Abstract
A total of 200 isolates of Haemophilus influenzae were analyzed by serotyping, biotyping, and pulsed-field gel electrophoresis (PFGE). A total of 178 epidemiologically unrelated strains of H. influenzae demonstrated a variety of genome patterns by PFGE, and 165 genotypes were thus obtained in this study. PFGE typing proved to have a much stronger discriminatory power than either serotyping or biotyping. Six serotype b strains were all classified into discrete genotypes. A PFGE analysis of 18 strains obtained from the nasopharynx, blood, and cerebrospinal fluid of patients with meningitis also supported the hypothesis that invasive H. influenzae disseminates from the nasopharynx to the bloodstream and then subsequently to other body sites. PFGE typing of 10 other strains isolated from household contacts of patients with H. influenzae infection revealed that the strain that caused the H. influenzae infection often colonized the nasopharynges of household contacts. Our findings suggest that PFGE analysis is useful for the epidemiological study of H. influenzae infection, even when the invasive disease is caused by serotype b strains.
Haemophilus influenzae is a pathogen exclusively found in humans, and it causes infections that range from asymptomatic colonization of the upper respiratory tract to serious invasive diseases, such as meningitis. Strains of H. influenzae are usually classified according to the antigenicities and compositions of their polysaccharide capsules. There are six structurally and antigenically distinct capsular types (serotypes), designated serotypes a to f (17). Among them, serotype b is a common cause of serious infections in younger children.
H. influenzae can also be divided into eight biotypes on the basis of three biochemical tests (13). However, this biotyping system is of limited use for epidemiological studies since the majority of clinical isolates are distributed into three biotypes (biotypes I, II, and III). More than 90% of invasive type b strains are of biotype I. The majority of strains isolated from the nasopharynges of healthy people are nonencapsulated and of biotypes II and III.
More useful subtyping procedures that use the outer membrane proteins (1, 15, 21), lipopolysaccharides (12), or isozymes (16) of H. influenzae have been developed. These methods demonstrated considerable heterogeneity among nontypeable strains; however, type b strains showed much less variation.
Typing based on the bacterial genomic DNA fingerprinting pattern obtained by pulsed-field gel electrophoresis (PFGE) has been reported to be a convenient tool for the epidemiological investigation of bacterial infections (18). In this study, we analyzed 200 clinical isolates of H. influenzae by PFGE and then compared the findings obtained by genome typing with those obtained by other typing methods (serotyping, biotyping).
MATERIALS AND METHODS
Bacterial strains.
From 1994 to 1996, 200 H. influenzae strains were collected: 160 from Fukuoka Children’s Hospital and Medical Center for Infectious Disease, Fukuoka, Japan; 36 from Kyushu University Hospital, Fukuoka, Japan; 3 from Kyushu Kousei-nenkin Hospital, Fukuoka, Japan; and 1 from Saga Prefectual Kouseikan Hospital, Saga, Japan. They were isolated from various sources including the nasopharynx (148 strains), sputum (19 strains), eye mucus from patients with conjunctivitis (4 strains), middle-ear mucus from patients with otitis media (8 strains), urine from patients with urinary tract infection (7 strains), vaginal mucus from a patient with vaginitis (1 strain), blood from patients with sepsis (5 strains), and cerebrospinal fluid (CSF) from patients with meningitis (8 strains) (see Table 1). Among the isolates, 178 strains were isolated from epidemiologically unrelated patients who happened to be admitted to the hospitals mentioned above. In six of eight patients with meningitis, cultures of nasopharyngeal or blood specimens were performed. In total, 12 H. influenzae strains were obtained from cultures of their nasopharyngeal or blood specimens. Ten other strains were isolated from household contacts of patients with an H. influenzae infection.
TABLE 1.
Distribution into serotypes and biotypes of the 200 H. influenzae strains in this study
| Source | No. of strains | No. of strains with the following serotype and biotype:
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| a
|
b
|
c, I | Nontypeable
|
|||||||||||
| I | V | I | II | I | II | III | IV | V | VI | VII | VIII | |||
| Nasopharynx | 148 | 2 | 1 | 4 | 1 | 2 | 16 | 63 | 42 | 5 | 1 | 1 | 10 | |
| Sputum | 19 | 1 | 8 | 7 | 2 | 1 | ||||||||
| Eye mucus | 4 | 1 | 2 | 1 | ||||||||||
| Middle ear mucus | 8 | 2 | 1 | 3 | 1 | 1 | ||||||||
| Urine | 7 | 3 | 3 | 1 | ||||||||||
| Vaginal mucus | 1 | 1 | ||||||||||||
| Blood | 5 | 2 | 2 | 1 | ||||||||||
| CSF | 8 | 4 | 2 | 1 | 1 | |||||||||
Bacteriological methods.
The isolates were identified as H. influenzae on the basis of the following biological characteristics: they were gram-negative small rods and required X and V factors for growth. The growth factor requirement was tested for with a small disc containing either X or V factor. All of the strains were serotyped by a slide agglutination assay with type a- to f-specific antiserum (Denka Seiken Co. Ltd., Tokyo, Japan) and were then biotyped on the basis of the findings of three biochemical tests (indole production, urease activity, ornithine decarboxylase activity) (13).
Preparation of chromosomal DNA.
The bacteria were suspended in 100 mM EDTA buffer (pH 8.0) at a concentration of 109 cells/ml. The suspension was mixed with melted low-melting-point agarose L (Wako Pure Chemical Industries Ltd., Osaka, Japan). After solidification the plugs were incubated in 100 mM EDTA buffer containing lysozyme (1 mg/ml) for 5 h at 37°C. Then the plugs were incubated in 250 mM EDTA containing 0.5 mg of proteinase K per ml and 1% sodium dodecyl sulfate overnight at 50°C (5, 8).
PFGE.
The DNA extracted from each agarose block prepared as described above was digested with the enzyme SmaI (Toyobo Co. Ltd., Osaka, Japan) (3, 4). Pulsed-field gel electrophoresis (PFGE) was performed in 0.5× TBE (Tris-borate-EDTA) buffer in a contour-clamped homogeneous electric field apparatus (CHEF-DR II apparatus; Bio-Rad, Richmond, Calif.). Portions of the agarose plugs containing SmaI-digested DNA were loaded directly into the wells of a 1% agarose gel. Electrophoresis was performed for 30 h at 12°C at 5.0 V/cm with a ramped pulse time of 1 to 26 s. After electrophoresis, the gels were stained for 50 min with 0.5 mg of ethidium bromide per liter. DNA bands were visualized on a UV transilluminator and were photographed through a red filter (3–5, 8).
ID.
A numerical index of discrimination (ID) for serotyping, biotyping, and PFGE typing was calculated by the method of Hunter and Gaston (9, 10), which is based on the probability that two unrelated strains sampled from the test population will be placed into different groups by the typing method. An ID of 1.0 indicates that the typing method is able to distinguish each strain from the test population. Conversely, an ID of 0.0 indicates that all strains of the test population have identical types.
RESULTS
Analysis of isolates by serotyping and biotyping.
A total of 200 isolates were serotyped and biotyped. The results are shown in Table 1. All of the serotype b and c strains were isolated from patients with meningitis. Serotype a and nontypeable strains were from various sources including one patient with meningitis. Serotype a strains were biotyped as biotype I or V. Serotype b strains were biotyped as type I or II. Serotype c strains belonged to biotype I. Nontypeable strains were biotyped into all eight types, and the majority of them belonged to biotype II (44%) or III (30%).
PFGE analysis.
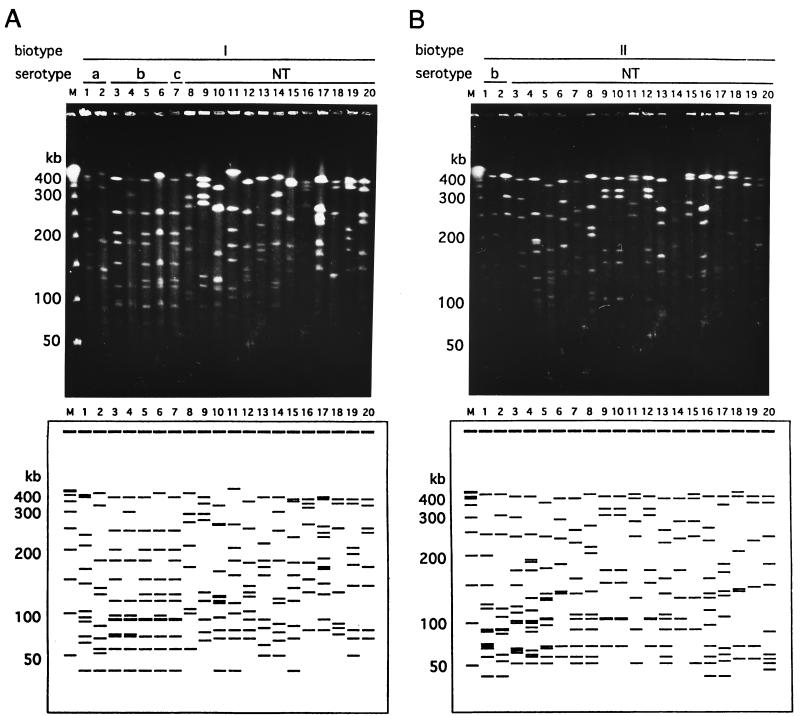
We compared the fingerprints of the genomic DNAs of all the strains which were obtained in this study. The strains belonging to the same biotype showed various distinctive PFGE patterns (Fig. 1). Twenty-seven distinctive restriction patterns were identified among the 28 epidemiologically unrelated strains which were biotyped as biotype I. Figure 1A shows examples of the PFGE patterns of the 20 strains which belonged to biotype I. It is noteworthy that these two strains, which belonged to different serotypes (lane 5, serotype b; lane 7, serotype c), had the same PFGE patterns. Sixty-eight restriction patterns were identified among 76 biotype II strains. Examples of the PFGE patterns for 20 strains are demonstrated in Fig. 1B. Forty-eight restriction patterns were identified among 52 biotype III strains. Figure 1C shows examples of the PFGE patterns of the 20 strains that belonged to biotype III. The strains which belonged to biotypes IV to VIII showed different genome patterns, as shown in Fig. 1D. The variations in the PFGE patterns for the H. influenzae strains examined in this study are summarized in Table 2. The genome patterns of the 178 epidemiologically unrelated isolates were classified into 165 groups. In our study, no group obtained by genotyping contained strains of more than one group obtained by biotyping.
FIG. 1.
Examples of PFGE patterns of chromosomal DNAs extracted from clinical H. influenzae isolates. Chromosomal DNA was digested with the SmaI restriction endonuclease. Lanes M, bacteriophage lambda concatamer molecular size marker. (A) Patterns of 20 strains of biotype I. Nineteen different patterns were observed, and a pair with the same pattern (lanes 5 and 7) was found. Lanes: 1 and 2, serotype a; 3 to 6, serotype b; 7, serotype c; 8 to 20, nontypeable (NT) strains (lanes 8 to 14, strains isolated from the nasopharynx; lane 15, strain isolated from sputum; lane 16, strain isolated from eye mucus; lanes 17 and 18, strains isolated from middle ear mucus; lanes 19 and 20, strains isolated from urine). (B) Patterns of 20 strains of biotype II. The strains in lanes 7 and 13 showed the same genome patterns, and the strains in lanes 9, 10, and 12 and in lanes 11 and 15 also showed the same genome patterns. Therefore, 16 different patterns are shown in panel B. Lanes: 1 and 2, serotype b; 3 to 20, nontypeable strains (lanes 3 to 10, strains isolated from the nasopharynx; lanes 11 to 14, strains isolated from sputum; lane 15, strain isolated from eye mucus; lane 16, strain isolated from middle-ear mucus; lanes 17 and 18, strain isolated from urine; lane 19, strain isolated from vaginal mucus; lane 20, strain isolated from CSF). (C) Patterns of 20 strains of biotype III. The strains in lanes 17 and 20 had the same genome patterns; therefore, 19 different patterns are shown in panel C. All strains were nontypeable by serotyping. Lanes: 1 to 11, strains isolated from nasopharynx; lanes 12 to 15, strains isolated from sputum; lane 16, strain isolated from eye mucus; 17 to 19, strains isolated from middle-ear mucus; 20, strain isolated from urine. (D) Patterns of 19 strains of biotypes IV to VIII. All strains except the one in lane 5 (serotype A) were nontypeable by serotyping. Lanes: 1 to 4, biotype IV strains isolated from the nasopharynx; 5 to 9, biotype V strains (lanes 5 and 6, strains isolated from the nasopharynx; lanes 7 and 8, strains isolated from sputum; 9, strains isolated from middle-ear mucus); 10, biotype VI strain isolated from nasopharynx; 11 to 18, biotype VII strains isolated from nasopharynx; 19, biotype VIII strain isolated from sputum.
TABLE 2.
Variations in PFGE patterns of the 178 epidemiologically unrelated strains of H. influenzae in this study
| Biotype | No. of strains | No. of different PFGE patterns |
|---|---|---|
| I | 28 | 27 |
| II | 76 | 68 |
| III | 52 | 48 |
| IV | 4 | 4 |
| V | 5 | 5 |
| VI | 1 | 1 |
| VII | 11 | 11 |
| VIII | 1 | 1 |
| Total | 178 | 165 |
PFGE analysis of isolates from patients with meningitis.
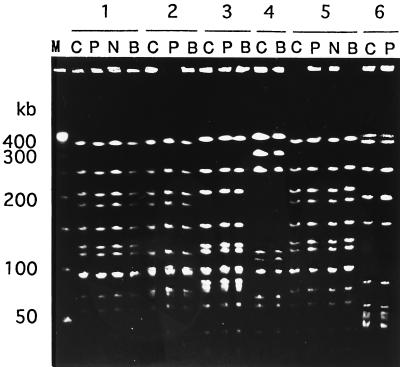
We obtained 12 isolates from either nasopharyngeal or blood samples, in addition to CSF, from six of eight patients with meningitis. These isolates were also analyzed by PFGE, and their PFGE patterns were then compared with the PFGE patterns of the strains isolated from the CSF of the individual patients. The results are shown in Fig. 2. For patients 1 (serotype b, biotype I), 2 (serotype b, biotype I), 3 (serotype b, biotype II), and 5 (serotype c, biotype I), H. influenzae strains were also obtained from both nasopharyngeal and blood samples, and PFGE analysis demonstrated that the genome patterns of the strains were the same as those of the isolates from CSF. We obtained an isolate (serotype b, biotype II) from a blood sample from patient 4 but not from his nasopharyngeal sample.
FIG. 2.
PFGE patterns of isolates from six patients (patients 1 to 6) with meningitis. The strains were isolated from CSF (lanes C), pharynx (lanes P), nasal cavity (lanes N), and blood (lanes B). Lane M, molecular size marker. Patients 1 and 2 had meningitis caused by serotype b, biotype I strains, patients 3 and 4 had meningitis caused by serotype b, biotype II strains, patient 5 had meningitis caused by a serotype c, biotype I strain, and patient 6 had meningitis caused by a nonencapsulated biotype II strain.
H. influenzae strains which were isolated from CSF and pharynx of patient 6 were nontypeable by serotyping and also had the same genome patterns. We could not isolate H. influenzae from a blood sample from this patient. After a detailed examination, we confirmed that this patient with meningitis had CSF leakage into his nasopharynx.
PFGE analysis of isolates from household contacts.
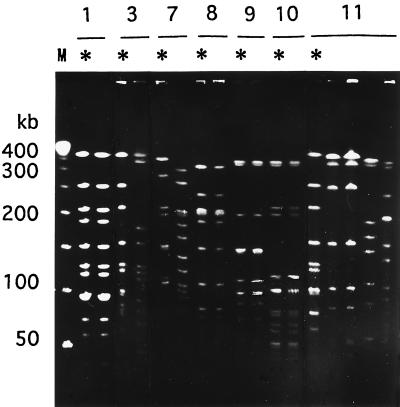
We obtained H. influenzae strains isolated from the nasopharynges of the family members of seven patients. The PFGE patterns of these isolates are shown in Fig. 3. For patients 1 (serotype b, biotype I), 8 (serotype NT [nontypeable], biotype III), 9 (serotype NT, biotype III), and 10 (serotype NT, biotype II), the strains from the patients had the same PFGE patterns as isolates that colonized the nasopharynges of their household contacts. For patients 3 (serotype b, biotype II), 7 (serotype NT, biotype III), and 11 (serotype b, biotype I), the strains obtained from the household contacts had PFGE patterns different from those of the patients with meningitis. For patient 11, strains of the same genotype colonized the nasopharynges of two household contacts, although the genotype of the isolates from the patient with meningitis was different from that of the strains colonizing the household contacts.
FIG. 3.
PFGE patterns of isolates from seven patients with H. influenzae infection (∗) and their household contacts (no mark). Patients 1, 3, and 11 had meningitis; patients 7, 8, 9, and 10 had respiratory tract infections. Patients 1 and 3 are the same as patients 1 and 3 in Fig. 2.
IDs for various typing methods.
The IDs for three typing methods (serotyping, biotyping, and PFGE typing) are shown in Table 3. An extremely high level of discrimination was observed by PFGE typing. Serotyping and biotyping alone allowed less discrimination than PFGE typing. The indices for various combinations of these typing methods are also shown in Table 3. The combination of serotyping and PFGE typing had the highest level of discrimination.
TABLE 3.
IDs of three typing methods for 178 epidemiologically unrelated strains of H. influenzae
| Method | No. of types | ID |
|---|---|---|
| Serotyping | 4 | 0.108360 |
| Biotyping | 8 | 0.706405 |
| PFGE typing | 165 | 0.999047 |
| Serotyping and biotyping | 13 | 0.726274 |
| Serotyping and PFGE typing | 166 | 0.999111 |
| Biotyping and PFGE typing | 165 | 0.999047 |
| Serotyping, biotyping, and PFGE typing | 166 | 0.999111 |
DISCUSSION
H. influenzae type b strains cause invasive disease in children, including sepsis and meningitis. However, the nonencapsulated strains of H. influenzae commonly colonize the upper respiratory tracts of healthy persons and are also implicated in such mucosal infections as otitis media, sinusitis, bronchitis, and conjunctivitis. Although various methods for the typing of clinical H. influenzae isolates have been reported in the past, no method has proven to be significantly useful in epidemiological investigations because of their poor discriminatory ability, particularly regarding type b strains.
In recent years, PFGE has become a useful tool for the typing and differentiation of strains for epidemiological studies with multiple bacterial species (5, 11, 14, 20). PFGE has been used for epidemiological studies of H. influenzae infections. Heath et al. (7) analyzed H. influenzae serotype b strains isolated from cultures of blood from two elderly nursing home residents by PFGE. The isolates from those patients were suggested to be related because their PFGE patterns were identical. Gazagne et al. (6) used PFGE to compare clinical ampicillin-resistant, non-beta-lactamase-producing H. influenzae strains. PFGE enabled the identification of 20 different patterns among 31 strains. They also studied the strains using other molecular biology tools, ribotyping and arbitrarily primed PCR with two primers, and each technique gave nearly the same number of different patterns as PFGE. However, ribotyping is more fastidious and time-consuming than arbitrarily primed PCR and PFGE (6).
The apparatuses for PFGE have recently come into widespread use in clinical laboratories. PFGE is considered a popular tool for epidemiological studies with various bacterial species. This study was carried out in order to determine whether the PFGE typing system could be useful as a tool for epidemiological studies of H. influenzae on the basis of the use of IDs.
The epidemiologically unrelated strains of H. influenzae which were collected in this study showed a variety of genome patterns by PFGE and also had high IDs. A total of 165 genotypes were obtained in this study, and a larger number of different genome patterns are also expected to be found if more strains are collected and analyzed by PFGE. It was noteworthy that six H. influenzae type b strains had separate PFGE patterns. These data confirm the fact that PFGE analysis is indeed useful even for epidemiological studies of invasive disease caused by H. influenzae type b. All serotype b strains were epidemiologically unrelated; however, the number of fragment differences among them was only three to nine. Serotype b strains are thus considered to be genetically related to each other. This phenomenon may explain why serotype b strains have been shown to have very few variations when they are typed by other subtyping methods. The guidelines for PFGE typing (19) indicated that variations in some bands were observed for strains of some species when they were cultured repeatedly over time. We subcultured three different strains of H. influenzae seven times and compared their PFGE types with those obtained before passage. The genome patterns of no strains changed after seven passages (data not shown). This result demonstrates the reproducibility of the PFGE typing technique with H. influenzae.
The present study also showed that strains of two different serotypes could be classified into the same group by genotyping. The degree of correlation between serotyping and PFGE typing was not significantly high. On the other hand, in our study no group delineated by genotyping contained strains of more than one group delineated by biotyping.
For five of eight patients with meningitis, H. influenzae strains were isolated from their blood samples, in addition to CSF, and the genome patterns of the strains were the same as those of isolates from CSF. It is generally known that invasive H. influenzae infections including meningitis are caused by the dissemination of bacteria, which are almost always of serotype b, from the nasopharynx to the bloodstream and subsequently to other body sites (22). All of the isolates simultaneously obtained from the nasopharynx, blood, and CSF of five patients with meningitis were thus considered to be the same strain. As expected, the isolates from each patient were classified into a single genotype. These results indicate that although PFGE demonstrated various genome patterns, identical strains were placed into the same group by genotyping. For the patient with meningitis caused by a nontypeable H. influenzae strain and with CSF leakage, no bacteria were isolated from his blood sample. This is probably because the bacteria disseminated from his nasopharynx to his spinal cord directly through CSF leakage. A previous study reported that anatomic disorders, including skull fractures, craniotomy, and CSF leakage, are the major predisposing factors for invasive disease caused by nontypeable H. influenzae strains in older children (2).
H. influenzae strains from four of seven patients had the same PFGE pattern as the isolates from the nasopharynges of household contacts. This finding strongly suggests that the same strain that caused H. influenzae serotype b infections (meningitis) also often colonized the nasopharynges of household contacts.
Our data therefore suggest that genotyping is a powerful tool for the epidemiological typing of H. influenzae strains.
ACKNOWLEDGMENTS
We gratefully acknowledge the following persons for supplying the strains used in this study: K. Takemori, Kyushu University Hospital, Fukuoka, Japan; T. Murao, K. Nakamura, T. Sakamoto, and K. Tsuzuki, Fukuoka Children’s Hospital and Medical Center for Infectious Disease; T. Ohno and H. Kariyazono, Kyushu Kousei-nenkin Hospital, Fukuoka, Japan; and E. Ishii, Saga Prefectual Kouseikan Hospital, Saga, Japan. We also thank K. Okada and K. Amako for helpful discussions and advice. We also express our appreciation to B. Quinn for valuable editorial advice on the manuscript.
REFERENCES
- 1.Barenkamp S J, Munson R S J, Granoff D M. Subtyping isolates of Haemophilus influenzae type b by outer-membrane protein profiles. J Infect Dis. 1981;143:668–676. doi: 10.1093/infdis/143.5.668. [DOI] [PubMed] [Google Scholar]
- 2.Bol P, Spanjaard L, van Alphen L, Zanen H C. Epidemiology of H. influenzae meningitis in patients more than 6 years of age. J Infect. 1987;15:81–94. doi: 10.1016/s0163-4453(87)91601-x. [DOI] [PubMed] [Google Scholar]
- 3.Butler P D, Moxon E R. A physical map of the genome of Haemophilus influenzae type b. J Gen Microbiol. 1990;136:2333–2342. doi: 10.1099/00221287-136-12-2333. [DOI] [PubMed] [Google Scholar]
- 4.Curran R, Hardie K R, Towner K J. Analysis by pulsed-field gel electrophoresis of insertion mutation in the transferrin-binding system of Haemophilus influenzae type b. J Med Microbiol. 1994;41:120–126. doi: 10.1099/00222615-41-2-120. [DOI] [PubMed] [Google Scholar]
- 5.Fujita M, Fujimoto S, Morooka T, Amako K. Analysis of strains of Campylobacter fetus by pulsed-field gel electrophoresis. J Clin Microbiol. 1995;33:1676–1678. doi: 10.1128/jcm.33.6.1676-1678.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gazagne L, Delmas C, Bingen E, Dabernat H. Molecular epidemiology of ampicillin-resistant non-beta-lactamase-producing Haemophilus influenzae. J Clin Microbiol. 1998;36:3629–3635. doi: 10.1128/jcm.36.12.3629-3635.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heath T C, Hewitt M C, Jalaludin B, Roberts C, Capon A G, Jelfs P, Gilbert G L. Invasive Haemophilus influenzae type b disease in elderly nursing home residents: two related cases. Emerg Infect Dis. 1997;3:179–181. doi: 10.3201/eid0302.970212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hu L, Umeda A, Kondo S, Amako K. Typing of Staphylococcus aureus colonising human nasal carriers by pulsed-field gel electrophoresis. J Med Microbiol. 1995;42:127–132. doi: 10.1099/00222615-42-2-127. [DOI] [PubMed] [Google Scholar]
- 9.Hunter P R. Reproducibility and indices of discriminatory power of microbial typing methods. J Clin Microbiol. 1990;28:1903–1905. doi: 10.1128/jcm.28.9.1903-1905.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hunter P R, Gaston M A. Numerical index of the discriminatory ability of typing system: an application of Simpson’s index of diversity. J Clin Microbiol. 1988;26:2465–2466. doi: 10.1128/jcm.26.11.2465-2466.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ichiyama S, Ohta M, Shimokata K, Kato N, Takeuchi J. Genomic DNA fingerprinting by pulsed-field gel electrophoresis as an epidemiological marker for study of nosocomial infections caused by methicillin-resistant Staphylococcus aureus. J Clin Microbiol. 1991;29:2690–2695. doi: 10.1128/jcm.29.12.2690-2695.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Inzana T. Electrophoretic heterogeneity and interstrain variation of the lipopolysaccharide of Haemophilus influenzae. J Infect Dis. 1983;148:492–499. doi: 10.1093/infdis/148.3.492. [DOI] [PubMed] [Google Scholar]
- 13.Kilian M. A taxonomic study of the genus Haemophilus with the proposal of a new species. J Gen Microbiol. 1976;93:9–62. doi: 10.1099/00221287-93-1-9. [DOI] [PubMed] [Google Scholar]
- 14.Kristjansson M, Samore M H, Gerding D N, Degirolami P C, Bettin K M, Karchmer A W, Arbeit R D. Comparison of restriction endonuclease analysis, ribotyping, and pulsed-field gel electrophoresis for molecular differentiation of Clostridium difficile strains. J Clin Microbiol. 1994;32:1963–1969. doi: 10.1128/jcm.32.8.1963-1969.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Loeb M R, Smith D H. Outer membrane protein composition in disease isolates of Haemophilus influenzae: pathogenic and epidemiological implications. Infect Immun. 1980;30:709–717. doi: 10.1128/iai.30.3.709-717.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Musser J M, Granoff D M, Pattison P E, Sellander R K. A population genetic framework for the study of invasive disease caused by serotype b strains of Haemophilus influenzae. Proc Natl Acad Sci USA. 1985;82:5078–5082. doi: 10.1073/pnas.82.15.5078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pittman M. Variation and type specificity in the bacterial species Haemophilus influenzae. J Exp Med. 1931;53:471–493. doi: 10.1084/jem.53.4.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smith C L, Canter C R. Purification, specific fragmentation, and separation of large DNA molecules. Methods Enzymol. 1987;155:449–467. doi: 10.1016/0076-6879(87)55030-3. [DOI] [PubMed] [Google Scholar]
- 19.Tenover F C, Arbeit R D, Goering R V, Mickelsen P A, Murray B A, Persing D H, Swaminathan B. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thong K, Ngeow Y, Altwegg M, Navaratnam P, Pang T. Molecular analysis of Salmonella enteritidis by pulsed-field gel electrophoresis and ribotyping. J Clin Microbiol. 1995;33:1070–1074. doi: 10.1128/jcm.33.5.1070-1074.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van Alphen L, Riemens T, Poolman J, Hopman C, Zanen H C. Homogeneity of cell envelope protein subtypes, lipopolysaccharide serotypes, and biotypes among Haemophilus influenzae type b from patients with meningitis in The Netherlands. J Infect Dis. 1983;148:75–81. doi: 10.1093/infdis/148.1.75. [DOI] [PubMed] [Google Scholar]
- 22.Ward J I, Lieberman J M, Cochi S L. Haemophilus influenzae vaccines. In: Plotkin S A, Mortimer E A, editors. Vaccines. Philadelphia, Pa: The W. B. Saunders Co.; 1994. pp. 337–386. [Google Scholar]