Abstract
3Y1 rat fibroblasts overexpressing the epidermal growth factor (EGF) receptor (EGFR cells) become transformed when treated with EGF. A common response to oncogenic and mitogenic stimuli is elevated phospholipase D (PLD) activity. RalA, a small GTPase that functions as a downstream effector molecule of Ras, exists in a complex with PLD1. In the EGFR cells, EGF induced a Ras-dependent activation of RalA. The activation of PLD by EGF in these cells was dependent upon both Ras and RalA. In contrast, EGF-induced activation of Erk1, Erk2, and Jun kinase was dependent on Ras but independent of RalA, indicating divergent pathways activated by EGF and mediated by Ras. The transformed phenotype induced by EGF in the EGFR cells was dependent upon both Ras and RalA. Importantly, overexpression of wild-type RalA or an activated RalA mutant increased PLD activity in the absence of EGF and transformed the EGFR cells. Although overexpression of PLD1 is generally toxic to cells, the EGFR cells not only tolerated PLD1 overexpression but also became transformed in the absence of EGF. These data demonstrate that either RalA or PLD1 can cooperate with EGF receptor to transform cells.
Overexpression of a tyrosine kinase is a common genetic defect in a variety of human tumors (21). The epidermal growth factor (EGF) receptor, which has an intrinsic tyrosine kinase that is activated in response to EGF, is frequently overexpressed in human breast and ovarian cancer (35). However, overexpression of a tyrosine kinase such as the EGF receptor is not sufficient for a fully transformed or cancerous phenotype. We recently demonstrated that downregulation of protein kinase C δ (PKC δ) transforms 3Y1 rat fibroblasts overexpressing either c-Src (28) or the EGF receptor (19). The EGF receptor-overexpressing cells (EGFR cells) could also be transformed when treated with EGF (19), suggesting that EGF could accomplish what PKC δ downregulation accomplished. Interestingly, downregulation of PKC δ also caused an increase in phospholipase D (PLD) activity (19, 38), which is commonly elevated in response to oncogenic and mitogenic stimuli (11, 41). Both EGF-induced increases in PLD activity and EGF-induced transformation were dependent upon the α isoform of PKC (19), suggesting that PLD may be an important component of the mitogenic and oncogenic properties of the EGF receptor.
We demonstrated previously (30) that PLD1 associates directly with the small GTPase RalA, a downstream target of Ras (13). RalA is required for PLD activation in response to v-Src and v-Ras (22). RalA has also been implicated in cell transformation (1, 39), indicating a possible role for PLD in mitogenic signaling. In this paper, we report that both RalA and PLD1 can cooperate with an overexpressed EGF receptor to transform cells.
MATERIALS AND METHODS
Cells and cell culture conditions.
Rat 3Y1 cells or rat 3Y1 cells expressing the EGF receptor were maintained in Dulbecco's modified Eagle's medium supplemented with 10% bovine calf serum (HyClone) as described previously (28, 29). The EGFR cells were constructed by transfecting into rat 3Y1 cells pPEGFr (6), which expresses the EGF receptor from the simian virus 40 promoter, as described previously (19). Cell cultures were made quiescent by being grown to confluence and then having the medium replaced with fresh medium containing 0.5% bovine calf serum for 1 day. For growth of cells in soft agar, 103 cells were suspended in top agar (Dulbecco's modified Eagle's medium, 20% calf serum, 0.38% agar) and overlaid onto hardened bottom agar (Dulbecco's modified Eagle's medium, 20% calf serum, 0.7% agar) as described previously (19, 28).
Transfection.
Cells were plated at a density of 105 cells/100-mm dish 18 h before transfection. Transfections were performed by using Lipofectamine reagent (GIBCO) as specified by the vendor. Transfected cultures were selected with either G418 (400 μg/ml), puromycin (5 μg/ml), or hygromycin (200 μg/ml) for 10 to 14 days at 37°C. At that time, antibiotic-resistant colonies were picked and expanded for further analysis under selective conditions.
Materials.
Monoclonal antibodies to the EGF receptor, Ras, Jun, and RalA were obtained from Transduction Laboratories; polyclonal PLD1 and anti-phospho–c-Jun antibodies were from Upstate Biotechnology; Erk1 and Erk2 polyclonal and anti-phospho–Erk1 and Erk2 antibodies were from Santa Cruz Biotechnology. pCEP4, which contains the hygromycin resistance gene, was obtained from Invitrogen.
Western analysis.
Proteins were extracted from cultured cells as previously described (28, 29). Equal amounts of protein were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis with an 8% acrylamide separating gel, transferred to nitrocellulose, and blocked overnight at 4°C with 5% nonfat dry milk isotonic phosphate-buffered saline (136 mM NaCl, 2.6 mM KCl, 1.4 mM KH2PO4, 4.2 mM Na2HPO4). The nitrocellulose filters were washed three times for 5 min each in phosphate-buffered saline and then incubated with antibodies as described below. Depending upon the origin of the primary antibodies, either anti-mouse or anti-rabbit immunoglobulin G was used for detection by the enhanced chemiluminescence system (Amersham).
PLD assays.
Confluent 35-mm culture dishes were prelabeled for 4 h with [3H]myristate (3 μCi [40 Ci/mmol]) in 3 ml of medium containing 0.5% newborn calf serum. PLD-catalyzed transphosphatidylation in the presence of 1% butanol was performed as described previously (37, 38). Extraction and characterization of lipids by thin-layer chromatography were performed as previously described (38).
RalA activation assay.
Activated RalA was detected as described by Wolthuis et al. (45, 46). The cells were first lysed with 15% glycerol–50 mM Tris-HCl (pH 7.4)–1% Nonidet P-40–200 nM NaCl–5 mM MgCl2–1 mM phenylmethylsulfonyl fluoride–1 μM leupeptin–0.1 μM aprotinin–10 μg of soybean trypsin inhibitor per ml. The lysates were then treated with glutathione S-transferase (GST)–Ral-BD fusion protein immobilized with glutathione-agarose beads prepared as described previously (31). Ral-BD is the Ral binding domain of Ral-BP that binds to activated GTP-bound Ral proteins (45). The activated Ral proteins were then recovered by centrifugation and subjected to Western blot analysis with an antibody raised against RalA (Transduction Laboratories).
hPLD1 expression.
A vector expressing Flu-tagged hPLD1 (pCGN-hPLD1) was generated by inserting the entire coding region of hPLD1 into pCGN as described previously (16). This gene was introduced into cells by cotransfection with pCEF4 (Invitrogen), which expresses a hygromycin resistance marker gene. PLD1 expression was detected by Western blot analysis with a monoclonal antibody raised against the Flu epitope (Santa Cruz Biotechnology).
RESULTS
Ras-dependent activation of RalA by EGF in 3Y1 cells overexpressing the EGF receptor.
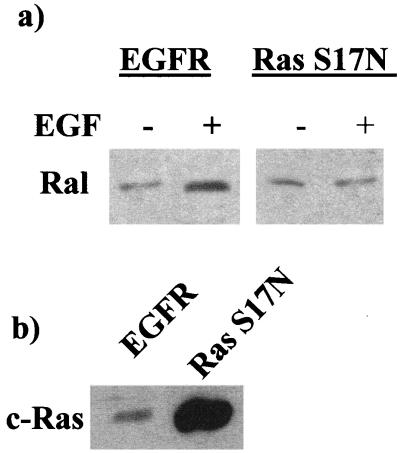
Upon activation, RalA binds GTP and associates with the downstream effector molecule Ral-BP1 (4). We took advantage of this by using the Ral binding domain (Ral-BD) of this protein fused to GST (GST–Ral-BD) to detect activated GTP-bound RalA as described by Wolthuis et al. (45, 46). 3Y1 rat fibroblasts overexpressing the EGF receptor (EGFR cells) (19) were treated with EGF, and cell lysates were prepared 10 min later and treated with GST–Ral-BD immobilized on glutathione-agarose beads. The GST–Ral-BD was recovered by centrifugation, and the pellets were subjected to Western blot analysis with an antibody raised against RalA. As shown in Fig. 1a, EGF treatment resulted in a substantial increase in the amount of RalA detected in the GST–Ral-BP1 precipitates. These data indicate that RalA is activated in response to EGF treatment in the EGFR cells. Ral-GDS, the GDP-GTP exchange factor for RalA, is a downstream effector molecule of Ras (17, 23, 40). However, RalA can be activated by Ras-independent mechanisms as well (18). We therefore wished to determine whether the activation of RalA by EGF was dependent upon Ras. To do this, we stably transfected a dominant negative Ras mutant (S17N) (12) into the EGFR cells and verified expression of the Ras mutant by Western blot analysis (Fig. 1b). We then investigated whether EGF was able to activate RalA in the cells expressing the dominant negative Ras; as shown in Fig. 1a, expression of the dominant negative Ras prevented the EGF-induced activation of RalA. Thus, EGF activates RalA in a Ras-dependent manner. These data are consistent with those reported previously by Wolthuis et al. (46), who demonstrated a Ras-dependent activation of Ral in rat fibroblasts expressing endogenous EGF receptor.
FIG. 1.
EGF activates RalA in a Ras-dependent manner. (a) EGFR cells and EGFR cells expressing the S17N dominant negative Ras mutant (Ras S17N) were treated with EGF (100 ng/ml) for 10 min. The cells were then lysed and treated with immobilized GST–Ral-BD as described in Materials and Methods. The GST–Ral-BD was recovered by centrifugation, and the precipitate was subjected to Western blot analysis with an antibody raised against RalA. (b) The parental EGFR cells and the EGFR cells stably transfected with the S17N dominant negative Ras mutant were examined for expression of Ras proteins by Western blot analysis.
EGF-induced PLD activity is dependent upon the Ras/RalA GTPase cascade.
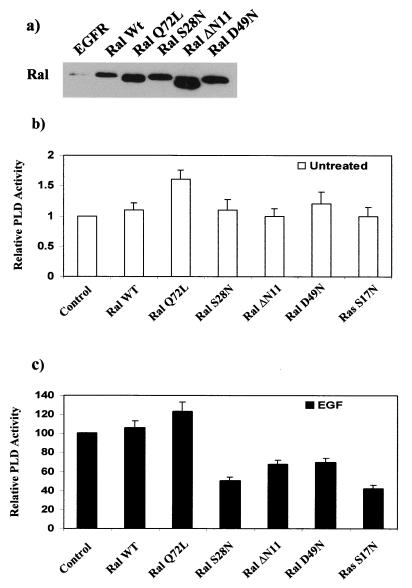
EGF induces an increase in PLD activity (19, 39, 47). PLD1 (16) associates directly with RalA (30). To investigate whether the Ras/RalA GTPase cascade played a role in the EGF-induced increase in PLD activity, we established several EGFR cell lines that stably expressed either wild-type or mutant RalA. The mutants of RalA used included an activated RalA (Q72L) and three inactivating RalA mutants: S28N, which is homologous to the mutant with the S17N mutation in Ras; D49N, which is an effector domain mutant and is defective in associating with Ral-BP1 (4); and ΔN11, which has an amino-terminal deletion of 11 amino-terminal amino acids unique to Ral GTPases. The ΔN11 mutant is defective in recruiting the PLD activator Arf into an active PLD complex (31). Expression of these RalA genes in the EGFR cells was verified by Western blot analysis, as shown in Fig. 2a. We then examined the effect of these RalA gene products upon PLD activity in the presence and absence of EGF. Overexpression of wild-type RalA and the activated Q72L RalA mutant induced a small but reproducible increase in the basal PLD activity of the unstimulated cells (Fig. 2b). Overexpression of all three defective RalA mutants inhibited EGF-induced PLD activity (Fig. 2c). The S28N mutant inhibited EGF-induced PLD activity the most efficiently and to the same extent as the S17N dominant negative Ras mutant (Fig. 2c). The reduced ability of the ΔN11 and D49N mutants to inhibit the EGF-induced PLD activity is probably due to the higher efficiency of the GDP-GTP mutants to act as dominant negative mutants (14). The defective RalA mutants had little or no effect upon the basal PLD activity (Fig. 2b). These data indicate that EGF-induced PLD activity is dependent upon RalA and that in cells overexpressing the EGF receptor, activated RalA can elevate PLD activity.
FIG. 2.
EGF-induced PLD activity is dependent upon the Ras/RalA GTPase cascade. EGFR cells were stably transfected with plasmid vectors expressing wild-type (wt) RalA; Q72L, an activated RalA mutant; S28N, an inactivating mutant that is homologous to the Ras S17N mutant; D49N, a RalA effector domain mutant; and ΔN11, which has an amino-terminal deletion of 11 amino acids. (a) Expression of these RalA genes was verified by Western blot analysis. (b and c) The PLD activity was then determined in untreated (b) and EGF-treated (100 ng/ml for 10 min) (c) EGFR cells and the EGFR cells expressing the RalA mutants and the S17N Ras dominant negative mutant described in Fig. 1. PLD activity was measured by the PLD-catalyzed transphosphatidylation of phosphatidylcholine to phosphatidylbutanol in the presence of 1% butanol as described in Materials and Methods. The relative PLD activity in the untreated cells was normalized to the PLD activity in the control EGFR cells, which was given a value of 1. The relative PLD activity in the EGF-treated cells was normalized to the PLD activity in the control EGFR cells, which was given a value of 100%. The PLD activity in the EGF-treated cells was approximately sixfold greater than that in the untreated cells (19). Error bars represent the standard deviation for three independent experiments performed in duplicate.
EGF-induced Erk1, Erk2, and Jun kinase activation is dependent upon Ras but independent of Ral.
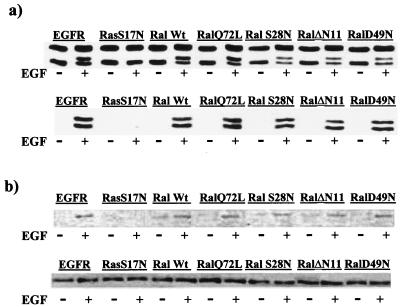
EGF treatment also activates Erk1, Erk2, and Jun kinases in a Ras-dependent manor (27). In addition to Ral-GDS, there are several other downstream effector molecules of Ras (32). EGF-induced activation of Erk1 and Erk2 is dependent on Raf (20, 36), and EGF-induced activation of Jun kinase is dependent on another Ras effector, phosphatidylinositol-3-kinase (27). To establish that the effect of the dominant negative RalA mutants was specific for the Ras/RalA pathway, we examined the ability of EGF to activate these kinases in the EGFR cells expressing the dominant negative RalA mutants. The activation of Erk1 and Erk2 (Fig. 3a) and Jun kinase (Fig. 3b) was inhibited by the dominant negative Ras but not by the dominant negative RalA mutants. These data indicate that the RalA mutants are affecting only the Ras/RalA pathway and not other signaling pathways mediated by the Ras downstream effector molecules Raf and phosphatidylinositol-3-kinase.
FIG. 3.
EGF-induced Erk1, Erk2, and Jun kinase activation is dependent upon Ras but independent of Ral. The EGFR cells and the EGFR cells expressing the dominant negative S17N Ras (RasS17N) and the various RalA genes described in the legend to Fig. 2 were treated with EGF (100 ng/ml for 10 min) as shown, and the activation of Erk1 and Erk2 (a) and Jun kinase (b) was determined. The activation of Erk1 and Erk2 was examined by subjecting cell lysates to Western blot analysis with an antibody that recognizes Erk1 and Erk2 and detects an electrophoretic mobility shift in Erk1 that occurs upon activation (a, top panel). We also performed Western blot analysis with an antibody that recognizes phosphorylated (activated) Erk1 and Erk2 (a, bottom panel). The top panel also indicates that the levels of Erk1 and Erk2 were not affected by the EGF treatment. The activation of Jun kinase was determined by subjecting cell lysates to Western blot analysis with an antibody specific for phosphorylated (activated) Jun (b, top panel) and an antibody to Jun that establishes that the levels of Jun were not altered by EGF treatment (b, bottom panel). Wt, wild type.
EGF-induced transformation is dependent upon Ral.
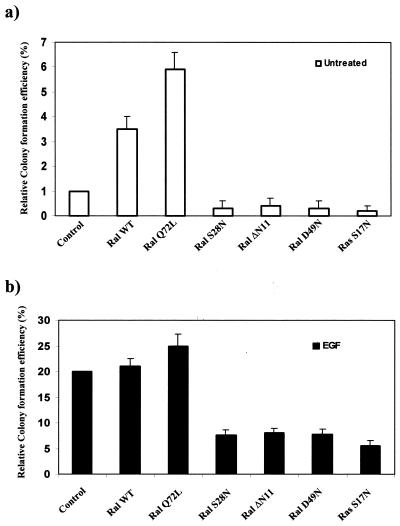
We reported previously that in response to EGF, the EGFR cells form colonies in soft agar (19). Moreover, the EGFR cells become transformed upon downregulation of PKC δ (19). Interestingly, the downregulation of PKC δ results in the elevation of PLD activity (19). We therefore examined the role of the Ras/RalA pathway on transformation in the EGFR cells. We investigated the ability of the EGFR cells expressing the S17N dominant negative Ras and the various RalA genes (Fig. 2) to form colonies in soft agar in the absence and presence of EGF. As shown in Fig. 4a, overexpression of wild-type RalA or the activated Q72L RalA mutant resulted in a substantial increase in colony-forming efficiency in the absence of EGF. Overexpression of wild-type RalA or the activated Q72L RalA mutant did not significantly increase colony-forming efficiency in the presence of EGF (Fig. 4b), and, as expected, the ability of EGFR cells to form colonies in soft agar was inhibited by expression of the dominant negative S17N Ras gene (Fig. 4b). Thus, RalA overexpression or activation can apparently result in at least partial transformation of cells overexpressing the EGF receptor. As observed for EGF-induced PLD activity, the corresponding S28N RalA mutant reduced the colony-forming efficiency to that observed with the S17N Ras mutant. The effector domain RalA mutant (S49N) and the amino-terminal deletion mutant (ΔN11) also reduced the colony-forming efficiency of the EGF-treated cells (Fig. 4b). The RalA mutants also reduced the basal colony number of the untreated EGFR cells (Fig. 4a). These data indicate that RalA is required for the EGF-induced transformation of the EGFR cells and that activated RalA is able to compensate, at least partially, for the effect of EGF.
FIG. 4.
EGF-induced transformation is dependent upon Ral. Anchorage-independent growth of the EGFR cells and the EGFR cells expressing the dominant negative S17N Ras (Ras S17N) and the various RalA genes described in the legend to Fig. 2 was examined in the absence (a) and presence (b) of EGF (100 ng/ml). EGF was replenished every 4 days. A total of 103 cells were suspended in soft agar, and the percentage of cells that formed colonies was determined 3 weeks later. The relative colony-forming efficiency in the untreated cells was normalized to that in the control EGFR cells, which was given a value of 1. The relative colony-forming efficiency in the EGF-treated cells was normalized to that in the control EGFR cells, which was given a value of 100%. The colony-forming efficiency was approximately 1% for the untreated EGFR cells and 20% for the EGF-treated cells, as reported previously (19). Error bars represent the standard deviation for three independent experiments performed in duplicate. WT, wild type.
Expression of PLD1 in EGFR cells increases colony-forming efficiency.
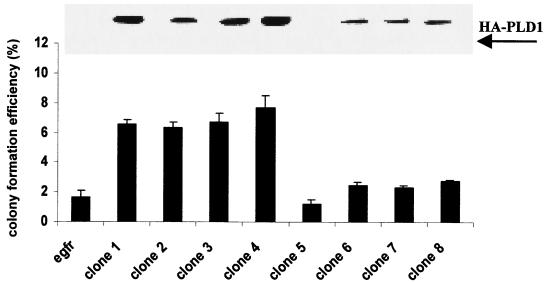
As discussed in the introduction, cells are very intolerant of PLD expression. We nevertheless attempted to express PLD1 in the EGFR cells and in the parental 3Y1 cells. A plasmid vector that expresses Flu-tagged hPLD1 (16) was cotransfected into the EGFR cells along with pCEF4 (Invitrogen), which expresses a hygromycin resistance marker gene. Transfection was attempted in both the EGFR and parental 3Y1 cells. Interestingly, hygromycin-resistant colonies were detected after 10 days only in the EGFR cells (58 hygromycin-resistant colonies were found); no hygromycin-resistant colonies were detected in the parental 3Y1 cells. This was not due to differences in transfection efficiency between the two cell lines, because transfection with pCEF4 alone gave very similar numbers of hygromycin-resistant colonies in both the EGFR and parental 3Y1 cells (97 and 110 hygromycin-resistant colonies, respectively). Thus, expression of PLD1 in the parental 3Y1 cells is apparently toxic to the parental 3Y1 cells, which is consistent with previous reports suggesting that a high level of PLD expression is difficult to obtain (19). The EGFR cells, however, are tolerant of higher levels of PLD1 expression. Several hygromycin colonies were expanded, and the level of PLD1 expression and their ability to form colonies in soft agar in the absence of EGF were examined. As shown in Fig. 5, several of the clones displayed an increased colony-forming efficiency, and, importantly, the ability to form colonies correlated with the level of hPLD1 expression (Fig. 5). Basal PLD activity in PLD1-transfected cells also correlated with PLD1 expression, with about a 2.5- to 3-fold increase in the number of cells with the highest levels of PLD1 expression (data not shown). In addition, the colonies formed in the PLD1-overexpressing cells were much larger than the background colonies formed by the EGFR cells. These data are consistent with the ability of overexpressed RalA to increase colony-forming efficiency in the EGFR cells, and they suggest that the effect of RalA is mediated by PLD. These data are also consistent with our previous data where we demonstrated that downregulation or inhibition of PKC δ elevates PLD activity in the EGFR cells and transforms them (19).
FIG. 5.
Overexpression of PLD1 in EGFR cells increases colony-forming efficiency in the absence of EGF. EGFR cells transfected with pCGN-hPLD1, which expresses Flu-tagged hPLD1 (13), were suspended in soft agar, and the ability to form colonies was determined as in the experiment in Fig. 4. Western blot analysis of the hPLD1-expressing and parental EGFR cells with monoclonal anti-Flu antibody is shown. Error bars represent the standard deviation for assays performed in triplicate.
DISCUSSION
Rat fibroblasts overexpressing the EGF receptor become transformed when treated with EGF. In the absence of EGF, they become transformed if PKC δ is downregulated (19). PKC δ downregulation leads to an elevation of PLD activity. Previous reports have indicated that the elevation of PLD activity in response to mitogenic signals is mediated by RalA (22), which interacts directly with PLD1 (30). In this report, we have demonstrated that RalA is required for the EGF-induced activation of PLD. The transformed phenotype induced by EGF on the EGFR cells was also dependent upon RalA, suggesting the possibility that an important component of EGF-induced cell division signals is the activation of PLD. Consistent with this hypothesis, overexpression of either RalA or PLD1 in EGFR cells led to the formation of colonies in soft agar in the absence of EGF.
The mitogenic effects of EGF probably involve multiple downstream effector molecules. Simply overexpressing the EGF receptor in some way is able to provide a partial mitogenic signal. It was previously reported that a kinase-defective EGF receptor could activate the Ras/Erk1/Erk2 pathway and allow cell survival but not proliferation in murine hematopoietic cells (44), indicating that the EGF receptor activates multiple downstream pathways to generate a complete mitogenic response. This is similar to the findings in the present study, where the overexpressed EGF receptor induces only a partially transformed phenotype, which can be complemented by overexpression of either RalA or PLD1. The dependence of EGF-induced transformation upon RalA also indicates that RalA is essential for mitogenic signaling mediated by an activated EGF receptor.
Overexpression of a tyrosine kinase is frequently observed in human cancers (21). However, tyrosine kinase overexpression is not sufficient convert a normal cell to a transformed one. Downregulation of PKC δ by tumor-promoting phorbol esters leads to the transformation of rat fibroblasts overexpressing c-Src (28). Similarly, inhibition of PKC δ transformed the EGFR cells used in this study (19). Downregulation of PKC δ elevates PLD activity (19), suggesting that tumor promotion may involve the activation of PLD. Interestingly, EGF leads to the tyrosine phosphorylation of PKC δ (9), which results in reduced PKC δ kinase activity (8, 48). Tumor promotion, which is brought about by compounds that stimulate cell division in partially cancerous cells, might therefore be achieved by substances that either downregulate PKC δ or elevate PLD activity. Since many substances activate PLD, this could play a significant role in the promotion phase of tumor progression for cells that have an overexpressed tyrosine kinase such as the EGF receptor.
Weinberg and colleagues characterized complementation groups of oncogenes that transform primary cells (10, 15, 26). In their model, a signaling oncogene such as Ras or Src will cooperate with Myc or large T antigen to cause transformation (15, 26). Interestingly, tumor-promoting phorbol esters also were able to complement the signaling oncogenes but not Myc (10). In this regard, RalA and PLD1 would appear to substitute for either Myc or large T antigen in the model for cooperating oncogenes (15, 26). This would indicate that RalA and PLD1 facilitate passage through the G1/S cell cycle checkpoint, since this is where large T antigen exerts its effects (34). Since constitutive PLD activity is not tolerated well by cells, it is not likely that a PLD gene will be found to be an oncogene in any tumors. However, substances that activate PLD activity could very well contribute to the promotion phase of tumor progression by pushing cells overexpressing a tyrosine kinase past the G1/S cell cycle checkpoint into S phase. Consistent with this hypothesis, inhibition of PKC δ results in an increase in DNA synthesis that can be detected with 2 h (28).
Three different defective RalA mutants blocked the transformed phenotype induced by EGF. The ΔN11 mutant is defective in recruiting the PLD activator protein Arf into a RalA-PLD complex (31). Thus, the ability of this mutant to block transformation suggests that Arf is required for the activation of PLD by EGF and further implicates Arf as a signaling molecule. The D49N effector domain RalA mutant also blocked the transformed phenotype induced by EGF. We demonstrated previously that an activated RalA was not sufficient to induce either PLD activity (22) or transformation (42) in nontransformed NIH 3T3 cells. Therefore, we were somewhat surprised that the D49N was as effective as the ΔN11 mutant in blocking the EGF-induced transformation. The D49N mutant is defective in binding to the RalA effector molecule Ral-BP1, a GTPase-activating protein for Rho family GTPases (4), which have also been implicated in the regulation of PLD activity (11). Thus, it is likely that the regulation of PLD activity in response to mitogenic signaling is complex and involves multiple small GTPases of the Ras, Ral, Arf, and Rho families.
The role that PLD plays in mitogenic signaling is not clear. PLD converts phosphatidylcholine to phosphatidic acid, which results in a significant change in the charge and pH at the membrane. PLD has been implicated in vesicle formation in the Golgi apparatus (3, 5, 25). Many signaling molecules including Ras, Src, and the EGF receptor localize to caveolin-enriched light membrane fractions (2), and we have found that there is an enrichment of RalA in these membrane microdomains (unpublished data). Similarly, both PLD1 and PLD2 are present in caveolae (7, 24). Thus, PLD may in some way regulate signaling molecules in this plasma membrane microdomain, where so many signaling molecules are localized. We speculated previously that PLD activity may contribute to the formation of “signaling vesicles” that are endocytosed from the plasma membrane (31). In this regard, it is interesting that RalA is required for EGF-induced receptor-mediated endocytosis (33) and that endocytosis is required for many of the signals generated in response to EGF (43). Thus, it is possible that the role PLD plays in the transduction of intracellular signals is similar to that proposed for protein trafficking in the Golgi apparatus (3, 5, 25), i.e., the generation of endocytic signaling vesicles. How such a vesicle might carry mitogenic signals remains to be determined.
ACKNOWLEDGMENTS
We thank Andrew Morris for the Flu-tagged hPLD1 expression vector (pCGN-hPLD1) and Johannes Bos for the GST-Ral-BD clone.
This investigation was supported by grants from the National Institutes of Health (CA46677) and the American Cancer Society (BE-243) (to D.A.F.), National Institutes of Health GM47707 and an American Cancer Society faculty research award (to L.A.F.), and a Research Centers in Minority Institutions (RCMI) award from the Division of Research Resources, National Institutes of Health (RR-03037), to Hunter College.
REFERENCES
- 1.Aguirre Ghiso J, Frankel P, Lu Z, Jiang H, Farías E, Olsen A, Feig L A, Bal de Kier Joffé E, Foster D A. RalA requirement for v-Src- and v-Ras-induced tumorigenicity and overproduction of urokinase-type plasminogen activator and metalloproteases. Oncogene. 1999;18:4718–4725. doi: 10.1038/sj.onc.1202850. [DOI] [PubMed] [Google Scholar]
- 2.Anderson R G W. The caveolae membrane system. Annu Rev Biochem. 1998;67:199–225. doi: 10.1146/annurev.biochem.67.1.199. [DOI] [PubMed] [Google Scholar]
- 3.Bi K, Roth M G, Ktistakis N T. Phosphatidic acid formation by phospholipase D is required for transport from the endoplasmic reticulum to the Golgi complex. Curr Biol. 1997;7:301–307. doi: 10.1016/s0960-9822(06)00153-9. [DOI] [PubMed] [Google Scholar]
- 4.Cantor S B, Urano T, Feig L A. Identification of Ral-binding protein 1, a potential downstream target of Ral GTPases. Mol Cell Biol. 1995;15:4578–4584. doi: 10.1128/mcb.15.8.4578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen Y-G, Siddhanta A, Austin C D, Hammond S M, Sung T-C, Frohman M A, Morris A J, Shields D. Phospholipase D stimulates release of nascent secretory vesicles from the trans-Golgi network. J Cell Biol. 1997;138:495–504. doi: 10.1083/jcb.138.3.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coppola D, Ferber A, Miura M, Sell C, D'Ambrosio C, Rubin R, Baserga R. A functional insulin-like growth factor I receptor is required for the mitogenic and transforming activities of the epidermal growth factor receptor. Mol Cell Biol. 1994;14:4588–4595. doi: 10.1128/mcb.14.7.4588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Czarny M, Lavie Y, Fiucci G, Liscovitch M. Localization of phospholipase D in detergent-insoluble, caveolin-rich membrane domains. Modulation by caveolin-1 expression and caveolin-182–101. J Biol Chem. 1999;274:2717–2724. doi: 10.1074/jbc.274.5.2717. [DOI] [PubMed] [Google Scholar]
- 8.Denning M F, Dlugosz A A, Howett M K, Yuspa S H. Expression of an oncogenic rasHa gene in murine keratinocytes induces tyrosine phosphorylation and reduced activity of protein kinase C δ. J Biol Chem. 1993;268:26079–26081. [PubMed] [Google Scholar]
- 9.Denning M F, Dlugosz A A, Threadgill D W, Magnuson T, Yuspa S H. Activation of the epidermal growth factor receptor signal transduction pathway stimulates tyrosine phosphorylation of protein kinase C δ. J Biol Chem. 1996;271:5325–5331. doi: 10.1074/jbc.271.10.5325. [DOI] [PubMed] [Google Scholar]
- 10.Dotto G P, Parada L F, Weinberg R A. Specific growth response of ras-transformed embryo fibroblasts to tumour promoters. Nature. 1985;318:472–475. doi: 10.1038/318472a0. [DOI] [PubMed] [Google Scholar]
- 11.Exton J H. Phospholipase D. Biochim Biophys Acta. 1998;1436:105–115. doi: 10.1016/s0005-2760(98)00124-6. [DOI] [PubMed] [Google Scholar]
- 12.Feig L A, Cooper G M. Inhibition of NIH 3T3 cell proliferation by a mutant ras protein with preferential affinity for GDP. Mol Cell Biol. 1988;8:3235–3243. doi: 10.1128/mcb.8.8.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Feig L A, Urano T, Cantor S. Evidence for a Ras/Ral signaling cascade. Trends Biochem Sci. 1996;21:438–441. doi: 10.1016/s0968-0004(96)10058-x. [DOI] [PubMed] [Google Scholar]
- 14.Feig L A. Tools of the trade: use of dominant inhibitory mutants of Ras-family GTPases. Nat Cell Biol. 1999;1:22–25. doi: 10.1038/10018. [DOI] [PubMed] [Google Scholar]
- 15.Hahn W C, Counter C M, Lundberg A S, Beijersbergen R L, Brooks M W, Weinberg R A. Creation of human tumour cells with defined genetic elements. Nature. 1999;400:464–468. doi: 10.1038/22780. [DOI] [PubMed] [Google Scholar]
- 16.Hammond S M, Altshuller Y M, Sung T C, Rudge S A, Ross K, Engebrecht J, Morris A J, Frohman M A. Human Arf-activated phosphatidylcholine-specific phospholipase D defines a new highly conserved gene family. J Biol Chem. 1995;270:29640–29643. doi: 10.1074/jbc.270.50.29640. [DOI] [PubMed] [Google Scholar]
- 17.Hofer F, Fields S, Schneider C, Martin G S. Activated Ras interacts with the Ral guanine nucleotide dissociation stimulator. Proc Natl Acad Sci USA. 1994;91:11089–11093. doi: 10.1073/pnas.91.23.11089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hofer F, Berdeaux R, Martin G S. Ras-independent activation of Ral by a Ca(2+)-dependent pathway. Curr Biol. 1998;8:839–842. doi: 10.1016/s0960-9822(98)70327-6. [DOI] [PubMed] [Google Scholar]
- 19.Hornia A, Lu Z, Sukezane T, Zhong M, Foster D A. Antagonistic effects of protein kinase C α and δ on both transformation and phospholipase D activity mediated by the EGF receptor. Mol Cell Biol. 1999;19:7672–7680. doi: 10.1128/mcb.19.11.7672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Howe L R, Leevers S J, Gomez N, Nakielny S, Cohen P, Marshall C J. Activation of the MAP kinase pathway by the protein kinase raf. Cell. 1992;71:335–342. doi: 10.1016/0092-8674(92)90361-f. [DOI] [PubMed] [Google Scholar]
- 21.Hunter T. The Croonian Lecture 1997. The phosphorylation of proteins on tyrosine: its role in cell growth and disease. Philos Trans R Soc London Ser B. 1998;353:583–605. doi: 10.1098/rstb.1998.0228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jiang H, Luo J-Q, Urano T, Lu Z, Foster D A, Feig L. Involvement of Ral GTPase in v-Src-induced phospholipase D activation. Nature. 1995;378:409–412. doi: 10.1038/378409a0. [DOI] [PubMed] [Google Scholar]
- 23.Kikuchi A, Demo S D, Ye Z, Chen Y, Williams L T. RalGDS family members interact with the effector loop of ras p21. Mol Cell Biol. 1994;14:7483–7491. doi: 10.1128/mcb.14.11.7483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim J H, Han J M, Lee S, Kim Y, Lee T G, Park J B, Lee S D, Suh P G, Ryu S H. Phospholipase D1 in caveolae: regulation by protein kinase Cα and caveolin-1. Biochemistry. 1999;38:3763–3769. doi: 10.1021/bi982478+. [DOI] [PubMed] [Google Scholar]
- 25.Ktistakis N T, Brown H A, Waters M G, Sternweis P C, Roth M G. Evidence that phospholipase D mediates ADP-ribosylation factor-dependent formation of coated vesicles. J Cell Biol. 1996;134:295–306. doi: 10.1083/jcb.134.2.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Land H, Parada L F, Weinberg R A. Tumorigenic conversion of primary embryo fibroblasts requires at least two cooperating oncogenes. Nature. 1983;304:596–602. doi: 10.1038/304596a0. [DOI] [PubMed] [Google Scholar]
- 27.Logan S K, Falasca M, Hu P, Schlessinger J. Phosphatidylinositol 3-kinase mediates epidermal growth factor-induced activation of the c-Jun N-terminal kinase signaling pathway. Mol Cell Biol. 1997;17:5784–5790. doi: 10.1128/mcb.17.10.5784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lu Z, Hornia A, Jiang Y-W, Frankel P, Zang Q, Foster D A. Tumor promotion by depleting cells of protein kinase C δ. Mol Cell Biol. 1997;17:3418–3428. doi: 10.1128/mcb.17.6.3418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lu Z, Liu D, Hornia A, Devonish W, Pagano M, Foster D A. Activation of protein kinase C triggers its ubiquitination and degradation. Mol Cell Biol. 1998;18:839–845. doi: 10.1128/mcb.18.2.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Luo J-Q, Liu X, Hammond S M, Colley W C, Feig L A, Frohman M A, Morris A J, Foster D A. Ral interacts directly with the Arf-responsive PIP2-dependent phospholipase D1. Biochem Biophys Res Commun. 1997;235:854–859. doi: 10.1006/bbrc.1997.6793. [DOI] [PubMed] [Google Scholar]
- 31.Luo J-Q, Liu X, Frankel P, Rotunda T, Ramos M, Flom J, Jiang H, Feig L A, Morris A J, Kahn R A, Foster D A. Functional association between RalA and Arf in active phospholipase D complexes. Proc Natl Acad Sci USA. 1998;95:3632–3637. doi: 10.1073/pnas.95.7.3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marshall C J. Ras effectors. Curr Opin Cell Biol. 1996;8:197–204. doi: 10.1016/s0955-0674(96)80066-4. [DOI] [PubMed] [Google Scholar]
- 33.Nakashima S, Morinaka K, Koyama S, Ikeda M, Kishida M, Okawa K, Iwamatsu A, Kishida S, Kikuchi A. Small G protein Ral and its downstream molecules regulate endocytosis of EGF and insulin receptors. EMBO J. 1999;18:3629–3642. doi: 10.1093/emboj/18.13.3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nevins J R. Disruption of cell-cycle control by viral oncoproteins. Biochem Soc Trans. 1993;21:935–938. doi: 10.1042/bst0210935. [DOI] [PubMed] [Google Scholar]
- 35.Reese D M, Slamon D J. HER-2/neu signal transduction in human breast and ovarian cancer. Stem Cells. 1997;15:1–8. doi: 10.1002/stem.150001. [DOI] [PubMed] [Google Scholar]
- 36.Schaap D, van der Wal J, Howe L R, Marshall C J, van Blitterswijk W J. A dominant-negative mutant of raf blocks mitogen-activated protein kinase activation by growth factors and oncogenic p21ras. J Biol Chem. 1993;268:20232–20236. [PubMed] [Google Scholar]
- 37.Song J, Pfeffer L M, Foster D A. v-Src increases diacylglycerol levels via a type D phospholipase-mediated hydrolysis of phosphatidylcholine. Mol Cell Biol. 1991;11:4903–4908. doi: 10.1128/mcb.11.10.4903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Song J, Foster D A. v-Src activates a phospholipase D activity that is distinguishable from phospholipase D activity activated by protein kinase C. Biochem J. 1993;294:711–717. doi: 10.1042/bj2940711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Song J, Jiang Y-W, Foster D A. EGF induces the production of biologically distinguishable diglyceride species from phosphatidylinositol and phosphatidylcholine: evidence for the independent activation of type C and type D phospholipases. Cell Growth Differ. 1994;5:79–85. [PubMed] [Google Scholar]
- 40.Spaargaren M, Bischoff J R. Identification of the guanine nucleotide dissociation stimulator for Ral as a putative effector molecule of R-ras, H-ras, K-ras and Rap. Proc Natl Acad Sci USA. 1994;91:12609–12613. doi: 10.1073/pnas.91.26.12609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Spiegel S, Foster D A, Kolesnick R. Signal transduction through lipid second messengers. Curr Opin Cell Biol. 1996;8:159–167. doi: 10.1016/s0955-0674(96)80061-5. [DOI] [PubMed] [Google Scholar]
- 42.Urano T, Emkey R, Feig L A. Ral GTPases mediate a distinct downstream signaling pathway from Ras that facilitates cellular transformation. EMBO J. 1996;16:810–816. [PMC free article] [PubMed] [Google Scholar]
- 43.Vieira A V, Lamaze C, Schmid S L. Control of EGF receptor signaling by clathrin-mediated endocytosis. Science. 1996;274:2086–2089. doi: 10.1126/science.274.5295.2086. [DOI] [PubMed] [Google Scholar]
- 44.Walker F, Kato A, Gonez L J, Hibbs M L, Pouliot N, Levitzki A, Burgess A W. Activation of the Ras/mitogen-activated protein kinase pathway by kinase-defective epidermal growth factor receptors results in cell survival but not proliferation. Mol Cell Biol. 1998;18:7192–7204. doi: 10.1128/mcb.18.12.7192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wolthuis R M F, Franke B, van Triest M, Bauer B, Cool R H, Camonis J H, Akkerman J W, Bos J L. Activation of the small GTPase Ral in platelets. Mol Cell Biol. 1998;18:2486–2491. doi: 10.1128/mcb.18.5.2486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wolthuis R M F, Zwartkruis F, Moen T C, Bos J L. Ras-dependent activation of the small GTPase Ral. Curr Biol. 1998;8:471–474. doi: 10.1016/s0960-9822(98)70183-6. [DOI] [PubMed] [Google Scholar]
- 47.Yeo E J, Exton J H. Stimulation of phospholipase D by epidermal growth factor requires protein kinase C activation in Swiss 3T3 cells. J Biol Chem. 1995;270:3980–3988. doi: 10.1074/jbc.270.8.3980. [DOI] [PubMed] [Google Scholar]
- 48.Zang Q, Lu Z, Curto M, Barile N, Shalloway D, Foster D A. Interaction between v-Src and protein kinase C δ in v-Src-transformed fibroblasts. J Biol Chem. 1997;272:13275–13280. doi: 10.1074/jbc.272.20.13275. [DOI] [PubMed] [Google Scholar]