DOI] [PubMed] [Google Scholar]