ABSTRACT
An investigation of members of the soil keratinophilic fungi community in China resulted in the identification of one new monotypic genus, Zongqia, and 10 new species, 2 of which are affiliated with Solomyces, 1 with the new genus Zongqia, 4 with Pseudogymnoascus, and 3 with Scedosporium. These novel taxa form an independent lineage distinct from other species, based on morphological and multilocus phylogenetic analyses. Descriptions, illustrations, and notes are provided for each taxon. These new taxa of the soil keratinophilic fungi add to the increasing number of fungi known from China, and it is now evident that numerous novel taxa are waiting to be described.
IMPORTANCE Keratinophilic fungi are a group that can degrade and utilize keratin-rich material. It is also because of this ability that many taxa can cause infections in animals or humans but remain poorly studied. In this study, we reported a novel genus and 10 novel species, 7 novel species belonging to the order Thelebolales and 3 to the genus Scedosporium, based on multilocus phylogenetic analyses combined with morphological characteristics. Our study significantly updates the taxonomy of Thelebolales and Scedosporium and enhances our understanding of this group of the keratin-degrading fungal community. The findings also encourage future studies on the artificially constructed keratin-degrading microbial consortia.
KEYWORDS: new taxa, keratinophilic fungi, Thelebolales, soil fungi, Zongqia, hair baiting technique
INTRODUCTION
Soil microbes are the richest component of terrestrial biodiversity, and among them, soil fungi play a major role in the ecosystem processes. To date, many studies have explored fungi in ocean, caves, forests (especially pristine rainforests), extreme environments, volcanoes, mountains, deserts, freshwater aquatic systems, lakes, grasslands, indoor environments, and many other habitats (1), and they have found that fungi in different habitats have very high species diversity. At the same time, many new fungal taxa have been reported, and they have shown potential high value in the industries of agriculture and medicine. However, as global urbanization continues to expand (2, 3), urban soil fungi, which are closely related to human health, have not been systematically investigated although they are a focal area for ecological and environmental issues. China has diverse urban soil types, diverse habitats, rapid urbanization, and high population mobility. Investigating the diversity of soil fungi in different cities in China will provide scientific data for understanding their ecological functions and maintaining public health safety and will enable the isolation of many new resources with potential applications.
The enrichment culture method using different substrates can often screen for the specific fungal consortium, so this method is often used for the isolation of fungal taxa with specific physiological functions. The distribution of keratinophilic fungi, as a special fungal consortium that can degrade and utilize keratin-rich materials, is greatly influenced by the activities of humans and animals, and the presence of such fungus is high in areas where humans and animals are frequently active, especially in urban parks, hospitals, and school campuses (4–7). According to the habitat, keratinophilic fungi can be broadly classified into three eco-types, anthropophilic, zoophilic, and geophilic species, and are mostly pathogenic or potentially pathogenic fungi. For human health and safety, their distribution should attract the attention of governments and scientists. Keratinophilic fungi have been reported in soils of different habitats in different geographic regions of the world, so the investigation of keratinophilic fungi has epidemiological significance (8).
Since the report of the degradable keratin of Onygena equina (9), new taxa of keratinophilic fungi and their applications have been reported. Keratinophilic fungi involve a large number of taxa belonging to several orders, families, and genera, including mainly dermatophytes and some saprophytic fungi, such as some species of Arthrodermataceae and Onygenaceae in the order Onygenales (10) and some members of the genera Geomyces and Pseudogymnoascus in the order Thelebolales (11). In addition, they contain a large number of common taxa, such as some species of the genera Aspergillus, Penicillium, and Trichoderma (12, 13). In the years since we investigated the members of keratin-degrading fungal communities in Chinese soils, several new taxa have been identified and reported (14–22). Here, we introduce one new genus, Zongqia (Thelebolales genera incertae sedis, Thelebolales), and 10 new species, 2 of which are affiliated with Solomyces, 1 with the new genus Zongqia, 4 with Pseudogymnoascus, and 3 with Scedosporium.
RESULTS
In this study, the internal transcribed spacer (ITS) regions of all isolates were sequenced, and all ITS sequences obtained were BLASTn searched in NCBI and assigned to potential genera and species. Then, strains belonging to Thelebolales and Scedosporium were screened and tested for further identification through morphological characterization and phylogenetic analyses.
Molecular phylogeny.
The first concatenated alignment (including Pseudogymnoascus and its related taxa) consisted of 2,806 nucleotides, including inserted gaps (ITS: 430 bp, large subunit ribosomal DNA [LSU]: 790 bp, minichromosomal maintenance protein 7 [MCM7]: 485 bp, RNA polymerase II subunit 2 [RPB2]: 467 bp, and elongation factor 1 alpha [EF1A]: 634 bp). The second concatenated data set (mainly involving the genera of Thelebolales) included 1,208 nucleotides, including inserted gaps (ITS: 433 bp; LSU: 775 bp). The third concatenated matrix (including Scedosporium and its related taxa) contained 964 nucleotides, including inserted gaps (ITS: 544 bp; beta-tubulin [BT2]: 420 bp). The best-fit evolutionary models of ML and BI analyses of each locus are listed in Table 1. The tree topology from both maximum likelihood (ML) and Bayesian interference (BI) analyses was almost identical.
TABLE 1.
The best-fit evolutionary models in our phylogenetic analyses
| Data set | Method | Model |
|||||
|---|---|---|---|---|---|---|---|
| ITS | BT2 | LSU | MCM7 | RPB2 | EF1A | ||
| First | ML | TIM2e + I + G4 | GTR + F + I | TVMe + I + G4 | K2P + I + G4 | SYM + R3 | |
| BI | SYM + I + G4 | TR + F + I | SYM + I + G4 | K2P + I + G4 | SYM + I + G4 | ||
| Second | ML | SYM + R3 | TIM + F + I + G4 | ||||
| BI | SYM + I + G4 | GTR + F + I + G4 | |||||
| Third | ML | TIM3 + F + R3 | HKY + F + R2 | ||||
| BI | HKY + F + I + G4 | HKY + F + G4 | |||||
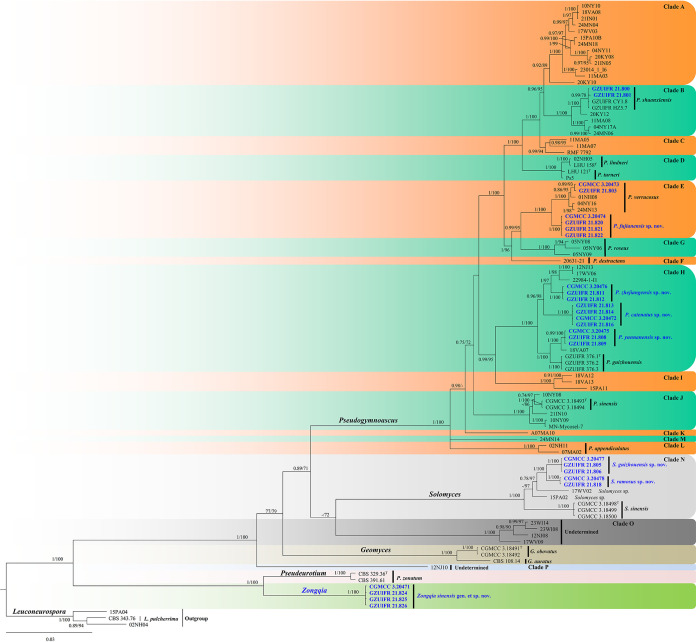
In the first phylogenetic tree (Fig. 1), the clades formed by each genus and by undetermined taxa had a high support rate: Pseudogymnoascus (1 posterior probability [PP]/100% bootstrap support [BS]), Solomyces (1 PP/100% BS), undetermined (clade O, 1 PP/100% BS), Geomyces (1 PP/100% BS), Pseudeurotium (1 PP/100% BS), and Zongqia gen. nov. (1 PP/100% BS). Our new species is divided into three genera. Eighteen of our new strains belong to three clades of genus Pseudogymnoascus, five are contained in genus Solomyces, and the remaining four are located within the new genus Zongqia.
FIG 1.
Bayesian inference strict consensus tree illustrating the phylogeny of new taxa and related species in Thelebolales based on a five-loci (ITS, LSU, MCM7, RPB2, EF1A) concatenated data set. Branches are labeled with Bayesian posterior probabilities of >0.70 and maximum likelihood bootstrap values of >70%. The new taxa and strains are in bold and blue. Clade names follow previous studies (21, 24).
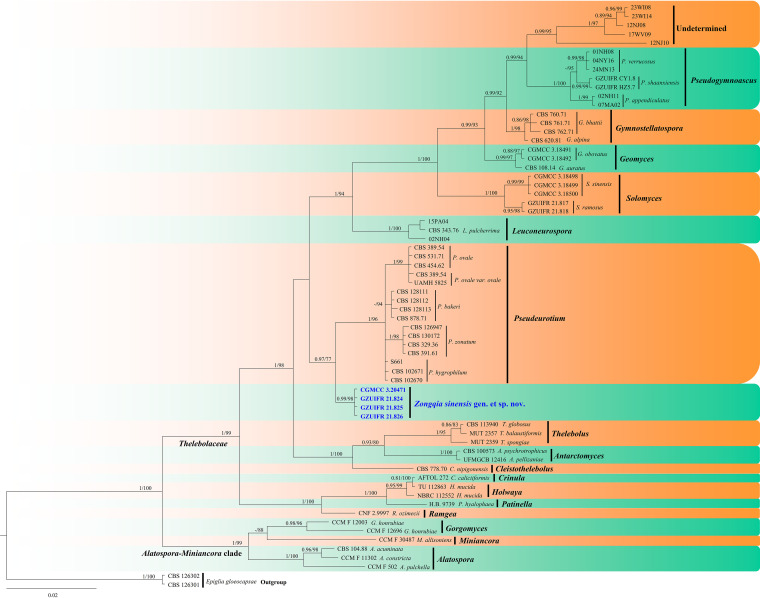
In the second phylogenetic tree (Fig. 2), each genus clusters into a monophyletic clade. The new genus Zongqia forms a well-supported (0.99 PP/98% BS) clade separated from other genera in Thelebolales.
FIG 2.
Bayesian inference strict consensus tree illustrating the phylogeny of genera in Thelebolales based on a two-loci (ITS and LSU) concatenated data set. Branches are labeled with Bayesian posterior probabilities of >0.70 and maximum likelihood bootstrap values of >70%. The new taxa and strains are in bold and blue.
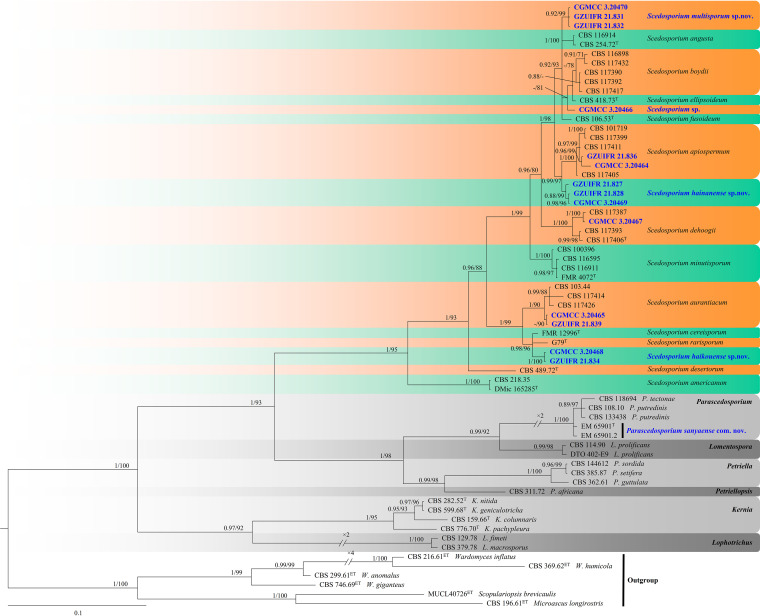
In the third phylogenetic tree (Fig. 3), the clades formed by each genus had a high support rate: Scedosporium (1 PP/95% BS), Parascedosporium (1 PP/100% BS), Lomentospora (0.99 PP/98% BS), Petriella (1 PP/100% BS), Kernia (1 PP/95% BS), and Lophotrichus (1 PP/100% BS). Our new species is nested in Scedosporium, and our strains are spread into five well-supported main clades, representing the species Scedosporium hunanense sp. nov. (0.92 PP/99% BS), Scedosporium apiospermum (1 PP/100% BS), Scedosporium hainanense sp. nov. (0.88 PP/99% BS), Scedosporium aurantiacum (1 PP/90% BS), and Scedosporium haikouense sp. nov. (1 PP/100% BS), except for CGMCC3.20466, which is associated with the species Scedosporium boydii and Scedosporium ellipsoideum.
FIG 3.
Phylogeny of Scedosporium and related species generated by BI analyses based on combined two-loci (ITS and BT2) sequences. Branches are labeled with Bayesian posterior probabilities of >0.70 and maximum likelihood bootstrap of >70%, respectively. New species and strains are indicated in bold and blue.
TAXONOMY
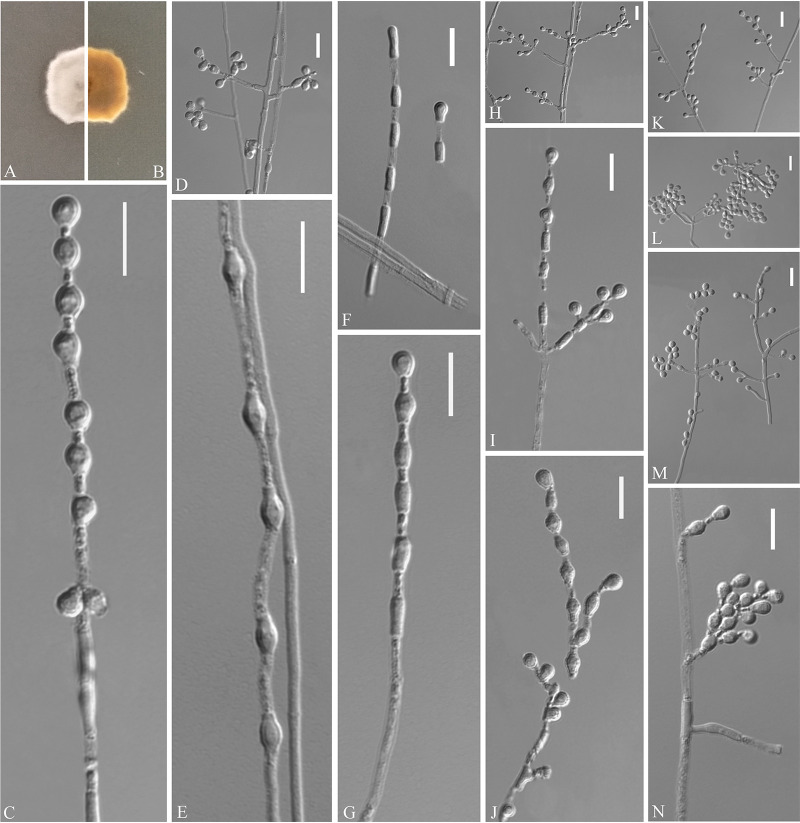
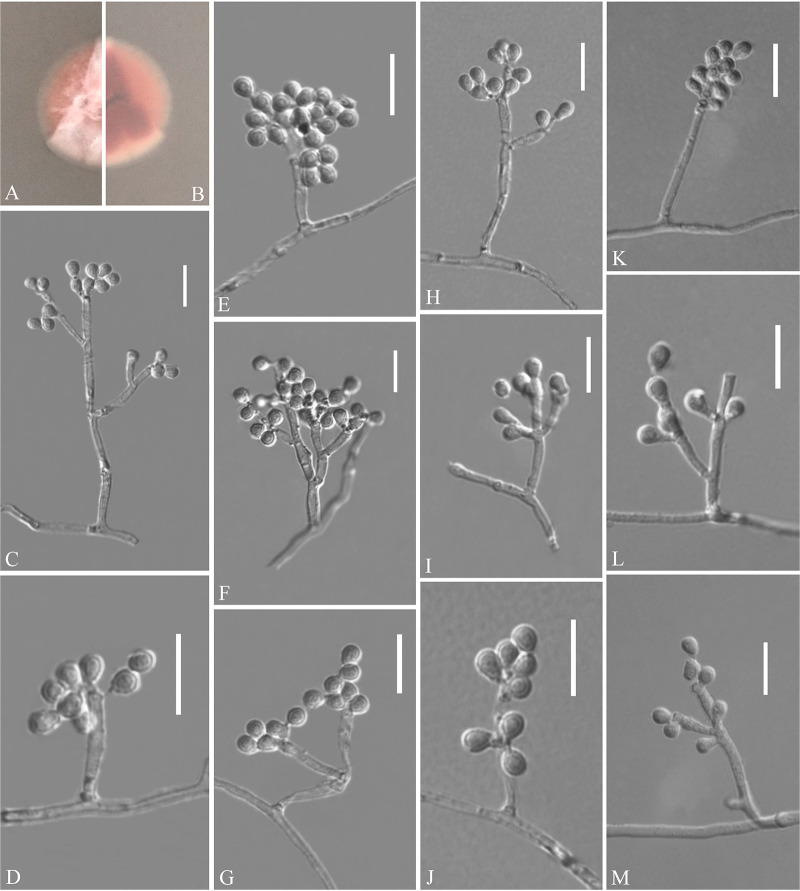
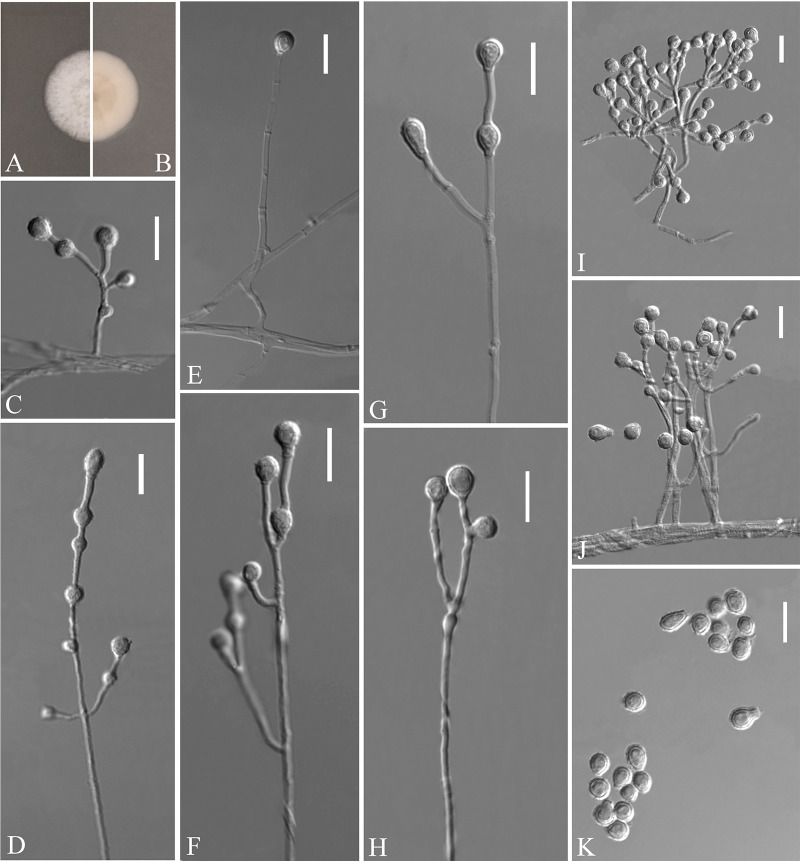
Pseudogymnoascus catenatus Zhang, Han, and Liang, sp. nov. (Fig. 4). MycoBank number: MB 840436. Etymology: referring to the catenation of its intercalary conidia. Diagnosis: similar to Pseudogymnoascus verrucosus but differs in obovoid conidia and intercalary conidia. Type: China, Fujian Province, Wuyishan City, Lie Ning Park, 27.758010N, 118.034403E, isolated from green belt soil, 18 August 2019, Z.Y. Zhang. (Holotype HMAS 350322, stored in a metabolically inactive state; ex-holotype culture CGMCC 3.20472 = GZUIFR 21.815, ibid., GZUIFR 21.816.) GenBank: MZ444080, MZ444081 (ITS); MZ444107, MZ444108 (LSU); MZ490762, MZ490763 (MCM7); MZ488545, MZ488546 (RPB2); MZ488522, MZ488523 (translation elongation factor [TEF]).
FIG 4.
Pseudogymnoascus catenatus (from ex-holotype CGMCC 3.20472). (A, B) Upper and reverse views of culture on PDA 14 days after inoculation; (C, E, G, I, J) intercalary conidia; (D, H, K to N) conidiophores and conidia; (F) arthroconidia. Scale bars (C to N), 10 μm.
Description. Sexual morph: not observed. Asexual morph: colonies on peptone-dextrose agar (PDA) slowly growing, attaining 6 to 10 mm diameter after 14 days at 25°C, velvety, short and fluffy, margins irregular, light gray to white, absent pigment and exudates; reverse brown. No growth at 37°C. Hyphae hyaline, branched, septate, smooth, 1 to 3 μm wide. Racquet hyphae absent. Conidiophores abundant, frequent branches, at acute angles, often 1 to 2 verticillate with 1 to 4 branches per whorl, secondary and tertiary branches can still branch again. Conidia abundant, normally borne terminally on verticillate branches or borne laterally and solitary on short protrusions or short side branches; subhyaline to hyaline, smooth-walled or rough; obovoid, sometimes subglobose, 3.0 to 6.0 by 3.0 to 4.0 μm (n = 50). Intercalary conidia are borne on the verticillate hyphae or hyphae, solitary or 1 to 6 in chains, smooth-walled or rough, obovoid, subglobose, fusiform, drum-shaped, truncated at both ends, 3.5 to 6.5 by 3.0 to 4.5 μm (n = 50), cylindrical, barrel-shaped, truncated at both ends, 5.5 to 6.5 by 2.5 to 3.5 μm (n = 50). Arthroconidia hyaline, cylindrical, sometimes obovoid, 3.0 to 6.0 by 2.0 to 3.5 μm (n = 50).
Substrate: soil. Distribution: Wuyishan City, Fujian Province; Ningbo City, Zhejiang Province, China. Material examined: China, Zhejiang Province, Ningbo City, Moon Lake, 29.870001N, 121.544021E, isolated from green belt soil, 16 August 2019, Z.Y. Zhang, GZUIFR 21.813, ibid., GZUIFR 21.814. GenBank: MZ444078, MZ444079 (ITS); MZ444105, MZ444106 (LSU); MZ490760, MZ490761 (MCM7); MZ488543, MZ488544 (RPB2); MZ488520, MZ488521 (TEF).
Notes. Morphologically, Pseudogymnoascus catenatus is similar to P. verrucosus in having arthroconidia but is clearly distinguished by the obovoid conidia and intercalary conidia (23). Phylogenetically, four isolates of P. catenatus formed a single clade separate from other species in Pseudogymnoascus (Fig. 1), which indicates that they are distinct species.
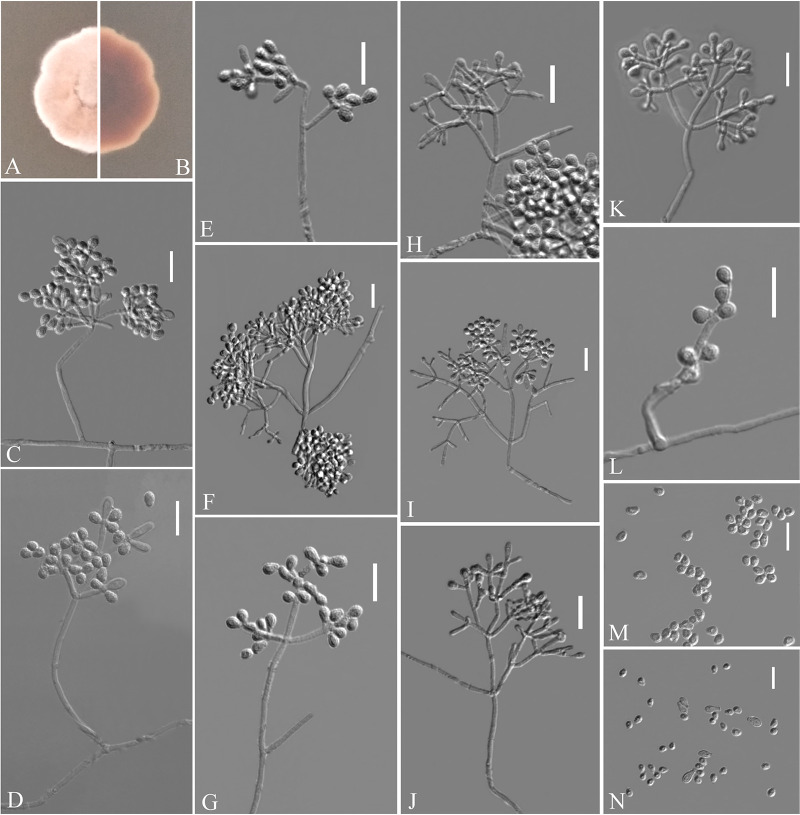
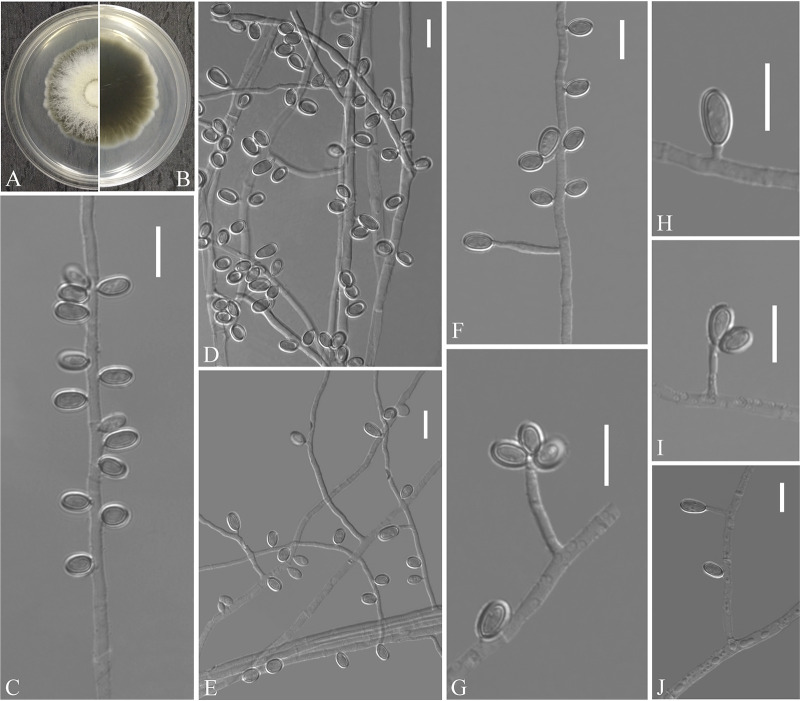
Pseudogymnoascus fujianensis Zhang, Han, and Liang, sp. nov. (Fig. 5). MycoBank number: MB 840437. Etymology: refers to the region from which the fungus was isolated. Diagnosis: similar to P. verrucosus, Pseudogymnoascus roseu, and Pseudogymnoascus destructans but differs in the presence of intercalary conidia and the absence of arthroconidia. Type: China, Fujian Province, Wuyishan City, Lie Ning Park, 27.758545N, 118.034134E, isolated from green belt soil, 18 August 2019, Z.Y. Zhang. (Holotype HMAS 350324, stored in a metabolically inactive state; ex-holotype culture CGMCC 3.20474 = GZUIFR 21.819, ibid., GZUIFR 21.820.) GenBank: MZ444084, MZ444085 (ITS); MZ444111, MZ444112 (LSU); MZ490766, MZ490767 (MCM7); MZ488549, MZ488550 (RPB2); MZ488526, MZ488527 (TEF).
FIG 5.
Pseudogymnoascus fujianensis (from ex-holotype CGMCC 3.20474). (A, B) Upper and reverse views of culture on PDA 14 days after inoculation; (C to M) conidiophores, conidia, and intercalary conidia. Scale bars (C to M), 10 μm.
Description. Sexual morph: not observed. Asexual morph: colonies on PDA attaining 19 to 20 mm diameter after 14 days at 25°C, flat, flocculent, sectorization, margin identified, white to pink, absent pigment and exudates; reverse brown. No growth at 37°C. Hyphae hyaline, branched, septate, smooth-walled, 0.5 to 3.5 μm wide. Racquet hyphae absent. Conidiophores abundant, branches, at acute angles, irregular, acyclic arrangement. Conidia abundant, mostly terminal or lateral, sessile or borne on hyphae, short protrusions or side branches; solitary, fasciation, or 2 in chains; hyaline, smooth-walled; obovoid, 2.5 to 5.5 by 2.5 to 4.0 μm (n = 50). Intercalary conidia abundant, normally chained with terminal conidia; solitary, smooth-walled or rough; obovoid, sometimes drum-shaped, 2.5 to 5.0 by 2.5 to 3.5 μm (n = 50).
Substrate: soil. Distribution: Wuyishan City, Fujian Province, China. Material examined: China, Fujian Province, Wuyishan City, Wuyi University, 27.728722N, 118.002862E, isolated from green belt soil, 18 August 2019, Z.Y. Zhang, GZUIFR 21.821, ibid., GZUIFR 21.822. GenBank: MZ444086, MZ444087 (ITS); MZ444113, MZ444114 (LSU); MZ490768, MZ490769 (MCM7); MZ488551, MZ488552 (RPB2); MZ488528, MZ488529 (TEF).
Notes. Morphological and phylogenetic analyses (Fig. 1) support our four strains as new species of Pseudogymnoascus fujianensis. P. fujianensis is phylogenetically closely related to P. verrucosus, P. roseu, and P. destructans. However, P. fujianensis is distinguished from other species of Pseudogymnoascus by the presence of intercalary conidia and the absence of arthroconidia (23–26).
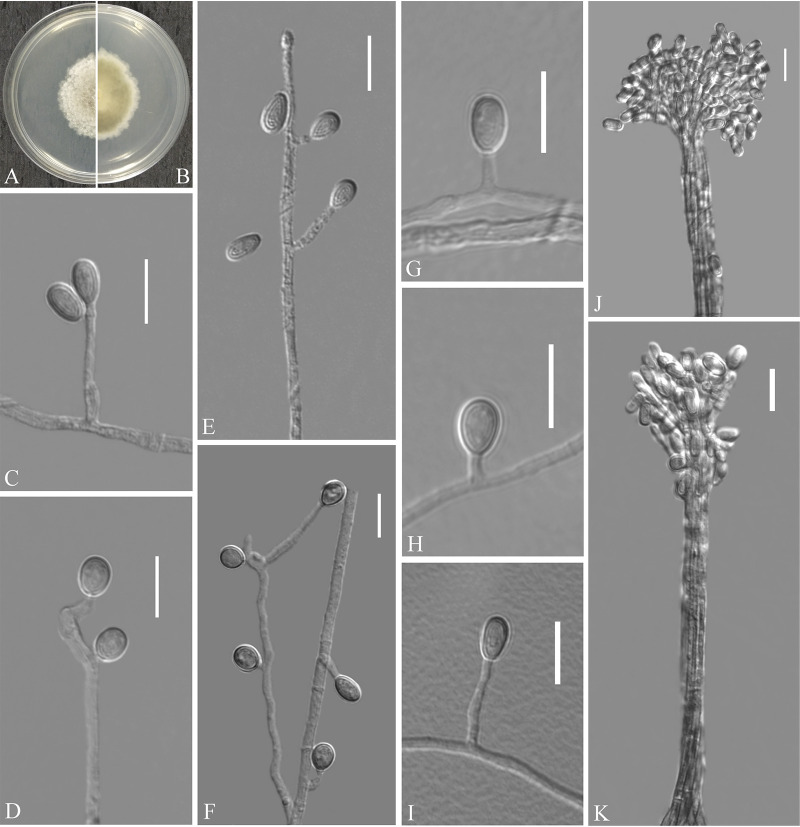
Pseudogymnoascus yunnanensis Zhang, Han, and Liang, sp. nov. (Fig. 6). MycoBank number: MB 840438. Etymology: refers to the region from which the fungus was isolated. Diagnosis: similar to Pseudogymnoas lindneri, Pseudogymnoas turneri, and Pseudogymnoas guizhouensis but differs in the clavate, fusiform with basal scars terminal conidia, and reniform, fusiform, truncated at both ends of intercalary conidia. Type: China, Yunnan Province, Dali City, Dali Bai Autonomous Prefecture People’s Hospital, 25.578478N, 100.222121E, isolated from green belt soil, 3 September 2019, Z.Y. Zhang. (Holotype HMAS 350320, stored in a metabolically inactive state; ex-holotype culture CGMCC 3.20475 = GZUIFR 21.807, ibid., GZUIFR 21.808.) GenBank: MZ444072, MZ444073 (ITS); MZ444099, MZ444100 (LSU); MZ490754, MZ490755 (MCM7); MZ488537, MZ488538 (RPB2); MZ488514, MZ488515 (TEF).
FIG 6.
Pseudogymnoascus yunnanensis (from ex-holotype CGMCC 3.20475). (A, B) Upper and reverse views of culture on PDA 14 days after inoculation; (C to L) conidiophores and conidia; (M to N) conidia. Scale bars (C to N), 10 μm.
Description. Sexual morph: not observed. Asexual morph: colonies on PDA attaining 23 to 25 mm diameter after 14 days at 25°C, velvety, powdery, margin identified, locally indented, pink, white at the edge, absent pigment and exudates; reverse brown. No growth at 37°C. Hyphae hyaline, branched, septate, smooth-walled, 1 to 3 μm wide. Racquet hyphae absent. Conidiophores abundant, frequent branches, at acute angles, often 2 to 3 verticillate with 1 to 4 branches per whorl, secondary and tertiary branches can still branch again. Conidia abundant, normally borne terminally on verticillate branches, or borne laterally and solitary on short protrusions or short side branches; subhyaline to hyaline, smooth-walled or echinulate; obovoid, subglobose to globose, sometimes pyriform, 2.5 to 4.5 by 2.5 to 3.5 μm (n = 50); sometimes terminal conidia clavate, fusiform with basal scars, 6.5 to 9.0 by 2.5 to 4.5 μm (n = 50). Intercalary conidia are borne on the outer branches of the hyphae or verticillate hyphae, solitary or two in chains, smooth-walled or rough, reniform and fusiform truncate at both ends, 2.5 to 5.5 by 2.5 to 4.0 μm (n = 50).
Substrate: soil. Distribution: Dali City, Yunnan Province, China. Material examined: China, Yunnan Province, Dali City, Dali University, 25.674141N, 100.154757E, isolated from green belt soil, 2 September 2019, Z.Y. Zhang, GZUIFR 21.809. GenBank: MZ444074 (ITS); MZ444101 (LSU); MZ490756 (MCM7); MZ488539 (RPB2); MZ488516 (TEF).
Notes. Morphologically, Pseudogymnoascus yunnanensis is similar to P. lindneri, P. turneri, and P. guizhouensis in having obovoid, globose conidia (27). However, P. yunnanensis can be distinguished from P. lindneri and P. turneri by the presence of its clavate, fusiform with basal scars terminal conidia and no observed sexual morph. P. yunnanensis differs from P. guizhouensis because it is reniform, fusiform, and truncated at both ends of intercalary conidia (22). Phylogenetically, three isolates of P. yunnanensis constitute a strongly supported subclade, sister to P. guizhouensis with high support values (Fig. 1), but they can be easily distinguished.
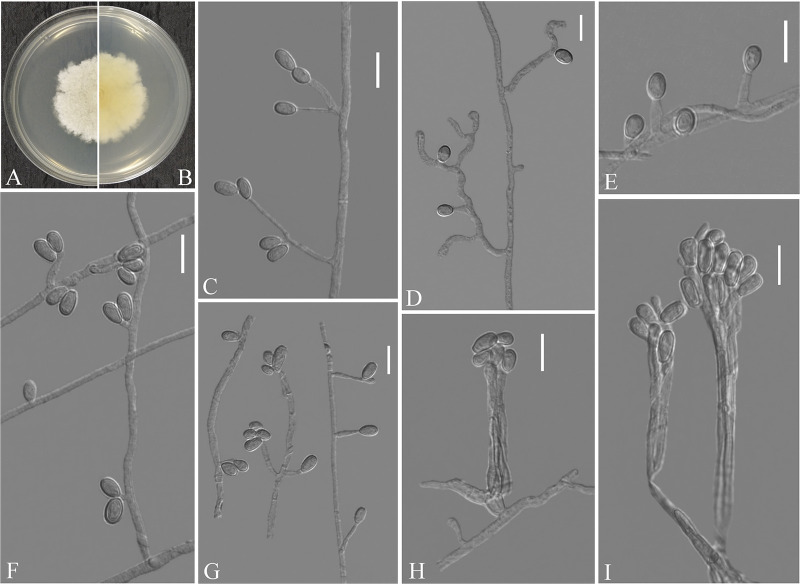
Pseudogymnoascus zhejiangensis Zhang, Han, and Liang, sp. nov. (Fig. 7). MycoBank number: MB 840439. Etymology: refers to the region from which the fungus was isolated. Diagnosis: similar to P. lindneri, P. turneri, and P. yunnanensis but differs in the obovoid, subglobose intercalary conidia. Type: China, Zhejiang Province, Ningbo City, Moon Lake, 29.871117N, 121.544218E, isolated from green belt soil, 16 August 2019, Z.Y. Zhang. (Holotype HMAS 350321, stored in a metabolically inactive state; ex-holotype culture CGMCC 3.20476 = GZUIFR 21.810, ibid., GZUIFR 21.811; ibid., GZUIFR 21.812.) GenBank: MZ444075, MZ444076, MZ444077 (ITS); MZ444102, MZ444103, MZ444104 (LSU); MZ490757, MZ490758, MZ490759 (MCM7); MZ488540, MZ488541, MZ488542 (RPB2); MZ488517, MZ488518, MZ488519 (TEF).
FIG 7.
Pseudogymnoascus zhejiangensis (from ex-holotype CGMCC 3.20476). (A, B) Upper and reverse views of culture on PDA 14 days after inoculation; (C to M) conidiophores, conidia, and intercalary conidia. Scale bars (C to M), 10 μm.
Description. Sexual morph: not observed. Asexual morph: colonies on PDA attaining 20 mm diameter after 14 days at 25°C, gradually increased from the edge to the center, velvety, floccose, margin entire, white, absent pigment and exudates; reverse pink, white at the edge. No growth at 37°C. Hyphae hyaline, branched, septate, smooth, 1 to 3 μm wide. Racquet hyphae absent. Conidiophores abundant, frequent branches, at acute angles, often 1 to 4 verticillate with 1 to 4 branches per whorl, secondary and tertiary branches can still branch again. Conidia abundant, normally borne terminally on verticillate branches or borne laterally and solitary on short protrusions or short side branches; subhyaline to hyaline, smooth-walled or rough; obovoid to globose, 2.5 to 4.5 by 2.5 to 4.0 μm (n = 50); clavate, long obovoid, 5 to 9 by 2.5 to 4 μm (n = 50). Intercalary conidia are borne on the verticillate hyphae or hyphae, solitary, smooth-walled or rough, obovoid, subglobose to globose, 3.5 to 4.5 by 3.0 to 4.0 μm (n = 50).
Substrate: Soil. Distribution: Ningbo City, Zhejiang Province, China.
Notes. Morphologically, Pseudogymnoascus zhejiangensis resembles P. lindneri, P. turneri, and P. yunnanensis because of the obovoid, globose conidia. However, P. zhejiangensis differs from P. lindneri, P. turneri, and P. yunnanensis in that it has obovoid, subglobose intercalary conidia (the intercalary conidia of P. linderi and P. turneri are globose to truncate, and those of P. yunnanensis are reniform, fusiform, and truncated at both ends) (27). Phylogenetically, three isolates of P. zhejiangensis formed one clade and share a sister relationship to three undescribed isolates (12NJ13, 17WV06, and 22984-1-I1) with high BS (Fig. 1). However, we did not compare morphological characteristics between P. zhejiangensis and another three isolates within Pseudogymnoascus because of the lack of morphological description of these three isolates (24).
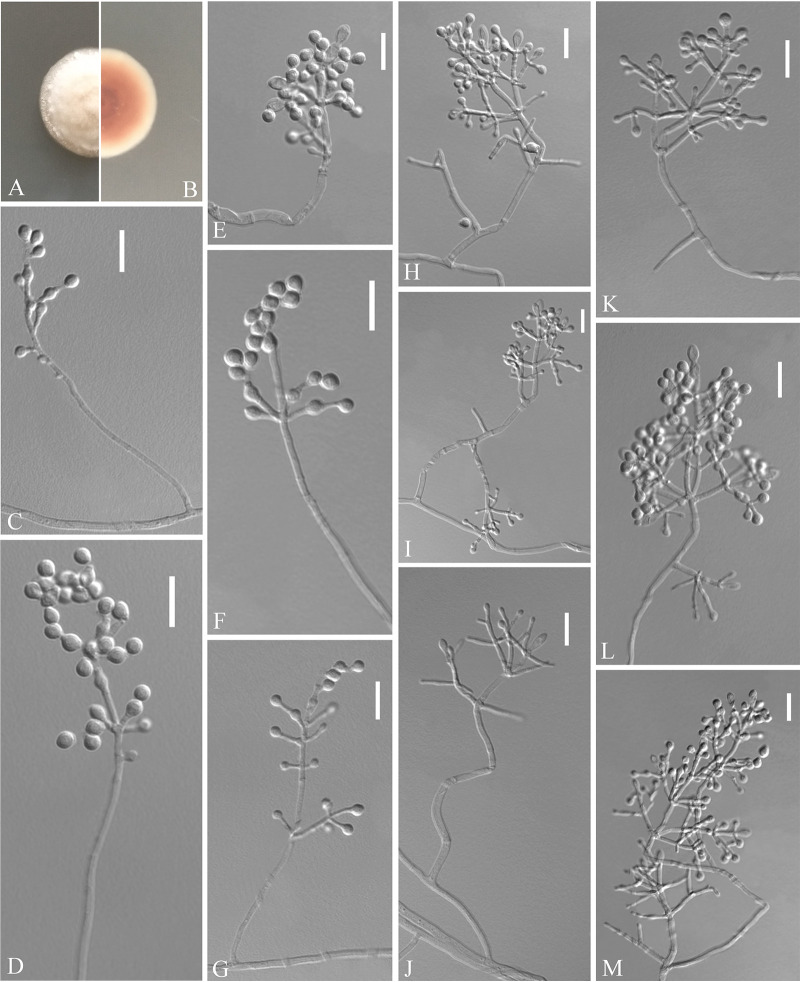
Solomyces guizhouensis Zhang, Han, and Liang, sp. nov. (Fig. 8). MycoBank number: MB 840440. Etymology: refers to Guizhou, the province where the isolate was collected. Diagnosis: Solomyces guizhouensis differs from other species by the presence of 2 to 3 conidia in chains and 2 to 3 intercalary conidia in chains. Type: China, Guizhou Province, Anshun City, Anshun University, 26.244748N, 105.898997E, isolated from green belt soil, 5 September 2019, Z.Y. Zhang. (Holotype HMAS 350319, stored in a metabolically inactive state; ex-holotype culture CGMCC 3.20477 = GZUIFR 21.804.) GenBank: MZ444069 (ITS); MZ444096 (LSU); MZ490751 (MCM7); MZ488534 (RPB2); MZ488511 (TEF).
FIG 8.
Solomyces guizhouensis (from ex-holotype CGMCC 3.20477). (A, B) Upper and reverse views of culture on PDA 14 days after inoculation; (C to M) terminal, lateral conidia, and intercalary conidia. Scale bars (C to M), 10 μm.
Description. Sexual morph: not observed. Asexual morph: colonies on PDA, reaching 16 to 17 mm diameter after 14 days at 25°C, floccose, margins regular, white, absent pigment and exudates; reverse white. No growth at 37°C. Hyphae abundant, smooth and thin-walled, septate, 1.5 to 3.0 μm wide. Conidia terminal and laterally borne on hyphae, short protrusions, or side branches; solitary, sometimes 2 to 3 in chains, hyaline, smooth or rough walled, obovoid, subglobose to globose, pyriform, 4.0 to 7.0 by 4.0 to 6.0 μm (n = 50). Intercalary conidia abundant, solitary or 2 to 3 in chains, hyaline, smooth or rough walled, olivary, subglobose to globose, 4.5 to 8.5 by 3.5 to 5.0 μm (n = 50). Lateral branches may emerge from intercalary conidia.
Substrate: soil. Distribution: Anshun City, Guizhou Province, China. Material examined: China, Guizhou Province, Anshun City, People’s Hospital of Anshun City Guizhou Province, 26.247091N, 105.967968E, isolated from green belt soil, 5 September 2019, Z.Y. Zhang, GZUIFR 21.805, ibid., GZUIFR 21.806. GenBank: MZ444070, MZ444071 (ITS); MZ444097, MZ444098 (LSU); MZ490752, MZ490753 (MCM7); MZ488535, MZ488536 (RPB2); MZ488512, MZ488513 (TEF).
Notes. Morphologically, Solomyces guizhouensis is distinguished from other species of Solomyces by the presence of 2 to 3 conidia in chains and 2 to 3 intercalary conidia in chains. Solomyces guizhouensis is phylogenetically allied to Solomyces ramosus (Fig. 1), but they can be easily distinguished (see notes on S. ramosus [22]).
Solomyces ramosus Zhang, Han, and Liang, sp. nov. (Fig. 9). MycoBank number: MB 840442. Etymology: referring to the ramose of its conidiophore. Diagnosis: Solomyces ramosus differ from other species by the presence of ramose conidiophores. Type: China, Shanghai City, Ruijin Hospital, Shanghai Jiao Tong University School of Medicine, 31.212090N, 121.467721E, isolated from green belt soil, 15 August 2019, Z.Y. Zhang. (Holotype HMAS 350323, stored in a metabolically inactive state; ex-holotype culture CGMCC 3.20478 = GZUIFR 21.817, ibid., GZUIFR 21.818.) GenBank: MZ444082, MZ444083 (ITS); MZ444109, MZ444110 (LSU); MZ490764, MZ490765 (MCM7); MZ488547, MZ488548 (RPB2); MZ488524, MZ488525 (TEF).
FIG 9.
Solomyces ramosus (from ex-holotype CGMCC 3.20478). (A, B) Upper and reverse views of culture on PDA 14 days after inoculation; (C to H) terminal, lateral conidia, and intercalary conidia; (I and J) ramose of conidiophore; (K) conidia. Scale bars (C to K), 10 μm.
Description. Sexual morph: not observed. Asexual morph: colonies on PDA reaching 17 mm diameter after 14 days at 25°C, slightly felty to floccose, margin identified, white; reverse white; absent pigment and exudates. No growth at 37°C. Hyphae abundant, smooth, hyaline, branched, septate, 1.0 to 3.5 μm wide. Conidiophores abundant, branches, at acute angles, 1 to 2 verticillate with 1 to 4 branches per whorl. Conidia terminal and laterally borne on hyphae, short protrusions, or side branches, solitary, hyaline, obovoid, subglobose, smooth or rough walled, 5 to 8.5 by 4.0 to 5.5 μm (n = 50). Intercalary conidia abundant, globose, olivary, subglobose to globose, 3.5 to 6.5 by 3.5 to 5.0 μm (n = 50).
Substrate: soil. Distribution: Shanghai City, China.
Notes. Morphologically, Solomyces ramosus is distinguished from other species of Solomyces by the presence of ramose conidiophores (22). Phylogenetically, our two new isolates of S. ramosus formed one clade and share a sister relationship to S. guizhouensis with high BS (Fig. 1), which indicates that they are distinct species.
Zongqia Zhang and Han, gen. nov. MycoBank number: MB 840447. Typification: Zongqia sinensis Zhang and Han. Etymology: in honor of Zong-Qi Liang, acknowledging his contributions to our group. Diagnosis: in addition to the phylogenetic distinctions (Fig. 1 to 2), Zongqia differs from Pseudeurotium by the presence of chains of conidia, conidiophores degenerated into conidiophore cells, clavate conidiophores cells.
Description. Saprobic on the soil. Sexual morph: not observed. Asexual morph: hyphae branched, septate, smooth. Conidiophores not observed and were degenerated into conidiophore cells. Conidiophores cells hyaline, cylindrical, clavate, occurring directly from the hyphae, smooth-walled, solitary. Conidia aseptate, smooth-walled, one-celled, solitary or chains, obovate, subglobose, fusiform, cylindrical, clavate. Chlamydospores not observed.
Notes. The new genus Zongqia is introduced here based on phylogeny and morphological evidence. Until now, the Thelebolales consisted of 23 genera (22, 28). In five-loci (ITS, LSU, MCM7, RPB2, and EF1A; Fig. 1) and two-loci (ITS and LSU; Fig. 2) phylogenetic analyses, Zongqia was related to Pseudeurotium with high support values (1 PP/100% BS). However, because no ITS, LSU, MCM7, RPB2, and EF1A sequence data were reported for Ascophanus, Ascozonus, Caccobius, Coprobolus, Leptokalpion, Neelakesa, and Pseudascozonus (22), we could not compare the phylogenetic relationships between these genera and Zongqia. Morphologically, because there is no record of the asexual stage of Ascophanus, Ascozonus, Caccobius, Coprobolus, Leptokalpion, Neelakesa, and Pseudascozonus in the literature (29), we could not compare the morphology between these genera and Zongqia. Of the remaining genera, Zongqia is similar to Pseudeurotium, but there are still noteworthy differences between them. Zongqia is distinguished from Pseudeurotium by the presence of chains of conidia, conidiophores degenerated into conidiophore cells, clavate conidiophores cells, and no observed sexual morph.
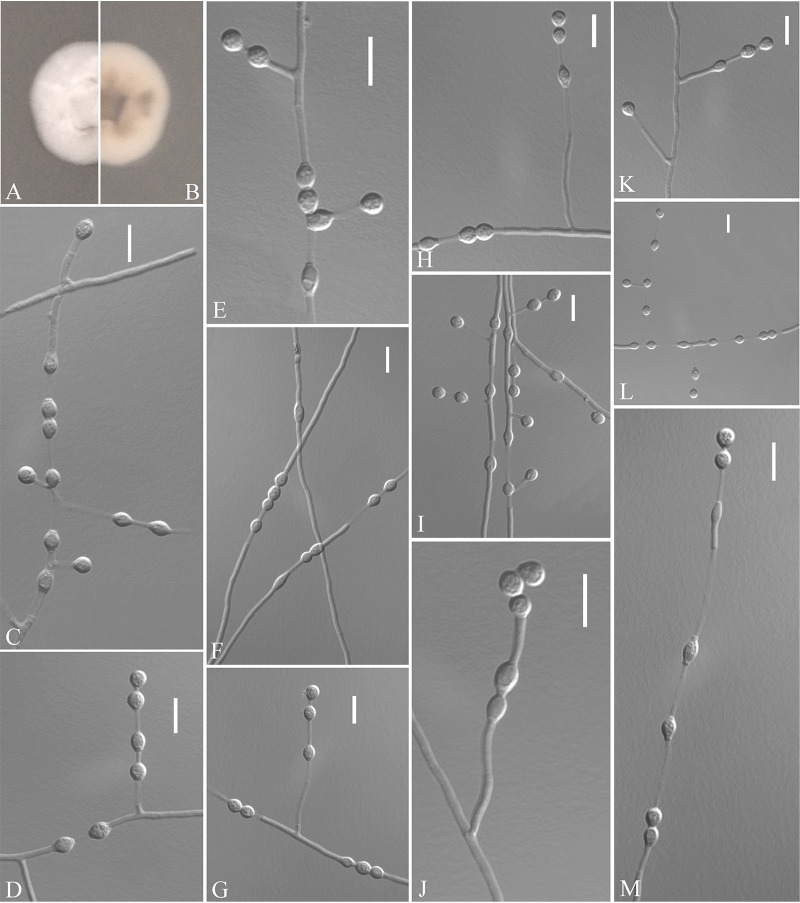
Zongqia sinensis Zhang and Han, sp. nov. (Fig. 10). MycoBank number: MB 840448. Etymology: named after China where the species is distributed. Diagnosis: the main diagnostic criteria of the species Zongqia sinensis are presence of chains of conidia, conidiophores degenerated into conidiophore cells, clavate conidiophores cells. Type: China, Guizhou Province, Guiyang, The Affiliated Hospital of Guizhou Medical University, 26.594218N, 106.713166E, isolated from green belt soil, 13 September 2019, Z.Y. Zhang. (Holotype HMAS 350325, stored in a metabolically inactive state; ex-holotype culture CGMCC 3.20471 = GZUIFR 21.823, ibid., GZUIFR 21.824.) GenBank: MZ444088, MZ444089 (ITS); MZ444115, MZ444116 (LSU); MZ490770, MZ490771 (MCM7); MZ488553, MZ488554 (RPB2).
FIG 10.
Zongqia sinensis (from ex-holotype CGMCC 3.20471). (A, B) Upper and reverse views of culture on PDA 14 days after inoculation; (C to E, H, I, L, M) conidia chains; (F) conidia borne on hyphae; (G) differentiation of conidiophore cells; (J) two conidia on the apex of conidiophore cells; (K) degenerated conidiophores; (N to P) solitary conidia. Scale bars (C to P), 10 μm.
Description. Sexual morph: not observed. Asexual morph: colonies grow slowly on PDA, reaching 11 to 13 mm diameter after 14 days at 25°C, suborbicular, white, floccose, margins regular; reverse white, no growth at 37°C. Hyphae hyaline, branched, septate, smooth, 1.5 to 3.5 μm wide. Conidiophores not observed but degenerated into conidiophore cells. Conidiophore cells hyaline, cylindrical, clavate, arising directly from the aerial hyphae, smooth-walled, solitary. Conidia aseptate, smooth-walled, one-celled, solitary, obovate to subobovoid, 5 to 9 by 3 to 5 μm (n = 50); or 2 to 20 in chains, obovate, subglobose, fusiform and obtuse at apex and base, sometimes cylindrical, clavate, 3.5 to 8.5 (to 12) by 2.5 to 4.5 μm (n = 50). Chlamydospores not observed.
Substrate: soil. Distribution: Guiyang City, Guizhou Province, China. Material examined: China, Guizhou Province, Guiyang, Guizhou University, 26.444504N, 106.669296E, isolated from green belt soil, 13 September 2019, Z.Y. Zhang, GZUIFR 21.825. GenBank: MZ444090 (ITS); MZ444117 (LSU); MZ490772 (MCM7); MZ488555 (RPB2). Guizhou Province, Guiyang, Qianlingshan Park, 26.592019N, 106.695434E, isolated from green belt soil, 13 September 2019, Z.Y. Zhang, GZUIFR 21.826. GenBank: MZ444091 (ITS); MZ444118 (LSU); MZ490773 (MCM7); MZ488556 (RPB2).
Notes. Based on multilocus phylogenetic analyses (Fig. 1 and 2) and similar morphological characteristics, the four strains are regarded as the same species, which cluster together very well and form a single clade separated from other species of Thelebolales. Morphologically, Zongqia sinensis is the only species that produces the conidia chains in this order. Therefore, based on both morphological and phylogenetic evidence, Z. sinensis is proposed as a novel species as a type of Zongqia.
Scedosporium haikouense Zhang, Han, and Liang, sp. nov. (Fig. 11). MycoBank number: MB 840443. Etymology: refers to Haikou, the city where the isolate was collected. Diagnosis: the main diagnostic criteria of the species Scedosporium haikouense are abundant ovoid, ellipsoidal, subcylindrical conidia, conidiogenous cells solitary or 2 to 3 fascicled conidia, and absent pigment and exudates and lack of synnemata. Type: China, Hainan Province, Haikou City, Hainan university Haidian Campus, 20.059602N, 110.330436E, isolated from green belt soil, 28 August 2019, Z.Y. Zhang. (Holotype HMAS 350313, stored in a metabolically inactive state; ex-holotype culture CGMCC 3.20468 = GZUIFR 21.833, ibid., GZUIFR 21.834.) GenBank: MZ469289, MZ469290 (ITS); MZ488563, MZ488564 (BT2).
FIG 11.
Scedosporium haikouense (from ex-holotype CGMCC 3.20468). (A, B) Upper and reverse views of culture on PDA 14 days after inoculation; (C to J) conidiogenous cells and conidia. Scale bars (C to J), 10 μm.
Description. Sexual morph: not observed. Asexual morph: colonies on PDA attaining 54 to 56 mm diameter after 5 days at 25°C, fluffy, flavescens to white, gray at margins, annular at the center, margin slightly undulate; reverse cream-yellow to black; absent pigment and exudates. Colonies on PDA attaining 68 to 70 mm diameter after 5 days at 37°C. Hyphae hyaline, branched, septate, smooth-walled, 0.5 to 5.5 μm wide. Conidiophores solitary, usually reduced to conidiogenous cells, arising terminally or laterally from hypha, hyaline, smooth-walled, cylindrical, 1.5 to 26.0 by 1.0 to 2.0 μm (n = 50). Conidia are borne on hyphae, short protrusions, or side branches, one-celled, solitary, or 2 to 3 fascicled, pale brown to brown, ovoid, ellipsoidal, subcylindrical and bilaterally compressed, rounded at the ends, 5.0 to 9.0 by 3.0 to 4.5 μm (n = 50). Synnemata not observed.
Substrate: soil. Distribution: Haikou City, Hainan Province, China.
Notes. Phylogenetically, Scedosporium haikouense is closely related to Scedosporium rarisporum, Scedosporium cereisporum, and S. aurantiacum. However, S. haikouense can be distinguished from S. rarisporum by the presence of abundant ovoid, ellipsoidal, subcylindrical conidia (30), from S. cereisporum by the solitary conidiogenous cells, solitary or 2 to 3 fascicled conidia (31), and from S. aurantiacum by the absent pigment and exudates and lack of synnemata (32).
Scedosporium hainanense Zhang, Han, and Liang, sp. nov. (Fig. 12). MycoBank number: MB 840445. Etymology: refers to Hainan, the province where the isolate was collected. Diagnosis: similar to S. apiospermum but differs in the ellipsoidal conidia. Type: China, Hainan Province, Sanya City, Hainan Tropical Ocean University, 18.311670N, 109.534152E, isolated from green belt soil, 26 August 2019, Z.Y. Zhang. (Holotype HMAS 350311, stored in a metabolically inactive state; ex-holotype culture CGMCC 3.20469 = GZUIFR 21.829.) GenBank: MZ469285 (ITS); MZ488559 (BT2).
FIG 12.
Scedosporium hainanense (from ex-holotype CGMCC 3.20469). (A, B) Upper and reverse views of culture on PDA 14 days after inoculation; (C to I) conidiogenous cells and conidia; (J and K) synnematous conidiomata. Scale bars (C to J), 10 μm.
Description. Sexual morph: not observed. Asexual morph: colonies on PDA attaining 38 to 43 mm diameter after 5 days at 25°C, cottony, floccose, light gray, margins irregular; reverse peltricolor, white to margins; absent pigment and exudates. Colonies on PDA attaining 64 to 66 mm diameter after 5 days at 37°C. Hyphae hyaline, branched, septate, smooth-walled, 0.5 to 4.5 μm wide. Conidiophores solitary, often consisting of a single conidiogenous cell, or arranged in whorls of 2 to 3 conidiogenous cells, arising terminally or laterally from hypha, undifferentiated hypha, short-stalked, or inside branches. Conidiogenous cells annellidic, hyaline, thin- and smooth-walled, lateral or terminal, cylindrical or slightly broad at the base, sometimes with several annellations at the top with the age, 2.5 to 33.0 by 1.0 to 2.5 μm (n = 50). Conidia are borne on hyphae, short protrusions, or side branches, one-celled, solitary, hyaline, ovoid, 5.0 to 8.0 by 2.5 to 6.0 μm (n = 50), ellipsoidal, 5.5 to 7.0 by 5.0 to 5.5 μm (n = 50). Conidiomata synnematous, erect, consisting of a cylindrical stipe, hyaline, smooth-walled; conidia cylindrical or claviform with a truncated base, 4.5 to 8.5 by 2.5 to 3.5 μm (n = 50).
Substrate: soil. Distribution: Sanya and Danzhou City, Hainan Province, China. Material examined: China, Hainan Province, Sanya City, Seaside parks, 18.272349N, 109.479274E, isolated from green belt soil, 26 August 2019, Z.Y. Zhang, GZUIFR 21.828. GenBank: MZ469284 (ITS); MZ488558 (BT2). Hainan Province, Danzhou City, Hainan University Danzhou Campus, 19.508080N, 109.494579E, isolated from green belt soil, 27 August 2019, Z.Y. Zhang, GZUIFR 21.827. GenBank: MZ469283 (ITS); MZ488557 (BT2).
Notes. Morphological and phylogenetic data (Fig. 3) support our strains as new species of Scedosporium hainanense. Scedosporium hainanense is phylogenetically closely related to S. apiospermum complex that comprises Scedosporium angustum, S. apiospermum, S. boydii, S. ellipsoideum, and Scedosporium fusarium. However, S. hainanense can be distinguished from S. apiospermum by the ellipsoidal conidia. We did not compare morphological characteristics between S. hainanense and the S. apiospermum complex (S. angustum, S. apiospermum, S. boydii, S. ellipsoideum, and Scedosporium fusarium) because of the lack of asexual morph descriptions of these species (33).
Scedosporium multisporum Zhang, Han, and Liang, sp. nov. (Fig. 13). MycoBank number: MB 840446. Etymology: referring to the 2 to 3 fascicled conidia. Diagnosis: similar to S. apiospermum complex but differs in the presence of 2 to 3 fascicled conidia, conidiomata synnematous. Type: China, Hunan Province, Huaihua City, Huaihua University, 27.572703N, 110.023832E, isolated from green belt soil, 12 August 2019, Z.Y. Zhang. (Holotype HMAS 350312, stored in a metabolically inactive state; ex-holotype culture CGMCC 3.20470 = GZUIFR 21.830, ibid., GZUIFR 21.831; ibid., GZUIFR 21.832.) GenBank: MZ469286, MZ469287, MZ469288 (ITS); MZ488560, MZ488561, MZ488562 (BT2).
FIG 13.
Scedosporium multisporum (from ex-holotype CGMCC 3.20470). (A, B) Upper and reverse views of culture on PDA 14 days after inoculation; (C to G) conidiogenous cells and conidia; (H and I) synnematous conidiomata. Scale bars (C to I), 10 μm.
Description. Sexual morph: not observed. Asexual morph: colonies on PDA attaining 45 to 50 mm diameter after 5 days at 25°C, cottony, powdery at the center; reverse white, light yellow at the center; absent pigment and exudates. Colonies on PDA attaining 70 to 73 mm diameter after 5 days at 37°C. Hyphae hyaline, branched, septate, smooth-walled, 1.0 to 4.0 μm wide. Conidiophores solitary, often consisting of a single conidiogenous cell, or arranged in whorls of 2 to 3 conidiogenous cells, arising terminally or laterally from hypha, undifferentiated hypha, short-stalked, or inside branches. Conidiogenous cells annellidic, hyaline, thin- and smooth-walled, lateral or terminal, cylindrical or slightly broad at the base, sometimes with several annellations at the top with the age, 0.5 to 16.0 by 1.0 to 3.5 μm (n = 50). Conidia are borne on hyphae, short protrusions, or side branches, one-celled, solitary, or 2 to 3 fascicled, hyaline, ovoid to subglobose, 3.0 to 7.5 by 3.0 to 5.0 μm (n = 50). Conidiomata synnematous, erect, consisting of a cylindrical stipe, hyaline, smooth-walled; conidia cylindrical, ovoid, long ovoid with a truncated base, 5.0 to 10.0 by 2.0 to 4.0 μm (n = 50).
Substrate: soil. Distribution: Huaihua City, Hunan Province, China.
Notes. Scedosporium multisporum is phylogenetically closely related to the S. apiospermum complex that comprises S. angustum, S. apiospermum, S. boydii, S. ellipsoideum, and S. fusarium. However, S. multisporum is distinguished from other species of Scedosporium by the presence of 2 to 3 fascicled conidia, conidiomata synnematous (33).
Parascedosporium sanyaense (Han, Zheng, Luo, Wang, and Liang 2017) Zhang, Han, and Liang 2021, comb. nov. MycoBank: MB 818105. Basionym: Scedosporium sanyaense (see reference 30).
Description: Y.F. Han, Huan Zheng, Y. Luo, Y.R. Wang, and Z.Q. Liang 2017.
Notes. In 2017, Han et al. introduced S. sanyaense to the genus Scedosporium, based on morphological and internal transcribed spacers (ITS) phylogenetic analysis (30). However, in our phylogenetic study, S. sanyaense is placed in the genus Parascedosporium. Therefore, we propose a new combination for that species.
DISCUSSION
The hair baiting technique was first used to isolate keratinophilic fungi from the soil by Vanbreuseghem (34) and has become applied widely. So far, the investigation of such resources is still dominated by traditional isolated cultures and baiting with materials of human or animal origin, such as feathers (35), horsehair (4), wool (36), human hair (37), and human nails (38). Only a small number of studies have used next-generation sequencing technologies (39).
Taxonomy and phylogenetic identification of fungi remain significant challenges (40). One of the main fundamental needs in fungal ecology is a strong taxonomic basis, which is dependent on advances in nucleic acid sequence technology. However, some researchers have relied too much on these techniques to the complete exclusion of fungal isolation and characterization using classical methods. While bacterial microbiome studies have relatively reliable taxonomic identification using 16S ribosomal DNA (rDNA) and even metagenome sequencing, mycobiome studies are still few and far between, with limited taxonomic interpretation capabilities. Indeed, phenotypic and culture-based studies remain an invaluable tool for fungal biology and ecology (41). The advantage of placing these organisms in pure culture is, of course, that almost all aspects of their biology can be studied, which may help to understand how they function in their natural ecological context. Thus, many challenges remain in studying the hundreds of niches on Earth that may be inhabited by fungi, not only to demonstrate their presence in these niches but also to culture them in pure form and store them properly for further study (42).
The ability of microorganisms to degrade recalcitrant materials has been widely explored for environmental remediation and industrial production. Significant success has been achieved with single strains, but the focus is now on the use of microbial consortia because of their functional stability and efficiency (43). The keratin degradation process requires the synergistic action of different enzymes, such as endoproteases, exoproteases, oligopeptidases, and disulfide reductases (44); thus, this process involves the synergistic cooperation of multiple species. We did not isolate purified fungal strains directly from feathers after enrichment using hair bating but did isolate members of the fungal community from the soil. Therefore, we could not determine whether the obtained strains are keratinophilic fungi and whether they are able to degrade and utilize keratin. However, numerous studies have shown that many members of Thelebolales and Scedosporium are indeed keratinophilic fungi (45–48). Hence, we think that our obtained strains are the keratinophilic fungi and should at least be constituent members of the keratin-degrading fungal consortia, although it is not clear what role they play in this consortium. In this study, 10 new species were identified and introduced, not only contributing to the further understanding of the keratin-degrading fungal community but also accumulating strains for future artificially constructed keratin-degrading microbial consortia.
MATERIALS AND METHODS
Sampling, fungal isolation, and morphology.
Soil samples were collected from Guizhou, Hunan, Zhejiang, Yunnan, Fujian, Hainan, Jiangxi, Guangdong, and Zhejiang provinces in southern China and transported to the laboratory in Ziploc plastic bags. The soil samples were processed using the method we described previously (22). Briefly, clean and sterile chicken feathers were placed in a sterile petri dish after the soil sample was added, wetted with distilled water, and incubated at room temperature for 1 month. Fungi were isolated using a conventional dilution technique based on Sabouraud’s dextrose agar (SDA; 10 g of peptone, 40 g of dextrose, 20 g of agar, 1 liter of ddH2O) supplemented with chloramphenicol and cycloheximide, and the purification of the strains was performed using potato dextrose agar (PDA; Shanghai Bio-way Technology Co., Ltd., China) (20, 22). Colonies on PDA were incubated after 14 days at 25°C, and the cultures were placed to slowly dry at 50°C to produce the holotype. Holotypes were deposited in the Mycological Herbarium of the Institute of Microbiology, Chinese Academy of Sciences, Beijing, China (HMAS). All strains were deposited in the Institute of Fungus Resources, Guizhou University (GZUIFR, the Herbarium of Guizhou Agricultural College, code GZAC), and the ex-type strains were also deposited in the China General Microbiological Culture Collection Center (CGMCC). The living cultures were stored in a metabolically inactive state, i.e., kept in sterile 30% glycerol in a −80°C freezer. Macroscopic and morphological characterization of the colonies was performed on PDA incubated for 14 days in the dark at 25°C. The characterization and measurement of fungal microscopic characteristics were performed in 25% lactic acid. Images were obtained using an optical microscope (OM; DM4 B, Leica, Germany) with differential interference contrast (DIC). Taxonomic descriptions and nomenclature were deposited at MycoBank (https://www.mycobank.org/).
DNA extraction, PCR amplification, and sequencing.
Total genomic DNA was extracted from fungal mycelia using the BioTeke fungus genomic DNA extraction kit (DP2032, BioTeke, Beijing, China) following the manufacturer’s instructions. Multiple loci were amplified and sequenced for each new isolate, and the primer sets are listed in Table 2. Amplification conditions were carried out as in the original literature where the primers were reported. The PCR thermal cycle programs for each locus amplification were performed as in the original literature where the primers were reported. The PCR products were sequenced with the amplified primers at a commercial sequencing service provider (Shanghai Sangon Biological Engineering Technology & Services Co., Shanghai, China) in an ABI 3730xl DNA analyzer using the Sanger method. The consensus sequences were obtained using the SeqMan software v. 7 (DNASTAR Lasergene, Madison, WI, USA).
TABLE 2.
Primers used in this study
| Locus | Primer | Primer sequence 5′ to 3′ | Orientation | Reference |
|---|---|---|---|---|
| ITS | ITS1 | TCCGTAGGTGAACCTGCGG | Forward | 56 |
| ITS4 | TCCTCCGCTTATTGATATGC | Reverse | 56 | |
| Beta-tubulin (BT2) | Bt2a | GGTAACCAAATCGGTGCTGCTTTC | Forward | 57 |
| Bt2b | ACCCTCAGTGTAGTGACCCTTGGC | Reverse | 57 | |
| Large subunit ribosomal DNA (LSU) | LROR | ACCCGCTGAACTTAAGC | Forward | 58 |
| LR7 | TACTACCACCAAGATCT | Reverse | 59 | |
| Translation elongation factor 1-alpha (TEF1-α) | 983F | GCYCCYGGHCAYCGTGAYTTYAT | Forward | 60 |
| EF1-2218R | ATGACACCRACRGCRACRGTYTG | Reverse | 60 | |
| RNA polymerase II subunit 2 (RPB2) | fRPB2-7cF | ATGGG[T/C]AA[A/G]CAAGC[T/C]ATGGG | Forward | 61 |
| RPB2-3053bR | TGRATYTTRTCRTCSACCAT | Reverse | 62 | |
| Minichromosomal maintenance protein 7 (MCM7) | MCM7-709 | ACIMGIGTITCVGAYGTHAARCC | Forward | 63 |
| MCM7-1348 | GAYTTDGCIACICCIGGRTCWCCCAT | Reverse | 63 |
Phylogenetic analysis.
The data sets were assembled based on the closest matches from the BLASTn search results and recently published data. Sequences generated from each locus were analyzed along with other sequences retrieved from GenBank. The individual loci matrix was aligned with MAFFT v7.037b (49) and was further edited manually, where necessary, using BioEdit v.7.0.9.0 (50). The best-fit model of maximum likelihood (ML) and Bayesian analyses of each locus were estimated using IQ-TREE’s ModelFinder function (51) using the Akaike Information Criterion (AIC).
Phylogenetic analyses of the combined aligned data were performed under ML and Bayesian inference (BI). ML analyses were performed with IQ-TREE v. 1.6.11 (52). Bootstrap analyses were performed using the ultrafast bootstrap approximation (53) with 10,000 replicates, and bootstrap support (BS) greater than 70% was considered significantly supported. The BI was conducted with MrBayes v. 3.2.6 (54). Four Markov chains were run for two runs from random starting trees for 5 million generations, and trees were sampled every 1,000 generations. The first 25% of the sampled trees were discarded as burn-in, and the remaining ones were used to reconstruct a majority rule consensus tree and calculate Bayesian posterior probabilities (BPP) of the clades. The above analyses were carried out in PhyloSuite v1.16 (55).
Data availability.
The sequences generated in this study can be found in GenBank. The accession numbers of the sequences deposited in GenBank are listed in Table 3.
TABLE 3.
List of GenBank accession numbers of sequences used in this studya
Accession numbers for these strains generated from this study.
ACKNOWLEDGMENTS
The work was supported by Key Areas of Research and Development Program of Guangdong Province (no. 2018B020205003), “Hundred” Talent Projects of Guizhou Province (Qian Ke He [2020] 6005), the National Natural Science Foundation of China (no. 32060011, 31860002), and Construction Program of Biology First-class Discipline in Guizhou (GNYL [2017] 009).
We declare no conflicts of interest.
Contributor Information
Yan-Feng Han, Email: swallow1128@126.com.
Soo Chan Lee, University of Texas at San Antonio.
REFERENCES
- 1.Hyde KD, Jeewon R, Chen YJ, Bhunjun CS, Calabon MS, Jiang HB, Lin CG, Norphanphoun C, Sysouphanthong P, Pem D, Tibpromma S, Zhang Q, Doilom M, Jayawardena RS, Liu JK, Maharachchikumbura SSN, Phukhamsakda C, Phookamsak R, Al-Sadi AM, Thongklang N, Wang Y, Gafforov Y, Jones EBG, Lumyong S. 2020. The numbers of fungi: is the descriptive curve flattening? Fungal Divers 103:219–271. doi: 10.1007/s13225-020-00458-2. [DOI] [Google Scholar]
- 2.Rydin Y, Bleahu A, Davies M, Dávila JD, Friel S, De Grandis G, Groce N, Hallal PC, Hamilton I, Howden-Chapman P, Lai K-M, Lim CJ, Martins J, Osrin D, Ridley I, Scott I, Taylor M, Wilkinson P, Wilson J. 2012. Shaping cities for health: complexity and the planning of urban environments in the 21st century. Lancet 379:2079–2108. doi: 10.1016/S0140-6736(12)60435-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nieuwenhuijsen MJ, Khreis H, Triguero-Mas M, Gascon M, Dadvand P. 2017. Fifty shades of green. Epidemiology 28:63–71. doi: 10.1097/EDE.0000000000000549. [DOI] [PubMed] [Google Scholar]
- 4.Shadzi S, Chadeganipour M, Alimoradi M. 2002. Isolation of keratinophilic fungi from elementary schools and public parks in Isfahan, Iran. Mycoses 45:496–499. doi: 10.1046/j.1439-0507.2002.00798.x. [DOI] [PubMed] [Google Scholar]
- 5.Vidyasagar GM, Hosmani N, Shivkumar D. 2005. Keratinophilic fungi isolated from hospital dust and soils of public places at Gulbarga, India. Mycopathologia 159:13–21. doi: 10.1007/s11046-004-9483-1. [DOI] [PubMed] [Google Scholar]
- 6.Maghraby TA, Gherbawy YAMH, Hussein MA. 2008. Keratinophilic fungi inhabiting floor dusts of student houses at the South Valley University in Egypt. Aerobiologia 24:99–106. doi: 10.1007/s10453-008-9089-z. [DOI] [Google Scholar]
- 7.Deshmukh SK, Verekar SA, Chavan YG. 2017. Incidence of keratinophilic fungi from the selected soils of Kaziranga National Park, Assam (India). Mycopathologia 182:371–377. doi: 10.1007/s11046-016-0083-7. [DOI] [PubMed] [Google Scholar]
- 8.Verekar SA, Deshmukh SK. 2017. Developments in fungal biology and applied mycology. Springer Press, Singapore. [Google Scholar]
- 9.Jain PC, Agrawal SC. 1980. A note on the keratin decomposing capability of some fungi. Trans Mycol Soc Jpn 21:513–517. [Google Scholar]
- 10.Sharma R, Shouche YS. 2020. Diversity of Onygenalean fungi in keratin-rich habitats of Maharashtra (India) and description of three novel taxa. Mycopathologia 185:67–85. doi: 10.1007/s11046-019-00346-7. [DOI] [PubMed] [Google Scholar]
- 11.Singh I, Kushwaha RKS. 2010. Dermatophytes and related keratinophilic fungi in soil of parks and agricultural fields of Uttar Pradesh, India. Indian J Dermatol 55:306–308. doi: 10.4103/0019-5154.70700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.El-Ghonemy DH, Ali TH. 2017. Optimization of physicochemical parameters for hyper keratinase production from a newly isolated Aspergillus sp. DHE7 using chicken feather as substrate - management of biowaste. J Appl Pharm Sci 7:171–178. doi: 10.7324/japs.2017.70923. [DOI] [Google Scholar]
- 13.Kumawat TK, Sharma A, Sharma V, Chandra S, Bhadauria S. 2020. A study on the prevalence of keratinophilic fungal biota of semi-arid region of Rajasthan, India. J King Saud Univ Sci 32:1014–1020. doi: 10.1016/j.jksus.2019.09.008. [DOI] [Google Scholar]
- 14.Zhang Y-W, Chen W, Zeng G, Zou XIAO, Wen T, Han Y, Qiu S-YI, Liang Z-QI. 2016. Two new Chrysosporium (Onygenaceae, Onygenales) from China. Phytotaxa 270:210–216. doi: 10.11646/phytotaxa.270.3.5. [DOI] [Google Scholar]
- 15.Chen WH, Zeng GP, Luo Y, Liang ZQ, Han YF. 2017. Morphological traits and molecular analysis for Geomyces fujianensis sp. nov. from China. Mycosphere 8:38–43. doi: 10.5943/mycosphere/8/1/5. [DOI] [Google Scholar]
- 16.Li Z, Zeng GP, Ren J, Zou X, Han YF. 2017. Chrysosporium leigongshanense sp. nov. from Guizhou Province, China. Mycosphere 8:1210–1216. doi: 10.5943/mycosphere/8/8/17. [DOI] [Google Scholar]
- 17.Zhang YW, Zeng GP, Zou X, Han YF, Liang ZQ, Qiu SY. 2017. Two new keratinophilic fungal species. Phytotaxa 303:173–180. doi: 10.11646/phytotaxa.303.2.7. [DOI] [Google Scholar]
- 18.Zhang ZY, Han YF, Chen WH, Liang ZQ. 2019. Phylogeny and taxonomy of three new Ctenomyces (Arthrodermataceae, Onygenales) species from China. MycoKeys 47:1–16. doi: 10.3897/mycokeys.47.30740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang ZY, Han YF, Chen WH, Liang ZQ. 2019. Gongronella sichuanensis (Cunninghamellaceae, Mucorales), a new species isolated from soil in China. Phytotaxa 416:167–174. doi: 10.11646/phytotaxa.416.2.4. [DOI] [Google Scholar]
- 20.Zhang ZY, Chen WH, Zou X, Han YF, Huang JZ, Liang ZQ, Deshmukh SK. 2019. Phylogeny and taxonomy of two new Plectosphaerella (Plectosphaerellaceae, Glomerellales) species from China. MycoKeys 57:47–60. doi: 10.3897/mycokeys.57.36628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang ZY, Zhao YX, Shen X, Chen WH, Han YF, Huang JZ, Liang ZQ. 2020. Molecular phylogeny and morphology of Cunninghamella guizhouensis sp. nov. (Cunninghamellaceae, Mucorales), from soil in Guizhou, China. Phytotaxa 455:31–39. doi: 10.11646/phytotaxa.455.1.4. [DOI] [Google Scholar]
- 22.Zhang Z, Dong C, Chen W, Mou Q, Lu X, Han Y, Huang J, Liang Z. 2020. The enigmatic Thelebolaceae (Thelebolales, Leotiomycetes): one new genus Solomyces and five new species. Front Microbiol 11:572596. doi: 10.3389/fmicb.2020.572596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rice AV, Currah RS. 2006. Two new species of Pseudogymnoascus with Geomyces anamorphs and their phylogenetic relationship with Gymnostellatospora. Mycologia 98:307–318. doi: 10.3852/mycologia.98.2.307. [DOI] [PubMed] [Google Scholar]
- 24.Minnis AM, Lindner DL. 2013. Phylogenetic evaluation of Geomyces and allies reveals no close relatives of Pseudogymnoascus destructans, comb. nov., in bat hibernacula of eastern North America. Fungal Biol 117:638–649. doi: 10.1016/j.funbio.2013.07.001. [DOI] [PubMed] [Google Scholar]
- 25.Samson RA. 1972. Notes on Pseudogymnoascus, Gymnoascus and related genera. Acta Bot Neerl 21:517–527. doi: 10.1111/j.1438-8677.1972.tb00804.x. [DOI] [Google Scholar]
- 26.Gargas A, Trest MT, Christensen M, Volk TJ, Blehert DS. 2009. Geomyces destructans sp. nov. associated with bat white-nose syndrome. Mycotaxon 108:147–154. doi: 10.5248/108.147. [DOI] [Google Scholar]
- 27.Crous PW, Wingfield MJ, Lombard L, Roets F, Swart WJ, Alvarado P, Carnegie AJ, Moreno G, Luangsaard J, Thangavel R, Alexandrova AV, Baseia IG, Bellanger J-M, Bessette AE, Bessette AR, De la Peña-Lastra S, García D, Gené J, Pham THG, Heykoop M, Malysheva E, Malysheva V, Martín MP, Morozova OV, Noisripoom W, Overton BE, Rea AE, Sewall BJ, Smith ME, Smyth CW, Tasanathai K, Visagie CM, Adamčík S, Alves A, Andrade JP, Aninat MJ, Araújo RVB, Bordallo JJ, Boufleur T, Baroncelli R, Barreto RW, Bolin J, Cabero J, Caboň M, Cafà G, Caffot MLH, Cai L, Carlavilla JR, Chávez R, de Castro RRL, et al. 2019. Fungal planet description sheets: 951–1041. Persoonia 43:223–425. doi: 10.3767/persoonia.2019.43.06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ekanayaka AH, Hyde KD, Gentekaki E, McKenzie EHC, Zhao Q, Bulgakov TS, Camporesi E. 2019. Preliminary classification of Leotiomycetes. Mycosphere 10:310–489. doi: 10.5943/mycosphere/10/1/7. [DOI] [Google Scholar]
- 29.Wijayawardene NN, Hyde KD, Rajeshkumar KC, Hawksworth DL, Madrid H, Kirk PM, Braun U, Singh RV, Crous PW, Kukwa M, Lücking R, Kurtzman CP, Yurkov A, Haelewaters D, Aptroot A, Lumbsch HT, Timdal E, Ertz D, Etayo J, Phillips AJL, Groenewald JZ, Papizadeh M, Selbmann L, Dayarathne MC, Weerakoon G, Jones EBG, Suetrong S, Tian Q, Castañeda-Ruiz RF, Bahkali AH, Pang KL, Tanaka K, Dai DQ, Sakayaroj J, Hujslová M, Lombard L, Shenoy BD, Suija A, Maharachchikumbura SSN, Thambugala KM, Wanasinghe DN, Sharma BO, Gaikwad S, Pandit G, Zucconi L, Onofri S, Egidi E, Raja HA, Kodsueb R, Cáceres MES, et al. 2017. Notes for genera: Ascomycota. Fungal Divers 86:1–594. doi: 10.1007/s13225-017-0386-0. [DOI] [Google Scholar]
- 30.Han YF, Zheng H, Zhang ZY, Chen WH, Luo Y, Wang YR, Liang ZQ. 2017. Two new Scedosporium species from Guangxi and Hainan. Mycosystema 36:145–153. [Google Scholar]
- 31.Crous PW, Wingfield MJ, Burgess TI, Hardy GESJ, Crane C, Barrett S, Cano-Lira JF, Le Roux JJ, Thangavel R, Guarro J, Stchigel AM, Martín MP, Alfredo DS, Barber PA, Barreto RW, Baseia IG, Cano-Canals J, Cheewangkoon R, Ferreira RJ, Gené J, Lechat C, Moreno G, Roets F, Shivas RG, Sousa JO, Tan YP, Wiederhold NP, Abell SE, Accioly T, Albizu JL, Alves JL, Antoniolli ZI, Aplin N, Araújo J, Arzanlou M, Bezerra JDP, Bouchara J-P, Carlavilla JR, Castillo A, Castroagudín VL, Ceresini PC, Claridge GF, Coelho G, Coimbra VRM, Costa LA, da Cunha KC, da Silva SS, Daniel R, de Beer ZW, Dueñas M, et al. 2016. Fungal planet description sheets: 469–557. Persoonia 37:218–403. doi: 10.3767/003158516X694499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gilgado F, Cano J, Gené J, Guarro J. 2005. Molecular phylogeny of the Pseudallescheria boydii species complex: proposal of two new species. J Clin Microbiol 43:4930–4942. doi: 10.1128/JCM.43.10.4930-4942.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.von Arx JA. 1973. The genera Petriellidium and Pithoascus. Persoonia 7:367–375. [Google Scholar]
- 34.Vanbreuseghem R. 1952. Technique biologique pour l’isolement des dermatophytes du sol. Ann Soc Belge Med Trop 32:173–178. [PubMed] [Google Scholar]
- 35.Mandeel Q, Nardoni S, Mancianti F. 2011. Keratinophilic fungi on feathers of common clinically healthy birds in Bahrain. Mycoses 54:71–77. doi: 10.1111/j.1439-0507.2009.01755.x. [DOI] [PubMed] [Google Scholar]
- 36.Muhsin TM, Hadi RB. 2002. Degradation of keratin substrates by fungi isolated from sewage sludge. Mycopathologia 154:185–189. doi: 10.1023/A:1016335623534. [DOI] [PubMed] [Google Scholar]
- 37.Malek E, Moosazadeh M, Hanafi P, Nejat ZA, Amini A, Mohammadi R, Kohsar F, Niknejad F. 2013. Isolation of keratinophilic fungi and aerobic actinomycetes from park soils in Gorgan, north of Iran. Jundishapur J Microbiol 6:e11250. doi: 10.5812/jjm.11250. [DOI] [Google Scholar]
- 38.Singh I, Kushwaha RKS. 2015. Keratinases and microbial degradation of keratin. Adv Appl Sci Res 6:74–82. [Google Scholar]
- 39.Hamm PS, Mueller RC, Kuske CR, Porras-Alfaro A. 2020. Keratinophilic fungi: specialized fungal communities in a desert ecosystem identified using cultured-based and Illumina sequencing approaches. Microbiol Res 239:126530. doi: 10.1016/j.micres.2020.126530. [DOI] [PubMed] [Google Scholar]
- 40.James TY, Stajich JE, Hittinger CT, Rokas A. 2020. Toward a fully resolved fungal tree of life. Annu Rev Microbiol 74:291–313. doi: 10.1146/annurev-micro-022020-051835. [DOI] [PubMed] [Google Scholar]
- 41.Walker LM, Cedeño-Sanchez M, Carbonero F, Herre EA, Turner BL, Wright SJ, Stephenson SL. 2019. The response of litter-associated Myxomycetes to long-term nutrient addition in a lowland tropical forest. J Eukaryot Microbiol 66:757–770. doi: 10.1111/jeu.12724. [DOI] [PubMed] [Google Scholar]
- 42.Carbonero F, Strobel G. 2021. Fungal ecology special issue: editorial. Microb Ecol 82:1–4. doi: 10.1007/s00248-021-01784-x. [DOI] [PubMed] [Google Scholar]
- 43.Kang D, Jacquiod S, Herschend J, Wei S, Nesme J, Sørensen SJ. 2019. Construction of simplified microbial consortia to degrade recalcitrant materials based on enrichment and dilution-to-extinction cultures. Front Microbiol 10:3010. doi: 10.3389/fmicb.2019.03010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lange L, Huang Y, Busk PK. 2016. Microbial decomposition of keratin in nature - a new hypothesis of industrial relevance. Appl Microbiol Biotechnol 100:2083–2096. doi: 10.1007/s00253-015-7262-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chaturvedi V, DeFiglio H, Chaturvedi S. 2018. Phenotype profiling of white-nose syndrome pathogen Pseudogymnoascus destructans and closely-related Pseudogymnoascus pannorum reveals metabolic differences underlying fungal lifestyles. F1000Res 7:665. doi: 10.12688/f1000research.15067.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pakshir K, Ghiasi MR, Zomorodian K, Gharavi AR. 2013. Isolation and molecular identification of keratinophilic fungi from public parks soil in Shiraz, Iran. Biomed Res Int 2013:619576. doi: 10.1155/2013/619576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Singh SM, Tsuji M, Gawas-Sakhalker P, Loonen MJJE, Hoshino T. 2016. Bird feather fungi from Svalbard Arctic. Polar Biol 39:523–532. doi: 10.1007/s00300-015-1804-y. [DOI] [Google Scholar]
- 48.Hayes MA. 2012. The Geomyces fungi: ecology and distribution. Bioscience 62:819–826. doi: 10.1525/bio.2012.62.9.7. [DOI] [Google Scholar]
- 49.Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and us-ability. Mol Biol Evol 30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hall TA. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41:95–98. [Google Scholar]
- 51.Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods 14:587–589. doi: 10.1038/nmeth.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol 32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Minh Q, Nguyen M, von Haeseler AA. 2013. Ultrafast approximation for phylogenetic bootstrap. Mol Biol Evol 30:1188–1195. doi: 10.1093/molbev/mst024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol 61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang D, Gao F, Jakovlić I, Zou H, Zhang J, Li WX, Wang GT. 2020. PhyloSuite: an integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol Ecol Resour 20:348–355. doi: 10.1111/1755-0998.13096. [DOI] [PubMed] [Google Scholar]
- 56.White TJ, Bruns T, Lee S, Taylor J. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics, p 315–322. In Innis MA, Gelfand DH, Sninsky JJ, White TJ (ed), PCR protocols, a guide to methods and applications. Academic Press, San Diego, CA, USA. doi: 10.1016/b978-0-12-372180-8.50042-1. [DOI] [Google Scholar]
- 57.Glass NL, Donaldson G. 1995. Development of primer sets designed for use with PCR to amplify conserved genes from filamentous ascomycetes. Appl Environ Microbiol 61:1323–1330. doi: 10.1128/aem.61.4.1323-1330.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rehner SA, Samuels GJ. 1994. Taxonomy and phylogeny of Gliocladium analyzed from nuclear large subunit ribosomal DNA sequences. Mycol Res 98:625–634. doi: 10.1016/S0953-7562(09)80409-7. [DOI] [Google Scholar]
- 59.Vilgalys R, Hester M. 1990. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J Bacteriol 172:4238–4246. doi: 10.1128/jb.172.8.4238-4246.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rehner SA, Buckley E. 2005. A Beauveria phylogeny inferred from nuclear ITS and EF1-alpha sequences: evidence for cryptic diversification and links to Cordyceps teleomorphs. Mycologia 97:84–98. doi: 10.3852/mycologia.97.1.84. [DOI] [PubMed] [Google Scholar]
- 61.Liu YJ, Whelen S, Hall BD. 1999. Phylogenetic relationships among ascomycetes: evidence from an RNA polymerase II subunit. Mol Biol Evol 16:1799–1808. doi: 10.1093/oxfordjournals.molbev.a026092. [DOI] [PubMed] [Google Scholar]
- 62.Reeb V, Lutzoni F, Roux C. 2004. Contribution of RPB2 to multilocus phylogenetic studies of the euascomycetes (Pezizomycotina, Fungi) with special emphasis on the lichen-forming Acarosporaceae and evolution of polyspory. Mol Phylogenet Evol 32:1036–1060. doi: 10.1016/j.ympev.2004.04.012. [DOI] [PubMed] [Google Scholar]
- 63.Schmitt I, Crespo A, Divakar PK, Fankhauser JD, Herman-Sackett E, Kalb K, Nelsen MP, Nelson NA, Rivas-Plata E, Shimp AD, Widhelm T, Lumbsch HT. 2009. New primers for promising single-copy genes in fungal phylogenetics and systematics. Persoonia 23:35–40. doi: 10.3767/003158509X470602. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The sequences generated in this study can be found in GenBank. The accession numbers of the sequences deposited in GenBank are listed in Table 3.
TABLE 3.
List of GenBank accession numbers of sequences used in this studya
Accession numbers for these strains generated from this study.