Abstract
The exosome is a protein complex consisting of a variety of 3′-to-5′ exonucleases that functions both in 3′-to-5′ trimming of rRNA precursors and in 3′-to-5′ degradation of mRNA. To determine additional exosome functions, we examined the processing of a variety of RNAs, including tRNAs, small nuclear RNAs (snRNAs), small nucleolar RNAs (snoRNAs), RNase P, RNase MRP, and SRP RNAs, and 5S rRNAs in mutants defective in either the core components of the exosome or in other proteins required for exosome function. These experiments led to three important conclusions. First, exosome mutants accumulate 3′-extended forms of the U4 snRNA and a wide variety of snoRNAs, including snoRNAs that are independently transcribed or intron derived. This finding suggests that the exosome functions in the 3′ end processing of these species. Second, in exosome mutants, transcripts for U4 snRNA and independently transcribed snoRNAs accumulate as 3′-extended polyadenylated species, suggesting that the exosome is required to process these 3′-extended transcripts. Third, processing of 5.8S rRNA, snRNA, and snoRNA by the exosome is affected by mutations of the nuclear proteins Rrp6p and Mtr4p, whereas mRNA degradation by the exosome required Ski2p and was not affected by mutations in RRP6 or MTR4. This finding suggests that the cytoplasmic and nuclear forms of the exosome represent two functionally different complexes involved in distinct 3′-to-5′ processing and degradation reactions.
The production of a wide variety of RNA species in eukaryotic cells requires specific 3′ trimming reactions. Such reactions occur in the processing of a diversity of primary transcripts, including those produced by RNA polymerase I, 5.8S rRNA, 18S rRNA, and 25S rRNA (reviewed in reference 35); those produced by RNA polymerase II, including both small nuclear RNAs (snRNAs) and small nucleolar RNAs (snoRNAs); and transcripts produced by RNA polymerase III, including tRNAs (reviewed in reference 10) and 5S rRNA (33). This diversity of substrates raises the issue of how each unique RNA is properly processed to generate a specific mature 3′ end. In addition, mRNAs are subjected to a variety of 3′ trimming events, including a specific-length trimming of the poly(A) tail (3), cytoplasmic deadenylation of the mRNA (reviewed in reference 14), and eventual 3′-to-5′ degradation of the body of the transcript in the cytoplasm (23, 24). An unresolved question is whether these different 3′ trimming reactions are carried out by a few relatively general 3′-to-5′ exonucleases, or if there are a large number of specific nucleases that each act on limited subsets of substrates.
Recent observations have suggested that at least two distinct 3′-to-5′ exonucleolytic processing events are performed by the same exonuclease complex. This is based on the identification of a multiprotein complex, termed the exosome, that contains at least 10 different polypeptides that have been shown to be 3′-to-5′ exonucleases or have sequence similarity to known 3′-to-5′ exonucleases (1, 22). More important, this complex has been shown to be required both for the 3′ processing of 5.8S rRNA in the nucleolus (22) and for the 3′-to-5′ pathway of mRNA degradation in the cytoplasm (13). Interestingly, these two different processing reactions appear to require additional proteins that are specific for each reaction. For example, although 3′-to-5′ degradation of mRNA requires Ski2p, Ski3p, and Ski8p, these Ski proteins are not required for 5.8S rRNA processing (13). Conversely, the Ski2p-related protein Mtr4p is known to be required for 5.8S rRNA processing (9). Since both Ski2p and Mtr4p are members of the DEVH box family of putative RNA helicases, an appealing model is that these proteins function in different exosome-mediated reactions, although it is not known whether Mtr4p also functions in cytoplasmic mRNA decay.
Since the components of the exosome are highly conserved and found in both the cytoplasm and the nucleus (1, 37), it was likely that this complex would have additional roles in 3′-to-5′ RNA processing and/or degradation. To identify such roles, we have systematically screened a number of Saccharomyces cerevisiae mutants defective in exosome function. Mutants impaired in the functioning of the nuclear exosome accumulate 3′-extended polyadenylated forms of U4 snRNA and independently transcribed snoRNAs and show alterations in snoRNAs produced from introns or polycistronic transcripts. This finding suggests that the exosome is involved in processing of these 3′-extended species. In addition, mutations in the nuclear proteins Rrp6p and Mtr4p affected processing of 5.8S rRNA (2, 9), snRNA, and snoRNA by the exosome. In contrast, mRNA degradation by the exosome required Ski2p, Ski3p, and Ski8p and was not affected by mutations in RRP6 or MTR4. This finding suggests that the cytoplasmic and nuclear forms of the exosome represent two functionally different complexes involved in 3′-to-5′ processing and degradation reactions.
MATERIALS AND METHODS
Strains and plasmids.
The genotypes of the S. cerevisiae strains used are given in Table 1. All but three strains are completely isogenic to each other; the exceptions are yRP1379, yRP1223, and yRP1381, which are partially isogenic as described below.
TABLE 1.
Strains used in this study
| Strain | Genotype | Reference |
|---|---|---|
| yRP686 | MATα leu2-3,112 lys2-201 trp1 ura3-52 | 11a |
| yRP687 | MATa leu2-3,112 his4-539 trp1 ura3-52 | 11a |
| yRP840 | MATa leu2-3,112 his4-539 trp1 ura3-52 cup1::LEU2/PGK1pG/MFA2pG | 11a |
| yRP841 | MATα leu2-3,112 lys2-201 trp1 ura3-52 cup1::LEU2/PGK1pG/MFA2pG | 11a |
| yRP1195 | MATa leu2-3,112 his4-539 trp1 ura3-52 cup1::LEU2/PGK1pG/MFA2pG ski2::LEU2 | 13 |
| yRP1196 | MATa leu2-3,112 his4-539 trp1 ura3-52 cup1::LEU2/PGK1pG/MFA2pG ski3::TRP1 | 13 |
| yRP1197 | MATa leu2-3,112 his4-539 lys2-201 trp1 ura3-52 cup1::LEU2/PGK1pG/MFA2pG ski8::URA3 | 13 |
| yRP1204 | MATα leu2-3,112 his4-539 lys2-201 trp1 ura3-52 cup1::LEU2/PGK1pG/MFA2pG ski6::URA3 [ski6-100/LYS2] | 13 |
| yRP1223 | MATα leu2 trp1 ura3 cup1::LEU2/PGK1pG/MFA2pG ade1-100 rrp4-1 | 13 |
| yRP1377 | MATα leu2-3,112 trp1 ura3-52 cup1::LEU2/PGK1pG/MFA2pG rrp6::URA3 | This study |
| yRP1378 | MATa leu2-3,112 trp1 ura3-52 cup1::LEU2/PGK1pG/MFA2pG mtr4::LEU2 [mtr4-1/URA3] | This study |
| yRP1379 | MATα leu2 his4-539 trp1 ura3-52 cup1::LEU2/PGK1pG/MFA2pG mtr3-1 | This study |
| yRP1380 | MATα leu2-3,112 lys2-201 trp1 ura3-52 cup1::LEU2/PGK1pG/MFA2pG mtr4::LEU2 [mtr4-1/URA3] | This study |
| yRP1381 | MATα leu2-3,112 ura3-52 cup1::LEU2/PGK1pG/MFA2pG rrp6::URA3 pap1-1 | This study |
| yRP1416 | MATα leu2-3,112 lys2-201 trp1 ura3-52 his4-539 cup1::LEU2/PGK1pG/MFA2pG | This study |
| yRP1417 | MATa leu2-3,112 lys2-201 trp1 ura3-52 his4-539 cup1::LEU2/PGK1pG/MFA2pG rrp6Δ::URA3 | This study |
| yRP1418 | MATa leu2-3,112 trp1 ura3-52 cup1::LEU2/PGK1pG/MFA2pG rnt1Δ::Neor rrp6Δ::URA3 | This study |
| yRP1419 | MATα leu2-3,112 trp1 ura3-52 cup1::LEU2/PGK1pG/MFA2pG rnt1Δ::Neor | This study |
The rrp6Δ strain was made by crossing yRP840 and yRP841. One copy of the RRP6 gene of the resulting diploid (yRP1375) was deleted and replaced by a URA3 marker by transformation with the SacI-to-ClaI fragment of pRP950, resulting in yRP1376. Successful gene disruption was confirmed by Southern analysis. Tetrads from yRP1376 were dissected to yield yRP1377. Spores were scored for rrp6Δ by their slow-growth phenotype and uracil autotrophy. pRP950 is a pBluescript SK+ derivative which contains positions −915 to −15 relative to the AUG codon of RRP6, the URA3 marker, and positions +82 to +727 relative to the stop codon of RRP6. This plasmid was constructed by amplifying these regions from total genomic DNA by using oligonucleotides containing restriction enzyme recognition sequences (XbaI, SacII, XhoI, and XmaI, respectively), and cloning the resulting PCR products into pRP312. pRP312 is a pBluescript SK+ derivative that contains the URA3 gene inserted into the BamHI site.
yRP1379 was made by crossing strain T208 (a generous gift from Alan M. Tartakoff) to yRP840. Spores were scored for the mtr3-1 mutation by their temperature sensitivity.
Strain yRP1380 was made by crossing yRP686 and yRP687. One copy of the MTR4 gene in the resulting diploid was knocked out by using plasmid pDM (19). Successful gene disruption was confirmed by Southern analysis. The resulting diploid was transformed with plasmid pRM(ts) (19), which carries the mtr4-1 allele on pRS316 (32). The resulting diploid was dissected, and spores were scored for mtr4::LEU2 by autotrophy and inability to grow on plates containing 5-fluoro-orotic acid, for mtr4-1 by the URA3 marker on the plasmid, by temperature-sensitive growth phenotype, and by 5.8S rRNA processing defect. The resulting strain (yRP1378) was crossed to yRP841, and spores were scored for mtr4-1 as before and for the CUP1::LEU2PM (11a) insert by Northern blotting.
Strain yRP1381 was made by crossing strain yRP1377 to yRP851. yRP851 was made by backcrossing the pap1-1 mutation (25) into the yRP840 background multiple times. The diploid was dissected, and the double mutant was scored by its uracil autotrophy and pap1-1 temperature-sensitive growth, which was partially suppressed by rrp6Δ as previously reported (2).
An rnt1Δ strain was constructed in the yrp840 strain background by amplifying an rnt1Δ::Neor cassette from a strain obtained from Research Genetics (record number 20825). The PCR product was used to transform the diploid strain yRP1376, which is heterozygous for rrp6Δ::URA3 (see above), and neomycin-resistant colonies were selected. Successful gene deletion was confirmed by Southern analysis. Tetrads from one such rnt1 deletion were dissected, and resulting strains were scored for neomycin resistance, uracil autotrophy, and RNA phenotypes. Two complete tetrads that contained all four possible combinations were analyzed. One of these tetrads yielded strains yRP1416 to yRP1419 and is shown.
RNA extraction and Northern blotting.
All yeast strains were grown in standard yeast extract-peptone (YEP) containing 2% galactose or 2% glucose. Strains carrying a deletion of a nonessential gene were grown at 30°C to an optical density at 600 nm of 0.3 to 0.5, and strains carrying a temperature-sensitive mutation were grown similarly at 24°C (a temperature permissive for all mutations used), followed by incubation for 1 h at 37°C (a temperature restrictive for all mutations used).
RNA was extracted, cleaved with RNase H, separated on agarose or polyacrylamide gels, transferred, and probed as described by Jacobs Anderson and Parker (13). Sequences of oligonucleotides used as probes are given in Table 2.
TABLE 2.
Oligonucleotides used in this study
| RNA species | snoRNA class | Name | Sequence |
|---|---|---|---|
| Independently transcribed snoRNAs | |||
| snR33 | H/ACA | oRP778 | AGGAACCGACTCAAACCGG (mature) |
| oRP880 | AAGTTTTGCAAATCGATTGTCC (3′ extended) | ||
| snR40 | C/D | oRP776 | ACCTGAGTACTTGTGGCATC (mature) |
| Intron-derived snoRNAs | |||
| U24 | C/D | oRP775 | CGGAACTCAAAGTTCCATCTG (mature) |
| snR44 | H/ACA | oRP785 | GTAAGAAGCATTTCCACATGGG (mature) |
| U18 | C/D | oRP774 | TCCCATCATAAACACGGACC (mature) |
| oRP | TACTCTGCTCTGTGCTATCG (3′ extended) | ||
| Polycistronic snoRNAs | |||
| snR190 | C/D | oRP772 | CCCTTGTCGTCATGGTCGA (mature) |
| U14 | C/D | oRP773 | TCCTACCGTGGAAACTGCG (mature) |
| oRP878 | GATACTACAGTATACGATCACTC (3′ extended) | ||
| snR72 | C/D | oRP852 | ATCAGACTGACGTGCAAATCAT (mature) |
| oRP882 | GAATTTTATATGGCTTGTGATCAG (3′ extended) | ||
| snR73 | C/D | oRP858 | AGCTCAGTACCACGCCCTG (mature) |
| oRP883 | GCTAAAAATATTAAAGCTCAGTAC (3′ extended) | ||
| snRNA | |||
| U4 | oRP756 | CGGACGAATCCTCACTGATA (mature) | |
| oRP768 | CAGTCCCTTTGAAAGAATGAAT (3′ extended) |
RESULTS
Experimental approach.
To identify additional functions for the exosome complex, we created a collection of eight strains carrying mutations in different components of the exosome, or in the proteins related to exosome function (Table 3), and then examined a diversity of 3′-to-5′ processing reactions in that collection of strains. Three strains that contained conditional temperature-sensitive alleles in core components of the exosome (i.e., rrp4-1, ski6-100, and mtr3-1) were used. These core components are thought to be subunits of both the nuclear and the cytoplasmic exosome (1) and are all essential. In addition, a strain carrying a deletion of RRP6 was examined. Rrp6p is an exonuclease associated with the exosome in nuclear fractions (1), and rrp6Δ strains have a defect in 5.8S rRNA maturation (2). A temperature-sensitive mtr4-1 strain was also included. Mtr4p is an essential protein required for 5.8S rRNA processing by the exosome (9). Last, strains with deletions of the nonessential SKI2, SKI3, and SKI8 genes were examined. These three genes are known to be required for exosome-mediated 3′-to-5′ degradation of mRNA (13).
TABLE 3.
Mutants used in this study
| Mutant | Protein | Protein required fora:
|
|||
|---|---|---|---|---|---|
| 5.8S rRNA processing | mRNA decay | snRNA/snoRNA processing | 5′ ETSb decay | ||
| rrp4-1 | Core exosome component | Yes (21) | Yes (13) | Yes (*) | Yes (9) |
| ski6-100 | Core exosome component | Yes (13) | Yes (13) | Yes (*) | Yes (9) |
| mtr3-1 | Core exosome component | Yes (16) | ND | Yes (*) | Yes (1) |
| rrp6Δ | Nuclear exosome component required for 3′ end processing of 5.8S rRNA | Yes (2) | No (*) | Yes (*) | Yes (1) |
| mtr4-1 | Nuclear DEVH box protein required for 3′ end processing of 5.8S rRNA | Yes (8) | No (*) | Yes (*) | Yes (9) |
| ski2Δ | DEVH box protein required for 3′-to-5′ degradation of mRNA | No (13) | Yes (13) | No (*) | ND |
| ski3Δ | Required for 3′-to-5′ degradation of mRNA | No (13) | Yes (13) | No (*) | ND |
| ski8Δ | Required for 3′-to-5′ degradation of mRNA | No (13) | Yes (13) | No (*) | ND |
Reference numbers are given in parentheses. (*), this study; ND, not determined.
External transcribed spaces of the pre-rRNA.
Yeast strains containing a deletion of a nonessential gene (SKI2, SKI3, SKI8, or RRP6) were grown at 30°C. Therefore, the pool of stable RNA in these strains represent a steady-state condition. In contrast, yeast strains containing conditional mutations in essential genes (RRP4, SKI6, MTR3, and MTR4) were grown at 24°C (a temperature permissive for growth) and shifted to 37°C (a temperature restrictive for growth) for 1 h. A relatively short shift to the restrictive temperature was used to minimize the occurrence of secondary effects. As a consequence, the pool of stable RNA present is a mixture of RNA synthesized during growth at the permissive temperature and RNA synthesized at the restrictive temperature. A wild-type strain was grown under both conditions. Total RNA from wild-type and mutant strains was analyzed by Northern blotting using oligonucleotide probes for a wide variety of RNA species. These included rRNA, tRNA, mRNA, snRNA, snoRNA, and the RNA subunits of SRP and RNases P and MRP. This collection of RNAs includes species transcribed by all three RNA polymerases and includes transcripts previously suggested to be processed from 3′-extended precursors, transcripts whose mature 3′ end appears to match the polymerase termination signal, and species for which little, if anything, is known about 3′ end formation.
Importantly, as detailed below, examination of U4 snRNA and snoRNAs revealed several alterations in these transcripts in the ski6-100, rrp4-1, rrp6Δ, mtr3-1, and mtr4-1 mutant strains but not in the ski2Δ, ski3Δ, or ski8Δ strains. In contrast, to date we have not found obvious defects in the processing of many other RNAs, including 5S rRNA, several tRNAs, and the RNA subunits of SRP and RNases MRP and P in any of the mutant strains examined (data not shown). This observation is consistent with these particular RNA species having mature 3′ ends that are formed by transcriptional termination, by processing by other 3′-to-5′ exonucleases (A. van Hoof, P. Lennertz, and R. Parker, unpublished data), or by redundancy of the exosome with other exonucleases for these functions. In either case, these results serve as negative controls that indicate that the defects described below are specific to U4 snRNA and snoRNAs.
Exosome mutants accumulate 3′-extended forms of snoRNAs and U4 snRNA. (i) The exosome is involved in the processing of independently transcribed snoRNAs and U4 snRNA.
snoRNAs are produced from independent transcripts, excised introns, or polycistronic transcripts (reviewed in references 20 and 34). To examine the role of the exosome in the processing of independently transcribed snoRNAs, we examined a member of each of the two major classes of snoRNAs (Table 2). snoRNAs can structurally and functionally be divided into C/D box-containing snoRNAs, required for methylation of the 2′ hydroxyl of RNA, and H/ACA box-containing snoRNAs, required for pseudouridyl formation in rRNA. For each class, we examined one snoRNA transcribed from an independent transcription unit. For the C/D box-containing snoRNAs, we examined snR40; for the H/ACA box-containing snoRNAs, we examined snR33.
Two sets of observations suggest that the exosome is involved in the processing of 3′-extended forms of snR33 and snR40. First, in rrp6Δ strains, the majority of snR33 and snR40 RNA species were longer by a few nucleotides (Fig. 1A and C, lower panels). This observation suggested that the exosome, and perhaps Rrp6p specifically, is involved in the removal of the last few 3′ nucleotides, to give rise to the mature 3′ end of these RNAs. The second alteration was that rrp6Δ, mtr4-1, ski6-100, mtr3-1, and rrp4-1 mutant strains all accumulated longer heterogeneous forms of the snR33 and snR40 transcripts (Fig. 1A and C, top panels). Hybridization of a Northern blot containing RNA from wild-type and rrp6Δ strains with a probe designed to hybridize 3′ of the mature snR33 showed that the heterogeneous population seen in Fig. 1A represents 3′-extended forms of snR33 (Fig. 1B). These observations suggest that snR33 and snR40 are made as 3′-extended forms that are then processed in a manner requiring the exosome (see below).
FIG. 1.
Exosome mutants have defects in processing of independently transcribed snoRNAs and U4 snRNA. Shown are two different exposures of the same Northern blots containing RNA from the indicated strains. All strains were grown in YEP–2% galactose to early to mid-log phase. The ski2Δ, ski3Δ, ski8Δ, and rrp6Δ strains were grown at 30°C. The ski6-100, rrp4-1, mtr3-1, and mtr4-1 strains were grown at 24°C and incubated for an additional hour at 37°C. Blots were probed for snR33 (A), 3′-extended snR33 (B), snR40 (C), U4 snRNA (D), or 3′-extended U4 snRNA (E). The species of 240 and 212 nt seen in panel C likely represent 5′ processing intermediates as recently proposed (5). wt, wild type. Positions of size markers (lanes M) are given in nucleotides.
Mutations affecting exosome function, especially mtr4-1, mtr3-1, ski6-100, and rrp4-1, led to only modest accumulation of the heterogeneous 3′-extended forms of snoRNAs. However, these defects are likely to be biologically significant for four reasons. First, snoRNAs are thought to be stable, and thus the pool of snoRNAs in a temperature-sensitive mutant after 1 h at the restrictive temperature reflects mostly RNA produced before the shift to the restrictive conditions. Second, the defects seen here are similar in strength to those seen in rRNA processing and mRNA degradation in the same mutants. For example, mutations in RRP4 or MTR4 led to only modest increases in 3′-extended 5.8S rRNA, which were detected only by long exposures or with use of probes designed not to hybridize to the mature RNA (9, 22). Third, the defects seen here are similar in strength to defects in snoRNA 5′ processing in a rat1-1 xrn1Δ double mutant. This mutant accumulates low levels of 5′-extended forms of snR190, U14, U24, and U18 snoRNAs after a 2-h incubation at the restrictive temperature (27), some of which were shown to exist only by using probes specific for 5′-extended forms. Fourth, reproducible accumulation of heterogeneous 3′-extended species were seen for six different snoRNAs from different classes (see below).
Analysis of the independently transcribed U4 snRNA revealed results similar to but distinct from those for snR33 and snR40. Wild-type strains accumulated a mature U4 species of 160 nucleotides (nt) and 3′-extended forms of between 270 and 300 nt. In addition, rrp6Δ, mtr3-1, mtr4-1, ski6-100, and rrp4-1 strains accumulated RNA species intermediate in size between the mature species and the 270- to 300-nt species. A heterogeneous population of transcripts was also seen in the rrp6Δ strain extending above the 270- to 300-nt precursor (Fig. 1D). The use of a specific oligonucleotide probe 3′ of the mature 3′ end of the U4 RNA showed that these species were extended on the 3′ side of the RNA (Fig. 1E). These data suggest that the exosome is involved in the processing of the 3′-extended forms of U4 snRNA.
(ii) The exosome is involved in the processing of intron-derived snoRNAs.
To determine if the exosome functions in the processing of intron-derived snoRNAs, we examined a member of each of the two major classes of snoRNAs. For the C/D box-containing snoRNAs, we examined U24; for the H/ACA box-containing snoRNAs, we examined snR44.
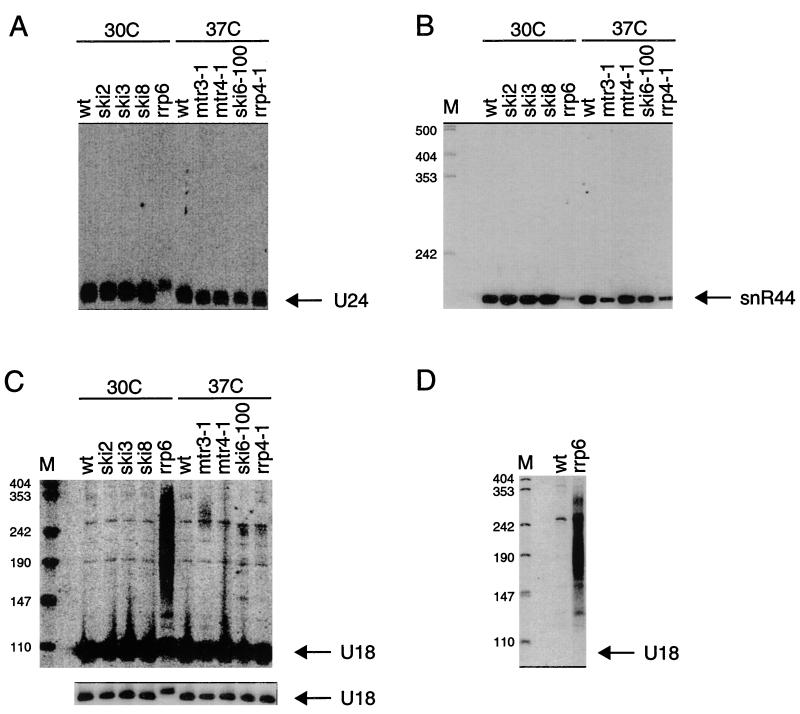
Intron-derived snoRNAs also showed altered patterns of accumulation in the various strains examined. First, for U24 in the rrp6Δ strain, the predominant RNA species was slightly larger than the normal RNA (Fig. 2A). This observation is similar to that noted above for the independently transcribed snoRNAs snR33 and snR40, and it suggests that the exosome, and perhaps Rrp6p specifically, is involved in the removal of the last few nucleotides to give rise to the mature 3′ end of these transcripts. Second, we observed a slight but reproducible reduction in the level of snR44 in the various mutants. The residual snR44 levels varied from 10% of the wild-type level in the rrp6Δ strain to 75% for the mtr4-1 strain (Fig. 2B). Less drastic reductions were seen for the level of U24 in various exosome mutants (Fig. 2A). One possible interpretation of these observations is that in the absence of proper 3′-to-5′ trimming, snR44 is degraded.
FIG. 2.
Exosome mutants have defects in processing of intron-derived snoRNAs. Shown are Northern blots containing RNA from the indicated strains. All strains were grown in YEP–2% galactose to early to mid-log phase. The ski2Δ, ski3Δ, ski8Δ, and rrp6Δ strains were grown at 30°C. The ski6-100, rrp4-1, mtr3-1, and mtr4-1 strains were grown at 24°C and incubated for an additional hour at 37°C. Blots were probed for U24 (A), snR44 (B), U18 (C), or 3′-extended U18 (D). In panel C, two different exposures of the same Northern blot are shown. wt, wild type. Positions of size markers (lanes M) are given in nucleotides.
We also examined the effects of exosome mutations on the processing of the U18 snoRNA. This snoRNA is embedded within the intron of the EFB1 gene, but it has been shown to be processed by two different pathways (36). In one pathway, it is processed from an intron released from the EFB1 pre-mRNA; in the other pathway, it is processed from the primary transcript in a manner that does not require splicing. Thus, this snoRNA does not fit neatly in either the intron-derived or the independently transcribed snoRNA class.
The defects seen for U18 snoRNA were similar to those seen for other snoRNAs. First, the rrp6Δ strain primarily accumulated a U18 RNA that was slightly larger than normal, as well as a heterogeneous population of larger species. A strain containing the mtr3-1 mutation (and possibly mtr4-1, ski6-100, and rrp4-1 strains) accumulated low levels of heterogeneous species of 200 to 300 nt (Fig. 2C). Probing of a Northern blot with RNA from wild-type and rrp6Δ strains with an oligonucleotide probe 3′ of the mature 3′ end of the U18 snoRNA showed that the heterogeneous species in rrp6Δ were extended on the 3′ side of the RNA (Fig. 2D). The observation that essentially no mature U18 accumulates in an rrp6Δ strain suggests that both pathways of U18 maturation are affected. Based on the defects seen in rrp6Δ, combined with the slight defects seen in other exosome mutants, we conclude that Rrp6p, and presumably also the core exosome, is involved in both pathways of U18 processing.
The exosome is involved in the processing of polycistronic snoRNAs.
To examine the role of the exosome in the processing of polycistronic snoRNAs, we examined two polycistronic precursors. One transcript examined contains snR190 and U14 C/D box-containing snoRNAs, with U14 being the 3′ snoRNA (6). The second polycistronic transcript examined contains seven C/D box-containing snoRNAs, in the 5′-to-3′ order of snR78, snR77, snR76, snR75, snR74, snR73, and snR72 (28). No H/ACA box-containing snoRNAs derived from polycistronic transcripts have been described to date.
Exosome mutants showed two distinct defects in the processing of snoRNAs from polycistronic precursors. Similar to what was seen with other snoRNAs, the rrp6Δ strain accumulated slightly larger than normal transcripts for U14, snR73, and snR72 (Fig. 3B to D, lower panels). No difference was seen for snR190 (Fig. 3A), but a difference of one or a few nucleotides might not be resolved, due to the relatively large size of this snoRNA. While snR33 is similar in size to snR190, we were able to detect an effect of rrp6Δ on snR33; thus, if rrp6Δ results in a larger snR190, this size difference must be smaller than the corresponding size difference for snR33. In addition to this effect, rrp6Δ, mtr3, mtr4, ski6, and rrp4 mutant strains accumulated heterogeneous populations of RNAs that were 3′-extended forms of U14, snR73, and possibly snR72 but not of snR190 (Fig. 3). These data, combined with the defects seen for independently transcribed and intron-derived snoRNAs, suggest that the exosome is involved in the processing of 3′-extended snoRNAs, irrespective of whether they are derived from independent transcripts, introns, or polycistronic transcripts.
FIG. 3.
Exosome mutants have defects in processing of polycistronic snoRNAs. Shown are two different exposures of the same Northern blots containing RNA from the indicated strains. All strains were grown in YEP–2% galactose to early to mid-log phase. The ski2Δ, ski3Δ, ski8Δ, and rrp6Δ strains were grown at 30°C. The ski6-100, rrp4-1, mtr3-1, and mtr4-1 strains were grown at 24°C and incubated for an additional hour at 37°C. Blots were probed for snR190 (A), U14 (B), snR73 (C), snR72 (D), 3′-extended U14 (E), 3′-extended snR73 (F), or 3′-extended snR72 (G). The species of 385 nt seen in panels A and B likely represents a dicistronic precursor, while the species of 184 nt seen in panel B likely represents U14 that contains a 5′ extension but carries the mature 3′ end. Based on known Rnt1p cleavage sites, the 238-nt species seen in panels C and F likely represents snR73 that has been released from its precursor but carries 5′ and 3′ extensions. wt, wild type. Positions of size markers (lanes M) are given in nucleotides.
Exosome mutants accumulate polyadenylated forms of U4 snRNA and some snoRNAs.
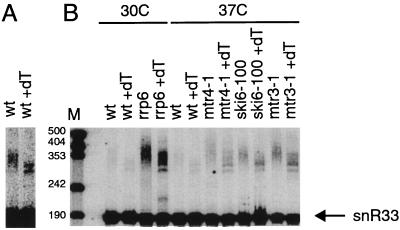
In the rrp6Δ mutant and in several of the other exosome mutant strains, we observed a larger heterogeneous population of transcripts derived from U4 snRNA and snR33, snR40, U18, U14, snR73, and snR72 snoRNAs. The simplest explanation is that this population represents a pool of polyadenylated RNAs with poly(A) tails varying in length. To examine this possibility, two experiments were performed. First, RNAs from different mutant strains and a wild-type strain were treated with RNase H in the presence of oligo(dT). RNase H cleaves RNA in a RNA-DNA duplex and is commonly used in combination with oligo(dT) to remove poly(A) tails in vitro. The products of these RNase H reactions were analyzed by probing a Northern blot for snR33. An important observation was that following treatment with RNase H and oligo(dT), the heterogeneous population migrated faster in the gel. Since the largest stretch of A's encoded in the corresponding region of the genome (three A's) is not long enough to efficiently hybridize to oligo(dT), this RNase H induced shift is not caused by a stretch of encoded A's and thus must be the result of a poly(A) tail. Removal of the poly(A) tail by RNase H treatment did not result in one discrete band, presumably because the poly(A) tail in individual molecules was added at different sites. This is similar to what occurs with mRNAs. For example, Graber et al. (11) recently analyzed 1,352 unique mRNA 3′ ends, which they found to be derived from 861 genes. Thus, even in this small sample, the average number of 3′ ends per gene was 1.6, indicating that many genes produce mRNAs with alternative 3′ ends. Our results indicate that the heterogeneous population seen in untreated samples corresponded to polyadenylated snoRNAs. These polyadenylated snR33 species are present at low levels in the wild type (Fig. 4A), and these levels are elevated in at least four strains carrying exosome mutations (Fig. 4B).
FIG. 4.
Wild-type and exosome mutant strains accumulate polyadenylated snR33. Shown is a Northern blot containing RNA from the indicated strains. All strains were grown in YEP–2% galactose to early to mid-log phase. The rrp6Δ strain was grown at 30°C. The ski6-100, mtr3-1, and mtr4-1 strains were grown at 24°C and incubated for an additional hour at 37°C. RNA was incubated with RNase H in the presence or absence of oligo(dT) as indicated. (A) Darker exposure of the wild-type (wt) lanes; (B) light exposure of the same gel. Positions of size markers (lane M) are given in nucleotides.
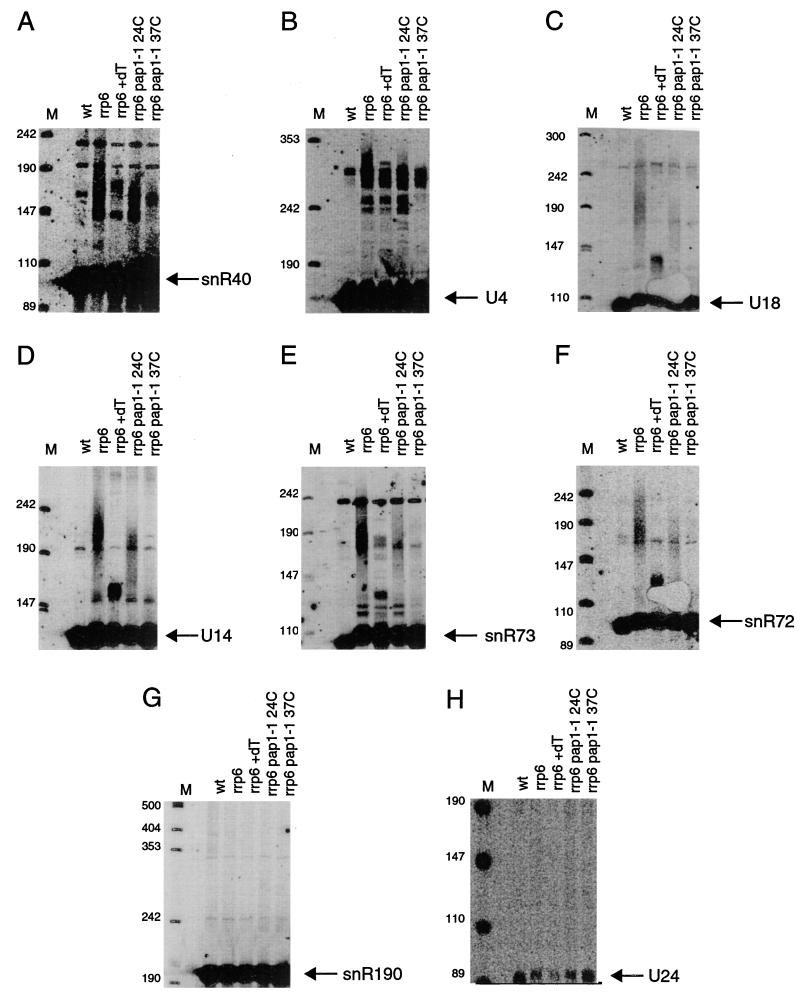
Similar experiments were also done to test whether the heterogeneous populations seen for snR40, U4, U18, U14, snR73, and snR72 in the rrp6Δ strain correspond to polyadenylated RNAs. Upon RNase H and oligo(dT) treatment, the heterogeneous population of 3′-extended species for U18, U14, snR73, and snR72 collapsed into a faster-migrating species (Fig. 5C to F, compare rrp6 lanes to rrp6 +dT lanes). While snR40 and U4 snRNA did not collapse into a single band upon RNase H and oligo(dT) treatment, the heterogeneous species for these two RNA migrate faster after this treatment, indicating that these species were also cleaved. Because the genomic regions just 3′ of these RNAs also do not contain long stretches of A's, this RNase H-induced shift must also be the result of a poly(A) tail. Thus, the heterogeneous species of all six RNAs are polyadenylated. As a negative control, the same analyses were done with snR190 and U24, which did not accumulate as heterogeneous populations. RNase H and oligo(dT) treatment did not result in a change in the pattern seen for either snR190 or U24 (Fig. 5G and H), suggesting that these two snoRNAs are not polyadenylated.
FIG. 5.
The rrp6Δ mutant accumulates polyadenylated forms of snR40, U4, U18, U14, snR73, and snR72 but not U24 or snR190. Shown are Northern blots containing RNA from the indicated strains. All strains were grown in YEP–2% galactose to early to mid-log phase at 24°C. The rrp6Δ pap1-1 double mutant was incubated for an additional hour at 37°C as indicated. RNA from the rrp6Δ strain was treated with RNase H and oligo(dT) as indicated. wt, wild type. Positions of size markers (lanes M) are given in nucleotides.
The second experiment to address whether the longer species were polyadenylated was to examine their presence in an rrp6Δ pap1-1 double-mutant strain. The PAP1 gene encodes the yeast poly(A) polymerase. The pap1-1 allele confers a temperature-sensitive defect in the addition of poly(A) tails to mRNA (25). Northern blot analysis of RNA from an rrp6Δ pap1-1 strain grown at 24°C showed that this strain accumulated the heterogeneous 3′-extended transcripts, but they were shorter than those in the rrp6Δ control strain (Fig. 5A to F, compare rrp6 lanes to rrp6 pap1-1 24C lanes). This indicates a partial defect in polyadenylation, even at the permissive temperature. More important, the heterogeneous population disappeared after a 1-h shift to 37°C (the restrictive temperature for pap1-1; Fig. 5A to F, compare rrp6 and rrp6 pap1-1 24C lanes to rrp6 pap1-1 37C lanes), indicating that Pap1p is required for the accumulation of this heterogeneous population.
We interpret the above data to suggest that at least a portion of the primary transcripts of snoRNAs are produced as polyadenylated species utilizing Pap1p and that the exosome functions to deadenylate these RNAs (see Discussion). Interestingly, the rrp6Δ pap1-1 double mutant did not accumulate abundant 3′-extended nonpolyadenylated snoRNAs that would comigrate with the species seen in the rrp6Δ +dT lanes. This finding indicates that while rrp6 mutants deadenylate these species slowly, further 3′ trimming is not as strongly affected. Finally, the polyadenylated snoRNA species that accumulated in the rrp6Δ pap1-1 mutant at 24°C disappeared within 1 h after inactivation of Pap1p. This observation suggests that this strain still contains an activity that can process, or degrade, the polyadenylated snoRNA species (see below for further discussion).
Rnt1p and the exosome act in the same pathway of U4 snRNA processing.
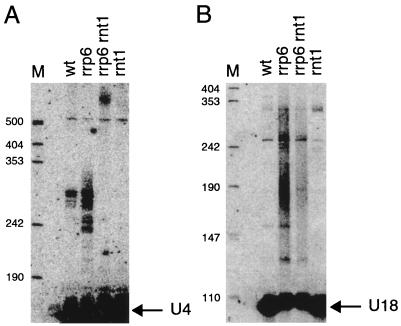
Recently it has been shown that Rnt1p, a double-strand-specific endoribonuclease, processes the U4 snRNA at two sites located 135 and 169 nt 3′ of the mature 3′ end, generating precursors of approximately 300 nt (1a). This is the same size as the 3′-extended U4 RNA that we detect in our Northern blots and that accumulates as a polyadenylated species in the rrp6Δ strain. These observations suggest two mechanisms for the formation of the polyadenylated U4 RNAs. In one model, the 3′-extended forms of the U4 snRNA would be made by the assembly of the normal mRNA polyadenylation machinery onto the nascent transcript, which would then lead to cleavage and polyadenylation. This would generate polyadenylated transcripts similar in size to the Rnt1p cleavage products but independently of Rnt1p. Alternatively, following Rnt1p cleavage, the 3′-extended U4 snRNA could be a substrate for polyadenylation by Pap1p. This would be striking since it would imply that the poly(A) polymerase can function on a second class of substrates cleaved by a distinct endonuclease. This latter hypothesis predicts that the formation of the polyadenylated U4 RNAs should be dependent on the Rnt1p.
To test this prediction, we examined the processing of the U4 snRNA in rnt1Δ and rnt1Δ rrp6Δ double-mutant strains. As expected, the rnt1Δ strain lacked the approximately 300-nt-long 3′-extended forms of U4 snRNA that are normally seen in wild-type cells. More important, in the rnt1Δ rrp6Δ double mutant strain the 3′-extended U4 snRNA species normally seen in rrp6 strains disappeared. Instead, a new heterogeneous species of >500 nt that may itself be polyadenylated appeared in the double mutant (Fig. 6A). These data strongly suggest that the polyadenylated U4 RNAs are produced by the addition of poly(A) tails to precursors that were cleaved by Rnt1p (see Discussion). For comparison, the processing of U18 snoRNA was analyzed in the rnt1Δ rrp6Δ strain. Rnt1p has no known role in U18 snoRNA processing, and the rnt1Δ mutation did not affect U18 processing (Fig. 6B) or prevent the accumulation of polyadenylated U18 species in the rrp6Δ strain, although the abundance of the U18 polyadenylated species was slightly reduced.
FIG. 6.
Rnt1p and Rrp6p act in the same pathway of U4 snRNA processing. Shown are Northern blots containing RNA from the indicated strains. All strains were grown in YEP–2% glucose to early to mid-log phase at 24°C. Blots were probed for U4 snRNA (A) or U18 snoRNA (B) as indicated. wt, wild type. Positions of size markers (lanes M) are given in nucleotides.
Effects of mutations in exosome components on mRNA degradation.
Since the exosome is involved in deadenylation of snoRNAs, as well as in 3′-to-5′ degradation of mRNA, the exosome might also function in the cytoplasmic deadenylation of mRNA. To examine this possibility, we measured the deadenylation rate of the MFA2pG and PGK1pG mRNAs (11a) by transcriptional pulse-chase analyses (8). In this analysis, a short pulse of transcription is used to produce mRNA with long poly(A) tails. The rate of deadenylation can then be determined in a chase period following transcription shutoff. This analysis indicated that ski2Δ, rrp6Δ, rrp4-1, ski6-100, and mtr3-1 mutants shortened the poly(A) tail on both PGK1 and MFA2 at rates indistinguishable from those for the wild type (data not shown). This finding is consistent with prior observations that overall mRNA decay rates are not substantially altered in exosome mutants (data not shown and reference 13) and argues that the exosome is not required for cytoplasmic deadenylation, although it might be functionally redundant with other 3′-to-5′ exonucleases in the cytoplasm.
We also determined the possible role of different exosome subunits in 3′-to-5′ mRNA degradation of the body of the transcript following deadenylation. A simple assay for 3′-to-5′ degradation of mRNA is to assess the levels and integrity of a poly(G)-to-3′-end fragment from the MFA2pG transcript (13). This fragment is produced by the 5′-to-3′ mRNA decay pathway. Because it is normally degraded by the exosome, it accumulates in mutants defective in 3′-to-5′ decay of mRNA (13). As described by Jacobs Anderson and Parker (13), ski2Δ, ski3Δ, ski8Δ, or ski6-100 resulted in the appearance of a ladder of partially degraded mRNA fragment (Fig. 7). In contrast, the rrp6Δ and mtr4-1 mutants accumulated MFA2pG mRNA degradation intermediates at levels similar to those for the wild type, indicating that Rrp6p and Mtr4p are not required for 3′-to-5′ degradation of mRNA.
FIG. 7.
mRNA degradation phenotypes of exosome mutants. Shown is a Northern blot containing RNA from the indicated strains. All strains were grown in YEP–2% galactose to early to mid-log phase. The ski2Δ, ski3Δ, ski8Δ, and rrp6Δ strains were grown at 30°C. The ski6-100 and mtr4-1 strains were grown at 24°C and incubated for an additional hour at 37°C. The blot was probed with oRP140, which hybridizes to the full-length MFA2pG mRNA and a degradation intermediate as indicated. wt, wild types. Positions of size markers (lane M) are given in nucleotides.
We also genetically tested the involvement of Rrp6p in 3′-to-5′ decay of mRNA. This test is based on the observation that mutations that block 5′-to-3′ decay of mRNA (such as deletion of the gene encoding the decapping enzyme, DCP1) are synthetically lethal with mutations that block the alternative 3′-to-5′ pathway (13, 15). A dcp1Δ strain was crossed with an rrp6Δ strain, and tetrads from the resulting diploid were dissected. Although both single-mutant strains grow slowly, the double mutant was recovered at the expected frequency and did not show an additional growth defect (data not shown). Similarly, an mtr4-1 mutant was crossed to a dcp1Δ strain. The double mutants were recovered at the expected frequency from this cross, and these double mutants did not show a more severe growth defect at any temperature tested (23 to 36°C). Thus, both genetic and molecular data indicate that the exosome and the Ski2p, Ski3p, and Ski8p are required for 3′-to-5′ degradation of mRNA but Rrp6p and Mtr4p are not (see Discussion).
DISCUSSION
The exosome functions in 3′-to-5′ processing of some snRNAs and snoRNAs.
Our data suggest that the exosome functions in the processing of U4 snRNA and snoRNA species in two manners. One role of the exosome is to deadenylate 3′-extended forms of these transcripts that contain poly(A) tails. The key observation is that exosome mutants, and particularly rrp6Δ strains, accumulate 3′-extended polyadenylated U4 snRNA and snoRNA species. Evidence that these heterogeneous populations are polyadenylated is that they are converted into shorter species upon treatment with RNase H in the presence of oligo(dT). In addition, these heterogeneous populations are shorter in an rrp6Δ pap1-1 double mutant strain at 24°C than in the rrp6Δ mutant and disappear after this double mutant has been shifted to the restrictive temperature of 37°C for 1 h. Since both Rrp6p and Mtr4p are involved in processing of U4 and snoRNAs and both of these proteins have been localized to the nucleus (1, 19), this processing most likely occurs in the nucleus.
Two sets of observations suggest that the exosome also functions in 3′-to-5′ trimming of U4 snRNA and snoRNA species in addition to the role in deadenylation. First, in various exosome mutants, longer discrete species accumulate for a variety of transcripts. For example, discrete 3′-extended U4 transcripts are detected in the mtr4-1, mtr3-1, rrp4-1, rrp6Δ, and ski6-100 mutants (Fig. 1). Second, the RNase H oligo(dT) treatment showed that the poly(A) tail of independently transcribed snoRNA species was added at a site (or sites) 3′ of the mature 3′ end (Fig. 4 and 5). If the poly(A) tail had been added to the mature 3′ end, the heterogeneous population would have collapsed into the mature RNA band. This result indicates a requirement for further 3′ trimming after removal of the poly(A) tail. This further trimming is not carried out correctly in an rrp6Δ strain, resulting in snoRNAs slightly (2- to 5-nt) larger than wild-type snoRNAs (Fig. 1 and 3). Similarly, the rrp6Δ strain accumulated snoRNA species that were slightly (∼3-nt) larger than wild type for the intron-derived U24 snoRNA, which apparently is not deadenylated by the exosome. We have not seen these slightly larger species in any of the conditional exosome alleles tested. It is not clear whether Rrp6p has a special role or whether the difference in this particular exosome phenotype seen between rrp6Δ and other mutant strains is caused by the specific alleles used (see below). The conclusion that the exosome functions in the 3′-to-5′ trimming of snoRNAs is also supported by similar results from Allmang et al. (1a).
Relationship of polyadenylation to 3′ processing of U4 snRNA and snoRNAs.
The accumulation of polyadenylated snRNAs and snoRNAs in the various mutant strains raises the related issues of the mechanism by which these poly(A) tails are produced and the relationship of the polyadenylation to the normal pathway of transcript maturation. Our data suggest that there are two mechanisms for the polyadenylation of these transcripts. In some cases, it appears that the polyadenylation of the transcript follows cleavage by the RNase III homolog, Rnt1p. The critical observation is that the polyadenylated U4 transcripts seen in an rrp6Δ strain are not produced in an rnt1Δ rrp6Δ double mutant (Fig. 6). Since the polyadenylation of the U4 transcripts also requires the poly(A) polymerase encoded by the PAP1 gene (Fig. 3), this suggests that the same polymerase that normally is directed to RNA substrates by the cleavage and polyadenylation machinery can also add poly(A) tails to 3′ ends generated by a different endonuclease.
A second possible mechanism for the synthesis of 3′-extended polyadenylated U4 snRNAs and snoRNAs would be by 3′ end formation and concurrent polyadenylation, similar to the formation of poly(A) tails on mRNA. This would be similar to the proposed role of polyadenylation in the biogenesis of the telomerase RNA subunit in yeast (7), another stable RNA. This possibility is supported by the observation that the U18 snoRNA receives a poly(A) tail in a manner that is largely Rnt1p independent (Fig. 6B). In addition, RNase H and oligo(dT) treatment roughly maps the polyadenylation site for snR73 to about position +30 relative to the mature 3′ end. This does not match the known Rnt1p cleavage site (at position +71). This mechanism of polyadenylation would also be consistent with the detection of polyadenylated snR33 transcripts in wild-type cells (Fig. 4) and with the detection of a longer heterogeneous U4 snRNA transcript in the rnt1Δ rrp6Δ double mutant (Fig. 6A).
A related issue is the relationship of the polyadenylation of the transcripts and their normal processing. For some snoRNAs (e.g., snR33), detection of the polyadenylated species at low levels in wild-type strains suggests that these are normal intermediates in the biogenesis of the transcript. For U4 snRNA and other snRNAs, there are two possible relationships between polyadenylation and normal processing. First, the polyadenylation of these transcripts could be a normal step in their biogenesis, and the polyadenylated species is normally rapidly processed by the Rrp6p and the exosome and is therefore not detectable in wild-type strains. Alternatively, these transcripts could become polyadenylated as a result of a block in their normal processing. For example, the U4 RNA could normally be processed by Rrp6p and the exosome following Rnt1p cleavage, but if this 3′ trimming is inhibited by the rrp6Δ or conditional exosome lesions, the Rnt1p cleavage product, or slightly trimmed versions of it, could be adenylated by Pap1p. This would be similar to what occurs in Escherichia coli, wherein RNAs resistant to degradation can become polyadenylated in order to promote further processing (18). At present, these two possibilities cannot be clearly distinguished. However, since unadenylated 3′-extended precursor of most snoRNAs do not accumulate in a pap1-1 rrp6Δ mutant strain, the addition of poly(A) tails is not a required step in the biogenesis of these RNAs.
In the cases of U18 and snR73, there may be two independent pathways for processing. One would be initiated by cleavage and polyadenylation of the nascent transcript relatively close to the mature 3′ end (at approximately +25 and +30, respectively), while the other would be initiated by cleavage and polyadenylation further downstream (downstream of the second EFB1 exon and snR72, respectively). An interesting implication of the possibility of 3′ end formation by different mechanisms is that it allows the production of different amounts of individual snoRNAs from one polycistronic gene or of intron-derived snoRNAs and mRNAs. This could simply be controlled by altering the relative frequency at which different cleavage and polyadenylation sites are used. The generation of snoRNA 3′ ends by two different mechanisms may be a general phenomenon of many snoRNA and snRNAs. For example, the U5 snRNA in yeast exists as a long and a short form (U5L and U5S), which are produced by alternative cleavage and processing pathways (4).
Implications of polyadenylated snoRNAs for the role of the nucleolus in mRNA transport.
The demonstration that several different snoRNAs accumulate as polyadenylated species in exosome mutants has implications for the interpretation of nuclear polyadenylated RNA accumulation in various mRNA export mutants. For example, mtr3-1 and mtr4-1 strains were initially isolated as mutants that accumulated polyadenylated RNA in the nucleus, based on in situ hybridization with labeled oligo(dT) (16). This was interpreted as a defect in mRNA export from the nucleus. Moreover, since the in situ localization of the polyadenylated species coincided with the nucleolus (17, 19), these observations have led to the hypothesis that mRNA may be transported through the nucleolus (26, 31). An alternative hypothesis is that this nucleolar polyadenylated RNA accumulation in the mtr3-1 and mtr4-1 mutants is due to the accumulation of polyadenylated snoRNAs we have described above. This hypothesis is supported by the observation that the poly(A) in situ signal in mtr3-1 and mtr4-1 strains colocalizes with Nop1p, a subunit of the C/D box snoRNA particle (17, 19). Moreover, snoRNAs may be sufficiently abundant to be detected by in situ hybridization. Thirty-eight independently transcribed snoRNAs and five polycistronic snoRNA genes have been characterized, and it appears likely that a significant number remain to be discovered. The abundance of each snoRNA in yeast cells is not well established, but published estimates vary between 10 to 500 and 200 to >1,000 molecules per snoRNA species (20, 29, 30). Considering that there are 15,000 mRNA molecules per yeast cell (12), snoRNAs may significantly contribute to the nucleolar poly(A) signal in mtr3 and mtr4 (and perhaps other) mutants.
Distinct phenotypes seen in rrp6Δ strains and other exosome mutants.
The different phenotypes for snoRNA processing seen in rrp6Δ strains, compared to other exosome mutants, may have implications for structure-function relationships of the exosome. For example, although an rrp6Δ strain accumulates mainly snoRNAs with short 3′ extensions, rrp4-1, ski6-100, mtr3-1, and mtr4-1 strains all accumulate mainly snoRNAs of wild-type size, with the accumulation of smaller amounts of larger species following a shift to the restrictive temperature. Interestingly, similar results have been obtained for 5.8S rRNA processing (data not shown; reference 1; compare data in reference 2 with those in references 9 and 21). In this case, rrp6 strains accumulate half of their 5.8S rRNA as a species that is approximately 30 nt larger than wild-type 5.8S rRNA (2), while other exosome mutants do not. In addition, rrp6 mutants accumulate significantly higher levels of polyadenylated U4 snRNA and snoRNAs than other exosome mutant strains.
At least four distinct hypotheses can explain the differences between rrp6Δ and other exosome mutants. First, the difference may simply be due to the fact that the rrp6Δ phenotype is a steady-state phenotype, while the other phenotypes are seen when the cells have been incubated at the restrictive temperature for a relatively short time. Second, the different phenotypes could reflect different catalytic roles of exosome subunits. In the case of U4, the phenotypes could result from preferred removal of the poly(A) tail by Rrp6p, followed by processing by any subunit of the exosome, snoRNAs would be processed by preferred removal of the poly(A) tail by Rrp6p and specific removal of the last few nucleotides by Rrp6p. Third, Rrp6p might act independently as a monomeric exonuclease in addition to being a part of the exosome. In rrp6Δ strains, the phenotype would then be more severe because these strains are missing not only a completely functional exosome but also the monomeric form of Rrp6p. Fourth, Rrp6p may not have a catalytic role at all, but in the absence of Rrp6p an altered exosome that does not fully carry out its normal function may assemble. These hypotheses could be tested by analyses of conditional alleles and/or catalytic site mutants of Rrp6p, but no such alleles exist at this time.
Two functionally distinct exosome complexes.
Several observations now suggest that the exosome is not a homogeneous complex, but that there are at least two functionally distinct exosome complexes in the cell that can be distinguished genetically. The key observation here is that while some mutations of exosome subunits (i.e., rrp4-1 and ski6-100) result in defects in mRNA degradation (13), rRNA processing (13, 22), snRNA processing, and snoRNA processing, there are two classes of mutations that specifically disrupt some but not other exosome functions. The first class includes rrp6Δ and mtr4-1, which cause accumulation of 3′-extended forms of 5.8S rRNA, U4 snRNA, and snoRNAs but have no effect on 3′-to-5′ mRNA degradation. In contrast, yeast strains containing ski2, ski3, or ski8 deletions are completely defective in exosome-mediated mRNA degradation but are indistinguishable from wild-type strains for all known other exosome functions.
Three points suggest that these two functionally distinct sets of processing reactions reflect differences between cytoplasmic and nuclear reactions. First, Rrp6p and Mtr4p are localized to the nucleus (1, 19), whereas Rrp4p is localized to both the nucleus and the cytoplasm (1). Second, the nature of the processing reactions is consistent with this distinction. For example, rRNA, snRNA, and snoRNA processing are thought to be nuclear events, whereas mRNA degradation is thought to be a cytoplasmic event. Third, purification of exosomes demonstrated that the Rrp6p was found only in association with a fraction of the exosome complex (1). It is notable that all of the nuclear events are processing reactions with specific endpoints and that the cytoplasmic mRNA decay is a complete digestion of the molecule. This suggests that there will be features, either of the substrate, or of the different exosome complexes, which dictate its endpoint in the 3′-to-5′ digestion. A detailed understanding of both the biochemical activities and structure-function relations of the exosome and its associated proteins is necessary to resolve these questions.
ACKNOWLEDGMENTS
We thank Alan M. Tartakoff for helpful comments on the manuscript, mtr3-1 and mtr4-1 strains, and mtr4 plasmids. We are grateful to members of the Parker laboratory and Harold E. Smith for helpful comments on the manuscript.
This work was supported by the Howard Hughes Medical Institute and NIH grant GM45443 to R.P.
REFERENCES
- 1.Allmang C, Petfalski E, Podtelejnikov A, Mann M, Tollervey D, Mitchell P. The yeast exosome and human PM-Scl are related complexes of 3′ to 5′ exonucleases. Genes Dev. 1999;13:2148–2158. doi: 10.1101/gad.13.16.2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1a.Allmang C, Kufel J, Chanfreau G, Mitchell P, Petfalski E, Tollervey D. Functions of the exosome in rRNA, snoRNA and snRNA synthesis. EMBO J. 1999;18:5399–5410. doi: 10.1093/emboj/18.19.5399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Briggs M W, Burkard K T, Butler J S. Rrp6p, the yeast homologue of the human PM-Scl 100-kDa autoantigen, is essential for efficient 5.8 S rRNA 3′ end formation. J Biol Chem. 1998;273:13255–13263. doi: 10.1074/jbc.273.21.13255. [DOI] [PubMed] [Google Scholar]
- 3.Brown C E, Sachs A B. Poly(A) tail length control in Saccharomyces cerevisiae occurs by message-specific deadenylation. Mol Cell Biol. 1998;18:6548–6559. doi: 10.1128/mcb.18.11.6548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chanfreau G, Elela S A, Ares M, Jr, Guthrie C. Alternative 3′-end processing of U5 snRNA by RNase III. Genes Dev. 1997;11:2741–2751. doi: 10.1101/gad.11.20.2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chanfreau G, Legrain P, Jacquier A. Yeast RNase III as a key processing enzyme in small nucleolar RNAs metabolism. J Mol Biol. 1998;284:975–988. doi: 10.1006/jmbi.1998.2237. [DOI] [PubMed] [Google Scholar]
- 6.Chanfreau G, Rotondo G, Legrain P, Jacquier A. Processing of a dicistronic small nucleolar RNA precursor by the RNA endonuclease Rnt1. EMBO J. 1998;17:3726–3737. doi: 10.1093/emboj/17.13.3726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chapon C, Cech T R, Zaug A J. Polyadenylation of telomerase RNA in budding yeast. RNA. 1997;3:1337–1351. [PMC free article] [PubMed] [Google Scholar]
- 8.Decker C J, Parker R. A turnover pathway for both stable and unstable mRNAs in yeast: evidence for a requirement for deadenylation. Genes Dev. 1993;7:1632–1643. doi: 10.1101/gad.7.8.1632. [DOI] [PubMed] [Google Scholar]
- 9.de la Cruz J, Kressler D, Tollervey D, Linder P. Dob1p (Mtr4p) is a putative ATP-dependent RNA helicase required for the 3′ end formation of 5.8S rRNA in Saccharomyces cerevisiae. EMBO J. 1998;17:1128–1140. doi: 10.1093/emboj/17.4.1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deutscher M P. Ribonucleases, tRNA nucleotidyltransferase and the 3′ processing of tRNA. Prog Nucleic Acids Res Mol Biol. 1990;39:209–240. doi: 10.1016/s0079-6603(08)60628-5. [DOI] [PubMed] [Google Scholar]
- 11.Graber J H, Cantor C R, Mohr S C, Smith T F. Genomic detection of new yeast pre-mRNA 3′-end-processing signals. Nucleic Acids Res. 1999;27:888–894. doi: 10.1093/nar/27.3.888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11a.Hatfield L, Beelman C A, Stevens A, Parker R. Mutations in trans-acting factors affecting mRNA decapping in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:5830–5838. doi: 10.1128/mcb.16.10.5830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hereford L M, Rosbash M. Number and distribution of polyadenylated RNA sequences in yeast. Cell. 1977;10:453–462. doi: 10.1016/0092-8674(77)90032-0. [DOI] [PubMed] [Google Scholar]
- 13.Jacobs Anderson J S, Parker R. The 3′ to 5′ degradation of yeast mRNAs is a general mechanism for mRNA turnover that requires the SKI2 DEVH box protein and 3′ to 5′ exonucleases of the exosome complex. EMBO J. 1998;17:1497–1506. doi: 10.1093/emboj/17.5.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jacobson A. Poly(A) metabolism and translation: the closed loop model. In: Hershey J W B, Mathews M B, Sonenberg N, editors. Translational control. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 451–480. [Google Scholar]
- 15.Johnson A W, Kolodner R D. Synthetic lethality of sep1 (xrn1) ski2 and sep1 (xrn1) ski3 mutants of Saccharomyces cerevisiae is independent of killer virus and suggests a general role for these genes in translation control. Mol Cell Biol. 1995;15:2719–2727. doi: 10.1128/mcb.15.5.2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kadowaki T, Chen S, Hitomi M, Jacobs E, Kumagai C, Liang S, Schneiter R, Singleton D, Wisniewska J, Tartakoff A M. Isolation and characterization of Saccharomyces cerevisiae mRNA transport-defective (mtr) mutants. J Cell Biol. 1994;126:649–659. doi: 10.1083/jcb.126.3.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kadowaki T, Schneiter R, Hitomi M, Tartakoff A M. Mutations in nucleolar proteins lead to nucleolar accumulation of polyA+ RNA in Saccharomyces cerevisiae. Mol Biol Cell. 1995;6:1103–1110. doi: 10.1091/mbc.6.9.1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li Z, Pandit S, Deutscher M P. Polyadenylation of stable RNA precursors in vivo. Proc Natl Acad Sci USA. 1998;95:12158–12162. doi: 10.1073/pnas.95.21.12158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liang S, Hitomi M, Hu Y H, Liu Y, Tartakoff A M. A DEAD-box-family protein is required for nucleocytoplasmic transport of yeast mRNA. Mol Cell Biol. 1996;16:5139–5146. doi: 10.1128/mcb.16.9.5139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maxwell E S, Fournier M J. The small nucleolar RNAs. Annu Rev Biochem. 1995;64:897–934. doi: 10.1146/annurev.bi.64.070195.004341. [DOI] [PubMed] [Google Scholar]
- 21.Mitchell P, Petfalski E, Tollervey D. The 3′ end of yeast 5.8S rRNA is generated by an exonuclease processing mechanism. Genes Dev. 1996;10:502–513. doi: 10.1101/gad.10.4.502. [DOI] [PubMed] [Google Scholar]
- 22.Mitchell P, Petfalski E, Shevchenko A, Mann M, Tollervey D. The exosome: a conserved eukaryotic RNA processing complex containing multiple 3′→5′ exoribonucleases. Cell. 1997;91:457–466. doi: 10.1016/s0092-8674(00)80432-8. [DOI] [PubMed] [Google Scholar]
- 23.Muhlrad D, Parker R. Premature translational termination triggers mRNA decapping. Nature. 1994;370:578–581. doi: 10.1038/370578a0. [DOI] [PubMed] [Google Scholar]
- 24.Muhlrad D, Decker C J, Parker R. Turnover mechanisms of the stable yeast PGK1 mRNA. Mol Cell Biol. 1995;15:2145–2156. doi: 10.1128/mcb.15.4.2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Patel D, Butler J S. Conditional defect in mRNA 3′ end processing caused by a mutation in the gene for poly(A) polymerase. Mol Cell Biol. 1992;12:3297–3304. doi: 10.1128/mcb.12.7.3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pederson T. The pluripotent nucleolus. Nucleic Acids Res. 1998;26:3871–3876. doi: 10.1093/nar/26.17.3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Petfalski E, Dandekar T, Henry Y, Tollervey D. Processing of the precursors to small nucleolar RNAs and rRNAs requires common components. Mol Cell Biol. 1998;18:1181–1189. doi: 10.1128/mcb.18.3.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Qu L H, Henras A, Lu Y J, Zhou H, Zhou W X, Zhu Y Q, Zhao J, Henry Y, Caizergues-Ferrer M, Bachellerie J P. Seven novel methylation guide small nucleolar RNAs are processed from a common polycistronic transcript by Rat1p and RNase III in yeast. Mol Cell Biol. 1999;19:1144–1158. doi: 10.1128/mcb.19.2.1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rasmussen T P, Culbertson M R. Analysis of yeast trimethylguanosine-capped RNAs by midwestern blotting. Gene. 1996;182:89–96. doi: 10.1016/s0378-1119(96)00519-7. [DOI] [PubMed] [Google Scholar]
- 30.Riedel N, Wise J A, Swerdlow H, Mak A, Guthrie C. Small nuclear RNAs from Saccharomyces cerevisiae: unexpected diversity in abundance, size, and molecular complexity. Proc Natl Acad Sci USA. 1986;83:8097–8101. doi: 10.1073/pnas.83.21.8097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schneiter R, Kadowaki T, Tartakoff A M. mRNA transport in yeast: time to reinvestigate the functions of the nucleus. Mol Biol Cell. 1995;6:357–370. doi: 10.1091/mbc.6.4.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sikorski R S, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tekamp P A, Garcea R L, Rutter W J. Transcription and in vitro processing of yeast 5 S rRNA. J Biol Chem. 1980;255:9501–9506. [PubMed] [Google Scholar]
- 34.Tollervey D, Kiss T. Function and synthesis of small nucleolar RNAs. Curr Opin Cell Biol. 1997;9:337–342. doi: 10.1016/s0955-0674(97)80005-1. [DOI] [PubMed] [Google Scholar]
- 35.Venema J, Tollervey D. Processing of pre-ribosomal RNA in Saccharomyces cerevisiae. Yeast. 1995;11:1629–1650. doi: 10.1002/yea.320111607. [DOI] [PubMed] [Google Scholar]
- 36.Villa T, Ceradini F, Presutti C, Bozzoni I. Processing of the intron-encoded U18 small nucleolar RNA in the yeast Saccharomyces cerevisiae relies on both exo- and endonucleolytic activities. Mol Cell Biol. 1998;18:3376–3383. doi: 10.1128/mcb.18.6.3376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zanchin N I, Goldfarb D S. Nip7p interacts with Nop8p, an essential nucleolar protein required for 60S ribosome biogenesis, and the exosome subunit Rrp43p. Mol Cell Biol. 1999;19:1518–1525. doi: 10.1128/mcb.19.2.1518. [DOI] [PMC free article] [PubMed] [Google Scholar]