Abstract
Introduction
We aimed to examine if severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) polymerase chain reaction (PCR) cycle quantification (Cq) value, as a surrogate for SARS-CoV-2 viral load, could predict hospitalisation and disease severity in adult patients with coronavirus disease 2019 (COVID-19).
Methods
We performed a prospective cohort study of adult patients with PCR positive SARS-CoV-2 airway samples including all out-patients registered at the Department of Infectious Diseases, Odense University Hospital (OUH) March 9-March 17 2020, and all hospitalised patients at OUH March 10-April 21 2020. To identify associations between Cq-values and a) hospital admission and b) a severe outcome, logistic regression analyses were used to compute odds ratios (OR) and 95% Confidence Intervals (CI), adjusting for confounding factors (aOR).
Results
We included 87 non-hospitalised and 82 hospitalised patients. The median baseline Cq-value was 25.5 (interquartile range 22.3–29.0). We found a significant association between increasing Cq-value and hospital-admission in univariate analysis (OR 1.11, 95% CI 1.04–1.19). However, this was due to an association between time from symptom onset to testing and Cq-values, and no association was found in the adjusted analysis (aOR 1.08, 95% CI 0.94–1.23). In hospitalised patients, a significant association between lower Cq-values and higher risk of severe disease was found (aOR 0.89, 95% CI 0.81–0.98), independent of timing of testing.
Conclusions
SARS-CoV-2 PCR Cq-values in outpatients correlated with time after symptom onset, but was not a predictor of hospitalisation. However, in hospitalised patients lower Cq-values were associated with higher risk of severe disease.
Introduction
As the novel Severe Acute Respiratory Syndrome Coronavirus-2 (SARS-CoV-2) sweeps through the world, detection of viral RNA by polymerase chain reaction (PCR) has become the gold standard for diagnosing coronavirus disease 2019 (COVID-19) [1, 2]. Nasopharyngeal or oropharyngeal swabs make up the majority of tests, since most patients are unable to produce sputum despite higher sensitivity of the latter [3].
In viral diseases, the PCR Quantification Cycle-value or Cycle threshold-value (Cq or Ct -value) can be used as a surrogate for viral load, with inverse correlation between the Cq-value and viral load. The use of Cq-value as a prognostic marker for disease severity in viral respiratory infections has been tested with varying results [4–6]. For coronaviruses, there is evidence from cohort studies supporting a correlation between Cq-values in upper airway samples and disease severity for human coronavirus in children, and between upper airway sample viral loads and disease severity for SARS-CoV-1 and Middle East Respiratory Syndrome (MERS) coronavirus infections in adults [7–9].
The most common symptoms of COVID-19 are fever, cough and dyspnea, and the course of disease can be complicated with Acute Respiratory Distress Syndrome (ARDS), respiratory failure and death [10–12]. There is limited data on whether viral load of SARS-CoV-2 correlate with disease severity. Two Chinese studies found that both in- and outpatients with COVID-19 had lower Cq-values indicating higher viral loads early in their disease course [13, 14]. A German study including hospitalised patients diagnosed with COVID-19 found that viral loads were high in the initial oropharyngeal samples and declining in 1–2 weeks [15]. Hospitalised patients in China with severe disease were found to have higher initial viral loads and prolonged time to reach PCR-negativity compared with patients with mild disease [16, 17].
With this study, we aimed to examine if baseline PCR Cq-values can identify 1) SARS-CoV-2 positive patients at increased risk of hospitalisation, and 2) hospitalised COVID-19 patients at increased risk of severe disease.
We hypothesized that the initial PCR Cq-values were lower among hospitalised patients, as a surrogate for higher viral loads, compared with non-hospitalised patients. We also hypothesized that due to a failure to reduce viral burden after the initial infection phase, lower PCR Cq-values were related to severe disease in hospitalised patients.
Materials and methods
Study setting and population
Odense University Hospital (OUH) serves as a tertiary hospital for the Region of Southern Denmark (approximately 1.2 million inhabitants) as well as a secondary hospital for the island of Funen (approximately 0.5 million inhabitants) [18]. The Danish public healthcare system supplies free, tax-funded healthcare for all residents.
Initially, the Danish national COVID-19 strategy was based on containment, where individuals who met the case definition were tested for SARS-CoV-2. The strategy later changed to mitigation, where only patients with symptoms of COVID-19 requiring hospital admission were tested for SARS-CoV-2.
Data sources
We used the unique 10-digit personal identification number assigned to all individuals in Denmark at birth or upon immigration to link the following two registries electronically with laboratory data:
The COVID-19 Hospital Cohort at OUH; a prospective hospital-based cohort of all adult (≥18 years old) COVID-19 patients admitted or referred to OUH since March 10, 2020. The cohort is ongoing and consecutively includes patients diagnosed with COVID-19. All patients admitted until April 21, with an available PCR Cq-value were included in this study. More details about this cohort is published elsewhere [11].
The COVID-19 Outpatient Cohort in the Region of Southern Denmark; a database of all adult COVID-19 patients from the Region of Southern Denmark tested positive for SARS-CoV-2 between March 9, 2020 and March 17, 2020, who had an available PCR Cq-value and were not admitted to a COVID-19 unit during their course of disease.
Data collection
For the hospital cohort, demography-, clinical-, laboratory-, management- and outcome data were gathered through review of medical records [11]. For the outpatient cohort, all eligible patients were invited to participate in an online survey two months after symptom onset. By signing an electronic consent form, a survey could be filled-out and electronically retrieved into a database. The data included information on demography, disease exposure, clinical symptoms of COVID-19, days until recovery and remaining symptoms (see S1 Appendix).
Data on PCR assays, type of airway samples (naso- and/or oropharyngeal swab, or sputum), PCR Cq-values and sample dates were collected from the Department of Clinical Microbiology, OUH and the Department of Clinical Microbiology, Lillebaelt Hospital.
SARS-CoV2 PCR assays
SARS-CoV-2 detection was established on three different analysis platforms—the fully automated high throughput Cobas 6800 (Roche), the commercially available kit RealStar® SARS-CoV-2 RT-PCR kit 1.0 (Altona Diagnostics) and a laboratory developed real-time (RT)-PCR.
On Cobas 6800 a 650 μl respiratory sample (oropharyngeal swab sample or sputum) was applied onto the system and subsequently RNA extraction, reverse transcription, PCR analysis and detection were performed. SARS-CoV-2 detection on Cobas 6800 included an internal RNA control, primers and probes targeting the ORF1a/b non-structural region that is unique for SARS-CoV-2 (target 1) and a conserved region in the structural protein envelope E gene that is shared by the Sarbecovirus subgenus (target 2).
RNA used for the RealStar® SARS-CoV-2 RT-PCR test (Altona Diagnostics) and for the laboratory developed test was either extracted from: 1) 500 μl respiratory sample material (oropharyngeal swab sample or sputum) using MagNA Pure 96 (Roche) with the extraction kit DNA and viral NA large volume kit (Roche) using the protocol Pathogen Universal, or 2) 300 μl respiratory sample material (naso- and oropharyngeal swab sample or sputum) using the Maxwell® 16 Viral Total Nucleic Acid Purification Kit (Promega) following the manufacturer’s protocol. RealStar® SARS-CoV-2 RT-PCR kit 1.0 included three PCR analyses for the qualitative detection of and differentiation of Sarbecovirus subgenus (E gene) and SARS-CoV-2 specific RNA (S gene) in addition to an internal control. The kit was used according to the manufacturer’s instructions with 30 μl reaction volume, and the 1-step RT-PCR was performed using Lightcycler 480 (Roche) or Stratagene Mx3005P (Agilent) in 96 well formats.
The laboratory developed real-time PCR E gene assay used for SARS-CoV-2 detection has been described previously [19]. This assay targeted a conserved sequence in the E gene region that is shared by the Sarbecovirus subgenus group (FP: ACAGGTACGTTAATAGTTAATAGCGT, RP: ATATTGCAGCAGTACGCACACA, Probe: FAM-ACACTAGCCATCCTTACTGCGCTTCG-BHQ1). Real-time PCR was performed in 15 μl reactions containing 3.75 μl TaqMan Fast Virus 1-Step master mix (ThermoFisher) with 1000 nM of each primer and 200 nM of the probe, and 5 μl RNA eluate. An internal RNA control (Newcastle disease virus vaccine strain; MSD) was added to the sample prior to RNA extraction (NDV-FWD-2: 5’-CACTGTCGGCATTATCGATGA-3’, NDV-REV: 5’-GAGCATCGCAGCGGAAA-3’, NDV-Probe: 5’-FAM-CCCAAGCGCGAGTTA-MGB-3’). Reverse transcription and amplification was performed using Lightcycler 480 (Roche) in 384 well formats. The cycling conditions were as follows: Reverse transcription at 50°C for 5 min, inactivation of RT/initial denaturation at 95° C for 20 sec, followed by 45 cycles of 95°C for 15 sec, 60°C for 1 min for amplification.
For all assays, the PCR Cq-value cut-off for a negative test was set at 40 cycles.
As for choice of baseline test assay when more than one test was available, in assays with both a target for pan-Sarbecovirus and Coronavirus SARS-CoV-2 (Cobas 6800 and RealStar® SARS-CoV-2 RT-PCR test), the Cq-value for the pan-Sarbecovirus was chosen if available, if not the Cq-value for Coronavirus SARS-CoV-2 target was used. If one sample was tested with multiple assays, the assays were prioritized in the following order; 1. Cobas 6800 (Roche), 2. The laboratory developed real-time PCR, and 3. RealStar SARS-CoV-2 RT-PCR test (Altona Diagnostics). The order was chosen by an experienced molecular biologist and a senior clinical microbiologist. For both naso/oropharyngeal swabs and sputum samples, the baseline PCR-sample for each patient was set to the first registered test for that patient (= day 0). If there were multiple tests for one patient within the first 3 days (Day 0, 1 and 2), the sample with the lowest Cq-value within this period was chosen.
Study design
We conducted a retrospective case-control study consisting of two different comparisons of sub-groups: 1) a case-control study with the hospital cohort as cases and the outpatient cohort as controls, and 2) a case-control study of our hospital cohort where the hospitalised patients with severe disease defined as ARDS, admittance to the Intensive Care Unit (ICU) and/or death during admission were included as cases, and the hospitalised patients with moderate disease (not fulfilling the definition of severe disease) were controls. The criteria for ARDS and grading of severity of ARDS were based on current international guidelines [20, 21].
Exposures
PCR Cq-values were used to estimate predictors for 1) hospital admission, and 2) ARDS, ICU admission and/or death.
Statistics
For baseline variables, descriptive statistics were reported as numbers and percentages for categorical variables and medians with interquartile ranges (IQR) for continuous variables. Chi-squared test or Fisher’s exact test were used to compare categorical variables between groups, student’s t-test and Wilcoxon Mann-Whitney test were used for parametric and non-parametric continuous variables, respectively.
We plotted the Cq-values according to days since symptom onset and examined a possible association using linear regression. To identify whether the Cq-value could predict 1) hospital admission and 2) ARDS, ICU admission and/or death, we used logistic regression to compute odds ratios (OR) and 95% confidence intervals (CI). Analyses were adjusted for potential confounding variables, which based on the current knowledge on COVID-19 was predetermined to be age, sex, comorbidities, Body Mass Index (BMI) and days from symptom debut to baseline PCR-sample (1). To reduce the risk of over-fitting, we only included confounders considered most important (sex, age) in the final multiple regression model (2).
Data on all patients were registered in a REDCap database hosted by Open Patient data Explorative Network (OPEN) [22]. STATA version 15 (StataCorp LP, Texas) was used for data processing and analyses.
Ethics approval
This study was registered as a quality development project at OUH, approved by the Danish Data Protection Agency (j.nr. 20/16169 and 20/20759) and the Danish Patient Safety Authority (Sagsnr. 31-1521-344). All data were handled in accordance with The General Data Protection Regulation (GDPR), the Danish Act on Data Protection, the Danish Act on Research Ethics Review of Health Research Projects and the Danish Health Act. The study adheres to the STROBE guidelines for observational studies. All patients gave informed consent for study participation prior to inclusion.
Results
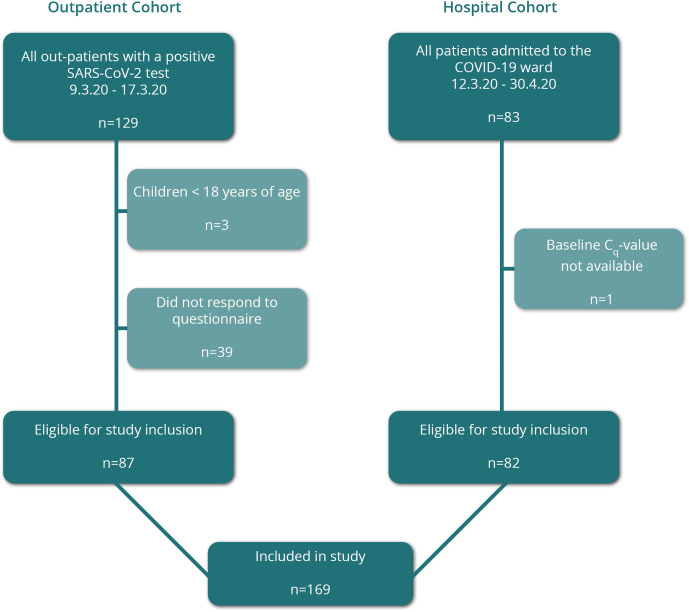
A total of 169 patients were included in the final cohort; 87 from the outpatient cohort (Fig 1).
Fig 1. Study inclusion of non-hospitalised and hospitalised patients into the Odense University Hospital COVID-19 cohort.
Patient characteristics
The baseline patient characteristics of the two cohorts are shown in Table 1.
Table 1. Characteristics and exposures in a Danish outpatients and hospitalised patients with COVID-19.
| Study population | All patients n = 169 | Outpatient cohort n = 87 | Hospital cohort n = 82 | p-value | |
|---|---|---|---|---|---|
| Age (years), median (IQR) | 54 (45–64) | 46 (36–54) | 63 (55–74) | <0.001 | |
| N = 169 | |||||
| Sex | Male (%) | 110 (65.1) | 59 (67.8) | 51 (62.2) | 0.44 |
| N = 169 | |||||
| BMI, median (IQR) | 25.6 (23.3–28.8) | 24.6 (23.1–27.2) | 26.5 (23.7–30.1) | 0.003 | |
| N = 167 | |||||
| Tobacco use (%) | 0.28a | ||||
| N = 167 | |||||
| Current smoker | 12 (7.2) | 7 (8.1) | 5 (6.2) | ||
| Former smoker | 54 (32.3) | 23 (26.7) | 31 (38.3) | ||
| Never smoker | 101 (60.5) | 56 (65.1) | 45 (55.6) | ||
| Alcohol consumption (units/week) (%) | 0.02a | ||||
| N = 166 | |||||
| >7 for women / >14 for men | 18 (10.8) | 14 (16.5) | 4 (4.9) | ||
| ≤7 for women / ≤14 for men | 148 (89.2) | 71 (83.5) | 77 (95.1) | ||
| Comorbidity (%) | |||||
| Cardiovascular disease n = 169 | 25 (14.8) | 4 (4.6) | 21 (25.6) | <0.001 | |
| Hypertension n = 168 | 50 (29.8) | 15(17.4) | 35 (42.7) | <0.001 | |
| Pulmonary disease n = 169 | 21 (12.4) | 8 (9.2) | 13 (15.9) | 0.19 | |
| Diabetes mellitus Type I+II n = 167 | 14 (8.4) | 1 (1.2) | 13 (15.9) | 0.001 | |
| Malignancy n = 167 | 19 (11.4) | 5 (5.9) | 14 (17.1) | 0.03 | |
| Health care worker (%) | 16 (10.1) | 11 (12.9) | 5 (6.8) | 0.29 | |
| N = 159 | |||||
| COVID-19 exposure | |||||
| Travel to high risk area (%) | 78 (56.1) | 65 (77.4) | 13 (23.6) | <0.001 | |
| N = 139 | |||||
| Austria (region of Tyrol) | 60 (76.9) | 58 (89.2) | 2 (15.4) | ||
| Italy | 5 (6.4) | 4 (6.2) | 1 (7.7) | ||
| Other | 13 (16.7) | 3 (4.6) | 10 (76.9) | ||
| Contact with suspected/confirmed COVID-19 case (%) | 80 (47.3) | 50 (57.5) | 30 (36.6) | 0.007 | |
| N = 169 | |||||
| Household | 26 (15.4) | 23 (26.4) | 3 (3.7) | <0.001 | |
| Colleague | 9 (5.3) | 7 (8.1) | 2 (2.4) | 0.17 | |
| Other | 37 (21.9) | 29 (33.3) | 8 (9.8) | <0.001 |
IQR = interquartile range; BMI = body mass index.
aAmong all groups.
The hospital cohort was significantly older (median age 63 years (IQR 55–74) vs 46 years (IQR 36–54), p<0.001), had higher weights (median BMI 26.5 (IQR 23.7–30.1) vs 24.6 (IQR 23.1–27.2), p = 0.003) and had a significantly higher proportion of all comorbidities except pulmonary diseases compared with the outpatient cohort.
COVID-19 exposure and symptoms
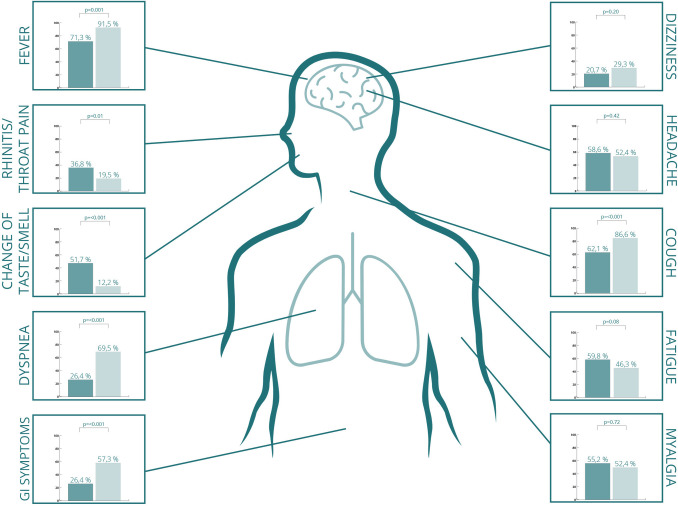
Compared with the hospital cohort, the outpatients had a significantly higher degree of known exposure to COVID-19 (Table 1). In the outpatient group, 65 patients (77.4%) had travelled to a COVID-19 hot-spot in the 14 days prior to symptom onset. Of these, 58 patients (66.7%) had been on skiing holidays in the Tyrol region of Austria. COVID-19 symptoms in the two cohorts are illustrated in Fig 2.
Fig 2. Number of patients displaying different symptoms among a non-hospitalised cohort (n = 87, displayed in dark green), and a hospitalised cohort (n = 82, displayed in light green) of adult COVID-19 patients.
Compared with the outpatient cohort, hospitalised patients more often had fever, cough, dyspnoea and gastrointestinal symptoms but less often rhinitis/throat pain and loss of smell/taste.
SARS-CoV-2 PCR Cq-value as a marker for hospital admission
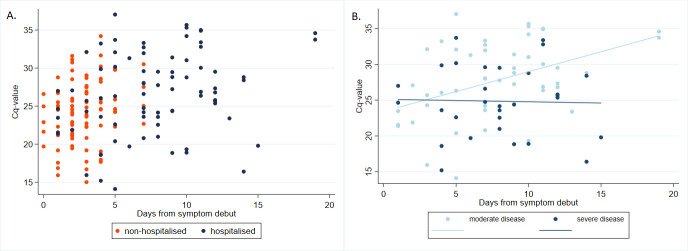
The median baseline SARS-CoV-2 PCR Cq-value for the entire study population was 25.5 (IQR 22.3–29.0). The outpatients had a significantly lower median baseline SARS-CoV-2 PCR Cq-value (24.6, IQR 21.8–27.5) compared with the hospitalised patients (median Cq-value 26.9, IQR 23.6–31.3), p = 0.001 (Fig 3A).
Fig 3.
SARS-CoV-2 PCR baseline Cq-values and days from symptom debut to baseline sample in non-hospitalised (n = 87, in orange) and hospitalised (n = 82, in blue) patients with COVID-19 disease (a), and of the admitted patients with moderate (n = 51, light blue) and severe (n = 31, dark blue) disease (b).
We found a statistically significant association between an increasing baseline Cq-value and higher risk of admission to hospital (OR 1.11, 95%CI 1.04–1.19, p = 0.002) when using unadjusted logistic regression (see S1 Table). However, this was mainly due to a strong association between time from symptom onset and Cq-value (coefficient 0.26, 95%CI 0.15–0.38, p<0.001), as the patients in the outpatient cohort were tested significantly earlier in their course of disease compared with the hospital cohort (median 3 days (IQR 2–4) vs. 8 days (IQR 5–11), p<0.001). When adjusting for this difference in timing of testing, we no longer found a significant association between Cq-values and admission (OR 1.00, 95%CI 0.91–1.09, p = 0.97), irrespective of further adjustment for confounding factors (OR 1.08, 95%CI 0.94–1.24 p = 0.27).
SARS-CoV-2 PCR Cq- values in different airway samples
We found no significant difference in median baseline Cq-values between naso-and/or-oropharyngeal swabs (143 patients; 13 naso-and-oropharyngeal and 130 oropharyngeal) and sputum samples (26 inpatients), with median Cq-values of 25.5 (IQR 22.3–28.8) and 24.4 (IQR 19.8–32.7), respectively (p = 0.61). Among the 165 patients with known symptom onset, we observed a significantly shorter time from symptom onset to first PCR sample in patients tested with naso-and/or-oropharyngeal swabs compared with patients tested with sputum samples (median 3 days (IQR 2–7) vs 8 days (IQR 6–11), p<0.001).
SARS-CoV-2 PCR Cq-value as a predictive marker for disease severity in hospitalised patients
A total of 31 of the 82 patients (38.0%) in the hospital cohort developed severe COVID-19 disease. Patients with moderate and severe disease did not differ with regards to sex, age, BMI, comorbidities, tobacco or alcohol consumption (see Table 2).
Table 2. Characteristics of 82 patients admitted to Odense University Hospital with COVID-19, of which 31 patients had severe disease defined as either Acute Respiratory Distress Syndrome (ARDS), admittance to intensive care unit and/or death during admission, and 51 patients had moderate disease.
| COVID-19 mild disease n = 51 | COVID-19 severe disease n = 31 | p-value | ||
|---|---|---|---|---|
| Age (years), median (IQR) | 61 (52–72) | 67 (58–78) | 0.09 | |
| N = 82 | ||||
| Sex | Male (%) | 28 (54.9) | 23 (74.2) | 0.08 |
| N = 82 | ||||
| BMI, median (IQR) | 26.1 (23.6–30.1) | 26.6 (23.7–32.2) | 0.73 | |
| N = 82 | ||||
| Tobacco use (%) | 0.24 | |||
| N = 81 | ||||
| Current smoker | 5 (9.8) | 0 (0.0) | ||
| Former smoker | 18 (35.3) | 13 (43.3) | ||
| Never smoker | 28 (54.9) | 17 (56.7) | ||
| Alcohol consumption (units/week) (%) | 0.50 | |||
| N = 81 | ||||
| >7 for women / >14 for men | 3 (6.0) | 1 (3.2) | ||
| ≤7 for women / ≤14 for men | 47 (94.0) | 30 (96.8) | ||
| Comorbidity (%) | ||||
| N = 82 | ||||
| Cardiovascular disease | 11 (21.6) | 10 (32.3) | 0.31 | |
| Hypertension | 18 (35.3) | 17 (54.8) | 0.08 | |
| Pulmonary disease | 8 (15.7) | 5 (16.1) | 0.96 | |
| Diabetes mellitus I+II | 7 (13.7) | 6 (19.4) | 0.54 | |
| Malignancy | 8 (15.7) | 6 (19.4) | 0.77 |
IQR = interquartile range; BMI = body mass index.
Patients with severe disease had significantly lower baseline Cq-values compared with patients with moderate disease (median 24.8 (IQR 21.0–28.8) vs 28.1 (IQR 24.3–33.2), p = 0.01). We found a statistically significant association between lower Cq-values and higher risk of severe disease (OR 0.89, 95%CI 0.81–0.98, p = 0.018). This association was independent of timing of the test in relation to symptom onset as well as presence of confounding factors including type of airway sample.
For patients with moderate disease, we found a direct linear association between the Cq-value and time of baseline test (Fig 3B). In contrast, we observed that patients with severe disease had a low baseline PCR Cq-value irrespective of time of testing. However, the regression coefficient between these two curves did not differ statistically (coef.-0.59 95%CI -1.20–0.02, p = 0.056) conferring to no significant interaction between Cq-value and time of test.
Course of disease
Median symptom duration in the out-patient cohort was 11 days (IQR 5–16) when excluding fatigue and loss of taste/smell, which persisted two months after onset of COVID-19 disease in 15 (17.2%) and 27 patients (31.0%), respectively. A SARS-CoV-2 PCR test was repeated in 17 patients after a median of 8 days (IQR 6–9); all Cq-values increased, and 6 patients were PCR negative (Fig 4A).
Fig 4.
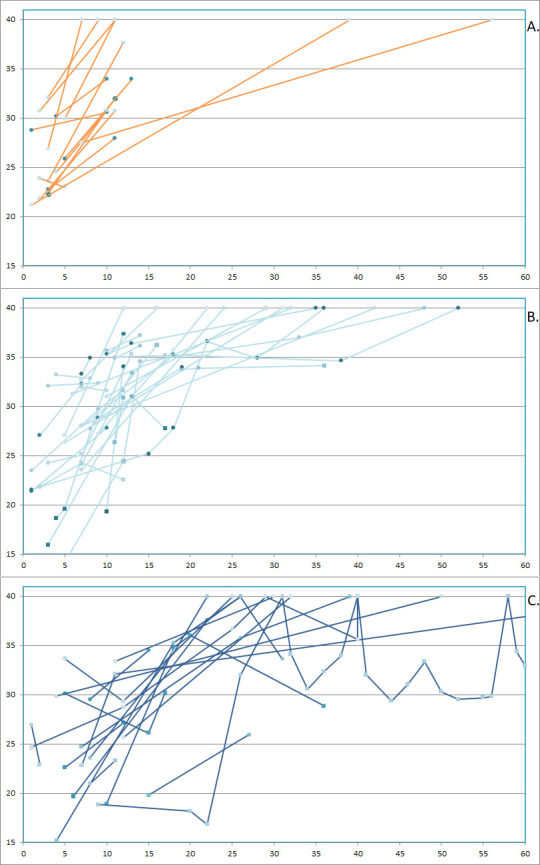
SARS-CoV-2 PCR Cq-values over time in 17 non-hospitalised patients (displayed in orange) (a), 33 hospitalised patients with moderate COVID-19 disease (displayed in light blue) (b) and 18 hospitalised patients with severe disease (displayed in dark blue) (c). The y-axis displays Cq-value and the x-axis displays days from symptom onset. Circle = naso-and/or-oropharyngeal swabs, squares = sputum samples.
In the hospital cohort, the median time from hospital admittance to either discharge (n = 78) or death (n = 4) was 7.5 days (IQR 3–11). Multiple PCR-samples were available in 33 patients with moderate disease and 18 patients with severe disease and showed a more complex pattern compared with the out-patient cohort. We observed a less linear increase in Cq-values, longer PCR positivity and several patients with subsequently decreasing Cq-values (Fig 4B+4C).
Discussion
To our knowledge, this prospective study is the first to compare SARS-CoV-2 PCR Cq-values between non-hospitalised and hospitalised patients. Our most important findings were the strong linear association between Cq-values and time of testing after symptom onset, the correlation between lower Cq-values and increased disease severity in hospitalised patients and the lack of association between Cq-values and risk of hospitalisation.
Our results of a linear association between Cq-values and timing of the test after symptom onset are in line with available data that suggest higher SARS-CoV-2 viral loads in airway samples at symptom presentation followed by a gradual decrease [13–15, 17]. In this way, the novel Coronavirus differs from SARS-CoV-1, where viral loads were found to increase in airway samples until day 12–14 after symptom onset before decreasing [23, 24].
In hospitalised patients, we found that a lower Cq-value was associated with a significantly higher risk of severe disease irrespective of time of sampling and confounding factors. These findings are in line with the initial Chinese studies by Zheng and Liu, where patients with clinically severe disease had lower Cq-values and were PCR positive longer than patients with mild disease [16, 17]. Due to the limited size of our population, a specific PCR Cq-cutoff-value for patients in high risk of severe disease could not be estimated. Other studies are needed to explore this further in order to use it in a prediction model.
We could not confirm our hypothesis of an association between lower baseline Cq-values and higher risk of hospital admission when adjusting for timing of the test and confounding factors. To our knowledge, there are no available studies that have investigated this possible correlation.
Two systematic literature reviews regarding the use of PCR Cq-values in SARS-CoV-2 have been published since we undertook our study [25, 26]. In accordance with our findings, both studies report evidence of increasing Cq-values in respiratory samples over time, and an association between Cq-values and disease severity in hospitalized patients. However, the evidence was not conclusive and more data is needed in this area.
Of symptoms of COVID-19, we found significantly more patients in the hospitalized cohort with cough, dyspnea, fever and gastrointestinal symptoms. On the other hand, significantly more non-hospitalized patients suffered from rhinitis/throat pain and change in taste and/or smell. Other studies have shown conflicting results regarding change in taste/smell and severity of disease [27, 28].
The main strength of this study is the well-described cohort with near-complete data of high quality for all patients as well as electronically retrieved Cq-values. Furthermore, patients in both cohorts have been tested for SARS-CoV-2 based on the national standardized guidelines.
Our study has some limitations. Available data show varying inter-test agreement between different SARS-CoV-2 assays, especially in samples with high Cq-values [29–32]. In our study, three different PCR assays were used. This may have affected the reproducibility of the results. The different assays used reflect time and availability of assays during the pandemic; in the beginning most patients were tested using the in-house Flow, which was later replaced by the Cobas 6800. We also included results from both naso- and/or-oropharyngeal swabs and sputum samples, the latter only from hospitalised patients. Though we did not find any significant difference between sputum and oropharyngeal baseline Cq-values, this could be explained by the time of sampling, as the sputum samples were generally tested later in the patients’ disease course. However, when including type of airway sample in our regression model for hospitalised patients, it did not alter the results. All airway-samples used in our study were sampled after clinical indication, and not as part of a research project. The airway swabs have therefore been sampled by different medical personnel. As this is an operator-dependent procedure, this lack of standardization may have affected the results. Whereas data on the hospital cohort was based on hospital files, data from the outpatient cohort was based on questionnaires filled out approximately two months after onset of disease. Therefore, recall bias cannot be excluded. Due to the small size of the two cohorts, we cannot exclude a risk of type 2 errors.
Finally, despite omission of confounding variables deemed not statistically significant, we cannot exclude some degree of over-fitting of the multivariate regression analyses. More research in this area is needed, and larger cohorts would be able to confirm our findings with greater certainty.
There are still questions that need to be enlightened regarding why some patients get severe COVID-19 disease and others do not. Our findings suggest that clinicians cannot use the baseline Cq-value in outpatients to predict risk of hospitalisation later in their disease course. However, treating physicians should be vigilant of admitted patients with initial low Cq-values in their airway samples. When interpreting Cq-values, time of symptom onset should be considered, and patients with continuously low Cq-values should be closely monitored.
In conclusion, SARS-CoV-2 PCR Cq-values correlated with time after onset of symptoms. Early in the disease course Cq-values were low as a sign of high viral loads. We did not find Cq-values to be a predictor for hospitalisation. However, in hospitalised patients lower Cq-values were found to be predictive of more severe disease.
Supporting information
(DOCX)
(DOCX)
Acknowledgments
For the laboratory-developed real time PCR, the primer and probe sequences targeting the internal control virus were kindly provided by Dr. Kurt Handberg, Department of Clinical Microbiology, Aarhus University Hospital. We thank Benedicte Christie Knudtzen for help with graphic design of Figs 1 and 2.
Data Availability
Due to The Danish General Data Protection Regulation (GDPR), the data used in this article is not publicly available. Researchers can request access to the data from the Danish Patient Safety Authority (https:/stps.dk/en or by email stps@stps.dk) and the Danish Data Protection Agency (https://www.datatilsynet.dk/english/).
Funding Statement
The authors received no specific funding for this work.
References
- 1.Wu F, Zhao S, Yu B, Chen YM, Wang W, Song ZG, et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579(7798):265–9. Epub 2020/02/06. doi: 10.1038/s41586-020-2008-3 ; PubMed Central PMCID: PMC7094943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.WHO. Laboratory testing for 2019 novel coronavirus (2019-CoV) in suspected human cases 2020 [updated 19 March 2020April 29th 2020]. Available from: https://www.who.int/publications-detail/laboratory-testing-for-2019-novel-coronavirus-in-suspected-human-cases-20200117.
- 3.Yu F, Yan L, Wang N, Yang S, Wang L, Tang Y, et al. Quantitative Detection and Viral Load Analysis of SARS-CoV-2 in Infected Patients. Clinical infectious diseases: an official publication of the Infectious Diseases Society of America. 2020. Epub 2020/03/30. doi: 10.1093/cid/ciaa345 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lalueza A, Folgueira D, Munoz-Gallego I, Trujillo H, Laureiro J, Hernandez-Jimenez P, et al. Influence of viral load in the outcome of hospitalized patients with influenza virus infection. European journal of clinical microbiology & infectious diseases: official publication of the European Society of Clinical Microbiology. 2019;38(4):667–73. Epub 2019/03/02. doi: 10.1007/s10096-019-03514-1 ; PubMed Central PMCID: PMC7102091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Spencer S, Chung J, Thompson M, Piedra PA, Jewell A, Avadhanula V, et al. Factors associated with real-time RT-PCR cycle threshold values among medically attended influenza episodes. Journal of medical virology. 2016;88(4):719–23. Epub 2015/09/04. doi: 10.1002/jmv.24373 . [DOI] [PubMed] [Google Scholar]
- 6.Vos LM, Bruyndonckx R, Zuithoff NPA, Little P, Oosterheert JJ, Broekhuizen BDL, et al. Lower respiratory tract infection in the community: associations between viral aetiology and illness course. Clin Microbiol Infect. 2020. Epub 2020/04/04. doi: 10.1016/j.cmi.2020.03.023 ; PubMed Central PMCID: PMC7118666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wishaupt JO, Ploeg TV, Smeets LC, Groot R, Versteegh FG, Hartwig NG. Pitfalls in interpretation of CT-values of RT-PCR in children with acute respiratory tract infections. Journal of clinical virology: the official publication of the Pan American Society for Clinical Virology. 2017;90:1–6. Epub 2017/03/06. doi: 10.1016/j.jcv.2017.02.010 ; PubMed Central PMCID: PMC7185604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hung IF, Cheng VC, Wu AK, Tang BS, Chan KH, Chu CM, et al. Viral loads in clinical specimens and SARS manifestations. Emerging infectious diseases. 2004;10(9):1550–7. Epub 2004/10/23. doi: 10.3201/eid1009.040058 ; PubMed Central PMCID: PMC3320271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Oh MD, Park WB, Choe PG, Choi SJ, Kim JI, Chae J, et al. Viral Load Kinetics of MERS Coronavirus Infection. The New England journal of medicine. 2016;375(13):1303–5. Epub 2016/09/30. doi: 10.1056/NEJMc1511695 . [DOI] [PubMed] [Google Scholar]
- 10.De Vito A, Fiore V, Princic E, Geremia N, Panu Napodano CM, Muredda AA, et al. Predictors of infection, symptoms development, and mortality in people with SARS-CoV-2 living in retirement nursing homes. PLoS One. 2021;16(3):e0248009. Epub 2021/03/17. doi: 10.1371/journal.pone.0248009 ; PubMed Central PMCID: PMC7963051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Madsen LW, Lindvig SO, Rasmussen LD, Knudtzen FC, Laursen CB, Øvrehus A, et al. Low mortality of hospitalised patients with COVID-19 in a tertiary Danish hospital setting. International journal of infectious diseases: IJID: official publication of the International Society for Infectious Diseases. 2021;102:212–9. Epub 2020/10/16. doi: 10.1016/j.ijid.2020.10.018 ; PubMed Central PMCID: PMC7550098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.De Vito A, Geremia N, Fiore V, Princic E, Babudieri S, Madeddu G. Clinical features, laboratory findings and predictors of death in hospitalized patients with COVID-19 in Sardinia, Italy. Eur Rev Med Pharmacol Sci. 2020;24(14):7861–8. Epub 2020/08/04. doi: 10.26355/eurrev_202007_22291 . [DOI] [PubMed] [Google Scholar]
- 13.Zou L, Ruan F, Huang M, Liang L, Huang H, Hong Z, et al. SARS-CoV-2 Viral Load in Upper Respiratory Specimens of Infected Patients. The New England journal of medicine. 2020;382(12):1177–9. Epub 2020/02/20. doi: 10.1056/NEJMc2001737 ; PubMed Central PMCID: PMC7121626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu T, Chen C, Zhu Z, Cui M, Chen C, Dai H, et al. Clinical features and dynamics of viral load in imported and non-imported patients with COVID-19. International journal of infectious diseases: IJID: official publication of the International Society for Infectious Diseases. 2020;94:68–71. Epub 2020/03/18. doi: 10.1016/j.ijid.2020.03.022 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wölfel R, Corman VM, Guggemos W, Seilmaier M, Zange S, Müller MA, et al. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581(7809):465–9. Epub 2020/04/03. doi: 10.1038/s41586-020-2196-x . [DOI] [PubMed] [Google Scholar]
- 16.Zheng S, Fan J, Yu F, Feng B, Lou B, Zou Q, et al. Viral load dynamics and disease severity in patients infected with SARS-CoV-2 in Zhejiang province, China, January-March 2020: retrospective cohort study. BMJ (Clinical research ed). 2020;369:m1443. Epub 2020/04/23. doi: 10.1136/bmj.m1443 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu Y, Yan LM, Wan L, Xiang TX, Le A, Liu JM, et al. Viral dynamics in mild and severe cases of COVID-19. The Lancet Infectious diseases. 2020;20(6):656–7. Epub 2020/03/23. doi: 10.1016/S1473-3099(20)30232-2 ; PubMed Central PMCID: PMC7158902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Statistik D. Statistikbanken Befolkning og valg Folketal 2020 [May 18th 2020]. Available from: https://www.statistikbanken.dk/statbank5a/default.asp?w=1280.
- 19.Corman VM, Landt O, Kaiser M, Molenkamp R, Meijer A, Chu DK, et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro surveillance: bulletin Europeen sur les maladies transmissibles = European communicable disease bulletin. 2020;25(3). Epub 2020/01/30. doi: 10.2807/1560-7917.Es.2020.25.3.2000045 ; PubMed Central PMCID: PMC6988269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ferguson ND, Fan E, Camporota L, Antonelli M, Anzueto A, Beale R, et al. The Berlin definition of ARDS: an expanded rationale, justification, and supplementary material. Intensive care medicine. 2012;38(10):1573–82. Epub 2012/08/29. doi: 10.1007/s00134-012-2682-1 . [DOI] [PubMed] [Google Scholar]
- 21.Griffiths MJD, McAuley DF, Perkins GD, Barrett N, Blackwood B, Boyle A, et al. Guidelines on the management of acute respiratory distress syndrome. BMJ open respiratory research. 2019;6(1):e000420. Epub 2019/07/02. doi: 10.1136/bmjresp-2019-000420 ; PubMed Central PMCID: PMC6561387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bennett John E DR, Blaser Martin J Encephalitis. In: J David Beckham KLT, editor. Mandell, Douglas, and Bennett’s Principles and Practice of Infectious Diseases. 8th Edition ed: Elsevier; 2014.
- 23.Cheng PK, Wong DA, Tong LK, Ip SM, Lo AC, Lau CS, et al. Viral shedding patterns of coronavirus in patients with probable severe acute respiratory syndrome. Lancet (London, England). 2004;363(9422):1699–700. Epub 2004/05/26. doi: 10.1016/S0140-6736(04)16255-7 ; PubMed Central PMCID: PMC7112423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chan PK, To WK, Ng KC, Lam RK, Ng TK, Chan RC, et al. Laboratory diagnosis of SARS. Emerging infectious diseases. 2004;10(5):825–31. Epub 2004/06/18. doi: 10.3201/eid1005.030682 ; PubMed Central PMCID: PMC3323215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Walsh KA, Jordan K, Clyne B, Rohde D, Drummond L, Byrne P, et al. SARS-CoV-2 detection, viral load and infectivity over the course of an infection. The Journal of infection. 2020;81(3):357–71. Epub 2020/07/03. doi: 10.1016/j.jinf.2020.06.067 ; PubMed Central PMCID: PMC7323671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rao SN, Manissero D, Steele VR, Pareja J. A Systematic Review of the Clinical Utility of Cycle Threshold Values in the Context of COVID-19. Infect Dis Ther. 2020;9(3):573–86. Epub 2020/07/30. doi: 10.1007/s40121-020-00324-3 ; PubMed Central PMCID: PMC7386165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vaira LA, Deiana G, Fois AG, Pirina P, Madeddu G, De Vito A, et al. Objective evaluation of anosmia and ageusia in COVID-19 patients: Single-center experience on 72 cases. Head Neck. 2020;42(6):1252–8. Epub 2020/04/29. doi: 10.1002/hed.26204 ; PubMed Central PMCID: PMC7267244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vaira LA, De Vito A, Deiana G, Pes C, Giovanditto F, Fiore V, et al. Systemic inflammatory markers and psychophysical olfactory scores in coronavirus disease 2019 patients: is there any correlation? J Laryngol Otol. 2021:1–6. Epub 2021/06/30. doi: 10.1017/S0022215121001651 ; PubMed Central PMCID: PMC8267245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smithgall MC, Scherberkova I, Whittier S, Green DA. Comparison of Cepheid Xpert Xpress and Abbott ID Now to Roche cobas for the Rapid Detection of SARS-CoV-2. Journal of clinical virology: the official publication of the Pan American Society for Clinical Virology. 2020;128:104428. Epub 2020/05/15. doi: 10.1016/j.jcv.2020.104428 ; PubMed Central PMCID: PMC7217789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pujadas E, Ibeh N, Hernandez MM, Waluszko A, Sidorenko T, Flores V, et al. Comparison of SARS-CoV-2 detection from nasopharyngeal swab samples by the Roche cobas 6800 SARS-CoV-2 test and a laboratory-developed real-time RT-PCR test. Journal of medical virology. 2020. Epub 2020/05/10. doi: 10.1002/jmv.25988 ; PubMed Central PMCID: PMC7267546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van Kasteren PB, van der Veer B, van den Brink S, Wijsman L, de Jonge J, van den Brandt A, et al. Comparison of seven commercial RT-PCR diagnostic kits for COVID-19. Journal of clinical virology: the official publication of the Pan American Society for Clinical Virology. 2020;128:104412. Epub 2020/05/18. doi: 10.1016/j.jcv.2020.104412 ; PubMed Central PMCID: PMC7206434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lieberman JA, Pepper G, Naccache SN, Huang ML, Jerome KR, Greninger AL. Comparison of Commercially Available and Laboratory-Developed Assays for In Vitro Detection of SARS-CoV-2 in Clinical Laboratories. Journal of clinical microbiology. 2020;58(8). Epub 2020/05/01. doi: 10.1128/JCM.00821-20 ; PubMed Central PMCID: PMC7383518. [DOI] [PMC free article] [PubMed] [Google Scholar]