Abstract
The mucus layer in the intestine plays a critical role in regulation of host–microbe interactions and maintaining homeostasis. Disruptions of the mucus layer due to genetic, environmental, or immune factors may lead to inflammatory bowel diseases (IBD). IBD frequently are accompanied with infections, and therefore are treated with antibiotics. Hence, it is important to evaluate risks of antibiotic treatment in individuals with vulnerable gut barrier and chronic inflammation. Mice with a knockout of the Muc2 gene, encoding the main glycoprotein component of the mucus, demonstrate a close contact of the microbes with the gut epithelium which leads to chronic inflammation resembling IBD. Here we demonstrate that the Muc2−/− mice harboring a gut protozoan infection Tritrichomonas sp. are susceptible to an antibiotic-induced depletion of the bacterial microbiota. Suppression of the protozoan infection with efficient metronidazole dosage or L-fucose administration resulted in amelioration of an illness observed in antibiotic-treated Muc2−/− mice. Fucose is a monosaccharide presented abundantly in gut glycoproteins, including Mucin2, and is known to be involved in host–microbe interactions, in particular in microbe adhesion. We suppose that further investigation of the role of fucose in protozoan adhesion to host cells may be of great value.
Keywords: fucose, protozoa, antibiotic, Mucin2, inflammation, microbiome
1. Introduction
The mammalian gut is inhabited by an enormous number of microorganisms referred to as the gut microbiota. The microbiota is known to contribute to host physiology, particularly the intestinal and systemic immunity [1]. In a healthy gut, the host–microbiota interactions are accurately regulated by the gut epithelial barrier. The barrier is comprised of enterocytes covered with a gel-like mucus layer formed substantially by glycoprotein Mucin2, antimicrobial peptides, and secretory immunoglobulins [2,3]. The barrier prevents close interaction between the gut microbiota and the mucosal immune system, thus precluding excessive immune activation. A breakdown of this barrier leads to a close interaction between the gut microbes and the immune system resulting in the disturbance of the intestinal homeostasis. This might be followed by different pathological conditions, such as IBD, which comprises Crohn’s disease (CD) and ulcerative colitis (UC) [4]. A disruption of the gut barrier may be caused by miscellaneous factors including genetic predisposition, environmental factors (such as nutrition, drug treatment, stress, etc.), status of the immune system and the microbiota [5]. Several genes are identified to be involved in IBD susceptibility including genes of the gut barrier, immune response and microbial defense pathways [6]. In patients with UC, the depletion of mucus thickness [7,8,9] and penetration of bacteria in the inner mucus layer [10] are observed. According to different studies, this may result from the defective differentiation of goblet cells and impaired mucus production [11,12,13,14,15], alteration in mucin glycosylation and sulfation [16,17,18,19] as well as increase of mucin degrading bacteria [20]. The precise etiology of such mucus disruptions is currently unknown and considered to be caused either by genetic factors, or inflammatory response and dysbiosis [14].
In addition to the intestinal barrier dysfunction, IBD are characterized by dysbiosis and concomitant infections. This provides a proinflammatory environment and aggravates inflammation in the gut [21,22]. Hence, one of the most used approaches of IBD treatment is antibiotic therapy. Antibiotics are used to decrease a bacterial load as well as to eliminate a particular infection. However, efficiency of antibiotic therapy is quite controversial and may be followed by several negative side effects [23]. Moreover, some data suggest that antibiotic uptake is a risk factor for IBD onset in childhood [24,25,26]. Studies of the gut microbiome in IBD patients in remission and acute phase demonstrate stronger dysbiosis in the latter [27], suggesting that microbiota may somehow be involved in IBD relapses. Furthermore, treatment of IBD can also be complicated by opportunistic microorganisms widely represented in the gut microflora. A disturbance of the microbiota may result in decrease of colonization resistance for pathogenic and opportunistic microorganisms which can provoke intestinal inflammation [28,29,30]. There are associations between antibiotic treatment and disorders of the gastrointestinal tract caused by opportunistic bacteria [31,32]. Apart from bacteria, the intestinal microbiota comprises fungi, archaea, protists. Less is known about the role of these microorganisms in host physiology and pathogenesis of IBD, but the questions are under investigation [33,34]. Therefore, it is important to understand risks of antibiotic usage, especially in the presence of concomitant infections, for treatment of individuals with defective gut barrier and chronic inflammation.
Animal models are very useful in studying genetically determined defects of the gut barrier and mechanisms of maintaining homeostasis and development of inflammation. Muc2 gene knockout mice (Muc2−/−) are widely used as a genetic animal model of IBD. Muc2−/− mice lack the main secreted glycoprotein Mucin2 forming mucus layer in the gut [35]. As a result, bacteria can directly contact the epithelial cells in the intestine of Muc2−/− mice [36]. This model is characterized by a chronic gut inflammation that can shift to spontaneous colitis, resembling UC, and colorectal cancer [35,37,38,39]. Muc2−/− mice are highly susceptible to enteric pathogens colonization [40,41] and the gut barrier disruption [37] displaying severe inflammation, the gut barrier dysfunction and mortality upon such conditions. Moreover, according to our data, antibiotic treatment for eradication of Helicobacter spp. infection in Mucin2 deficient mice (Muc2−/−; Kaiso−/− double knockout) did not result in clearance of the pathogen, but led to mortality of the animals [42,43]. Thus, due to the sensitivity to changes in the intestinal environment, Mucin2 knockout model can be used for studying the effects of dysbiosis upon chronic inflammation. In the current investigation, we used Muc2−/− mice harboring both Helicobacter spp. infection and intestinal protozoa Tritrichomonas sp. to reveal the effect of antibiotic treatment on the host health.
The microbiota is highly involved in maintaining homeostasis in the gut providing resistance to pathogens [44]. Therefore, development of new approaches for microbiota modulation is of great value, this includes modulation of microbiota during antibiotic therapy. Oligo- and polysaccharides are substances widely investigated as microbiota modulators [45]. In particular, fucose-containing saccharides are under consideration [46]. Fucose is a monosaccharide abundant in the intestinal glycoproteins, including mucin2 [47,48], it is presented in the terminal position of mucin2 polysaccharide chains, thus it is available to bacteria in the gut lumen. Fucose is known to mediate interactions between microbes and host: it serves as a nutrient for microbes [49], can affect virulence of pathogens (demonstrated for E. coli [50]), is involved in pathogen adhesion [49,51]. Defective fucosylation in the gut results in alteration of the microbiota [52]. Resident microbiota can stimulate fucosylation in the intestine, thus protecting against inflammation and pathogenic infection [53] Several studies have shown that fucose-containing substances are able to modulate the gut microbiota [54,55,56] and ameliorate antibiotic-induced dysbiosis [57]. Recently we have demonstrated that fucose contributes to amelioration of colitis-associated dysbiosis [58]. Lack of the glycoprotein mucin2 in Muc2−/− mice is likely result in decrease of the amount of fucose in the intestine. Thus, the protective properties of fucose against infection and inflammation might be reduced in this mouse model. Considering the beneficial effect of fucose on the gut microbiota, in the current study we investigated if supplementation of antibiotic treatment with fucose contributes to the susceptibility to the antibiotic-induced mortality of Muc2−/− mice.
2. Results
2.1. Antibiotics-Induced Mortality of Muc2−/− Mice Is Associated with Presence of Tritrichomonas sp.
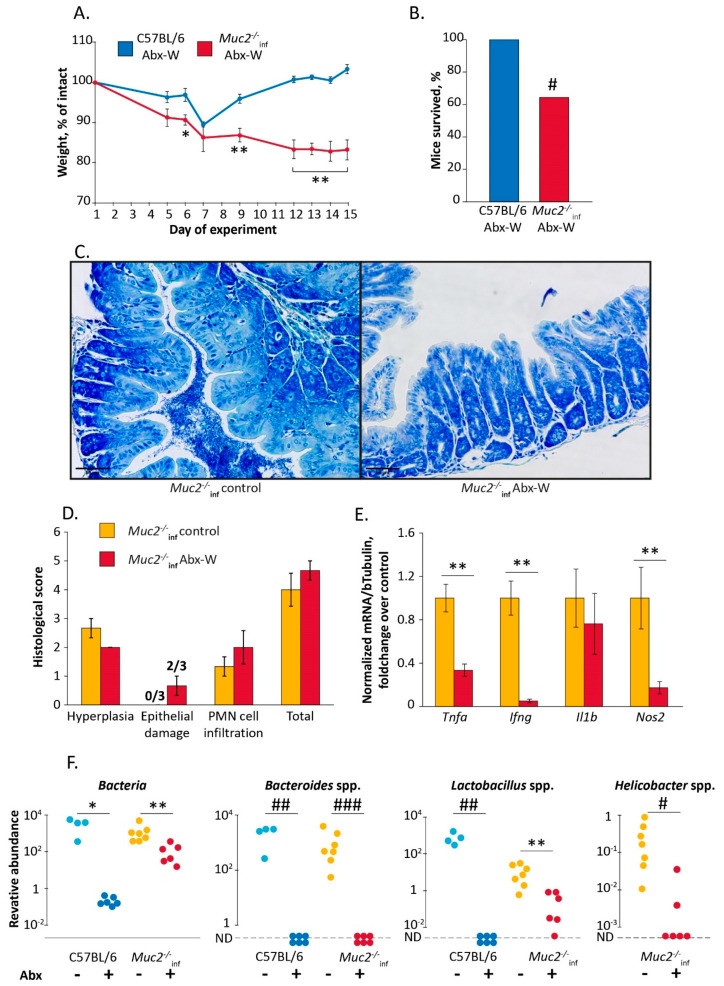
First, we tested the effect of the gut microbiota depletion on the health of Muc2−/− mice. The mice were treated with a cocktail of broad-spectrum antibiotics (clarithromycin, amoxicillin and metronidazole) in drinking water. According to our data, this treatment eradicates Helicobacter spp. in C57BL/6 mice [42]. The treatment with the antibiotics resulted in death of 35% of Muc2−/− mice harboring Helicobacter spp. (Hspp) (9 of 14 mice survived, group “Muc2−/−inf Abx-W”). The same treatment did not affect C57BL/6 mice free of infections (12 of 12 mice survived, group “C57BL/6 Abx-W”) (Fisher Exact test vs. “Muc2−/−inf Abx-W”: p < 0.05) (Figure 1B). We observed significant changes in body weight during the experiment in both “C57BL/6 Abx-W” and “Muc2−/−inf Abx-W” groups (Friedman chi-squared 46.6 and 21.6, df = 7, p < 0.001 and p < 0.01 respectively). The body weight loss in “Muc2−/−inf Abx-W” was greater than in “C57BL/6 Abx-W” on the days 6, 9, and 12–15 (Mann–Whitney u-test: Z = 2.31, p < 0.05; Z = 2.94; Z = 3.36; Z = 3.24; Z = 3.1, p < 0.01 respectively) (Figure 1A). After the two-week antibiotic treatment, survived Muc2−/− mice demonstrated severe weight loss compared to C57BL/6, which recovered by the end of the experiment (Figure 1A). To reveal if the mortality of mutant mice was associated with increased inflammation severity, we performed histological and gene expression analysis in colon. We did not observe significant effects of the antibiotic treatment on histological scores; however, 2 of 3 Muc2−/− mice demonstrated epithelial damage indicated by intensified desquamation (Figure 1C). The antibiotic treatment resulted in decreased expression of proinflammatory genes Tnfa, Ifng and Nos2 (Mann–Whitney u-test compared to control group: Z = 3.06; Z = 3.18; Z = 2.72; p < 0.01 respectively) and did not affect Il1b (Figure 1E) Thus, the antibiotic-induced mortality was not associated with aggravation of inflammation.
Figure 1.
The antibiotic treatment induced mortality of Muc2−/− mice. (A) Body weight dynamics during the antibiotic treatment. *, **—“C57BL/6 Abx-W” vs. “Muc2−/−inf Abx-W”, p < 0.05, p < 0.01, Mann–Whitney u-test. (B) Survival rate. #—“C57BL/6 Abx-W” vs. “Muc2−/−inf Abx-W”, p < 0.05, Fisher Exact test. (C) Azur-II-eosin-stained colonic sections of Muc2−/− mice (treated with the antibiotics and control group). Scale bar = 50µm. (D) Histological score of inflammation in colon of Muc2−/− mice. (E) Gene expression in colon tissue normalized on beta Tubulin (Tubb5) gene. **—differences between groups p < 0.01, Mann–Whitney u-test. (F) Relative abundance of the gut bacteria in feces on the day 15, normalized on Mus musculus 28S rRNA DNA. *, **—differences between groups p < 0.05, p < 0.01, Mann–Whitney u-test. #, ##, ###—differences between groups p < 0.05, p < 0.01, p < 0.001, Fisher Exact test. ND—not detected. Experimental groups: “Abx-W”—treatment with the antibiotics by adding in drinking water.
Muc2−/− mice used in the experiment harbored Helicobacter spp. infection, so we suggested that the pathogen might be implicated in the mortality of mutant mice. Next, we analyzed fecal microbiota by real-time PCR. The antibiotics decreased Helicobacter spp. infection in Muc2−/− mice (Fisher Exact test: p < 0.05, Figure 1F). As expected, the antibiotic treatment resulted in decrease of total gut bacteria determined by measuring the amount of bacterial 16S rRNA gene in feces of Muc2−/− and C57BL/6 mice (effect of the experimental group by Kruskal–Wallis test: H (3,23) = 18.64, p < 0.001, Figure 1F). Gut symbiotic bacteria Bacteroides spp. were undetectable after the treatment in mice of both genotypes (Fisher Exact test: p < 0.01 for C57BL/6 and p < 0.001 for Muc2−/−inf, Figure 1F). Lactobacillus spp. also decreased after the antibiotic treatment in Muc2−/− mice (Mann–Whitney u-test: Z = 2.64, p < 0.01, Figure 1F) and were undetectable in C57BL/6 (Fisher Exact test: p < 0.01, Figure 1F). Thus, we supposed that the mortality observed was not caused by Helicobacter spp. infection, but depletion of the intestinal microbiota was critical for viability of Muc2−/− mice.
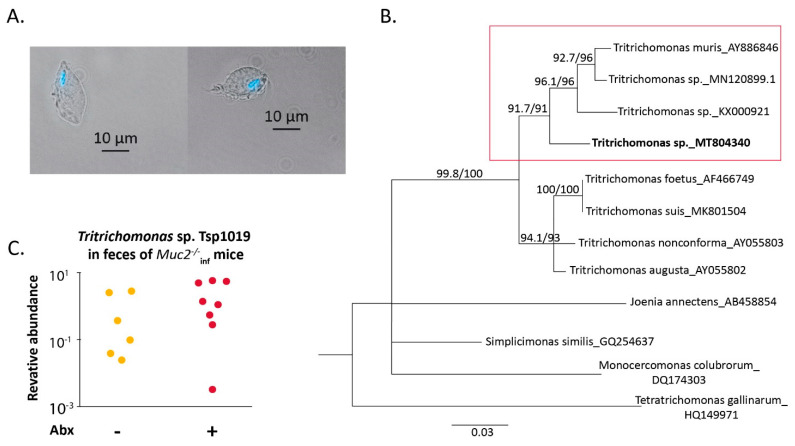
Interestingly, microscopic investigation of intestinal contents of Muc2−/− mice revealed a protozoan microorganism that morphologically resembled Tritrichomonas species (Figure 2A). To determine the microorganism, we performed sequencing of 18S rRNA gene and phylogenetic analysis as described previously [59]. We found that the detected protozoan clone Tsp1019 (Tsp) (deposed in GenBank, accession number MT804340) was phylogenetically related to Tritrichomonas species found in the gut of rodents (T. muris clone 1-6 AY886846, Tritrichomonas sp. strain LL5 MN120899.1 and Tritrichomonas sp. MEG-2016a KX000921—referred to as T. musculis, Figure 2B). Next, using the obtained sequence we designed specific primers for quantitative detection of Tritrichomonas sp. clone1019. Importantly, amount of Tritrichomonas sp. clone Tsp1019 in feces of Muc2−/− mice treated with the antibiotics did not differ from level observed in control Muc2−/− mice (Figure 2C). Thus, the antibiotic treatment did not affect the protozoa, suggesting that the microorganism might be involved in the antibiotic-induced mortality upon bacterial microbiota depletion.
Figure 2.
Protozoan microorganism Tritrichomonas sp. clone Tsp1019 was detected in the intestine of Muc2−/− mice. (A) Micrograph demonstrates microorganism purified from cecal content stained with Hoechst 33258 (magnification ×400), the bright field and Hoechst 33258 are merged. Scale bar = 10 µm. (B) Phylogenetic analysis of 18S rRNA DNA sequence obtained from gut content of Muc2−/− mice. (C) Relative abundance of Tritrichomonas sp. Tsp1019 DNA in feces of Muc2−/− mice on the day 15, normalized on Mus musculus 28S rRNA DNA.
2.2. Muc2−/− Mice Free of Infections Recover after Antibiotic Treatment
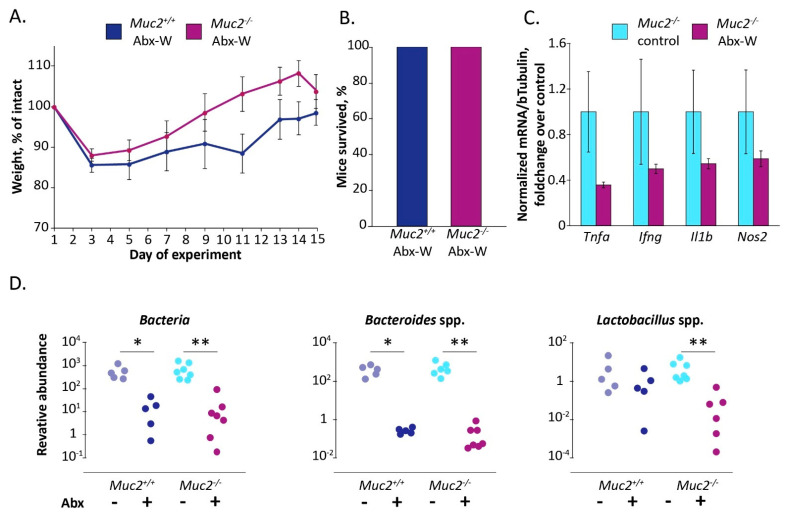
To test if the antibiotic-induced death of Muc2−/− mice resulted from the presence of the protozoan infection, we performed an experiment using Muc2−/− mice free of infections (including Tsp), Muc2+/+ littermates were used as a control. In contrast to the previous experiment, the antibiotic treatment did not result in severe weight loss and mortality of infection-free Muc2−/− mice (100% of mice survived) (Figure 3A,B). Moreover, we did not observe significant effects on expression of the proinflammatory genes after the antibiotics administration (Figure 3C). Thus, infection-free mutant mice were less sensitive to antibiotic treatment than infected mice. Therefore, we questioned, if the treatment decreased intestinal bacteria in infection-free mice. In accordance with the previous experiment, the antibiotics decreased the amount of total bacterial 16S rRNA copy number as well as Bacteroides and Lactobacillus spp. (the effect of the experimental group by Kruskal–Wallis test: H (3,24) = 17.34 for Bacteria, H (3,24) = 17.57 for Bacteroides spp., H (3,24) = 13.23 for Lactobacillus spp. p < 0.001, Figure 3D).
Figure 3.
Muc2−/− mice free of the infections survive after the antibiotic treatment. (A) Body weight dynamics upon antibiotic treatment. (B) Survival rate. (C) Gene expression in colon tissue normalized on beta Tubulin (Tubb5) gene. (D) Relative abundance of the gut bacteria in feces on the day 15, normalized on Mus musculus 28S rRNA gene. *, **—differences between groups p < 0.05, p < 0.01, Mann–Whitney u-test. Experimental groups: “Abx-W”—treatment with the antibiotics by adding in drinking water.
Therefore, the antibiotics did not induce mortality in Muc2−/− mice free of infections. We assumed that the lethal effect indeed resulted from the presence of Tsp infection upon depletion of the gut microbiota. Further we tested if suppression of the infection contributes to the amelioration of the antibiotic-induced lethal effect in Muc2−/− mice.
2.3. Elimination of Tritrichomonas sp. upon Antibiotic Treatment Results in Recovery of Infected Muc2−/− Mice
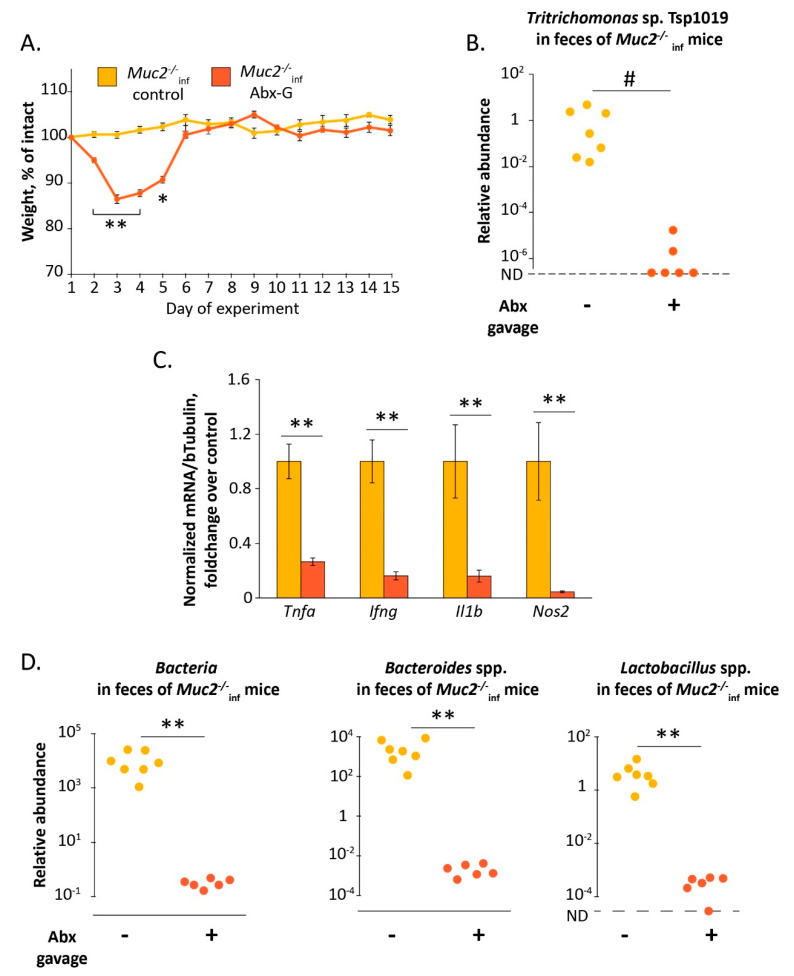
Furthermore, to suppress Tsp infection upon the gut microbiota depletion in Muc2−/− mice, we treated the mice with the antibiotics by intragastric gavage for two weeks (group “Muc2−/−inf Abx-G”). According to several studies, this way of antibiotic treatment is effective and has less adverse effects [60,61]. The daily doses of the antibiotics used were the same as those being used during treatment via drinking water in our previous experiments. The dose of metronidazole used was proportionate to those reported to be effective for suppression of Tritrichomonas spp. infections [62]. We assumed that treatment by gavage may be of more advantage due to administration of the daily dose in a one-time manner. Indeed, Tsp infection significantly decreased in the group “Muc2−/−inf Abx-G” compared to the control group “Muc2−/−inf Cont” (gavaged with drinking water) after two weeks of the treatment (Fisher Exact test: p < 0.05, Figure 4B). We observed a significant effect on body weight during the experiment in both groups (Friedman chi-squared 52.7 and 62.8 df = 14, p < 0.001 respectively for “Muc2−/−inf Cont” and “Muc2−/−inf Abx-G” groups, Figure 4A). The antibiotic treatment resulted in significant decrease of body weight in Muc2−/− mice in comparison to the control mice. The body weight loss in “Muc2−/−inf Abx-G” group was greater than in “Muc2−/−inf Cont” from the day 2 to the day 5 (Mann–Whitney u-test: Z = 3, p < 0.01; Z = 2, p < 0.05), but by the day 6 the mice recovered (Figure 4A). Similar to the previous experiments antibiotic treatment via gavage caused decrease of total bacteria, Bacteroides spp. and Lactobacillus spp. (Mann–Whitney u-test: Z = 2.08; p < 0.01; Figure 4D). Thus, a suppression of Tsp upon an alteration of the gut microbiota was associated with the recovery of the mutant mice after the antibiotic treatment. Moreover, in accordance with the fist experiment (Figure 1E) the treatment of the infected mice with the antibiotics resulted in decreased expression of the proinflammatory genes Tnfa, Nos2, Ifng, Il1b, (Mann–Whitney u-test: Z = 2.92; Z = 2.64; Z = 2.78; p < 0.01; Figure 4C). Therefore, we assumed that the antibiotic-induced death of the Muc2−/− mice observed previously resulted from insufficient suppression of Tsp infection upon the bacterial microbiota depletion. Next, we attempted to improve antibiotic-induced mortality by supplementation with the monosaccharide L-fucose.
Figure 4.
Suppression of Tritrichomonas sp. by antibiotic treatment by gavage results in the recovery of the infected Muc2−/− mice. (A) Body weight dynamics upon antibiotic treatment. *, **—“Muc2−/−inf control” vs. “Muc2−/−inf Abx-G”, p < 0.05, p < 0.01, Mann–Whitney u-test. (B) Relative abundance of Tritrichomonas sp. T.sp1019 in feces on the day 15, normalized on Mus musculus 28S rRNA gene. #—differences between groups p < 0.05, Fisher Exact test. ND—not detected. (C) Gene expression in colon tissue normalized on beta Tubulin (Tubb5) gene. **—differences between groups p < 0.01, Mann–Whitney u-test. (D) Relative abundance of the gut bacteria in feces on the day 15, normalized on Mus musculus 28S rRNA gene. **—differences between groups p < 0.01, Mann–Whitney u-test. ND—not detected. Experimental groups: “control”—gavage with drinking water; “Abx-G”—gavage with the antibiotics.
2.4. L-Fucose Prevents an Expansion of Tritrichomonas sp. and Ameliorates Antibiotic-Induced Emaciation in Infected Muc2−/− Mice
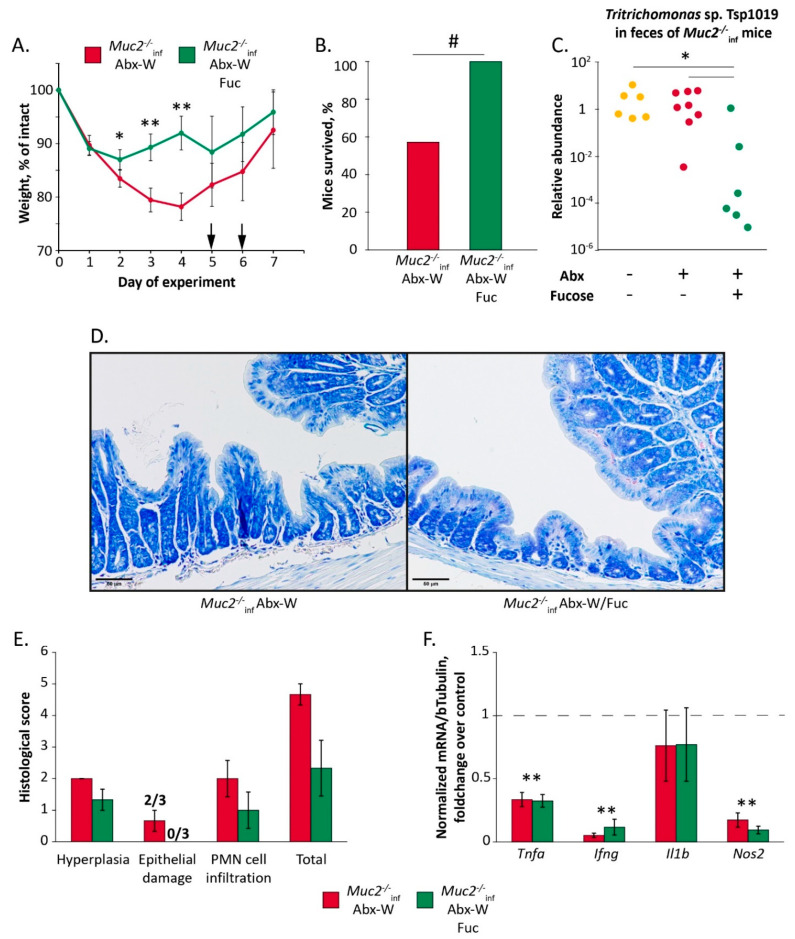
In this experiment, the mice were treated with the antibiotics via drinking water in dosage same to the previous experiments (“Muc2−/−inf Abx-W” group) or with the antibiotics supplemented with 0.1% L-fucose (“Muc2−/−inf Abx-W/Fuc” group) for 7 days. Similar to the previous experiments, the antibiotic treatment resulted in a severe weight loss and death of the mutant mice (8 of 14 mice survived in “Muc2−/−inf Abx-W” group). The mortality of the mice was observed on the days 5 and 6 (indicated with arrows in Figure 5A). Interestingly, supplementation of the antibiotic treatment with L-fucose resulted in 100% survival of the Muc2−/− mice (12 of 12 mice). The difference in survival rate was of significance with p < 0.05 (Fisher Exact test, Figure 5B). The both treatments significantly affected body weight (Friedman chi-squared 13.7, df = 6, p < 0.05 for “Muc2−/−inf Abx-W” group and 18.4, df = 6, p < 0.01 for “Muc2−/−inf Abx-W/Fuc” group). However, the body weight loss in “Muc2−/−inf Abx-W” was greater than in “Muc2−/−inf Abx-W/Fuc” on the days 2, 3 and 4 (Mann–Whitney u-test: Z = 2.05, p < 0.05; Z = 3.18; Z = 3.13, p < 0.01 respectively) (Figure 5A).
Figure 5.
L-fucose suppresses Tritrichomonas sp. and ameliorates antibiotic-induced emaciation in infected Muc2−/− mice. (A) Body weight dynamics upon treatment with antibiotics and L-fucose. *, **—p < 0.05 and p < 0.01, Mann–Whitney u-test. Arrows indicate the days when death was observed. (B) Survival rate. #—p < 0.05, Fisher Exact test. (C) Relative abundance of Tritrichomonas sp. in feces normalized on Mus musculus 28S rRNA DNA. *—differences between groups p < 0.05, Mann–Whitney u-test. (D) Azur-II-eosin-stained colonic sections of Muc2−/− mice (treated with the antibiotics and control group). Scale bar = 50µm. (E) Histological score of inflammation in colon of Muc2−/− mice. (F) Gene expression in colon tissue normalized on beta Tubulin (Tubb5) gene. **—differences compared to mRNA level in control group indicated by dotted line, p < 0.01, Mann–Whitney u-test. Experimental groups: “Abx-W”—treatment with the antibiotics via drinking water for 7 days; “Abx-W/Fuc”—treatment with antibiotics with L-fucose via drinking water for 7 days.
In accordance with the previous results, the antibiotics provided in drinking water did not affect relative abundance of the Tsp in feces, but supplementation of the treatment with L-fucose resulted in decrease of Tsp DNA in 13 times (Mann–Whitney u-test: Z = 2.5 for “Muc2−/−inf Abx-W/Fuc” vs. “Muc2−/−inf Abx-W” p < 0.05, Figure 5C). Thus, recovery of Muc2−/− mice after the antibiotic treatment was associated with suppression of Tsp. infection by L-fucose.
In the previous experiment we did not observe significant histological changes in colon of Muc2−/− mice after the antibiotics, with the exception of epithelial damage scores that had tended to increase (Figure 1D). Supplementation of the antibiotics with L-fucose did not induce significant changes in colon histology, but abrogated slightly increased epithelial damage score (Figure 5D,E). The antibiotic treatment resulted in a decrease of proinflammatory Tnf, Ifng and Nos2 genes expression (Mann–Whitney u-test compared to control group indicated by dotted line: Z = 3.06; Z = 3.18; Z = 2.72; p < 0.01 respectively Figure 5F) and L-fucose did not ameliorate the mRNA level of Tnf, Ifng, and Nos2 genes (Mann–Whitney test over control group indicated by dotted line: Z = 2.93, Z = 2.78; Z = 2.78; p < 0.01 respectively Figure 5F). Therefore, recovery of the mice and suppression of Tsp. infection were not accompanied with amelioration of the immune suppression.
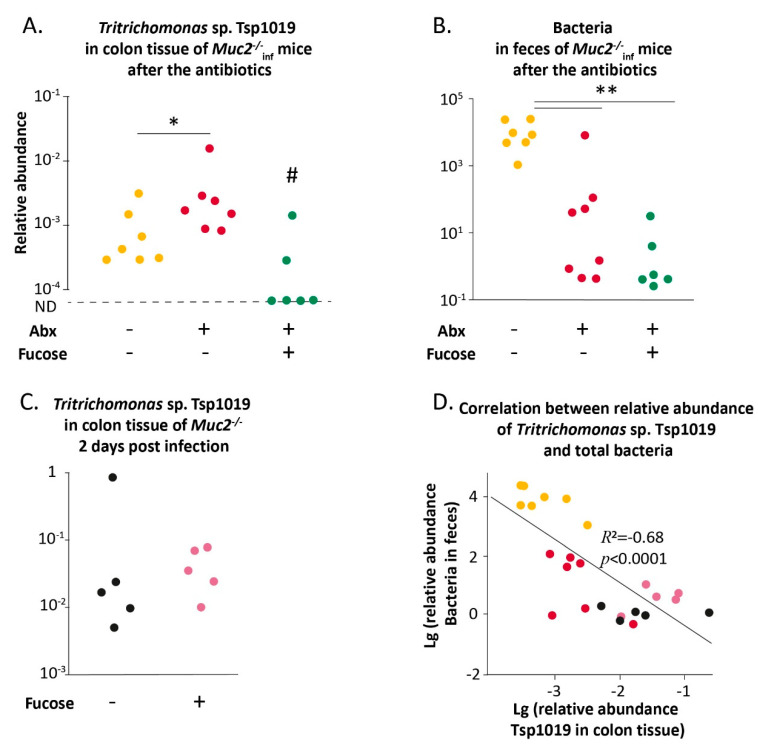
Tritrichomonas species attach to the epithelial cells, which is critical for establishment of the infection [63]. In mice lacking mucin2 intestinal epithelial cells are more vulnerable to contact with microbes, so next we assessed the amount of Tsp DNA in the colon tissue of Muc2−/− mice. Remarkably, treatment with the antibiotics resulted in an increase of Tsp in the colon tissue (“Muc2−/−inf control” vs. “Muc2−/−inf Abx-W” Mann–Whitney u-test: Z = 2.04 p < 0.05, Figure 6A). L-fucose promoted decrease of the microorganism in the colon tissue, Tsp DNA was detected only in 2 of 6 mice (Fisher Exact test: p < 0.05 for “Muc2−/−inf Abx-W/Fuc” vs. “Muc2−/−inf control” and “Muc2−/−inf Abx-W/Fuc” vs. “Muc2−/−inf Abx-W”, Figure 6A). In accordance with the previous results, antibiotics caused decrease of fecal bacteria (Mann–Whitney u-test: Z = 2.83, p < 0.01, Figure 6B). We assumed that bacterial microbiota depletion might contribute to Tsp. expansion and L-fucose might provide partial microbiota reconstitution, thus promoting recovery of Muc2−/− mice after the antibiotic treatment. However, L-fucose did not affect the amount of total bacterial DNA after the antibiotics in Muc2−/− mice (Mann–Whitney u-test: Z = 2.93, p < 0.01, Figure 6B).
Figure 6.
L-fucose prevents an expansion of Tritrichomonas sp. in colon of Muc2−/− mice upon antibiotic-induced microbiota depletion. (A) Relative abundance of Tritrichomonas sp. in colon tissue of Muc2−/− mice after treatment with antibiotics and antibiotics supplemented with L-fucose, normalized on Mus musculus 28S rRNA DNA. *—differences between groups p < 0.05, Mann–Whitney u-test. #—p < 0.05, Fisher Exact test. ND—not detected. (B) Relative abundance of the gut bacteria in feces of Muc2−/− mice after treatment with antibiotics and antibiotics supplemented with L-fucose normalized on Mus musculus 28S rRNA gene. **—differences between groups p < 0.01, Mann–Whitney u-test. (C) Relative abundance of Tritrichomonas sp. in colon tissue of Muc2−/− mice after infection via gavage. (D) Spearman correlation between relative abundance of Tsp1019 in colon tissue and bacteria in feces.
Therefore, L-fucose either interfered with the attachment of Tsp to colon cells or negatively affected the viability of Tsp in the intestinal lumen. To test this, Muc2−/− mice with intact microbiota were infected with Tsp. via gavage of fecal suspension containing Tritrichomonas sp. The mice were provided with drinking water supplemented with L-fucose for two days post-inoculation and then were tested for Tsp. DNA in colon tissue. The gavage resulted in infection with Tritrichomonas sp. that was detected in all inoculated mice. Supplementation with L-fucose did not affect amount of Tsp. DNA in colon tissue (Figure 6C). Thus, L-fucose did not alter the viability of Tritrichomonas sp. in the intestine. It should be noted that the mice infected with Tritrichomonas sp. (Figure 6C) did not demonstrate deterioration of well-being despite the detected level of Tsp in intestinal tissue being higher to that observed after the antibiotics (Figure 6B) (Mann–Whitney u-test: Z = 3.07, p < 0.01). Therefore, intact bacterial microbiota seemed to be crucial factor for outcomes of Tsp. infection in Muc2−/− mice. In fact, we observed negative correlation between bacteria in feces and Tsp. in colon tissue (Spearman correlation: R = −0.68, p < 0.0001, Figure 6D). Thus, normal bacterial microbiota might affect colonization of Tritrichomonas sp. and upon depletion of the microbiota L-fucose is able to protect from expansion of Tsp in the colon.
3. Discussion
Here we investigated the effect of the gut microbiota and the presence of the infections on susceptibility to antibiotic treatment in a host with a disturbed gut barrier. We made use of Muc2−/− mice lacking main intestinal secreted mucin, so the gut microbiota closely contacts the intestinal epithelia and the immune cells [36]. Previously, we have shown that Muc2−/−Kaiso−/− double knockout mice are extremely sensitive to antibiotics used for eradication of Helicobacter spp. infection. The treatment not only was ineffective for elimination of the pathogen, but also resulted in mortality of the animals [42,43]. Moreover, Tadesse and colleagues mentioned the rapid death of Muc2−/− mice upon antibiotic treatment in the discussion [64], but to our knowledge, the data have not been published. In the current study we demonstrated that antibiotics caused dramatic weight loss (Figure 1A) and mortality (Figure 1B) in Muc2−/− mice, which was accompanied by depletion of the symbiotic microbiota as well as a decrease of Helicobacter spp. infection (Figure 1F). The interesting finding was that the Muc2−/− mice also harbored protozoan microorganisms identified as Tritrichomonas sp. T.sp1019. Phylogenetic analysis revealed that the microorganisms are related to the common murine protozoans T. muris and T. musculis (Figure 2B). Moreover, the infection was also detected in the mutant mice after the antibiotic treatment (Figure 2C). Tritrichomonas spp. are generally considered to be non-pathogenic commensal microbiota of mice, but monitoring of the microorganisms in SPF-facilities is under discussion [65]. Tritrichomonas spp. are known to affect host physiology by changing the gut proteome [66] and triggering immune responses in the intestine [67,68]. In the previous studies, the mice were not tested for presence of protozoan microorganisms [42,43], so we assumed Tsp. probably being involved in the antibiotic-induced mortality of Muc2−/− mice.
We tested if the antibiotic treatment has such a negative effect on Muc2−/− mice free of the infections (including Tsp). Turned out that the mice without infections did not demonstrate mortality and recovered by the end of two-week treatment with the antibiotics (Figure 3). Thus, depletion of the gut microbiota by itself did not cause the adverse outcome observed. Then we examined if suppression of Tritrichomonas sp. infection upon microbiota depletion ameliorates the antibiotic-induced emaciation in Muc2−/− mice. We did not have isolated cultures of Tritrichomonas sp., we infected mice through intragastric gavage with fecal microbiota from mice positive for Tritrichomonas sp. Tsp1019. To avoid the effect of the infection inoculation we used these infected mice to generate offspring being born with the infection. The infected mice were treated with the antibiotics by intragastric gavage, this approach was reported to be effective and less harmful [60,61]. The treatment resulted in a decrease of Tritrichomonas sp. in Muc2−/− mice (Figure 4B), probably due to appropriate concentration of metronidazole in antibiotic cocktail. Tsp. suppression was associated with complete body weight recovery by the 6th day of the treatment (Figure 4A). Thus, we supposed that the antibiotic-associated body weight loss in mice lacking mucin2 resulted from the presence of Tritrichomonas sp. in context of the bacterial microbiota depletion.
The gut bacterial microbiota is known to promote resistance to pathogens [44] and to affect outcomes of various protozoan infections [69]. Some symbiotic bacteria seem to have inhibitory effects on several protozoan parasites [70], thus protecting the host organism. During the infection, Tritrichomonas and Trichomonas spp. attach to the host epithelial cells leading to the barrier dysfunction and cell death [63]. Gut bacteria are known to mediate epithelium turnover and repair. It was shown that antibiotic-treated mice demonstrate decreased turnover of the intestinal epithelial cells [71] and mortality induced by DSS-treatment [72]. Thus, we hypothesized that reconstitution of the bacterial microflora may ameliorate the antibiotic-associated emaciation in Muc2−/− mice with Tritrichomonas sp. infection.
We attempted to modulate the microbiota depletion by supplementing the antibiotic treatment with fucose upon insufficient suppression of Tritrichomonas sp. Mucin2 is a heavily O-glycosylated glycoprotein that serves not only as the main component of host protecting mucus, but also as a nutritional niche for commensal bacteria in the gut. Fucose is a terminal monosaccharide of Mucin2 glycoprotein, it contributes to host-microbiome interaction [73]. Moreover, fucose-containing substances are known to modulate the gut microbiota [54,55,56], fucose monosaccharide contributes to amelioration of colitis-associated dysbiosis [58]. Furthermore, it was shown that fucose-containing polysaccharide fucoidan improved antibiotic-induced dysbiosis and promoted the recovery of gut microbiome in mice [57]. Contrary to our assumption, L-fucose did not affect the number of total bacteria in the gut upon the antibiotic treatment (Figure 6B), but it did ameliorate the antibiotic-induced weight loss in the mutant mice (Figure 5A,B). Interestingly, the observed recovery of the mice was associated with suppression of Tsp infection in colon by L-fucose (Figure 6A). At the same time, treatment with the antibiotics did not cause mortality in Muc2−/− mice free of the infections and 100% of the animals recovered by the end of the experiment (Figure 3). Altogether our results imply that the antibiotic-induced emaciation resulted from the presence of Tritrichomonas sp. infection upon the microbiota depletion in the host with a compromised barrier in the gut.
Antibiotic treatment is known to suppress immune function. For instance, broad-spectrum antibiotics impair leukocytes count by suppression of hematopoiesis [74]. We observed inhibition of the proinflammatory genes expression in the colon tissue (Figure 1E). Thus, the immune suppression in mice lacking Mucin2 in combination with increase of Tritrichomonas sp. in colon, probably, was critical for the health of mutant mice. L-fucose did not modulate suppressed immune function (Figure 5F). However, L-fucose, as well as intact bacterial microbiota, prevented Tritrichomonas sp. expansion, we assume that the monosaccharide or intestinal bacteria affect the infection through modulation of adhesion in mice lacking mucin2. When attached to the epithelial cells, trichomonads become cytotoxic causing cell damage, loss of function and cell death [63]. Thus, Tritrichomonas spp. colonization may result in intoxication and alteration of nutrition and energy uptake. Mechanisms of the adhesion are not completely understood, the most explored are Tritrichomonas foetus and Trichomonas vaginalis. The studies indicate several surface molecules mediating membrane-membrane interactions of the parasites with host epithelia. These include sialic acid-binding lectins, adhesins, lipophosphoglycan (LPG) and cysteine proteases [63,75]. According to our analysis T. foetus is phylogenetically close to Tritrichomonas sp. T.sp1019, so we suppose that the adhesion mechanisms of the microorganisms are likely to be similar. Saccharides on the parasite and host cells are involved in cell-cell interaction and adhesion. For instance, sialic acid is known to inhibit T. foetus adhesion in vitro [76,77] due to presence of sialic acid-binding lectin in the parasite membrane [78]. Thus, T. foetus can use sialic acid on the host cells for adhesion. Membrane of T. foetus also contains LPG that binds to host lectins promoting the adhesion [79]. Several studies show that LPG of T. foetus contains several saccharides with fucose and mannose being prevalent ones [80], but the surface saccharide patterns in different isolates may vary [81]. Thus, it is possible that fucose can be involved in Tritrichomonas spp. pathogenesis, but this needs further investigations. Yet, it was shown that L-fucose inhibits phagocytosis in T. vaginalis, probably by blocking mannose receptors on the microorganism [82]. It should be noted that the mechanisms described were explored mostly in vitro, using cells from different hosts, which can differ by glycosylation patterns. Therefore, the effects obtained in one cell culture may not be reproduced in the other. Moreover, to our knowledge, the mechanisms have not been verified in vivo.
Currently, oligosaccharides, including those containing fucose, are considered to be prospective compounds for microbiota modulation, particularly by blocking the microbe adhesion to host cells [83]. Therefore, understanding the mechanisms of protozoa adhesion to the host cells will promote development of new therapeutic approaches for urogenital and gastrointestinal diseases treatment. The human gastrointestinal tract is inhabited by several protozoa, including members of trichomonads—Dientamoeba fragilis and Pentatrichomonas hominis known to cause gastrointestinal symptoms [84]. Here we demonstrate that treatment with broad-spectrum antibiotics, aimed to eradicate Helicobacter spp., led to emaciation and mortality in mice with deficient barrier in the gut due to blooming of Tritrichomonas sp. T.sp1019. The effective suppression of the protozoa by the antibiotic gavage and L-fucose supplementation promoted recovery in the mice. Current approaches being used for IBD treatment often do not consider the variety of the microorganisms inhabiting the gastrointestinal tract [85,86], including protozoa. Our results imply that this may result in health deterioration in individuals defective in gut barrier due to overgrowth of non-bacterial agents. Thus, when choosing a therapy, it is important to consider a potential protozoa infection, monitor the microorganisms and, if necessary, make use of drugs for protozoa elimination. One of the successful ways to treat a protozoa infection may be preventing the adhesion to the epithelial cells. Thus, understanding the mechanisms of adhesion and developing drugs affecting these mechanisms are of great perspective.
4. Materials and Methods
4.1. Mice and Housing Conditions
The study was conducted using the equipment of the Center for Genetic Resources of Laboratory Animals at the Institute of Cytology and Genetics of the Siberian Branch of the Russian Academy of Sciences (ICG SB RAS), supported by the Ministry of Education and Science of the Russian Federation (Unique identifier of the project RFMEFI62117X0015). All experimental procedures were carried out according to Russian legislation in agreement with the Good Laboratory Practice standards (directive #267 from 19.06.2003 of the Ministry of Health of the Russian Federation, Moscow, Russian Federation) and the European Convention for the protection of vertebrate animals used for experimental and other scientific purposes; all procedures were approved by the inter-institutional bioethical committee, protocol #28 (19 June 2015).
Two animal colonies were used in the experiments. The one was positive for Helicobacter spp. (Hspp) and negative for all other pathogens listed in the annual FELASA 2014 recommendations [87]. The second was negative for all listed pathogens, further on referred to as mice free of infections. In accordance with the FELASA 2014 recommendations for surveillance, pathogens testing was conducted quarterly in sentinel mice that received both dirty bedding and water from colony mice for a 3-month period. During the experiments protozoan microorganisms were detected in the first colony of Muc2−/− mice harboring Helicobacter spp. The microorganisms were determined as Tritrichomonas sp. (Tsp), which is not listed in the annual FELASA 2014 recommendations. In all experiments this microorganism was detected only in Muc2−/− and Muc2+/+ littermates within “infected” groups.
All animals were housed in single-sex groups of 3-6 mice in individually ventilated caging systems (Optimice®, Animal Care Systems, Centennial, CO, USA). The light/dark photoperiod was 14 h/10 h (light off at 16:00 h), the temperature was 22–24 °C, the humidity was 30%–60%, the air exchange was 7–10 volumes per hour. The mice were provided with sterile food (Mouse Maintenance autoclavable, V1534-300, Sniff, Spezialdiäten, GmbH, Spezialdiäten, Germany) and sterile water ad libitum. All experimental groups comprised 8-10-week-old female mice. We used female mice, as male mice obtained from the breeding were used in another investigation. The first experiment was carried out using Muc2−/− mice backcrossed on C57BL/6JNskrc substrain in our facility and C57BL/6JNskrc mice as a control. Muc2−/− mice harbored both Tsp and Hspp infections and control C57BL/6JNskrc were free of the infections. All other experiments were conducted using Muc2−/− and Muc2+/+ littermates harboring Tsp or free of infections.
4.2. Tritrichomonas sp. Colonization
To obtain mice infected with Tsp., 5 female and 5 male heterozygous (Muc2+/−) mice free of infections were gavaged intragastrically with fecal suspension from infected animals. To prepare the suspension 3 fecal boli were homogenized in 1 mL of drinking water, filtered through 70 µm cell-strainer (Corning, New York, NY, USA) and diluted up to 1.5 mL. Mice were gavaged with 100 µL of the suspension for three days. In 14 days after infection all mice were positive for Tsp. detected in feces using primers designed specific to Tritrichomonas sp. clone Tsp1019 sequence (see below). These infected mice were crossed to obtain infected Muc2+/− used for subsequent breeding (infection and breeding strategies are pictured in Supplementary Materials Figure S1).
4.3. Antibiotic and L-Fucose Treatment
In the study a mix of three antibiotics (clarithromycin, amoxicillin, and metronidazole) was used. Antibiotics concentrations were 0.2 mg/mL for clarithromycin and metronidazole, 0.6 mg/mL for amoxicillin when added in drinking water in accordance with previously used protocol [42]. The dosage for intragastric gavage was 25 mg/kg for clarithromycin and metronidazole, 75 mg/kg for amoxicillin. The dosage of metronidazole was proportionate to that being used for treatment of Trichomonas spp. in mice [62]. L-fucose was added in the drinking water in the final concentration of 0.1% as reported previously [58].
4.4. Experimental Design
In the first (Figure 1 and Figure 2) and second (Figure 3) experiments mice were treated with the antibiotics by adding in drinking water for 14 days (“Abx-W” groups), control mice obtained the drinking water (“control” groups). During the experiment mice were weighted. Some mutant mice with the infections died on the 13 and 14 days of the experiment. After two-week treatment fecal samples were collected from survived mice and the mice were euthanized using CO2.
In the third experiment (Figure 4) mice were treated with the antibiotics by intragastric gavage for 14 days (“Abx-G” group). To estimate if the gavage procedure affected weight loss, control mice were gavaged with drinking water in parallel with the antibiotic treatment (“control” group). During the experiment mice were weighted, after two-week treatment fecal samples were collected and the mice were euthanized using CO2.
In the fourth experiment (Figure 5) mice were treated with the antibiotics (“Abx-W” group) and antibiotics supplemented with L-fucose (“Abx-W/Fuc” group) via drinking water for 7 days. Some mice from the “Abx-W” group died on the 5 and 6 days of the experiment. Mice were weighed, fecal samples were collected from survived mice. Mice were euthanized using CO2.
In the fifth experiment (Figure 6) Muc2−/− mice free of infections were inoculated with Tsp infection via gavage. For two days after the inoculation mice were provided with 0.1% L-fucose (“fucose” group) or drinking water (“control” group). Mice were euthanized in two days post-infection using CO2.
For microbiota analysis fecal and colon tissue samples were stored at −20 °C until the analysis. Colon tissue samples for gene expression were frozen in liquid nitrogen and stored at −70 °C until the extraction. Colon tissues for histological analysis were fixed in 4% paraformaldehyde.
4.5. Fecal and Colon DNA Extraction and Bacteria Real-Time PCR
Fecal and colon tissue DNA was extracted using QIAamp Fast DNA Stool Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. Briefly, samples stored at −20 °C were homogenized and lysed in Inhibitex Buffer at 70 °C for 30 min and centrifuged at 10,000 rpm for 5 min. Supernatant was placed in a clean tube and mixed with Proteinase K and AL Buffer and incubated at 70 °C for 10 min. Then 96% ethanol was added, samples were mixed, applied to spin columns and washed with AW1 and AW2 buffers. Purified DNA was eluted, measured with a NanoDrop 2000 spectrophotometer (ThermoScientific, Waltham, MA, USA) and stored at −20 °C until the analysis.
Bacteria (total bacteria, Bacteroides spp., Lactobacillus spp. and Helicobacter spp.) in fecal samples were evaluated by the amount of bacterial 16S rRNA DNA normalized to Mus musculus 28S rRNA DNA. PCR contained PCR Master Mix HS-qPCR SYBR Blue (Biolabmix, Novosibirisk, Russian Federation), specific primers with final concentration of 300 nM (all primer sequences are presented in Supplementary Materials Table S1), and 25–100 ng of DNA. Real-time PCR was performed in a CFX96 real-time PCR Detection System (BioRad Laboratories, Hercules, CA, USA) according to the following protocol: 95 °C for 3 min; 40 cycles of: 95 °C for 15 s, 62 °C for 25 s, 72 °C for 25 s; melt curve 65–95 °C. Relative amount of bacterial DNA was calculated by formula: 2[Ct(28S rRNA)-Ct(16S rRNA)].
4.6. Sequencing of Tritrichomonas sp. DNA, Phylogenetic Analysis, and Tritrichomonas sp., Clone Tsp1019 Real-Time PCR
To taxonomically determine the detected protozoa we performed Sanger sequencing of. 18S rRNA DNA in intestinal content of Muc2−/− mice using previously described primer sets [59]. Sequencing was performed using BigDye™ Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems™, Warrington, Cheshire, UK) according to the manufacturer’s instructions. The reaction protocol was as follows: 96 °C for 60 s; 35 cycles of: 96 °C for 30 s, 56 °C for 15 s, 60 °C for 4 min. Then samples were purified by precipitation in 64% isopropanol (+4 °C) for 20 min at room temperature with following centrifugation at 14,000 rpm for 20 min. Invisible pallets were washed with 70% ethanol (+4 °C) and dried using vacuum concentrator. Samples were sequenced at the “Molecular and Cellular Biology” core facility of the IMCB SB RAS (Novosibirsk, Russian Federation).
Sequences obtained were analyzed using Unipro UGENE software [88], the assembly resulted in 1355 bp DNA sequence clone T.sp1019 (deposited in GenBank with accession number MT804340). The DNA sequence was analyzed using BLAST algorithm [89], for further analysis 11 sequences were picked (listed in Supplementary Materials Table S2). The sequences were aligned with MUSCLE algorithm using UGENE software and phylogenetic analysis was performed using IQ-tree web server [90] and visualized using FigTree software.
To further analyze the amount of this specific Tritrichomonas sp. Tsp1019 DNA in gut of mice we designed specific primers to the obtained sequence MT804340 (listed in Supplementary Materials Table S1), to verify unique specificity BLAST search was used. PCR analysis with fecal and colonic tissue DNA was performed as described above, according to the following protocol: 95 °C for 3 min; 40 cycles of: 95 °C for 15 s, 57 °C for 25 s, 72 °C for 25 s; melt curve 65–95 °C. Relative amount of protozoan DNA was calculated by formula: 2[Ct(28S rRNA)-Ct(T.sp)].
4.7. RNA Extraction and Gene Expression Analysis
The gene expression analysis was performed according to our previously described protocol [58] with following modifications. Total RNA was purified from colon tissue samples using TRIzol reagent (Invitrogen, Waltham, MA, USA) and genomic DNA was removed by DNaseI (Roche, Mannheim, Germany) according to recommendations of the manufacturers. RNA concentration was determined using a NanoDrop 2000 spectrophotometer (ThermoScientific, Waltham, MA, USA). Reverse transcription reaction was performed using 7–10 µg of RNA, mix of random hexa-deoxyribonucleotide and Oligo-dT primers and M-MuLV reverse transcriptase (SibEnzyme, Novosibirsk, Russian Federation) according to the manufacturer’s recommendations. The cDNA obtained was five times diluted with deionized water and used for real-time PCR. Real-time PCR was performed using PCR Master Mix HS-qPCR SYBR Blue (Biolabmix, Novosibirisk, Russian Federation), 5 µL of cDNA, and 250 nM specific primers (listed in Supplementary Materials Table S3). The reaction was performed in a CFX96 real-time PCR Detection System (BioRad Laboratories, Hercules, CA, USA) according to the following protocol: 95 °C for 3 min; 40 cycles of: 95 °C for 15 s, 62 °C for 25 s, 72 °C for 25 s; melt curve 65–95 °C. Expression of the target gene was normalized to Tubb5 level using the formula: 2[Ct(Tubb5 mRNA)-Ct(gene of interest mRNA)].
4.8. Tritrichomonas sp. Microscopy
Tritrichomonas sp. microorganisms were harvested from cecum contents of mice using protocol previously described [64]. Briefly, cecum content was collected in 25 mL of ice-cold PBS, thoroughly resuspended, passed through a 70 µm cell-strainer and centrifuged at 1000 rpm for 5 min. A pallet was washed with ice-cold PBS, centrifuged as described above and resuspended in PBS. Then percoll 40/80% gradient was performed at 1000× g centrifugation for 15 min with breaks off and microorganisms were collected from the percoll 40/80 interface, centrifuged at 1750 rpm for 10 min and washed with ice-cold PBS. The microorganisms obtained were stained with Hoechst 33258 (Sigma-Aldrich, Darmstadt, Germany), mounted on a slide and examined under the fluorescent microscope (Zeiss Axio Imager M2, Carl Zeiss, Oberkochen, Germany) with ×100 magnification.
4.9. Histological Analysis
Colon tissue fixed in 4% paraformaldehyde were embedded in paraffin, n = 3 per group. Paraffin sections (4 µm) were stained with azur-II-eosin, the sections were examined in a blinded manner. Images were taken with an AxioImager.M2 microscope using an Axiocam 305 color camera (Zeiss, Oberkochen, Germany). Hyperplasia was defined as percentage of crypt elongation above the mean crypt length counted in C57BL/6 sections. Crypt length was determined using ImageJ software. Epithelial damage was defined as distortion of epithelial cell layer. PMN cells per section were counted on ×1000 magnification.
Hyperplasia: 0: ≤10%, 1: 0–50%, 2: 50–100%, 3: ≥100%
Epithelial damage: 0—no pathological changes detectable, 1—epithelial desquamation, 2—erosion of the epithelial surface (gaps of 1 to 10 epithelial cells/lesion), 3—epithelial ulceration (gaps of > 10 epithelial cells)
PMN cell infiltration: 0: 1–30 per section, 1: 31–60 per section, 2: 61–90 per section, 3: ≥90 per section.
4.10. Statistical Analysis
The data were tested for normality with the Kolmogorov–Smirnov test, all data had abnormal distributions. All data are presented as mean ± Standard Error of Mean (SEM), except for abundance of bacteria and Tsp1019 DNA, which are shown as actual values. The effect on body weight dynamics within the group was analyzed using Friedman test and differences between the groups were analyzed with Mann–Whitney u-test. All other data were analyzed using Kruskal–Wallis test and Mann–Whitney u-test. Within several data sets obtained for bacteria and Tsp1019 abundance some values were under detection limit, in that case Fisher Exact test was used.
5. Conclusions
Here we have demonstrated that the antibiotic treatment resulting in the bacterial microbiota depletion leads to dramatic weight loss and mortality in mice lacking mucin2 (Muc2−/− mice). This effect is associated with the increase of Tritrichomonas sp. in colon. Elimination of Tritrichomonas sp. using both an effective scheme of the antibiotic treatment and supplementation of the antibiotics with L-fucose, promotes survival of the mice. L-fucose does not affect viability and colonization of Tritrichomonas sp. infection in Muc2−/− mice with intact microbiota.
Acknowledgments
We thank Galina Kalmykova and Nadezhda Akulova from Siberian Federal Scientific Centre of Agro-BioTechnologies of the Russian Academy of Sciences for kindly providing the microscopy of protozoa in gut content.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/ijms221910699/s1, Figure S1: Infection and breeding strategies being used to obtain Muc2+/+ and Muc2−/− littermates infected with Tritrichomonas sp. (T.sp), Table S1: Primer sets used for microbial DNA PCR, Table S2: Sequences obtained by BLAST search and used for phylogenetic analysis, Table S3: Primer sets used for gene expression analysis.
Author Contributions
Conceptualization, K.M.A. and E.A.L.; methodology, E.N.K., K.M.A. and M.A.B.; formal analysis, E.A.L. and K.M.A.; investigation, K.M.A. and M.A.B.; data curation, E.A.L. and K.M.A.; writing—original draft preparation, E.A.L. and K.M.A.; writing—review and editing, E.A.L., E.N.K., K.M.A. and M.A.B.; visualization, K.M.A.; project administration, E.A.L. and K.M.A.; funding acquisition, E.A.L. and E.N.K. All authors have read and agreed to the published version of the manuscript.
Funding
Laboratory animals were obtained with the support of the Budgetary funding for basic scientific research # 0533-2019-0003 and # АААА-А21-121011990039-2. Reagents for manipulation with microflora were supported by the Russian Foundation for Basic Research (RFBR) grant # 19-015-00169. Bacterial analysis and sequencing were supported by the Russian Science Foundation (RSF) grant # 20-64-47020. Gene expression analyses were supported by the RSF grant # 20-74-10022.
Institutional Review Board Statement
The study was conducted according to the guidelines of the Declaration of Helsinki, and approved by the Inter-institutional bioethical committee at ICG SB RAS, protocol #28 (19 June 2015).
Data Availability Statement
The data presented in this study are available on request from the corresponding author.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Grigg J.B., Sonnenberg G.F. Host-Microbiota Interactions Shape Local and Systemic Inflammatory Diseases. J. Immunol. 2017;198:564–571. doi: 10.4049/jimmunol.1601621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McGuckin M.A., Linden S.K., Sutton P., Florin T.H. Mucin dynamics and enteric pathogens. Nat. Rev. Microbiol. 2011;9:265–278. doi: 10.1038/nrmicro2538. [DOI] [PubMed] [Google Scholar]
- 3.Okumura R., Takeda K. Maintenance of intestinal homeostasis by mucosal barriers. Inflamm. Regen. 2018;38:5. doi: 10.1186/s41232-018-0063-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Martini E., Krug S.M., Siegmund B., Neurath M.F., Becker C. Mend Your Fences: The Epithelial Barrier and Its Relationship with Mucosal Immunity in Inflammatory Bowel Disease. Cell. Mol. Gastroenterol. Hepatol. 2017;4:33–46. doi: 10.1016/j.jcmgh.2017.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ananthakrishnan A.N. Epidemiology and risk factors for IBD. Nat. Rev. Gastroenterol. Hepatol. 2015;12:205–217. doi: 10.1038/nrgastro.2015.34. [DOI] [PubMed] [Google Scholar]
- 6.Khor B., Gardet A., Xavier R.J. Genetics and pathogenesis of inflammatory bowel disease. Nature. 2011;474:307–317. doi: 10.1038/nature10209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pullan R.D., Thomas G.A., Rhodes M., Newcombe R.G., Williams G.T., Allen A., Rhodes J. Thickness of adherent mucus gel on colonic mucosa in humans and its relevance to colitis. Gut. 1994;35:353–359. doi: 10.1136/gut.35.3.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Swidsinski A., Loening-Baucke V., Theissig F., Engelhardt H., Bengmark S., Koch S., Lochs H., Dörffel Y. Comparative study of the intestinal mucus barrier in normal and inflamed colon. Gut. 2007;56:343–350. doi: 10.1136/gut.2006.098160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Strugala V., Dettmar P.W., Pearson J.P. Thickness and continuity of the adherent colonic mucus barrier in active and quiescent ulcerative colitis and Crohn’s disease. Int. J. Clin. Pract. 2008;62:762–769. doi: 10.1111/j.1742-1241.2007.01665.x. [DOI] [PubMed] [Google Scholar]
- 10.Johansson M.E., Gustafsson J.K., Holmén-Larsson J., Jabbar K.S., Xia L., Xu H., Ghishan F.K., Carvalho F.A., Gewirtz A.T., Sjövall H., et al. Bacteria penetrate the normally impenetrable inner colon mucus layer in both murine colitis models and patients with ulcerative colitis. Gut. 2014;63:281–291. doi: 10.1136/gutjnl-2012-303207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Alipour M., Zaidi D., Valcheva R., Jovel J., Martínez I., Sergi C., Walter J., Mason A.L., Wong G.K., Dieleman L.A., et al. Mucosal Barrier Depletion and Loss of Bacterial Diversity are Primary Abnormalities in Paediatric Ulcerative Colitis. J. Crohn’s Colitis. 2016;10:462–471. doi: 10.1093/ecco-jcc/jjv223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gersemann M., Becker S., Kübler I., Koslowski M., Wang G., Herrlinger K.R., Griger J., Fritz P., Fellermann K., Schwab M., et al. Differences in goblet cell differentiation between Crohn’s disease and ulcerative colitis. Differentiation. 2009;77:84–94. doi: 10.1016/j.diff.2008.09.008. [DOI] [PubMed] [Google Scholar]
- 13.Dorofeyev A.E., Vasilenko I.V., Rassokhina O.A., Kondratiuk R.B. Mucosal barrier in ulcerative colitis and Crohn’s disease. Gastroenterol. Res. Pract. 2013;2013:431231. doi: 10.1155/2013/431231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sheng Y.H., Hasnain S.Z., Florin T.H., McGuckin M.A. Mucins in inflammatory bowel diseases and colorectal cancer. J. Gastroenterol. Hepatol. 2012;27:28–38. doi: 10.1111/j.1440-1746.2011.06909.x. [DOI] [PubMed] [Google Scholar]
- 15.Tytgat K.M., van der Wal J.W., Einerhand A.W., Büller H.A., Dekker J. Quantitative analysis of MUC2 synthesis in ulcerative colitis. Biochem. Biophys. Res. Commun. 1996;224:397–405. doi: 10.1006/bbrc.1996.1039. [DOI] [PubMed] [Google Scholar]
- 16.Hanski C., Born M., Foss H.D., Marowski B., Mansmann U., Arastéh K., Bachler B., Papenfuss M., Niedobitek F. Defective post-transcriptional processing of MUC2 mucin in ulcerative colitis and in Crohn’s disease increases detectability of the MUC2 protein core. J. Pathol. 1999;188:304–311. doi: 10.1002/(SICI)1096-9896(199907)188:3<304::AID-PATH375>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 17.Larsson J.M., Karlsson H., Crespo J.G., Johansson M.E., Eklund L., Sjövall H., Hansson G.C. Altered O-glycosylation profile of MUC2 mucin occurs in active ulcerative colitis and is associated with increased inflammation. Inflamm. Bowel Dis. 2011;17:2299–2307. doi: 10.1002/ibd.21625. [DOI] [PubMed] [Google Scholar]
- 18.Fu J., Wei B., Wen T., Johansson M.E., Liu X., Bradford E., Thomsson K.A., McGee S., Mansour L., Tong M., et al. Loss of intestinal core 1-derived O-glycans causes spontaneous colitis in mice. J. Clin. Investig. 2011;121:1657–1666. doi: 10.1172/JCI45538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shirazi T., Longman R.J., Corfield A.P., Probert C.S. Mucins and inflammatory bowel disease. Postgrad. Med. J. 2000;76:473–478. doi: 10.1136/pmj.76.898.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Png C.W., Lindén S.K., Gilshenan K.S., Zoetendal E.G., McSweeney C.S., Sly L.I., McGuckin M.A., Florin T.H. Mucolytic bacteria with increased prevalence in IBD mucosa augment in vitro utilization of mucin by other bacteria. Am. J. Gastroenterol. 2010;105:2420–2428. doi: 10.1038/ajg.2010.281. [DOI] [PubMed] [Google Scholar]
- 21.Trifan A., Stanciu C., Stoica O., Girleanu I., Cojocariu C. Impact of Clostridium difficile infection on inflammatory bowel disease outcome: A review. World J. Gastroenterol. 2014;20:11736–11742. doi: 10.3748/wjg.v20.i33.11736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shen Z.H., Zhu C.X., Quan Y.S., Yang Z.Y., Wu S., Luo W.W., Tan B., Wang X.Y. Relationship between intestinal microbiota and ulcerative colitis: Mechanisms and clinical application of probiotics and fecal microbiota transplantation. World J. Gastroenterol. 2018;24:5–14. doi: 10.3748/wjg.v24.i1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nitzan O., Elias M., Peretz A., Saliba W. Role of antibiotics for treatment of inflammatory bowel disease. World J. Gastroenterol. 2016;22:1078–1087. doi: 10.3748/wjg.v22.i3.1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kronman M.P., Zaoutis T.E., Haynes K., Feng R., Coffin S.E. Antibiotic exposure and IBD development among children: A population-based cohort study. Pediatrics. 2012;130:e794–e803. doi: 10.1542/peds.2011-3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Virta L., Auvinen A., Helenius H., Huovinen P., Kolho K.L. Association of repeated exposure to antibiotics with the development of pediatric Crohn’s disease—A nationwide, register-based Finnish case-control study. Am. J. Epidemiol. 2012;175:775–784. doi: 10.1093/aje/kwr400. [DOI] [PubMed] [Google Scholar]
- 26.Shaw S.Y., Blanchard J.F., Bernstein C.N. Association between the use of antibiotics in the first year of life and pediatric inflammatory bowel disease. Am. J. Gastroenterol. 2010;105:2687–2692. doi: 10.1038/ajg.2010.398. [DOI] [PubMed] [Google Scholar]
- 27.Duboc H., Rajca S., Rainteau D., Benarous D., Maubert M.A., Quervain E., Thomas G., Barbu V., Humbert L., Despras G., et al. Connecting dysbiosis, bile-acid dysmetabolism and gut inflammation in inflammatory bowel diseases. Gut. 2013;62:531–539. doi: 10.1136/gutjnl-2012-302578. [DOI] [PubMed] [Google Scholar]
- 28.Sekirov I., Tam N.M., Jogova M., Robertson M.L., Li Y., Lupp C., Finlay B.B. Antibiotic-induced perturbations of the intestinal microbiota alter host susceptibility to enteric infection. Infect. Immun. 2008;76:4726–4736. doi: 10.1128/IAI.00319-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wlodarska M., Willing B., Keeney K.M., Menendez A., Bergstrom K.S., Gill N., Russell S.L., Vallance B.A., Finlay B.B. Antibiotic treatment alters the colonic mucus layer and predisposes the host to exacerbated Citrobacter rodentium-induced colitis. Infect. Immun. 2011;79:1536–1545. doi: 10.1128/IAI.01104-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lewis B.B., Buffie C.G., Carter R.A., Leiner I., Toussaint N.C., Miller L.C., Gobourne A., Ling L., Pamer E.G. Loss of Microbiota-Mediated Colonization Resistance to Clostridium difficile Infection with Oral Vancomycin Compared with Metronidazole. J. Infect. Dis. 2015;212:1656–1665. doi: 10.1093/infdis/jiv256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Högenauer C., Hammer H.F., Krejs G.J., Reisinger E.C. Mechanisms and management of antibiotic-associated diarrhea. Clin. Infect. Dis. 1998;27:702–710. doi: 10.1086/514958. [DOI] [PubMed] [Google Scholar]
- 32.Wiström J., Norrby S.R., Myhre E.B., Eriksson S., Granström G., Lagergren L., Englund G., Nord C.E., Svenungsson B. Frequency of antibiotic-associated diarrhoea in 2462 antibiotic-treated hospitalized patients: A prospective study. J. Antimicrob. Chemother. 2001;47:43–50. doi: 10.1093/jac/47.1.43. [DOI] [PubMed] [Google Scholar]
- 33.Sartor R.B., Wu G.D. Roles for Intestinal Bacteria, Viruses, and Fungi in Pathogenesis of Inflammatory Bowel Diseases and Therapeutic Approaches. Gastroenterology. 2017;152:327–339.e4. doi: 10.1053/j.gastro.2016.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yamamoto-Furusho J.K., Torijano-Carrera E. Intestinal protozoa infections among patients with ulcerative colitis: Prevalence and impact on clinical disease course. Digestion. 2010;82:18–23. doi: 10.1159/000273871. [DOI] [PubMed] [Google Scholar]
- 35.Velcich A., Yang W., Heyer J., Fragale A., Nicholas C., Viani S., Kucherlapati R., Lipkin M., Yang K., Augenlicht L. Colorectal cancer in mice genetically deficient in the mucin Muc2. Science. 2002;295:1726–1729. doi: 10.1126/science.1069094. [DOI] [PubMed] [Google Scholar]
- 36.Johansson M.E., Phillipson M., Petersson J., Velcich A., Holm L., Hansson G.C. The inner of the two Muc2 mucin-dependent mucus layers in colon is devoid of bacteria. Proc. Natl. Acad. Sci. USA. 2008;105:15064–15069. doi: 10.1073/pnas.0803124105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Van der Sluis M., De Koning B.A., De Bruijn A.C., Velcich A., Meijerink J.P., Van Goudoever J.B., Büller H.A., Dekker J., Van Seuningen I., Renes I.B., et al. Muc2-deficient mice spontaneously develop colitis, indicating that MUC2 is critical for colonic protection. Gastroenterology. 2006;131:117–129. doi: 10.1053/j.gastro.2006.04.020. [DOI] [PubMed] [Google Scholar]
- 38.Wenzel U.A., Magnusson M.K., Rydström A., Jonstrand C., Hengst J., Johansson M.E., Velcich A., Öhman L., Strid H., Sjövall H., et al. Spontaneous colitis in Muc2-deficient mice reflects clinical and cellular features of active ulcerative colitis. PLoS ONE. 2014;9:e100217. doi: 10.1371/journal.pone.0100217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bao Y., Guo Y., Li Z., Fang W., Yang Y., Li X., Li Z., Xiong B., Chen Z., Wang J., et al. MicroRNA profiling in Muc2 knockout mice of colitis-associated cancer model reveals epigenetic alterations during chronic colitis malignant transformation. PLoS ONE. 2014;9:e99132. doi: 10.1371/journal.pone.0099132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bergstrom K.S., Kissoon-Singh V., Gibson D.L., Ma C., Montero M., Sham H.P., Ryz N., Huang T., Velcich A., Finlay B.B., et al. Muc2 protects against lethal infectious colitis by disassociating pathogenic and commensal bacteria from the colonic mucosa. PLoS Pathog. 2010;6:e1000902. doi: 10.1371/journal.ppat.1000902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zarepour M., Bhullar K., Montero M., Ma C., Huang T., Velcich A., Xia L., Vallance B.A. The mucin Muc2 limits pathogen burdens and epithelial barrier dysfunction during Salmonella enterica serovar Typhimurium colitis. Infect. Immun. 2013;81:3672–3683. doi: 10.1128/IAI.00854-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Litvinova E.A., Kozhevnikova E.N., Achasova K.M., Kontsevaya G.V., Moshkin M.P. Eradication of Helicobacter spp. in mucin2-deficient mice. Lab. Anim. 2017;51:311–314. doi: 10.1177/0023677216670687. [DOI] [PubMed] [Google Scholar]
- 43.Litvinova E.A., Belyaev M.D., Prokhortchouk A.V., Korostina V.S., Prokhortchouk E.B., Kozhevnikova E.N. Role of intestinal mucin-2 in the effectiveness of the treatment of Helicobacter spp. infection in laboratory mice. Vavilov J. Genet. Breed. 2015;19:494–498. doi: 10.18699/VJ15.066. [DOI] [Google Scholar]
- 44.Kamada N., Chen G.Y., Inohara N., Núñez G. Control of pathogens and pathobionts by the gut microbiota. Nat. Immunol. 2013;14:685–690. doi: 10.1038/ni.2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Markowiak P., Śliżewska K. Effects of Probiotics, Prebiotics, and Synbiotics on Human Health. Nutrients. 2017;9:1021. doi: 10.3390/nu9091021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Garber J.M., Hennet T., Szymanski C.M. Significance of fucose in intestinal health and disease. Mol. Microbiol. 2021;115:1086–1093. doi: 10.1111/mmi.14681. [DOI] [PubMed] [Google Scholar]
- 47.Pickard J.M., Chervonsky A.V. Intestinal fucose as a mediator of host-microbe symbiosis. J. Immunol. 2015;194:5588–5593. doi: 10.4049/jimmunol.1500395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bergstrom K.S., Xia L. Mucin-type O-glycans and their roles in intestinal homeostasis. Glycobiology. 2013;23:1026–1037. doi: 10.1093/glycob/cwt045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sicard J.F., Le Bihan G., Vogeleer P., Jacques M., Harel J. Interactions of Intestinal Bacteria with Components of the Intestinal Mucus. Front. Cell. Infect. Microbiol. 2017;7:387. doi: 10.3389/fcimb.2017.00387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pacheco A.R., Curtis M.M., Ritchie J.M., Munera D., Waldor M.K., Moreira C.G., Sperandio V. Fucose sensing regulates bacterial intestinal colonization. Nature. 2012;492:113–117. doi: 10.1038/nature11623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Suwandi A., Galeev A., Riedel R., Sharma S., Seeger K., Sterzenbach T., García Pastor L., Boyle E.C., Gal-Mor O., Hensel M., et al. Std fimbriae-fucose interaction increases Salmonella-induced intestinal inflammation and prolongs colonization. PLoS Pathog. 2019;15:e1007915. doi: 10.1371/journal.ppat.1007915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kashyap P.C., Marcobal A., Ursell L.K., Smits S.A., Sonnenburg E.D., Costello E.K., Higginbottom S.K., Domino S.E., Holmes S.P., Relman D.A., et al. Genetically dictated change in host mucus carbohydrate landscape exerts a diet-dependent effect on the gut microbiota. Proc. Natl. Acad. Sci. USA. 2013;110:17059–17064. doi: 10.1073/pnas.1306070110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pickard J.M., Maurice C.F., Kinnebrew M.A., Abt M.C., Schenten D., Golovkina T.V., Bogatyrev S.R., Ismagilov R.F., Pamer E.G., Turnbaugh P.J., et al. Rapid fucosylation of intestinal epithelium sustains host-commensal symbiosis in sickness. Nature. 2014;514:638–641. doi: 10.1038/nature13823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pham T.A., Clare S., Goulding D., Arasteh J.M., Stares M.D., Browne H.P., Keane J.A., Page A.J., Kumasaka N., Kane L., et al. Epithelial IL-22RA1-mediated fucosylation promotes intestinal colonization resistance to an opportunistic pathogen. Cell Host Microbe. 2014;16:504–516. doi: 10.1016/j.chom.2014.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shi H., Chang Y., Gao Y., Wang X., Chen X., Wang Y., Xue C., Tang Q. Dietary fucoidan of Acaudina molpadioides alters gut microbiota and mitigates intestinal mucosal injury induced by cyclophosphamide. Food Funct. 2017;8:3383–3393. doi: 10.1039/C7FO00932A. [DOI] [PubMed] [Google Scholar]
- 56.Liu M., Ma L., Chen Q., Zhang P., Chen C., Jia L., Li H. Fucoidan alleviates dyslipidemia and modulates gut microbiota in high-fat diet-induced mice. J. Funct. Foods. 2018;48:220–227. doi: 10.1016/j.jff.2018.07.006. [DOI] [Google Scholar]
- 57.Wang L., Ai C., Wen C., Qin Y., Liu Z., Wang L., Gong Y., Su C., Wang Z., Song S. Fucoidan isolated from Ascophyllum nodosum alleviates gut microbiota dysbiosis and colonic inflammation in antibiotic-treated mice. Food Funct. 2020;11:5595–5606. doi: 10.1039/D0FO00668H. [DOI] [PubMed] [Google Scholar]
- 58.Borisova M.A., Snytnikova O.A., Litvinova E.A., Achasova K.M., Babochkina T.I., Pindyurin A.V., Tsentalovich Y.P., Kozhevnikova E.N. Fucose Ameliorates Tryptophan Metabolism and Behavioral Abnormalities in a Mouse Model of Chronic Colitis. Nutrients. 2020;12:445. doi: 10.3390/nu12020445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dimasuay K.G., Rivera W.L. Molecular characterization of trichomonads isolated from animal hosts in the Philippines. Vet. Parasitol. 2013;196:289–295. doi: 10.1016/j.vetpar.2013.03.019. [DOI] [PubMed] [Google Scholar]
- 60.Hill D.A., Hoffmann C., Abt M.C., Du Y., Kobuley D., Kirn T.J., Bushman F.D., Artis D. Metagenomic analyses reveal antibiotic-induced temporal and spatial changes in intestinal microbiota with associated alterations in immune cell homeostasis. Mucosal Immunol. 2010;3:148–158. doi: 10.1038/mi.2009.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Reikvam D.H., Erofeev A., Sandvik A., Grcic V., Jahnsen F.L., Gaustad P., McCoy K.D., Macpherson A.J., Meza-Zepeda L.A., Johansen F.-E. Depletion of Murine Intestinal Microbiota: Effects on Gut Mucosa and Epithelial Gene Expression. PLoS ONE. 2011;6:e17996. doi: 10.1371/journal.pone.0017996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cobo E.R., Eckmann L., Corbeil L.B. Murine models of vaginal trichomonad infections. Am. J. Trop. Med. Hyg. 2011;85:667–673. doi: 10.4269/ajtmh.2011.11-0123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tolbert M.K., Gookin J.L. Mechanisms of Tritrichomonas foetus Pathogenicity in Cats with Insights from Venereal Trichomonosis. J. Vet. Intern. Med. 2016;30:516–526. doi: 10.1111/jvim.13920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tadesse S., Corner G., Dhima E., Houston M., Guha C., Augenlicht L., Velcich A. MUC2 mucin deficiency alters inflammatory and metabolic pathways in the mouse intestinal mucosa. Oncotarget. 2017;8:71456–71470. doi: 10.18632/oncotarget.16886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Da Costa A.S., Graham T.M., Duncan J.A., Pillai S.P.S., Lund J.M. Detection and treatment strategy for Tritrichomonas muris in the common laboratory mouse. bioRxiv. 2019:827055. doi: 10.1101/827055. [DOI] [Google Scholar]
- 66.Kashiwagi A., Kurosaki H., Luo H., Yamamoto H., Oshimura M., Shibahara T. Effects of Tritrichomonas muris on the mouse intestine: A proteomic analysis. Exp. Anim. 2009;58:537–542. doi: 10.1538/expanim.58.537. [DOI] [PubMed] [Google Scholar]
- 67.Chudnovskiy A., Mortha A., Kana V., Kennard A., Ramirez J.D., Rahman A., Remark R., Mogno I., Ng R., Gnjatic S., et al. Host-Protozoan Interactions Protect from Mucosal Infections through Activation of the Inflammasome. Cell. 2016;167:444–456.e14. doi: 10.1016/j.cell.2016.08.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Escalante N.K., Lemire P., Cruz Tleugabulova M., Prescott D., Mortha A., Streutker C.J., Girardin S.E., Philpott D.J., Mallevaey T. The common mouse protozoa Tritrichomonas muris alters mucosal T cell homeostasis and colitis susceptibility. J. Exp. Med. 2016;213:2841–2850. doi: 10.1084/jem.20161776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bär A.-K., Phukan N., Pinheiro J., Simoes-Barbosa A. The Interplay of Host Microbiota and Parasitic Protozoans at Mucosal Interfaces: Implications for the Outcomes of Infections and Diseases. PLoS Negl. Trop. Dis. 2015;9:e0004176. doi: 10.1371/journal.pntd.0004176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Travers M.-A., Florent I., Kohl L., Grellier P. Probiotics for the Control of Parasites: An Overview. J. Parasitol. Res. 2011;2011:610769. doi: 10.1155/2011/610769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Park J.-H., Kotani T., Konno T., Setiawan J., Kitamura Y., Imada S., Usui Y., Hatano N., Shinohara M., Saito Y., et al. Promotion of Intestinal Epithelial Cell Turnover by Commensal Bacteria: Role of Short-Chain Fatty Acids. PLoS ONE. 2016;11:e0156334. doi: 10.1371/journal.pone.0156334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rakoff-Nahoum S., Paglino J., Eslami-Varzaneh F., Edberg S., Medzhitov R. Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell. 2004;118:229–241. doi: 10.1016/j.cell.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 73.Tailford L.E., Crost E.H., Kavanaugh D., Juge N. Mucin glycan foraging in the human gut microbiome. Front. Genet. 2015;6:81. doi: 10.3389/fgene.2015.00081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Josefsdottir K.S., Baldridge M.T., Kadmon C.S., King K.Y. Antibiotics impair murine hematopoiesis by depleting the intestinal microbiota. Blood. 2017;129:729–739. doi: 10.1182/blood-2016-03-708594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Petrin D., Delgaty K., Bhatt R., Garber G. Clinical and microbiological aspects of Trichomonas vaginalis. Clin. Microbiol. Rev. 1998;11:300–317. doi: 10.1128/CMR.11.2.300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bonilha V.L., Ciavaglia M.d.C., de Souza W., e Silva Filho F.C. The involvement of terminal carbohydrates of the mammalian cell surface in the cytoadhesion of trichomonads. Parasitol. Res. 1995;81:121–126. doi: 10.1007/BF00931616. [DOI] [PubMed] [Google Scholar]
- 77.Babál P., Russell L.C. Sialic Acid-Specific Lectin-Mediated Adhesion of Tritrichomonas foetus and Tritrichomonas mobilensis. J. Parasitol. 1999;85:33–40. doi: 10.2307/3285696. [DOI] [PubMed] [Google Scholar]
- 78.Babál P., Pindak F.F., Russell L.C., Gardner W.A., Jr. Sialic acid-specific lectin from Tritrichomonas foetus. Biochim. Biophys. Acta. 1999;1428:106–116. doi: 10.1016/S0304-4165(99)00062-8. [DOI] [PubMed] [Google Scholar]
- 79.Singh B.N., Lucas J.J., Beach D.H., Shin S.T., Gilbert R.O. Adhesion of Tritrichomonas foetus to bovine vaginal epithelial cells. Infect. Immun. 1999;67:3847–3854. doi: 10.1128/IAI.67.8.3847-3854.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Singh B.N. Lipophosphoglycan-like glycoconjugate of Tritrichomonas foetus and Trichomonas vaginalis. Mol. Biochem. Parasitol. 1993;57:281–294. doi: 10.1016/0166-6851(93)90204-B. [DOI] [PubMed] [Google Scholar]
- 81.Doumecq M.L., Soto P., Casalini M.B., Gimeno E.J., Barbeito C.G., Monteavaro C.E. Variation in the saccharide lectin binding pattern from different isolates of Tritrichomonas foetus. Exp. Parasitol. 2014;147:48–53. doi: 10.1016/j.exppara.2014.09.009. [DOI] [PubMed] [Google Scholar]
- 82.Pereira-Neves A., Benchimol M. Phagocytosis by Trichomonas vaginalis: New insights. Biol. Cell. 2007;99:87–101. doi: 10.1042/BC20060084. [DOI] [PubMed] [Google Scholar]
- 83.Wiciński M., Sawicka E., Gębalski J., Kubiak K., Malinowski B. Human Milk Oligosaccharides: Health Benefits, Potential Applications in Infant Formulas, and Pharmacology. Nutrients. 2020;12:266. doi: 10.3390/nu12010266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Maritz J.M., Land K.M., Carlton J.M., Hirt R.P. What is the importance of zoonotic trichomonads for human health? Trends Parasitol. 2014;30:333–341. doi: 10.1016/j.pt.2014.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Toruner M., Loftus E.V., Jr., Harmsen W.S., Zinsmeister A.R., Orenstein R., Sandborn W.J., Colombel J.F., Egan L.J. Risk factors for opportunistic infections in patients with inflammatory bowel disease. Gastroenterology. 2008;134:929–936. doi: 10.1053/j.gastro.2008.01.012. [DOI] [PubMed] [Google Scholar]
- 86.Inflammatory Bowel Disease Group, Chinese Society of Gastroenterology, Chinese Medical Association Evidence-based consensus on opportunistic infections in inflammatory bowel disease (republication) Intest. Res. 2018;16:178–193. doi: 10.5217/ir.2018.16.2.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Mähler Convenor M., Berard M., Feinstein R., Gallagher A., Illgen-Wilcke B., Pritchett-Corning K., Raspa M. FELASA recommendations for the health monitoring of mouse, rat, hamster, guinea pig and rabbit colonies in breeding and experimental units. Lab. Anim. 2014;48:178–192. doi: 10.1177/0023677213516312. [DOI] [PubMed] [Google Scholar]
- 88.Okonechnikov K., Golosova O., Fursov M. Unipro UGENE: A unified bioinformatics toolkit. Bioinformatics. 2012;28:1166–1167. doi: 10.1093/bioinformatics/bts091. [DOI] [PubMed] [Google Scholar]
- 89.Zhang Z., Schwartz S., Wagner L., Miller W. A greedy algorithm for aligning DNA sequences. J. Comput. Biol. 2000;7:203–214. doi: 10.1089/10665270050081478. [DOI] [PubMed] [Google Scholar]
- 90.Trifinopoulos J., Nguyen L.T., von Haeseler A., Minh B.Q. W-IQ-TREE: A fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 2016;44:W232–W235. doi: 10.1093/nar/gkw256. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data presented in this study are available on request from the corresponding author.