The helix-loop-helix (HLH) family of transcriptional regulatory proteins are key players in a wide array of developmental processes. Over 240 HLH proteins have been identified to date in organisms ranging from the yeast Saccharomyces cerevisiae to humans (6). Studies in Xenopus laevis, Drosophila melanogaster, and mice have convincingly demonstrated that HLH proteins are intimately involved in developmental events such as cellular differentiation, lineage commitment, and sex determination. In yeast, HLH proteins regulate several important metabolic pathways, including phosphate uptake and phospholipid biosynthesis (19, 67, 112). In multicellular organisms, HLH factors are required for a multitude of important developmental processes, including neurogenesis, myogenesis, hematopoiesis, and pancreatic development (12, 86, 127, 179). The purpose of this review is to examine the structure and functional properties of HLH proteins.
E-box sites: elements mediating cell-type-specific gene transcription.
Gene transcription of the immunoglobulin heavy-chain (IgH) gene has long been known to be regulated, in part, by a cis-acting DNA element known as the IgH intronic enhancer (109, 156). By in vivo methylation protection assays, a number of sites were identified in both the IgH and the kappa light-chain gene enhancers which were specifically protected in B cells but not in nonlymphoid cells (41). These elements shared a signature motif which consisted of the core hexanucleotide sequence, CANNTG, and were subsequently dubbed E boxes (41). A total of five E-box elements are present in the IgH gene enhancer: μE1, μE2, μE3, μE4, and μE5. The Ig kappa enhancer also contains three cannonical E boxes, designated κE1, κE2, and κE3. E-box sites have been subsequently found in B-cell-specific promoter and enhancer elements, including a subset of Ig light-chain gene promoters, the IgH and Ig light-chain 3′ enhancers, and, more recently, the λ5 promoter (110, 118, 156).
E-box elements have also been identified in promoter and enhancer elements that regulate muscle-, neuron-, and pancreas-specific gene expression. For example, in muscle, the muscle creatine kinase gene, acetylcholine receptor genes α and δ, and the myosin light-chain gene all require E-box elements for full activity (27, 51, 85). A number of genes whose expression is limited to the pancreas also require E-box sites for proper expression. The insulin and somatostatin genes, for example, contain E-box sites that, when multimerized, are sufficient to regulate pancreatic β-cell-specific gene expression (168). More recently, E-box regulatory sites have been identified in a number of neuron-specific genes, including the opsin, hippocalcin, beta 2 subunit of the neuronal nicotinic acetylcholine receptor, and muscarinic acetylcholine receptor genes (1, 21, 52, 125).
E-box sites: cognate recognition sequence for HLH proteins.
Two proteins, termed E12 and E47, were originally identified as binding to the κE2/μE5 site (65, 102). They have a region of homology with the Drosophila Daughterless protein, the myogenic differentiation factor MyoD, members of the achaete-scute gene complex, and the Myc family of transcription factors (102). This stretch of conserved residues, known as the Myc homology region, appeared to be critical for the DNA binding properties of E12 and E47 (102). The E12 and E47 proteins, which differ only within this Myc homology region, arise by alternative splicing of the E2A gene (157). This conserved sequence, which was modeled as two amphipathic alpha helices separated by a flexible loop structure, was named the HLH motif and shown to function as a dimerization domain.
The HLH structure.
The solution structure of the basic HLH (bHLH)-leucine zipper (LZ) factor Max first confirmed the existence of the HLH motif (44). Subsequently, the three-dimensional structure of the E47 bHLH polypeptide bound to its E-box recognition site, CACCTG, has been solved at 2.8-Å resolution (38). A number of interesting features were revealed from analysis of the E47 crystal structure. The E47 dimer forms a parallel, four-helix bundle which allows the basic region to contact the major groove (38). In addition to the basic region, residues in the loop and helix 2 also make contact with DNA (38). Stable interaction of the HLH domain is favored by van der Waals interactions between conserved hydrophobic residues (38). The E47 dimer is centered over the E box, with each monomer interacting with either a CAC or CAG half-site. A glutamate present in the basic region of each subunit makes contact with the cytosine and adenine bases in the E-box half-site. An adjacent arginine residue stabilizes the position of the glutamate by direct interaction with these nucleotides and additionally the phosphodiester backbone. Both the glutamate and the arginine residues are conserved in most bHLH proteins, consistent with a role in specific DNA binding (6, 38, 102).
Classification of the HLH proteins.
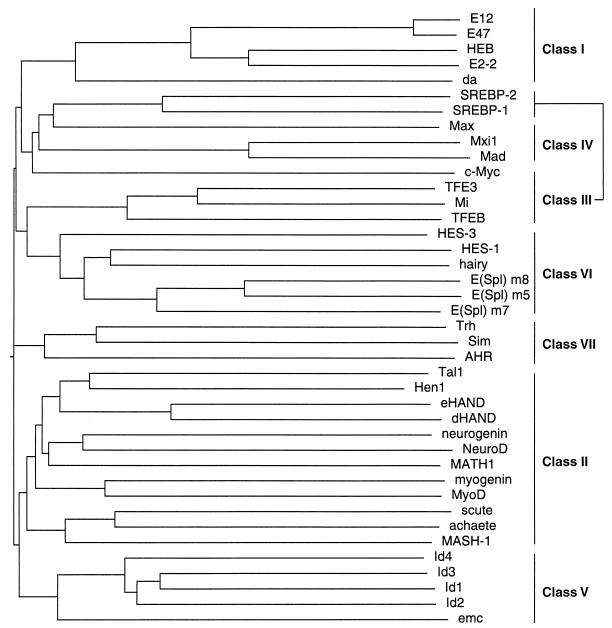
Owing to the large number of HLH proteins that have been described, a classification scheme that was based upon tissue distribution, dimerization capabilities, and DNA-binding specificities was devised (Fig. 1) (101). Class I HLH proteins, also known as the E proteins, include E12, E47, HEB, E2-2, and Daughterless. These proteins are expressed in many tissues and capable of forming either homo- or heterodimers (103). The DNA-binding specificity of class I proteins is limited to the E-box site. Class II HLH proteins, which include members such as MyoD, myogenin, Atonal, NeuroD/BETA2, and the achaete-scute complex, show a tissue-restricted pattern of expression. With few exceptions, they are incapable of forming homodimers and preferentially heterodimerize with the E proteins. Class I-class II heterodimers can bind both canonical and noncanonical E-box sites (103). Class III HLH proteins include the Myc family of transcription factors, TFE3, SREBP-1, and the microphthalmia-associated transcription factor, Mi. Proteins of this class contain an LZ adjacent to the HLH motif (66, 177). Class IV HLH proteins define a family of molecules, including Mad, Max, and Mxi, that are capable of dimerizing with the Myc proteins or with one another (7, 22, 174). A group of HLH proteins that lack a basic region, including Id and emc, define the class V HLH proteins (18, 39, 47). Class V members are negative regulators of class I and class II HLH proteins (18, 39, 47). Class VI HLH proteins have as their defining feature a proline in their basic region. This group includes the Drosophila proteins Hairy and Enhancer of split (76, 141). Finally, the class VII HLH proteins are categorized by the presence of the bHLH-PAS domain and include members such as the aromatic hydrocarbon receptor (AHR), the AHR nuclear-translocator (Arnt), hypoxia-inducible factor 1α, and the Drosophila Single-minded and Period proteins (34).
FIG. 1.
Multiple sequence alignment and classification of some representative members of the HLH family of transcription factors. Shown is a dendrogram created by aligning the sequences of the indicated HLH proteins by the Clustal W algorithm (160).
Recently, another classification method of HLH proteins has been described (6). Based on the amino acid sequences of 242 HLH proteins, a phylogenetic tree was created to group family members according to evolutionary relationships (6). Four major groups, A through D, which comprise more than 24 protein families were identified (6). The groupings were based upon DNA-binding specificity as well as conservation of amino acids at certain positions (6). As the number of HLH proteins continues to grow, this evolutionary or “natural” classification may provide a more accurate and convenient means of categorization.
THE HLH PROTEINS
Yeast.
Genetic studies in S. cerevisiae have identified a number of HLH factors that are important for regulating the transcription of genes involved in different metabolic pathways. The INO2 and INO4 genes encode HLH proteins that function together as a heterodimer to activate transcription of the fatty acid synthase genes required for the biosynthesis of phospholipids (67, 112). The HLH protein Pho4 plays an essential role in the regulation of phosphate uptake in yeast (see below). The yeast HLH protein CBF1 has a dual role in regulating methionine biosynthesis and controlling chromosomal integrity (28). Under conditions of mitochondrial stress, an isoform of citrate synthase encoded by the nuclear gene CIT2 is transcriptionally induced (90). Two HLH proteins, Rtg1p and Rtg3p, have been shown to be required for both the basal and induced expression of CIT2 (89, 137). Rtg1p and Rtg3p form a heterodimeric complex that binds to two sequence elements, each containing an E-box half-site present in the promoter of the CIT2 gene (73).
Caenorhabditis elegans.
Recent analysis of open reading frames present in the C. elegans genome has revealed 24 putative HLH proteins (142). A number of these genes have been identified previously in genetic screens. For example, the bHLH protein lin-32 is required for proper formation of the male tail rays, which are peripheral sense organs important for mating (176). A class VI HLH protein, lin-22, is important for proper nervous system patterning (169). A Daughterless ortholog has also been cloned, demonstrating that both the ubiquitous and cell-type-specific HLH proteins are present in nematodes (81). Surprisingly, however, like S. cerevisiae, the C. elegans genome lacks HLH proteins of the Id class (142). This suggests that inhibition of HLH activity through the formation of non-DNA-binding complexes was a strategy that evolved later in evolution.
Sex determination in Drosophila.
The decision of what sex to become is regulated in Drosophila primarily by the ratio of X chromosomes to sets of autosomes (122). A number of genes, including daughterless (da), sisterless-a (sis-a), and sisterless-b (sis-b), that are involved in sex determination have been identified (122). The da gene product, a bHLH protein, is required maternally and is present in equal amounts in both male and female embryos (35, 122). Mothers of the da genotype fail to produce female, but not male, offspring (35). sis-b, which also encodes a bHLH protein, is located on the X chromosome and is required zygotically for proper sex determination. Both genes have been shown to be required for activation of the Sxl gene promoter (23, 75). Since females are 2X, they make twice as much sis-b as males, and thus only females make sufficient concentrations of da–sis-b heterodimers to activate Sxl expression (32). In support of this model, da–sis-b heterodimers have been shown to bind E-box sites in vitro and can activate transcription in S. cerevisiae (4, 161). The choice between male and female in flies is thus dependent upon the concentration of active bHLH heterodimers.
Myogenesis.
Perhaps the best understood of the bHLH proteins are the myogenic regulatory factors (MRFs). Four genes whose protein products regulate both establishment and differentiation of the myogenic lineage have been identified. These proteins, named MyoD, MRF-4, Myf-5, and myogenin, are approximately 80% similar in their bHLH regions (115, 167). In addition, the MRFs have homology outside the bHLH domain, including a cysteine-histidine-rich stretch adjacent to the basic region and a serine-threonine-rich region at the carboxyl terminus (115). All MRFs are capable of converting the mesodermal cell line C3H10T1/2 into muscle progenitor cells known as myoblasts (115). This provided support for the existence of a muscle-specific master regulatory gene(s) (167). All MRFs are expressed solely in skeletal muscle, as determined by Northern blot analysis (164). In situ hybridization studies of the developing mouse embryo have shown that the MRFs are expressed in the somites and limb buds, consistent with a role in skeletal muscle development (164).
Gene targeting experiments in mice have demonstrated that the MRFs are important for different stages of muscle development. For example, both MyoD and Myf-5 null mutant mice show normal skeletal muscle development, suggesting a redundancy in MRF function (25, 138). Mice containing a homozygous deletion of both MyoD and Myf-5, however, die at birth and lack skeletal muscle (139). The myogenin gene product is required at a later stage of muscle development, specifically during the terminal differentiation of myoblasts to myotubes (61, 107). Three groups have independently generated mice homozygously null for MRF-4 (24, 124, 175). Although all used a similar gene targeting strategy, the severity of the phenotype was dependent upon the knockout allele used (116). Interestingly, the phenotypic variation in MRF-4−/− mice was due to an unanticipated inhibitory effect on the expression of the neighboring Myf-5 allele caused by disruption of the MRF-4 locus (116). These studies ultimately concluded, however, that MRF-4 is not required for the formation of skeletal muscle.
Heart development.
Two class II HLH proteins have been shown to be essential for proper heart morphogenesis. In situ hybridization studies in the mouse and chick have shown that two closely related HLH genes, dHAND and eHAND, are expressed in the developing heart (154). Inhibition of both dHAND and eHAND expression with antisense oligonucleotides resulted in arrested development at the cardiac looping stage (154). Consistent with these findings, both dHAND and eHAND null mutant mice exhibit pronounced defects in heart formation and die as embryos (45, 135, 155).
Pancreatic development.
As discussed above, a number of pancreas-specific genes contain E-box elements that regulate their tissue-restricted expression. α and β cells express both HEB and E2A and heterodimerize with a class II HLH protein, BETA2 (49, 111, 119, 150). Surprisingly, BETA2 turned out to be identical to NeuroD, a factor involved in neurogenic differentiation (86). Recently, mice that carry a targeted deletion of the BETA2/NeuroD gene have been generated (108). BETA2/NeuroD knockout mice exhibit no obvious neuronal phenotype but show marked pancreatic defects (108). These animals are severely diabetic (BETA2−/− mice have blood glucose levels fourfold higher than those in wild-type littermate controls) and die several days after birth (108). Normal morphogenesis of the pancreas is disrupted in BETA2 null mutant mice, and there is a dramatic reduction in insulin-producing β cells (108).
Neurogenesis.
A number of HLH proteins have been implicated in the control of vertebrate neurogenesis. Many of these factors were identified by a degenerate PCR strategy based on the sequences of Drosophila genes which regulate neurogenesis. For example, the mammalian achaete-scute homolog, MASH-1, plays an essential role in the generation of both autonomic and olfactory neurons (55). Recent gene targeting experiments in mice have demonstrated that Math1, the murine homolog of Atonal, is required for the development of granule neurons and the external germinal layer of the cerebellum (17). Interestingly, Math1 is also required for the formation of hair cells in the inner ear (20). Injection of NeuroD-related bHLH factor neurogenin 1 (ngn1) mRNA into Xenopus embryos induces ectopic neurogenesis and the expression of NeuroD (94). Although expression of NeuroD also promotes the generation of ectopic neurons in Xenopus, it does not activate ngn1 expression (86, 94). These data suggest that NeuroD is a downstream target of ngn1 (94). Consistent with these findings, ngn1 null mutant mice fail to activate NeuroD expression and show defects in cranial sensory neuron development (93).
The HLH proteins described above act as transcriptional activators. However, known HLH repressors also function in neurogenesis. Mice lacking the HLH repressor hairy and Enhancer of split homolog, HES-1, display a marked neural tube defect that may be the result of premature neurogenesis (70). The Id family of inhibitory HLH factors also play key roles in neurogenesis. All four mammalian Id genes are expressed in the developing brain and spinal cord (72). The unique expression patterns of the Id genes suggest that each is performing a specialized or nonredundant function (72). Retrovirus-mediated overexpression studies in chick embryos have demonstrated that Id2 can promote premature neurogenesis (95). Additionally, ectopic Id2 expression was able to convert ectoderm into neural crest cells, suggesting that the relative level of active bHLH proteins in these tissues helps determine whether cells will adopt a neural or epidermal fate (95).
Hematopoiesis.
HLH factors play significant roles in hematopoiesis. The bHLH protein SCL/Tal1, originally discovered by virtue of its involvement in a T-cell acute lymphoblastic leukemia (T-ALL)-specific translocation, has been shown to be essential for the development of all hematopoietic cell lineages (127). Disruption of the SCL/tal1 gene in mice leads to early embryonic lethality and a complete absence of blood (151). Specifically, immature erythrocytes are lacking in the embryo, placenta, or yolk sac of tal1-deficient mice, indicating a key role for Tal1 in erythropoiesis (151). Further studies indicated that the Tal1 protein is also required for proper B- and T-lineage development (127).
E2A: an essential regulator of B- and T-lymphocyte development.
Class II tissue-specific bHLH proteins are required for a number of developmental processes in organisms as diverse as flies and mammals. This, however, does not seem to be the case for the regulation of B-cell development, as homodimers of E47, a widely expressed E protein, appear to be the relevant DNA-binding species (11, 104, 149). These results put to rest, temporarily, the notion that B-cell-specific gene expression could be regulated by a heterodimer composed of E47 and a B-cell-restricted HLH protein (149). Recently, however, a B-cell-restricted bHLH protein, ABF-1, which is capable of binding DNA as a heterodimer with E2A has been identified (98). We note that ABF-1 expression has been detected only in human activated B cells and its function in B lineage development remains to be clarified.
Ectopic expression of E12 and E47 in various cell types is sufficient to activate B-lineage-specific gene expression. Overexpression of E47 in a pre-T-cell line leads to Ig germ line transcription and promotes IgH DJ gene rearrangements (146). Ectopic expression of E12 in a macrophage cell line leads to activation of λ5, RAG-1, early B-cell factor (EBF), and Pax-5 transcription (74, 152). In summary, these data support a model whereby E12 and E47 homodimers activate B-lineage-specific gene expression (101, 104).
Targeted deletion studies in mice have since demonstrated that the E2A gene is absolutely required for B-cell development (12, 179). E2A null mutant mice lack pre-B and mature B lymphocytes and contain significantly reduced numbers of B220+ CD43+ B-cell progenitors (12, 179). Additionally, thymocyte development is also severely perturbed. Both E2A- and HEB-deficient mice show abnormalities during the early stages of thymocyte differentiation (10, 178). Furthermore, ectopic expression of Id3 in human fetal thymic organ cultures showed a complete block in thymocyte development at a stage similar to that seen in E2A-deficient bone marrow cells (62). The E2A proteins are also required for proper γδ T-lineage development (13). Specifically, in the absence of E2A, γδ T-cell receptor (TCR) V(D)J recombination is severely perturbed (13). Interestingly, the dosage of E2A proteins was shown to regulate site-specific recombination of these loci. These data suggest that the concentration of E2A proteins in developing thymocytes may be rate limiting for the recombination reaction, preventing the rearrangement of two alleles at a given time.
Potential role for E2A proteins as tumor suppressors.
E2A-deficient mice revealed novel aspects for E2A function in addition to the role of E2A proteins in lymphocyte development. Specifically, E2A-deficient mice rapidly develop T-cell lymphomas (10). The tumors are monoclonal in origin, are of an immature T-cell phenotype, and will form tumors when injected into nude mice (10). Ectopic expression of Id2 in developing thymocytes also leads to the rapid development of lymphomas, consistent with these observations (100). However, it is remarkable that in these mice polyclonal tumors develop rather than the monoclonal populations observed in E2A-deficient thymocytes. These seemingly contradictory results may be explained by assuming that ectopic expression of Id2 blocks both E2A and HEB DNA binding activity, thus leading to a more severe phenotype compared to the E2A deficiency alone.
It is conceivable that loss of E2A homodimers, through targeted gene disruption or through ectopic expression of other bHLH proteins such as Lyl1 or Tal1, is an important event driving the development of T-cell leukemias (10). Thus, a function for E2A as a tumor suppressor in human malignancies is a reasonable possibility. Ectopic expression of E2A in E2A-deficient lymphomas rapidly leads to the induction of apoptosis, suggesting that indeed E2A is acting as a tumor suppressor (40). Similarly, overexpression of E2A in human T-ALL-derived cells results in cell death, providing further evidence for a key role of these proteins in lymphomagenesis (120).
E2A proteins also contribute to the regulation of cell cycle progression. Specifically, ectopic expression of E2A in fibroblasts blocks cells in the G1 phase (126). Consistent with a role for the E proteins in growth control, it was recently shown that Id2 is overexpressed in human pancreatic cancer (77). It was demonstrated previously that Id2 can reverse retinoblastoma protein (pRb)-mediated growth arrest in Saos-2 cells (69). Remarkably, ectopic expression of Id1 in primary keratinocytes leads to immortalization (2). Furthermore, deregulated expression of Id1 alone activates telomerase activity and promotes pRb phosphorylation in keratinocytes (2). How pRb phosphorylation is regulated by Id1 remains to be determined. Interestingly, both MyoD and E2A have the ability to regulate cyclin-dependent kinase inhibitor p21 expression (56, 128). It is conceivable, therefore, that Id1 controls pRb phosphorylation by indirectly controlling the expression of cyclin inhibitors. We propose that since the E proteins are targets of Id family members in these cells, they may contribute, directly or indirectly, to the regulation of telomerase activity. It will be important to dissect the role of E proteins in cell cycle progression, cell survival, and immortalization. Taken together, these observations suggest that E proteins regulate a wide variety of genes that are involved in many aspects of cellular differentiation and homeostasis.
TRANSCRIPTIONAL REGULATION BY THE HLH PROTEINS
The E proteins: activators of transcription.
Consistent with their role in activating the expression of B-lineage-specific genes, the E proteins, which include E2A (E47 and E12), E2-2, and HEB, have been shown to function as transcriptional activators (65). Recent studies have demonstrated that the activation domains of E2A are required for proper regulation of target genes in vivo. Removal of the N-terminal transactivation domains renders E12 incapable of inducing B-lineage-specific gene expression in a macrophage cell line (74). The transcriptional activity mapped to two domains, AD1 and AD2, located in the N-terminal half of the E2A protein (5, 65, 97, 132).
Interestingly, mutations that affected the transcriptional activity of the AD1 and AD2 domains in mammalian cells showed the same effects in the yeast S. cerevisiae, suggesting that the targets of these domains are highly conserved (97, 132). Two recent reports have shed some light on the transactivation properties of the E proteins. The coactivator p300, originally identified by virtue of its ability to interact with the E1A oncoprotein, has been shown to interact with E12 when bound to DNA (37). p300 has been shown to contain histone acetyltransferase (HAT) activity, thus raising the possibility that the E proteins recruit enzymes involved in chromatin modification (14, 37, 114). The interaction between p300 and E12 was mapped to the bHLH region of E12 and did not require the presence of the transactivation domains (37). These data are somewhat surprising since the bHLH domain does not contain intrinsic transactivation potential. p300 also has the ability to potentiate both AD1- and AD2-mediated transcription (130). More recently, studies have indicated that a conserved motif, LDFS, present in the AD1 domain directly interacts with a distinct nuclear HAT complex, termed SAGA (Fig. 2) (96). Many of the subunits that make up the SAGA complex are conserved from yeast to mammals. This includes a subset of the TATA binding protein-associated factors (TAFs), the HAT Gcn5, the Ada and Spt proteins, and an ATM family member called Tra1 (53, 54, 113). Specifically, amino acid substitutions within a conserved region of AD1 that disrupts SAGA binding in vitro abrogate transcriptional activation in vivo (96). Furthermore, AD1 cannot activate transcription in yeast strains lacking functional SAGA complexes (96). Taken together, these findings suggest that the E proteins may stimulate target gene transcription through interaction of their conserved activation domains with protein complexes that exhibit HAT activity. We note that it remains to be determined whether solely the SAGA complex or p300 or both are essential to mediate E-protein-mediated transcription.
FIG. 2.
Models for transcriptional activation and repression by the HLH proteins. Shown is a schematic depiction of an HLH target gene containing an E-box element (green box) (or an N box for Hairy [H]). (A) Cell-type-specific activators (A) can directly interact with the p300/CBP HAT (37, 105, 129, 130, 145) when bound as a heterodimer with an E protein (E). This interaction may be via independent domains present in the HLH activator. (B) Two models for activation by the E proteins. (Upper panel) Recruitment of p300/CBP may be directly mediated by the bHLH and/or activation domains (37, 130). Alternatively, the association with p300/CBP could be bridged by another factor (X). (Lower panel) In the second model, DNA-bound E-protein homodimers directly recruit the SAGA HAT complex via a conserved transactivation domain (96). Note that these models are not mutually exclusive. The wavy arrow represents transcriptional activation, and Ac denotes acetylase activity of the indicated protein complex. (C) A model for transcriptional activation by c-Myc. Once bound to DNA as a heterodimer with Max, Myc can recruit HAT complexes (such as SAGA) which contain the ATM family member TRRAP. This recruitment is mediated by a direct interaction of the N terminus of c-Myc with TRRAP (indicated in red type) (54, 99). Myc has also been shown to interact with hSNF5, a component of the ATP-dependent SWI-SNF chromatin-remodeling complex (30). (D) Repression by the HLH proteins. The top two diagrams indicate two forms of passive (indirect) repression. A bHLH repressor (R) may heterodimerize with a bHLH activator and inhibit its ability to bind an E box, thus preventing transactivation of the target gene (58, 87). The formation of non-DNA-binding Id–E-protein heterodimers can inhibit bHLH activator function by titrating away the available pool of E proteins. The bottom three illustrations show active forms of repression that necessitate the recruitment of transcriptional corepressor molecules. An HLH repressor may homodimerize or form a heterodimer with an E protein and repress transcription through E-box (or N-box) elements (87, 98). It is also possible that these heterodimers occupy E-box elements but are transcriptionally inert (121). The deacetylase activity of Rpd3 is indicated in the figure as dAc. It is thought that once brought to DNA, deacetylase activity induces localized nucleosomal condensation and thus promotes a transcriptionally repressed state. Transcriptional repression by Max-Mad (Mxi1) heterodimers is mediated by recruitment of the Sin3–N-CoR–HDAC complex. The blackened region of Mad represents the Sin3 interaction domain.
The E proteins and control of DNA rearrangement.
In addition to controlling Ig gene transcription, the Ig enhancers function to promote site-specific recombination. Since E2A binding sites are required for Ig enhancer activity, the potential role for these proteins in V(D)J recombination has been extensively studied. When E47 is expressed in a pre-T-cell line, it is capable of activating germ line transcription from the IgH enhancer and enhancing IgH DJ gene rearrangements (146). Overexpression of E47 activates IgH germ line transcription (31). More recently, E2A has been shown to be required for proper V(D)J recombination of certain TCR γ and δ loci (13). Remarkably, ectopic expression of E2A in a nonlymphoid line activates rearrangement of the Ig kappa light-chain gene (B. Romanow et al., submitted for publication). More specifically, overexpression of E12/E47 and RAG1 and RAG2 in embryonic kidney cells promotes endogenous Ig kappa VJ recombination utilizing variable regions that are interspersed over large segments of DNA (Romanow et al., submitted).
E2A proteins also are involved in DNA rearrangement at a later stage of B-cell development, class switch recombination (CSR). Immunohistochemical staining of human lymph nodes, spleens, and appendixes with an anti-E2A antibody showed that E2A protein was present in the dark zone of the germinal centers (50). This region is known to contain activated B cells that are actively undergoing CSR and somatic mutation (16). This is consistent with earlier reports demonstrating that high levels of E2A transcripts are present in splenic germinal centers (136). Similarly, in vitro evidence indicates that E2A proteins levels increase significantly upon activation with various mitogens (131). A role for E2A-like proteins in isotype switch recombination was demonstrated both in cell lines and in primary B-lineage cells (50). Interestingly, E2A-deficient B-lineage cells were not able to promote class switch recombination as detected by genomic rearrangements (131). Thus, E2A proteins are required for proper Ig isotype switching. These data open the intriguing possibility that E2A is directly involved in regulating various aspects of DNA rearrangement of both TCR and Ig loci.
How might E2A control DNA recombination in lymphocytes? It is conceivable that E2A directly recruits a recombination factor(s) to specific sites within the target gene. In this model, the ability of E2A to regulate recombination may be distinct from its transcriptional activation capability. While such a scenario cannot be excluded, we favor a mechanism in which E2A proteins promote accessibility to target gene loci. E2A binding sites are present in, for example, both the promoter and enhancer elements of the Ig kappa locus. E2A could recruit a nuclear HAT activity such as p300/CREB binding protein (CBP) or a mammalian SAGA complex (PCAF or human GCN5 [hGCN5] complexes) to the target gene, which could alter the acetylation state of nucleosomal histones (113). Covalent modification could destabilize the target nucleosomes, perhaps by weakening contact between adjacent nucleosomes (91). This, in turn, could lead to a localized opening of chromatin and allow access by the recombination machinery.
Activation by class II HLH proteins.
MyoD and BETA2 also have the ability to bind p300/CBP (37, 105, 130). Interestingly, the p300/CBP-associated factor PCAF also directly interacts with MyoD in vitro and has the ability to enhance MyoD-directed transcription (129). The HAT activity of PCAF but not that of p300 was required for coactivation of p21 (129). Furthermore, when myotube cell extracts were affinity purified with an E-box oligonucleotide, MyoD, p300, and PCAF were specifically retained (129). What are the functional consequences of the MyoD-p300 and MyoD-PCAF interactions on muscle differentiation? Microinjection studies demonstrated that myogenic differentiation was inhibited the addition of either anti-PCAF or anti-p300 antibodies (129). Conversely, overexpression of PCAF enhanced the differentiation of myoblasts to myotubes and cooperated with MyoD to promote increased conversion of fibroblasts into muscle cells (129). Hence, the physical association of MyoD with nuclear HATs is essential for its ability to direct skeletal muscle differentiation. What are the effects on chromatin structure on HLH target genes in vivo? A recent study has provided insight into how MyoD activates transcription by altering chromatin structure (48). Expression of a hormone-inducible form of MyoD activates transcription of myogenin, and MyoD was shown to remodel chromatin structure at several muscle-lineage specific genes, including MCK, myoD, and myogenin (48). These findings suggest that a MyoD-HAT complex may be recruited to promote accessibility to the transcription machinery, similar to that described above for the E2A proteins (Fig. 2).
Activation by class III proteins.
Sterol-responsive element-binding proteins 1 and 2 (SREBP-1 and -2) are bHLH-LZ transcription factors that are important for chloresterol-mediated induction of the low-density lipoprotein receptor (26, 143). These proteins, which are normally anchored in the endoplasmic reticulum membrane, are released by a chloresterol-induced protease and translocate to the nucleus, where they activate, in concert with Sp1, low-density lipoprotein receptor transcription (26, 143). The transcriptional synergy between SREBP-1a and Sp1 was not observed on naked DNA but required templates to be packaged into chromatin (106). More detailed biochemical analysis revealed that, in addition to the TAFs, a novel CBP-containing protein complex was required for transcriptional synergy in vitro (106). This complex contains at least 10 polypeptides in addition to CBP and, not surprisingly, exhibits HAT activity.
The Myc family of proteins has been shown to regulate a wide range of processes, including oncogenic transformation, apoptosis, and cellular differentiation (reviewed in references 42 and 64). Significant progress into the mechanism of transcriptional regulation by c-Myc has been achieved. A member of the SWI-SNF chromatin remodeling complex, hSNF5 (Ini1), interacts with the bHLH-LZ domain of c-Myc (30). Expression of a dominant-negative form of BRG1, another member of the SWI-SNF complex, blocks c-Myc transcriptional activation (30). Collectively, these data suggest that direct recruitment of SWI-SNF complexes by c-Myc is required for its transactivation function (Fig. 2).
The N terminus of c-Myc is required for transactivation and is essential for mediating its oncogenic effects (33). A novel ATM-related factor, TRRAP, interacts with the N terminus of c-Myc (99). TRAPP is closely related to Tra1, a yeast homolog that is present in several biochemically distinct HAT complexes, including SAGA (54). Furthermore, TRRAP has been shown to be a component of two mammalian SAGA-related activities: the PCAF and hGCN5 complexes (162). Interestingly, overexpression of dominant-negative forms of TRRAP blocked the ability of c-Myc to transform, in conjunction with activated ras, rat embryo fibroblasts (99). Taken together, these data indicate that c-Myc may be regulating gene expression, at least in part, by recruiting distinct HAT complexes through the association with its cofactor, TRRAP (Fig. 2).
Inducible gene expression by HLH proteins and chromatin remodeling.
AHR and Arnt are ligand-inducible mammalian HLH transcription factors that function together to activate gene transcription in response to the environmental toxin dioxin (reviewed in reference 59). AHR normally resides in the cytoplasm but translocates to the nucleus when liganded and forms dimeric complex with Arnt. AHR-Arnt heterodimers recognize sequence elements known as dioxin response elements present in a number of genes which regulate hydrocarbon detoxification and activate their transcription. Both AHR and Arnt contain glutamine-rich transactivation domains in their C termini (71). Using a DNase I digestion–ligation-mediated PCR assay, Ko et al. have assessed effects on the chromatin structure on the promoter and enhancer regions of the CYP1A1 gene, a direct target of the AHR-Arnt heterodimer (78, 79). Upon agonist treatment, cells expressing wild-type AHR, but not a carboxyl-terminal deletion mutant, showed increased DNase I hypersensitivity at the promoter element (78). Interestingly, the CYP1A1 promoter does not contain any known AHR-Arnt binding sites, so the effect is likely mediated by AHR-Arnt bound at the upstream enhancer. The Q-rich activation domain present in AHR is thus required for altering chromatin structure 1 kb away (78). Interestingly, other types of transactivation domains can functionally substitute for the Q-rich activator of AHR. Chimeric AHR proteins containing acidic activation domains derived from VP16 or Arnt were capable of activating CPY1A1 transcription in a dioxin-inducible manner (79). Furthermore, both AHR activator chimeras were shown to facilitate nucleosome disruption and allow occupancy of the TATA box and NF1 site at the CYP1A1 promoter (79).
Elegant studies focused on the control of phosphate uptake in the yeast S. cerevisiae also have provided key insights into how HLH proteins regulate gene expression. When exposed to limiting phosphate conditions, transcriptional activation of the PHO5 gene, which encodes a secreted acid phosphatase, is induced (88). Two upstream activating sequences (UAS) called UASp1 and UASp2 are present within PHO5 and are required for its transcriptional induction (140). The HLH protein Pho4, which is absolutely essential for PHO5 activation, binds both UASp1 and UASp2 (15). Under high-phosphate conditions, when PHO5 transcription is repressed, an ordered series of four nucleosomes that occludes the UASp2 site is present in the PHO5 promoter (163). The UASp1 site, however, lies in between nucleosomes in a DNase I hypersensitive region. When phosphate becomes limiting, disruption of the four nucleosomes occurs and Pho4 binds to both UASp1 and UASp2 (158). Transcriptional induction of PHO5 requires the integrity of the Pho4 transcriptional activation domain (159). Interestingly, when the PHO5 TATA box is deleted, Pho4-dependent chromatin remodeling can still occur (43, 158). Thus, PHO5 transcription per se is not required for chromatin remodeling to occur.
Recently, Komeili and O'Shea have provided further insight into how Pho4 is regulated by differential modification (80). First, when the Pho80/Pho85 cyclin-dependent kinase becomes activated under conditions of adequate phosphate, five sites within Pho4 become phosphorylated, inactivating its ability to induce transcription of phosphate-responsive genes (80). Second, phosphorylation of residues present in the N-terminal portion of Pho4 promotes nuclear export by facilitating an interaction with the nuclear export factor Msn5 (80). Third, in vitro binding studies demonstrated that phosphorylation of a residue present in the nuclear localization signal of Pho4 disrupts its ability to bind the nuclear import protein Pse1 (80). Interestingly, Pho4 mutants that were constitutively nuclear were still capable of inducing acid phosphatase when grown under limiting phosphate conditions, suggesting yet another mechanism for Pho4 regulation. Indeed, phosphorylation of yet another residue was shown to disrupt binding to Pho2, another transcription factor required for expression of PHO5 (80). Thus, posttranslational modification of Pho4 regulates both its intracellular localization and its ability to cooperate with other transcriptional regulators.
Transcriptional repression by HLH proteins.
Much progress has been made towards the understanding of how the class VI HLH proteins, including Enhancer of split and Hairy, function as repressors and has been the subject of several recent reviews (46, 123). Once bound to DNA, Hairy recruits a corepressor known as Groucho (Fig. 2). This interaction is mediated through a conserved tetrapeptide sequence, WRPW/Y, present at the extreme C terminus of all HES family members. A recent study has shown that Groucho can functionally interact with the histone deacetylase (HDAC) Rpd3 (29). In vitro binding assays demonstrated that the interaction with Rpd3 is direct and is mediated by the glycine-proline-rich domain of Groucho (29). These data indicate that at least one mechanism by which Groucho can repress transcription is through the recruitment of a chromatin-modifying activity to target genes (Fig. 2).
The class IV proteins Mad and Mxi1 function as repressors when bound to E-box elements as a heterodimer with Max (148). Over the last several years, significant progress has been made toward the understanding of how Mad proteins repress transcription. A breakthrough came with the identification of mSin3A and mSin3B, mammalian homologs of the yeast repressor Sin3, as factors that could directly interact with the Mad repression domain (SID) (8, 147). Subsequently, a number of groups identified a protein complex containing Sin3, the nuclear receptor corepressor N-CoR, and HDAC (3, 63, 82). Thus, Mad regulates repression through a SID-dependent association with a complex containing HDAC activity (Fig. 2).
Class II HLH proteins such as Tal1, ABF-1, and Mist1 have been shown to function as transcriptional repressors as well (Fig. 2) (87, 98, 121). Tal1 and ABF-1 inhibit E47-dependent activation presumably through the formation of heterodimers which are incapable of activating transcription through E-box elements (98, 121). Both ABF-1 and Mist1 can repress transcription in mammalian cells when expressed as GAL4 DNA-binding domain fusions (87, 98). These data suggest that ABF-1–E2A and Mist1-E2A heterodimers can function to actively repress E-box-containing target genes. Another model that has been proposed to explain how Mist1 inhibits transactivation by MyoD suggests that Mist1 forms transcriptionally inactive heterodimers with MyoD (87). How ABF-1 and Mist repress transcription remains to be determined.
Regulation of HLH activity by the Id protein family.
The class V HLH proteins, which include emc and Id, inhibit HLH activators through passive repression. These proteins contain a highly conserved HLH motif but lack an adjacent basic region necessary for DNA binding. Heterodimerization with these dominant-negative factors results in a form that is incapable of binding DNA (Fig. 2). Four known vertebrate Id gene products have been identified (reviewed in reference 93). The Id1 to Id4 gene products are closely related in their HLH regions and show similar affinities for the various E proteins (83). However, they differ in their expression patterns (72, 133, 134). The function of Id proteins has recently been addressed by using a gene targeting approach in mice. Whereas Id1 null mutant mice do not exhibit abnormalities, Id2−/− mice lack lymph nodes and the development of the natural killer (NK) cell lineage is severely perturbed (171, 173). On the other hand, we note that in the absence of E2A activity, NK cell development is significantly enhanced (62). Taken together, the data suggest that during early thymocyte differentiation the relative dose of E proteins and Id2 levels may ultimately determine cell fate, NK versus T-lineage cells. Intriguing data have recently revealed a key role for Id1 and Id3 in angiogenesis. Specifically, distinct tumors failed to grow and metastasize in Id1+/− Id3−/− null mutant mice, suggesting that these Id proteins are required for the invasiveness of vasculature structures (92).
How Id proteins are regulated is largely unknown. Recent studies have indicated that cell cycle-regulated phosphorylation of Id2 and Id3 by Cdk2 inhibits their ability to dimerize with the E proteins (36, 60). Ultimately it will be important to determine precisely how Id and E-protein levels are regulated by signals emanating from the cell surface and how their relative levels contribute to determine cell fate.
A new twist on transcriptional inhibition.
The class II HLH protein Twist was originally identified as a factor required for proper gastrulation and mesoderm formation in Drosophila. A number of reports have demonstrated that Twist has the ability to inhibit the differentiation of multiple cell types, including muscle and neurons. Twist appears to be able to inhibit myogenic differentiation by interfering with the activity of the myogenic transcription factors MyoD and MEF2. Several independent mechanisms have been proposed to explain the inhibitory effects of Twist on the muscle-specific transcriptional program. Twist, like Id, has the ability to repress MyoD function by titrating away E proteins, thus preventing the formation of functional MyoD–E-protein heterodimers (153). These authors further demonstrated that Twist inhibits muscle-specific gene activation by interfering with MEF2-mediated transactivation (153). This dually inhibitory mechanism of Twist may interfere with synergistic transactivation of target genes by the myogenic HLH proteins and MEF2 (153). Based on the observation that overexpression of E proteins and MEF2 does not relieve the inhibitory effect of Twist on MyoD activity, another inhibitory mechanism has been proposed (58). Twist was shown to directly interact with MyoD, MRF4, Myf-5, and myogenin. Surprisingly, MyoD and Twist associated via their basic regions (58). This unusual dimerization with Twist thus sequesters the myogenic HLH proteins in a transcriptionally inactive form.
Twist may also play a more general role in negative regulation of transcription. The amino terminus of Twist has recently been shown to mediate a direct interaction with the HAT domains of two different coactivators which are known to associate in vivo: p300 and PCAF (57, 172). In an in vitro HAT assay to monitor acetylation of purified histones, HAT activity of both p300 and PCAF was inhibited upon interaction with Twist (57). Overexpression of Twist in cell lines blocked PCAF-enhanced p300 autoacetylation, suggesting that, in vivo, HAT activity is impaired (57). Transient-transfection studies showed that the activity of several p300-dependent acidic activators could be dramatically reduced when Twist was coexpressed (57). These data demonstrate that p300- and PCAF-dependent gene expression may be downregulated in cells that express Twist, by directly inhibiting their acetyltransferase activity and possibly by preventing the interaction between PCAF and p300 (57). This novel mechanism of Twist-mediated transcriptional inhibition may help to explain its ability to interfere with the differentiation of diverse cell types.
Functional collaboration of HLH proteins with other classes of transcription factors.
Tal1 is a class II HLH protein that was originally identified as a result of a chromosomal translocation found in a subset of T-ALLs (9). Lmo2 is a member of the LIM-only family of transcription factors that is inappropriately expressed in T-ALL due to a chromosomal translocation (144). Targeted deletion of these genes in mice has demonstrated that both Tal1 and Lmo2 are required for erythroid development (127, 151, 166, 170). Studies in transgenic mice have shown that Tal1 and Lmo2 can collaborate to induce T-cell leukemia (84). By a binding-site selection strategy, a novel DNA-binding protein complex that contains the HLH proteins E47, Tal-1, zinc finger transcription factor GATA-1, and Lmo2 and Ldb1, two LIM domain-containing proteins, has recently been identified in erythroid cells (165). These five proteins bind to a bipartite sequence element composed of an E box and a GATA site. Complex binding required the integrity of both binding sites and was sensitive to the spacing between them. Reconstitution of this oligomeric complex by cotransfection experiments in COS cells demonstrated that it was capable of activating transcription of a reporter gene containing two tandem copies of the E box-GATA element. A protein complex present in a T-ALL line which is composed of Tal-1, Lmo, and GATA-3 was recently characterized (117). This complex has the ability to synergistically activate the RALDH2 gene in an E-box-independent fashion (117). Thus, HLH proteins may be associated with other transcription factors to regulate cell-type-specific gene transcription and tumorigenesis in a manner that may not require direct binding to E boxes.
HLH proteins are also known to be important for inducible expression of genes downstream of signal transduction pathways. Recently, a novel cDNA expression cloning technique based on retroviral gene transfer has been used to identify proteins involved in transforming growth factor β (TGF-β)-inducible expression of plasminogen activator inhibitor 1 (PAI-1), an extracellular matrix protein involved in cell adhesion and blood coagulation (68). A cDNA encoding the HLH protein TFE3 was isolated and found to be important for inducible expression of the PAI-1 promoter by TGF-β (68). An E-box element that binds TFE3 in vitro was identified in the PAI-1 promoter and was shown to be required for TGF-β-induced transcription in vivo (68). Transient-transfection analysis in a hepatocellular carcinoma cell line demonstrated that two transcription factors in the TGF-β signalling pathway, Smad3 and Smad4, synergized with TFE3 to activate PAI-1 in a TGF-β-dependent manner (68). The functional collaboration between TFE3 and the Smads does not appear to involve a direct protein-protein interaction, but gel shift analysis has shown that they can bind to adjacent sites on the PAI-1 promoter (68).
CONCLUDING REMARKS
Studies of developmental pathways, cell growth, and cell death have in part become studies of transcriptional regulators. Efforts to identify the nature of these regulators have revealed families of proteins that are characterized by highly conserved DNA binding, dimerization, and transactivation and repression motifs. One of these includes the HLH family. Despite the advances in understanding HLH structure and function, how they regulate gene expression and recombination is largely unknown. What is the mechanism by which this particular class of regulators influences RNA polymerase or recombinase function? Also, much is to be learned about how these proteins act in synergy with other families of transcriptional regulators to control gene expression and how they modulate chromatin structure. Finally, we will need to learn more about how these proteins are regulated at the levels of synthesis, at both the transcriptional and posttranscriptional levels, by signals emanating from cell surface receptors.
ACKNOWLEDGMENTS
We thank the reviewers for their helpful comments and suggestions which improved the quality of the manuscript.
M.E.M. is supported by a postdoctoral Cancer Biology training grant from the National Institutes of Health. C.M. is supported by grants from the National Institutes of Health.
REFERENCES
- 1.Ahmad I. Mash-1 is expressed during ROD photoreceptor differentiation and binds an E-box, E opsin-1 in the rat opsin gene. Brain Res Dev Brain Res. 1995;90:184–189. doi: 10.1016/0165-3806(96)83500-0. [DOI] [PubMed] [Google Scholar]
- 2.Alani R, Hasskarl J, Grace M, Hernandez M-C, Israel M, Munger K. Immortalization of primary human keratinocytes by the helix-loop-helix protein, Id-1. Proc Natl Acad Sci USA. 1999;96:9637–9641. doi: 10.1073/pnas.96.17.9637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alland L, Muhle R, Hou H, Jr, Potes J, Chin L, Schreiber-Agus N, DePinho R A. Role for N-CoR and histone deacetylase in Sin3-mediated transcriptional repression. Nature. 1997;387:49–55. doi: 10.1038/387049a0. [DOI] [PubMed] [Google Scholar]
- 4.Alonso M, Cabrera C. The Achaete-Scute gene complex of Drosophila melanogaster comprises four homologous genes. EMBO. 1988;7:2585–2591. doi: 10.1002/j.1460-2075.1988.tb03108.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aronheim A, Shiran R, Rosen A, Walker M D. The E2A gene product contains two separable and functionally distinct transcription activation domains. Proc Natl Acad Sci USA. 1993;90:8063–8067. doi: 10.1073/pnas.90.17.8063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Atchley W, Fitch W. A natural classification of the basic helix-loop-helix class of transcription factors. Proc Natl Acad Sci USA. 1997;94:5172–5176. doi: 10.1073/pnas.94.10.5172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ayer D, Kretzner L, Eisenman R. Mad: a heterodimeric partner for Max that antagonizes Myc transcriptional activity. Cell. 1993;72:211–222. doi: 10.1016/0092-8674(93)90661-9. [DOI] [PubMed] [Google Scholar]
- 8.Ayer D, Lawrence Q, Eisenman R. Mad-Max transcriptional repression is mediated by ternary complex formation with mammalian homologs of yeast repressor Sin3. Cell. 1995;80:767–776. doi: 10.1016/0092-8674(95)90355-0. [DOI] [PubMed] [Google Scholar]
- 9.Baer R. TAL1, TAL2 and LYL1: a family of basic helix-loop-helix proteins implicated in T cell acute leukaemia. Semin Cancer Biol. 1993;4:341–347. [PubMed] [Google Scholar]
- 10.Bain G, Engel I, Maandag E R, te Riele H, Voland J, Sharp L, Chun J, Huey B, Pinkel D, Murre C. E2A deficiency leads to abnormalities in αβ T-cell development and to the rapid development of T-cell lymphomas. Mol Cell Biol. 1997;17:4782–4791. doi: 10.1128/mcb.17.8.4782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bain G, Gruenwald S, Murre C. E2A and E2-2 are subunits of B-cell-specific E2-box DNA-binding proteins. Mol Cell Biol. 1993;13:3522–3529. doi: 10.1128/mcb.13.6.3522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bain G, Maandag E, Izon D, Amsen D, Kruisbeek A, Weintraub B, Krop I, Schlissel M, Feeney A, van Roon M, van der Valk M, te Riele H, Berns A, Murre C. E2A proteins are required for proper B cell development and initiation of immunoglobulin gene rearrangements. Cell. 1994;79:885–892. doi: 10.1016/0092-8674(94)90077-9. [DOI] [PubMed] [Google Scholar]
- 13.Bain G, Romanow W, Albers K, Havran W, Murre C. Positive and negative regulation of V(D)J recombination by the E2A proteins. J Exp Med. 1999;189:289–300. doi: 10.1084/jem.189.2.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bannister A, Kouzarides T. The CBP co-activator is a histone acetyltransferase. Nature. 1996;384:641–643. doi: 10.1038/384641a0. [DOI] [PubMed] [Google Scholar]
- 15.Barbaric S, Munsterkotter M, Goding C, Horz W. Cooperative Pho2-Pho4 interactions at the PHO5 promoter are critical for binding of Pho4 to UASp1 and for efficient transactivation by Pho4 at UASp2. Mol Cell Biol. 1998;18:2629–2639. doi: 10.1128/mcb.18.5.2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barek C. The development of B cells and the B cell repertoire in the microenvironment of the germinal center. Immunol Rev. 1992;126:7–18. doi: 10.1111/j.1600-065x.1992.tb00628.x. [DOI] [PubMed] [Google Scholar]
- 17.Ben-Arie N, Bellen H J, Armstrong D L, McCall A E, Gordadze P R, Guo Q, Matzuk M M, Zoghbi H Y. Math1 is essential for genesis of cerebellar granule neurons. Nature. 1997;390:169–172. doi: 10.1038/36579. [DOI] [PubMed] [Google Scholar]
- 18.Benezra R, Davis R, Lockshon D, Turner D, Weintraub H. The protein Id—a negative regulator of helix-loop-helix DNA binding proteins. Cell. 1990;61:49–59. doi: 10.1016/0092-8674(90)90214-y. [DOI] [PubMed] [Google Scholar]
- 19.Berben G, Legrain M, Gilliquet V, Hilger F. The yeast regulatory gene PHO4 encodes a helix-loop-helix motif. Yeast. 1990;6:451–454. doi: 10.1002/yea.320060510. [DOI] [PubMed] [Google Scholar]
- 20.Bermingham N, Hassan B, Price S, Vollrath M, Ben-Arie N, Eatock R, Bellen H, Lysakowski A, Zoghbi H. Math1: an essential gene for the generation of inner ear hair cells. Science. 1999;284:1837–1842. doi: 10.1126/science.284.5421.1837. [DOI] [PubMed] [Google Scholar]
- 21.Bessis A, Salmon A, Zoli M, Le Novere N, Picciotto M, Changeux J. Promoter elements conferring neuron-specific expression of the beta 2-subunit of the neuronal nicotinic acetylcholine receptor studied in vitro and in transgenic mice. Neuroscience. 1995;69:807–819. doi: 10.1016/0306-4522(95)00303-z. [DOI] [PubMed] [Google Scholar]
- 22.Blackwood E M, Eisenman R N. Max: a helix-loop-helix zipper protein that forms a sequence-specific DNA-binding complex with Myc. Science. 1991;251:1211–1217. doi: 10.1126/science.2006410. [DOI] [PubMed] [Google Scholar]
- 23.Bopp D, Bell L, Cline T, Schedl P. Developmental distribution of female-specific Sex-lethal proteins in Drosophila melanogaster. Genes Dev. 1991;5:403–415. doi: 10.1101/gad.5.3.403. [DOI] [PubMed] [Google Scholar]
- 24.Braun T, Arnold H H. Inactivation of Myf-6 and Myf-5 genes in mice leads to alterations in skeletal muscle development. EMBO J. 1995;14:1176–1186. doi: 10.1002/j.1460-2075.1995.tb07101.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Braun T, Rudnicki M A, Arnold H H, Jaenisch R. Targeted inactivation of the muscle regulatory gene Myf-5 results in abnormal rib development and perinatal death. Cell. 1992;71:369–382. doi: 10.1016/0092-8674(92)90507-9. [DOI] [PubMed] [Google Scholar]
- 26.Brown M S, Goldstein J L. The SREBP pathway: regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell. 1997;89:331–340. doi: 10.1016/s0092-8674(00)80213-5. [DOI] [PubMed] [Google Scholar]
- 27.Buskin J N, Hauschka S D. Identification of a myocyte nuclear factor which binds to the muscle-specific enhancer of the mouse muscle creatine kinase gene. Mol Cell Biol. 1989;9:2627–2640. doi: 10.1128/mcb.9.6.2627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cai M, Davis R. Yeast centromere binding protein CBF1, of the helix-loop-helix protein family, is required for chromosome stability and methionine prototrophy. Cell. 1990;61:437–446. doi: 10.1016/0092-8674(90)90525-j. [DOI] [PubMed] [Google Scholar]
- 29.Chen G, Fernandez J, Mische S, Courey A. A functional interaction between the histone deacetylase Rpd3 and the corepressor Groucho in Drosophila development. Genes Dev. 1999;13:2218–2230. doi: 10.1101/gad.13.17.2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cheng S W, Davies K P, Yung E, Beltran R J, Yu J, Kalpana G V. c-MYC interacts with INI1/hSNF5 and requires the SWI/SNF complex for transactivation function. Nat Genet. 1999;22:102–105. doi: 10.1038/8811. [DOI] [PubMed] [Google Scholar]
- 31.Choi J, Shen C-P, Radomska H, Eckhardt L, Kadesch T. E47 activates the Ig-heavy chain and TdT loci in non-B cells. EMBO J. 1996;15:5014–5021. [PMC free article] [PubMed] [Google Scholar]
- 32.Cline T W. The affairs of daughterless and the promiscuity of developmental regulators. Cell. 1989;59:231–234. doi: 10.1016/0092-8674(89)90280-8. [DOI] [PubMed] [Google Scholar]
- 33.Cole M. The myc oncogene: its role in transformation and differentiation. Annu Rev Gen. 1986;20:361–385. doi: 10.1146/annurev.ge.20.120186.002045. [DOI] [PubMed] [Google Scholar]
- 34.Crews S. Control of cell lineage-specific development and transcription by bHLH-PAS proteins. Genes Dev. 1998;12:607–620. doi: 10.1101/gad.12.5.607. [DOI] [PubMed] [Google Scholar]
- 35.Cronmiller C, Cline T W. The sex determination gene daughterless has different functions in the germ line versus the soma. Cell. 1987;48:479–487. doi: 10.1016/0092-8674(87)90198-x. [DOI] [PubMed] [Google Scholar]
- 36.Deed R, Hara E, Atherton G, Peters G, Norton J. Regulation of Id3 cell cycle function by Cdk-2-dependent phosphorylation. Mol Cell Biol. 1997;17:6815–6821. doi: 10.1128/mcb.17.12.6815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Eckner R, Yao T-P, Oldread E, Livingston D. Interaction and functional collaboration of p300/CBP and bHLH proteins in muscle and B-cell differentiation. Genes Dev. 1996;10:2478–2490. doi: 10.1101/gad.10.19.2478. [DOI] [PubMed] [Google Scholar]
- 38.Ellenberger T, Fass D, Arnaud M, Harrison S. Crystal structure of transcription factor E47: E-box recognition by a basic region helix-loop-helix dimer. Genes Dev. 1994;8:970–980. doi: 10.1101/gad.8.8.970. [DOI] [PubMed] [Google Scholar]
- 39.Ellis H M, Spann D R, Posakony J W. extramacrochaetae, a negative regulator of sensory organ development in Drosophila, defines a new class of helix-loop-helix proteins. Cell. 1990;61:27–38. doi: 10.1016/0092-8674(90)90212-w. [DOI] [PubMed] [Google Scholar]
- 40.Engel I, Murre C. Ectopic expression of E47 or E12 promotes the death of E2A-deficient lymphomas. Proc Natl Acad Sci USA. 1999;96:996–1001. doi: 10.1073/pnas.96.3.996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ephrussi A, Church G M, Tonegawa S, Gilbert W. B-lineage-specific interactions of an immunoglobulin enhancer with cellular factors in vivo. Science. 1985;227:134–140. doi: 10.1126/science.3917574. [DOI] [PubMed] [Google Scholar]
- 42.Facchini L, Penn L. The molecular role of Myc in growth and transformation: recent discoveries lead to new insights. FASEB J. 1998;12:633–651. [PubMed] [Google Scholar]
- 43.Fascher K-D, Schmitz J, Horz W. Structural and functional requirements for the chromatin transition at the PHO5 promoter in Saccharomyces cerevisiae upon PHO5 activation. J Mol Biol. 1993;231:658–667. doi: 10.1006/jmbi.1993.1317. [DOI] [PubMed] [Google Scholar]
- 44.Ferre-D'Amare A, Prendergast G, Ziff E, Burley S. Recognition by Max of its cognate DNA through a dimeric b/HLH/Z domain. Nature. 1993;363:38–45. doi: 10.1038/363038a0. [DOI] [PubMed] [Google Scholar]
- 45.Firulli A B, McFadden D G, Lin Q, Srivastava D, Olson E N. Heart and extra-embryonic mesodermal defects in mouse embryos lacking the bHLH transcription factor Hand1. Nat Genet. 1998;18:266–270. doi: 10.1038/ng0398-266. [DOI] [PubMed] [Google Scholar]
- 46.Fisher A L, Caudy M. Groucho proteins: transcriptional corepressors for specific subsets of DNA-binding transcription factors in vertebrates and invertebrates. Genes Dev. 1998;12:1931–1940. doi: 10.1101/gad.12.13.1931. [DOI] [PubMed] [Google Scholar]
- 47.Garrell J, Modolel J. The Drosophila extramacrochaetae locus, an antagonist of proneural genes that like these genes, encodes a helix-loop-helix protein. Cell. 1990;61:39–48. doi: 10.1016/0092-8674(90)90213-x. [DOI] [PubMed] [Google Scholar]
- 48.Gerber A, Klesert T, Bergstrom D, Tapscott S. Two domains of MyoD mediate transcriptional activation of genes in repressive chromatin: a mechanism for lineage determination in myogenesis. Genes Dev. 1997;11:436–450. doi: 10.1101/gad.11.4.436. [DOI] [PubMed] [Google Scholar]
- 49.German M, Blanar M, Nelson C, Moss L, Rutter W. Two related helix-loop-helix proteins participate in separate cell-specific complexes that bind the insulin enhancer. Mol Endocrinol. 1991;5:292–299. doi: 10.1210/mend-5-2-292. [DOI] [PubMed] [Google Scholar]
- 50.Goldfarb A, Flores J, Lewandowska K. Involvement of the E2A basic helix-loop-helix protein in immunoglobulin heavy chain class switching. Mol Immunol. 1996;33:947–956. doi: 10.1016/s0161-5890(96)00047-8. [DOI] [PubMed] [Google Scholar]
- 51.Gossett L, Kelvin D, Sternberg E, Olsen E. A new myocyte-specific enhancer binding factor that recognizes a conserved element associated with multiple muscle-specific genes. Mol Cell Biol. 1989;9:5022–5033. doi: 10.1128/mcb.9.11.5022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Grant A, Jones A, Thomas K, Wisden W. Characterization of the rat hippocalcin gene: the 5′ flanking region directs expression to the hippocampus. Neuroscience. 1996;75:1099–1115. doi: 10.1016/0306-4522(96)00344-2. [DOI] [PubMed] [Google Scholar]
- 53.Grant P, Schieltz D, Pray-Grant M, Steger D, Reese J, Yates III J, Workman J. A subset of TAFIIs are integral components of the SAGA complex required for nucleosomal acetylation and transcription stimulation. Cell. 1998;94:45–53. doi: 10.1016/s0092-8674(00)81220-9. [DOI] [PubMed] [Google Scholar]
- 54.Grant P, Schieltz D, Pray-Grant M, Yates III J, Workman J. The ATM-related cofactor Tra1 is a component of the purified SAGA complex. Mol Cell. 1998;2:863–867. doi: 10.1016/s1097-2765(00)80300-7. [DOI] [PubMed] [Google Scholar]
- 55.Guillemot F, Lo L-C, Johnson J, Auerbach A, Anderson D, Joyner A. Mammalian achaete-scute homolog 1 is required for the early development of olfactory and autonomic neurons. Cell. 1993;75:463–476. doi: 10.1016/0092-8674(93)90381-y. [DOI] [PubMed] [Google Scholar]
- 56.Halevy O, Novitch B, Spicer D, Skapek S, Rhee J, Hannon G, Beach D, Lassar A. Correlation of terminal cell cycle arrest of skeletal muscle with induction of p21 by MyoD. Science. 1995;267:1018–1021. doi: 10.1126/science.7863327. [DOI] [PubMed] [Google Scholar]
- 57.Hamamori Y, Sartorelli V, Ogryzko V, Puri P, Wu H-Y, Wang J, Nakatani Y, Kedes L. Regulation of histone acetyltransferases p300 and PCAF by the bHLH protein twist and adenoviral oncoprotein E1A. Cell. 1999;96:405–413. doi: 10.1016/s0092-8674(00)80553-x. [DOI] [PubMed] [Google Scholar]
- 58.Hamamori Y, Wu H-Y, Sartorelli V, Kedes L. The basic domain of myogenic basic helix-loop-helix (bHLH) proteins is the novel target for direct inhibition by another bHLH protein, Twist. Mol Cell Biol. 1997;17:6563–6573. doi: 10.1128/mcb.17.11.6563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hankinson O. The aryl hydrocarbon receptor complex. Annu Rev Pharmacol Toxicol. 1995;35:307–340. doi: 10.1146/annurev.pa.35.040195.001515. [DOI] [PubMed] [Google Scholar]
- 60.Hara E, Hall M, Peters G. Cdk2-dependent phosphorylation of Id2 modulates activity of E2A-related transcription factors. EMBO J. 1997;16:101–110. doi: 10.1093/emboj/16.2.332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hasty P, Bradley A, Morris J H, Edmondson D G, Venuti J M, Olson E N, Klein W H. Muscle deficiency and neonatal death in mice with a targeted mutation in the myogenin gene. Nature. 1993;364:501–506. doi: 10.1038/364501a0. [DOI] [PubMed] [Google Scholar]
- 62.Heemskerk M H, Blom B, Nolan G, Stegmann A P, Bakker A Q, Weijer K, Res P C, Spits H. Inhibition of T cell and promotion of natural killer cell development by the dominant negative helix loop helix factor Id3. J Exp Med. 1997;186:1597–1602. doi: 10.1084/jem.186.9.1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Heinzel T, Lavinsky R M, Mullen T M, Söderstrom M, Laherty C D, Torchia J, Yang W M, Brard G, Ngo S D, Davie J R, Seto E, Eisenman R N, Rose D W, Glass C K, Rosenfeld M G. A complex containing N-CoR, mSin3 and histone deacetylase mediates transcriptional repression. Nature. 1997;387:43–48. doi: 10.1038/387043a0. [DOI] [PubMed] [Google Scholar]
- 64.Henriksson M, Luscher B. Proteins of the Myc network: essential regulators of cell growth and differentiation. Adv Cancer Res. 1996;68:109–182. doi: 10.1016/s0065-230x(08)60353-x. [DOI] [PubMed] [Google Scholar]
- 65.Henthorn P, Kiledjian M, Kadesch T. Two distinct transcription factors that bind the immunoglobulin enhancer μE5/ κE2 motif. Science. 1990;247:467–470. doi: 10.1126/science.2105528. [DOI] [PubMed] [Google Scholar]
- 66.Henthorn P, Stewart C, Kadesch T, Puck J. The gene encoding human TFE3, a transcription factor that binds the immunoglobulin heavy-chain enhancer, maps to xp11;22. Genomics. 1991;11:374–378. doi: 10.1016/0888-7543(91)90145-5. [DOI] [PubMed] [Google Scholar]
- 67.Hoshizaki D, Hill J, Henry S. The Saccharomyces cerevisiae INO4 gene encodes a small, highly basic protein required for derepression of phospholipid biosynthetic enzymes. J Biol Chem. 1990;265:3320–3328. [PubMed] [Google Scholar]
- 68.Hua X, Liu X, Ansari D, Lodish H. Synergistic cooperation of TFE3 and Smad proteins in TGF-β-induced transcription of the plasminogen activator inhibitor-1 gene. Genes Dev. 1998;12:3084–3095. doi: 10.1101/gad.12.19.3084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Iavarone A, Garg P, Lasorella A, Hsu J, Israel M. The helix-loop-helix protein Id-2 enhances cell proliferation and binds to the retinoblastoma protein. Genes Dev. 1994;8:1270–1284. doi: 10.1101/gad.8.11.1270. [DOI] [PubMed] [Google Scholar]
- 70.Ishibashi M, Ang S L, Shiota K, Nakanishi S, Kageyama R, Guillemot F. Targeted disruption of mammalian hairy and Enhancer of split homolog-1 (HES-1) leads to up-regulation of neural helix-loop-helix factors, premature neurogenesis, and severe neural tube defects. Genes Dev. 1995;9:3136–3148. doi: 10.1101/gad.9.24.3136. [DOI] [PubMed] [Google Scholar]
- 71.Jain S, Dolwick K, Schmidt J, Bradfield C. Potent transactivation domains of the Ah receptor and the Ah receptor nuclear translocator map to their carboxy termini. J Biol Chem. 1994;269:31518–31524. [PubMed] [Google Scholar]
- 72.Jen Y, Manova K, Benezra R. Each member of the Id gene family exhibits a unique expression pattern in mouse gastrulation and neurogenesis. Dev Dyn. 1997;208:92–106. doi: 10.1002/(SICI)1097-0177(199701)208:1<92::AID-AJA9>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 73.Jia Y, Rothermel B, Thornton J, Butow R. A basic helix-loop-helix-leucine zipper transcription complex in yeast functions in a signaling pathway from mitochondria to the nucleus. Mol Cell Biol. 1997;17:1110–1117. doi: 10.1128/mcb.17.3.1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kee B, Murre C. Induction of early B cell factor (EBF) and multiple B lineage genes by the basic helix-loop-helix transcription factor E12. J Exp Med. 1998;188:699–713. doi: 10.1084/jem.188.4.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Keyes L, Cline T, Schedl P. The primary sex determination signal of Drosophila acts at the level of transcription. Cell. 1992;68:933–943. doi: 10.1016/0092-8674(92)90036-c. [DOI] [PubMed] [Google Scholar]
- 76.Klambt C, Knust E, Tietze K, Campos-Ortega J A. Closely related transcripts encoded by the neurogenic gene complex enhancer of split of Drosophila melanogaster. EMBO J. 1989;8:203–210. doi: 10.1002/j.1460-2075.1989.tb03365.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kleeff J, Ishiwata T, Friess H, Buchler M, Israel M, Korc M. The helix-loop-helix protein Id2 is overexpressed in human pancreatic cancer. Cancer Res. 1998;58:3769–3772. [PubMed] [Google Scholar]
- 78.Ko H, Okino S, Ma Q, Whitlock J., Jr Dioxin-induced CYP1A1 transcription in vivo: the aromatic hydrocarbon receptor mediates transactivation, enhancer-promoter communication, and changes in chromatin structure. Mol Cell Biol. 1996;16:430–436. doi: 10.1128/mcb.16.1.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ko H, Okino S, Ma Q, Whitlock J., Jr Transactivation domains facilitate promoter occupancy for the dioxin-inducible CYP1A1 gene in vivo. Mol Cell Biol. 1997;17:3497–3507. doi: 10.1128/mcb.17.7.3497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Komeili A, O'Shea E. Roles of phosphorylation sites in regulating activity of the transcription factor Pho4. Science. 1999;284:977–980. doi: 10.1126/science.284.5416.977. [DOI] [PubMed] [Google Scholar]
- 81.Krause M, Park M, Zhang J M, Yuan J, Harfe B, Xu S Q, Greenwald I, Cole M, Paterson B, Fire A. A C. elegans E/Daughterless bHLH protein marks neuronal but not striated muscle development. Development. 1997;124:2179–89. doi: 10.1242/dev.124.11.2179. [DOI] [PubMed] [Google Scholar]
- 82.Laherty C D, Yang W M, Sun J M, Davie J R, Seto E, Eisenman R N. Histone deacetylases associated with the mSin3 corepressor mediate mad transcriptional repression. Cell. 1997;89:349–356. doi: 10.1016/s0092-8674(00)80215-9. [DOI] [PubMed] [Google Scholar]
- 83.Langlands K, Yin X, Anand G, Prochownik E V. Differential interactions of Id proteins with basic-helix-loop-helix transcription factors. J Biol Chem. 1997;272:19785–19793. doi: 10.1074/jbc.272.32.19785. [DOI] [PubMed] [Google Scholar]
- 84.Larson R C, Lavenir I, Larson T A, Baer R, Warren A J, Wadman I, Nottage K, Rabbitts T H. Protein dimerization between Lmo2(Rbtn2) and Tal1 alters thymocyte development and potentiates T cell tumorigenesis in transgenic mice. EMBO J. 1996;15:1021–1027. [PMC free article] [PubMed] [Google Scholar]
- 85.Lassar A B, Buskin J N, Lockshon D, Davis R L, Apone S, Hauschka S D, Weintraub H. MyoD is a sequence-specific DNA binding protein requiring a region of myc homology to bind to the muscle creatine kinase enhancer. Cell. 1989;58:823–831. doi: 10.1016/0092-8674(89)90935-5. [DOI] [PubMed] [Google Scholar]
- 86.Lee J E, Hollenberg S M, Snider L, Turner D L, Lipnick N, Weintraub H. Conversion of Xenopus ectoderm into neurons by neuroD, a basic helix-loop-helix protein. Science. 1995;268:836–844. doi: 10.1126/science.7754368. [DOI] [PubMed] [Google Scholar]
- 87.Lemercier C, To R Q, Carrasco R A, Konieczny S F. The basic helix-loop-helix transcription factor Mist1 functions as a transcriptional repressor of MyoD. EMBO J. 1998;17:1412–1422. doi: 10.1093/emboj/17.5.1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lenburg M E, O'Shea E K. Signaling phosphate starvation. Trends Biochem Sci. 1996;21:383–387. [PubMed] [Google Scholar]
- 89.Liao X, Butow R. RTG1 and RTG2: two yeast genes required for a novel path of communication from mitochondria to the nucleus. Cell. 1993;72:61–71. doi: 10.1016/0092-8674(93)90050-z. [DOI] [PubMed] [Google Scholar]
- 90.Liao X, Small W, Srere P, Butow R. Intramitochondrial functions regulate nonmitochondrial citrate synthase (CIT2) expression in Saccharomyces cerevisiae. Mol Cell Biol. 1991;11:38–46. doi: 10.1128/mcb.11.1.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Luger K, Mader A, Richmond R, Sargent D, Richmond T. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 92.Lyden D, Young A, Zagzag D, Yan W, Gerald W, O'Reilly R, Bader B, Hynes R, Zhuang Y, Manova K, Benezra R. Id1 and Id3 are required for neurogenesis, angiogenesis and vascularization of tumor xenografts. Nature. 1999;401:670–677. doi: 10.1038/44334. [DOI] [PubMed] [Google Scholar]
- 93.Ma Q, Chen Z, del Barco Barrantes I, de la Pompa J L, Anderson D J. Neurogenin1 is essential for the determination of neuronal precursors for proximal cranial sensory ganglia. Neuron. 1998;20:469–482. doi: 10.1016/s0896-6273(00)80988-5. [DOI] [PubMed] [Google Scholar]
- 94.Ma Q, Kintner C, Anderson D J. Identification of neurogenin, a vertebrate neuronal determination gene. Cell. 1996;87:43–52. doi: 10.1016/s0092-8674(00)81321-5. [DOI] [PubMed] [Google Scholar]
- 95.Martinsen B J, Bronner-Fraser M. Neural crest specification regulated by the helix-loop-helix repressor Id2. Science. 1998;281:988–991. doi: 10.1126/science.281.5379.988. [DOI] [PubMed] [Google Scholar]
- 96.Massari M, Grant P, Pray-Grant M, Berger S, Workman J, Murre C. A conserved motif present in a class of helix-loop-helix proteins activates transcription by direct recruitment of the SAGA complex. Mol Cell. 1999;4:63–73. doi: 10.1016/s1097-2765(00)80188-4. [DOI] [PubMed] [Google Scholar]
- 97.Massari M, Jennings P, Murre C. The AD1 transactivation domain of E2A contains a highly conserved helix which is required for its activity in both Saccharomyces cerevisiae and mammalian cells. Mol Cell Biol. 1996;16:121–129. doi: 10.1128/mcb.16.1.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Massari M, Rivera R, Voland J, Quong M, Breit T, van Dongen J, de Smit O, Murre C. Characterization of ABF-1, a novel basic helix-loop-helix transcription factor expressed in activated B lymphocytes. Mol Cell Biol. 1998;18:3130–3139. doi: 10.1128/mcb.18.6.3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.McMahon S B, Van Buskirk H A, Dugan K A, Copeland T D, Cole M D. The novel ATM-related protein TRRAP is an essential cofactor for the c-Myc and E2F oncoproteins. Cell. 1998;94:363–374. doi: 10.1016/s0092-8674(00)81479-8. [DOI] [PubMed] [Google Scholar]
- 100.Morrow, M., E. Mayer, C. Perez, M. Adlam, and G. Siu. Overexpression of the helix-loop-helix protein Id2 blocks T cell development at multiple stages. Mol. Immunol., in press. [DOI] [PubMed]
- 101.Murre C, Bain G, Dijk M A v, Engel I, Furnari B A, Massari M E, Matthews J R, Quong M W, Rivera R R, Stuiver M H. Structure and function of helix-loop-helix proteins. Biochim Biophys Acta. 1994;1218:129–135. doi: 10.1016/0167-4781(94)90001-9. [DOI] [PubMed] [Google Scholar]
- 102.Murre C, McCaw P S, Baltimore D. A new DNA binding and dimerization motif in immunoglobulin enhancer binding, daughterless, MyoD, and myc proteins. Cell. 1989;56:777–783. doi: 10.1016/0092-8674(89)90682-x. [DOI] [PubMed] [Google Scholar]
- 103.Murre C, Mccaw P S, Vaessin H, Caudy M, Jan L Y, Jan Y N, Cabrera C V, Buskin J N, Hauschka S D, Lassar A B, Weintraub H, Baltimore D. Interactions between heterologous helix-loop-helix proteins generate complexes that bind specifically to a common DNA sequence. Cell. 1989;58:537–544. doi: 10.1016/0092-8674(89)90434-0. [DOI] [PubMed] [Google Scholar]
- 104.Murre C, Voronova A, Baltimore D. B-cell- and myocyte-specific E2-box-binding factors contain E12/E47-like subunits. Mol Cell Biol. 1991;11:1156–1160. doi: 10.1128/mcb.11.2.1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Mutoh H, Naya F, Tsai M-J, Leiter A. The basic helix-loop-helix protein BETA2 interacts with p300 to coordinate differentiation of secretin-expressing enteroendocrine cells. Genes Dev. 1998;12:820–830. doi: 10.1101/gad.12.6.820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Naar A, Beaurang P, Robinson K, Oliner J, Avizonis D, Scheek S, Zwicker J, Kadonaga J, Tjian R. Chromatin, TAFs, and a novel multiprotein coactivator are required for synergic activation by Sp1 and SREBP-1a in vitro. Genes Dev. 1998;12:3020–3031. doi: 10.1101/gad.12.19.3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Nabeshima Y, Hanaoka K, Hayasaka M, Esumi E, Li S, Nonaka I, Nabeshima Y. Myogenin gene disruption results in perinatal lethality because of severe muscle defects. Nature. 1993;364:532–535. doi: 10.1038/364532a0. [DOI] [PubMed] [Google Scholar]
- 108.Naya F, Huang H-P, Qiu Y, Mutoh H, DeMayo F, Leiter A, Tsai M-J. Diabetes, defective pancreatic morphogenesis, and abnormal enteroendocrine differentiation in BETA2/NeuroD-deficient mice. Genes Dev. 1997;11:2323–2334. doi: 10.1101/gad.11.18.2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Nelsen B, Sen R. Regulation of immunoglobulin gene transcription. Int Rev Cytol. 1992;133:121–149. doi: 10.1016/s0074-7696(08)61859-8. [DOI] [PubMed] [Google Scholar]
- 110.Nelsen B, Tian G, Erman B, Gregoire J, Maki R, Graves B, Sen R. Regulation of the lymphoid-specific immunoglobulin mu heavy chain gene enhancer by the ETS-domain proteins. Science. 1993;261:82–86. doi: 10.1126/science.8316859. [DOI] [PubMed] [Google Scholar]
- 111.Nelson C, Shen L P, Meister A, Fodor E, Rutter W J. Pan: a transcriptional regulator that binds chymotrypsin, insulin and AP-4 enhancer motifs. Genes Dev. 1990;4:1035–1044. doi: 10.1101/gad.4.6.1035. [DOI] [PubMed] [Google Scholar]
- 112.Nikoloff D, McGraw P, Henry S. The INO2 gene of Saccharomyces cerevisiae encodes a helix-loop-helix protein that is required for activation of phospholipid synthesis. Nucleic Acids Res. 1992;20:3253. doi: 10.1093/nar/20.12.3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Ogryzko V, Kotani T, Zhang X, Schlitz R, Howard T, Yang X, Howard B, Qin J, Nakatani Y. Histone-like TAFs within the PCAF histone acetylase complex. Cell. 1998;94:35–44. doi: 10.1016/s0092-8674(00)81219-2. [DOI] [PubMed] [Google Scholar]
- 114.Ogryzko V, Schiltz R, Russanova V, Howard B, Nakatani Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 115.Olson E. MyoD family: a paradigm for development? Genes Dev. 1990;4:1454–1461. doi: 10.1101/gad.4.9.1454. [DOI] [PubMed] [Google Scholar]
- 116.Olson E N, Arnold H H, Rigby P W, Wold B J. Know your neighbors: three phenotypes in null mutants of the myogenic bHLH gene MRF4. Cell. 1996;85:1–4. doi: 10.1016/s0092-8674(00)81073-9. [DOI] [PubMed] [Google Scholar]
- 117.Ono Y, Fukuhara N, Yoshie O. TAL1 and LIM-only proteins synergistically induce retinaldehyde dehydrogenase 2 expression in T-cell acute lymphoblastic leukemia by acting as cofactors for GATA3. Mol Cell Biol. 1998;18:6939–6950. doi: 10.1128/mcb.18.12.6939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.O'Riordan M, Grosschedl R. EBF and E47 collaborate to induce expression of the endogenous immunoglobulin surrogate light chain genes. Immunity. 1997;7:25–36. doi: 10.1016/s1074-7613(00)80507-5. [DOI] [PubMed] [Google Scholar]
- 119.Park C, Walker M. Subunit structure of cell-specific E-box binding proteins analyzed by quantitation of electrophoretic mobility shift. J Biol Chem. 1992;267:15642–15649. [PubMed] [Google Scholar]
- 120.Park S T, Nolan G P, Sun X H. Growth inhibition and apoptosis due to restoration of E2A activity in T cell acute lymphoblastic leukemia cells. J Exp Med. 1999;189:501–508. doi: 10.1084/jem.189.3.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Park S T, Sun X H. The Tal1 oncoprotein inhibits E47-mediated transcription. Mechanism of inhibition. J Biol Chem. 1998;273:7030–7037. doi: 10.1074/jbc.273.12.7030. [DOI] [PubMed] [Google Scholar]
- 122.Parkhurst S, Meneely P. Sex determination and dosage compensation: lessons from flies and worms. Science. 1994;264:924–932. doi: 10.1126/science.8178152. [DOI] [PubMed] [Google Scholar]
- 123.Parkhurst S M. Groucho: making its Marx as a transcriptional co-repressor. Trends Genet. 1998;14:130–132. doi: 10.1016/s0168-9525(98)01407-3. [DOI] [PubMed] [Google Scholar]
- 124.Patapoutian A, Yoon J K, Miner J H, Wang S, Stark K, Wold B. Disruption of the mouse MRF4 gene identifies multiple waves of myogenesis in the myotome. Development. 1995;121:3347–3358. doi: 10.1242/dev.121.10.3347. [DOI] [PubMed] [Google Scholar]
- 125.Pepitoni S, Wood I, Buckley N. Structure of the m1 muscarinic acetylcholine receptor gene and its promoter. J Biol Chem. 1997;272:17112–17117. doi: 10.1074/jbc.272.27.17112. [DOI] [PubMed] [Google Scholar]
- 126.Peverali F, Ramqvist T, Saffrich R, Pepperkok R, Barone M, Philipson L. Regulation of G1 progression by E2A and Id helix-loop-helix proteins. EMBO J. 1994;13:4291–4301. doi: 10.1002/j.1460-2075.1994.tb06749.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Porcher C, Swat W, Rockwell K, Fujiwara Y, Alt F, Orkin S. The T cell leukemia oncoprotein SCL/tal-1 is essential for development of all hematopoietic lineages. Cell. 1996;86:47–57. doi: 10.1016/s0092-8674(00)80076-8. [DOI] [PubMed] [Google Scholar]
- 128.Prabhu S, Ignatova A, Park S, Sun X-H. Regulation of the expression of cyclin-dependent kinase inhibitor p21 by E2A and Id proteins. Mol Cell Biol. 1997;17:5888–5896. doi: 10.1128/mcb.17.10.5888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Puri P, Sartorelli V, Yang X-J, Hamamori Y, Ogryzko V, Howard B, Kedes L, Wang J, Graessmann A, Nakatani Y, Levrero M. Differential roles of p300 and PCAF acetyltransferases in muscle differentiation. Mol Cell. 1997;1:35–45. doi: 10.1016/s1097-2765(00)80005-2. [DOI] [PubMed] [Google Scholar]
- 130.Qiu Y, Sharma A, Stein R. p300 mediates transcriptional stimulation by the basic helix-loop-helix activators of the insulin gene. Mol Cell Biol. 1998;18:2957–2964. doi: 10.1128/mcb.18.5.2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Quong M, Harris D, Swain S, Murre C. E2A activity is induced during B-cell activation to promote immunoglobulin class switch recombination. EMBO J. 1999;18:6307–6318. doi: 10.1093/emboj/18.22.6307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Quong M W, Massari M E, Zwart R, Murre C. A new transcriptional activation motif restricted to a class of helix-loop-helix proteins is functionally conserved in both yeast and mammalian cells. Mol Cell Biol. 1993;13:792–800. doi: 10.1128/mcb.13.2.792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Riechmann V, Cruchten I v, Sablitzky F. The expression pattern of Id4, a novel dominant negative helix-loop-helix protein, is distinct from Id1, Id2 and Id3. Nucl Acids Res. 1994;22:749–755. doi: 10.1093/nar/22.5.749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Riechmann V, Sablitzky F. Mutually exclusive expression of two dominant-negative helix-loop-helix (dnHLH) genes, Id4 and Id3, in the developing brain of the mouse suggests distinct regulatory roles of these dnHLH proteins during cellular proliferation and differentiation of the nervous system. Cell Growth and Differentiation. 1995;6:837–43. [PubMed] [Google Scholar]
- 135.Riley P, Anson-Cartwright L, Cross J C. The Hand1 bHLH transcription factor is essential for placentation and cardiac morphogenesis. Nature Genetics. 1998;18:271–5. doi: 10.1038/ng0398-271. [DOI] [PubMed] [Google Scholar]
- 136.Roberts V J, Steenbergen R, Murre C. Localization of E2A mRNA expression in developing and adult rat tissues. Proc Natl Acad Sci USA. 1993;90:7583–7587. doi: 10.1073/pnas.90.16.7583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Rothermel B, Thornton J, Butow R. Rtg3p, a basic helix-loop-helix/leucine zipper protein that functions in mitochondrial-induced changes in gene expression, contains independent activation domains. J Biol Chem. 1997;272:19801–19807. doi: 10.1074/jbc.272.32.19801. [DOI] [PubMed] [Google Scholar]
- 138.Rudnicki M, Braun T, Hinuma S, Jaenisch R. Inactivation of MyoD in mice leads to an up-regulation of the myogenic HLH gene Myf5 and results in apparently normal muscle development. Cell. 1992;71:383–390. doi: 10.1016/0092-8674(92)90508-a. [DOI] [PubMed] [Google Scholar]
- 139.Rudnicki M, Schnegelsbery P, Stead R, Braun T, Arnold H, Jaenisch R. MyoD or Myf5 is required for the formation of skeletal muscle. Cell. 1993;75:1351–1359. doi: 10.1016/0092-8674(93)90621-v. [DOI] [PubMed] [Google Scholar]
- 140.Rudolph H, Hinnen A. The yeast PHO5 promoter: phosphate-control elements and sequences mediating mRNA start-site selection. Proc Natl Acad Sci USA. 1987;84:1340–1344. doi: 10.1073/pnas.84.5.1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Rushlow C A, Hogan A, Pinchin S M, Howe K M, Lardelli M, Ish-Horowicz D. The Drosophila hairy protein acts in both segmentation and bristle patterning and shows homology to N-myc. EMBO J. 1989;8:3095–3103. doi: 10.1002/j.1460-2075.1989.tb08461.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Ruvkun G, Hobert O. The taxonomy of developmental control in Caenorhabditis elegans. Science. 1998;282:2033–2041. doi: 10.1126/science.282.5396.2033. [DOI] [PubMed] [Google Scholar]
- 143.Sanchez H B, Yieh L, Osborne T F. Cooperation by sterol regulatory element-binding protein and Sp1 in sterol regulation of low density lipoprotein receptor gene. J Biol Chem. 1995;270:1161–1169. doi: 10.1074/jbc.270.3.1161. [DOI] [PubMed] [Google Scholar]
- 144.Sánchez-García I, Rabbitts T. LIM domain proteins in leukaemia and development. Semin Cancer Biol. 1993;4:349–358. [PubMed] [Google Scholar]
- 145.Sartorelli V, Huang J, Hamamori Y, Kedes L. Molecular mechanisms of myogenic coactivation by p300: direct interaction with the activation domain of MyoD and with the MADS box of MEF2C. Mol Cell Biol. 1997;17:1010–1026. doi: 10.1128/mcb.17.2.1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Schlissel M, Voronova A, Baltimore D. Helix loop helix transcription factor-E47 activates germ-line immunoglobulin heavy-chain gene transcription and rearrangement in a pre-T-cell line. Genes Dev. 1991;5:1367–1376. doi: 10.1101/gad.5.8.1367. [DOI] [PubMed] [Google Scholar]
- 147.Schreiber-Argus N, Chin L, Chen K, Torres R, Rao G, Guida P, Skoultchi A, DePinho R. An amino-terminal domain of Mxi1 mediates anti-Myc oncogenic activity and interacts with a homolog of the yeast transcriptional repressor SIN3. Cell. 1995;80:777–786. doi: 10.1016/0092-8674(95)90356-9. [DOI] [PubMed] [Google Scholar]
- 148.Schreiber-Argus N, DePinho R. Repression by the Mad(Mxi1)-Sin3 complex. Bioessays. 1998;20:808–818. doi: 10.1002/(SICI)1521-1878(199810)20:10<808::AID-BIES6>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 149.Shen C P, Kadesch T. B-cell-specific DNA binding by an E47 homodimer. Mol Cell Biol. 1995;15:4518–4524. doi: 10.1128/mcb.15.8.4518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Shieh S-Y, Tsai M-J. Cell-specific and ubiquitous factors are responsible for the enhancer activity of the rat insulin II gene. J Biol Chem. 1991;266:16707–16714. [PubMed] [Google Scholar]
- 151.Shivdasani R, Mayer E, Orkin S. Absence of blood formation in mice lacking the T-cell leukaemia oncoprotein tal-1/SCL. Nature. 1995;373:432–434. doi: 10.1038/373432a0. [DOI] [PubMed] [Google Scholar]
- 152.Sigvardsson M, O'Riordan M, Grosschedl R. EBF and E47 collaborate to induce expression of the endogenous immunoglobulin surrogate light chain genes. Immunity. 1997;7:25–36. doi: 10.1016/s1074-7613(00)80507-5. [DOI] [PubMed] [Google Scholar]
- 153.Spicer D, Rhee J, Cheung W, Lassar A. Inhibition of myogenic bHLH and MEF2 transcription factors by the bHLH protein twist. Science. 1996;272:1476–1480. doi: 10.1126/science.272.5267.1476. [DOI] [PubMed] [Google Scholar]
- 154.Srivastava D, Cserjesi P, Olson E N. A subclass of bHLH proteins required for cardiac morphogenesis. Science. 1995;270:1995–1999. doi: 10.1126/science.270.5244.1995. [DOI] [PubMed] [Google Scholar]
- 155.Srivastava D, Thomas T, Lin Q, Kirby M L, Brown D, Olson E N. Regulation of cardiac mesodermal and neural crest development by the bHLH transcription factor, dHAND. Nat Genet. 1997;16:154–160. doi: 10.1038/ng0697-154. [DOI] [PubMed] [Google Scholar]
- 156.Staudt L, Lenardo M. Immunoglobulin gene transcription. Annu Rev Immunol. 1991;9:373–398. doi: 10.1146/annurev.iy.09.040191.002105. [DOI] [PubMed] [Google Scholar]
- 157.Sun X-H, Baltimore D. An inhibitory domain of E12 transcription factor prevents DNA binding in E12 homodimers but not in E12 heterodimers. Cell. 1991;64:459–470. doi: 10.1016/0092-8674(91)90653-g. [DOI] [PubMed] [Google Scholar]
- 158.Svaren J, Hörz W. Transcription factors vs nucleosomes: regulation of the PHO5 promoter in yeast. Trends Biochem Sci. 1997;22:93–97. doi: 10.1016/s0968-0004(97)01001-3. [DOI] [PubMed] [Google Scholar]
- 159.Svaren J, Schmitz J, Horz W. The transactivation domain of Pho4 is required for nucleosome disruption at the PHO5 promoter. EMBO J. 1994;13:4856–4862. doi: 10.1002/j.1460-2075.1994.tb06812.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Thompson J, Higgins D, Gibson T. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Van Doren M, Ellis H, Posakony J. The Drosophila extramacrochaete protein antogonizes sequence-specific DNA binding by daughterless/achaete-scute protein complexes. Development. 1991;113:245–255. doi: 10.1242/dev.113.1.245. [DOI] [PubMed] [Google Scholar]
- 162.Vassilev A, Yamauchi J, Kotani T, Prives C, Avantaggiati M L, Qin J, Nakatani Y. The 400 kDa subunit of the PCAF histone acetylase complex belongs to the ATM superfamily. Mol Cell. 1998;2:869–875. doi: 10.1016/s1097-2765(00)80301-9. [DOI] [PubMed] [Google Scholar]
- 163.Venter U, Svaren J, Schmitz J, Schmid A, Horz W. A nucleosome precludes binding of the transcription factor Pho4 in vivo to a critical target site in the PHO5 promoter. EMBO J. 1994;13:4848–4855. doi: 10.1002/j.1460-2075.1994.tb06811.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Venuti J, Cserjesi P. Molecular embryology of skeletal myogenesis. Curr Topics Dev Biol. 1996;34:169–206. doi: 10.1016/s0070-2153(08)60711-5. [DOI] [PubMed] [Google Scholar]
- 165.Wadman I A, Osada H, Grütz G G, Agulnick A D, Westphal H, Forster A, Rabbitts T H. The LIM-only protein Lmo2 is a bridging molecule assembling an erythroid, DNA-binding complex which includes the TAL1, E47, GATA-1 and Ldb1/NLI proteins. EMBO J. 1997;16:3145–3157. doi: 10.1093/emboj/16.11.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Warren A, Colledge W, Carlton M, Evans M, Smith A, Rabbitts T. The oncogenic cysteine-rich LIM domain protein rbtn2 is essential for erythroid development. Cell. 1994;78:45–57. doi: 10.1016/0092-8674(94)90571-1. [DOI] [PubMed] [Google Scholar]
- 167.Weintraub H, Dwarki V, Verma I, Davis R, Hollenberg S, Snider L, Lassar A, Tapscott S. Muscle-specific transcriptional activation by MyoD. Genes Dev. 1991;5:1377–1386. doi: 10.1101/gad.5.8.1377. [DOI] [PubMed] [Google Scholar]
- 168.Whelan J, Cordle S R, Henderson E, Weil P A, Stein R. Identification of a pancreatic β-cell insulin gene transcription factor that binds to and appears to activate cell-type-specific gene expression: its possible relationship to other cellular factors that bind to a common insulin gene sequence. Mol Cell Biol. 1990;10:1564–1572. doi: 10.1128/mcb.10.4.1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Wrischnik L A, Kenyon C J. The role of lin-22, a hairy/enhancer of split homolog, in patterning the peripheral nervous system of C. elegans. Development. 1997;124:2875–2888. doi: 10.1242/dev.124.15.2875. [DOI] [PubMed] [Google Scholar]
- 170.Yamada Y, Warren A J, Dobson C, Forster A, Pannell R, Rabbitts T H. The T cell leukemia LIM protein Lmo2 is necessary for adult mouse hematopoiesis. Proc Natl Acad Sci USA. 1998;95:3890–3895. doi: 10.1073/pnas.95.7.3890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Yan W, Young A Z, Soares V C, Kelley R, Benezra R, Zhuang Y. High incidence of T-cell tumors in E2A-null mice and E2A/Id1 double-knockout mice. Mol Cell Biol. 1997;17:7317–7327. doi: 10.1128/mcb.17.12.7317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Yang X-J, Ogryzko V, Nishikawa J, Howard B, Nakatani Y. A p300/CBP-associated factor that competes with the adenoviral oncoprotein E1A. Nature. 1996;382:319–324. doi: 10.1038/382319a0. [DOI] [PubMed] [Google Scholar]
- 173.Yokota Y, Mansouri A, Mori S, Sugawara S, Adachi S, Nishikawa S, Gruss P. Development of peripheral lymphoid organs and natural killer cells depends on the helix-loop-helix inhibitor Id2. Nature. 1999;397:702–706. doi: 10.1038/17812. [DOI] [PubMed] [Google Scholar]
- 174.Zervos A, Gyuris J, Brent R. Mxi1, a protein that specifically interacts with Max to bind Myc-Max recognition sites. Cell. 1994;79:388–398. [PubMed] [Google Scholar]
- 175.Zhang W, Behringer R R, Olson E N. Inactivation of the myogenic bHLH gene MRF4 results in up-regulation of myogenin and rib anomalies. Genes Dev. 1995;9:1388–1399. doi: 10.1101/gad.9.11.1388. [DOI] [PubMed] [Google Scholar]
- 176.Zhao C, Emmons S W. A transcription factor controlling development of peripheral sense organs in C. elegans. Nature. 1995;373:74–78. doi: 10.1038/373074a0. [DOI] [PubMed] [Google Scholar]
- 177.Zhao Q, Zhou X, Mattei M. TFEC, a basic helix-loop-helix protein, forms heterodimers with TFE3 and inhibits TFE3-dependent transcription activation. Mol Cell Biol. 1993;13:4505–4512. doi: 10.1128/mcb.13.8.4505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Zhuang Y, Cheng P, Weintraub H. B-lymphocyte development is regulated by the combined dosage of three basic helix-loop-helix genes, E2A, E2-2, and HEB. Mol Cell Biol. 1996;16:2898–2905. doi: 10.1128/mcb.16.6.2898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Zhuang Y, Soriano P, Weintraub H. The helix-loop-helix gene E2A is required for B cell formation. Cell. 1994;79:875–884. doi: 10.1016/0092-8674(94)90076-0. [DOI] [PubMed] [Google Scholar]