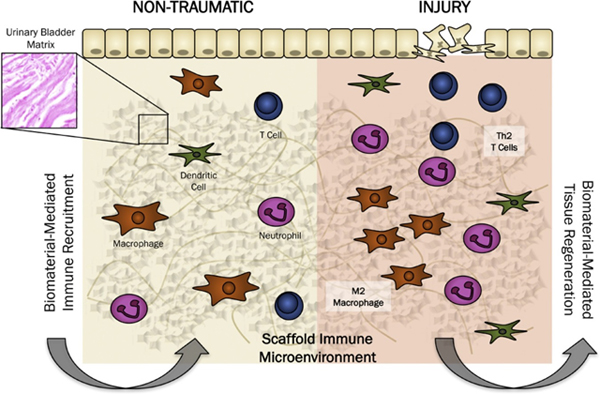
Abstract
Urinary bladder matrix (UBM) is used clinically for management of wounds and reinforcement of surgical soft tissue repair, among other applications. UBM consists of the lamina propria and basal lamina of the porcine urinary bladder, and is decellularized as part of the process to manufacture the medical device. UBM is composed mainly of Collagen I, but also contains a wide variety of fibrillar and basement membrane collagens, glycoproteins, proteoglycans and ECM-associated factors. Upon application of the biomaterial in a traumatic or nontraumatic setting in a mouse model, there is a cascade of immune cells that respond to the damaged tissue and biomaterial. Here, through the use of multicolor flow cytometry, we describe the various cells that infiltrate the UBM scaffold in a subcutaneous and volumetric muscle injury model. A wide variety of immune cells are found in the UBM scaffold immune microenvironment (SIM) including F4/80+ macrophages, CD11c+ dendritic cells, CD3+ T cells and CD19+ B cells. A systemic IL-4 upregulation and a local M2-macrophage response were observed in the proximity of the implanted UBM. The recruitment and activation of these cells is dependent upon signals from the scaffold and communication between the different cell types present.
Keywords: Biomaterials, Immunology, T cells, Macrophages
Graphical Abstract
1. Introduction
Urinary bladder matrix (UBM), a decellularized extracellular matrix (ECM), is used clinically in a variety of applications [1–5]. Clinical indications of the commercialized UBM include reinforcement of abdominal wall repair [6,7], management of diabetic ulcers [8], gastrointestinal tissue reinforcement [9], urologic and gynecologic surgical reinforcement, skin wounding [10], and management of deep, partial thickness burns [11]. Off-label, the material, and other ECM-derived scaffolds, have been used in treatment of larger soft tissue defects such as skeletal muscle repair [5,12,13], breast reconstruction [14], dural repair, subcutaneous injection with or without PRP (platelet-rich plasma) to induce hair growth [15], and tendon repair. These varied applications capitalize on a pro-regenerative host remodeling response that is induced when ECM scaffolds such as UBM are implanted within the body, although the determinants of this response are still being investigated.
ECM scaffolds are created by treating native tissue, often porcine or human, with a variety of acids and detergents to remove the majority of cellular components and leave behind a complex structural and signaling scaffold [4,16]. A large array of tissues have been decellularized for tissue engineering applications including but not limited to, urinary bladder, small intestinal submucosa, cardiac muscle, demineralized bone, and amnion. Urinary bladder matrix in particular can be synthesized either as a sheet (used for large surfaces) or particulate (used in skin wound management) and are currently being investigated in other forms such as hydrogels [3,17–19]. As they are derived from native tissue, ECM scaffolds such as UBM carry a structural and biochemical complexity that cannot be mimicked synthetically, and are likely an important factor in successful scaffold remodeling.
Current evaluations of UBM composition have shown that is composed primarily of collagens, interspersed with proteoglycans ECM sequestered growth factors, and cytokines [18,20,21]. While the constituents of these scaffolds can greatly affect their biologic activity and performance in the clinical setting, comprehensive characterization of their composition is incomplete. Currently, the primary methods used for determining components of ECM scaffolds are through histological staining, chemical assays, and enzyme-linked immunosorption assays (ELISA) of target proteins such as growth factors. Further understanding of the components of these scaffolds can be achieved through proteomic analysis [22], but the chemical nature of ECM proteins have made such characterizations challenging and incomplete. Contrary to typical substrates for proteomic analysis, ECM scaffolds are highly hydrophobic, difficult to solubilize, and are protease resistant, requiring more stringent protocols to prepare fragments that can be detected by mass-spectrometry. However, with a distinct profile of the proteins found in ECM scaffolds, we can better characterize the results seen in the clinic with the scaffold composition.
The proteomic composition of a material can be correlated with alterations in cell function, including immune responses [23–25]. Immune cells are the first responders to injury and biomaterial implantation. The immune microenvironment created by a scaffold will alter the presence of various cytokines and growth factors that can contribute to stem cell differentiation and tissue regeneration [26]. UBM materials are clinically implanted in areas of acute injury (i.e. an inflammatory environment) and in areas of chronic injury or to reinforce tissue (i.e. a more homeostatic environment). Each of these represent very different immune milieus that may influence the host response to the implanted UBM.
A wide variety of immune cells have been implicated in regeneration of murine muscle, liver, and salamander limbs [27–29]. In muscle tissue, we have shown the importance of Th2 polarized T cells [30], and Heredia et al. described the importance of eosinophils in muscle recovery after cardiotoxin injury [27]. In both cases, if elements of the type-2 immune response were depleted, there was a reduction in functional muscle fiber formation, replaced with fibrosis, ectopic adipogenesis and small irregularly shaped muscle fibers. Eosinophils have also been implicated as a major mediator of liver regeneration [28]. In the salamander, depletion of macrophages results in full inhibition of limb regeneration and deposition of a thick collagenous extracellular matrix at the limb bud [29]. Characterizing the scaffold immune microenvironment (SIM) of clinical grade materials would allow insight into the cell dynamics that could be expected in a clinical setting. In the context of extracellular matrix scaffolds, studies have demonstrated that these materials induce a pro-regenerative M2-macrophage phenotype that is dependent upon signaling from Th2 T cells [17,30,31]. The present study provides a detailed characterization of the proteomic composition of clinically utilized UBM-ECM scaffolds and of immune cell recruitment/polarization in sites of injury vs homeostasis.
2. Materials and methods
2.1. Mechanical testing and scanning electron microscopy
Sterile UBM particulate (MicroMatrix®) was obtained from ACell, Inc. (Columbia, MD). Rheology was performed with an Ares G2 rheometer (TA Instruments New Castle, DE). Experiments were conducted at physiological temperatures (37 °C) and performed in sequence. Freshly made samples of different UBM concentrations were loaded in 400 mg quantities on the rheometer stage. Experiments were conducted at a rheometer gap of 1.5 mm, using a 25 mm parallel plate geometry. Determination of linear viscoelastic region by a frequency sweep conducted from 0.1 rad/s to 15 rad/s at a constant oscillation strain of 0.5%, followed by a strain sweep at constant oscillation frequency 15 rad/s was performed. Finally, storage and lost moduli were observed over time using constant oscillation frequency 15 rad/s and oscillation strain 0.5%. Three samples per group were tested under the same rheological protocol. Samples were kept hydrated during these rheology experiments using a solvent trap.
Collagen from bovine tendon (Sigma) was obtained from a commercial supplier to use as a material control. Collagen and UBM samples were scanned using a double furnace, power compensation differential scanning calorimeter (DSC 8500, Perkin Elmer) fitted with an intracooler. Samples were individually weighed, placed in, and crimpsealed in manufacturer supplied aluminum pans and lids. Sample pans were placed in the DSC and heated at 5 °C per minute from 20 °C to 90 °C. An empty pan was used as reference.
The structure and surface topography of UBM particles was evaluated using scanning electron microscopy (SEM). Particles were adhered to aluminum stubs with copper tape and sputter coated with 20 nm Au/Pd. SEM was performed using LEO 1530 Field Emission Scanning Electron Microscope (Carl Zeiss, Jena, Germany) operating at 1–20 kV.
2.2. Proteomics
UBM proteomic composition was determined by mass spectometry of tryptic peptides derived from the matrix components of three separate manufacturing lots. UBM particles were suspended in 9 M urea (pH 8, 30 mM HEPES buffer) at a concentration of 2.5 mg ECM/ml, vortexed for 60 s, reduced with dithiothreitol (DTT, 5 mM final concentration), and sonicated (20 W output, 5 s on 20 s off for 12 cycles). Further reduction was performed by heating samples to 60 °C for 20 min followed by alkylation with iodoacetic acid (10 mM final concentration). Sample urea concentration was adjusted to 8 M with HEPES buffer for digestion with Lys-c (Wako Chemicals) for 4 h at 37 °C with agitation (1:110 ECM:Lys-c dry wt. ratio). Sample urea concentration was then further reduced to 2.5 M with HEPES buffer for digestion with trypsin (Promega Gold) overnight at 37 °C with agitation (1:50 trypsin:ECM dry wt. ratio). Small aliquots of samples were collected before and after enzymatic digestion for size evaluation by SDS-gel electrophoresis. Following digestion, samples were centrifuged for 10 min at 16,000 × g to confirm the absence of remaining insoluble material, and the supernatant collected.
Tryptic peptides for each lot of material were analyzed in triplicate runs using an Orbitrap Fusion Lumos mass spectrometer (Thermo Scientific). Peptides were separated by on-line reverse phase chromatography consisting of EASY-Spray analytical column (2 μm, 50 cm, Thermo Scientific) over a 150 min gradient. The data was acquired in data dependent manner in ‘top speed’ mode over 3 s. MS1 scans were acquired in Orbitrap analyzer at 120,000 resolution followed by MS2 scans also acquired in the Orbitrap analyzer at 60,000 resolution. Peptides were fragmented in HCD mode at 35% collision energy. Dynamic exclusion of 30 s was included in the method.
Peptide search, protein identification and label free quantification was carried out using MaxQuant software (Supplementary Table S1). Peptide search parameters were as follows: precursor mass error of 5 PPM and fragment mass error of 0.05 Da was allowed. Cysteine car-bamidomethylation was used as static modification while methionine oxidation and acetylation of protein n terminal were used as dynamic modifications. Refseq73 protein database for the species Sus scrofa with common contaminant proteins was used for peptide search. 1% FDR rate at PSM level and protein level was permitted. Replicate runs for each lot were combined for label free quantification using the intensity-based absolute quantification (iBAQ) method. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD005400 [32]. via. Gene level annotation was applied for each protein match and gene symbols beginning with LOC were manually matched to current gene annotations if available. Protein gene products were then categorized according to their function or homology in the ECM compartment according to a previously curated list [33], and are provided in Supplementary Table S1. These include the core ECM compartment consisting of collagens, proteoglycans, and glycoproteins, and the ECM associated compartment consisting of ECM affiliated proteins, ECM regulators, and secreted factors. All identified proteins not listed in these categories were characterized as non-ECM. Each lot of material was averaged for compositional analysis of the most abundant ECM components. Repeatability from lot-to-lot was calculated by weighted iBAQ intensity, which shows that >99% of the overall signal detected is derived from proteins identified in all 3 lots.
2.3. Cell culture
Hematopoetic progenitors were isolated from the bone marrow of wild type C57BL/6 mice. These cells were then cultured with 100 ng/ml M-CSF (BioLegend) in DMEM-F12 supplemented with 10% FBS and 1.0% penicillin-streptomycin. After 4 days, media and M-CSF was refreshed after washing away non-adherent cells with 1X PBS. At 7 days post-seeding, cells (bone marrow derived macrophages, BMDMs) were harvested with trypsin and plated onto ECM coated plates.
Cell culture dishes were coated with UBM particles by creating a 4–5 mg/ml suspension of particles in sterile distilled water. One ml of this suspension was added per well in a 6 well culture dish and allowed to dry overnight. Plates were UV sterilized for 1 h on each side, then washed with 1X PBS to remove non-adhered particles prior to cell seeding. M0 medium contained 100 ng/ml M-CSF in supplemented DMEM-F12. M1 polarizing media consisted of M0 media supplemented with 200 ng/ml bacterial lipopolysaccharide (LPS; 055:B5, Sigma) and 20 ng/ml interferon gamma (IFNg; Peprotech). The M2 polarizing medium was supplemented with 20 ng/ml interleukin 4 (IL-4; Peprotech).
2.4. Subcutaneous implantation
Particulate UBM was hydrated with PBS to 300 mg/ml. Hydrated scaffold was injected subcutaneously (0.2 ml) into the dorsal region of female wild type C57BL/6 mice at 6–7 weeks of age. Mice were anesthestized under 3% isoflurane and maintained at 2% isoflurane and oxygen for the duration of the surgery. Prior to injection, the dorsal side of the mouse was shaved and sterilized with 70% ethanol. After 1 and 3 weeks post-injection, animals were sacrificed and implants and surrounding tissue were dissected with or without the skin for histology and flow cytometry, respectively.
2.5. Volumetric muscle loss surgery
Six week old wild type female C57BL/6 mice (Charles River Laboratories) were used in all studies. Mice were anesthetized under 3.0% isoflurane and maintained at 2.0% isoflurane for the duration of the surgery. Hair was removed from both hindlimbs through the use of an electric razor. Skin was sanitized with 70% ethanol before making a 1 cm incision in the skin above the quadriceps muscle group. Using surgical scissors, a 3 mm × 4 mm wound was created and filled with 50 ul of a 300 mg/ml UBM particulate paste. The wound was stapled closed and procedure was repeated on the contralateral leg. After surgery, mice were injected subcutaneously with 5 mg/kg Rimadyl for pain relief and monitored until amubulatory. All procedures were done in accordance with guidelines provided by the Johns Hopkins University Animal Care and Use Committee.
2.6. Histology & immunofluorescent staining
Subcutaneous implants were incubated in 10% Formalin overnight prior to dehydration and mounting in paraffin. Five (5) micron sections were taken on a microtome (Leica) and stained with hematoxylin and eosin (Sigma).
For immunofluorescent staining of 1 week post-injection implant, 5 μm sections were re-hydrated and processed for antigen retrieval by treatment with a citrate antigen retrieval buffer (10 mM Citrate, pH 6) via microwave treatment. Sections were sequentially stained for B220 (rat monoclonal RA3–6B2, 1:5000 dilution, Biolegend), F4/80 (rat monoclonal BM8, 1:500 dilution, Biolegend), and CD3 (rabbit monoclonal SP7, 1:1000 dilution, abcam) using the tyramide signal amplification (TSA) system with Opal dye reagents (Perkin-Elmer). Each labeling step consisted of the following at room temperature: blocking for 30 min at (4% normal goat serum, 1% BSA, 0.05% Tween in TBS), incubation with primary antibody diluted in blocking buffer for 30 min (except for F4/80 which was incubated overnight at 4 °C), incubation in anti-rat or -rabbit HRP polymer conjugated secondary antibody (Biocare medical) for 10 min, TSA amplification with Opal reagent (Opal-520/B220, Opal-570/F/480, Opal-650/CD3), and finally antibody stripping via microwave treatment in citrate buffer. Extensive washing in TBS/0.05% Tween was performed following each antibody. After coverslipping, slides were imaged at 50X for mosaic generation and at 200X at the host-implant interface along dorsal, ventral, and edge surfaces and within the center of the implant.
2.7. Flow cytometry
Subcutaneous implants and muscle samples were diced finely with a scalpel before incubation in 0.5 mg/ml Liberase TL (Roche) and 0.2 mg/ml DNase I (Roche) in RPMI media for 45 min with agitation at 37 °C. Resultant material was filtered through a 100 μm cell strainer and washed twice with 1X PBS. To remove debris the suspension was subjected to density separation with Lympholyte-M (CedarLane Labs) as per manufacturer’s instructions. Interphase was washed twice with PBS before staining with the following panels. Cell population analysis: CD3 Alexa Fluor-488 (BioLegend), CD19 BrilliantViolet-421 (BioLegend), CD11c APC/Cy7 (BD Biosciences), F4/80 PE/Cy7 (BioLegend), CD34 Per/CP-Cy5.5 (BioLegend), CD86 Alexa Fluor-700 (BioLegend), CD206 APC (BioLegend) and Viability Aqua (Thermo). Cells were then fixed with BD Cytofix prior to analysis on a LSRII flow cytometer (BD). For T Cell analysis, cells were stained with CD3 Alexa Fluor-488 (BioLegend), CD8 Alexa Fluor 700 (BioLegend), CD4 PE/Cy7 (BioLegend), and Viability Aqua (Thermo); then, the cells were fixed as per previously described and permeabilized with the Cytofix/Perm reagent for staining with FoxP3 Pacific Blue (BioLegend) prior to analysis on a LSRII flow cytometer. For analysis of MHCII expression on myeloid cells, samples were stained with F4/80 PE/Cy7 (BioLegend), CD206 PE (BioLegend), and MHCII AlexaFluor 488 (BioLegend) and Viability eFluor 780 (eBioscience) prior to analysis as stated above.
2.8. Gene expression analysis
Cell culture samples were harvested in 1 ml TRIzol, and mixed with 200 μl Chloroform before spinning at 12,000 × g for 15 min. The aqueous phase was added to 500 ul 70% ethanol and run through an RNeasy Spin column. The RNA was cleaned and eluted as per manufacturer’s instructions. cDNA was synthesized using SuperScript RTIII reagents. Gene expression was determined using the following primers with a SYBR Green reporter dye: B2m forward CTC GGT GAC CCT GGT CTT TC, B2m reverse GGA TTT CAA TGT GAG GCG GG; Tnfa forward GTC CAT TCC TGA GTT CTG, Tnfa reverse GAA AGG TCT GAA GGT AGG; Il1b forward GTA TGG GCT GGA CTG TTT C, Il1b reverse GCT GTC TGC TCA TTC ACG; Arg1 forward CAG AAG AAT GGA AGA GTC AG, Arg1 reverse CAG ATA TGC AGG GAG TCA CC; Retnla forward CTT TCC TGA GAT TCT GCC CCA G, Retnla reverse CAC AAG CAC ACC CAG TAG CA; Il10 forward TCT CAC CCA GGG AAT TCA AA, Il10 reverse AAG TGA TGC CCC AGG CA; Inos forward GAC GAG ACG GAT AGG CAG AG, Inos reverse GTG GGG TTG TTG CTG AAC TT.
2.9. Statistics
One-way ANOVA and Students T-test were performed in GraphPad Prism operating with a P value < 0.05.
3. Results
3.1. Mechanical properties and proteomic analysis of UBM scaffolds
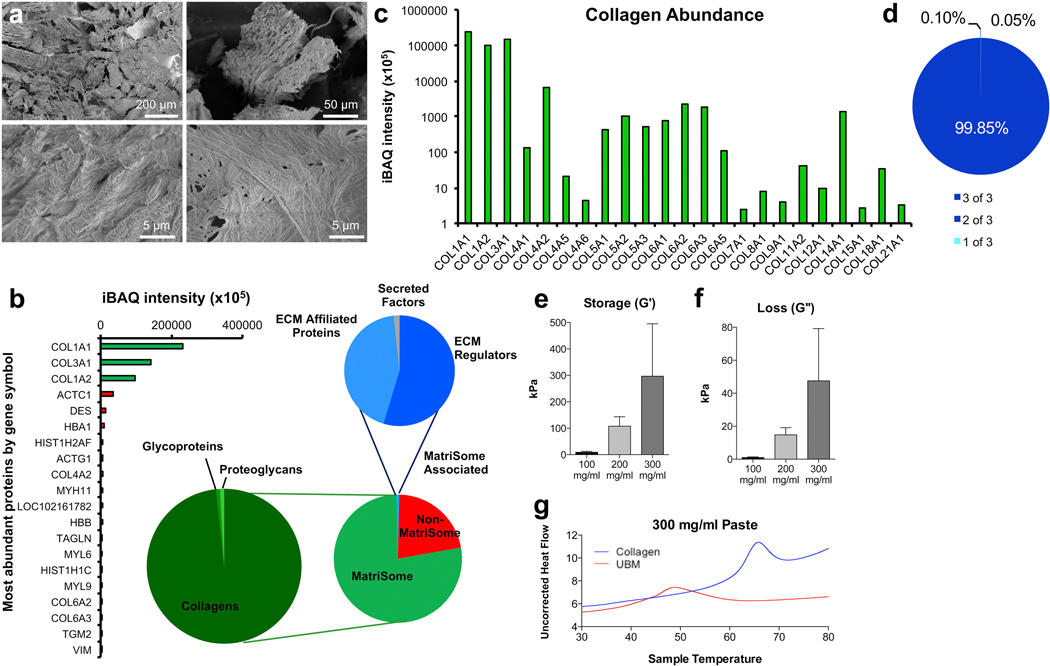
As urinary bladder matrix (UBM) is synthesized from native tissue, we analyzed the physical and mechanical properties, along with the proteomic composition, to determine its macro- and micro-architecture. SEM analysis showed that UBM particulates were composed of both fibrous strands and sheet-like connective tissue components characteristic of collagen strands and basement membrane components, respectively (Fig. 1a). Proteomic analysis was conducted to determine the protein components within the UBM scaffold (Fig. 1b–d). Over 500 proteins were identified via mass spectrometry, with 78% identified as MatriSome and MatriSome-associated proteins (Fig. 1b). Within the MatriSome category of proteins (77% of total), 98% were collagens, 1% ECM glycoproteins, and 1% proteoglycans. Within the MatriSome-Associated fraction (1% of total) 55% were ECM regulators, 44% ECM affiliated proteins, and 2% secreted factors. The most abundant nonMatriSome proteins included actin, desmin, and hemoglobin, all of which are present in high abundance in the native bladder tissue [34]. As collagens represented the majority of MatriSome proteins, we further categorized the different collagen types present in UBM (Fig. 1c). High-abundance collagens, such as Type I Collagen and Type III Collagen, were detected at the highest frequency. Additionally, we detected basement membrane-specific Type IV collagen at high levels, characteristic of the natural pre-processing bladder matrix. These protein identifications were highly reproducible, with 99.85% of proteins (by iBAQ intensity) being common to all 3 lots tested, and only 0.15% present in either 1 of 3 or 2 of 3 lots (Fig. 1d).
Fig. 1.
UBM materials characterization. (a) SEM of particulate UBM. (b) Proteomic evaluation of the proteins with highest abundance in UBM and categories of proteins detected. (c) Collagen abundance in UBM (d) Repeatability of iBAQ detection of peptides in 3 different lots of UBM. (e and f) Storage and Loss moduli of 100, 200 and 300 mg/ml ECM pastes. (g) Differential scanning calorimetry hydrated 300 mg/ml paste UBM compared to a milled collagen I control. For rheology, data are means ± SEM, n = 3 samples per group. For proteomics, data are means of 3 lots tested in 3 mass-spectrometry runs per lot.
Rheological characterization of the UBM pastes was performed to determine the relationship between total ECM content and mechanics. Storage modulus (G’) increased non-linearly with increasing UBM concentrations. A near ten-fold increase in storage modulus was observed between UBM pastes of 100 mg/ml compared to 200 mg/ml; correspondingly, 300 mg/ml UBM materials also demonstrated higher storage moduli, roughly three times stronger (G’ = 300 kPa) than 200 mg/ml materials (Fig. 1e). Higher UBM concentrations also increase viscosity of the paste resulting in an increase in loss modulus (Fig. 1f).
Differential Scanning Calorimetry analyses demonstrated significant differences between bovine tendon collagen and UBM, in both denaturation temperatures and enthalpies of denaturation. Thermographs obtained from hydrated samples revealed a lower melting point of 48.56 °C in 300 mg/ml UBM samples as compared to 65.65 °C for type I collagen samples at the same concentration (Fig. 1g). Lower denaturation temperature of the UBM may be attributed to disruption of the overall ECM structure during their processing and due to the complex composition of UBM. The denaturation peaks in UBM samples were broader and the enthalpy of denaturation was higher in UBM (8.93 J/g) as compared to collagen (5.88 J/g), expected in part due to interactions between various protein and proteoglycan components of the UBM.
3.2. Immunomodulation of macrophage behavior by UBM scaffolds
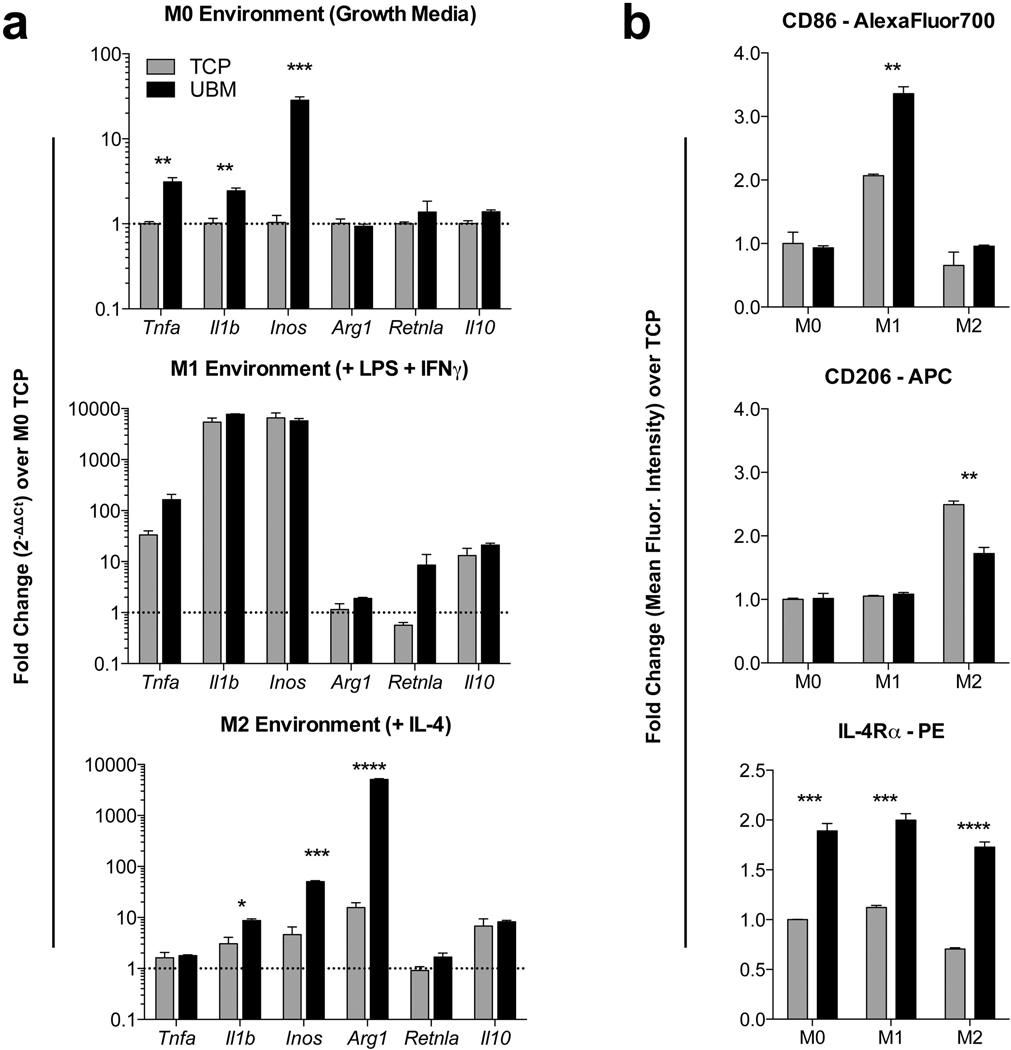
Macrophages are phagocytic cells of the innate immune system that respond to stimuli in their environment such as pathogen presence or wounding. Additionally, these cells play a key role in the foreign body response [35,36]. As macrophages form part of the body’s first line of defense, we cultured murine bone marrow derived macrophages (BMDM) on particulate UBM to determine if there is a direct effect of the scaffold on macrophage behavior. Macrophages were cultured on UBM-coated tissue culture plastic for 24 h prior to gene expression analysis via RT-PCR. We analyzed genes associated with M1 (inflammatory, classical activation) and M2 (anti-inflammatory, alternative activation) polarization phenotypes, Tnfa, Il1b and Inos or Arg1, Retnla and Il10, respectively (Fig. 2). In M0, unstimulated conditions, UBM upregulated M1 inflammatory genes, but did not alter M2 gene expression. However, in M2 media conditions (+IL-4) UBM induced a higher expression of Arg1, a canonical M2 marker, suggesting a mixed phenotype that does not fit into a specific M1- or M2-promoting material (Fig. 2a). To further test macrophage response to UBM, we analyzed the expression of CD86 (M1) and CD206 via flow cytometry. As with RT-PCR, in vitro, UBM promotes expression of CD86, an M1 marker, and decreases expression of CD206, an M2 marker. However, UBM did up-regulate expression of IL-4Rα in all 3 media conditions, again confirming the mixed M1/M2 phenotype (Fig. 2b).
Fig. 2.
Particulate UBM induces alterations in macrophage phenotype in vitro. (a) RT-PCR analysis of M1 (Tnfa, Il1b, Inos) and M2 (Arg1, Retnla, Il10) genes in bone-marrow derived macrophages cultured on UBM for 24 h in M0 (growth, unstimulated), M1 (inflammatory, LPS+IFNg) or M2 (anti-inflammatory, IL-4) media conditions. (b) Flow cytometric analysis of CD86 (M1), CD206 (M2) and IL4ra (M2) on macrophages at 24 h post-seeding on UBM coated tissue culture plastic (TCP). Data are means ± SEM, n = 4 mice per group.
3.3. Scaffold immune microenvironment of subcutaneous UBM
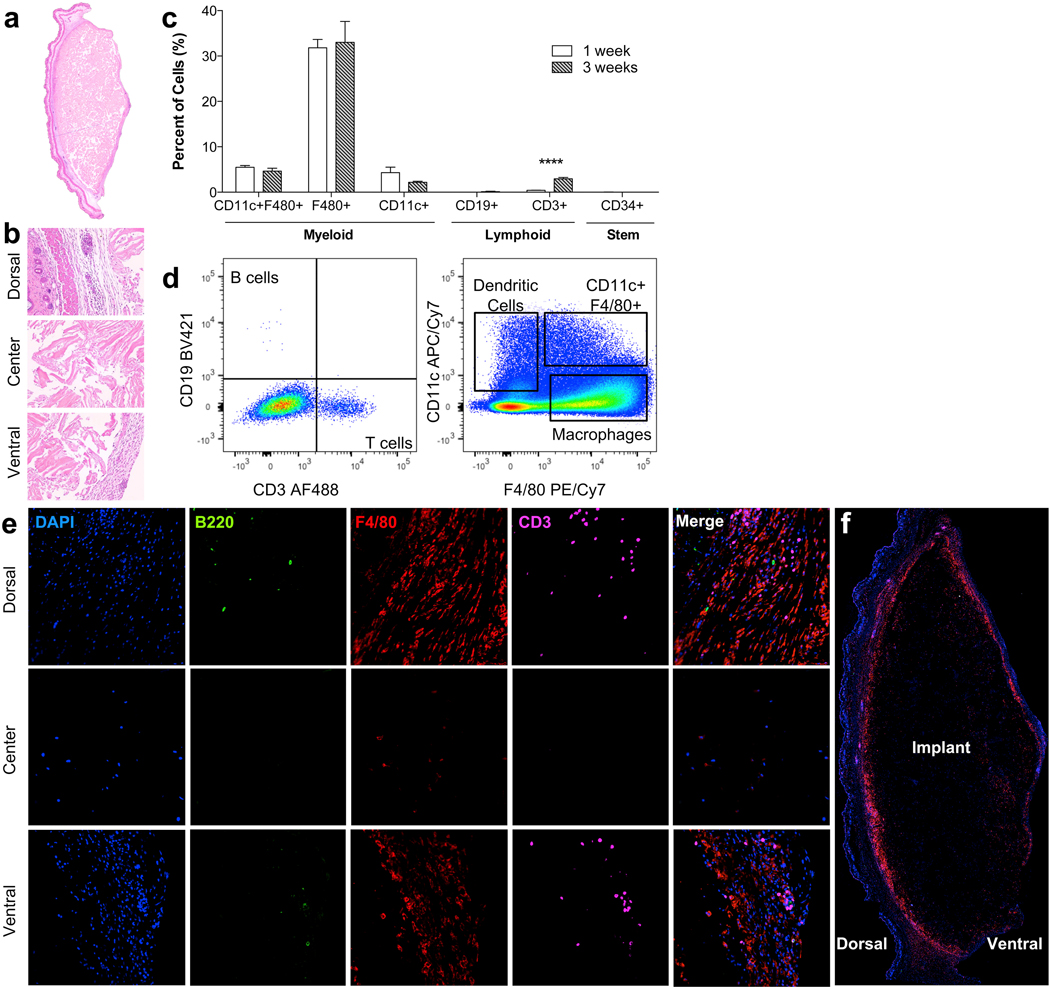
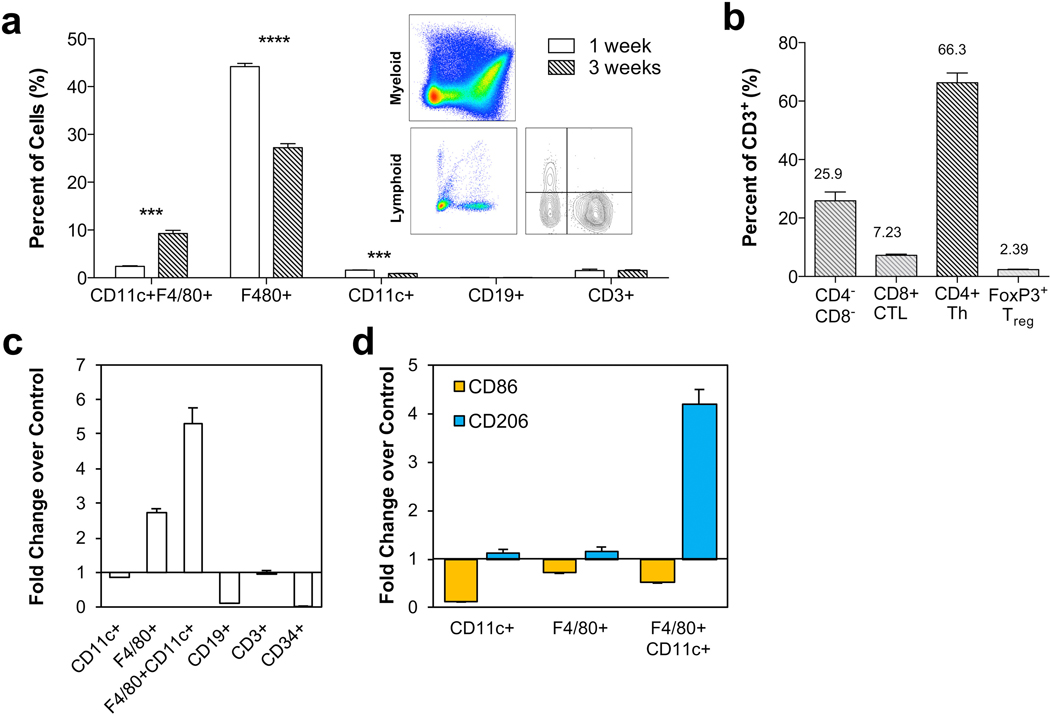
The immune response is a complex coordination of local and systemic effects from multiple cell types involving many signaling molecules. Therefore, it is hard to recapitulate in an in vitro setting using isolated cell types. To define the scaffold immune microenvironment (SIM) of UBM in non-injury setting, we injected 0.2 cc of a 300 mg/ml particulate UBM paste in the subcutaneous space of C57BL/6 WT mice (Figs. 3 and 4). Histologically, by 1 week post-injection a thin capsule had formed around the implant adhering it tightly to the underside of the skin (Fig. 3a). Cellular infiltration was present through the center of the scaffold by 3 weeks post-injection (Fig. 3b). Using flow cytometry, we were able to identify these cells that formed the UBM-SIM. After mechanical and enzymatic separation, a single cell suspension was stained for the following markers: CD3 (T cells), CD19 (B cells), CD34 (progenitor and endothelial cells), CD11c (dendritic cells), F4/80 (macrophages), CD86 (M1 macrophages) and CD206 (M2 macrophages). We detected a high proportion of F4/80+CD11c+/− macrophages in the UBM-SIM at 1 and 3 weeks post-injection (Fig. 3c and d). Low numbers of CD11c+F4/80− dendritic cells were also detected. In addition to macrophages and dendritic cells (cells of the innate immune system) we detected both CD3+ T cells and CD19+ B cells suggesting activation of the adaptive immune response (Fig. 3c and d). We confirmed the presence of Macrophages (F4/80+, red), T cells (CD3+, magenta) and B cells (B220+, green) through immunofluorescence staining of the subcutaneous implant at 1 week post-injection (Fig. 3e and f). B cells and T cells were found exclusively in the capsule, often in cellular clusters as seen in the H & E staining of the dorsal surface of the implant. F4/80hi macrophages were present around the implant, with low numbers of F4/80+ cells within the implant, and notably lower expression levels of F4/80. As with flow cytometric analysis, macrophages were most abundant, followed by T cells and very few B cells.
Fig. 3.
The scaffold immune microenvironment of UBM. C57BL/6 mice received 200 ul of a 300 mg/ml subcutaneous UBM implant. (a) Cross-section of subcutaneous UBM implant at 1 week post-injection. (b) Dorsal (skin), center, and ventral (capsule) sections of UBM implant at 1 week post-injection. (c) FACS analysis of resident immune cells at 1 and 3 weeks post-injection showing high presence of F4/80+ macrophages and an increase in CD3+ T cells over time. (d) Representative FACS plots from data quantified in (c). (e) Immunofluorescence (IF) staining of implant at 1 week post-injection showing localization of B cells (B220, green), macrophages (F4/80, Red), and T cells (CD3, magenta). (f) Composite cross-section of IF-stained implant at 1 week post-injection.
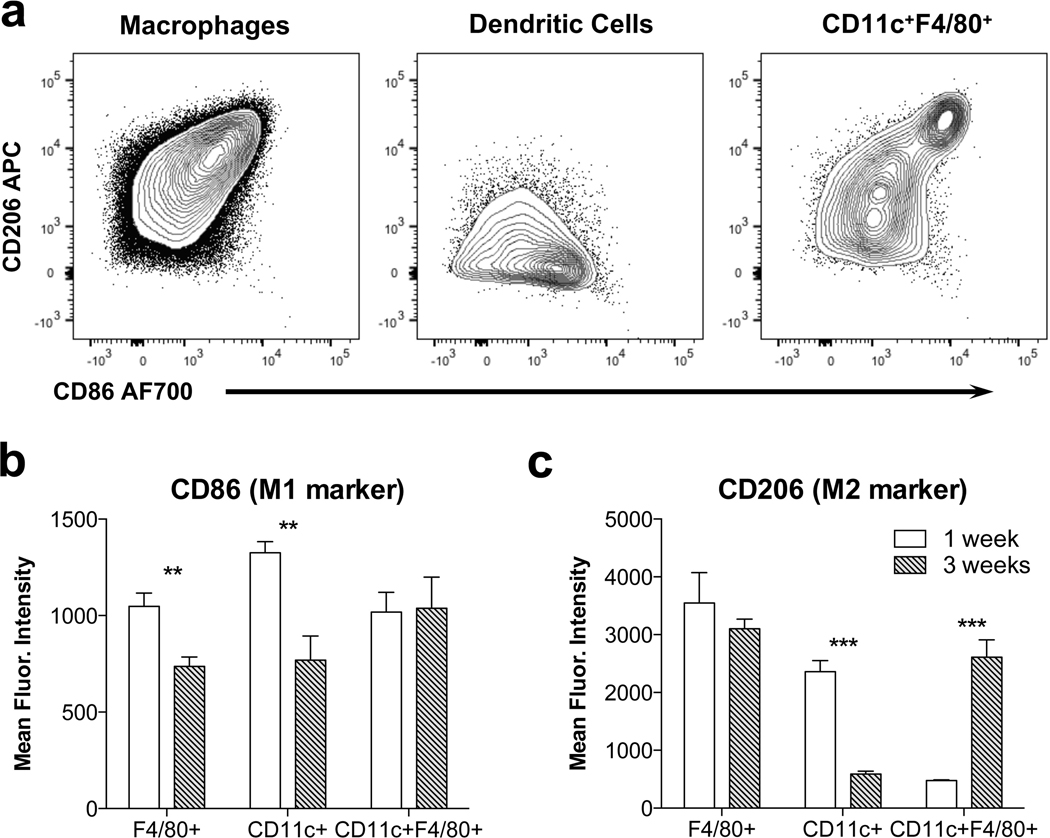
Fig. 4.
UBM promotes an M2-macrophage phenotype that matures over time. (a) CD206 and CD86 expression on 3 myeloid subtypes detected in the implant at 3 weeks post-injection. (b) CD86 expression quantified as mean fluorescence intensity (MFI) at 1 and 3 weeks post-injection. (c) CD206 expression quantified as MFI. Data are means ± SEM, n = 4 mice per group.
To further evaluate the UBM-SIM, we analyzed the expression of CD86 and CD206 on myeloid cells (Fig. 4). F4/80+ macrophages expressed intermediate levels of CD206, with more mature macrophages (F4/80hi) expressing higher levels of CD206 (CD206hi). These macrophages also expressed CD86, an M1 marker, which decreased over time (Fig. 4b). Expression of the M2 and M1 macrophage markers was not mutually exclusive suggesting a complex phenotype that does not neatly categorize into a distinct “type-1” or “type-2” macrophage. Additionally, CD11c+ dendritic cells were detected with low CD206 expression and high CD86 expression. CD11c+F4/80+ macrophages also expressed both CD86 and CD206.
3.4. UBM macrophage and T cell recruitment in traumatic muscle injury
UBM is clinically applied in cases of tumor resection and soft tissue loss, which often follow traumatic injury, either from the surgical removal of cancerous tissue or from wounding. To model the traumatic environment of these clinical applications, we applied the UBM scaffold to a murine model of volumetric muscle loss (VML). A 3 mm × 4 mm portion of the quadriceps muscle was removed using surgical scissors, and the resulting defect was filled with 0.05 cc of 300 mg/ml UBM. After 1 and 3 weeks post-injury, the scaffold and surrounding area were harvested for analysis of the SIM via flow cytometry. As with the subcutaneous implantation, response to the UBM scaffold was dominated by F4/80+CD11c+/− macrophages, followed by CD11c+ dendritic cells, CD3+ T cells and CD19+ B cells (Fig. 5a). There was a peak level of macrophage infiltration at 1 week post-injury which decreased by 3 weeks post-injury. CD3+ T cells were present at 1 week post-injury, and persisted after 3 weeks. Of these CD3+ T cells, the majority were CD4+ helper T cells (Fig. 5b, 66.3%) compared to CD8+ cytotoxic T lymphocytes (7.23%) and double negative cells (25.9%, mainly natural killer T cells). Additionally, CD4+FoxP3+ Tregs were present in low numbers (2.39% of total CD3+ cells). Compared to a saline-treated control wound, UBM induced higher levels of F4/80+ and F4/80+CD11c+ macrophages at 3 weeks post-injury (Fig. 5c). Additionally, UBM promoted lower levels of CD86 expression and higher levels of CD206 expression, suggesting a more M2-like phenotype compared to a saline-treated control (Fig. 5d).
Fig. 5.
UBM-treated muscle wounds recruit a diverse immune cell repertoire. C57BL/6 WT mice received a bilateral 3 mm × 4 mm muscle removal from their quadriceps muscle group which was back filled with 50 ul of a 300 mg/ml UBM paste and analyzed via flow cytometry. (a) Immune cell populations at 1 and 3 weeks post-injury. (b) CD4:CD8 ratio at 3 weeks post-injury. (c) Comparison of cell populations in UBM-treated wound versus saline-treated control at 3 weeks post-injury. Data displayed as fold change over control. (d) Comparison of CD86 (M1 marker) and CD206 (M2 marker) expression in UBM-treated wound versus saline-treated control at 3 weeks post-injury. Data displayed as fold change over control. Data are means ± SEM, n = 4 mice per group.
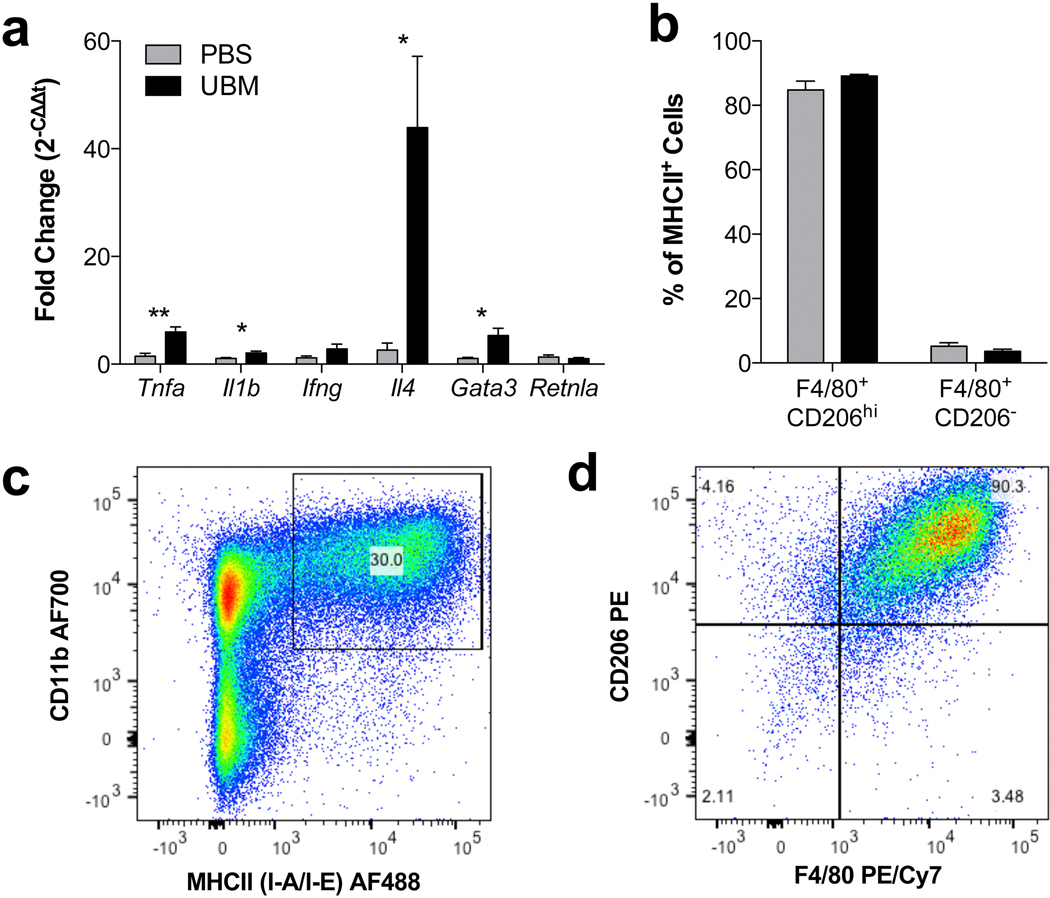
As the SIM recruited high numbers of CD3+ T cells, we measured gene expression in the distal (axillary/brachial) lymph nodes to determine if there was a systemic activation of the immune system by the UBM scaffold (Fig. 6a). Compared to the saline-treated control, mice treated with UBM induced a systemic up-regulation of Th2-associated genes, Il4 and Gata3, in the distal lymph nodes. Genes associated with Th1 T cells, such as Tnfa and Il1b were also up-regulated, but not to the same extent as IL-4, suggesting an activation and polarization of T cells towards a Th2 lineage. To probe the activation of the adaptive immune system, we evaluated the expression levels of MHCII (I-A/I-E), a protein complex that is responsible for antigen presentation to CD4+ T cells (Fig. 6b–d). In the UBM-SIM, macrophages expressed higher levels of MHCII than the canonical CD11c+ dendritic cells, suggesting communication between the local macrophages and lymph node Th2T cells.
Fig. 6.
UBM induces a systemic IL-4 upregulation correlated with local antigen-presenting M2-macrophages. (a) RT-PCR of distal lymph nodes (axillary/brachial) at 3 weeks post-injury displayed as RQ to saline treated VML control. (b) Percent (%) of MHCII+ cells that are present in the scaffold immune microenvironment at 1 week post-injury. (c) MHCII+ expression is selectively detected on CD11b+ myeloid cells. (d) MHCII+ cells are mainly F4/80+CD206+ M2 macrophages, representative FACS plot of data quantified in (b). Data are means ± SEM, n = 4 mice per group.
4. Discussion
Clinically used biomaterials were historically designed in the context of a foreign body response by the immune system. The foreign body response, extensively characterized by James Anderson in the 1980’s, described immune infiltration by neutrophils and macrophages resulting in oxidation and degradation of the biomaterial, foreign body giant cell formation, tissue-damaging inflammation, and fibrous encapsulation [35–37]. However, biomaterials are being leveraged in many forms to modulate the immune system. For example, synthetic nanoparticles are used as delivery vehicles for drugs and peptides that can alter the immune response, especially in the case of cancer immunotherapy [38–40]. Macroscopically, solid scaffolds are being investigated with applications in vaccines in oncology, acting as peripheral hubs of activation and polarization that educate the immune system [41–43]. New techniques to screen the immune response to biomaterials are being developed to provide a baseline of activity onto which immunomodulatory signals can be integrated [44]. The role of the immune system in tissue repair is providing new insights on key parameters in biomaterials design for regenerative medicine. Unexpected immune cell types such as eosinophils have even been implicated in tissue repair [28]. Specific to ECM-derived biological scaffolds, research over the past 10 years has correlated specific macrophage polarization and phenotypes with regenerative efficacy of the implants [7,45–47]. The intersection of immunomodulatory biomaterials research in device compatibility, cancer, vaccines and now regeneration will lead to a new generation of materials development.
The inherent biocompatibility described by ECM scaffolds is hypothesized to be due in part to maintenance of the natural ECM structure and composition. UBM particulate in the present study was found to consistently possess a multilayer sheet-like morphology indicating preservation of this structure. Likewise, proteomic analysis showed a highly complex compositional profile. The structural Type I and Type III collagens were most abundant, however numerous other collagen types were identified. These include basement membrane specific Collagen IV at high levels, and Collagen VII, albeit at low levels, as well as laminins, consistent with immunohistochemical studies of UBM [48,49]. This suggests, that ECM components survived decellularization during manufacturing. Numerous non-ECM components were also detected. Although proteomics is a comprehensive characterization technique, a potential limitation of this analysis is the poor solubility of ECM components and dearth of unique fragments for identification compared to intracellular and membrane components. Additional processing considerations are necessary for mass spectrometry analysis, and previous attempts to characterize UBM may have been biased towards the more easily solubilized and digested non-ECM components [50]. Non-ECM components were also found in this analysis, and may contribute to the immune modulating milieu. Smooth muscle actin and desmin are likely residuals from the smooth muscle layer of the bladder, and can act as damage associated molecular patterns (DAMPs) activating wound-healing and anti-inflammatory immune responses by signaling a type-2 polarization of recruited immune cells such as M2 macrophages and Th2T cells [51]. However, mass spectrometry only characterizes primary structure and does not infer protein functionality, and future studies are necessary to determine which components are directly responsible for observed responses.
In our in vitro studies, we noted a preference towards an M1 polarization when bone marrow-derived macrophages were exposed to particulate UBM. This is in contrast to previous studies that have used pepsin-digested UBM. As the material is being presented to the macrophages in a different form (solid particulates as opposed to protease digested soluble material) this could account for the differences noted in our study. Additionally, we have previously shown a dependence of macrophages upon the adaptive immune system, specifically Th2 T cells, to polarize towards an M2-phenotype [30,52]. In the absence of T cells, macrophages substantially decreased several markers of the M2-phenotype including surface CD206 expression, and transcript levels of Arg1, Retnla, and Cebpb.
Both subcutaneous and muscle-injury applications of UBM recruited large numbers of F4/80+ macrophages, that were CD86+CD206hi. Additionally, UBM recruited cells of the adaptive immune system including T cells and B cells, predominately CD4+ T cells. In addition to the local M2 macrophage phenotype, there was a systemic up-regulation of IL-4, a canonical Th2 cytokine, in the distal lymph nodes. The local M2 macrophages expressed high levels of MHCII, and were the dominant antigen-presenting cell in the UBM-SIM. Normally, M1 macrophages are more associated with high levels of MHCII, however in the UBM-SIM, there is an up-regulation of M2-macrophage communication with the adaptive immune system, which correlates with the systemic IL-4 detection. This suggests a macrophage-dependent T cell activation that in turn feeds back onto the macrophage population to further promote a pro-regenerative niche.
Formulated into different concentrations, UBM paste adopts dose-dependent rheological properties. These differing mechanical properties could endow the material with different immunological properties. Further research would involve determining the immunologic effects of scaffold modification, from differing concentrations of material, to physical modifications such as gelation [18] or combination with synthetics [53]. Understanding the structure-function relationship of scaffold modification could allow researchers to leverage material delivery and structural properties with immune response to determine the optimal configuration for the desired application.
In summary, a clinical formulation of UBM was found to possess a highly diverse repertoire of ECM and non-ECM components. This UBM was found to recruit high proportions of M2 macrophages when implanted in injured and uninjured tissues. Additionally, adaptive immune T and B cells were also detected suggesting a role for antigen specificity to the remodeling response. Further investigation is necessary to determine whether specific ECM antigen recognition plays a role in ECM induced immune responses.
Supplementary Material
Acknowledgments
The authors would like to acknowledge ACell Inc. for providing UBM material and funding for this study. The Sidney Kimmel Comprehensive Cancer Center Flow Cytometry Core, Ada Tam, and Lee Blosser for guidance in flow cytometric studies. The Michael Delannoy and Barbara Smith at the Institute for Basic Biomedical Sciences microscope facility for assistance in electron microscopy. Jeeyoen Sohn for assistance with qRT-PCR studies. We acknowledge support of the Center for Proteomics Discovery at the Johns Hopkins University and shared instrumentation grant S10OD021844. M.T.W. would like to acknowledge support from the Hartwell Foundation Postdoctoral Fellowship.
Footnotes
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.smim.2017.05.002.
References
- [1].Badylak SF, Freytes DO, Gilbert TW, Extracellular matrix as a biological scaffold material: structure and function, Acta Biomater. 5 (1) (2009) 1–13. [DOI] [PubMed] [Google Scholar]
- [2].Gilbert TW, Stolz DB, Biancaniello F, Simmons-Byrd A, Badylak SF, Production and characterization of ECM powder: implications for tissue engineering applications, Biomaterials 26 (12) (2005) 1431–1435. [DOI] [PubMed] [Google Scholar]
- [3].Zhang L, Zhang F, Weng Z, Brown BN, Yan H, Ma XM, Vosler PS, Badylak SF, Dixon CE, Cui XT, Chen J, Effect of an inductive hydrogel composed of urinary bladder matrix upon functional recovery following traumatic brain injury, Tissue Eng. Part A 19 (17–18) (2013) 1909–1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Gilbert TW, Sellaro TL, Badylak SF, Decellularization of tissues and organs, Biomaterials 27 (19) (2006) 3675–3683. [DOI] [PubMed] [Google Scholar]
- [5].Sicari BM, Rubin JP, Dearth CL, Wolf MT, Ambrosio F, Boninger M, Turner NJ, Weber DJ, Simpson TW, Wyse A, Brown EH, Dziki JL, Fisher LE, Brown S, Badylak SF, An acellular biologic scaffold promotes skeletal muscle formation in mice and humans with volumetric muscle loss, Sci. Transl. Med 6 (234) (2014) 234ra58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Agrawal V, Almond PS, Reyna R, Emran MA, Successful three stage repair of a large congenital abdominal region defect, J. Pediatr. Surg. Case Rep 3 (6) (2015) 230–233. [Google Scholar]
- [7].Brown BN, Londono R, Tottey S, Zhang L, Kukla KA, Wolf MT, Daly KA, Reing JE, Badylak SF, Macrophage phenotype as a predictor of constructive remodeling following the implantation of biologically derived surgical mesh materials, Acta Biomater. 8 (3) (2012) 978–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Frykberg RG, Cazzell SM, Arroyo-Rivera J, Tallis A, Reyzelman AM, Saba F, Warren L, Stouch BC, Gilbert TW, Evaluation of tissue engineering products for the management of neuropathic diabetic foot ulcers: an interim analysis, J. Wound Care 25 (Suppl. 7) (2016) S18–25. [DOI] [PubMed] [Google Scholar]
- [9].Afaneh C, Abelson J, Schattner M, Janjigian YY, Ilson D, Yoon SS, Strong VE, Esophageal reinforcement with an extracellular scaffold during total gastrectomy for gastric cancer, Ann. Surg. Oncol 22 (4) (2015) 1252–1257. [DOI] [PubMed] [Google Scholar]
- [10].Lanteri PA, Abernathie B, Datiashvili R, The use of urinary bladder matrix in the treatment of complicated open wounds, Wounds 26 (7) (2014) 189–196. [PubMed] [Google Scholar]
- [11].Kim JS, Kaminsky AJ, Summitt JB, Thayer WP, New innovations for deep partial-thickness burn treatment with ACell MatriStem Matrix, Adv. Wound Care 5 (12) (2016) 546–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Mase VJ Jr., Hsu JR, Wolf SE, Wenke JC, Baer DG, Owens J, Badylak SF, Walters TJ, Clinical application of an acellular biologic scaffold for surgical repair of a large, traumatic quadriceps femoris muscle defect, Orthopedics 33 (7) (2010) 511. [DOI] [PubMed] [Google Scholar]
- [13].Dziki J, Badylak S, Yabroudi M, Sicari B, Ambrosio F, Stearns K, Turner N, Wyse A, Boninger ML, Brown EH, An acellular biologic scaffold treatment for volumetric muscle loss: results of a 13-patient cohort study, npj Regen. Med 1 (2016) 16008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Salzberg CA, Nonexpansive immediate breast reconstruction using human acellular tissue matrix graft (AlloDerm), Ann. Plast. Surg 57 (1) (2006) 1–5. [DOI] [PubMed] [Google Scholar]
- [15].Rose PT, Hair restoration surgery: challenges and solutions, Clin. Cosmet. Invest. Dermatol 8 (2015) 361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Crapo PM, Gilbert TW, Badylak SF, An overview of tissue and whole organ decellularization processes, Biomaterials 32 (12) (2011) 3233–3243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Faulk DM, Londono R, Wolf MT, Ranallo CA, Carruthers CA, Wildemann JD, Dearth CL, Badylak SF, ECM hydrogel coating mitigates the chronic inflammatory response to polypropylene mesh, Biomaterials 35 (30) (2014) 8585–8595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Freytes DO, Martin J, Velankar SS, Lee AS, Badylak SF, Preparation and rheological characterization of a gel form of the porcine urinary bladder matrix, Biomaterials 29 (11) (2008) 1630–1637. [DOI] [PubMed] [Google Scholar]
- [19].Wolf MT, Carruthers CA, Dearth CL, Crapo PM, Huber A, Burnsed OA, Londono R, Johnson SA, Daly KA, Stahl EC, Freund JM, Medberry CJ, Carey LE, Nieponice A, Amoroso NJ, Badylak SF, Polypropylene surgical mesh coated with extracellular matrix mitigates the host foreign body response, J. Biomed. Mater. Res Part A 102 (1) (2014) 234–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Dahms S, Piechota H, Dahiya R, Lue T, Tanagho E, Composition and biomechanical properties of the bladder acellular matrix graft: comparative analysis in rat, pig and human, Br. J. Urol 82 (1998) 411–419. [DOI] [PubMed] [Google Scholar]
- [21].Wilson CB, Leopard J, Cheresh DA, Nakamura RM, Extracellular matrix and integrin composition of the normal bladder wall, World J. Urol 14 (1) (1996) S30–S37. [DOI] [PubMed] [Google Scholar]
- [22].Naba A, Clauser KR, Hoersch S, Liu H, Carr SA, Hynes RO, The matrisome: in silico definition and in vivo characterization by proteomics of normal and tumor extracellular matrices, Mol. Cell. Proteom. MCP 11 (4) (2012) M111.014647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Watt FM, Huck WT, Role of the extracellular matrix in regulating stem cell fate, Nat. Rev. Mol. Cell Biol 14 (8) (2013) 467–473. [DOI] [PubMed] [Google Scholar]
- [24].Kleinman HK, Philp D, Hoffman MP, Role of the extracellular matrix in morphogenesis, Curr. Opin. Biotechnol 14 (5) (2003) 526–532. [DOI] [PubMed] [Google Scholar]
- [25].Beachley VZ, Wolf MT, Sadtler K, Manda SS, Jacobs H, Blatchley MR, Bader JS, Pandey A, Pardoll D, Elisseeff JH, Tissue matrix arrays for high-throughput screening and systems analysis of cell function, Nat. Methods 12 (12) (2015) 1197–1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Sadtler K, Singh A, Wolf MT, Wang X, Pardoll DM, Elisseeff JH, Design, clinical translation and immunological response of biomaterials in regenerative medicine, Nat. Rev. Mater 1 (2016) 16040. [Google Scholar]
- [27].Heredia JE, Mukundan L, Chen FM, Mueller AA, Deo RC, Locksley RM, Rando TA, Chawla A, Type 2 innate signals stimulate fibro/adipogenic progenitors to facilitate muscle regeneration, Cell 153 (2) (2013) 376–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Goh YP, Henderson NC, Heredia JE, Red Eagle A, Odegaard JI, Lehwald N, Nguyen KD, Sheppard D, Mukundan L, Locksley RM, Chawla A, Eosinophils secrete IL-4 to facilitate liver regeneration, Proc. Natl. Acad. Sci. U. S. A 110 (24) (2013) 9914–9919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Godwin JW, Pinto AR, Rosenthal NA, Macrophages are required for adult salamander limb regeneration, Proc. Natl. Acad. Sci. U. S. A 110 (23) (2013) 9415–9420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Sadtler K, Estrellas K, Allen BW, Wolf MT, Fan H, Tam AJ, Patel CH, Luber BS, Wang H, Wagner KR, Powell JD, Housseau F, Pardoll DM, Elisseeff JH, Developing a pro-regenerative biomaterial scaffold microenvironment requires T helper 2 cells, Science (New York, N.Y.) 352 (6283) (2016) 366–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Sicari BM, Dziki JL, Siu BF, Medberry CJ, Dearth CL, Badylak SF, The promotion of a constructive macrophage phenotype by solubilized extracellular matrix, Biomaterials 35 (30) (2014) 8605–8612. [DOI] [PubMed] [Google Scholar]
- [32].Vizcaíno JA, Csordas A, del-Toro N, Dianes JA, Griss J, Lavidas I, Mayer G, Perez-Riverol Y, Reisinger F, Ternent T, 2016 update of the PRIDE database and its related tools, Nucleic Acids Res. 44 (D1) (2016) D447–D456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Naba A, Clauser KR, Hoersch S, Liu H, Carr SA, Hynes RO, The matrisome: in silico definition and in vivo characterization by proteomics of normal and tumor extracellular matrices, Mol. Cell. Proteom 11 (4) (2011) M111.014647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Kim M-S, Pinto SM, Getnet D, Nirujogi RS, Manda SS, Chaerkady R, Madugundu AK, Kelkar DS, Isserlin R, Jain S, Thomas JK, Muthusamy B, Leal-Rojas P, Kumar P, Sahasrabuddhe NA, Balakrishnan L, Advani J, George B, Renuse S, Selvan LDN, Patil AH, Nanjappa V, Radhakrishnan A, Prasad S, Subbannayya T, Raju R, Kumar M, Sreenivasamurthy SK, Marimuthu A, Sathe GJ, Chavan S, Datta KK, Subbannayya Y, Sahu A, Yelamanchi SD, Jayaram S, Rajagopalan P, Sharma J, Murthy KR, Syed N, Goel R, Khan AA, Ahmad S, Dey G, Mudgal K, Chatterjee A, Huang T-C, Zhong J, Wu X, Shaw PG, Freed D, Zahari MS, Mukherjee KK, Shankar S, Mahadevan A, Lam H, Mitchell CJ, Shankar SK, Satishchandra P, Schroeder JT, Sirdeshmukh R, Maitra A, Leach SD, Drake CG, Halushka MK, Prasad TSK, Hruban RH, Kerr CL, Bader GD, Iacobuzio-Donahue CA, Gowda H, Pandey A, A draft map of the human proteome, Nature 509 (7502) (2014) 575–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Anderson JM, Biological responses to materials, Annu. Rev. Mater. Res 31 (1) (2001) 81–110. [Google Scholar]
- [36].Anderson JM, Miller KM, Biomaterial biocompatibility and the macrophage, Biomaterials 5 (1) (1984) 5–10. [DOI] [PubMed] [Google Scholar]
- [37].Anderson JM, Inflammatory response to implants, ASAIO J. 34 (2) (1988) 101–107. [DOI] [PubMed] [Google Scholar]
- [38].Irvine DJ, Hanson MC, Rakhra K, Tokatlian T, Synthetic nanoparticles for vaccines and immunotherapy, Chem. Rev 115 (19) (2015) 11109–11146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Leleux J, Roy K, Micro and nanoparticle-based delivery systems for vaccine immunotherapy: an immunological and materials perspective, Adv. Healthc. Mater 2 (1) (2013) 72–94. [DOI] [PubMed] [Google Scholar]
- [40].Rosenberg SA, Yang JC, Restifo NP, Cancer immunotherapy: moving beyond current vaccines, Nat. Med 10 (9) (2004) 909–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Ali OA, Tayalia P, Shvartsman D, Lewin S, Mooney DJ, Inflammatory cytokines presented from polymer matrices differentially generate and activate DCs in situ, Adv. Funct. Mater 23 (36) (2013) 4621–4628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Ali OA, Verbeke C, Johnson C, Sands RW, Lewin SA, White D, Doherty E, Dranoff G, Mooney DJ, Identification of immune factors regulating antitumor immunity using polymeric vaccines with multiple adjuvants, Cancer Res. 74 (6) (2014) 1670–1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Kim J, Mooney DJ, In vivo modulation of dendritic cells by engineered materials: towards new cancer vaccines, Nano Today 6 (5) (2011) 466–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Vegas AJ, Veiseh O, Doloff JC, Ma M, Tam HH, Bratlie K, Li J, Bader AR, Langan E, Olejnik K, Fenton P, Kang JW, Hollister-Locke J, Bochenek MA, Chiu A, Siebert S, Tang K, Jhunjhunwala S, Aresta-Dasilva S, Dholakia N, Thakrar T. Vietti, M. Chen, J. Cohen, K. Siniakowicz, M. Qi, J. McGarrigle, Lyle, D.M. Harlan, D.L. Greiner, J. Oberholzer, G.C. Weir, R. Langer,D.G. Anderson, Combinatorial hydrogel library enables identification of materials that mitigate the foreign body response in primates, Nat. Biotechnol 34 (3) (2016) 345–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Badylak SF, Valentin JE, Ravindra AK, McCabe GP, Stewart-Akers AM, Macrophage phenotype as a determinant of biologic scaffold remodeling, Tissue Eng. Part A 14 (11) (2008) 1835–1842. [DOI] [PubMed] [Google Scholar]
- [46].Brown BN, Ratner BD, Goodman SB, Amar S, Badylak SF, Macrophage polarization: an opportunity for improved outcomes in biomaterials and regenerative medicine, Biomaterials 33 (15) (2012) 3792–3802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Brown BN, Sicari BM, Badylak SF, Rethinking regenerative medicine: a macrophage-centered approach, Front. Immunol 5 (2014) 510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Brown B, Lindberg K, Reing J, Stolz DB, Badylak SF, The basement membrane component of biologic scaffolds derived from extracellular matrix, Tissue Eng. 12 (3) (2006) 519–526. [DOI] [PubMed] [Google Scholar]
- [49].Wetzels RH, Robben HC, Leigh IM, Schaafsma HE, Vooijs GP, Ramaekers FC, Distribution patterns of type VII collagen in normal and malignant human tissues, Am. J. Pathol 139 (2) (1991) 451–459. [PMC free article] [PubMed] [Google Scholar]
- [50].Marçal H, Ahmed T, Badylak SF, Tottey S, Foster LJR, A comprehensive protein expression profile of extracellular matrix biomaterial derived from porcine urinary bladder, Regen. Med 7 (2) (2012) 159–166. [DOI] [PubMed] [Google Scholar]
- [51].Zhang JG, Czabotar PE, Policheni AN, Caminschi I, Wan SS, Kitsoulis S, Tullett KM, Robin AY, Brammananth R, van Delft MF, Lu J, O’Reilly LA, Josefsson EC, Kile BT, Chin WJ, Mintern JD, Olshina MA, Wong W, Baum J, Wright MD, Huang DC, Mohandas N, Coppel RL, Colman PM, Nicola NA, Shortman K, Lahoud MH, The dendritic cell receptor Clec9A binds damaged cells via exposed actin filaments, Immunity 36 (4) (2012) 646–657. [DOI] [PubMed] [Google Scholar]
- [52].Sadtler K, Allen BW, Estrellas K, Housseau F, Pardoll DM, Elisseeff JH, The scaffold immune microenvironment: biomaterial-mediated immune polarization in traumatic and nontraumatic applications, Tissue Eng. Part A (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Stankus JJ, Freytes DO, Badylak SF, Wagner WR, Hybrid nanofibrous scaffolds from electrospinning of a synthetic biodegradable elastomer and urinary bladder matrix, J. Biomater. Sci. Polym Ed. 19 (5) (2008) 635–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.