Abstract
INTRODUCTION:
The brains of extant animals have evolved over hundreds of millions of years from simple circuits. Cell types diversified, connections elaborated, and new brain regions emerged. Models for brain region evolution range from duplication of existing regions to splitting of previously multifunctional regions and de novo assembly from existing cell types. These models, however, have not been demonstrated in vertebrate brains at cell-type resolution.
RATIONALE:
We investigated brain region evolution using the cerebellar nuclei as a model system. The cerebellum is a major hindbrain structure in jawed vertebrates, comprising the cerebellar cortex and cerebellar nuclei. It is thought to act as a feedforward model for motor control and cognitive processes. The cerebellar cortex receives and processes inputs and sends outputs to the cerebellar nuclei, which route the results of cerebellar computations to the rest of the brain. Whereas the cerebellar cortex is well conserved across vertebrates, the cerebellar nuclei vary in number, with none in jawless vertebrates, one pair in cartilaginous fishes and amphibians, two pairs in reptiles and birds, and three pairs in mammals. This pattern suggests that extant cerebellar nuclei evolved from a single ancestral nucleus. Cerebellar nuclei thus provide a good model to interrogate brain region evolution.
RESULTS:
We characterized the cerebellar nuclei in mice, chickens, and humans using whole-brain and spinal cord projection mapping in cleared samples, single-nucleus RNA sequencing (snRNAseq), and spatially resolved transcript amplicon readout mapping (STARmap) analysis. We first compared the projection patterns of the three cerebellar nuclei of mice. Our data reveal broad projections of all nuclei, which in common target regions are shifted relative to each other. To understand the transcriptomic differences that underlie these shifting projections, we produced a cell-type atlas of the mouse cerebellar nuclei using snRNAseq. We discovered three region-invariant inhibitory cell classes and 15 region-specific excitatory cell types. Excitatory cell types fall into two classes with distinct gene expression and electrophysiological properties. Members of each class are present in every nucleus and are putative sister cell types. STARmap analysis in mice revealed that the organizational unit of the cerebellar nuclei is cytoarchitectonically distinguishable subnuclei, each of which contains the three inhibitory and two excitatory classes.
To test whether this archetypal subnucleus is also the evolutionary unit of the cerebellar nuclei, we performed snRNAseq and STARmap on the chicken cerebellar nuclei. We identified four subnuclei, three of which had direct orthologs in mice. Each chicken subnucleus contained the same cell-type set of three inhibitory and two excitatory classes already identified in mice, confirming our hypothesis.
Cerebellar nuclei vary in size across vertebrates. In particular, the human lateral nucleus is markedly expanded. To understand this expansion, we performed snRNAseq in humans. We found that the medial and interposed nuclei maintained the archetypal cerebellar nuclei composition. However, the lateral nucleus expanded one excitatory cell class at the expense of the other. Conditional tracing in the mouse lateral nucleus revealed that the cell class expanded in humans preferentially accesses lateral frontal cortices via specific intermediate thalamic nuclei.
CONCLUSION:
We identified a conserved cell-type set that forms an archetypal cerebellar nucleus as the unit of cerebellar nuclei organization and evolution. We propose that this archetypal nucleus was repeatedly duplicated during evolution, accompanied primarily by transcriptomic divergence of excitatory neurons and shifts in their projection patterns. Our data support a model of duplication-and-divergence of entire cell-type sets for brain region evolution.
Graphical Abstract
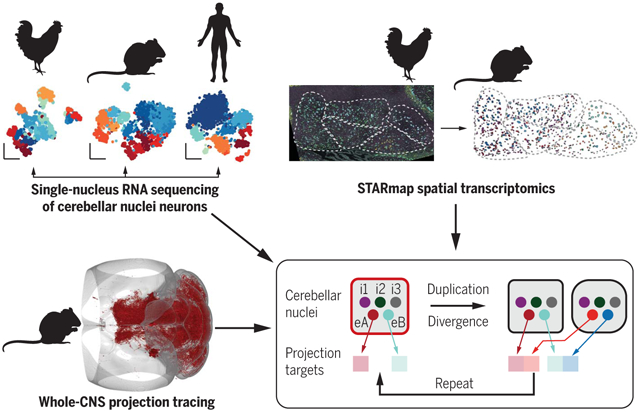
Evolution of the cerebellar nuclei. Comparative single-cell transcriptomics in mice, chickens, and humans (top left; neurons are color-coded by type), spatial transcriptomic analyses in mice and chickens (top right; neurons are color-coded by type in raw and processed data), and central nervous system (CNS)–wide projection mapping in mice (bottom left; axons in red in a three-dimensional mouse brain) revealed the unit of cerebellar nuclei organization and evolution. This unit (red box) comprises three inhibitory and two excitatory neuron classes (each colored circle indicates a neuron class). Extant cerebellar nuclei likely derived from the duplication and divergence of this unit, with more dynamic gene expression in excitatory neurons (changed color hues), along with projection target shifts.
How have complex brains evolved from simple circuits? Here we investigated brain region evolution at cell-type resolution in the cerebellar nuclei, the output structures of the cerebellum. Using single-nucleus RNA sequencing in mice, chickens, and humans, as well as STARmap spatial transcriptomic analysis and whole–central nervous system projection tracing, we identified a conserved cell-type set containing two region-specific excitatory neuron classes and three region-invariant inhibitory neuron classes. This set constitutes an archetypal cerebellar nucleus that was repeatedly duplicated to form new regions. The excitatory cell class that preferentially funnels information to lateral frontal cortices in mice becomes predominant in the massively expanded human lateral nucleus. Our data suggest a model of brain region evolution by duplication and divergence of entire cell-type sets.
The brains of extant animals are products of hundreds of millions of years of evolution. Over time, cell types diversified (1) and new brain regions appeared, giving rise to complex vertebrate brains today. Various models of brain region evolution have been proposed (2-5). These include the duplication of entire regions followed by either divergence (neofunctionalization, supporting new functions) or maintenance (isofunctionalization, supporting more of the same function) of the duplicated products. Brain regions could also arise by splitting previously multifunctional regions into more specialized ones (subfunctionalization) or might evolve from de novo generation and combination of cell types. To our knowledge, however, none of these processes have been demonstrated in vertebrate brain evolution at cell-type resolution. Doing so requires a comprehensive comparison of cell types across regions (6) and species (7-11) in a system that contains different numbers of homologous regions in different species.
The cerebellar nuclei are well suited for investigating brain region evolution. The cerebellum, consisting of the cerebellar cortex and cerebellar nuclei, is an ancient hindbrain structure present in all jawed vertebrates (12) and is involved in motor and cognitive functions (13, 14). The cerebellum sends its output through the cerebellar nuclei to a large number of target regions (15, 16) (Fig. 1A). Whereas the cerebellar cortex has expanded across evolution while maintaining a constant circuit motif (12), the cerebellar nuclei have been more plastic. Jawless vertebrates have cerebellum-like structures considered to be precursors to the cerebellar cortex but lack cerebellar nuclei. By contrast, a single pair of cerebellar nuclei can be recognized in cartilaginous fishes and amphibians, two pairs in reptiles and birds, and three pairs in mammals (12). These findings suggest that the last common ancestor of jawed vertebrates had a single pair of cerebellar nuclei and that nuclei numbers have increased in amniotes to expand the cerebellar output channels (Fig. 1B). The lateral nucleus in humans expanded to be 17 times larger than the medial or interposed nucleus (17), concomitant with the expansion of the prefrontal cortex that preferentially communicates with the lateral cerebellum (18).
Fig. 1. Brain-wide projections of mouse cerebellar nuclei (CN).
(A) Schematic of the cerebellar circuit. Information enters the cerebellar cortex through mossy fibers (MF) and climbing fibers (CF). Purkinje cells (PC) send cerebellar cortex output to the CN, which project to many brain regions. PN, pontine nuclei; Thal, thalamus; VN, vestibular nuclei; RN, red nucleus; SC, superior colliculus; IO, inferior olive; GC, granule cells. (B) Vertebrate cladogram, annotated with the number of CN pairs. (C) Schematic of the medial, interposed, and lateral CN in mice. (D) Schematic of experimental workflow. Anterograde tracers were injected into individual nuclei. Brains were cleared and imaged, and images were registered, showing a dorsal view of a representative brain volume with axons in red. Asterisk (*), tracer injection site. Dashed line denotes the midline. (E) Heat maps showing the mean projection strengths to the top innervated brain regions of each injection site. For abbreviations, see table S5. (F) Dendrogram showing hierarchical clustering of axon projections from 23 brains with indicated injection sites. Medial CN is most distinct from the other CN. Line color and gray numbers indicate bootstrapping-based branch confidence. Values >40 indicate good support. (G) Coronal heat maps of axonal innervation from the three mouse CN, with Allen compartments in background. Heat maps were derived from N = 5, 5, 6, and 7 anterior medial, posterior medial, interposed, and lateral CN injections, respectively. Asterisk (*), average tracer injection sites. Arrowheads and insets show shifted projections in contralateral thalamus [G(i)], ipsilateral brainstem [G(ii)], and cerebellar cortex [(G), middle panel]. (H) Sagittal heat map, showing shifted projection patterns in the contralateral superior colliculus. Scale bar: main panel, 1 mm; inset, 500 μm. In this and all subsequent figures: A, anterior; P, posterior; D, dorsal; V, ventral; M, medial; L, lateral.
Despite their obvious importance in cerebellar function, the cerebellar nuclei are poorly understood. Their transcriptomic cell types have not been identified in any species, beyond a basic division into glutamatergic, γ-aminobutyric acid–mediated (GABAergic), and glycinergic neurons in rodents (19). There have not been quantitative brain-wide comparisons of projection patterns of different cerebellar nuclei in any species (15, 16, 20, 21), and few cerebellar nuclei injections are available in the Allen Connectivity Atlas. Here, we characterize the transcriptomic cell types, spatial organization, and central nervous system (CNS)–wide projections of the three mouse cerebellar nuclei and compare these data to transcriptomic cell types and spatial organization of the two nuclei of chickens, as well as to the cell types in the three nuclei of humans. We identify an archetypal cerebellar nucleus—comprising a deeply conserved, stereotyped cell-type set—as the unit of cerebellar nuclei organization and evolution.
Results
CNS-wide projection mapping across mouse cerebellar nuclei
Mouse cerebellar nuclei are divided into three regions: medial (fastigial), interposed, and lateral (dentate) nuclei (Fig. 1C; see table S1 for nomenclature). The medial nucleus is considered to be phylogenetically the oldest, and the lateral nucleus the youngest (12). These three nuclei differ in their axonal projection patterns (16) and, potentially, gene expression (22). To comprehensively characterize the differences between the individual nuclei, we began by comparing their projection patterns. We performed CNS-wide anterograde tracing of each nucleus, followed by brain and spinal cord clearing and imaging (Fig. 1, D to H, and fig. S1 to S10). We aligned all brain volumes to the Allen Common Coordinate Framework reference brain, detected axons using a custom classification pipeline (materials and methods, Fig. 1D, and fig. S1), and quantified axonal innervation into ipsi- and contralateral brain regions (Fig. 1E, fig. S8 to S10, and table S2).
We traced 23 brains from four injection sites (anterior medial, posterior medial, interposed, and lateral nuclei). All three nuclei projected extensively to both hemispheres (16), innervating 125 ± 34 and 140 ± 32 (mean ± SD) ipsi- and contralateral brain regions, respectively. Medial and interposed nuclei also projected primarily to the contralateral cervical spinal cord (fig. S7). The brain-wide projection patterns of the medial and, particularly, anterior medial nuclei (which only has weak thalamic projections; fig. S3) were most distinct, whereas projections of the putatively more recently diverged interposed and lateral nuclei were comparatively more similar (Fig. 1F and fig. S8).
Closer inspection of projection patterns revealed cases wherein the three nuclei innervated adjacent brain regions with axons shifted relative to each other (Fig. 1, G and H); such shifts likely underestimated actual shift because of the spread of anterograde tracers at injection sites. Shifts were apparent in the ipsilateral cerebellar cortex (Fig. 1G), where medial, interposed, and lateral nuclei innervated the vermis, paravermis, and hemisphere, respectively (20), and in the anterior contralateral thalamus [Fig. 1G(i)], where interposed nucleus-innervated regions shifted dorsolaterally relative to lateral nucleus (21) and medial nucleus–innervated regions shifted ventromedially (23). Other shifts were observed in the ipsilateral brainstem [Fig. 1G(ii)], where the three nuclei innervated adjacent parasagittal stripes, and in the contralateral superior colliculus (Fig. 1H), where the interposed nucleus innervated more posterior regions than the lateral nucleus.
In summary, with the exception that the lateral nucleus does not appear to innervate the spinal cord, all mouse cerebellar nuclei innervate large portions of ipsi- and contralateral CNS. Different nuclei innervate grossly similar regions in the thalamus, midbrain, and hindbrain. Their projections, however, are often shifted relative to each other such that different nuclei innervate adjacent volumes within or across brain region boundaries. Interposed and lateral nuclei projections are more similar to each other than to medial nucleus projections.
Cell-type composition of mouse cerebellar nuclei
To investigate the molecular basis of the projection differences, we next used single-cell transcriptomics to determine the cell-type composition of the cerebellar nuclei. We separately dissected the three nuclei in each experiment and sorted NeuN+ neuronal nuclei into 384-well plates for high-depth, full-length single-nucleus RNA sequencing (snRNAseq; ~1 million aligned reads per cell) (Fig. 2A), yielding 4605 high-quality neurons. snRNAseq ensured relatively unbiased sampling of cerebellar nuclei neuronal types and is directly transferable to frozen brain samples from other species, owing to the conservation of NeuN (24).
Fig. 2. Mouse cerebellar nuclei cell types.
(A) Workflow of snRNAseq. The three regions were dissected separately, and cell nuclei were liberated, sorted for NeuN expression, and sequenced. (B) Marker expression for all neurons. Dashed line divides rhombic lip (RL)– and ventricular zone (VZ)-derived cells. N = 6 rounds of FACS using nine mice each. (C) Representative image of permanently labeled RL-derived cells probed for endogenous marker expression. Arrow, excitatory neuron; arrowhead, inhibitory neuron. Asterisk (*) in (B) and (C) labels Slc6a5+ RL-derived cluster e9*. Scale bar, 50 μm. N = 2 sections. (D and E) Clustering results for VZ- and RL-derived cells, labeled by clustering result (top) and CN dissection (middle), with marker expression at the bottom. Dissection labels are imperfect owing to close apposition of individual cerebellar nuclei in space. (F) Marker expression for all cell types. (G) Hierarchical clustering of excitatory cell types in the space of differentially expressed genes, using a correlation-based distance metric. Line color and gray numbers indicate bootstrapping-based branch confidence as in Fig. 1F. Class-A and Class-B neurons are color-coded with red and blue hues, respectively, in this and subsequent figures.
Overall, mouse cerebellar nuclei neurons separated into four broad clusters. Three were Gadl+ (encoding glutamic acid decarboxylase) inhibitory neurons. The remaining one was largely Slc17a6+ (encoding vesicular glutamate transporter 2) excitatory neurons; however, a small group of neurons within the Slc17a6+ cluster was Slc17a6− but Slc6a5+ (encoding glycine transporter 2) and likely glycinergic (Fig. 2B). We speculated that these broad divisions are driven by the developmental origins of excitatory and inhibitory neurons from the rhombic lip and ventricular zone, respectively (25, 26) (fig. S11A). To test this, we permanently labeled rhombic lip–derived neurons with tdTomato using Atoh1-Cre and performed spatially resolved transcript amplicon readout mapping (STARmap) in situ sequencing (27) on adult animals to quantify mRNA of various endogenous marker genes and tdTomato (Fig. 2C and fig. S11). All excitatory neurons were tdTomato+ and therefore derived from the rhombic lip. By contrast, all neurons falling into the three Gad1+ clusters were tdTomato− and thus ventricular zone-derived (Fig. 2C and fig. S11). The exception was a small cluster of Slc6a5+ neurons within the Slc17a6+ transcriptomic cluster (Fig. 2B, asterisk), which were tdTomato+ and therefore rhombic lip–derived (Fig. 2C and fig. S11E). On the basis of their large size and location in the lateral part of the medial nucleus (fig. S11B), these cells likely correspond to the previously described large glycinergic projection neurons (28). For simplicity, we hereafter refer to rhombic lip–and ventricular zone–derived cells as “excitatory” and “inhibitory,” respectively, despite the exception that a small cluster of rhombic lip–derived neurons are likely glycinergic inhibitory neurons.
To understand how neuronal types differ across nuclei, we separately clustered inhibitory and excitatory neurons (Fig. 2, D and E, and fig. S12 and S13). Inhibitory neurons showed relatively low diversity and formed three classes (table S1). Classes 1 and 3 each comprised a single transcriptomic cell type (i1, i3; referred to as “cell type” hereafter), whereas class 2 comprised one major (i2.1) and two minor (i2.2, i2.3) cell types. All cell types were represented across three nuclei without discernible nucleus-specific changes (Fig. 2D and fig. S12, A to C). i1 neurons were Gad1+Slc6a5− and likely correspond to inferior olive–projecting inhibitory neurons (29). i2.1 and i3 were Slc6a5+ glycinergic neurons. In contrast to the relatively low diversity of inhibitory neurons, excitatory neurons formed 15 distinct cell types, each specific to a single nucleus (Fig. 2E and fig. S12, D and E). Medial nucleus cell types were most distinct, whereas interposed and lateral nuclei cell types were more similar to each other (Fig. 2E), mirroring the projection data (Fig. 1F). Although we could map some of these cell types to previously described cell types (fig. S14), the diversity uncovered from our study far exceeds that of previous reports.
In summary, mouse cerebellar nuclei contain five nucleus-invariant inhibitory cell types in three classes and 15 nucleus-specific excitatory cell types, all of which can be distinguished by specific marker genes (Fig. 2F and fig. S15).
Excitatory neurons belong to two classes
If the three mouse cerebellar nuclei arose from a single ancestral nucleus, cell types with a common evolutionary origin might exist in the different nuclei (1). Such “sibling cell types” should share gene expression signatures that form an axis of variation independent of nucleus-specific changes. The nucleus-invariant inhibitory cell types found in each nucleus fulfill these requirements.
To investigate whether sibling cell types for excitatory neurons also exist, we hierarchically clustered all excitatory cell types in the space of differentially expressed genes between them (Fig. 2G). This analysis revealed a split of excitatory cell types into two classes, hereafter termed “Class-A” and “Class-B” (table S1). On average, more genes were detected in Class-B neurons than in Class-A neurons, hinting that Class-B neurons might be larger than Class-A neurons (fig. S12, G and H). Further, a large number of genes were differentially expressed in Class-A and -B neurons (fig. S16 and S17), including those with cell adhesion (fig. S16B) and ion channel activity (fig. S16C) that might contribute to different physiological properties of neurons in the two classes. Gene regulatory network analysis revealed regulons strongly associated with Class-A and Class-B (fig. S17). Using whole-cell patch-clamp recordings combined with RNAseq (Patch-Seq), we further showed that lateral nucleus Class-A and Class-B neurons are electrophysiologically distinct: Class-B neurons were larger, showed significantly higher spontaneous firing rates, and exhibited shorter, lower-amplitude action potentials than Class-A neurons (fig. S14, A to J). Notably, both Class-A and Class-B neurons were represented in each nucleus with one to three types each. Thus, the excitatory cell types within each class are putative sibling cell types to each other.
Each subnucleus contains a stereotyped cell-type set
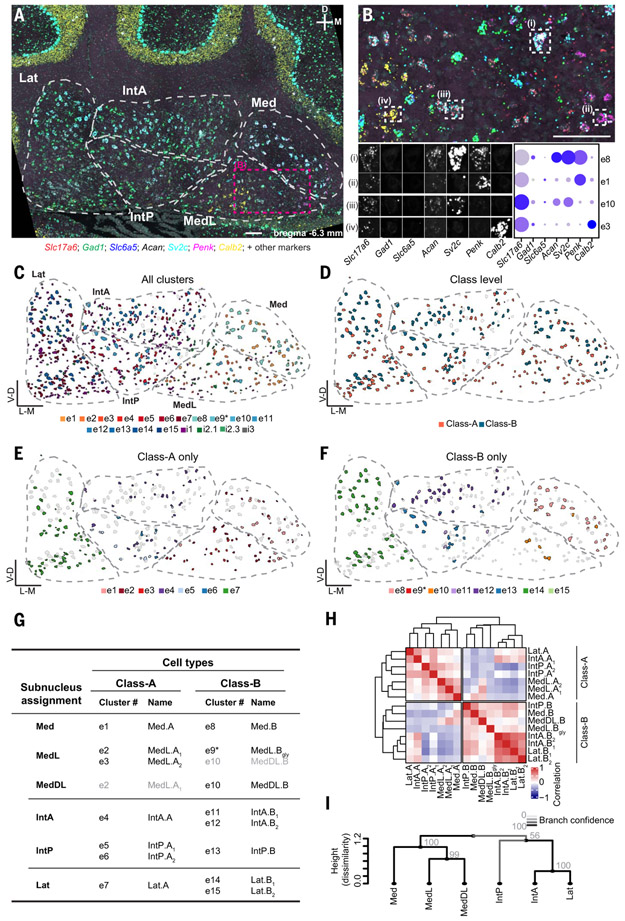
The existence of both Class-A and Class-B sibling cell types in each nucleus indicates that the cerebellar nuclei might have evolved through duplication. The finding of more than one Class-A or Class-B cell type within an individual nucleus, however, suggested that the cerebellar nuclei are evolutionarily organized into units smaller than individual nuclei. Indeed, mouse cerebellar nuclei can be divided into subnuclei on the basis of their cytoarchitecture (30) (table S1). To identify the relationship between subnuclei and cell types, we used sequential STARmap in situ sequencing (27). We detected up to 20 marker genes (materials and methods and table S3) chosen to distinguish all cell types within each nucleus on coronal sections spanning the anterior–posterior axis of the cerebellar nuclei (Fig. 3, A to F, and figs. S18 to S21). We then classified neurons by cell type on the basis of binarized marker gene expression (Fig. 3, A and B) and inspected their location. We found that individual excitatory cell types were largely confined to cytoarchitecturally defined subnuclei: The medial nucleus split into Med, MedL, and MedDL; the interposed nucleus split into IntA and IntP; and the lateral nucleus (Lat) remained unsplit (table S1). Within each subnucleus, Class-A and Class-B neurons were intermingled, albeit with local density differences (Fig. 3, C to F, and figs. S18 to S19). Most subnuclei contained only a single excitatory cell type per class; if two cell types from the same class were present in a subnucleus, they were often spatially segregated.
Fig. 3. Spatial organization of mouse cell types.
(A) A STARmap coronal section of the cerebellar nuclei, showing seven markers for illustration; representative of two animals, each including two hemispheres of three to six coronal sections spanning the anterior–posterior axis of the cerebellar nuclei. Cytoarchitectonic subnuclei boundaries are indicated. Scale bar, 100 μm. (B) Enlargement of the area marked in (A). Scale bar, 100 μm. Four excitatory cells are marked and decomposed into the seven illustrated STARmap channels. Comparison to snRNAseq data (dot plot) yields the classification of the cells into transcriptomic cell types. (C to F) Classification results of the same section shown in (A). All excitatory and inhibitory neurons are colored by their assigned transcriptomic cell type in Fig. 2C; excitatory neurons only colored by class (D); Class-A–only (E) and Class-B–only (F) excitatory neurons colored by their transcriptomic cluster showing subnuclei specificity. Unassigned neurons are in gray. (G) Summary of STARmap results for all excitatory cell types, noting the location of each cell type and new cell type names. Gray entries signify minor contributions to the indicated subnuclei. (H) Correlation matrix of all excitatory cell types annotated by subnuclei location. IntA correlates well with Lat in both Class-A and Class-B, whereas IntP is more similar to medial nucleus cell types. (I) Hierarchical clustering of subnuclei. Line color and numbers indicate bootstrapping-based branch confidence.
As an example, consider the interposed nucleus (Fig. 3, C to F, and fig. S18 to S19). Among Class-B cell types, e13 was restricted to IntP, whereas e11 and e12 were both located in IntA only. However, e11 was located only in anterior-most IntA, and e12 was located in posterior-most IntA. Likewise, among Class-A cell types, e5 and e6 were restricted to IntP—with e6 located more laterally than e5—and e4 was confined to IntA. To reflect these findings, we renamed excitatory cell types to indicate both their subnucleus location and class (Fig. 3G and table S1).
Both pairwise correlations of the average transcriptomes of excitatory cell types (Fig. 3H) and hierarchical clustering of excitatory neurons averaged at the level of subnuclei (Fig. 3I) revealed consistent relations between subnuclei within and across classes. Medial subnuclei grouped with each other. IntP grouped with the medial nucleus in Class-B, whereas IntA was more closely related to Lat. Inspection of differentially expressed genes across subnuclei revealed both class-specific, subnucleus-independent (fig. S16 and S17) and class-independent, subnucleus-specific gene sets (fig. S22, A and B). To explicitly account for these two independent axes of variation, we developed a region-aware neighbor-joining algorithm that allows both the duplication of cell types within a region and the duplication of regions themselves (materials and methods). The root of the resulting tree is formed by a single region containing both Class-A and Class-B cell types (fig. S22C), suggesting that the ancestral cerebellar nucleus contained both Class-A and Class-B neurons.
In contrast to the subnucleus specificity of excitatory neurons, inhibitory neurons were broadly distributed across subnuclei (figs. S20 and S21B). The only exception was reduced numbers of i1 neurons and increased numbers of i3 neurons in the medial nucleus, mirroring our snRNAseq data (fig. S12C).
In summary, spatial transcriptomic analysis indicated a simple organizing principle for the cerebellar nuclei. Subnuclei are the repeating units that form the cerebellar output channels. Each subnucleus contains a stereotyped cell-type set: one or two types each of Class-A and Class-B excitatory neurons that are subnucleus-specific, and three inhibitory classes that are subnucleus-invariant.
Subnuclei as units of evolutionary duplication
Our mouse data suggest a model of cerebellar nucleus evolution wherein a stereotyped cell-type set is duplicated over the course of evolution to form a new subnucleus (fig. S22C), accompanied by changes in gene expression and shifts of projection targets for the new subnucleus relative to old ones. To test this model, we investigated the transcriptomic cell types of chicken cerebellar nuclei.
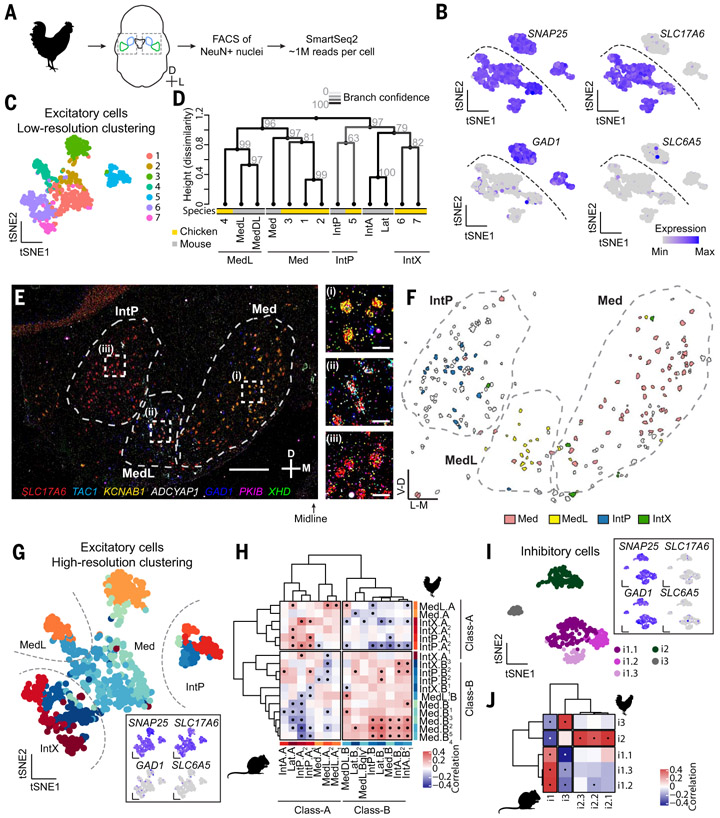
Chickens are thought to have two pairs of cerebellar nuclei without the equivalent of the mammalian lateral nucleus (Fig. 1B) (12, 31). We dissected the entire chicken cerebellar nuclei together for snRNAseq (Fig. 4A) and retained 1238 high-quality neurons. These cells split into major groups in a pattern comparable to that of mouse cells, with one broad group of excitatory neurons and two major groups of inhibitory neurons (Fig. 4B and figs. S23 and S24). SLC6A5 expression was sparse, indicating few glycinergic cells in the chicken cerebellar nuclei.
Fig. 4. Cerebellar nuclei cell types in the chicken.
(A) Workflow for chicken snRNAseq. The entire cerebellar nuclei were dissected together from frozen tissue. (B) Marker expression in all neurons. Dashed line divides excitatory and inhibitory neurons. N = 3 chickens. (C) Coarse clustering result of all excitatory neurons. (D) Dendrogram showing hierarchical clustering of coarse excitatory chicken clusters and mouse excitatory neurons averaged by subnuclei. Line color and gray numbers indicate bootstrapping-based branch confidence. (E) A STARmap coronal section of the chicken cerebellar nuclei, representative of N = 7 sections from three animals. Scale bar: main panel, 500 μm; inset, 50 μm. (F) Classification results of the excitatory cells shown in (E) into subnuclei inferred in (D). The subnuclei form spatially distinct structures. Unassigned neurons are in gray. (G) High-resolution clustering results of chicken excitatory neurons. Inset shows marker expression. (H) Correlation matrix between mouse and chicken excitatory cell types. A division of chicken excitatory neuron types into Class-A and Class-B is apparent. Dots indicate significant correlations. (I) Clustering results of inhibitory neurons. Inset shows marker expression. (J) Correlation matrix between mouse and chicken inhibitory neurons. Dots indicate significant correlations.
To understand cerebellar nuclei evolution at the level of subnuclei, we first focused on the excitatory chicken neurons and coarsely clustered them (Fig. 4C) on the basis of the observation that mouse excitatory cells clustered coarsely by subnuclei (Figs. 2E and 3H). We then built a joint phylogenetic tree of these coarse chicken clusters and mouse subnuclei in the space of differentially expressed genes shared across species (Fig. 4D and materials and methods). Mouse subnuclei intermingled with chicken clusters, indicating that chicken cerebellar nuclei contained regions homologous to mouse Med, MedL/MedDL, and IntP, but not IntA and Lat. The chicken cerebellar nuclei also included an additional region that fell within the same clade as the mouse interposed and lateral nuclei. We call this region IntX.
To confirm that the chicken subnuclei indeed form spatially distinct structures, we applied STARmap in situ sequencing to the chicken cerebellar nuclei (Fig. 4, E and F, and fig. S25). Probing for subnuclei marker genes, we identified Med, MedL, and IntP as separate structures spanning the mediolateral axis of the cerebellar nuclei (Fig. 4, E and F). IntX occupied the rostral end of the cerebellar nuclei (fig. S25A). The identification of shared and new regions in the chicken and mouse supports the notion that the cerebellar nuclei number increased by the duplication and divergence of subnuclei.
Conserved neuronal classes across amniotes
Next, we sought to determine if the above model held at the resolution of cell types; specifically, is the distinction between Class-A and Class-B excitatory neurons in the mouse conserved in the chicken? We clustered the chicken excitatory neurons at a higher resolution (Fig. 4G and fig. S23, A and D), aiming to match clustering resolution between mouse and chicken data, and compared them to the mouse excitatory cell types. Correlational analysis between mouse and chicken cell types in the space of shared differentially expressed genes revealed both Class-A and Class-B excitatory neurons in the chicken, with good correspondence to the mouse cell types (Fig. 4H). Notably, each subnucleus contained representatives of both Class-A and Class-B neurons, which was confirmed by STARmap in situ sequencing (fig. S25). We named the chicken cell types according to the mouse convention to reflect their subnuclei (Fig. 4, D to F) and class membership (Fig. 4H). Comparison of mouse and chicken data at single-cell resolution confirmed the results of our correlational analysis (figs. S26 and S27). Application of region-aware neighbor-joining algorithm to the chicken excitatory cell types revealed that, as in the mouse, the root of the tree is formed by a single region containing a Class-A and a Class-B cell type (fig. S23H). All chicken excitatory cell types could be robustly distinguished by differentially expressed genes (figs. S23G and S24).
Analysis of chicken inhibitory neurons revealed five cell types that, like mouse inhibitory neurons, fell into three classes (Fig. 4I and fig. S23, B to D). Correlation analysis to the mouse data showed a perfect match between the species at the class level (Fig. 4J). At a finer resolution, our data indicated independent cell-type diversification or loss of ancestral diversity in chickens and mice in classes i1 and i2, respectively. Whereas the putatively inferior olive-projecting i1 class comprised three cell types in chickens, only a single cell type was found in mice. Conversely, whereas class i2 contained three cell types in mice, it contained only a single type in chickens.
Taken together, our chicken data indicate the conservation of the previously identified archetypal subnuclei in both excitatory and inhibitory cell classes. Our findings thus support the proposal that amniote cerebellar nuclei evolved by repeatedly duplicating an archetypal subnucleus composed of a deeply conserved cell-type set (Fig. 5K, left).
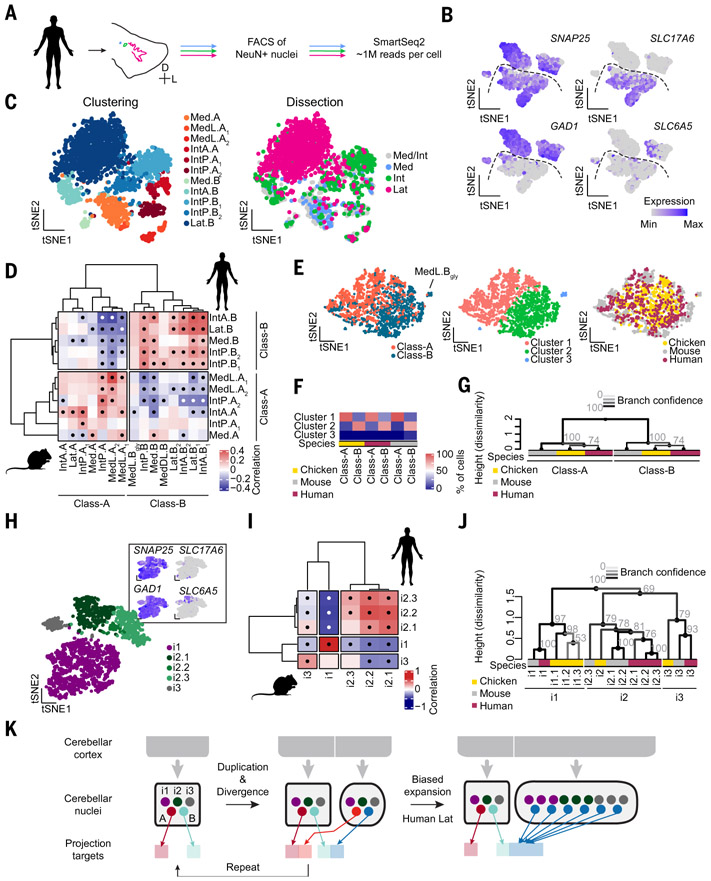
Fig. 5. Class-B neurons expanded in human lateral nucleus.
(A) Workflow for human snRNAseq. The three cerebellar nuclei are separately dissected from frozen tissue. (B) Marker expression for all neurons. N = 3 donors. Dashed line divides excitatory and inhibitory neurons. (C) Clustering results of human excitatory neurons, colored by cluster assignment and dissection. Dissection labels are imperfect, owing to close apposition of individual cerebellar nuclei. Med/Int indicates a mixed dissection. (D) Correlation matrix of mouse and human excitatory cell types. Medial and interposed nuclei contain cell types that correlate with both mouse Class-A and Class-B cell types. Lateral nucleus neurons only correlate with Class-B neurons. Dots indicate significant correlations. (E) Seurat integration of excitatory neurons from three species, colored by class (left), clustering results in integrated space (middle), and species (right). (F) Quantification of membership to the integrated clusters of Class-A and Class-B cells. Across species, Class-A and Class-B cells fall into the same clusters. (G) Hierarchical clustering of excitatory neurons averaged by class, showing conservation of excitatory cell classes across amniotes. Grayscales of line and numbers indicate bootstrapping-based branch confidence. (H) Clustering results of human inhibitory neurons. Cells are colored by cluster assignment. Marker expression is indicated in the inset. (I) Correlation matrix of mouse and human inhibitory neurons, showing one-to-one correspondences. Dots indicate significant correlations. (J) Hierarchical clustering of inhibitory cell types across all three species (color coded), showing conservation of three inhibitory classes across amniotes. Grayscales of line and numbers as above. (K) Schematic illustrating the proposed model of subnucleus duplication-and-divergence (left) and biased expansion of Class-B excitatory neurons in human lateral nucleus (right).
Class-B expanded in the human lateral nucleus
Cerebellar nuclei differ not only in number across vertebrates but also in size of individual nuclei. The marked expansion of the human lateral nucleus is a prime example. This expansion could be the result of an even increase in neuron numbers across all cell types, the formation of new subnuclei within the lateral nucleus by duplication-and-divergence, or the formation of many de novo subnuclei within the lateral nucleus. To distinguish these possibilities, we determined the transcriptomic cell types of the human medial, interposed, and lateral nuclei. We separately dissected the three nuclei from postmortem human cerebella and processed them for snRNAseq (Fig. 5A). The 4722 high-quality neurons clustered into four major groups, as in the mouse (Fig. 5B and figs. S28 and S29).
Human excitatory neurons readily separated by dissection labels (Fig. 5C and fig. S28A), mirroring the nucleus specificity observed in the mouse. The medial and interposed nuclei formed four and five distinct cell types, respectively. Unexpectedly, lateral nucleus neurons, although contributing almost half of all excitatory neurons in our dataset (1042 out of 2340 neurons), formed only a single cluster. We then compared excitatory cell types from mice and humans using correlation analysis (Fig. 5D). Whereas the human medial and interposed nuclei contained Class-A and Class-B neurons, the lateral nucleus contained only Class-B neurons.
Despite this variation on the archetypal subnucleus, Seurat integration of mouse, chicken, and human excitatory neurons resulted in Class-A and Class-B clusters that were conserved across all species (Fig. 5, E and F). Similarly, hierarchical clustering of excitatory neurons averaged at the level of classes in the three species supports the conservation of Class-A and Class-B across amniotes (Fig. 5G).
Clustering the inhibitory neurons revealed five nucleus-invariant cell types in three classes (Fig. 5H and fig. S28, B and C) with perfect correspondence to the mouse inhibitory classes (Fig. 5I). The Slc6a5− i2.3 cell type, which is rare in mice, is much more abundant in humans, reducing the overall abundance of Slc6a5+ cells in human cerebellar nuclei (fig. S28E). Taken together with the absence of SLC6A5+ neurons in the chicken, this suggests that glycinergic neurons became abundant in the clade leading to the mouse after the divergence of rodents and primates. Hierarchical clustering of all inhibitory cell types in chickens, mice, and humans confirms the results of the pairwise comparisons (Figs. 4J and 5I) and supports the classification of inhibitory neurons into three conserved classes (Fig. 5J).
In summary, the human medial and interposed nuclei follow the cell-type composition of the archetypal cerebellar nuclei. However, in the human lateral nucleus, Class-B neurons are expanded at the expense of Class-A neurons, suggesting that evolution tuned relative abundance of cell types within the framework of duplicating a stereotyped cell-type set (Fig. 5K, right).
Connectivity differences of Class-A and Class-B neurons
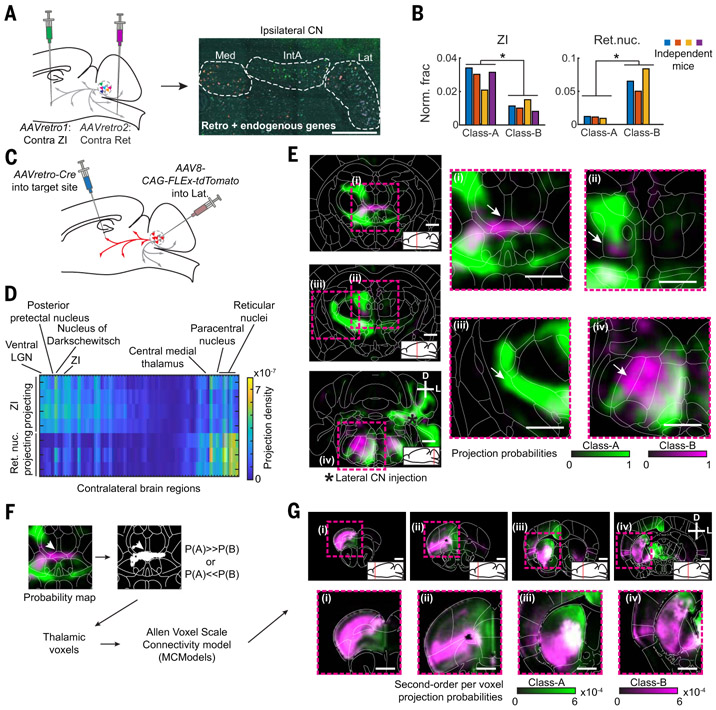
To investigate the implication of the selective Class-B neuron expansion in the human lateral nucleus, we sought to determine how Class-A and Class-B neurons differ in brain-wide projection patterns. As cell type–specific tracing is not feasible in humans, we performed this analysis in mice, where the lateral nucleus contains both Class-A and Class-B neurons. Double retrograde tracing combined with STARmap in situ sequencing to identify projection targets of either class (Fig. 6A) revealed that most target regions labeled both Class-A and Class-B neurons roughly equally. Consistently, collateralization mapping indicated broad projection patterns of cerebellar nuclei neurons (fig. S30). However, contralateral zona incerta (ZI) injections preferentially labeled Class-A neurons in the lateral nucleus, whereas contralateral brainstem reticular nucleus (Ret) injections primarily labeled Class-B neurons (Fig. 6B).
Fig. 6. Differential projections of lateral nucleus Class-A and Class-B neurons in mice.
(A) Schematic of retrograde tracing and STARmap identification of Class-A and Class-B neurons in the lateral nucleus. Contralateral zona incerta (ZI) and contralateral parvocellular reticular nucleus were injected with different AAVretro tracers. Gene expression was then measured by STARmap in the ipsilateral lateral nucleus. Scale bar, 500 μm. (B) Quantification of retrograde tracing results across N = 3 or 4 independent mice in the lateral nucleus at class resolution. *p < 0.05, paired t test without corrections for multiple comparisons. (C) Schematic of collateralization mapping experiments. (D) Heat map showing all differentially innervated contralateral regions (p < 0.05, no multiple comparison correction) from Ret-projecting (N = 3) and ZI-projecting (N = 4) lateral nucleus cells. Brain regions are sorted by mean innervation difference. (E) Probability maps of Class-A and Class-B projection patterns as computed from ZI and Ret collateralization patterns. Regions of differential intralaminar thalamus innervation ((i) and (ii)) and of the AAVretro-Cre injection sites ((iii) and (iv)) by Class-A and Class-B neurons are highlighted. Scale bar, 1 mm. (F) Workflow for in silico tracing of second-order projections from preferentially Class-A– or Class-B-innervated thalamic voxels. Starting voxels are identified and fed into a brain-wide voxel scale connectivity model (33). (G) Coronal sections showing brain-wide normalized projection probabilities from thalamic voxels preferentially innervated by Class-A (green) or Class-B (magenta). Scale bar, 1 mm.
To investigate which other brain regions are differentially innervated by Class-A and Class-B neurons, we performed whole-brain collateralization mapping (32) initiated at ZI and Ret (Fig. 6C). Ret injections labeled a smaller set of lateral nucleus neurons, with a more restricted projection pattern than ZI-projecting neurons. In many brain regions, projections of the Ret-projecting neurons and ZI-projecting neurons overlapped (figs. S31 to S33 and table S4). However, several regions of the contralateral intralaminar nuclei of the thalamus—including paracentral nucleus and central medial nucleus—were more innervated by Ret-projecting than ZI-projecting neurons (Fig. 6D and fig. S31, C and D).
Although ZI- and Ret-projecting neurons did not perfectly correspond to Class-A and Class-B, respectively (Fig. 6B), knowing the ratio of Class-A versus Class-B labeling from retrograde tracing allowed us to estimate the underlying projection probability maps for Class-A and Class-B neurons (materials and methods). The resulting computed maps reinforced the previous results of Class-B projections to the intralaminar thalamus (Fig. 6E, fig. S34) but also highlighted intralaminar regions innervated by nonoverlapping projections of both Class-A and Class-B neurons [Fig. 6E(ii)].
To investigate the relevance of these finer differences in Class-A and Class-B projection patterns, we first identified the thalamic voxels more likely to be innervated by Class-A than Class-B neurons, and vice versa. We then used these voxels as starting points for in silico anterograde tracing using the Allen Atlas voxel scale connectivity model (33) (Fig. 6F). The resulting projection probability maps revealed specific projections from primarily Class-B–innervated thalamic voxels to a lateral network of frontal association, ventral orbital, and insular cortices (30), as well as ventrolateral striatum (Fig. 6G and fig. S35, C and D). Conversely, in silico tracing from primarily Class-A neuron–innervated thalamic voxels revealed relatively broader projections to frontal cortical regions, but with a strong bias toward a medial network, including medial prefrontal cortex and anterior cingulate cortex, as well as dorsomedial striatum (Figs. 6G and fig. S35, A and B). We obtained similar results when we performed the same analysis based directly on ZI- and Ret-initiated collateralization maps (fig. S35, E to H), indicating that our results were not an artifact of our inferred class-level projection maps. Class-A and Class-B cerebellar nuclei neurons, therefore, funnel information through the thalamus to different prefrontal networks in the mouse. Given the expansion of Class-B in the human lateral nucleus, and assuming conservation of the discovered projection networks, these results suggest that cerebellar connectivity to the lateral prefrontal network is preferentially expanded in humans.
Discussion
Here we present comprehensive datasets describing cerebellar nuclei transcriptomic cell types and brain-wide projections in mice, as well as transcriptomic cell types in chickens and humans. These data reveal a conserved cell-type set that makes up an archetypal subnucleus, which we propose is effectively duplicated during evolution to increase the number of cerebellar subnuclei and thus the number of cerebellar output channels. In addition, the predominance of Class-B excitatory neurons in human lateral nuclei indicates that the archetypal composition can be modified by varying the relative abundance of constituent cell types (Fig. 5K).
Subnuclei are the repeating units
At the outset, we took advantage of the variations of the cerebellar nuclei number in different species to investigate brain region evolution (Fig. 1B). We discovered instead that the fundamental repeating units in the mouse cerebellar nuclei are the subnuclei, each of which is formed by the same stereotyped cell-type set (Fig. 3). This set contains one or two subnucleus-specific Class-A and Class-B excitatory neurons each and the three classes of subnucleus-invariant inhibitory cell types.
Comparisons of excitatory and inhibitory neurons across neocortical regions also suggest a region-specific set of excitatory cell types accompanied by a region-invariant set of inhibitory cell types (6). Developmentally, neocortical excitatory neurons derive from the ventricular zone through local radial migration, whereas inhibitory neurons originate from the ventral forebrain through long-distance tangential migration (34). Thus, despite the opposite migratory paths giving rise to excitatory and inhibitory neurons, the cerebellar nuclei and neocortex share a similar feature: region-specific excitatory cell types and region-invariant inhibitory cell types.
Brain region evolution by duplication and divergence
Comparison between mice and chickens revealed that the stereotyped cell-type set in subnuclei is deeply conserved across amniotes (Fig. 4) and thus likely describes an archetypic cell-type composition of the cerebellar nuclei in the last common ancestor of birds and mammals 320 million years ago. Our data suggest a model wherein cerebellar subnuclei increased in number by repeatedly duplicating the entire cell-type set—likely achieved by a coordinated expansion of cell numbers within all cell types followed by anatomical regionalization. Such duplications were accompanied by divergence in gene expression in the excitatory but not inhibitory neurons (Figs. 2 to 5), and in projection patterns (Fig. 1). Overall, cerebellar nuclei evolution is therefore best described as region-level duplication-and-divergence (Fig. 5K, left). At finer resolution, however, duplication-and-divergence (neofunctionalization) is restricted to rhombic lip–derived excitatory neurons, and duplication-and-maintenance (isofunctionalization) appears to govern the evolution of ventricular zone–derived inhibitory neurons.
The developmental implementation of such regional “duplications” of a cell-type set could take a multitude of paths. These include duplication of an early multipotent progenitor (l) or establishment of a new region-defining morphogen gradient (35). Analysis of the cerebellar nuclei in more species and detailed developmental investigations are needed to distinguish these possibilities. We expect that differences in the two developmental sources of cerebellar nuclei neurons will explain the divergence versus maintenance of transcriptomic state observed for excitatory and inhibitory neurons, respectively.
Functionally, the duplication-and-divergence model of cerebellar nuclei evolution implies that each subnucleus should be considered as an output node of the cerebellum. Together with evidence of topographic projections from Purkinje cells to the cerebellar nuclei (36) and the crystalline cerebellar motif (37), our finding suggests that specific regions of the cerebellar cortex and their connected subnuclei act as a functional module in parallel with other such modules (38). Increased functionality of the cerebellum across evolution might be implemented by the addition of such cerebellar cortex–nuclei modules to brain-wide circuits. The control of cerebellar cortex size by the number of excitatory cerebellar nuclei neurons (39,40) provides a simple mechanism for coordinated evolutionary expansion of cerebellar cortex and nuclei.
Brain region evolution by duplication-and-divergence naturally favors the evolution of modular neuronal circuits, with dense connections within the duplicated unit and comparatively sparse connections between units. This built-in modularity may speed up the rate of evolution (41) and explain the modular nature of brain networks.
Variations within the duplication-and-divergence framework
There is considerable variation in the brain region duplication-and-divergence framework proposed above. The existence of several representatives of Class-A or Class-B cell types in individual cerebellar subnuclei suggests within-subnucleus cell-type diversification (Fig. 3G). Conversely, varying numbers of cell types per inhibitory cell class i1 and i2 in mammals and chickens (Figs. 4J and 5I) highlight the possibility of gain of new diversity or loss of ancestral diversity that uniformly affects all regions. Individuation of cell types after region-level duplication, moreover, can be substantial, as illustrated by the apparent neurotransmitter switch in the rhombic lip–derived, Slcl7a6−/Slc6a5+ MedL.Bgly cell type in mice (Fig. 2C).
The biased expansion of the human lateral nucleus illustrates the possibility of drastic changes in relative cell type abundance within the archetypal set. In the mouse, Class-B neurons of the lateral nucleus preferentially funnel information into frontal association cortex and lateral orbital and insular regions via the thalamus, whereas Class-A neurons preferentially access a medial network, including medial prefrontal and anterior cingulate cortex (Fig. 6). The human lateral nucleus is greatly expanded relative to the other nuclei, likely owing to temporal expansion of the rhombic lip progenitor zones (42), but it has largely lost the Class-A neurons (Fig. 5). The expansion of the human lateral nucleus in general, and Class-B neurons within it, might have occurred in concert with the expansion of the human frontal cortical regions, and potentially elaboration of the thalamic intermediaries. We thus predict that the homolog to mouse lateral frontal cortex is expanded in humans. The precise evolutionary relationships between mouse and human frontal cortical regions, however, are currently unclear (43). Future comparative transcriptomic and connectomic work on mammalian frontal cortex evolution will shed more light on this question.
In conclusion, our studies of the cerebellar nuclei evolution suggest a duplication-and-divergence framework for brain region evolution at cell-type resolution. Investigations of other brain regions using approaches outlined here may provide insight into how generalizable this framework is and will deepen our understanding of how brains changed over the course of evolution.
Materials and methods summary
Animal procedures were approved by the Stanford University or the University of California Davis Animal Care and Use Committee and were carried out in accordance with National Institutes of Health standards. We performed all experiments in adult male mice and chickens, and human donors of both sexes.
For unconditional anterograde tracing (Fig. 1), we injected AAV8-CAG-tdTomato virus into one cerebellar nucleus per animal. In conditional tracing experiments (Fig. 6), we injected AAVretro-Ef1a-cre into a target region and AAV8-CAG-FLEx-tdTomato virus into the lateral nucleus. After a minimum of 3 weeks of expression, we perfused the mice with paraformaldehyde, dissected the brain and spinal cord, and subjected these tissues to brain clearing. Briefly, we dilipidated the whole-mount samples, stained them with antibodies against red fluorescent protein and Alexa-647-conjugated secondary antibody, and chemically cleared the samples. We then imaged the cleared brains using light-sheet microscopy and quantified axonal innervation. We traced second-order projections (Fig. 6) in silico on the basis of a voxel-level mouse brain connectivity matrix from the Allen Brain Institute.
For single-nucleus RNA sequencing experiments (Figs. 2, 4, and 5), we dissected the cerebellar nuclei from acute (mouse) or frozen sections (chicken, human), liberated cellular nuclei by mechanical force, and stained samples for NeuN using anti-NeuN antibody and phycoerythrin (PE)–conjugated secondary antibody. We then used fluorescence-activated cell sorting to select NeuN+ nuclei and sorted them into individual wells of 384-well plates. In each well, we performed a customized SmartSeq2 protocol. Briefly, we produced double-stranded cDNA by template switching, preamplified the cDNA using polymerase chain reaction (PCR), fragmented the amplified cDNA using Tn5 enzyme loaded with Nextera adapters, and finally produced barcoded Illumina libraries for each well by performing PCR on the fragments with primers containing i5 and i7 sample barcodes. We then pooled size-selected libraries and sequenced them on Illumina Novaseq machines with PE100. We aligned demultiplexed sequencing data to the respective genome (all from Ensembl). For each gene, we counted all reads that mapped to exons or introns. We then analyzed the count tables in Seurat. We performed cross-species comparisons using cluster-level gene correlations, using the intersection of within-species differentially expressed genes as a basis and considering only 1:1 orthologous genes. We also used the data integration tools of Seurat and CONOS for cross-species comparisons at the single-cell level.
For STARmap experiments (Figs. 2, 3, 4, and 6), we flash froze mouse or chicken brains and cryosectioned them into 16-μm sections. We then annealed gene-specific SNAIL probes, ligated them, and amplified the correctly annealed probes using rolling circle amplification. We gel-embedded these amplified probes and read out gene identify using sequential SEDAL sequencing on a five-color spinning disk confocal microscope. Using a modified spacetx STARfish pipeline, we detected signals and then analyzed data in Matlab.
Detailed descriptions of all experimental protocols and analyses are provided in the supplementary materials.
Supplementary Material
ACKNOWLEDGMENTS
We thank L. O’Connell, H. Fraser, J. Lui, C. McLaughlin, J. Li, and H. Li for comments on the manuscript; members of the Luo lab for support and advice; and F. Zanini, F. Horns, G. Stanley, X. Wang, J. Chang, and S. Holmes for technical advice. We thank the cooperation of Donor Network West and all of the organ and tissue donors and their families, for giving the gift of life and the gift of knowledge, by their generous donation.
Funding:
This work was supported by NIH R01-NS080835 (L.L.), NS104698 (L.L.), RM1-HG007735 (H.Y.C.), and NS091144 (J.B.D.); NSF NeuroNex (K.D., L.L.); Stanford Wu Tsai Neuroscience Institute Neuro-omics Initiative (L.L., S.R.Q.); DARE Fellowship (E.A.); and Jane Coffin Childs Memorial Fund postdoctoral fellowship (J.M.K.). H.Y.C., K.D., and L.L. are Howard Hughes Medical Institute investigators. S.R.Q. is a Chan Zuckerberg investigator.
Footnotes
Competing interests: The authors declare no competing interests.
Data and materials availability: The sequencing datasets generated in this study are available in the NCBI Gene Expression Omnibus under accession GSE160471. Custom analysis code and interactively browsable sequencing data are available at https://github.com/justuskebschull/CNcode_finalor Zenodo (44) unless otherwise indicated. All other data are available in the main paper or supplementary materials. All materials are available through requests to the corresponding authors.
REFERENCES AND NOTES
- 1.Arendt D et al. , The origin and evolution of cell types. Nat Rev. Genet 17, 744–757 (2016). doi: 10.1038/nrg.2016.127; pmid: 27818507 [DOI] [PubMed] [Google Scholar]
- 2.Tosches MA, Developmental and genetic mechanisms of neural circuit evolution. Dev. Biol 431, 16–25 (2017). doi: 10.1016/j.ydbio.2017.06.016; pmid: 28645748 [DOI] [PubMed] [Google Scholar]
- 3.Chakraborty M, Jarvis ED, Brain evolution by brain pathway duplication. Philos. Trans. R. Soc. Lond. B Biol. Sci 370, 20150056 (2015). doi: 10.1098/rstb.2015.0056; pmid: 26554045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grillner S, Robertson B, The Basal Ganglia Over 500 Million Years. Curr. Biol 26, R1088–R1100 (2016). doi: 10.1016/j.cub.2016.06.041; pmid: 27780050 [DOI] [PubMed] [Google Scholar]
- 5.Frangeul L et al. , A cross-modal genetic framework for the development and plasticity of sensory pathways. Nature 538, 96–98 (2016). doi: 10.1038/nature19770; pmid: 27669022 [DOI] [PubMed] [Google Scholar]
- 6.Yao Z et al. , A taxonomy of transcriptomic cell types across the isocortex and hippocampal formation. bioRxiv 2020.03.30.015214 [Preprint] (2020). 10.1101/2020.03.30.015214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hodge RD et al. , Conserved cell types with divergent features in human versus mouse cortex. Nature 573, 61–68 (2019). doi: 10.1038/s41586-019-1506-7; pmid: 31435019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bakken TE et al. , Evolution of cellular diversity in primary motor cortex of human, marmoset monkey, and mouse. bioRxiv 2020.03.31.016972 [Preprint]. (2020). 10.1101/2020.03.31.016972. [DOI] [Google Scholar]
- 9.Tosches MA et al. , Evolution of pallium, hippocampus, and cortical cell types revealed by single-cell transcriptomics in reptiles. Science 360, 881–888 (2018). doi: 10.1126/science.aar4237; pmid: 29724907 [DOI] [PubMed] [Google Scholar]
- 10.Krienen FM et al. , Innovations in Primate Interneuron Repertoire. bioRxiv 709501 [Preprint] (2019). 10.1101/709501. [DOI] [Google Scholar]
- 11.Peng YR et al. , Molecular Classification and Comparative Taxonomics of Foveal and Peripheral Cells in Primate Retina. Cell 176, 1222–1237.e22 (2019). doi: 10.1016/j.cell.2019.01.004; pmid: 30712875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yopak KE, Pakan JMP, Wylie D, in Evolution of Nervous Systems, Kaas JH, Ed. (Academic Press, ed. 2, 2016), vols. 1–4, pp. 373–385. [Google Scholar]
- 13.Buckner RL, The cerebellum and cognitive function: 25 years of insight from anatomy and neuroimaging. Neuron 80, 807–815 (2013). doi: 10.1016/j.neuron.2013.10.044; pmid: 24183029 [DOI] [PubMed] [Google Scholar]
- 14.Wagner MJ, Luo L, Neocortex-Cerebellum Circuits for Cognitive Processing. Trends Neurosci. 43, 42–54 (2020). doi: 10.1016/j.tins.2019.11.002; pmid: 31787351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chan-Palay V, in Cerebellar Dentate Nucleus (Springer, 1977). [Google Scholar]
- 16.Teune TM, van der Burg J, van der Moer J, Voogd J, Ruigrok TJH, Topography of cerebellar nuclear projections to the brain stem in the rat. Prog. Brain Res 124, 141–172 (2000). doi: 10.1016/S0079-6123(00)24014-4; pmid: 10943123 [DOI] [PubMed] [Google Scholar]
- 17.Tellmann S et al. , Cytoarchitectonic mapping of the human brain cerebellar nuclei in stereotaxic space and delineation of their co-activation patterns. Front. Neuroanat. 9, 54 (2015). doi: 10.3389/fnana.2015.00054; pmid: 26029057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bostan AC, Dum RP, Strick PL, Cerebellar networks with the cerebral cortex and basal ganglia. Trends Cogn. Sci 17, 241–254 (2013). doi: 10.1016/j.tics.2013.03.003; pmid: 23579055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Uusisaari MY, Knöpfel T, in Handbook of the Cerebellum and Cerebellar Disorders (Springer, 2013), pp. 1101–1110. [Google Scholar]
- 20.Gould BB, The organization of afferents to the cerebellar cortex in the cat: Projections from the deep cerebellar nuclei. J. Comp. Neurol 184, 27–42 (1979). doi: 10.1002/cne.901840103; pmid: 762281 [DOI] [PubMed] [Google Scholar]
- 21.Aumann TD, Rawson JA, Finkelstein DI, Horne MK, Projections from the lateral and interposed cerebellar nuclei to the thalamus of the rat: A light and electron microscopic study using single and double anterograde labelling. J. Comp. Neurol 349, 165–181 (1994). doi: 10.1002/cne.903490202; pmid: 7860776 [DOI] [PubMed] [Google Scholar]
- 22.Chung S-H, Marzban H, Hawkes R, Compartmentation of the cerebellar nuclei of the mouse. Neuroscience 161, 123–138 (2009). doi: 10.1016/j.neuroscience.2009.03.037; pmid: 19306913 [DOI] [PubMed] [Google Scholar]
- 23.Gao Z et al. , A cortico-cerebellar loop for motor planning. Nature 563, 113–116 (2018). doi: 10.1038/s41586-018-0633-x; pmid: 30333626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim KK, Adelstein RS, Kawamoto S, Identification of neuronal nuclei (NeuN) as Fox-3, a new member of the Fox-1 gene family of splicing factors. J. Biol. Chem 284, 31052–31061 (2009). doi: 10.1074/jbc.M109.052969; pmid: 19713214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Elsen GE, Juric-Sekhar G, Daza RAM, Hevner RF, in Handbook of the Cerebellum and Cerebellar Disorders (Springer, 2013), pp. 179–206. [Google Scholar]
- 26.Fink AJ et al. , Development of the deep cerebellar nuclei: Transcription factors and cell migration from the rhombic lip. J. Neurosci 26, 3066–3076 (2006). doi: 10.1523/JNEUROSCI.5203-05.2006; pmid: 16540585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang X et al. , Three-dimensional intact-tissue sequencing of single-cell transcriptional states. Science 361, eaat5691 (2018). doi: 10.1126/science.aat5691; pmid: 29930089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bagnall MW et al. , Glycinergic projection neurons of the cerebellum. J. Neurosci 29, 10104–10110 (2009). doi: 10.1523/JNEUROSCI.2087-09.2009; pmid: 19675244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Prekop HT et al. , Sox14 is required for a specific subset of cerebello–olivary projections. J. Neurosci 38, 9539–9550 (2018). doi: 10.1523/JNEUROSCI.1456-18.2018; pmid: 30242051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Paxinos G, Franklin KBJ, Paxinos and Franklin’s the Mouse Brain in Stereotaxic Coordinates (Academic Press, ed. 4, 2011). [Google Scholar]
- 31.Feirabend HKP, Voogd J, Myeloarchitecture of the cerebellum of the chicken (Gallus domesticus): An atlas of the compartmental subdivision of the cerebellar white matter. J. Comp. Neurol 251, 44–66 (1986). doi: 10.1002/cne.902510104; pmid: 3760258 [DOI] [PubMed] [Google Scholar]
- 32.Schwarz LA et al. , Viral-genetic tracing of the input-output organization of a central noradrenaline circuit. Nature 524, 88–92 (2015). doi: 10.1038/nature14600; pmid: 26131933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Knox JE et al. , High-resolution data-driven model of the mouse connectome. Netw. Neurosci 3, 217–236 (2018). doi: 10.1162/netn_a_00066; pmid: 30793081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marín O, Rubenstein JLR, Cell migration in the forebrain. Annu. Rev. Neurosci 26, 441–483 (2003). doi: 10.1146/annurev.neuro.26.041002.131058; pmid: 12626695 [DOI] [PubMed] [Google Scholar]
- 35.O’Leary DDM, Chou SJ, Sahara S, Area patterning of the mammalian cortex. Neuron 56, 252–269 (2007). doi: 10.1016/j.neuron.2007.10.010; pmid: 17964244 [DOI] [PubMed] [Google Scholar]
- 36.Sugihara I, Shinoda Y, Molecular, topographic, and functional organization of the cerebellar nuclei: Analysis by three-dimensional mapping of the olivonuclear projection and aldolase C labeling. J. Neurosci. 27, 9696–9710 (2007). doi: 10.1523/JNEUROSCI.1579-07.2007; pmid: 17804630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ito M, Cerebellar circuitry as a neuronal machine. Prog. Neurobiol 78, 272–303 (2006). doi: 10.1016/j.pneurobio.2006.02.006; pmid: 16759785 [DOI] [PubMed] [Google Scholar]
- 38.Ekerot CF, Jörntell H, Garwicz M, Functional relation between corticonuclear input and movements evoked on microstimulation in cerebellar nucleus interpositus anterior in the cat. Exp. Brain Res 106, 365–376 (1995). doi: 10.1007/BF00231060; pmid: 8983981 [DOI] [PubMed] [Google Scholar]
- 39.Willett RT et al. , Cerebellar nuclei excitatory neurons regulate developmental scaling of presynaptic Purkinje cell number and organ growth. eLife 8, e50617 (2019). doi: 10.7554/eLife.50617; pmid: 31742552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fleming J, Chiang C, The Purkinje neuron: A central orchestrator of cerebellar neurogenesis. Neurogenesis 2, e1025940 (2015). doi: 10.1080/23262133.2015.1025940; pmid: 27604220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Alon U, An Introduction to Systems Biology (CRC Press, 2020). [Google Scholar]
- 42.Haldipur P et al. , Spatiotemporal expansion of primary progenitor zones in the developing human cerebellum. Science 366,454–460 (2019). doi: 10.1126/science.aax7526;pmid: 31624095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Carlén M, What constitutes the prefrontal cortex? Science 358, 478–482 (2017). doi: 10.1126/science.aan8868; pmid: 29074767 [DOI] [PubMed] [Google Scholar]
- 44.Kebschull JM et al. , Code accompanying Kebschull et al. 2020 “Cerebellar nuclei evolved by repeatedly duplicating a conserved cell-type set,” Version v0.1). Zenodo; 10.5281/zenodo.4124155. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.