Abstract
HAV-infected Ifnar1-/- mice recapitulate many of the cardinal features of hepatitis A in humans, including serum alanine aminotransferase (ALT) elevation, hepatocellular apoptosis, and liver inflammation. Previous studies implicate MAVS-IRF3 signaling in pathogenesis, but leave unresolved the role of IRF3-mediated transcription versus the non-transcriptional, pro-apoptotic activity of ubiquitylated IRF3. Here, we compare the intrahepatic transcriptomes of infected versus naïve Mavs-/- and Ifnar1-/- mice using high-throughput sequencing, and identify IRF3-mediated transcriptional responses associated with hepatocyte apoptosis and liver inflammation. Infection was transcriptionally silent in Mavs-/- mice, in which HAV replicates robustly within the liver without inducing inflammation or hepatocellular apoptosis. By contrast, infection resulted in the upregulation of hundreds of genes in Ifnar1-/- mice that develop acute hepatitis closely modeling human disease. Upregulated genes included pattern recognition receptors, interferons, chemokines, cytokines and other interferon-stimulated genes. Compared with Ifnar1-/- mice, HAV-induced inflammation was markedly attenuated and there were few apoptotic hepatocytes in livers of infected Irf3S1/S1Ifnar1-/- mice in which IRF3 is transcriptionally-inactive due to alanine substitutions at Ser-388 and Ser-390. Although transcriptome profiling revealed remarkably similar sets of genes induced in Irf3S1/S1Ifnar1-/- and Ifnar1-/- mice, a subset of genes was differentially expressed in relation to the severity of the liver injury. Prominent among these were both type 1 and type III interferons and interferon-responsive genes associated previously with apoptosis, including multiple members of the ISG12 and 2’-5’ oligoadenylate synthetase families. Ifnl3 and Ifnl2 transcript abundance correlated strongly with disease severity, but mice with dual type 1 and type III interferon receptor deficiency remained fully susceptible to liver injury. Collectively, our data show that IRF3-mediated transcription is required for HAV-induced liver injury in mice and identify key IRF3-responsive genes associated with pathogenicity, providing a clear distinction from the transcription-independent role of IRF3 in liver injury following binge exposure to alcohol.
Author summary
Hepatitis A is a common and potentially serious disease involving inflammation and liver cell death resulting from infection with the picornavirus, hepatitis A virus (HAV). The pathogenesis of the disease is incompletely understood. Here, we have profiled changes in the RNA transcriptome of livers from mice with various genetic deficiencies in the innate immune response to HAV. We show that the liver injury associated with HAV infection in these mice results from the induction of genes under transcriptional control of interferon regulatory factor 3 (IRF3). We use high-throughput RNA sequencing to identify sets of genes induced in mice with wild-type versus transcriptionally-incompetent IRF3, rule out roles for type III interferons and IFIT proteins in disease pathogenesis, and identify genes with intrahepatic expression correlating closely with HAV-mediated liver pathology.
Introduction
Although viral hepatitis is an important cause of human morbidity and mortality worldwide, the absence of tractable small animal models for hepatotropic human viruses has handicapped efforts to understand anti-viral immunity and inflammatory responses within the liver [1,2]. Recent studies have shown, however, that Ifnar1-/- mice lacking expression of the type I interferon (IFN) receptor are highly permissive for infection with hepatitis A virus (HAV), the causative agent of type A hepatitis in humans [3–5]. These mice experience an hepatotropic HAV infection associated with elevated levels of liver-specific enzymes in the serum, fecal shedding of virus through the biliary track, and mixed inflammatory cell infiltrates surrounding apoptotic infected hepatocytes in the liver. The clinical course of HAV infection in these mice and histopathologic evidence of HAV-induced hepatocellular apoptosis closely mirror HAV infections in humans and experimentally-infected chimpanzees [6,7]. These mice thus offer unique opportunities for investigating pathogenic mechanisms underlying liver injury in acute viral hepatitis.
The inflammatory liver injury observed in HAV-infected Ifnar1-/- mice appears to result from cell-intrinsic, innate immune responses to the virus induced via RIG-I-like pattern recognition receptors (RLRs) [3]. Mavs-/- and Irf3-/-Irf7-/- mice, both of which lack the signaling downstream of RLRs required for virus-induced IFN synthesis, are just as permissive for infection as Ifnar1-/- mice, but they do not develop liver disease when infected with HAV. Neither Mavs-/- mice nor Irf3-/-Irf7-/- mice develop elevated serum alanine aminotransferase (ALT) activities or hepatic inflammation when infected, despite having equal or greater quantities of virus within the liver [3]. Numerous apoptotic hepatocytes are present in infected Ifnar1-/- liver, but few if any are found in infected Mavs-/- tissue [3]. Consistent with the notion that hepatitis results from cell-intrinsic responses, prior depletion of CD4+ or CD8+ T cells, NK/NK-T cells, or phagocytic macrophages has no impact on the development of hepatitis following intravenous challenge of Ifnar1-/- mice with HAV [3].
Cell-intrinsic responses to HAV mediated by mitochondrial antiviral signaling protein (MAVS) and interferon regulatory factor (IRF3) result in the expression of multiple ‘interferon-stimulated genes’ (ISGs) in infected Ifnar1-/- mice, despite the absence of paracrine IFN-signaling through the type I IFN receptor [3]. These genes are likely induced directly by transcriptionally-active, phosphorylated IRF3 [8]. Phosphorylated IRF3 can be detected in livers of infected Ifnar1-/- mice [3], as can transcripts of IRF3-responsive ISGs encoding potentially pro-apoptotic proteins such as phorbol-12-myristate-13-acetate-induced protein 1 (PMAIP, a.k.a Noxa) [3]. However, activated IRF3 can also induce apoptosis directly through a transcription-independent pathway involving its direct interaction with mitochondrial Bax [9]. Thus, either mechanism of IRF3-mediated apoptosis could be important for HAV induced liver disease.
There is increasing evidence that IRF3 plays a pivotal role in liver injury resulting from a variety of causes, including chemical and viral exposures and non-alcoholic fatty liver disease (NAFLD) [10–12]. Hepatocytes are highly sensitive to Fas ligation, which induces apoptosis and the expression of chemokines and other inflammatory mediators [13]. Deleting Irf3 protects against Fas-mediated liver injury as well as hepatocellular apoptosis resulting from acute ethanol or CCl4 exposure in mice [10,14]. IRF3 mediates pathology independently of downstream IFN signaling, as Ifnar1-/- mice are not protected against ethanol-related liver injury [10,15] or HAV-induced liver disease [3]. Acute alcohol exposure induces IRF3 to interact with mitochondrial Bax [15], which suggests that ethanol-induced hepatocyte apoptosis might result from the transcription-independent activity of ubiquitylated IRF3 [9]. Recent studies are consistent with this hypothesis, as Irf3S1/S1 mice, with Ser-to-Ala substitutions rendering IRF3 transcriptionally incompetent, remain susceptible to liver injury following acute-on-chronic exposure to ethanol, whereas Irf3-/- mice completely deficient in IRF3 are protected from liver injury [16].
Here, we report a series of experiments aimed at further elucidating the mechanism of liver injury in HAV-infected Ifnar1-/-mice. We profile changes in the intrahepatic transcriptomes of HAV-infected Mavs-/- and Ifnar1-/- mice, and investigate the role played by transcriptional activation of IRF3 in the pathogenesis of acute hepatitis A. We demonstrate that HAV-mediated hepatocellular apoptosis requires transcriptional activation of IRF3 by phosphorylation at Ser388/399, distinguishing IRF3-dependent HAV liver injury from IRF3-mediated alcohol-related disease, and identify IRF3-responsive genes for which the level of intrahepatic expression correlates closely with the severity of the liver injury.
Results
Transcriptional profiling of HAV-infected Mavs-/- and Ifnar1-/- mice
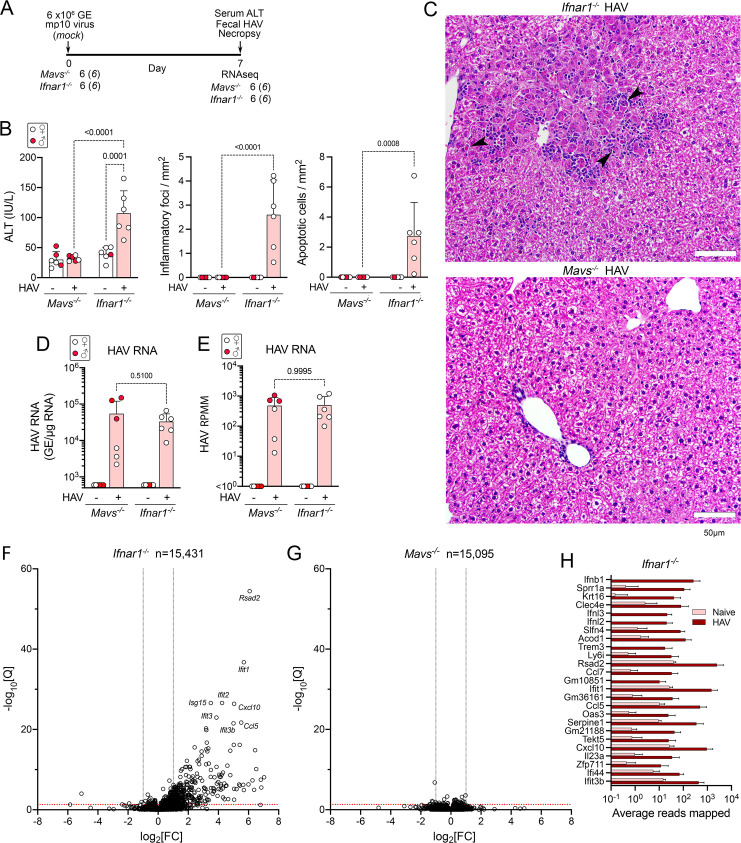
Cohorts of Mavs-/- and Ifnar1-/- mice (6 each) were inoculated intravenously with liver homogenate from an infected Mavs-/- mouse containing 6 x106 genome equivalents (GE) of 10th mouse-passage virus (HM175-mp10 inoculum), or mock-infected by inoculation of an equal volume of PBS (Fig 1A). HAV-infected and mock-infected animals were drawn from the same litters. Mice were necropsied 7 days post-inoculation (dpi). Serum ALT levels were modestly elevated at necropsy in infected Ifnar1-/- mice (mean 107 IU/L ± 38 s.d., compared with a mean 39 IU/L ± 11 s.d. in uninfected Ifnar1-/- mice, p = 0.0001) (Fig 1B). Apoptotic hepatocytes and scattered inflammatory cell infiltrates were present in H&E-stained liver sections from these animals (Fig 1B and 1C). By contrast, neither ALT elevation nor histopathologic changes were observed in the infected Mavs-/- mice (Fig 1B and 1C), despite similar intrahepatic viral load (Fig 1D). These findings recapitulate earlier observations of HAV infections in these genetically-modified mice [3].
Fig 1. Infection-induced changes in the intrahepatic transcriptome of HAV-infected Mavs-/- and Ifnar1-/- mice.
(A) Experimental scheme showing numbers of mice inoculated with 6 x106 genome equivalents (GE) of HM175-mp10 virus. (B) (left) Serum alanine transaminase (ALT) activities in sera of HAV-infected and mock-infected (naïve) mice 7 days after inoculation (dpi); (center) inflammatory foci; and, (right) apoptotic cells identified in H&E-stained liver sections from infected (+) and HAV-naive (-) mice. Liver sections were reviewed by a veterinary hepatic pathologist who was blinded to the inoculum for each mouse. Histopathology results are shown as the mean number of inflammatory foci or apoptotic cells per mm2 area derived from scoring of 20 random 40x microscopic fields of view. In this and all other panels, male animals are indicated by solid red symbols, and female animals by open symbols. (C) Representative H&E stained liver sections. Arrowheads indicate apoptotic hepatocytes surrounded by inflammatory immune cells. (D) HAV RNA abundance in liver tissue determined by RT-qPCR. (E) HAV RNA transcript abundance determined by high-throughput RNA sequencing. RPMM = reads per million reads mapped. (F) Volcano plot showing fold-change (FC) and significance of changes in intrahepatic transcripts of genes with at least 10 reads mapped in RNA from naïve versus HAV-infected Ifnar-/- mice following conditioning for sex and RNAseq batch. Q = adjusted p-value. Red horizontal line indicates Q = 0.05. (G) Similar plot of fold-change in transcript abundance in infected Mavs-/- mice. See also S1D Fig. (H) Mean reads mapped in liver tissue from naïve and HAV-infected female Ifnar-/- mice for the 25 differentially expressed transcripts with the greatest fold-change in abundance. Genes are rank-ordered according to the fold-change between naïve and infected. Error bars in all panels indicate s.d.; p-values are shown for 2-way ANOVA with Šidák’s test for multiple comparisons.
To characterize changes in the intrahepatic transcriptome associated with HAV infection, RNA extracted from the livers of these mice was subjected to high-throughput sequencing. A mean of 5.09 x107 individual reads (representing 61–80% of all reads) in each sample mapped uniquely to the mouse genome (S1A Table). Transcripts were identified from a total of 15,095 host genes in Mavs-/- mice and 15,431 genes in Ifnar1-/- mice. Comparable numbers of reads mapped to the HAV genome in samples from infected Mavs-/- and Ifnar1-/- mice (Fig 1E). Principal component analysis (PCA) identified sex and sequencing batch as the major contributors to variance in transcript abundance (samples were sequenced in two separate batches) (S1A Fig). Infection status contributed only ~9% to the overall variance, with PCA failing to distinguish infected from uninfected Mavs-/- mice (all of the infected Ifnar1-/- mice were females). While surprising, previous studies have shown that there is extensive sexual dimorphism in the intrahepatic transcriptome, driven in part by sex differences in growth hormone-regulated STAT5 and the hepatic estrogen receptor [17,18]. Nonetheless, after conditioning for sex and sequencing batch, a total of 376 genes were significantly differentially expressed (>2-fold change with p-value <0.05 after adjustment by the Benjamini-Hochberg method) in infected versus uninfected Ifnar1-/- mice (Table 1). Of these genes, 372 were upregulated and only 4 downregulated (Figs 1F, S1B, and S1C). By contrast, in infected Mavs-/- mice, there were no genes comparably upregulated, and 6 genes downregulated (Figs 1G and S1D and Table 1).
Table 1. High-throughput sequencing of intrahepatic RNA from HAV-infected mice.
| Sequencing batch run | Mouse genotype | n* | Virus Inoculum | Days p.i. | Mean HAV RNA† | ALT‡ | Differentially-expressed transcripts¶ | |||
|---|---|---|---|---|---|---|---|---|---|---|
| 2x ↑ | 2x ↓ | 4x ↑ | 4x ↓ | |||||||
| 1,2 | Mavs -/- | 6 | mp10 | 7 | 5.4 x104 | 32 | 0 | 6 | 0 | 0 |
| 1,2 | Ifnar1 -/- | 6 | mp10 | 7 | 3.3 x104 | 107 | 372 | 4 | 145 | 2 |
| 3 | Ifnar1 -/- | 4 | mp6 | 14 | 3.5 x105 | 195 | 1028 | 74 | 380 | 10 |
| 3 |
Irf3
S1/S1
Ifnar1 -/- |
4 | mp6 | 14 | 4.6 x105 | 26 | 182 | 24 | 66 | 3 |
*Number of mice in each naïve and infected cohort
†Intrahepatic viral RNA (genome equivalents/μg total RNA)
‡Serum ALT (IU/L) on day 7 p.i.
¶HAV-infected versus naïve animals with Q <0.
Genes encoding interferons (Ifnb, Ifnl2 and Ifnl3), chemokines (Ccl7, Ccl5, and Cxcl10), and ISGs (Ifit1, Ifit2, Ifit3b, Oas3, Ifi44, Rsad2, and Isg15, among others) were prominently represented among those induced by infection in Ifnar1-/- mice (Fig 1F and 1H and S1B Table). None of these were upregulated in Mavs-/- mice (S1E Fig and S1C Table). Consistent with earlier studies [3], fluorescent in situ hybridization of liver tissue indicated that Ccl5 transcripts were expressed primarily within HAV-infected hepatocytes, which were easily distinguished from infiltrating lymphocytes and macrophages by their generous cytoplasm (S2 Fig). HAV RNA was readily detected in infected liver from both Ifnar1-/- and Mavs-/- mice, but present in a patchy distribution involving less than 10–25% of hepatocytes (S2 Fig). Geneset Enrichment Analysis (GSEA) was consistent with a profound activation of the innate immune system, including a highly significant match with the hallmark complement component gene set (S1F Fig). As suggested previously, this striking transcriptional response to HAV infection in Ifnar1-/- mice likely arises from MAVS-dependent transcriptional activation of IRF3 [3].
HAV infection in Ifnar1-/- mice with locus-wide knockout of Ifit genes
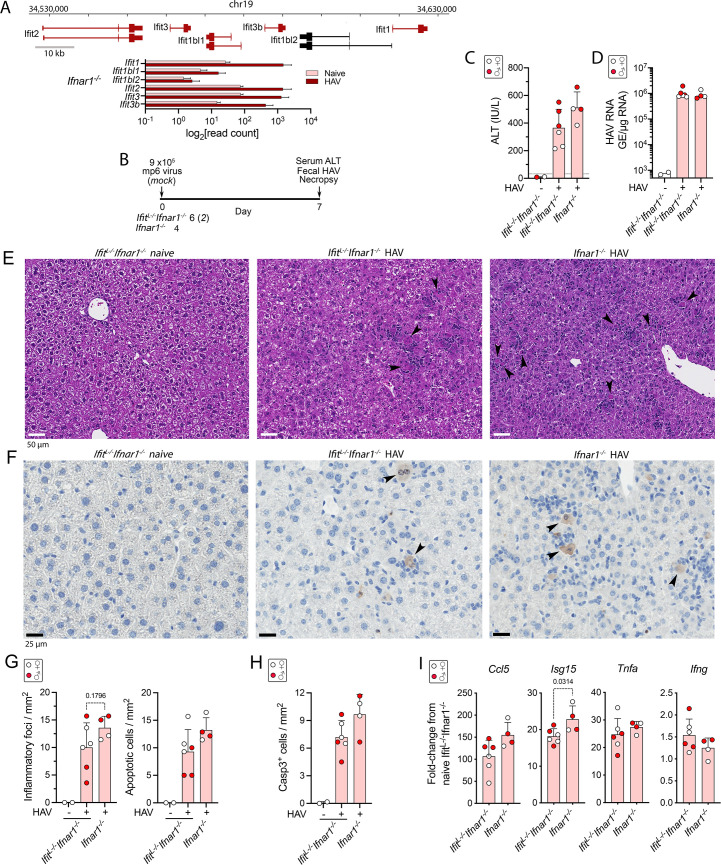
Multiple members of the IFIT (interferon-induced protein with tetratricopeptide repests) family of ISGs (Ifit1, Ifit2, Ifit3, and Ifit3b were among those most highly induced in the infected Ifnar1-/- mice (Fig 1F). This family is comprised of 6 proteins with tetratricopeptide repeats encoded by genes clustered in chromosome 19, 4 of which were induced over 10-fold (Fig 2A and 2B). These ISGs are diverse in sequence and exert a variety of antiviral activities by interacting with both viral RNAs and proteins [19]. The antiviral activity of IFITs against HAV has not been studied previously, and little is known about their activity within the murine liver. Although distinct in sequence from its murine ortholog, human IFIT2 promotes Bcl family member (Bax and Bak) dependent mitochondrial apoptosis when expressed in murine cells [20]. To determine whether the induction of IFITs contribute to hepatocellular apoptosis and inflammation in infected Ifnar1-/- mice [3], we challenged mice crossed with a genetic knockout of the entire chromosome 19 IFIT locus (IfitL-/- mice) [21].
Fig 2. HAV infection of Ifit locus knockout IfitL-/-Ifnar1-/- and Ifnar1-/- mice.
(A) Ifit gene locus in chromosome 19, showing exons and introns related to major transcripts encoding IFIT proteins. Transcripts identified by high-throughput sequencing in naïve and HAV-infected mouse liver are colored red. At the bottom are shown the normalized Ifit locus transcript read counts in livers of naïve and HAV-infected female Ifnar1-/- mice (n = 4). (B) Experimental scheme showing numbers of HAV- and mock-infected IfitL-/-Ifnar1-/- and Ifnar1-/- mice. Mice were necropsied 7 days after i.v. inoculation (dpi) of 9 x105 genome equivalents (GE) of HM175-mp6 virus. (C) Serum ALT activities in mock-infected and HAV-infected IfitL-/-Ifnar1-/- mice versus HAV-infected Ifnar1-/- mice. In this and other panels, male animals are indicated by solid red symbols, and female animals by open symbols. (D) Intrahepatic HAV RNA abundance 7 dpi determined by RT-qPCR GE = genome equivalents. (E) Representative H&E stained liver sections from naïve and HAV-infected IfitL-/-Ifnar1-/- and Ifnar1-/- mice. (F) Representative immunohistochemically stained liver sections demonstrating cells with intracellular cleaved caspase 3 (arrows). (G) Foci of inflammation (left) and apoptotic hepatocytes (right) observed by a blinded liver pathologist in H&E-stained sections of liver collected at necropsy of naïve and infected mice. (H) Mean number of hepatocytes staining positively for cleaved caspase 3 per mm2 in liver tissue collected at necropsy. (I) Cytokine transcript abundance in liver tissue collected at necropsy. Error bars in all panels indicate s.d.; p-values are shown only when <0.05 in 2-way comparisons by two-sided t test or ANOVA with Šidák’s test for multiple comparisons.
Cohorts of 4–6 IfitL-/-Ifnar1-/- or Ifnar1-/- mice were inoculated intravenously with 9 x105 GE of 6th mouse-passage virus (HM175-mp6) and necropsied 7 days later. This inoculum is notably more virulent than the HM175-mp10 inoculum used to infect mice in the experiment shown in Fig 1, and caused substantially higher serum ALT elevations in Ifnar1-/- mice on day 7 post infection (p.i.) (Fig 2C). Although ALT levels tended to be higher in infected Ifnar1-/- versus IfitL-/-Ifnar1-/- mice, this difference did not achieve statistical significance. The intrahepatic viral load was similar at necropsy on day 14 (Fig 2D), and liver sections from both IfitL-/-Ifnar1-/- and Ifnar1-/- mice demonstrated moderate inflammatory cell infiltrates with apoptotic hepatocytes scattered throughout the parenchyma (Fig 2E). Immunohistochemical staining for cleaved caspase 3 revealed numerous apoptotic hepatocytes in both types of mice (Fig 2F). While the histopathologic changes tended to be milder in the IfitL-/-Ifnar1-/- animals, there was clear overlap with Ifnar1-/- mice and this difference did not achieve statistical significance (Fig 2G and 2H). Similar trends existed in Ccl5 and Isg15 (p = 0.0314) transcript levels (Fig 2I), suggesting less IRF3 activation in IfitL-/-Ifnar1-/- mice despite similar intrahepatic viral loads (Fig 2D). Tnfa and Ifng responses, both of which tend to peak later in the infection [3], were similar in IfitL-/-Ifnar1-/- and Ifnar1-/- mice. We conclude that IFITs make no more than a minor contribution to the liver injury, including hepatocellular apoptosis, associated with HAV infection in Ifnar1-/- mice, and although robustly induced have no anti-HAV activity in the murine liver.
HAV pathogenesis in mice with transcriptionally-incompetent IRF3
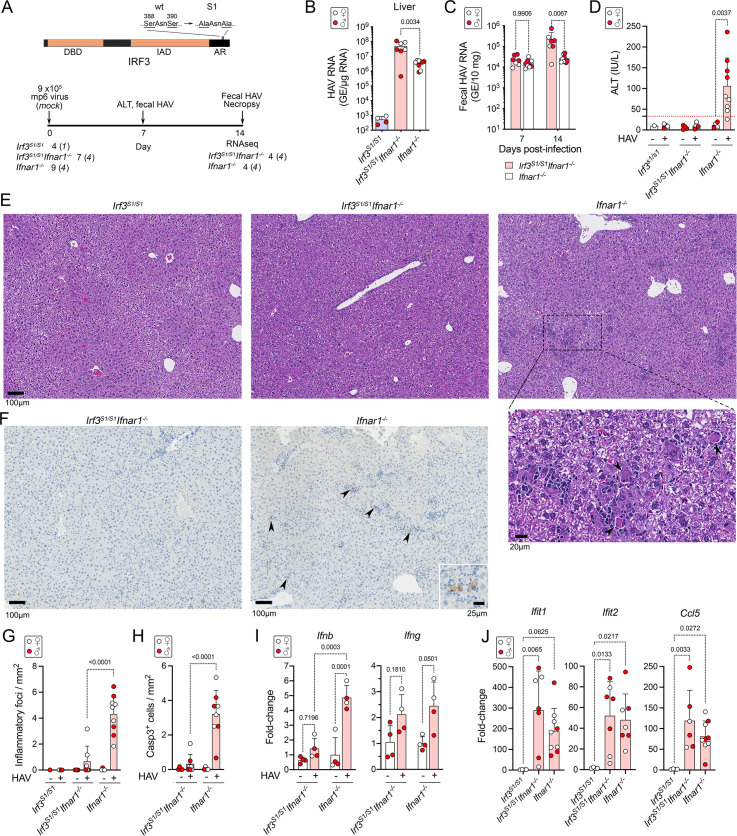
The MAVS-dependent phosphorylation of IRF3 in HAV-infected Ifnar1-/- mice induces pro-apoptotic and pro-inflammatory gene expression [3] (Fig 1F and S1B Table). However, RIG-I-like receptor (RLR)-induced signaling through MAVS and TRAF2/3/6 can also lead to the linear ubiquitylation of IRF3, which then interacts with mitochondrial Bax to induce apoptosis [9,22]. This alternative IRF3-mediated mechanism of apoptosis, known as the “RLR-induced IRF3-mediated pathway of apoptosis” (RIPA), is not dependent upon the phosphorylation of IRF3 at serine residues near its C-terminus (Ser-388 and Ser-390) that activate it transcriptionally. RIPA has been shown to provide protection against Sendai virus infection in genetically modified Irf3S1/S1 mice, in which IRF3 is transcriptionally incompetent due to alanine substitutions at Ser-388 and Ser-390 [9], and has also been shown to promote liver injury in alcohol-exposed mice [16] (Fig 3A). We challenged Irf3S1/S1 mice with HAV to ascertain whether hepatocellular apoptosis and liver injury associated with HAV infection requires IRF3-directed transcription, or alternatively might result from RIPA mediated by the direct interaction of ubiquitylated IRF3 with Bax.
Fig 3. Infectious HAV challenge of Irf3S1/S1, Irf3S1/S1Ifnar1-/-, and Ifnar1-/- mice.
(A) (top) IRF3 protein structure showing location of Ser-to-Ala substitutions in Irf3S1/S1 mice that render the protein transcriptionally inactive. DBD, DNA-binding domain; IAD, IRF-association domain; AR, autoinhibitory region. (bottom) Experimental scheme. A total of 4 Irf3SI/SI mice, 7 Irf3S1/S1Ifnar1-/- mice, and 9 Ifnar1-/- mice were infected with 9 x105 HM175-mp6 virus in two independent experiments. Liver tissue collected at necropsy 14 days later from representative infected and mock-infected Irf3S1/S1Ifnar1-/- and Ifnar1-/- mice (4 each) were selected for high-throughput RNA sequencing. Irf3SI/SI mice showed no evidence of infection and these livers were not subjected to sequencing. (B) HAV RNA in livers of mice determined by RT-qPCR 14 days post-inoculation (dpi). In all panels, male animals are indicated by solid red symbols, and female animals by open symbols. (C) Fecal HAV RNA 7 and 14 dpi. (D) Serum ALT in Irf3S1/S1, Irf3S1/S1Ifnar1-/-, and Ifnar1-/- mice 7 dpi. (E) Representative H&E-stained sections of (left to right) Irf3S1/S1, Irf3S1/S1Ifnar1-/-, and Ifnar1-/- mouse liver 14 dpi. Arrowheads in the enlarged image of Ifnar1-/- liver indicate apoptotic hepatocytes. (F) Representative immunohistochemically stained liver sections showing detection of cleaved caspase 3 in liver from (left) Irf3S1/S1Ifnar1-/- and (right) Ifnar1-/- mice 14 dpi. (G) Number of inflammatory foci per mm2 in H&E-stained liver sections. (H) Mean number of hepatocytes staining positively for cleaved caspase 3 (Casp3) per mm2 in liver sections by immunohistochemistry. (I) RT-qPCR determination of the fold-change in intrahepatic Ifnb and Ifng transcript abundance in groups of infected and mock-infected Irf3S1/S1Ifnar1-/- and Ifnar1-/- mice selected for high-throughput sequencing (n = 4 in each group). (J) RT-qPCR determination of infection-induced changes in intrahepatic Ifit1, Ifit2 and Ccl5 transcript levels in mice from both experiments. Error bars in all panels indicate s.d.; p-values are shown for 2-way comparisons by two-way ANOVA with Šidák’s test for multiple comparisons.
Cohorts of 4–9 Irf3S1/S1, Irf3S1/S1Ifnar1-/- or Ifnar1-/- mice were inoculated i.v. with 9 x105 GE HM175-mp6 virus or sham control. HAV failed to establish infection in Irf3S1/S1 mice, indicating that eliminating the transcriptional activity of IRF3 does not by itself render mice permissive for the virus (Fig 3B). These results mirror previous studies in which Irf3-/- mice lacking IRF3 expression were not susceptible to infection [3]. By contrast, Irf3S1/S1Ifnar1-/- mice were permissive for infection and shed more virus in feces than Ifnar1-/- mice on days 7 and 14 p.i. (Fig 3C). Viral load was also higher in the liver of Irf3S1/S1Ifnar1-/- mice on day 14 (Fig 3B). Thus, although loss of IRF3 transcription does not render mice permissive for infection, it does result in reduced virus control in mice lacking the type I IFN receptor.
In sharp contrast to the greater intrahepatic viral load in Irf3S1/S1Ifnar1-/- mice, there was no elevation of serum ALT levels in these mice (Fig 3D). Consistent with this, only rare inflammatory cell infiltrates and cleaved caspase 3-positive apoptotic hepatocytes were observed in livers from infected Irf3S1/S1Ifnar1-/- mice, whereas both were abundant in infected Ifnar1-/- mice (Fig 3E–3H). IFNβ (Ifnb) transcripts were minimally induced in the liver of Irf3S1/S1Ifnar1-/- mice compared with Ifnar1-/- mice, whereas there was no difference in IFNγ (Ifng) transcripts which were minimally elevated (about 2-fold) in both (Fig 3I). Thus, hepatocyte apoptosis, hepatic inflammation and HAV-induced IFNβ expression are highly dependent upon the phosphorylation of IRF3 in HAV-infected mice. Despite these differences, and the marked difference in liver injury, transcripts for chemokines and ISGs such as Ccl5, Ifit1, and Ifit2 were equivalently induced in Irf3S1/S1Ifnar1-/- and Ifnar1-/- mice (Fig 3J). These genes were not transcriptionally induced in Irf3S1/S1 mice.
Transcriptional profiling of HAV-infected Irf3S1/S1Ifnar1-/- mice
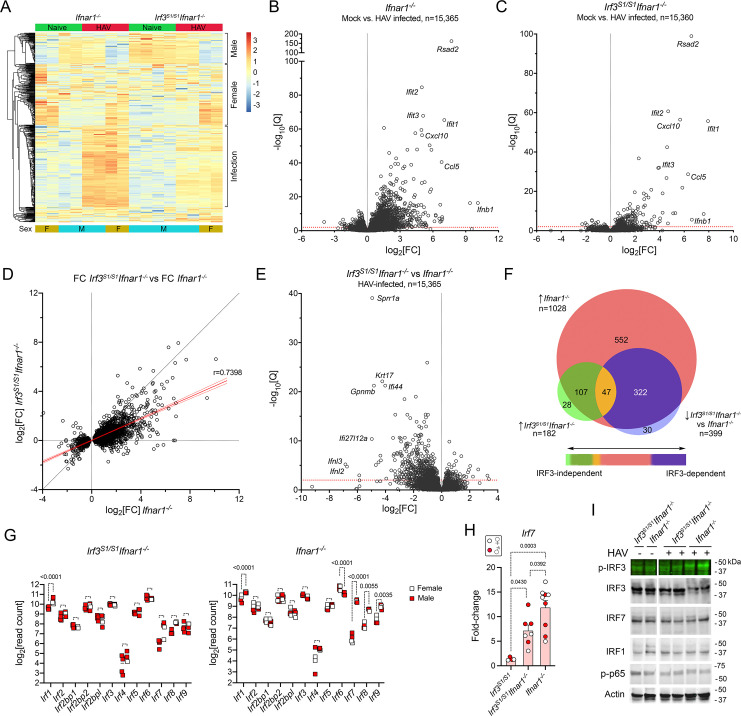
To better understand differences in the transcriptional response to HAV infection in Irf3S1/S1Ifnar1-/- and Ifnar1-/- mice, we sequenced RNA isolated from the livers of infected and naive animals of each genotype (n = 4 each) (Fig 4A and Table 1). Alignment statistics for the derived RNA reads (S1D Table) were similar to those described above, with a greater percentage of reads mapping to the HAV genome in livers from infected Irf3S1/S1Ifnar1-/- than Ifnar1-/- mice (S3A Fig). PCA suggested sex contributed ~57% of the variance in gene expression, with infection status contributing ~22% (S3B Fig). After conditioning for sex, a total of 1028 genes were significantly upregulated, and 74 down-regulated, by infection in Ifnar1-/- mice (Tables 1 and S1E). By contrast, only 182 genes were upregulated and 24 down-regulated in Irf3S1/S1Ifnar1-/- mice (Tables 1 and S1F). The greater number of genes that were significantly induced in the Ifnar1-/- mice in this second transcriptome study compared to that shown in Fig 1 reflects the use of a more potent viral inoculum (HM175-mp6 vs. HM-175-mp10), a greater length of time since infection (14 versus 7 days), and consequently more severe liver injury (mean serum ALT 195 IU/L versus 107 IU/L). These differences notwithstanding, changes in the expression levels of individual upregulated genes correlated well in the two experiments (S3C Fig). There was little difference in the transcriptomes of uninfected Irf3S1/S1Ifnar1-/- and Ifnar1-/- mice (S3D Fig).
Fig 4. Changes in the intrahepatic transcriptome of HAV-infected Irf3S1/S1Ifnar1-/- mice.
(A) Heat map showing changes in abundance of the top 500 differentially-expressed genes (ranked by variance of standardized read counts) in naïve and HAV-infected Irf3S1/S1Ifnar1-/- and Ifnar1-/- mice; n = 4 in each group selected for high-throughput RNA sequencing. Transcripts differentially expressed in relation to sex or HAV infection are noted on the right. The scale on the right represents z-score. (B, C) Volcano plots showing fold-change (fc) and significance of changes in transcripts of genes with at least 10 reads mapped in RNA from naïve versus HAV-infected liver tissue from (B) Ifnar1-/- and (C) Irf3S1/S1Ifnar1-/- mice. Q = adjusted p-value. (D) Correlation of fold-change (log2[FC]) observed in abundance of 2,503 differentially expressed transcripts in HAV-infected Irf3S1/S1Ifnar1-/- mice versus Ifnar1-/- mice. Red line = simple linear regression, ±95% C.I.; slope = 0.4415; Spearman r = 0.7398, p<0.0001. (E) Volcano plot showing fold-change and significance of infection-induced differences in intrahepatic RNA transcripts in Irf3S1/S1Ifnar1-/- versus Ifnar1-/- mice. (F) Venn diagram showing overlap of genes upregulated by infection in Ifnar1-/- and Irf3S1/S1Ifnar1-/- mice. (G) Normalized read counts of interferon regulatory factor transcripts in mock versus HAV-infected (left) Irf3S1/S1Ifnar1-/- and (right) Ifnar1-/- mice. Significance determined by two-way ANOVA with Šídák’s multiple comparisons test. (H) Infection induced fold-change in Irf7 transcript abundance determined by RT-qPCR. Fold increase for Irf3S1/S1 was 1.38 ± 0.31 s.d. Solid red symbols represent male animals. (I) Immunoblots of interferon regulatory factors, including phosphorylated IRF3 (p-IRF3, Ser396), and phosphorylated RELA (p-p65) in extracts of liver tissue from naïve and HAV-infected (left) Irf3S1/S1Ifnar1-/- and (right) Ifnar1-/- mice.
The genes induced by HAV infection were remarkably similar in Irf3S1/S1Ifnar1-/- and Ifnar1-/- mice, with Rsad2, Ifit1, Ifit2, Ifit3 and Cxcl10 among those most prominently upregulated (Fig 4B and 4C). Strikingly, Rsad2 (viperin) was the most highly induced gene in both studies of Ifnar1-/- mice (Figs 1F and 4B), and also in Irf3S1/S1Ifnar1-/- mice (Fig 4C). A strong positive correlation existed between the fold-change of individual transcripts in Irf3S1/S1Ifnar1-/- and Ifnar1-/- mice (Spearman r = 0.7398, p<0.0001) (Fig 4D). Nonetheless, a comparison of infected mice identified 399 genes that were >2-fold less expressed in Irf3S1/S1Ifnar1-/- compared with Ifnar1-/- mice (Fig 4E and S1G Table). These included 47 genes that were significantly induced by infection in both Irf3S1/S1Ifnar1-/- and Ifnar1-/- mice, and 322 induced only in Ifnar1-/- animals (Figs 4F and S3E). Particularly notable among genes that were significantly less induced in the Irf3S1/S1Ifnar1-/- mice were those encoding IFNλ2 (Ifnl2) and IFNλ3 (Ifnl3) (Fig 4E). These results thus establish sets of genes that are transcriptionally induced by HAV infection largely independent of IRF3 phosphorylation at Ser-388 or Ser-399, and others that are highly dependent upon phosphorylation of these IRF3 residues.
To gain insight into transcription factor(s) driving gene expression in infected Irf3S1/S1Ifnar1-/- and Ifnar1-/- mice, we sought evidence for transcriptional induction of members of the IRF family. Baseline expression prior to infection was similar in the two types of mice (S3F Fig). Irf1, Irf7, Irf8, and Irf9 transcripts were significantly induced by infection in Ifnar1-/- mice, whereas Irf6 transcripts (abundant in uninfected mice) were significantly down-regulated (Fig 4G). The mechanism underlying the decrease in Irf6 transcripts is uncertain, but IRF6 is known to function primarily in maintaining epidermal barrier function, not host responses to viral infection [23]. Irf1 was significantly upregulated in infected Irf3S1/S1Ifnar1-/- mice (Fig 4G). While Irf7 transcript abundance was much lower than Irf1, Irf7 was also upregulated when measured by RT-qPCR in a larger number of animals (Fig 4H). suggesting IRF1 may drive most ISG expression in these animals. Immunoblots of liver tissues confirmed phosphorylation of IRF3 at Ser396 in infected Ifnar1-/- mice, and the absence of this in infected Irf3S1/S1Ifnar1-/- animals (Fig 4I). Efforts to document the presence of ubiquitylated IRF3 were unsuccessful. We observed no changes in the abundance or molecular mass of IRF7 or IRF1 in either type of mouse. Similarly, efforts to detect phosphorylated IRF7 were unsuccessful. Although IRF1 is regulated transcriptionally by NF-κB [24], there was no detectable increase in phospho-p65 (Fig 4I). Efforts to document these changes in transcription factors by immunoblotting were likely limited by technical difficulties quantifying these proteins in liver extracts, as well as the low proportion of hepatocytes infected with virus (<25%) (S2B Fig).
Gene expression and liver histopathology in HAV-infected mice
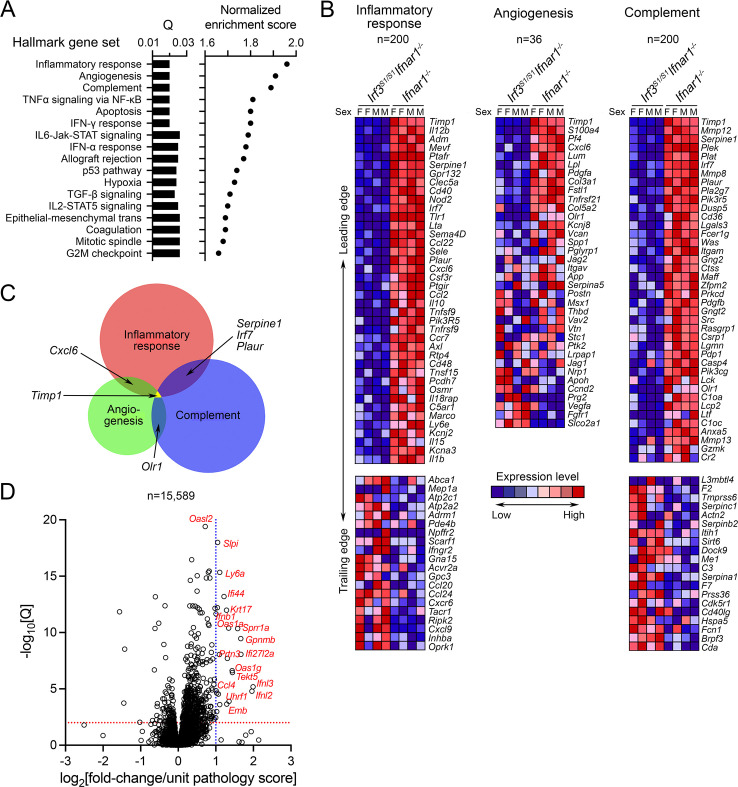
GSEA revealed a strong concordance between the IRF3-regulated genes that were significantly downregulated in infected Irf3S1/S1Ifnar1-/- mice versus Ifnar1-/- mice and established hallmark gene sets related to inflammatory response, angiogenesis, and complement [25] (Fig 5A and 5B). However, there was surprisingly little overlap in the leading edge genes in these three gene sets, suggesting that the enrichment scores for each are driven primarily by different genes (Fig 5C). Normalized enrichment scores were lower, yet still significant with hallmark gene sets for TNF-α signaling via NF-κB and apoptosis (Figs 5A and S4A). These two hallmark gene sets show greater overlap with the inflammatory response gene set, with Il1b (IL-1β) notably present among the top 40 leading edge genes down-regulated in Irf3S1/S1Ifnar1-/- mice in each gene set (S4B Fig). Thus, despite the substantial overlap in genes induced by HAV in Irf3S1/S1Ifnar1-/- and Ifnar1-/- mice (Fig 4D and 4F), differences in the innate immune responses elicited by infection in these two types of mice involve distinct sets of genes linked to each of the well-defined biological states represented by these specific hallmark gene sets.
Fig 5. Differentially expressed intrahepatic transcripts in HAV-infected Irf3S1/S1Ifnar1-/- versus Ifnar1-/- mice.
(A) Gene Set Enrichment analysis showing false discovery rate (Q) and normalized enrichment scores of hallmark gene sets most enriched for differentially expressed genes. (B) Heat maps reflecting transcript abundance of the top 40 genes in the leading edges, and bottom 20 genes in the trailing edges, of the hallmark “inflammatory response” and “complement” gene sets, and all genes in the hallmark “angiogenesis” gene set. The number of genes in each gene set is shown at the top. (C) Venn diagram showing overlap between genes in the leading edges of the hallmark gene sets shown in panel B. (D) Volcano plot showing differential expression of genes in HAV-infected Irf3S1/S1Ifnar1-/- and Ifnar1-/- mice relative to the liver tissue pathology score as a continuous variable (see S1H in S1 Table). Data were conditioned for sex prior to analysis.
The substantial overlap evident in genes induced by HAV in Irf3S1/S1Ifnar1-/- and Ifnar1-/- mice (Fig 4D and 4F) contrasts also with the absence of hepatocellular apoptosis and inflammation in the former (Fig 3G and 3H). While this could reflect the globally lower fold-induction of these genes in Irf3S1/S1Ifnar1-/- mice, the GSEA suggests it results from differences in the induction of specific genes. Thus, we sought to identify genes with a strong positive correlation between expression level and liver histopathology score in the combined cohort of HAV-infected and naïve Irf3S1/S1Ifnar1-/- and Ifnar1-/- mice. To accomplish this, liver tissue from each of the 8 infected Irf3S1/S1Ifnar1-/- and Ifnar1-/- mice was scored for histopathologic changes on a 0–4 scale by a veterinary hepatic pathologist based on the intensity of inflammatory infiltrates and numbers of apoptotic hepatocytes. We then analyzed the data from the these mice, first conditioning for sex, and then identifying genes that were differentially expressed using the pathology score as a continuous variable. This revealed 29 genes for which there was >1.0 log2-fold increase in expression, and 63 genes with >0.8 log2-fold increase in expression for each 1 unit increase in the recorded pathology score with Q <0.05 (Fig 5D and S1H Table). These included transcripts encoding both type 1 and type III IFNs (Ifnb, Ifna4, Ifnl3, Ifnl2) and multiple interferon-responsive genes, chemokines, and cytokines (see Discussion).
HAV challenge of Ifnlr1-/-Ifnar1-/-double receptor knockout mice
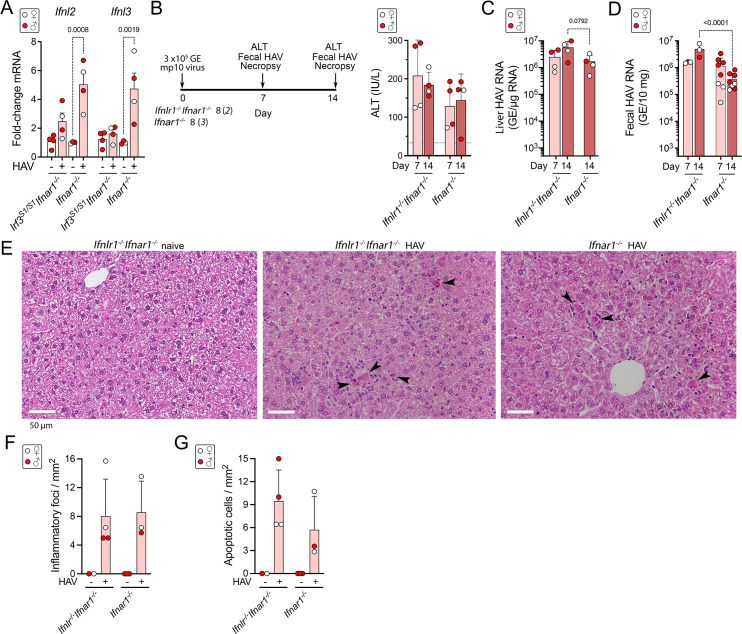
It was surprising to find a strong correlation between Ifnl3 and Ifnl2 expression levels and histopathology (Fig 5D), as murine hepatocytes are nonresponsive to type III IFN [26,27]. Nonetheless, RT-qPCR confirmed that these transcripts were minimally induced in infected Irf3S1/S1Ifnar1-/- mice compared with Ifnar1-/- mice (Fig 6A). Thus, to determine whether type III IFN expression contributes directly to HAV-induced liver injury, we challenged mice with dual deficiency of both type I and type III IFN receptors. Groups of 4 Ifnlr1-/-Ifnar1-/- and Ifnar1-/- mice were inoculated intravenously with 3 x105 GE mp10 virus. Serum ALT elevations were comparable in the two types of mice at days 7 and 14 p.i. (Fig 6B), but HAV RNA levels in both liver and feces were significantly greater in the Ifnlr1-/-Ifnar1-/- mice than Ifnar1-/- mice on day 14 post-infection (Fig 6C and 6D). Histopathologic examination revealed similar numbers of apoptotic hepatocytes and similar inflammatory infiltrates in the infected Ifnlr1-/-Ifnar1-/- and Ifnar1-/- mice (Fig 6E and 6G). Thus, while the additional absence of IFNλ signaling renders Ifnar1-/- mice somewhat more permissive for HAV infection, it does not reduce liver injury.
Fig 6. HAV infection in Ifnlr1-/-Ifnar1-/- double-knockout mice lacking receptors to both type I and III interferon.
(A) Infection-induced changes in Ifnl2 and Ifnl3 transcript abundance in Irf3S1/S1Ifnar1-/- and Ifnar1-/- mice determined by RT-qPCR in the experiment outlined in Fig 3A. In all panels, male animals are indicated by solid red symbols, and female animals by open symbols. (B) (left) Experimental scheme and (right) mean serum alanine transaminase (ALT) activities in sera of Ifnlr1-/-Ifnar1-/- and Ifnar1-/- mice 7 and 14 days after i.v. inoculation (dpi) of the 3 x105 GE (genome equivalents) HM175-mp10 virus. In each group, 4 infected animals were necropsied on day 7, and 4 on day 14 dpi. (C) Mean HAV RNA abundance in liver tissue and (D) feces determined by RT-qPCR. Fecal samples from additional Ifnar1-/- mice infected with the same inoculum were included to demonstrate the range of fecal shedding in these animals. GE = genome equivalents. (E) Representative H&E stained sections of liver from HAV-naïve and infected mice 14 dpi. Arrowheads indicate apoptotic hepatocytes, many surrounded by inflammatory immune cells. (F) Number of inflammatory foci and (G) apoptotic hepatocytes per mm2 in H&E-stained liver sections at 14 dpi. Error bars in all panels are s.d.
Discussion
Although liver injury in acute hepatitis A is considered generally to result from immune responses to the infection, it remains incompletely understood. The cytotoxic activity of virus-specific CD8+ T cells has been suggested to mediate HAV-induced liver disease [28,29], but more recent studies cast some doubt on this. Virus-specific CD4+ helper cells dominate the adaptive cellular response to HAV in experimentally-infected chimpanzees, whereas CD8+ T cell responses are generally weaker and correlate poorly with both liver injury and virus control [30]. Interleukin 15 (IL-15)-induced bystander activation of CD8+ memory T cells targeting viruses other than HAV has been shown to result in innate-like cytotoxic activity in HAV-infected humans [31]. The cytotoxic activity of bystander-activated T cells correlated better with serum ALT elevations than the cytotoxic activity of virus-specific CD8+ T cells in hospitalized patients with severe disease [31], but it is not known whether this mechanism of liver injury is active in individuals with less severe and more typical infection. Mucosal-associated invariant T (MAIT) cells have also been shown to be activated, and to exert innate-like granzyme B-dependent cytotoxicity in patients with severe disease [32]. In contrast to these proposed cell-mediated mechanisms of liver injury, hepatitis in infected Ifnar1-/- mice results from cell-intrinsic innate immune responses to the virus [3]. It is possible that no one mechanism accounts for the liver injury in infected humans, and that each of these mechanisms may have a role in producing the spectrum of disease observed in HAV infection.
HAV induces neither hepatocyte apoptosis nor hepatic inflammation in Mavs-/- mice, despite the quantity of virus in the liver being equal to or greater than that in infected Ifnar1-/- mice [3] (Fig 1C and 1D). The absence of liver injury in Mavs-/- mice shows that HAV replication is noncytopathic, and that hepatitis in Ifnar1-/- mice results from the host response to the infection. Ifnar1-/- mice recapitulate many of the cardinal features of hepatitis A in humans, including hepatocyte apoptosis and elevated serum ALT levels [3,7]. Apoptotic hepatocytes are surrounded by inflammatory cells, and CD4+ and CD8+ T cells, NK cells, and F4/80+CD11+ macrophages are all increased in number in the liver [3,33]. However, antibody depletion of CD4+ or CD8+ T cells or NK/NKT cells, or clodronate-depletion of phagocytic cells prior to virus challenge does not prevent hepatocellular apoptosis or liver inflammation [3]. Moreover, depleting T cells in mice with established infection leads to increased hepatocyte apoptosis, greater serum ALT elevation, and enhanced inflammation [34]. Like Mavs-/- mice, Irf3-/-Irf7-/- mice also show no evidence of liver injury when infected with HAV [3]. Pathogenesis is thus dependent upon signaling through the MAVS-IRF3 axis, likely following sensing of the virus by RLRs including RIG-1 [35].
Recent studies have revealed a key role for IRF3 activation in the pathogenesis of chemically-induced liver disease [10,15]. Following exposure of mice to ethanol or CCl4, IRF3 is phosphorylated by TANK-binding kinase 1 (TBK1) secondary to signaling from cyclic GMP-AMP synthase (cGAS) and stimulator of interferon genes protein (STING) [10,14]. Irf3-/- mice are protected against liver injury following binge exposure to ethanol, whereas Irf3S1/S1 mice with transcriptionally-incompetent IRF3 develop steatosis, ALT elevation and inflammatory cytokine expression [16]. The IRF3 S1 mutant is ubiquitylated and interacts with mitochondrial Bax, inducing apoptosis and liver injury independently of IRF3-mediated transcription [9,16]. In contrast, we found here that HAV-induced inflammation was markedly attenuated in Irf3S1/S1Ifnar1-/- mice compared to Ifnar1-/- mice expressing wild-type IRF3 (Fig 3E–3H). We cannot rule out a contribution from non-transcriptional IRF3 activity, but our results show that IRF3-mediated transcription is required for liver injury in HAV-infected mice, providing a clear distinction from IRF3-mediated alcohol liver injury.
The contrast between hepatic inflammation in infected Ifnar1-/- mice and its near complete absence in Irf3S1/S1Ifnar1-/- mice could reflect quantitatively greater induction of qualitatively similar innate immune response programs including multiple pro-apoptotic and pro-inflammatory mediators (Figs 4C and S4C). Alternatively, the liver injury in Ifnar1-/- mice could result from qualitative differences in the host response, with induction of specific genes uniquely dependent upon IRF3 phosphorylation and minimally induced by IRF1 and other transcription factors activated in Irf3S1/S1Ifnar1-/- mice (Fig 4E and 4F). We identified 63 genes for which the level of expression correlated well with liver pathology read blindly by an experienced veterinary hepatic pathologist (S1H Table). Many of these genes were minimally or not induced at all in infected Irf3S1/S1Ifnar1-/- mice. These included genes encoding IFNλ3 and IFNλ2 (S5B Fig). However, like type I IFN signaling, type III IFN signaling is not required for hepatocellular apoptosis or liver inflammation, as both occur in infected mice lacking IFNλ receptors (Fig 6).
No one gene is likely to be responsible for the liver injury associated with HAV infection in Ifnar1-/- mice, and many of the genes we found expressed in a manner correlating with liver pathology have been associated previously with apoptotic signaling. These include in particular Ifi27l2a and Ifi27l2b, which encode the BH3-like IFNα-inducible proteins 27-like 2A/B, also known as ISG12 (Fig 5D and S1H Table) [36,37]. There are three ISG12 family members expressed in mice, ISG12A (Ifi27), ISG12 (Ifi27l2a), and ISG12b2 (Ifi27l2b), each of which is significantly induced by HAV infection in Ifnar1-/- mice (S5A and S5B Fig). The physiological functions of these proteins are poorly understood, but both murine and human ISG12 family members are associated with mitochondria and have been implicated in regulating IFN-induced apoptosis [36,38,39]. Human ISG12A (which shares 75% identity with mouse ISG12A) has been linked to the induction of apoptosis in hepatitis C virus (HCV) infected human hepatocytes [37]. Other genes associated with apoptosis include Lta (lymphotoxin-alpha), Tnfrsf9 (TNF receptor superfamily member 9, or CD137), and Oas1g, Oas1a, and other members of the 2’-5’ oligoadenylate synthetase family.
Finally, it should be noted that previous HAV challenge experiments using Irf3-/-Irf7-/- mice [3] were potentially confounded by an unintended deletion of a pro-apoptotic gene, Bcl2l12, closely juxtaposed to Irf3 [40]. The Bcl2l12 gene is intact and normally expressed in Irf3S1/S1Ifnar1-/- mice (S5C Fig). Thus, the absence of liver injury in infected Irf3S1/S1Ifnar1-/- mice (Fig 4E) confirms that the lack of liver inflammation in earlier studies of infected Irf3-/-Irf7-/- mice [3] resulted from the deletion of Irf3, not Bcl2l12.
In summary, the studies described here show that IRF3-dependent gene expression promotes hepatocyte apoptosis and liver inflammation during acute HAV infection in mice, and identify sets of IRF3-responsive genes that act together to promote pathogenesis. Similar pathogenetic processes likely drive inflammation of the liver in many humans with acute hepatitis A, and may contribute to both acute and chronic liver injury associated with other hepatotropic viruses.
Materials and methods
Ethics statement
The care and use of mice in the experiments described in this report were reviewed and approved by the Animal Care and Use Committee of The University of North Carolina at Chapel Hill, Chapel Hill, North Carolina, or by the Animal Care and Use Committee of the National Institute of Infectious Diseases, Tokyo, Japan.
Hepatitis A Virus (HAV)
Infections were carried out with mouse-passaged HM175 virus [3]. Inocula were prepared from lysates of liver from infected Mavs-/- mice after a total of 6 (HM175-mp6) or 10 (HM175-mp10) mouse-to-mouse passages as described previously [3].
Mice
Mice were bred and housed at the University of North Carolina at Chapel Hill in accordance with the polices and guidelines of the UNC Institutional Animal Care and Use Committee, or at the National Institute of Infectious Diseases in accordance with the policies and guidelines of the NIID Animal Care and Use Committee. C57BL/6 mice were purchased from the Jackson Laboratory. Ifnar1-/- mice were provided by J. Sprent of the Scripps Research Institute, or by S. Morikawa of the National Institute of Infectious Diseases, Japan. Mavs-/- mice were provided by M. Gale Jr. of the University of Washington, USA. IfitL-/- mice with deletion of the complete Ifit gene locus [21] and Irf3S1/S1 mice with transcriptionally-incompetent IRF3 [9] have been described previously. Ifnar1-/-Ifnlr-/- mice [27] were provided by P. Staeheli, Institute for Virology, Universitätsklinikum Freiburg. All were bred on a C57BL/6 background for at least 10 generations. Ifit1L-/-Ifnar1-/- and Irf3S1/S1Ifnar1-/- mice were crosses between Ifnar1-/- mice and IfitL-/- mice [21] and Irf3S1/S1 mice [9], respectively. All mouse genotypes were confirmed by PCR-based genotyping.
HAV infectious challenge
Mice were intravenously inoculated with the indicated virus inocula at 6–8 weeks of age. Feces were collected from mice housed in individual cages, and serum samples were collected periodically. Tissues were harvested at necropsy and snap frozen on dry ice, stored in RNAlater (Thermo Fisher Scientific) and kept at -80°C, or fixed in 10% neutral phosphate-buffered formalin for 48 hours and stored subsequently in 70% ethanol until processed for histology. All two-way comparisons of infection outcomes in different mouse genotypes were based on use of the same inoculum, identical infection schedules, and data collected from mice infected contemporaneously in the same experiment.
Alanine Aminotransferase assay
Serum alanine aminotransferase (ALT) activity was measured using the Alanine Aminotransferase Activity Assay Kit (Reitman-Frankel Method, Elabscience) or Alanine Transaminase Colorimetric Activity Assay Kit (Cayman Chemical, Ann Arbor, MI), according to the manufacturers’ recommended procedures.
Histopathology and immunohistochemistry
Formalin-fixed paraffin-embedded (FFPE) livers were sectioned at 4 μm thickness for histopathology and immunohistochemistry. Sections were stained with hematoxylin and eosin (H&E) and scanned using a Leica Biosystems ScanScope AT2 brightfield line scanner within Translational Pathology Laboratory at University of North Carolina at Chapel Hill. Cleaved caspase 3 staining was accomplished using Dako Autostainer (Dako, Carpinteria, CA). Tissue sections were de-paraffinized and rehydrated prior to antigen retrieval in Pascal presser cooker (Dako) for 30 sec at 123°C in Tris-EDTA buffer. Slides were cooled, rinsed and then treated sequentially with endogenous peroxidase blocker and universal block prior to incubation for 2 hr at room temperature with antibody to cleaved caspase 3 (Cell Signaling, 9661S) diluted 1:100, and 30 min at room temperature with secondary antibody followed by development with 3,3′-diaminobenzidine and counterstain with hematoxylin. Slides were then dehydrated and mounted. Antibodies used in this study are listed in Supporting Information (S2 Table).
Histopathology scoring
Liver sections were reviewed and scored for pathological changes by a board-certified veterinary pathologist who was blinded to infection status and mouse genotype. Foci of inflammation or hepatocellular apoptosis were enumerated microscopically in 10–20 successive random fields of view using a 40X objective and a 10X ocular (FN22), or in similarly sized fields from high-resolution scanned digital images. Numbers of apoptotic cells, identified by acidophilic cytoplasmic staining and disrupted nuclear morphology in H&E-stained sections or by immunohistochemical staining for cleaved caspase 3, and foci of inflammatory cells were normalized to the area scanned and are reported as foci, or cells, per mm2. To assess the differential expression of genes based on pathology as a continuous variable, liver tissues were assigned a pathology score on a 0–4 scale by a veterinary hepatic pathologist based on the intensity of inflammatory infiltrates and numbers of apoptotic hepatocytes.
In situ Hybridization (ISH)
Fluorescent in situ hybridization of formalin-fixed paraffin imbedded tissue sections was carried out using the QuantiGene ViewRNA FFPE Assay (Affymetrix, Santa Clara, CA) as described previously [3].
High-throughput RNA sequencing and data analysis
RNA was extracted from mouse liver using the mirVana miRNA Isolation Kit (Thermo Fisher, AM1560). Libraries were prepared using the TruSeq Stranded Total RNA Library Prep Gold Kit (Illumina) and sequenced on an Illumina HiSeq 4000 sequencer with pair-end 50 bp setting within the High-Throughput Sequencing Facility of the University of North Carolina at Chapel Hill. Sequencing reads were aligned to the mouse genome mm10 with STAR 2.6 and transcripts quantified using Salmon 0.7 and Gencode vM17. Differential expression analysis was carried out using default settings in the DESeq2 package in R3.6. Transcript counts were normalized using the DESeq2 internal median of ratios method. Genes with base mean read counts <10 were excluded from analysis. Principal component analysis plots were generated by the DESeq2 plotPCA routine after variance-stabilizing transformation of the read counts. Conditioning of sex and batch effect were carried out in DESeq2 by introducing sex and batch as independent co-variants in the design of the experiment as: “design = ~ sex+ batch + condition”. Significance was adjusted for multiple comparisons using the Benjamini-Hochberg method. Gene set enrichment analysis was carried out using normalized read counts for all genes and the GSEA software package: https://www.gsea-msigdb.org/gsea/index.jsp [41,42].
Quantitative RT-qPCR
Total RNA was extracted from liver using Trizol (Invitrogen), and from feces using the QIAamp Viral RNA Mini Kit (Qiagen). First-strand cDNA was synthesized with Superscript III reverse transcriptase (Invitrogen), and quantitative polymerase chain reaction carried out using Universal SYBR Supermix (Bio-Rad) as described [3]. HAV RNA was quantified against a synthetic RNA standard. Primer sequences are listed in Key Resources.
Immunoblots
Frozen mouse liver samples were homogenized in the presence of lysis buffer (15% w/v, Invitrogen) containing protease inhibitors (Sigma-Aldrich), 1mM Na3VO4, and 50mM NaF, clarified by centrifugation twice at 12,000 g for 30 min at 4°C, and stored at -80°C. The samples were subjected to SDS-PAGE and immunoblotting using standard methods. Blots were blocked with Odyssey blocking buffer (LI-COR Bioscience) and probed with the following primary antibodies: IRF1 (8478, 1:500, Cell Signaling), IRF3 (4302, 1:400, Cell Signaling), phospho-IRF3 (Ser396) (4947, 1:200, Cell Signaling), IRF5 (20261S, 1:200, Cell Signaling), β-actin (3700, 1:5000, Cell Signaling). IRDye 680 and 800 secondary antibodies (LI-COR Bioscience) were used to visualize protein bands with an Odyssey two-color detection imaging system (LI-COR, Bioscience).
Statistical analysis
Statistical tests were carried out using GraphPad Prism 9.0.1 for Mac OS X software. Unless otherwise noted, comparisons between groups used the non-parametric Mann-Whitney test, and one-way or two-way ANOVA. Details of specific statistical tests and experimental design are given in the relevant figure captions. Statistical significance was defined as p<0.05 with appropriate adjustment for multiple tests.
Supporting information
(A) Principal component analysis of RNAseq data from 24 mice in groups of 6 infected and 6 uninfected of each genetic knock-out. First and second components (left) represent variance due to differences in sex (42%) and sequencing run (batch, 18%); RNAseq reads from noninfected and infected Ifnar1-/- mice cluster separately from naïve Mavs-/- mice based on the 3rd and 4th components (combined contribution to variance = 15%). (B,C) Heat maps reflecting (B) differential expression of the 617 genes which were significantly changed (padj<0.05) following infection in either Mavs-/- or Ifnar1-/- mice, and (C) differentially expressed genes in liver tissue from (left) Mavs-/- (n = 40 genes) and (right) Ifnar1-/- mice (n = 579 genes). Scales on the right represent z-score. (D) Expanded volcano plot showing fold-change (fc) and significance (padj = adjusted p-value) of transcripts with significant change in abundance in liver tissue from Mavs-/- mice (see Fig 1G in the main manuscript). (E) Average reads mapped in liver tissue from naïve and HAV-infected Mavs-/- mice for those 25 transcripts with the greatest fold-change in abundance in infected Ifnar-/- mice (see Fig 1F in the main manuscript). (F) Gene set enrichment analysis of transcripts with significant change in abundance in Ifnar-/- mice. No significant hallmark signatures were identified for tissue from Mavs-/- mice.
(TIF)
HAV RNA and Ccl5 (RANTES) mRNA were probed for in liver tissue from (top to bottom) uninfected Ifnar1-/- mice and HAV-infected Mavs-/- and Ifnar-/- mice. Merged fluorescence images are shown on the left, and single-channel images on the right.
(TIF)
Liver tissue was harvested 7 days post-inoculation of HM175-mp6 virus. (A) Normalized HAV RNA read counts. RPMM = reads per million reads mapped. (B) Principal component analysis showing variance related to sex, knockout type, and HAV infection status of animals. (C) Correlation plot of fold-change induction of individual genes in Ifnar1-/- mice infected with HM175-mp10 virus (see Fig 1) versus induction in Ifnar1-/- mice infected with HM175-mp6 virus (see Fig 4). R2 determined by linear regression analysis for all transcripts upregulated >2-fold. (D) Volcano plot showing difference (fold-change, fc) and significance of differences in transcript abundance in livers of naive Irf3S1/S1Ifnar1-/- and Ifnar1-/- mice. padj = adjusted p-value. (E) Venn diagram showing overlap in transcripts downregulated following HAV infection of Ifnar1-/- and Irf3S1/S1Ifnar1-/- mice. (F). Normalized read counts observed for interferon regulatory factor (IRF) transcripts in livers of naïve Irf3S1/S1Ifnar1-/- and Ifnar1-/- mice.
(TIF)
(A) Heat maps reflecting transcript abundance of top 40 genes in the leading edges, and bottom 20 genes in the trailing edges, of hallmark TNF-α signaling via NF-κB and apoptosis gene sets. The number of genes in each set is shown at the top. (B) Venn diagrams showing overlap between the genes in the (top) leading edges and (bottom) trailing edges of gene sets shown in panel A and the hallmark inflammatory response gene set in Fig 5 in the main manuscript.
(TIF)
Intrahepatic abundance of transcripts encoding ISG12a family members and BCL2L12 in naïve and HAV-infected Irf3S1/S1Ifnar1-/- and Ifnar1-/- mice (n = 4 each) 7 days after infection with HM175-mp6 virus. (A) Normalized Ifi27 gene family member read counts from high throughput sequencing. (B) Relative transcript abundance determined by RT-qPCR, showing fold-change from uninfected mouse liver. (C) Normalized Bcl2l12 gene family member read counts from high throughput sequencing. Error bars in all panels represent s.d. Significance determined by unpaired t-test.
(TIF)
(XLSX)
(PDF)
(XLSX)
Acknowledgments
We thank Sara R. Selitsky for helpful discussions and critical review of this manuscript.
Data Availability
Datasets generated and analyzed during this study are included with this publication or have been deposited with Gene Expression Omnibus (GEO), accession numbers GSE166353: GSM5068950-GSM5068989.
Funding Statement
This work was supported in part by grants from the National Institute of Allergy and Infectious Diseases of the U.S. National Institutes of Health: R01-AI131685 (SML and JKW), R01-AI103083, R01-AI150095 (SML), R01-AI143894, R01-AI138337 (JKW), R01-AA027456 and RO1-CA068782 (GCS); the Japanese Society for the Promotion of Science KAKENHI, JP18K05987 (AHY); and Japan Agency for Medical Research and Development, JP20fk0108102j0102 and JP20fk0210062j0302 (AHY). The UNC Animal Histopathology & Laboratory Medicine Core and UNC High-Throughput Sequencing Facility were supported in part by an NCI Center Core Support Grant, P30CA016086 to the Lineberger Comprehensive Cancer Center. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Protzer U, Maini MK, Knolle PA. Living in the liver: hepatic infections. Nat Rev Immunol. 2012;12(3):201–13. Epub 2012/03/01. doi: 10.1038/nri3169 [DOI] [PubMed] [Google Scholar]
- 2.Crispe IN. The liver as a lymphoid organ. Annu Rev Immunol. 2009;27:147–63. Epub 2009/03/24. doi: 10.1146/annurev.immunol.021908.132629 [DOI] [PubMed] [Google Scholar]
- 3.Hirai-Yuki A, Hensley L, McGivern DR, González-López O, Das A, Feng H, et al. MAVS-dependent host species range and pathogenicity of human hepatitis A virus. Science (New York, NY). 2016;353(6307):1541–5. Epub 2016/09/17. doi: 10.1126/science.aaf8325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hirai-Yuki A, Whitmire JK, Joyce M, Tyrrell DL, Lemon SM. Murine Models of Hepatitis A Virus Infection. Cold Spring Harbor perspectives in medicine. 2019;9(1). Epub 2018/04/18. doi: 10.1101/cshperspect.a031674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hirai-Yuki A, Hensley L, Whitmire JK, Lemon SM. Biliary secretion of quasi-enveloped human hepatitis A virus. mBio. 2016;7:e01998–16. doi: 10.1128/mBio.01998-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lanford RE, Feng Z, Chavez D, Guerra B, Brasky KM, Zhou Y, et al. Acute hepatitis A virus infection is associated with a limited type I interferon response and persistence of intrahepatic viral RNA. Proc Nat’l Acad Sci USA. 2011;108:11223–8. Epub 2011/06/22. doi: 10.1073/pnas.1101939108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lemon SM, Ott JJ, Van Damme P, Shouval D. Type A viral hepatitis: A summary and update on the molecular virology, epidemiology, pathogenesis and prevention. J Hepatol. 2018;68:167–84. Epub 2017/09/10. doi: 10.1016/j.jhep.2017.08.034 [DOI] [PubMed] [Google Scholar]
- 8.Grandvaux N, Servant MJ, tenOever B, Sen GC, Balachandran S, Barber GN, et al. Transcriptional profiling of interferon regulatory factor 3 target genes: direct involvement in the regulation of interferon-stimulated genes. J Virol. 2002;76(11):5532–9. doi: 10.1128/jvi.76.11.5532-5539.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chattopadhyay S, Kuzmanovic T, Zhang Y, Wetzel JL, Sen GC. Ubiquitination of the transcription factor IRF-3 activates RIPA, the apoptotic pathway that protects mice from viral pathogenesis. Immunity. 2016;44(5):1151–61. Epub 2016/05/15. doi: 10.1016/j.immuni.2016.04.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Petrasek J, Dolganiuc A, Csak T, Nath B, Hritz I, Kodys K, et al. Interferon regulatory factor 3 and type I interferons are protective in alcoholic liver injury in mice by way of crosstalk of parenchymal and myeloid cells. Hepatology. 2011;53(2):649–60. Epub 2011/01/29. doi: 10.1002/hep.24059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu LM, Tu WJ, Zhu T, Wang XT, Tan ZL, Zhong H, et al. IRF3 is an important molecule in the UII/UT system and mediates immune inflammatory injury in acute liver failure. Oncotarget. 2016;7(31):49027–41. Epub 2016/07/28. doi: 10.18632/oncotarget.10717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Qiao JT, Cui C, Qing L, Wang LS, He TY, Yan F, et al. Activation of the STING-IRF3 pathway promotes hepatocyte inflammation, apoptosis and induces metabolic disorders in nonalcoholic fatty liver disease. Metabolism: clinical and experimental. 2018;81:13–24. Epub 2017/11/07. doi: 10.1016/j.metabol.2017.09.010 [DOI] [PubMed] [Google Scholar]
- 13.Faouzi S, Burckhardt BE, Hanson JC, Campe CB, Schrum LW, Rippe RA, et al. Anti-Fas induces hepatic chemokines and promotes inflammation by an NF-kappa B-independent, caspase-3-dependent pathway. J Biol Chem. 2001;276(52):49077–82. Epub 2001/10/17. doi: 10.1074/jbc.M109791200 [DOI] [PubMed] [Google Scholar]
- 14.Iracheta-Vellve A, Petrasek J, Gyongyosi B, Satishchandran A, Lowe P, Kodys K, et al. Endoplasmic reticulum stress-induced hepatocellular death pathways mediate liver injury and fibrosis via stimulator of interferon genes. J Biol Chem. 2016;291(52):26794–805. Epub 2016/11/05. doi: 10.1074/jbc.M116.736991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Petrasek J, Iracheta-Vellve A, Csak T, Satishchandran A, Kodys K, Kurt-Jones EA, et al. STING-IRF3 pathway links endoplasmic reticulum stress with hepatocyte apoptosis in early alcoholic liver disease. Proc Natl Acad Sci U S A. 2013;110(41):16544–9. Epub 2013/09/21. doi: 10.1073/pnas.1308331110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sanz-Garcia C, Poulsen KL, Bellos D, Wang H, McMullen MR, Li X, et al. The non-transcriptional activity of IRF3 modulates hepatic immune cell populations in acute-on-chronic ethanol administration in mice. J Hepatol. 2019;70(5):974–84. Epub 2019/02/03. doi: 10.1016/j.jhep.2019.01.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Clodfelter KH, Holloway MG, Hodor P, Park SH, Ray WJ, Waxman DJ. Sex-dependent liver gene expression is extensive and largely dependent upon signal transducer and activator of transcription 5b (STAT5b): STAT5b-dependent activation of male genes and repression of female genes revealed by microarray analysis. Mol Endocrinol. 2006;20(6):1333–51. Epub 2006/02/14. doi: 10.1210/me.2005-0489 [DOI] [PubMed] [Google Scholar]
- 18.Melia T, Waxman DJ. Genetic factors contributing to extensive variability of sex-specific hepatic gene expression in Diversity Outbred mice. PLoS One. 2020;15(12):e0242665. Epub 2020/12/03. doi: 10.1371/journal.pone.0242665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fensterl V, Sen GC. Interferon-induced Ifit proteins: their role in viral pathogenesis. J Virol. 2015;89(5):2462–8. Epub 2014/11/28. doi: 10.1128/JVI.02744-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stawowczyk M, Van Scoy S, Kumar KP, Reich NC. The interferon stimulated gene 54 promotes apoptosis. J Biol Chem. 2011;286(9):7257–66. Epub 2010/12/31. doi: 10.1074/jbc.M110.207068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kimura T, Flynn CT, Alirezaei M, Sen GC, Whitton JL. Biphasic and cardiomyocyte-specific IFIT activity protects cardiomyocytes from enteroviral infection. PLoS Pathog. 2019;15(4):e1007674. Epub 2019/04/09. doi: 10.1371/journal.ppat.1007674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chattopadhyay S, Marques JT, Yamashita M, Peters KL, Smith K, Desai A, et al. Viral apoptosis is induced by IRF-3-mediated activation of Bax. The EMBO journal. 2010;29(10):1762–73. Epub 2010/04/03. doi: 10.1038/emboj.2010.50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oberbeck N, Pham VC, Webster JD, Reja R, Huang CS, Zhang Y, et al. The RIPK4-IRF6 signalling axis safeguards epidermal differentiation and barrier function. Nature. 2019;574(7777):249–53. Epub 2019/10/04. doi: 10.1038/s41586-019-1615-3 [DOI] [PubMed] [Google Scholar]
- 24.Feng H, Zhang Y-B, Gui J-F, Lemon SM, Yamane D. Interferon regulatory factor 1 (IRF1) and anti-pathogen innate immune responses. PLoS Pathog. 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liberzon A, Birger C, Thorvaldsdóttir H, Ghandi M, Mesirov JP, Tamayo P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell systems. 2015;1(6):417–25. Epub 2016/01/16. doi: 10.1016/j.cels.2015.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hermant P, Demarez C, Mahlakoiv T, Staeheli P, Meuleman P, Michiels T. Human but not mouse hepatocytes respond to interferon-lambda in vivo. PLoS One. 2014;9(1):e87906. Epub 2014/02/06. doi: 10.1371/journal.pone.0087906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mordstein M, Kochs G, Dumoutier L, Renauld JC, Paludan SR, Klucher K, et al. Interferon-lambda contributes to innate immunity of mice against influenza A virus but not against hepatotropic viruses. PLoS Pathog. 2008;4(9):e1000151. Epub 2008/09/13. doi: 10.1371/journal.ppat.1000151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vallbracht A, Maier K, Stierhof YD, Wiedmann KH, Flehmig B, Fleischer B. Liver-derived cytotoxic T cells in hepatitis A virus infection. J Infect Dis. 1989;160:209–17. doi: 10.1093/infdis/160.2.209 [DOI] [PubMed] [Google Scholar]
- 29.Schulte I, Hitziger T, Giugliano S, Timm J, Gold H, Heinemann FM, et al. Characterization of CD8+ T-cell response in acute and resolved hepatitis A virus infection. J Hepatol. 2010;54:201–08. Epub 2010/11/09. S0168-8278(10)00782-8 [pii] doi: 10.1016/j.jhep.2010.07.010 [DOI] [PubMed] [Google Scholar]
- 30.Zhou Y, Callendret B, Xu D, Brasky KM, Feng Z, Hensley LL, et al. Dominance of the CD4+ T helper cell response during acute resolving hepatitis A virus infection. J Exp Med. 2012;209:1481–92. Epub 2012/07/02. jem.20111906 [pii]doi: 10.1084/jem.20111906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim J, Chang DY, Lee HW, Lee H, Kim JH, Sung PS, et al. Innate-like cytotoxic function of bystander-activated CD8(+) T Cells is associated with liver injury in acute hepatitis A. Immunity. 2018;48(1):161–73.e5. Epub 2018/01/07. doi: 10.1016/j.immuni.2017.11.025 [DOI] [PubMed] [Google Scholar]
- 32.Rha MS, Han JW, Kim JH, Koh JY, Park HJ, Kim SI, et al. Human liver CD8(+) MAIT cells exert TCR/MR1-independent innate-like cytotoxicity in response to IL-15. J Hepatol. 2020;73(3):640–50. Epub 2020/04/06. doi: 10.1016/j.jhep.2020.03.033 [DOI] [PubMed] [Google Scholar]
- 33.Misumi I, Li Z, Sun L, Das A, Shiota T, Cullen J, et al. Iminosugar glucosidase inhibitors reduce hepatic inflammation in HAV-infected Ifnar1(-/-) mice. J Virol. 2021. Epub 2021/03/12. doi: 10.1128/JVI.00058-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Misumi I, Mitchell JE, Lund MM, Cullen J, Lemon SM, Whitmire JK. T cells protect against hepatitis A virus infection and limit infection-induced liver injury. J Hepatol. 2021;In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yamane D, Feng H, Rivera-Serrano EE, Selitsky SR, Hirai-Yuki A, Das A, et al. Constitutive expression of interferon regulatory factor 1 drives intrinsic hepatocyte resistance to multiple RNA viruses. Nat Microbiol 2019;4(7):1096–104. doi: 10.1038/s41564-019-0425-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gytz H, Hansen MF, Skovbjerg S, Kristensen AC, Hørlyck S, Jensen MB, et al. Apoptotic properties of the type 1 interferon induced family of human mitochondrial membrane ISG12 proteins. Biol Cell 2017;109(2):94–112. Epub 2016/10/27. doi: 10.1111/boc.201600034 [DOI] [PubMed] [Google Scholar]
- 37.Yang D, Meng X, Xue B, Liu N, Wang X, Zhu H. MiR-942 mediates hepatitis C virus-induced apoptosis via regulation of ISG12a. PLoS One. 2014;9(4):e94501. Epub 2014/04/15. doi: 10.1371/journal.pone.0094501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cheriyath V, Leaman DW, Borden EC. Emerging roles of FAM14 family members (G1P3/ISG 6–16 and ISG12/IFI27) in innate immunity and cancer. J Interferon Cytokine Res. 2011;31(1):173–81. Epub 2010/10/14. doi: 10.1089/jir.2010.0105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lu MY, Liao F. Interferon-stimulated gene ISG12b2 is localized to the inner mitochondrial membrane and mediates virus-induced cell death. Cell Death Differ. 2011;18(6):925–36. Epub 2010/12/15. doi: 10.1038/cdd.2010.160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nakajima A, Nishimura K, Nakaima Y, Oh T, Noguchi S, Taniguchi T, et al. Cell type-dependent proapoptotic role of Bcl2L12 revealed by a mutation concomitant with the disruption of the juxtaposed Irf3 gene. Proc Natl Acad Sci U S A. 2009;106(30):12448–52. Epub 2009/07/21. doi: 10.1073/pnas.0905702106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–50. Epub 2005/10/04. doi: 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genetics. 2003;34(3):267–73. Epub 2003/06/17. doi: 10.1038/ng1180 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Principal component analysis of RNAseq data from 24 mice in groups of 6 infected and 6 uninfected of each genetic knock-out. First and second components (left) represent variance due to differences in sex (42%) and sequencing run (batch, 18%); RNAseq reads from noninfected and infected Ifnar1-/- mice cluster separately from naïve Mavs-/- mice based on the 3rd and 4th components (combined contribution to variance = 15%). (B,C) Heat maps reflecting (B) differential expression of the 617 genes which were significantly changed (padj<0.05) following infection in either Mavs-/- or Ifnar1-/- mice, and (C) differentially expressed genes in liver tissue from (left) Mavs-/- (n = 40 genes) and (right) Ifnar1-/- mice (n = 579 genes). Scales on the right represent z-score. (D) Expanded volcano plot showing fold-change (fc) and significance (padj = adjusted p-value) of transcripts with significant change in abundance in liver tissue from Mavs-/- mice (see Fig 1G in the main manuscript). (E) Average reads mapped in liver tissue from naïve and HAV-infected Mavs-/- mice for those 25 transcripts with the greatest fold-change in abundance in infected Ifnar-/- mice (see Fig 1F in the main manuscript). (F) Gene set enrichment analysis of transcripts with significant change in abundance in Ifnar-/- mice. No significant hallmark signatures were identified for tissue from Mavs-/- mice.
(TIF)
HAV RNA and Ccl5 (RANTES) mRNA were probed for in liver tissue from (top to bottom) uninfected Ifnar1-/- mice and HAV-infected Mavs-/- and Ifnar-/- mice. Merged fluorescence images are shown on the left, and single-channel images on the right.
(TIF)
Liver tissue was harvested 7 days post-inoculation of HM175-mp6 virus. (A) Normalized HAV RNA read counts. RPMM = reads per million reads mapped. (B) Principal component analysis showing variance related to sex, knockout type, and HAV infection status of animals. (C) Correlation plot of fold-change induction of individual genes in Ifnar1-/- mice infected with HM175-mp10 virus (see Fig 1) versus induction in Ifnar1-/- mice infected with HM175-mp6 virus (see Fig 4). R2 determined by linear regression analysis for all transcripts upregulated >2-fold. (D) Volcano plot showing difference (fold-change, fc) and significance of differences in transcript abundance in livers of naive Irf3S1/S1Ifnar1-/- and Ifnar1-/- mice. padj = adjusted p-value. (E) Venn diagram showing overlap in transcripts downregulated following HAV infection of Ifnar1-/- and Irf3S1/S1Ifnar1-/- mice. (F). Normalized read counts observed for interferon regulatory factor (IRF) transcripts in livers of naïve Irf3S1/S1Ifnar1-/- and Ifnar1-/- mice.
(TIF)
(A) Heat maps reflecting transcript abundance of top 40 genes in the leading edges, and bottom 20 genes in the trailing edges, of hallmark TNF-α signaling via NF-κB and apoptosis gene sets. The number of genes in each set is shown at the top. (B) Venn diagrams showing overlap between the genes in the (top) leading edges and (bottom) trailing edges of gene sets shown in panel A and the hallmark inflammatory response gene set in Fig 5 in the main manuscript.
(TIF)
Intrahepatic abundance of transcripts encoding ISG12a family members and BCL2L12 in naïve and HAV-infected Irf3S1/S1Ifnar1-/- and Ifnar1-/- mice (n = 4 each) 7 days after infection with HM175-mp6 virus. (A) Normalized Ifi27 gene family member read counts from high throughput sequencing. (B) Relative transcript abundance determined by RT-qPCR, showing fold-change from uninfected mouse liver. (C) Normalized Bcl2l12 gene family member read counts from high throughput sequencing. Error bars in all panels represent s.d. Significance determined by unpaired t-test.
(TIF)
(XLSX)
(PDF)
(XLSX)
Data Availability Statement
Datasets generated and analyzed during this study are included with this publication or have been deposited with Gene Expression Omnibus (GEO), accession numbers GSE166353: GSM5068950-GSM5068989.