Abstract
Bad is a critical regulatory component of the intrinsic cell death machinery that exerts its death-promoting effect upon heterodimerization with the antiapoptotic proteins Bcl-2 and Bcl-xL. Growth factors promote cell survival through phosphorylation of Bad, resulting in its dissociation from Bcl-2 and Bcl-xL and its association with 14-3-3τ. Survival of interleukin 3 (IL-3)-dependent FL5.12 lymphoid progenitor cells is attenuated upon treatment with the Rho GTPase-inactivating toxin B from Clostridium difficile. p21-activated kinase 1 (PAK1) is activated by IL-3 in FL5.12 cells, and this activation is reduced by the phosphatidylinositol 3-kinase inhibitor LY294002. Overexpression of a constitutively active PAK mutant (PAK1-T423E) promoted cell survival of FL5.12 and NIH 3T3 cells, while overexpression of the autoinhibitory domain of PAK (amino acids 83 to 149) enhanced apoptosis. PAK phosphorylates Bad in vitro and in vivo on Ser112 and Ser136, resulting in a markedly reduced interaction between Bad and Bcl-2 or Bcl-xL and the increased association of Bad with 14-3-3τ. Our findings indicate that PAK inhibits the proapoptotic effects of Bad by direct phosphorylation and that PAK may play an important role in cell survival pathways.
The ability of multicellular organisms to maintain cellular homeostasis is critically dependent on a balance between cell survival and cell death (apoptosis). The responsiveness of individual cells to death signals can vary greatly depending on the presence of continuous survival cues from the extracellular environment. The perturbation of normal cell survival mechanisms, leading to an increase in cell death or cell survival, plays an important role in the development of a number of disease states, including cancer (26, 55).
Members of the Bcl-2 family are intracellular proteins that can either promote survival (Bcl-2, Bcl-xL, Mcl-1, and A1) or augment cell death (Bad, Bax, Bak, and Bcl-xS) (38, 41). Bcl-2 family proteins homo- and heterodimerize, and it has been suggested that susceptibility to cell death is dictated by the relative levels and interactions of Bcl family members (57). Bad, for example, has been shown to dimerize with Bcl-2 or Bcl-xL. This complex formation inhibits the ability of Bcl-2 and Bcl-xL to block the release of cytochrome c from mitochondria, a critical step in the activation of the downstream caspase protease cascade (17, 19, 20).
A number of growth factors, including insulin-like growth factor 1, platelet-derived growth factor, and nerve growth factor (46, 51), and many cytokines (such as interleukin 3 [IL-3]) promote cell survival through pathways requiring the activity of phosphatidylinositol 3-kinase (PI 3-kinase) (8, 11, 60). The lipid products of PI 3-kinase (phosphatidylinositol-3,4-P2 and phosphatidylinositol-3,4,5-P3) act as second messengers to stimulate the activity of the protein Ser/Thr kinase Akt (16). Activated Akt has been shown to phosphorylate the proapoptotic Bad protein on serine residue 136, resulting in its dissociation from complexes with Bcl-2/Bcl-xL and its subsequent association with the cytosolic adapter protein 14-3-3τ (7, 8). The uncomplexed Bcl-xL is then capable of suppressing cell death responses by blocking the release of mitochondrial cytochrome c (24). It is not yet clear if the binding of phosphorylated Bad to 14-3-3τ also plays an active role in promoting cell survival.
FL5.12 lymphoid progenitor cells die in the absence of the cytokine IL-3. Previous studies (60) have established that IL-3 induces the phosphorylation of Bad at serine residues 112 and 136. Phosphorylation of these sites is critical for cell survival signaling in a number of systems (7, 8, 11); after mutation of these phosphorylation sites, Bad effectively induces cell death which can no longer be antagonized by IL-3 (60). The Akt/PI 3-kinase-dependent phosphorylation of Ser136 on Bad cannot fully account for cell survival mediated by IL-3, however, as phosphorylation at Ser112 also antagonizes Bad activity (7). The kinases that catalyze this phosphorylation reaction have not been well characterized. A recent study identified mitochondrion-associated protein kinase A as a Bad Ser112-specific kinase (22). Raf-1, through interactions with Bcl-2, can induce phosphorylation of Bad in IL-3-dependent cell lines as well, although the sites of phosphorylation are distinct from Ser112 or Ser136 and remain unknown (53). A calcium-inducible apoptosis was found to occur through the dephosphorylation of Bad by the calcium-activated protein phosphatase calcineurin (54).
The p21-activated protein kinases (PAK1 to -3) are closely related serine/threonine kinases activated by the GTPases Rac and Cdc42 (32, 47) and by sphingosine (5). PAKs are implicated in the regulation of a number of cellular processes, including rearrangement of the actin-myosin cytoskeleton (47, 49), mitogen-activated protein kinase (MAPK) signaling pathways (61), growth factor-induced neurite outgrowth (6), and control of phagocyte NADPH oxidase (28). PAK2 is proteolytically cleaved in apoptotic cells by DEVD-sensitive caspases. This cleavage generates an active PAK2 COOH-terminal fragment, which has been implicated in the regulation of morphological changes occurring during the late stages of the apoptotic response (31, 42). In some cells sensitive to signaling through the c-Jun N-terminal kinase (JNK) and p38 MAPK pathways, PAK2 can be proapoptotic (43). However, there is also evidence that members of the PAK family play a role in antiapoptotic pathways. Faure et al. (14) demonstrated that Xenopus PAK is involved in arrest of oocytes at G2/prophase and prevention of apoptosis induced by progesterone withdrawal.
In this study we show that PAK1 is activated by IL-3 in FL5.12 cells and that active PAK1 protects FL5.12 cells from apoptosis induced by deprivation of IL-3. PAK1 phosphorylates Bad at both Ser112 and Ser136, leading to a disruption of the interaction between Bad and Bcl-2/Bcl-xL. PAK thus regulates cell survival, and this is mediated, at least in part, through the suppression of the proapoptotic activity of Bad.
MATERIALS AND METHODS
Plasmids.
For in vitro translation experiments we used pRC-CMV-Bcl-2, pBluescript II SK-BclxL (from B. C. Chang and Craig Thompson, University of Chicago), and pBluskriptIIKS-14-3-3τ (from S. J. Korsmeyer, Washington University School of Medicine, St. Louis, Mo.). To generate recombinant glutathione S-transferase (GST) fusion proteins of Bad, pGEX-4T-1-mBad-wt (full length), pGEX-4T-1-mBad-wt-aa 104-141, and pGEX-4T1-mBadS112, 136A-aa 104-141 (from H. Harada and S. J. Korsmeyer) were used. The cDNA expression plasmids pcDNA3-HA-mBad-wt and pcNDA-HA-mBad-S112/136A were a gift from M. E. Greenberg (Harvard Medical School, Boston, Mass.). PAK constructs used in these studies have been previously described (47, 49). Clostridium difficile toxin B was a kind gift from Klaus Aktories.
Antibodies.
The antibodies used in this study include rabbit polyclonal antibodies specific to the 20 C-terminal amino acids of Bad [Bad(C20); sc-943; for immunoprecipitation]; the 20 N-terminal amino acids of Bad [Bad(N20); sc-941; for Western blotting), and Bcl-2(ΔC21] (sc-784) (all from Santa Cruz Biotechnology, Santa Cruz, Calif.), rabbit polyclonal antibodies specific to phospho-Bad-Ser112 and phospho-Bad-Ser136 (both from New England BioLabs, Beverly, Mass.), and mouse monoclonal anti-hemagglutinin epitope (HA) antibody (BAbCo, Richmond, Calif.). The PAK1 rabbit polyclonal antibody R2124 is essentially the same as anti-PAK1 antibody R626, described in reference 28.
Green fluorescent protein (GFP)-annexin V was generously provided by Joel Ernst (University of California, San Francisco), and its use has been previously described (12).
In-gel kinase assay.
A 400-μg aliquot of lysate of FL5.12 cells was immunoprecipitated with PAK1 polyclonal antibody R2124, and samples were separated on 7% polyacrylamide gels containing 0.375 mg of p47phox peptide (amino acids [aa] 297 to 331) per ml. Gels were washed in 20% 2-propanol–50 mM Tris (pH 7.5) and then in 50 mM Tris (pH 7.5)–5 mM β-mercaptoethanol before a denaturation step in 5 mM Tris (pH 7.5)–6 M guanidine-HCl as described previously (10, 42). For renaturation of proteins, gels were washed extensively in 50 mM Tris (pH 7.5)–0.04% Tween 40–5 mM β-mercaptoethanol and in PAK kinase buffer (50 mM HEPES [pH 7.5], 10 mM MgCl2, 2 mM MnCl2, 1 mM dithiothreitol). In-gel phosphorylation was done in PAK kinase buffer containing 10 μCi of [γ-32P]ATP/ml (4,500 Ci/mmol; ICN) and 25 μM ATP for 1 h at 30°C. Gels were washed several times in 5% (wt/vol) trichloroacetic acid–1% sodium pyrophosphate, stained with Coomassie blue, and subjected to autoradiography.
In vitro kinase assay.
Bad was phosphorylated in vitro in 50 mM HEPES (pH 7.5)–10 mM MgCl2–2 mM MnCl2–0.2 mM dithiothreitol–25 μM ATP–5 μCi of [γ-32P]ATP (4,500 Ci/mmol; ICN) by 1 μg of His-PAK1 plus 400 mM sphingosine in a total volume of 50 μl for 30 min at 30°C. In some experiments, PAK1 was activated by the addition of 1 μg of Cdc42-GTPγS. Incubations were stopped in Laemmli sample buffer and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) in 12% gels.
In vitro protein interaction assay.
Purified GST fused to wild-type Bad (GST-Bad-wt) or to a Bad construct consisting of aa 104 to 141 [GST-Bad(104-141)WT or S112/136A] were phosphorylated by Cdc42-GTPγS-activated His-PAK1 for 30 min at 30°C. GST fusion proteins were then incubated with glutathione beads for 1 h at 4°C, and the beads were washed four times in 50 mM Tris (pH 7.5)–1 mM EDTA–100 mM NaCl–0.2% NP-40. GST-Bad-loaded beads (∼1 μg of protein on 20 μl of beads) were incubated with 5 μl of reticulocyte lysate (TNT-T7 lysates; Promega, Inc.) containing in vitro-translated [35S]methionine-labeled Bcl-2, Bcl-xL, or 14-3-3τ for 2 h at 4°C. After extensive washing in 50 mM Tris (pH 7.5)–1 mM EDTA–500 mM NaCl–1% NP-40, beads were boiled in Laemmli sample buffer; eluted proteins were analyzed by SDS-PAGE (12% gel) and detected by autoradiography.
Cell lines and transfections.
NIH 3T3 cell lines expressing PAK1-wt, PAK1-T423E (constitutively active), and PAK1-K299R under the control of a tetracycline-regulated repressor were used for these studies (48). Cells were cultured in Dulbecco's modified Eagle's medium with 5% calf serum, 5% newborn calf serum, 2.5 mM histidinol, 2 μg of puromycin/ml, and 0.5 μg of tetracycline/ml; 106 NIH 3T3 cells were transfected with 10 μg of pcDNA3-HAmBad-wt or pcDNA3-HAmBad-S112/136A by using LipofectAMINE (Life Technology, Inc.) and grown in the presence or absence of tetracycline for additional 36 h. FL5.12 cells were cultured as previously described (34) in Iscove's modified Dulbecco's medium (IMDM) with 10% fetal calf serum–10% WEHI-3 supernatant containing IL-3. For infection of FL5.12 cells, cDNAs encoding Myc-tagged PAK-T42E (mycPAK-T423E), mycPAK-K299R, or mycPAK83-149 were subcloned into the Semliki Forest vector pSFV3. In vitro transcription of linearized pSFV3 constructs and pSFV-Helper2 was performed with SP6 RNA polymerase. RNA transfection of BHK-21 cells was done by electroporation (33), yielding recombinant viral stocks of approximately 107 PFU/ml; 2 × 105 FL5.12 cells were seeded on fibronectin 2 days before infection. Cells were washed twice with IMDM and then incubated with 100 μl of activated virus for 2 h and for an additional 4 h without virus in IMDM plus 150 ng of recombinant mouse IL-3/ml at 37°C. Cells were again washed twice with IMDM and incubated with or without IL-3 for the indicated times before viability of the cells was assessed.
Immunoprecipitation.
Transfected NIH 3T3 cells were solubilized in 20 mM Tris (pH 8.0)–37 mM NaCl–1.5 mM MgCl2–1 mM EDTA–50 mM NaF–0.5% NP-40 containing 0.15 U of aprotinin/ml, 20 mM leupeptin, and 1 mM phenylmethylsulfonyl fluoride and incubated with a 1:200 dilution (as instructed by the manufacturer) of Bad(C20), phospho-Bad-S112, or phospho-Bad-S136 antibody for 2 h at 4°C. The antibody complexes were captured with protein A beads for 1 h; and the immunoprecipitate was washed with lysis buffer, resuspended in Laemmli sample buffer, and separated by SDS-PAGE. The gels were transferred to a nitrocellulose membrane for further immunoblot analysis.
RESULTS
Active PAK protects FL5.12 cells from apoptosis induced by IL-3 deprivation.
FL5.12 cells, lymphoid progenitor cells that die via apoptosis in the absence of IL-3, represent a system commonly used to investigate the regulation of cell survival. We observed (Fig. 1) that the ability of IL-3 to promote FL5.12 cell survival was abrogated by treatment of the cells with Clostridium toxin B, an inactivator of Rho GTPase function (1). These results suggested that Rho GTPases were necessary, at least to some extent, for IL-3 to generate cell survival signals. We therefore investigated whether the Rac and Cdc42 effector PAK1 might suppress (or enhance) the apoptosis induced by IL-3 withdrawal. Parental FL5.12 cells were infected with Semliki Forest virus encoding LacZ (Co) or mycPAK-T423E (a constitutively active mutant of PAK1). The kinetics of cell death were explored in these cells starting 6 h after infection, when we determined that 50 to 70% of the cells were expressing protein (Fig. 2B). A small percentage of cells (∼15 to 20%) were rendered nonviable by the virus infection. In the presence of IL-3, FL5.12 cells infected with the PAK-T423E construct or control virus survive over a time period of 24 h (Fig. 2A). However, after deprivation of IL-3, the control cells or cells transfected with a kinase-dead PAK-K299R mutant (not shown) undergo substantial apoptotic death, detectable by trypan blue exclusion within 3 h after IL-3 depletion. After 24 h of IL-3 deprivation, we detected only 37.7% ± 4.5% living cells after infection with LacZ; a similar decrease, to 26.3% ± 11.7% living cells, was observed after infection with mycPAK-K299R (not shown). In contrast, cells expressing mycPAK-T423E exhibited a dramatic delay in cell death in the absence of IL-3, with 61.5% ± 2.4% viable cells remaining at 24 h (Fig. 2A).
FIG. 1.
Inhibition of Rho GTPases prevents IL-3-dependent cell survival. FL5.12 cells were incubated in the presence or absence of the Rho GTPase-inactivating C. difficile (C. Diff) toxin B, as indicated. The cells were then assessed for viability by trypan blue exclusion as described in Materials and Methods.
FIG. 2.
Effect of PAK-T423E and PAK83-149 on apoptosis in FL5.12 cells induced by IL-3 deprivation. (A) Kinetics of cell viability. FL5.12 cells were infected with Semliki Forest virus encoding LacZ (Co) or mycPAK1-T423E (a constitutively active mutant of PAK1) as described in Materials and Methods. Cells were cultured in the presence or absence of IL-3 (150 ng/ml) for various times, and the fraction of viable cells was assessed by trypan blue exclusion. (B) Detection of apoptotic FL5.12 cells by GFP-annexin V. FL5.12 cells were transfected and treated as described above; 8 h after incubation of FL5.12 cells in the presence or absence of IL-3, cells were stained with GFP-annexin V and the fraction of apoptotic cells (nonviable) was assessed by counting fluorescent cells. Expression of the PAK1-T423E construct as detected by immunoblotting with the Myc epitope antibody 9E10 is shown at the right. (C) PAK1 autoinhibitory domain enhances death of FL5.12 cells. FL5.12 cells were infected with Semliki Forest virus encoding LacZ, mycPAK1-T423E, or mycPAK183-149 (the autoinhibitory domain of PAK1) and cultured in the presence or absence of IL-3 (150 ng/ml) for 8 h. Fraction of viable cells at 8 h was assessed by trypan blue exclusion; similar results were obtained with GFP-annexin V. All results shown represent 150 to 300 cells counted per data point in each experiment. All experimental data points are expressed as the mean ± standard error of the mean (n = 3).
Although it has been previously established that FL5.12 cells undergo apoptotic death upon withdrawal of IL-3, we confirmed that the cell death we were measuring represented apoptotic cell death by staining of dead cells with GFP-annexin V (Fig. 2B). Annexin V binds to phosphatidylserine exposed on the extracellular leaflet of the plasma membrane of cells undergoing apoptosis (12, 29). Using the annexin V method, in the presence of IL-3 only 15 to 20% dead cells were detected when LacZ, mycPAK-T423E, or mycPAK-K299R was expressed. In contrast, 3 h after deprivation of IL-3, 60% ± 4.2% of control cells and 66% ± 5.5% of cells expressing mycPAK-K299R were stained with GFP-annexin V, while only 29.1% ± 4.6% of cells expressing mycPAK-T423E were labeled with GFP-annexin V.
It has been established that a 67-residue polypeptide representing PAK83-149 serves as an autoinhibitory domain, capable of suppressing the kinase activity of PAK (59, 62). We tested whether overexpression of this autoinhibitory domain would enhance the level of cell death by inhibiting endogenous PAK activity. As shown in Fig. 2C, overexpression of PAK83-149 induced an increased level of cell death in the presence of IL-3 compared to vector control cells. We observed that 38.1% ± 1.3% of the cells died, as opposed to only about half as many (∼20%) in control cells and cells infected with mycPAK-T423E. This was verified to be apoptotic death by GFP-annexin V staining. PAK activation by IL-3 was not detectable in the presence of the autoinhibitory domain (not shown). These data suggest that PAK is likely to be a component of the cell survival signaling pathways normally induced by IL-3.
PAK is activated by IL-3 in FL5.12 cells.
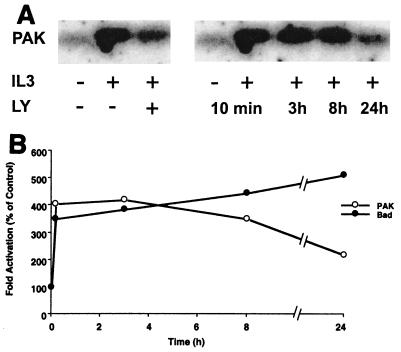
The antiapoptotic effect of activated PAK expressed in FL5.12 cells in the absence of IL-3, coupled with the partial suppression of the antiapoptotic of IL-3 in these cells by the PAK autoinhibitory domain, raised the question of whether PAK is activated by IL-3 and plays a role in the IL-3-dependent survival pathway. Immunoblot analysis with PAK1- and PAK2-specific antisera revealed that FL5.12 cells contained primarily a PAK isoform immunoreactive with the PAK1-reactive antibody R2124 (not shown). The cells were serum starved and incubated with or without recombinant IL-3 for 10 min, 3 h, 8 h, and 24 h, and PAK1 activity was analyzed after immunoprecipitation from these cells by using the R2124 antibody. In-gel kinase assays were performed with the selective p47phox peptide substrate (10), and PAK1 kinase activity was detected after stimulation of FL5.12 cells with IL-3 (Fig. 3). Activation of PAK1 was substantial as early as 10 min after stimulation with IL-3, and the activation was stable for more than 8 h but had decreased by 24 h. Interestingly, PAK activation correlated well with the IL-3-induced phosphorylation of Bad at Ser112 (Fig. 3B).
FIG. 3.
PAK is activated by IL-3 in FL5.12 cells. (A) FL5.12 cells were starved for 2 h and incubated with or without IL-3 (150 ng/ml) for 10 min, 3 h, 8 h, or 24 h. One set of cells was preincubated with 30 μM LY294002 (LY) for 15 min, as indicated, and then stimulated with IL-3 (150 ng/ml) for 10 min. PAK was immunoprecipitated from lysates with the PAK1 polyclonal rabbit antibody R2124. Kinase activity of PAK was assessed by an in-gel kinase assay with p47phox peptide as the substrate (see Materials and Methods. Quantitation by phosphorimager analysis indicated PAK1 activity was enhanced by 4.0-fold at 10 min, 4.4-fold at 3 and 8 h, and 1.9-fold at 24 h after IL-3 stimulation. Similarly, the 4.0-fold stimulation of PAK at 10 min was reduced to 2.4-fold in the presence of LY294002. (B) The time course of PAK1 activation in the presence of IL3 was quantified by phosphorimager analysis and compared with the time course of IL-3-stimulated phosphorylation of Bad at Ser112 determined as described in Materials and Methods.
PI 3-kinase has been shown to be activated by IL-3, and this activation is known to be critical for stimulation of Akt activity and cell survival signaling via Bad (8, 60). PI 3-kinase can also stimulate the activation of the small GTPase Rac, probably via regulation of various GDP-GTP exchange factors, resulting in the subsequent activation of PAK (23). We determined whether the activity of PAK1 induced by IL-3 in FL5.12 cells was sensitive to the PI 3-kinase specific inhibitor LY294002 (11, 52). The addition of 30 μM LY294002 partially blocked the IL-3-dependent PAK1 activation, and this level of inhibition was not increased at higher drug doses. Thus, the 4.0-fold activation of PAK1 by IL-3 was reduced to a 2.4-fold stimulation in the presence of LY294002 (Fig. 3A). These data show that PAK1 is at least partially activated by IL-3 via a PI 3-kinase dependent pathway and suggest that PAK1 could play a role in both PI 3-kinase-dependent and -independent survival signaling by IL-3.
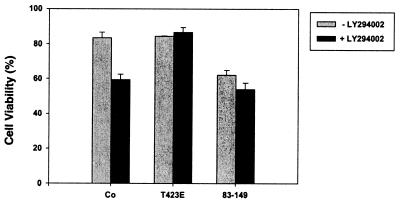
To rule out the possibility that the antiapoptotic effect of PAK-T423E was somehow mediated through activation of PI 3-kinase, we infected FL5.12 cells with Semliki Forest virus encoding LacZ, mycPAK-T423E, or mycPAK83-149 and examined cell death in the absence or presence of LY294002. Treatment of the FL5.12 cells cultured in the presence of IL-3 with LY294002 resulted in apoptosis of ∼25% more cells compared to untreated control cells (Fig. 4). In contrast, LY294002 treatment of FL5.12 cells infected with Semliki Forest virus encoding mycPAK-T423E did not result in increased apoptosis compared to cells cultured without LY294002 in the presence (or absence [not shown]) of IL-3 (Fig. 4). Cells expressing the PAK autoinhibitory domain (aa 83 to 149) showed enhanced cell death, and this was not changed significantly in the presence of the PI 3-kinase inhibitor. These data indicate that active PAK mediates its antiapoptotic effect by acting downstream or independently of PI 3-kinase.
FIG. 4.
The PI 3-kinase inhibitor LY294002 does not block the antiapoptotic effect of PAK-T423E. FL5.12 cells were infected with Semliki Forest virus encoding LacZ, mycPAK1-T423E, or mycPAK183-149 and cultured in the presence of IL-3 (150 ng/ml), without or with 30 μM LY294002, for 6 h. Fraction of viable cells was assessed by trypan blue exclusion. Results shown are the mean ± standard error of the mean n = 3).
Bad is phosphorylated by PAK1 in vitro.
Stimulation of FL5.12 cells with IL3 results in the phosphorylation of Bad on at least two sites, Ser112 and Ser136, thereby initiating its dissociation from Bcl-2 and Bcl-xL and inactivation of its proapoptotic effect (57, 60). The activation of Akt leads to the phosphorylation of Bad on Ser136, at least in some cells, while protein kinase A (PKA) can phosphorylate mitochondrion- localized Bad on Ser112. However, there is evidence that Akt activity is not always correlated with survival pathways acting at the level of Bad and that other pathways must exist for the phosphorylation of these sites (25). To test whether PAK1 is able to phosphorylate Bad in vitro, recombinant GST fusion proteins of human Bad (hBad), mouse Bad (mBad), mBad (104-141) containing the Ser112 and Ser136 phosphorylation sites but lacking the BH1 and BH2 domains, and the mBad(104-114)S112/136A mutant were incubated with activated PAK1 in the presence of [γ-32P]ATP. The kinase used for this experiment was His-PAK1 expressed from a recombinant baculovirus in Sf9 cells; this enzyme is partially active but can be stimulated in vitro with the activator Cdc42-GTPγS or sphingosine. As shown in Fig. 5A (lanes 1 to 4), both hBad and mBad proteins were heavily phosphorylated in vitro by sphingosine-activated PAK1. Sphingosine itself in the absence of PAK was without effect (lane 9). Similar results were obtained with Cdc42-GTPγS-activated His-PAK1 (lanes 10 and 11), as well as with a constitutively activated PAK2 (data not shown).
FIG. 5.
In vitro phosphorylation of Bad by PAK. (A) One microgram of recombinant GST fused to hBad-wt, mBad-wt, mBad(104–141)wt(Bad104-141wt), or mBad(104-141)-S112/136A (Bad104-141mt) was subjected to phosphorylation conditions (see Materials and Methods) with 1 μg of recombinant His-Pak1 (except for lane 9) in the presence or absence of 400 μM sphingosine or 1 μg of Cdc42-GTPγS, as indicated. Samples were separated by SDS-PAGE and analyzed by autoradiography. (B and C) GST-mBad(104-141)wt (Bad104-141wt) and GST-mBad(104-141)S112/136A (Bad104-141mt) were phosphorylated by 1 μg of recombinant His-Pak1 in the presence or absence of 400 μM sphingosine and 500 μM ATP. Samples were separated on SDS-PAGE and analyzed by Western blotting using the phospho-Bad(S112) (B) or phospho-Bad(S136) (C) antibody.
We evaluated whether PAK1 phosphorylated the critical Ser112 and Ser136 regulatory sites in Bad by incubating the recombinant GST-Bad(104-141)wt and the GST-Bad(104-141)S112/136A mutant with sphingosine-activated His-PAK in the presence of unlabeled ATP and then detecting phosphorylation of Ser112 or Ser136 immunochemically with specific phospho-Bad antibodies. Both antibodies detected phosphorylated Bad after incubation of Bad-wt with His-PAK1 activated by sphingosine or Cdc42 (data not shown). In contrast to Bad-wt, the Bad-S112/136A mutant did not show any signal, consistent with the mutation of these specific serine residues in this polypeptide (Fig. 5B and C). PAK1 therefore phosphorylates Bad on both residues Ser112 and Ser136 in vitro.
Although PAK1 was still capable of phosphorylating the Bad(104-141)S112/136A double mutant, as shown in Fig. 5A (lanes 5 to 8), we observed that the level of phosphorylation was consistently lower than in the wild-type fragment. Incorporation of 32P was quantified after precipitation with trichloroacetic acid and filtration. The wild-type fragment incorporated an average of 1.32 pmol of 32P/pmol of protein (range = 1.28 to 1.36 pmol, n = 2), while the mutant peptide incorporated only an average of 0.38 pmol of 32P/pmol of protein (range = 0.33 to 0.43 pmol; n = 2). These data indicate that the primary sites for phosphorylation by PAK are S112 and S136, although an additional (minor) site(s) also exists within the 104–141 segment.
Bad is phosphorylated by PAK in vivo.
IL-3-dependent FL5.12 cell survival is dependent on intact Rho GTPase function (Fig. 1), and PAK activity is regulated by the Rho family GTPases Rac and Cdc42. We examined whether expression of constitutively active or dominant negative forms of Cdc42 were capable of modulating Bad phosphorylation by PAK on the critical Ser112 site in vivo. NIH 3T3 cells stably expressing PAK1-wt in a tetracycline-dependent manner were transiently cotransfected with HA-hBad and either active Cdc42-Q61L or dominant negative Cdc42-T17N. Coexpression of PAK1 and Cdc42-Q61L substantially increased the phosphorylation of Bad on Ser112, while coexpression with Cdc42-T17N decreased the basal level of Bad phosphorylation at this site (Fig. 6A). Since PAK1-wt itself has little effect on Bad phosphorylation (see below), these data are consistent with the activation of Pak1 by coexpressed Cdc42-Q61L and the subsequent phosphorylation of Bad.
FIG. 6.
In vivo phosphorylation of Bad by PAK and/or Cdc42. (A) NIH 3T3 cells in which expression of PAK-wt was repressed in the presence of tetracycline (−) or induced by withdrawal of tetracycline (+) were transiently transfected with pcDNA3-HA-hBad and pRK5M-Cdc42-T17N or pRK5M-Cdc42-Q61L as described in Materials and Methods. (Upper panel) Bad proteins phosphorylated on Ser112 were immunoprecipitated by phospho-Bad-Ser112 antibody and analyzed by Western blotting using Bad(N20) antibody as described in Materials and Methods. (Lower panel) Lysates of the cells expressing PAK-wt, Bad, and Cdc42 were analyzed by Western blotting using the HA antibody, Bad(N20) antibody, and 9E10 Myc antibody 9E10, respectively, to verify the protein expression levels. The results are representative of three independent experiments. (B) NIH 3T3 cells in which expression of PAK-T432E or PAK-K299R was repressed in the presence of tetracycline (−) or induced by withdrawal of tetracycline (+) were transiently transfected with pcDNA3-HA-hBad and pRC-CMV-Bcl-2 as described in Materials and Methods. Co, vector-only control. We observed no difference in Bad phosphorylation with or without wortmannin in the presence or absence of tetracycline in these cells. Samples were treated with 20 nM wortmannin (WT) for 24 h as indicated. (Upper panel) Bad proteins phosphorylated on Ser112 were immunoprecipitated by phospho-Bad-Ser112 antibody and analyzed by Western blots using the Bad(N20) antibody. Note that the basal level of Bad phosphorylation varied between cell lines. (Lower panel) Lysates of NIH 3T3 cells coexpressing Bad and PAK were analyzed immunochemically, using the Bad(N20) antibody to verify that protein expression levels were essentially equal under each experimental condition. The results are representative of three independent experiments.
We investigated whether Bad is phosphorylated by PAK1 in vivo. NIH 3T3 cells stably expressing a PAK1 constitutively active mutant (PAK-T423E) or a kinase-dead mutant (PAK-K299R) in a tetracycline-regulated manner were transiently transfected with HA-hBad. To maintain cell viability in the presence of the expressed hBad, cells were cotransfected with Bcl-2. At 36 h after transfection, Bad was immunoprecipitated with an antibody specific to phosphorylated Ser112. (i.e., a phospho-specific antibody). In the NIH 3T3 cells expressing Bad-wt, in the absence of PAK, we observed a basal level of phosphorylated Ser112 as determined by immunoprecipitation with the anti-Ser112 antibody (Fig. 6B). This basal level of Bad phosphorylation was observed to vary in each individual cell line. However, it was readily evident that the level of Bad phosphorylation was significantly enhanced by expression of PAK-T423E but not by expression of kinase-inactive PAK-K299R. These data establish that PAK phosphorylates Bad on Ser112 in vivo. We obtained similar results using the Ser136 phospho-specific antibody for immunoprecipitation, as well as after immunoprecipitation with a Bad protein antibody followed by immunoblotting with phospho-specific antibodies (data not shown), indicating that both Ser112 and Ser136 are phosphorylated by PAK in vivo. Treatment of cells with the PI 3-kinase inhibitor wortmannin decreased the phosphorylation of Bad in control cells (stably transfected expressing empty vector), suggesting that the basal phosphorylation was mediated by the PI 3-kinase pathway. In contrast, wortmannin did not block the PAK-T423E-induced phosphorylation of Bad, demonstrating that constitutively active PAK-T423E phosphorylates Bad on residue Ser112 in vivo in a PI 3-kinase-independent manner (Fig. 6B). We also established that there was no increase in endogenous Akt activity in cells expressing PAK-T423E (data not shown).
Phosphorylation of Bad by PAK1 alters its interactions with Bcl-2, Bcl-xL, and 14-3-3τ.
Since the phosphorylation of Bad by Akt results in its dissociation from Bcl-2 and Bcl-xL complexes and in its subsequent association with 14-3-3τ, we examined whether phosphorylation by PAK1 affects the capacity of Bad to interact with Bcl-2, Bcl-xL, and 14-3-3τ. In vitro-translated [35S]methionine-labeled Bcl-2, Bcl-xL, and 14-3-3τ were allowed to interact with different GST-Bad constructs preincubated with or without activated PAK1. As shown in Fig. 7A, nonphosphorylated GST-Bad-wt bound effectively to [35S]methionine-labeled Bcl-2 and Bcl-xL. This interaction was markedly reduced after phosphorylation of Bad by PAK1. Bad(104-141)wt or the Bad(104-141)S112/136A mutant did not bind to Bcl-2 or Bcl-xL, consistent with the absence of the BH1 and BH2 domains that are necessary for dimerization with other members of the Bcl-2 family. GST-Bad-wt and Bad(104-141)wt interacted with [35S]methionine-labeled 14-3-3τ only after phosphorylation by PAK; no interaction was detected between unphosphorylated Bad and 14-3-3τ.
FIG. 7.
Phosphorylation of Bad by PAK regulates its interaction with Bcl-2, Bcl-xL, and 14-3-3τ. (A) Recombinant GST-hBad, (Bad-wt), GST-mBad(104-141)wt (Bad-104-141wt), and GST-mBad(104-141)S112/136A (Bad104-141mt) were phosphorylated by Cdc42-GTPγS activated recombinant His-Pak1, immobilized on glutathione-Sepharose, and then incubated with in vitro translated [35S]methionine-labeled Bcl-2 (upper panel), Bcl-xL (middle panel), or 14-3-3τ (lower panel). Bound proteins were analyzed by SDS-PAGE and autoradiography. An amount of in vitro-translated protein (IVT) equivalent to that in each binding reaction was run directly in the gel (far right lane in each panel). (B) The in vivo interaction of Bad with Bcl-2 decreased after overexpression of PAK-T423E and increased after overexpression of PAK-K299R. (Top) Coimmunoprecipitation experiments were performed with lysates of NIH 3T3 cells coexpressing Bad-wt (wt) or Bad-S112/136A (mt), Bcl-2, and PAK-T423E or PAK-K299R as described for Fig. 6 with Bad(C20) antibody. Western blots were developed with Bcl-2(ΔC21) antibody. (Bottom) Lysates of NIH-3T3 cells coexpressing PAK, Bad, and Bcl-2 were analyzed immunochemically by using the HA Bad(N20) and Bcl-2 (ΔC21) antibodies, respectively to verify the expression levels. The results shown are representative of three independent experiments.
In a second set of experiments we determined whether the phosphorylation of Bad by PAK also alters the interaction with Bcl-2 in vivo. Plasmids encoding Bad-wt or Bad-S112/136A were transfected together with Bcl-2 in the tetracycline-dependent NIH 3T3 cells stably expressing PAK-T423E, PAK-K299R, or PAK-wt. The levels of expression of PAK, Bad, and Bcl-2 were determined 36 h after transfection by Western blot analysis, and cell lysates expressing comparable amounts of PAK, Bad, and Bcl-2 (Fig. 7B, lower panel) were used for analysis of the formation of heterodimers in vivo. Bad was immunoprecipitated with the polyclonal Bad(C20) antibody, and coimmunoprecipitated Bcl-2 was detected by Western blotting with the polyclonal Bcl-2(ΔC21) antibody. After induction of PAK-T423E expression, the amount of Bcl-2 coimmunoprecipitated with Bad-wt was consistently decreased. Interestingly, we also observed a decrease in the amount of Bcl-2 associated with Bad-S112/136A after induction of PAK-T423E. This suggests that the phosphorylation of additional sites on Bad by PAK (Fig. 5) is also capable of attenuating the interaction between Bad and Bcl-2 in vivo. After induction of PAK-K299R expression, the interaction between Bad and Bcl-2 was increased, suggesting that this kinase-dead mutant may act to block some basal level of endogenous PAK activity which phosphorylates Bad. However, we could not detect a significant decrease in Bad phosphorylation at Ser112 when PAK-K299R was expressed (Fig. 6B). The expression of PAK-wt, which is typically not activated when expressed alone, did not affect the amount of Bcl-2 coimmunoprecipitated with the Bad antibody. Thus, based on both the in vitro protein binding assays and the in vivo coimmunoprecipitations, we observed that the phosphorylation of Bad by PAK alters the heterodimerization of Bad with Bcl-2 and 14-3-3τ in a manner consistent with promoting cell survival.
Active PAK protects NIH 3T3 cells from apoptoisis induced by overexpression of Bad or treatment with C2-ceramide.
Cellular overexpression of Bad induces apoptosis because of the consequent dimerization of Bad with Bcl-2 and/or Bcl-xL, resulting in the release of cytochrome c from mitochrondria (17) (Fig. 8A). Since the proapoptotic effect of Bad is blocked by its phosphorylation, activated PAK should therefore suppress apoptosis induced by overexpression of Bad. Transfection of plasmids encoding HA-hBad in NIH 3T3 cells in which expression of PAK was suppressed by the presence of tetracycline resulted in apoptosis of approximately one-third of the cells. Control cells expressing Bad transiently show no difference in apoptotic events in the presence or absence of tetracycline (data not shown), indicating that tetracycline itself did not influence the apoptotic or antiapoptotic pathway in NIH 3T3 cells. After induction of PAK-T423E expression, apoptosis in response to Bad was totally blocked, as determined both by trypan blue staining (not shown) and by GFP-annexin V staining (Fig. 8B). In contrast, induction of the expression of catalytically inactive PAK-K299R resulted in a slight increase in the percentage of dead cells in both the presence and absence of Bad (Fig. 8C).
FIG. 8.
Active PAK has an antiapoptotic effect in NIH 3T3 cells overexpressing Bad. NIH-3T3 cells stably expressing PAK-T423E or PAK-K299R in a tetracycline-dependent manner, or vector control cells (Co), were transiently transfected with pcDNA-HA-hBad; 16 h after transfection, cells were starved for 6 h, and both floating and attached cells were collected and stained with GFP-annexin V. The results represent mean ± standard error of the mean (n = 3).
It has been established that ceramide induces apoptosis through a mechanism inhibitable by antiapoptotic Bcl-2 family members (2, 13, 50). We incubated NIH 3T3 cells with 50 μM C2-ceramide and compared the apoptotic responses of cells in the presence or absence of PAK-T423E, PAK-K299R, or vector control (Fig. 9). Addition of C2-ceramide induced apoptosis in 42 to 50% of cells in which expression of PAK was suppressed. Control cells grown in the presence or absence of tetracycline exhibited only slight differences in the level of dead cells. However, in cells overexpressing PAK-T423E, the percentage of cells that died after treatment with C2-ceramide was markedly reduced (42.6% ± 7.7% without PAK, in comparison with 27.6% ± 2.6% for cells overexpressing PAK-T423E). Conversely, after overexpression of PAK-K299R, the level of dead cells was slightly increased. These data show that active PAK promotes cell survival in NIH 3T3 cells after induction of apoptosis either by direct overexpression of Bad or by treatment with C2-ceramide.
FIG. 9.
Active PAK protects NIH 3T3 cells from apoptosis induced by ceramide. NIH 3T3 cells stably expressing PAK-T423E or PAK-K299R in a tetracycline-dependent manner, or vector control cells (Co), were starved for 6 h and incubated with or without 50 M C2-ceramide for 6 h. Floating and attached cells were collected and stained with trypan blue. Results shown are the mean ± standard error of the mean (n = 3).
DISCUSSION
The requirement for PI 3-kinase activity in cell survival signaling in many systems is due, at least partially, to the role of PI 3-lipids to regulate activation of the Akt Ser/Thr kinase (15). Akt has been shown to phosphorylate Bad on Ser136, thereby reversing dimerization with and inhibition of Bcl-xL and/or Bcl-2 (7, 8). A number of survival factors, including IL-3, insulin-like growth factor 1 and Kit, induce the phosphorylation of Bad on both Ser112 and Ser136 (4, 30, 60). These factors act, at least in some situations, via the PI 3-kinase/Akt pathway to phosphorylate Ser136. In the FL5.12 IL-3-dependent cell line, survival signaling is also partially sensitive to inhibitors of PI 3-kinase (7, 8), and Akt is rapidly activated in response to IL-3 and phosphorylates Bad on Ser136 (7). The inability of Akt to phosphorylate Bad at Ser112, the inability of PI 3-kinase inhibitors to fully block survival signals in some systems, and studies demonstrating a dissociation between Akt activation and survival signaling all indicate that other kinase-dependent cell survival pathways are likely to exist. The findings reported here identify the Ser/Thr kinase PAK1 as one such alternate survival pathway.
We used Clostridium toxin B, which glucosylates and inactivates Rho family GTPases, to establish a requirement for Rho GTPase activity in IL-3-dependent survival of FL5.12 lymphoid progenitor cells (Fig. 1). This is consistent with recent reports that Rac GTPase exerts antiapoptotic effects in BaF3 cells upon IL-3 withdrawal (36) and in the presence of oncogenic Ras (27). We next established that the downstream effector of both Rac and Cdc42, PAK1, is rapidly activated by IL-3 in FL5.12 cells, and the time course of activation parallels the time course of IL-3-induced Bad phosphorylation (Fig. 3B). PAK1 phosphorylates the proapoptotic protein Bad in vitro and in vivo on both Ser112 and Ser136, leading to dissociation from Bcl-xL/Bcl-2 and binding of Bad to 14-3-3τ. PAK1-mediated phosphorylation of Bad effectively blocks Bad-induced cell death. The observations that a constitutively active PAK1 protects FL5.12 cells (Fig. 2A and B) and NIH 3T3 cells (Fig. 8) from apoptosis and that the autoinhibitory domain of PAK increases cell death in FL5.12 cells (Fig. 2C) indicate that PAK is likely to contribute to cell survival signaling by the IL-3 receptor. Interestingly, the activation of PAK1 by IL-3 in FL5.12 cells is partially blocked by the PI 3-kinase inhibitor LY294002. Since PAK can be activated by the small GTPases Rac and Cdc42, and activation of either GTPase can occur in response to PI 3-kinase-dependent guanine nucleotide exchange factors (3, 21, 23, 37), this may account for the partial sensitivity to PI 3-kinase inhibition. The data suggest that some part of the PI 3-kinase-dependent survival signals generated by IL-3 could involve PAK1 activation in addition to Akt. PAK1 may contribute to PI 3-kinase-independent survival signaling as well, since the activation of PAK1 by IL-3 is not totally blocked by LY294002. It is likely that PAK1 is also involved in the PI 3-kinase-independent survival pathways described for other cells (4, 25, 30, 39).
PAK activity alone was sufficient to phosphorylate Bad in vitro, and Bad phosphorylation by constitutive active PAK1 was independent of PI-3 kinase activity in vivo (Fig. 6B). Consistent with a direct effect of PAK, we measured Akt activity in FL5.12 cells expressing PAK1-T423E and did not detect any stimulation of Akt by Pak (not shown). Quantitative analysis indicates that PAK1 phosphorylates Bad (Fig. 5 and 6B) primarily on Ser112 and Ser136 within the span of aa 104 to 141. Another site (or sites) was phosphorylated within this region, albeit quantitatively to a lesser extent. There are additional serine residues at positions 108, 111, 128, and 134 that could serve as potential PAK phosphorylation sites. The data in Fig. 7B, showing that the coexpression of constitutive active PAK with Bad-S112/136A led to a decrease in the interaction between the Bad mutant and Bcl-2, suggests that the additional site(s) could be functional, i.e., that regulation at these sites adjacent to the Bcl protein-binding BH3 domain could also reduce interactions with Bcl-xL or Bcl-2. However, since IL-3 has not been reported to induce phosphorylation of Bad at residues apart from Ser112 and Ser136, the physiological significance of this observation is questionable. Indeed, no interaction between Bad-S112/136A and 14-3-3τ was detectable after phosphorylation of this Bad mutant by PAK (Fig. 8A). This is consistent with the findings of Zha et al. (60) and Datta et al. (7) that only the phosphorylation of Ser112 and/or Ser136 supports complexation of Bad with 14-3-3τ.
In addition to Akt, a number of other kinase signaling pathways have been identified as potential mediators of survival stimuli, including PKA, components of the Ras-MAPK-p90RSK pathway, the Raf-1 kinase, the calcium/calmodulin-dependent kinase, Bcr/Abl, and heart muscle kinase (a form of PKA) (35, 44, 45, 53, 58, 60). Only PKA has been shown to phosphorylate mitochondrion-associated Bad on Ser112 in vivo (22). There is no evidence that Rho GTPases or PAK are able to modulate the activity of PKA. Calcium/calmodulin-dependent kinase activates Akt directly, resulting in phosphorylation of Bad on Ser136 (58). Our studies clearly show that PAK phosphorylates Bad on both Ser112 and Ser136 in vitro and in vivo, suggesting that PAK promotes cell survival through an independent pathway distinct from the PI 3-kinase/Akt pathway. However, these results do not rule out the possibility that PAK promotes cell survival by other mechanisms in addition to that mediated by phosphorylation of Bad.
The activation of full-length PAKs (PAK1 or PAK2) is known to occur in response to activation of various cytokine and growth factor receptors (references 9, 18, and 61 and this report) as well as in response to cell adhesion (40). These stimuli are also known to generate effective cell survival signals, which our present data suggest may include the activation of PAKs. Certainly, the effect of PAK activity on cell survival will depend on the cellular context in which it becomes activated, as the mechanism that we have defined here would be relevant only in cells in which the Bad-Bcl2 mechanism is functioning to regulate cell death. In systems like the Jurkat cell where the Bad-Bcl2 mechanism is not operative, PAKs would not be expected to antagonize cell death by phosphorylating Bad.
Indeed, we have previously demonstrated that overexpression of the constitutively active PAK2 COOH-terminal fragment formed by caspase-mediated proteolysis of PAK2 can be proapoptotic in Jurkat T cells (42, 43). This effect is likely due to the ability of PAK to induce the activation of JNK and p38 kinases, which effectively induce cell death in the Jurkat system. In order to promote cell survival, activation of PAKs (either PAK1 or PAK2) by growth factors, etc., must occur prior to the initiation of the final effector phase of cell death, i.e., prior to the point when cytochrome c release has taken place and caspase 3 has been activated. After this stage, phosphorylation of Bad will not be able to prevent cell death. Since proteolytic cleavage and activation of PAK2 (but not PAK1) by caspase 3 occurs late in the apoptotic cascade, this is already too late for PAK2 activity to affect the death response by modulating Bad function. It has been shown that the survival kinase Akt is also proteolytically cleaved by caspase 3, but this is not likely to influence the subsequent death response at this stage. Proteolytic activation of PAK2 subsequent to caspase 3 activation appears to be primarily involved in cytoskeletal remodeling and/or JNK/p38 kinase activation.
We have identified a mechanism by which PAK kinase activity promotes cell survival. We suggest that it is likely that both PAK1 and PAK2 can regulate the Bad pathway, as our in vitro studies have shown that full-length PAK2 and PAK2 with a constitutively active C terminus are both as able as full-length PAK1 to catalyze the phosphorylation of Bad. Future studies will be directed at determining the contribution of PAK1 and PAK2 activity to other cell survival signaling pathways and the possible relevance of PAK activity to diseases in which apoptotic responsiveness has been reduced, including cancer.
ACKNOWLEDGMENTS
We acknowledge the expert technical assistance of Benjamin P. Bohl (TSRI), the support of Jon Chernoff (Fox Chase Cancer Center) and Klaus M. Hahn (TSRI), and secretarial assistance by Antonette Lestelle. Charles C. King provided much appreciated assistance with figure preparation. We thank S. J. Korsmeyer, M. E. Greenberg, and C. Thompson for providing reagents used in these studies.
This work was supported by Deutsche Forschungs Gemeinschaft fellowship Schu750/2-1 to A.S., U.S. Army grant DAMD 17-97-1-7230 to L.C.S. NIH grants CA-69381 (to J.C.R.) and AG15430 (to K. Hahn), and California Breast Cancer Research Program Award 3PB-0062 (to G.M.B.).
Footnotes
Manuscript no. 12215-IMM of The Scripps Research Institute.
REFERENCES
- 1.Aktories K, Just I. Monoglucosylation of low-molecular-mass GTP-binding Rho proteins by clostridial cytotoxins. Trends Cell Biol. 1995;5:441–443. doi: 10.1016/s0962-8924(00)89107-2. [DOI] [PubMed] [Google Scholar]
- 2.Basu S, Bayoumy S, Zhang Y, Lozano J, Kolesnick R. BAD enables ceramide to signal apoptosis via Ras and Raf-1. J Biol Chem. 1998;273:30419–30426. doi: 10.1074/jbc.273.46.30419. [DOI] [PubMed] [Google Scholar]
- 3.Benard V, Bohl B, Bokoch G. Characterization of Rac and Cdc42 activation in chemoattractant-stimulated human neutrophils using a novel assay for active GTPases. J Biol Chem. 1999;274:13198–13204. doi: 10.1074/jbc.274.19.13198. [DOI] [PubMed] [Google Scholar]
- 4.Blume-Jensen P, Janknecht R, Hunter T. The Kit receptor promotes cell survival via activation of PI 3-kinase and subsequent Akt-mediated phosphorylation of Bad on Ser136. Curr Biol. 1998;8:779–782. doi: 10.1016/s0960-9822(98)70302-1. [DOI] [PubMed] [Google Scholar]
- 5.Bokoch G M, Reilly A M, Daniels R H, King C C, Olivera A, Spiegel S, Knaus U G. A GTPase-independent mechanisms of p21-activated kinase activation. J Biol Chem. 1998;723:8137–8144. doi: 10.1074/jbc.273.14.8137. [DOI] [PubMed] [Google Scholar]
- 6.Daniels R H, Hall P S, Bokoch G M. Membrane targeting of p21-activated kinase (PAK1) induces neurite outgrowth from PC12 cells. EMBO J. 1998;17:754–764. doi: 10.1093/emboj/17.3.754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Datta S R, Dudek H, Tao X, Masters S, Fu H, Gotoh Y, Greenberg M E. Akt phosphorylation of Bad couples survival signals to the cell-intrinsic death machinery. Cell. 1997;91:231–241. doi: 10.1016/s0092-8674(00)80405-5. [DOI] [PubMed] [Google Scholar]
- 8.del Peso L, Gonzalez-Garcia M, Page C, Herrera R, Nunez G. Interleukin-3-induced phosphorylation of BAD through the protein kinase Akt. Science. 1997;278:687–689. doi: 10.1126/science.278.5338.687. [DOI] [PubMed] [Google Scholar]
- 9.Dharmawardhane S, Sanders L C, Martin S, Daniels R H, Bokoch G M. Localization of p21-activated kinase 1 (PAK1) to pinocytic vesicles and cortical actin structures in stimulated cells. J Cell Biol. 1997;138:1265–1278. doi: 10.1083/jcb.138.6.1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ding J, Knaus U G, Lian J P, Bokoch G M, Badwey J A. The renaturable 69- and 63-kDa protein kinases that undergo rapid activation in chemoattractant-stimulated guinea pig neutrophils are p21-activated kinases. J Biol Chem. 1996;271:24869–24873. doi: 10.1074/jbc.271.40.24869. [DOI] [PubMed] [Google Scholar]
- 11.Dudek H, Datta S R, Franke T F, Birnbaum M J, Yao R, Cooper G M, Segal R A, Kaplan D R, Greenberg M E. Regulation of neuronal survival by the serine-threonine protein kinase Akt. Science. 1997;275:661–665. doi: 10.1126/science.275.5300.661. [DOI] [PubMed] [Google Scholar]
- 12.Ernst J D, Yang L, Rosales J L, Broaddus V C. Preparation and characterization of an endogenously fluorescent annexin for detection of apoptotic cells. Anal Biochem. 1998;260:18–23. doi: 10.1006/abio.1998.2677. [DOI] [PubMed] [Google Scholar]
- 13.Farschon D M, Couture C, Mustelin T, Newmeyer D D. Temporal phases in apoptosis defined by the actions of Src homology 2 domains, ceramide, Bcl-2, interleukin-1 beta converting enzyme family proteases, and a dense membrane fraction. J Cell Biol. 1997;137:1117–1125. doi: 10.1083/jcb.137.5.1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Faure S, Vigneron S, Dorree M, Morin N. A member of the Ste20/PAK family of protein kinases is involved in both arrest in Xenopus oocytes and G2/prophase of the first meiotic cell cycle and prevention of apoptosis. EMBO J. 1997;16:5550–5561. doi: 10.1093/emboj/16.18.5550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Franke T F, Kaplan D R, Cantley L C. PI3K: downstream AKTion blocks apoptosis. Cell. 1997;88:435–437. doi: 10.1016/s0092-8674(00)81883-8. [DOI] [PubMed] [Google Scholar]
- 16.Franke T F, Kaplan D R, Cantley L C, Toker A. Direct regulation of the Akt proto-oncogene product by phosphatidylinositol-3,4-bisphosphate. Science. 1997;275:665–668. doi: 10.1126/science.275.5300.665. [DOI] [PubMed] [Google Scholar]
- 17.Gajewski T F, Thompson C B. Apoptosis meets signal transduction: elimination of BAD influence. Cell. 1996;87:589–592. doi: 10.1016/s0092-8674(00)81377-x. [DOI] [PubMed] [Google Scholar]
- 18.Galisteo M, Chernoff J, Su Y-C, Skolnik E Y, Schlessinger J. The adaptor protein Nck links receptor tyrosine kinases with the serine-threonine kinase Pak1. J Biol Chem. 1996;271:20997–21000. doi: 10.1074/jbc.271.35.20997. [DOI] [PubMed] [Google Scholar]
- 19.Golstein P. Controlling cell death. Science. 1997;275:1081–1082. doi: 10.1126/science.275.5303.1081. [DOI] [PubMed] [Google Scholar]
- 20.Green D, Reed J C. Mitochondria and apoptosis. Science. 1998;281:1309–1312. doi: 10.1126/science.281.5381.1309. [DOI] [PubMed] [Google Scholar]
- 21.Han J, Luby-Phelps K, Das B, Shu X, Xia Y, Mosteller R D, Krishna U M, Falk J R, White M A, Broek D. Role of substrates and products of PI 3-kinase in regulating activation of Rac-related guanosine triphosphates by Vav. Science. 1998;279:558–560. doi: 10.1126/science.279.5350.558. [DOI] [PubMed] [Google Scholar]
- 22.Harada H, Becknell B, Wilm M, Mann M, Huang L, Taylor S, Scott J, Korsmeyer S. Phosphorylation and inactivation of BAD by mitochondria-anchored protein kinase A. Mol Cell. 1999;3:413–422. doi: 10.1016/s1097-2765(00)80469-4. [DOI] [PubMed] [Google Scholar]
- 23.Hawkins P T, Eguinoa A, Qiu R G, Stokoe D, Cooke F T, Walters R, Wennstrom S, Claesson-Welsh L, Evans T, Symons M, et al. PDGF stimulates an increase in GTP-Rac via activation of phosphoinositide 3-kinase. Curr Biol. 1995;5:393–403. doi: 10.1016/s0960-9822(95)00080-7. [DOI] [PubMed] [Google Scholar]
- 24.Hengartner M O. Apoptosis. CED-4 is a stranger no more. Nature. 1997;397:714–715. doi: 10.1038/41873. [DOI] [PubMed] [Google Scholar]
- 25.Hinton H, Welham M. Cytokine-induced protein kinase B activation and Bad phosphorylation do not correlate with cell survival of hemopoietic cells. J Immunol. 1999;162:7002–7009. [PubMed] [Google Scholar]
- 26.Jacobsen M D, Weil M, Raff M C. Programmed cell death in animal development. Cell. 1997;88:347–354. doi: 10.1016/s0092-8674(00)81873-5. [DOI] [PubMed] [Google Scholar]
- 27.Joneson T, Bar-Sagi D. Suppression of Rad-induced apoptosis by the Rac GTPase. Mol Cell Biol. 1999;19:5892–5901. doi: 10.1128/mcb.19.9.5892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Knaus U G, Morris S, Dong H, Chernoff J, Bokoch G M. Regulation of human leukocyte p21-activated kinases through G protein-coupled receptors. Science. 1995;269:221–223. doi: 10.1126/science.7618083. [DOI] [PubMed] [Google Scholar]
- 29.Koopman G, Reutelingsperger C, Kuijten G, Keehen R, Pals S, van Oers M. Annexin V for flow cytometric detection of phosphatidylserine expression of B cells undergoing apoptosis. Blood. 1994;84:1415–1420. [PubMed] [Google Scholar]
- 30.Kulik G, Klippel A, Weber M J. Antiapoptotic signaling by the insulin-like growth factor I receptor, phosphatidylinositol 3-kinase, and Akt. Mol Cell Biol. 1997;17:1595–1606. doi: 10.1128/mcb.17.3.1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee N, MacDonald H, Reinhard C, Halenbeck R, Roulston A, Shi T, Williams L T. Activation of hPAK65 by caspase cleavage induces some of the morphological and biochemical changes of apoptosis. Proc Natl Acad Sci USA. 1997;94:13642–13647. doi: 10.1073/pnas.94.25.13642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lim L, Manser E, Leung T, Hall C. Regulation of phosphorylation pathways by p21 GTPases. The p21 Ras-related Rho subfamily and its role in phosphorylation signaling pathways. Eur J Biochem. 1996;242:171–185. doi: 10.1111/j.1432-1033.1996.0171r.x. [DOI] [PubMed] [Google Scholar]
- 33.Lundstrom K, Mills A, Buell G, Allet E, Adamis N, Liljestrom P. High-level expression of the human neurokinin-1 receptor in mammalian cell lines using the Semliki Forest virus expression system. Eur J Biochem. 1994;224:917–921. doi: 10.1111/j.1432-1033.1994.00917.x. [DOI] [PubMed] [Google Scholar]
- 34.McKearn J P, McCurbey J, Fagg B. Enrichment of hematopoetic precursor cells and cloning of multipotential B-lymphocyte precursors. Proc Natl Acad Sci USA. 1995;82:7414–7418. doi: 10.1073/pnas.82.21.7414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Neshat, M. S., A. B. Raitano, H. G. Wang, J. C. Reed, and C. L. Sawyers. Phosphorylation of the proapoptotic protein Bad in cells expressing the Bcr-Abl oncogene through phosphatidylinositol 3-kinase and Raf-dependent signaling pathways. Submitted for publication.
- 36.Nishida K, Kaziro Y, Satoh T. Anti-apoptotic function of Rac in hematopoietic cells. Oncogene. 1999;18:407–415. doi: 10.1038/sj.onc.1202301. [DOI] [PubMed] [Google Scholar]
- 37.Numnual A S, Yatsula B A, Bar-Sagi D. Coupling of Ras and Rac guanosine triphosphatases through the Ras exchanger Sos. Science. 1998;279:560–563. doi: 10.1126/science.279.5350.560. [DOI] [PubMed] [Google Scholar]
- 38.Oltvai Z N, Korsmeyer S J. Checkpoints of dueling dimers foil death wishes. Cell. 1994;79:189–192. doi: 10.1016/0092-8674(94)90188-0. [DOI] [PubMed] [Google Scholar]
- 39.Philpott K L, McCarthy M J, Klippel A, Rubin L L. Activated phosphatidylinositol 3-kinase and Akt kinase promote survival of superior cervical neurons. J Cell Biol. 1997;139:809–815. doi: 10.1083/jcb.139.3.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Price L S, Leng J, Schwartz M A, Bokoch G M. Activation of Rac and Cdc42 by integrins mediates cell spreading. Mol Biol Cell. 1998;9:1863–1871. doi: 10.1091/mbc.9.7.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reed J C. Bcl-2 family proteins. Oncogene. 1998;17:3225–3236. doi: 10.1038/sj.onc.1202591. [DOI] [PubMed] [Google Scholar]
- 42.Rudel T, Bokoch G M. Membrane and morphological changes in apoptotic cells regulated by caspase-mediated activation of PAK2. Science. 1997;276:1571–1574. doi: 10.1126/science.276.5318.1571. [DOI] [PubMed] [Google Scholar]
- 43.Rudel T, Zenke F T, Chuang T-H, Bokoch G M. p21-activated kinase (PAK) is required for Fas-induced JNK activation in Jurkat cells. J Immunol. 1998;160:7–11. [PubMed] [Google Scholar]
- 44.Rukenstein A, Rydel R E, Greene L A. Multiple agents rescue PC12 cells from serum-free cell death by translation and transcription-independent mechanisms. J Neurosci. 1991;11:2552–2563. doi: 10.1523/JNEUROSCI.11-08-02552.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Salomoni P, Wasik M A, Riedel R F, Reiss K, Choi J K, Skorski T, Calabretta B. Expression of constitutively active Raf-1 in the mitochondria restores antiapoptotic and leukemogenic potential of a transformation-deficient BCR/ABL mutant. J Exp Med. 1998;187:1995–2007. doi: 10.1084/jem.187.12.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Segal R A, Greenberg M E. Intracellular signaling pathways activated by neurotrophic factors. Annu Rev Neurosci. 1996;19:463–489. doi: 10.1146/annurev.ne.19.030196.002335. [DOI] [PubMed] [Google Scholar]
- 47.Sells M A, Chernoff J. Emerging from the Pak: the p21-activated protein kinase family. Trends Cell Biol. 1997;7:162–167. doi: 10.1016/S0962-8924(97)01003-9. [DOI] [PubMed] [Google Scholar]
- 48.Sells M A, Boyd J T, Chernoff J. p21-activated kinase 1 (PAK1) regulates cell motility in mammalian fibroblasts. J Cell Biol. 1999;145:837–849. doi: 10.1083/jcb.145.4.837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sells M A, Knaus U G, Bagrodia S, Ambrose D M, Bokoch G M, Chernoff J. Human p21-activated kinase (Pak1) regulates actin organization in mammalian cells. Curr Biol. 1997;7:202–210. doi: 10.1016/s0960-9822(97)70091-5. [DOI] [PubMed] [Google Scholar]
- 50.Smyth M J, Perry D K, Zhang J, Poirier G G, Hannun Y A, Obeid L M. prICE: a downstream target for ceramide-induced apoptosis and for the inhibitory action of Bcl-2. Biochem J. 1996;316:25–28. doi: 10.1042/bj3160025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stewart C E, Rotwein P. Growth, differentiation, and survival: multiple physiological functions for insulin-like growth factors. Physiol Rev. 1996;76:1005–1026. doi: 10.1152/physrev.1996.76.4.1005. [DOI] [PubMed] [Google Scholar]
- 52.Vlahos C J, Matter W F, Hui K Y, Brown R F. A specific inhibitor of phosphatidylinositol 3-kinase 2-(4-morpholinyl)-8-phenyl-4H-1-benzopyran-4-one (LY294002) J Biol Chem. 1994;269:5241–5248. [PubMed] [Google Scholar]
- 53.Wang H-G, Rapp U R, Reed J C. Bcl-2 targets the protein kinase Raf-1 to mitochondria. Cell. 1996;87:629–638. doi: 10.1016/s0092-8674(00)81383-5. [DOI] [PubMed] [Google Scholar]
- 54.Wang H-G, Pathan N, Ethell I M, Krajewski S, Yamaguchi Y, Shibasaki F, McKeon F, Bobo T, Franke T F, Reed J C. Ca2+-induced apoptosis through calcineurin dephosphorylation of BAD. Science. 1999;284:339–343. doi: 10.1126/science.284.5412.339. [DOI] [PubMed] [Google Scholar]
- 55.Wyllie H A, editor. Apoptosis. British Medical Bulletin 53. Dorchester, England: The Dorset Press; 1997. [Google Scholar]
- 56.Xia Z, Dickens M, Raingeaud J, Davis R J, Greenberg M E. Opposing effects of ERK and JNK-p38-MAP kinases on apoptosis. Science. 1995;270:1326–1331. doi: 10.1126/science.270.5240.1326. [DOI] [PubMed] [Google Scholar]
- 57.Yang E, Zha J, Jockel J, Boise L H, Thompson C B, Korsmeyer S J. Bad, a heterodimeric partner for Bcl-XL and Bcl-2, displaces Bax and promotes cell death. Cell. 1995;80:285–291. doi: 10.1016/0092-8674(95)90411-5. [DOI] [PubMed] [Google Scholar]
- 58.Yano S, Tokumitsu H, Soderling T R. Calcium promotes cell survival through CaM-K kinase activation of protein-kinase-B-pathway. Nature. 1998;396:584–587. doi: 10.1038/25147. [DOI] [PubMed] [Google Scholar]
- 59.Zenke F, King C, Bohl B, Bokoch G. Identification of a central phosphorylation site in p21-activated kinase regulating autoinhibition and kinase activity. J Biol Chem. 1999;274:32565–32573. doi: 10.1074/jbc.274.46.32565. [DOI] [PubMed] [Google Scholar]
- 60.Zha J, Harada H, Yang E, Jockel J, Korsmeyer S J. Serine phosphorylation of death agonist Bad in response to survival factor results in binding to 14-3-3 not Bcl-xL. Cell. 1996;87:619–628. doi: 10.1016/s0092-8674(00)81382-3. [DOI] [PubMed] [Google Scholar]
- 61.Zhang S, Han J, Sells M A, Chernoff J, Knaus U G, Ulevitch R J, Bokoch G M. Rho family GTPases regulated p38 mitogen-activated protein kinase through the downstream mediator Pak1. J Biol Chem. 1995;270:23934–23936. doi: 10.1074/jbc.270.41.23934. [DOI] [PubMed] [Google Scholar]
- 62.Zhao Z S, Chen X Q, Chong C, Leung T, Lim L. A conserved negative regulatory region in αPAK: inhibition of PAK kinases reveals their morphological roles downstream of Cdc42 and Rac1. Mol Cell Biol. 1998;18:2153–2163. doi: 10.1128/mcb.18.4.2153. [DOI] [PMC free article] [PubMed] [Google Scholar]