ABSTRACT
In this article, we argue that a careful examination of human microbiome science’s relationship with race and racism is necessary to foster equitable social and ecological relations in the field. We point to the origins and evolution of the problematic use of race in microbiome literature by demonstrating the increased usage of race both explicitly and implicitly in and beyond the human microbiome sciences. We demonstrate how these uses limit the future of rigorous and just microbiome research. We conclude with an outline of alternative actionable ways to build a more effective, antiracist microbiome science.
KEYWORDS: antiracist, ethnicity, justice, microbiome, microbiota, race, racism
PERSPECTIVE
INTRODUCTION: RACE AND MICROBIOME SCIENCE
We are a transdisciplinary group of researchers—a microbiologist, a geographer, an anthropologist, and an evolutionary and microbial ecologist—who are committed to antiracist scholarship and to the effective and ethical future of microbiome science. In this article, we argue that the use of race and other racial proxies as “ghost variables” in most current human microbiome research is problematic and requires interrogation. We examine how racial categories are used in the microbiome literature and conclude with alternative, actionable ways to build equitable and antiracist microbiome science.
Race has no coherent basis in biology. Human groups have extensively shared genetics over time and space through migration and forced displacement. As a result, while there are some geographic signatures of genetic variation, human populations are highly interconnected (1). There is more genetic diversity within racial categories than between “races” (2–4). Thus, genetics do not clearly map onto ideas of race. Racial categories vary across cultural and historical contexts and are socially determined (5). Nevertheless, the harmful effects of historical and contemporary racism have real impacts on people’s lives, bodies, and environments (6). As Amutah et al. argue, “Race is not a biologic category based on innate differences that produce unequal health outcomes. Rather, it is a social category that reflects the impact of unequal social experiences on health” (7). These uneven experiences must be studied in relation to the disease, environmental, and socioeconomic burdens of Black, Indigenous, and People of Color (BIPOC). Categories of race in the microbiome sciences must be used intentionally and with care, and race must be studied in relation to racism.
The microbiome sciences are often complicit in contributing to racial disparities by attributing findings to racial or ethnic differences without referencing racism or by using ghost variables of race. By ghost variables, we are referring to complex, historically loaded racial categories used in microbiome research without explicitly naming race (8). Studies use imprecise labels that inaccurately conflate race with other variables, present racial or ethnic differences in disease or environmental burden without context, or link racial groups with particular diseases or increased disease burden (6). In continuing to use the category of race as a determining variable in research design and analysis, human microbiome research has come to explicitly or implicitly rely on race as holding biological truth independent of social forces. Race, as Kozik demonstrates, serves as a “conveniently measurable proxy” without attending to the confounding structure of racism and its many effects on people and environments (9, 10).
In this article, we combine Ruth Wilson Gilmore’s definition of racism, the production and exploitation of “group-differentiated vulnerability to premature death,” with Paul Farmer’s use of structural violence, the normalized, interacting political and social structures that cause injury, injustice, and oppression, to examine race in studies of the microbiome (11, 12). If the microbiome sciences continue to explicitly and implicitly deploy the variable of race in research without accounting for the relevant forms of racism that impact people and environments, microbiome research will continually provide data and recommendations in support of a system based on inequities and harm.
RACE IN STUDIES OF THE HUMAN MICROBIOME
How did race come to occupy an important role in microbiome research? The advancement of genomics along with the initial findings of the Human Genome Project stoked enthusiasm for personalized medicine and the idea of a postracial science. Findings suggest that humans are more genetically similar than historically assumed, making biological justifications for racial categories untenable. Yet, biological understandings of race reemerged in studies of genetic ancestry (13–15). For the microbiome sciences, this resurgence exerted itself in subtle but powerful ways. For example, a primary goal of the Human Microbiome Project was to characterize the “healthy” microbiome as a critical first step in determining how deviation from a baseline state contributes to human disease (16). From this early work, major innovations included new sequence databases, laboratory methods and technologies, and bioinformatic tools. These advancements triggered a deluge of correlative microbiome studies in and beyond the biomedical sciences. While there have been advancements in mechanistic understandings of the microbiome, the definition of a “healthy microbiome” remains ambiguous (17). In this context characterized by uncertainty, race emerged as a common variable used to make deterministic and comparative claims about the microbiome and to explain health disparities among racial or ethnic groups (8, 18–21).
Among the human microbiome literature archived in PubMed Central (PMC) between 2000 and 2020, there are 14,103 results that mention race or ethnicity. Representative examples of this type of widely cited scholarship describe race as one of the strongest host phenotypes associated with the microbiome (19, 22–24). While a growing body of peer-reviewed human microbiome research exists mentioning race or ethnicity (Fig. 1), it is worrisome that only 114 of the 14,103 PMC-archived human microbiome articles also mention racism. Further review of those results reveals that only 8 of the 114 articles have engaged with the specificity of how racism is embedded in microbiome research; many of them are rarely cited (9, 25–31). These results suggest that current microbiome research risks using race or ethnicity as an explanatory factor—determinant or correlative—of the microbiome. Therefore, the field has limited engagement with the direct effects of racism on human physiology, which include but are not limited to increased trauma and stress, lack of access to quality health care, and historical and current discrimination in treatment within health care systems.
FIG 1.
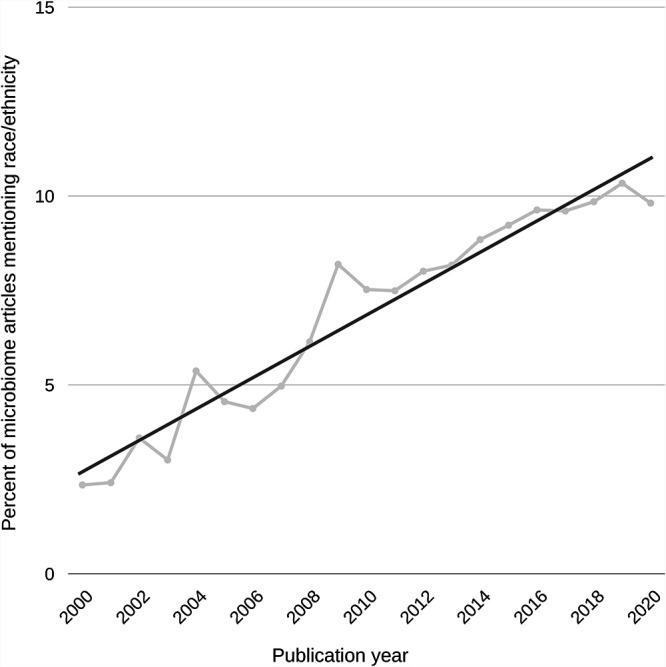
The use of race/ethnicity among human microbiome research has steadily increased since 2000. Data represent Boolean search results of the PMC archive using rentrez in R (69, 70). Search terms representing the human microbiome included microbiome, microbiota, or 16S rRNA and human, patient, subject, volunteer, or participant. Search terms representing race or ethnicity included race, racial, or ethnic.
RACE AS A GHOST VARIABLE IN AND BEYOND THE HUMAN MICROBIOME
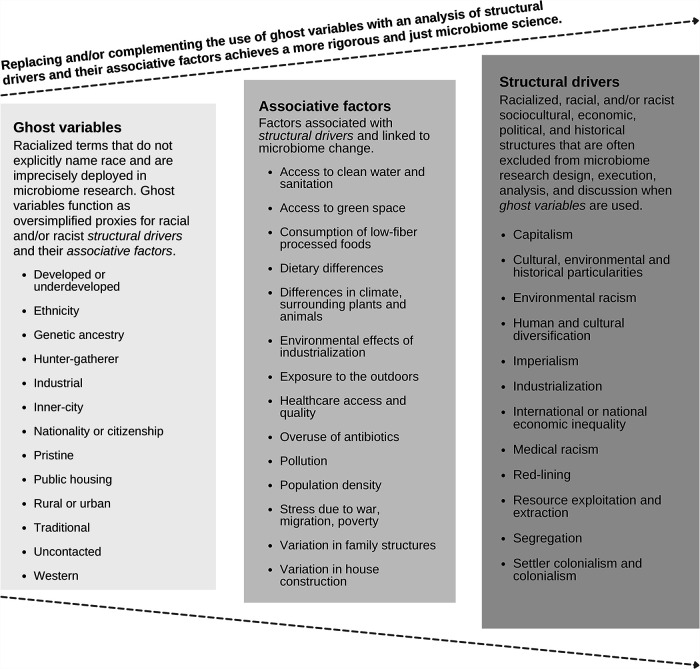
With increased recognition of the problems associated with using race as a category of analysis in microbiome science, we must also examine how often race is used as a ghost variable. Human microbiomes are categorized as “Western,” “industrialized,” or belonging to “Europeans” and “Americans” and compared against the microbiomes of racialized Indigenous and global South populations whose microbiomes are presupposed to be “underdeveloped,” “modernizing,” and closer to “pure” or “natural” states (32–36). Assuming that the microbiomes of Indigenous groups are a baseline somehow ancestral to all humans is a notion that ignores the complexity of genetic, ecological, and cultural divergence in human populations and fails to account for the rapid ecological and evolutionary changes in the microbiome (Fig. 2).
FIG 2.
Replacing and/or complementing the use of ghost variables with an analysis of structural drivers and their associative factors achieves a more rigorous and equitable microbiome science. This figure is not exhaustive but is intended to assist researchers in determining study groups during the early stages of research design or to help analyze existing studies’ use of race or ghost variables.
Throughout this kind of microbiome research is the assertion that BIPOC populations are less developed and have “wild” or “natural” microbiota and that “modern,” “disturbed,” “Western/white” microbiomes must be “rewilded.” In some of these studies, racial categories are implemented to address differences in health outcomes, but they fail to acknowledge social history and complexity, or the ecological intricacy of the microbiome itself. For example, studies looking at racial differences in vaginal microbiomes briefly consider correlated socioeconomic differences, but they do not account for the effects of stress and toxic exposures associated with structural racism (18, 20). Furthermore, this work fails to address BIPOC medical distrust and how that affects reporting and data. Similarly, studies of the effects of migration on the human microbiome often look at immigration to “Western” and “industrialized” countries but rarely at migration in the opposite direction to understand the effects of the new environment and diet (37).
Race as a ghost variable also extends beyond the human microbiome. In studies of the microbiome of the built environment, spaces are racialized but analyzed without explicit mention of race or structural racism. Studies investigate urban built environments and draw connections between microbiota and higher rates of asthma, inflammatory bowel diseases, and even affective and anxiety disorders—but race or ethnicity is rarely mentioned (38, 39). The omission of race elides effects of structural racism such as histories of segregation, red-lining, and enduring environmental injustice that creates spatial divisions in cities. Furthermore, in environmental microbiome studies, landscapes that have been long tended by Indigenous populations are often described as “pristine” or “undisturbed” (40–42), erasing the contributions of Indigenous peoples to these ecosystems and microbiomes (43, 44).
This article is focused on how race is used as an explicit or implicit category in microbiome research. But race is a “ghost” in other important ways—a lack of diversity and representation in researchers and study populations and a lack of meaningful community research engagement prevent important questions from being asked.
CONCLUSION: TOWARD AN ANTIRACIST MICROBIOME SCIENCE
We have traced how race in many iterations has been operationalized in microbiome science. Historically and presently, microbiome sciences either harmfully ignore systemic racism and its effects or unreflectively reproduce racial thinking—with far-reaching implications for the study of human, environmental, animal, and plant microbiota.
From the global pandemic to uprisings against racial injustice, this is a transformative moment. Microbiome researchers have the opportunity to make significant changes to the ways science addresses race and simultaneously improve the quality and precision of this important work. An emerging literature is calling for microbiome science to address racial disparities (8, 45, 46), and increasingly, human microbiome studies are directly addressing socioenvironmental aspects causing differences in microbiome composition and health outcomes (47–49). Unlike race or ethnicity, there are clear mechanisms linking these variables with microbiome composition; interventions on these environmental variables are possible and can directly address environmental and health inequalities. However, work must be done across microbiome science to connect differences in microbiota to health disparities caused by structural inequities (9, 50, 51).
We propose an integrated three-part approach to create antiracist microbiome science. We build upon work in Indigenous science and technology studies and literature on race and genomics to suggest actionable solutions for microbiome science (52–61). First, institutional changes must be made in funding, publishing, hiring, and recruiting practices. Second, transdisciplinary collaboration across the biological and social sciences must be established as essential and customary. Third, study populations and BIPOC communities must be engaged in the research and empowered through the science. These suggestions are ordered by their relative feasibility: we see the first as the most feasible and the third as the most challenging to implement but most impactful.
-
1.
STEM fields and microbiome sciences in particular continue to lack representational diversity (62, 63). BIPOC, people with disabilities, and gender-diverse students and scientists should be sought out and supported for academic research, teaching jobs, funding, and publication. Publishing and funding work by researchers from different backgrounds reduce bias and prevent omissions in data (64). Funders and journal reviewers must be diversified and can be trained to look critically at how concepts of race are being utilized in proposed work. We suggest that funding agencies and editors pledge to interrogate papers and grant proposals that use race without accounting for or referencing racism. This is the future gold standard for educating junior scientists, and thoughtful antiracist thinking should be a requirement for funding and publishing.
-
2.
As we endeavor to challenge racism, we assert that social science-microbiome science partnerships are central to this work. We use “transdisciplinary” to highlight the need to work across disciplines, types of knowledge, and expertise, integrating natural, social, and health sciences and transcending the boundaries of those fields (65). Anthropologists, geographers, social scientists, epidemiologists, and public health experts can contribute to the analysis of variables that have real explanatory power and can examine the ways sociopolitical systems interact with the microbial world, thus enriching and improving the science. Ultimately, scientific research can no longer in good faith use race as an inadequate and misleading proxy. More precise variables (such as food, environment, infrastructure, social relationships, and structural racism) need to be studied. Race is not a valid biological category, but racism has a consequential effect on biology.
-
3.As mentioned above, microbiome science has failed to attend to an “ethics of care” in regard to marginalized people and environments (66). To work toward antiracist research, we propose that microbiome researchers look to current science that intentionally engages with the problem of racism (please see work by the following: Dr. Rosie Alegado, University of Hawaiʻi at Mānoa; Dr. Katherine Amato, Northwestern University; Dr. Marie-Claire Arrieta, University of Calgary; Dr. Erin Eggleston, Middlebury College; Dr. Keolu Fox, UC San Diego; Dr. Sue Ishaq, University of Maine; Dr. Michael D. L. Johnson, University of Arizona; Drs. Cecil M. Lewis, Jr., and Paul Spicer, University of Oklahoma; Dr. Max Liboiron, Memorial University; Dr. Kat Milligan-Myhre, University of Connecticut) and consider the following conditions when working with BIPOC subjects and communities:
- Prioritize community engagement and community-led research that utilizes local knowledge in research design from the conception of each project.
- Solicit community and individual input on health and environmental priorities.
- Secure formal consent from each community/tribe/sovereignty/nation for sampling, land use, and community access.
- Participate in socially responsible sampling, management, and fair ownership of data (67).
- Actively account for social determinants of human and environmental health in scientific data, which could include systemic barriers like poor access to health care, jobs, housing, and education.
- Ensure equitable benefit-sharing of translational interventions developed from the scientific research for research participants and their communities.
- Legally protect subjects, their samples, data, and land against commercial, scientific, medical, and cultural exploitation (68).
We are very aware that what we are proposing is extremely difficult and that the practices we have outlined are not currently supported by funding structures, study designs, or institutional hierarchies. Which is precisely why, for instance, future human microbiome grants must add field-inclusive funding strategies that support equity initiatives. We challenge microbiome researchers using racial categories to ask themselves, what is the function of race in my study? What am I using it for, and is there something more precise and equitable (Fig. 2)? This article is just the barest of beginnings; developing an antiracist microbiome science requires investment by all to determine what the field standards should be and how to deploy all types of expertise and knowledge systems to address systemic drivers of microbial difference. Very few have started to put this work into action yet, but we have cited some researchers who can serve as methodological inspirations. There is no guidebook, but with determined commitments to equity, collaboration, and better science, it is a goal worth striving toward.
ACKNOWLEDGMENTS
We thank Dean Giustini (ORCID: 0000-0002-6197-8788) for their helpful conversation and guidance with library resources and Christopher Reimer for their work which improved the article figures.
T.J.D.W. was supported by the Michael Smith Foundation for Health Research Trainee award (RT-2020-04-64). M.R.A. was supported by the Wall Scholar fellowship granted by the Peter Wall Institute for Advanced Studies at the University of British Columbia. M.R.G. acknowledges support from the School of Biological Sciences and the Department of Ecology and Evolutionary Biology at University of California, Irvine.
Biographies
Travis J. De Wolfe is a Michael Smith Foundation for Health Research-funded Postdoctoral Fellow with the Department of Pediatrics at the University of British Columbia and BC Children’s Hospital. His research interests include Clostridioides difficile infection, colonization resistance, inflammatory bowel diseases, host-microbe interactions, and ecology of the gut microbiome.
Mohammed Rafi Arefin is an Assistant Professor in the Department of Geography at the University of British Columbia. His research and teaching are focused on urban environmental politics with a particular focus on sanitation, health, and environmental justice in the Middle East and North America.
Amber Benezra is an Assistant Professor of Science and Technology Studies at Stevens Institute of Technology. She is a sociocultural anthropologist researching how studies of the human microbiome intersect with biomedical ethics, public health/technological infrastructures, and care. In partnership with human microbial ecologists, she is developing an “anthropology of microbes” to address global health problems across disciplines.
María Rebolleda Gómez is an Assistant Professor in the Department of Ecology and Evolutionary Biology at the University of California, Irvine. Her research explores ecological and evolutionary dynamics in model microbial communities. She is also interested in Environmental History, Philosophy, and History of Science. María was born in Mexico City.
Contributor Information
Travis J. De Wolfe, Email: tjdewolfe@gmail.com.
Mohammed Rafi Arefin, Email: rafi.arefin@ubc.ca.
Amber Benezra, Email: abenezra@stevens.edu.
María Rebolleda Gómez, Email: mreboll1@uci.edu.
Kathryn C. Milligan-Myhre, University of Connecticut
REFERENCES
- 1.Mathias RA, Taub MA, Gignoux CR, Fu W, Musharoff S, O’Connor TD, Vergara C, Torgerson DG, Pino-Yanes M, Shringarpure SS, Huang L, Rafaels N, Boorgula MP, Johnston HR, Ortega VE, Levin AM, Song W, Torres R, Padhukasahasram B, Eng C, Mejia-Mejia D-A, Ferguson T, Qin ZS, Scott AF, Yazdanbakhsh M, Wilson JG, Marrugo J, Lange LA, Kumar R, Avila PC, Williams LK, Watson H, Ware LB, Olopade C, Olopade O, Oliveira R, Ober C, Nicolae DL, Meyers D, Mayorga A, Knight-Madden J, Hartert T, Hansel NN, Foreman MG, Ford JG, Faruque MU, Dunston GM, Caraballo L, Burchard EG, Bleecker E, Araujo MI, Herrera-Paz EF, Gietzen K, Grus WE, Barnshad M, Bustamante CD, Kenny EE, Hernandez RD, Beaty TH, Ruczinski I, Akey J, CAAPA, Barnes KC. 2016. A continuum of admixture in the Western Hemisphere revealed by the African Diaspora genome. Nat Commun 7:12522. doi: 10.1038/ncomms12522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lewontin RC. 1972. The apportionment of human diversity, p 381–398. In Dobzhansky T, Hecht MK, Steere WC (ed), Evolutionary biology. Springer US, New York, NY. [Google Scholar]
- 3.Yu N, Chen F-C, Ota S, Jorde LB, Pamilo P, Patthy L, Ramsay M, Jenkins T, Shyue S-K, Li W-H. 2002. Larger genetic differences within Africans than between Africans and Eurasians. Genetics 161:269–274. doi: 10.1093/genetics/161.1.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jorde LB, Wooding SP. 2004. Genetic variation, classification and ‘race’. Nat Genet 36:S28–S33. doi: 10.1038/ng1435. [DOI] [PubMed] [Google Scholar]
- 5.Yudell M, Roberts D, DeSalle R, Tishkoff S. 2016. Science and society. Taking race out of human genetics. Science 351:564–565. doi: 10.1126/science.aac4951. [DOI] [PubMed] [Google Scholar]
- 6.Fuentes A, Ackermann RR, Athreya S, Bolnick D, Lasisi T, Lee S-H, McLean S-A, Nelson R. 2019. AAPA statement on race and racism. Am J Phys Anthropol 169:400–402. doi: 10.1002/ajpa.23882. [DOI] [PubMed] [Google Scholar]
- 7.Amutah C, Greenidge K, Mante A, Munyikwa M, Surya SL, Higginbotham E, Jones DS, Lavizzo-Mourey R, Roberts D, Tsai J, Aysola J. 2021. Misrepresenting race - the role of medical schools in propagating physician bias. N Engl J Med 384:872–878. doi: 10.1056/NEJMms2025768. [DOI] [PubMed] [Google Scholar]
- 8.Benezra A. 2020. Race in the microbiome. Sci Technol Human Values 45:877–902. doi: 10.1177/0162243920911998. [DOI] [Google Scholar]
- 9.Kozik AJ. 2020. mSphere of Influence: frameshift—a vision for human microbiome research. mSphere 5:e00944-20. doi: 10.1128/mSphere.00944-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Taylor K. 2020. mSphere of Influence: that’s racist—COVID-19, biological determinism, and the limits of hypotheses. mSphere 5:e00945-20. doi: 10.1128/mSphere.00945-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gilmore RW. 2007. Golden gulag: prisons, surplus, crisis, and opposition in globalizing California. University of California Press, Oakland, CA. [Google Scholar]
- 12.Farmer PE, Nizeye B, Stulac S, Keshavjee S. 2006. Structural violence and clinical medicine. PLoS Med 3:e449. doi: 10.1371/journal.pmed.0030449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Benn Torres J. 2020. Anthropological perspectives on genomic data, genetic ancestry, and race. Am J Phys Anthropol 171(Suppl 70):74–86. doi: 10.1002/ajpa.23979. [DOI] [PubMed] [Google Scholar]
- 14.Fujimura JH, Rajagopalan R. 2011. Different differences: the use of ‘genetic ancestry’ versus race in biomedical human genetic research. Soc Stud Sci 41:5–30. doi: 10.1177/0306312710379170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fitzgerald KJ. 2014. The continuing significance of race: racial genomics in a postracial era. Humanity Soc 38:49–66. doi: 10.1177/0160597613519231. [DOI] [Google Scholar]
- 16.Turnbaugh PJ, Ley RE, Hamady M, Fraser-Liggett CM, Knight R, Gordon JI. 2007. The human microbiome project. Nature 449:804–810. doi: 10.1038/nature06244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yong E. 1 November 2014. There is no ‘healthy’ microbiome. New York Times. https://www.nytimes.com/2014/11/02/opinion/sunday/there-is-no-healthy-microbiome.html.
- 18.Zhou X, Brown CJ, Abdo Z, Davis CC, Hansmann MA, Joyce P, Foster JA, Forney LJ. 2007. Differences in the composition of vaginal microbial communities found in healthy Caucasian and black women. ISME J 1:121–133. doi: 10.1038/ismej.2007.12. [DOI] [PubMed] [Google Scholar]
- 19.Ravel J, Gajer P, Abdo Z, Schneider GM, Koenig SSK, McCulle SL, Karlebach S, Gorle R, Russell J, Tacket CO, Brotman RM, Davis CC, Ault K, Peralta L, Forney LJ. 2011. Vaginal microbiome of reproductive-age women. Proc Natl Acad Sci USA 108:4680–4687. doi: 10.1073/pnas.1002611107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fettweis JM, Brooks JP, Serrano MG, Sheth NU, Girerd PH, Edwards DJ, Strauss JF, The Vaginal Microbiome Consortium, Jefferson KK, Buck GA. 2014. Differences in vaginal microbiome in African American women versus women of European ancestry. Microbiology (Reading) 160:2272–2282. doi: 10.1099/mic.0.081034-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Srinivasan S, Hoffman NG, Morgan MT, Matsen FA, Fiedler TL, Hall RW, Ross FJ, McCoy CO, Bumgarner R, Marrazzo JM, Fredricks DN. 2012. Bacterial communities in women with bacterial vaginosis: high resolution phylogenetic analyses reveal relationships of microbiota to clinical criteria. PLoS One 7:e37818. doi: 10.1371/journal.pone.0037818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Human Microbiome Project Consortium. 2012. Structure, function and diversity of the healthy human microbiome. Nature 486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aagaard K, Ma J, Antony KM, Ganu R, Petrosino J, Versalovic J. 2014. The placenta harbors a unique microbiome. Sci Transl Med 6:237ra65. doi: 10.1126/scitranslmed.3008599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brooks AW, Priya S, Blekhman R, Bordenstein SR. 2018. Gut microbiota diversity across ethnicities in the United States. PLoS Biol 16:e2006842. doi: 10.1371/journal.pbio.2006842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johnson MDL. 2020. mSphere highlights Black In Microbiology Week. mSphere 5:e00966-20. doi: 10.1128/mSphere.00966-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Duggan CP, Kurpad A, Stanford FC, Sunguya B, Wells JC. 2020. Race, ethnicity, and racism in the nutrition literature: an update for 2020. Am J Clin Nutr 112:1409–1414. doi: 10.1093/ajcn/nqaa341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Handsley-Davis M, Jamieson L, Kapellas K, Hedges J, Weyrich LS. 2020. The role of the oral microbiota in chronic non-communicable disease and its relevance to the Indigenous health gap in Australia. BMC Oral Health 20:327. doi: 10.1186/s12903-020-01308-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aronsen GP, Fehren-Schmitz L, Krigbaum J, Kamenov GD, Conlogue GJ, Warinner C, Ozga AT, Sankaranarayanan K, Griego A, DeLuca DW, Eckels HT, Byczkiewicz RK, Grgurich T, Pelletier NA, Brownlee SA, Marichal A, Williamson K, Tonoike Y, Bellantoni NF. 2019. “The dead shall be raised”: multidisciplinary analysis of human skeletons reveals complexity in 19th century immigrant socioeconomic history and identity in New Haven, Connecticut. PLoS One 14:e0219279. doi: 10.1371/journal.pone.0219279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Agnello M, Marques J, Cen L, Mittermuller B, Huang A, Chaichanasakul Tran N, Shi W, He X, Schroth RJ. 2017. Microbiome associated with severe caries in Canadian First Nations children. J Dent Res 96:1378–1385. doi: 10.1177/0022034517718819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shamarina D, Stoyantcheva I, Mason CE, Bibby K, Elhaik E. 2017. Communicating the promise, risks, and ethics of large-scale, open space microbiome and metagenome research. Microbiome 5:132. doi: 10.1186/s40168-017-0349-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lincoln KD. 2020. Race, obesity, and mental health among older adults in the United States: a literature review. Innov Aging 4:igaa031. doi: 10.1093/geroni/igaa031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fragiadakis GK, Smits SA, Sonnenburg ED, Van Treuren W, Reid G, Knight R, Manjurano A, Changalucha J, Dominguez-Bello MG, Leach J, Sonnenburg JL. 2019. Links between environment, diet, and the hunter-gatherer microbiome. Gut Microbes 10:216–227. doi: 10.1080/19490976.2018.1494103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gomez A, Petrzelkova KJ, Burns MB, Yeoman CJ, Amato KR, Vlckova K, Modry D, Todd A, Jost Robinson CA, Remis MJ, Torralba MG, Morton E, Umaña JD, Carbonero F, Gaskins HR, Nelson KE, Wilson BA, Stumpf RM, White BA, Leigh SR, Blekhman R. 2016. Gut microbiome of coexisting BaAka pygmies and Bantu reflects gradients of traditional subsistence patterns. Cell Rep 14:2142–2153. doi: 10.1016/j.celrep.2016.02.013. [DOI] [PubMed] [Google Scholar]
- 34.Obregon-Tito AJ, Tito RY, Metcalf J, Sankaranarayanan K, Clemente JC, Ursell LK, Zech Xu Z, Van Treuren W, Knight R, Gaffney PM, Spicer P, Lawson P, Marin-Reyes L, Trujillo-Villarroel O, Foster M, Guija-Poma E, Troncoso-Corzo L, Warinner C, Ozga AT, Lewis CM. 2015. Subsistence strategies in traditional societies distinguish gut microbiomes. Nat Commun 6:6505. doi: 10.1038/ncomms7505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rampelli S, Schnorr SL, Consolandi C, Turroni S, Severgnini M, Peano C, Brigidi P, Crittenden AN, Henry AG, Candela M. 2015. Metagenome sequencing of the Hadza hunter-gatherer gut microbiota. Curr Biol 25:1682–1693. doi: 10.1016/j.cub.2015.04.055. [DOI] [PubMed] [Google Scholar]
- 36.Sonnenburg ED, Sonnenburg JL. 2019. The ancestral and industrialized gut microbiota and implications for human health. Nat Rev Microbiol 17:383–390. doi: 10.1038/s41579-019-0191-8. [DOI] [PubMed] [Google Scholar]
- 37.Vangay P, Johnson AJ, Ward TL, Al-Ghalith GA, Shields-Cutler RR, Hillmann BM, Lucas SK, Beura LK, Thompson EA, Till LM, Batres R, Paw B, Pergament SL, Saenyakul P, Xiong M, Kim AD, Kim G, Masopust D, Martens EC, Angkurawaranon C, McGready R, Kashyap PC, Culhane-Pera KA, Knights D. 2018. US immigration westernizes the human gut microbiome. Cell 175:962–972.e10. doi: 10.1016/j.cell.2018.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stamper CE, Hoisington AJ, Gomez OM, Halweg-Edwards AL, Smith DG, Bates KL, Kinney KA, Postolache TT, Brenner LA, Rook GAW, Lowry CA. 2016. The microbiome of the built environment and human behavior: implications for emotional health and well-being in postmodern Western societies. Int Rev Neurobiol 131:289–323. doi: 10.1016/bs.irn.2016.07.006. [DOI] [PubMed] [Google Scholar]
- 39.Gilbert JA, Stephens B. 2018. Microbiology of the built environment. Nat Rev Microbiol 16:661–670. doi: 10.1038/s41579-018-0065-5. [DOI] [PubMed] [Google Scholar]
- 40.Cadena M, Durso LM, Miller DN, Waldrip HM, Castleberry BL, Drijber RA, Wortmann C. 2018. Tetracycline and sulfonamide antibiotic resistance genes in soils from Nebraska organic farming operations. Front Microbiol 9:1283. doi: 10.3389/fmicb.2018.01283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fonseca JP, Hoffmann L, Cabral BCA, Dias VHG, Miranda MR, de Azevedo Martins AC, Boschiero C, Bastos WR, Silva R. 2018. Contrasting the microbiomes from forest rhizosphere and deeper bulk soil from an Amazon rainforest reserve. Gene 642:389–397. doi: 10.1016/j.gene.2017.11.039. [DOI] [PubMed] [Google Scholar]
- 42.Delavaux CS, Schemanski JL, House GL, Tipton AG, Sikes B, Bever JD. 2021. Root pathogen diversity and composition varies with climate in undisturbed grasslands, but less so in anthropogenically disturbed grasslands. ISME J 15:304–317. doi: 10.1038/s41396-020-00783-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Roos CI, Zedeño MN, Hollenback KL, Erlick MMH. 2018. Indigenous impacts on North American Great Plains fire regimes of the past millennium. Proc Natl Acad Sci USA 115:8143–8148. doi: 10.1073/pnas.1805259115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Heckenberger MJ, Kuikuro A, Kuikuro UT, Russell JC, Schmidt M, Fausto C, Franchetto B. 2003. Amazonia 1492: pristine forest or cultural parkland? Science 301:1710–1714. doi: 10.1126/science.1086112. [DOI] [PubMed] [Google Scholar]
- 45.Fortenberry JD. 2013. The uses of race and ethnicity in human microbiome research. Trends Microbiol 21:165–166. doi: 10.1016/j.tim.2013.01.001. [DOI] [PubMed] [Google Scholar]
- 46.Findley K, Williams DR, Grice EA, Bonham VL. 2016. Health disparities and the microbiome. Trends Microbiol 24:847–850. doi: 10.1016/j.tim.2016.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, Ling AV, Devlin AS, Varma Y, Fischbach MA, Biddinger SB, Dutton RJ, Turnbaugh PJ. 2014. Diet rapidly and reproducibly alters the human gut microbiome. Nature 505:559–563. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dannemiller KC, Gent JF, Leaderer BP, Peccia J. 2016. Indoor microbial communities: influence on asthma severity in atopic and nonatopic children. J Allergy Clin Immunol 138:76–83.e1. doi: 10.1016/j.jaci.2015.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fouladi F, Bailey MJ, Patterson WB, Sioda M, Blakley IC, Fodor AA, Jones RB, Chen Z, Kim JS, Lurmann F, Martino C, Knight R, Gilliland FD, Alderete TL. 2020. Air pollution exposure is associated with the gut microbiome as revealed by shotgun metagenomic sequencing. Environ Int 138:105604. doi: 10.1016/j.envint.2020.105604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Amato KR, Arrieta MC, Azad MB, Bailey MT, Broussard JL, Bruggeling CE, Claud EC, Costello EK, Davenport ER, Dutilh BE, Ewald HAS, Ewald P, Hanlon EC, Julion W, Keshavarzian A, Maurice CF, Miller GE, Preidis GA, Segurel L, Singer B, Subramanian S, Zhao L, Kuzawa CW. 2021. The human gut microbiome and health inequities. Proc Natl Acad Sci USA 118:e2017947118. doi: 10.1073/pnas.2017947118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ishaq SL, Parada FJ, Wolf PG, Bonilla CY, Carney MA, Benezra A, Wissel E, Friedman M, DeAngelis KM, Robinson JM, Fahimipour AK, Manus MB, Grieneisen L, Dietz LG, Pathak A, Chauhan A, Kuthyar S, Stewart JD, Dasari MR, Nonnamaker E, Choudoir M, Horve PF, Zimmerman NB, Kozik AJ, Darling KW, Romero-Olivares AL, Hariharan J, Farmer N, Maki KA, Collier JL, O’Doherty KC, Letourneau J, Kline J, Moses PL, Morar N. 2021. Introducing the Microbes and Social Equity Working Group: considering the microbial components of social, environmental, and health justice. mSystems 6:e00471-21. doi: 10.1128/mSystems.00471-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fox K. 2020. The illusion of inclusion - the “All of Us” research program and indigenous peoples’ DNA. N Engl J Med 383:411–413. doi: 10.1056/NEJMp1915987. [DOI] [PubMed] [Google Scholar]
- 53.Garrison NA. 2018. Genetic ancestry testing with tribes: ethics, identity & health implications. Daedalus 147:60–69. doi: 10.1162/DAED_a_00490. [DOI] [Google Scholar]
- 54.TallBear K. 2013. Native American DNA: tribal belonging and the false promise of genetic science. University of Minnesota Press, Minneapolis, MN. [Google Scholar]
- 55.Tsosie KS, Yracheta JM, Kolopenuk JA, Geary J. 2021. We have “gifted” enough: indigenous genomic data sovereignty in precision medicine. Am J Bioeth 21:72–75. doi: 10.1080/15265161.2021.1891347. [DOI] [PubMed] [Google Scholar]
- 56.Benjamin R. 2019. Assessing risk, automating racism. Science 366:421–422. doi: 10.1126/science.aaz3873. [DOI] [PubMed] [Google Scholar]
- 57.Fullwiley D. 2007. The molecularization of race: institutionalizing human difference in pharmacogenetics practice. Sci Cult 16:1–30. doi: 10.1080/09505430601180847. [DOI] [Google Scholar]
- 58.Morning A. 2007. “Everyone knows it’s a social construct”: contemporary science and the nature of race. Sociol Focus 40:436–454. doi: 10.1080/00380237.2007.10571319. [DOI] [Google Scholar]
- 59.Nelson A. 2016. The social life of DNA: race, reparations, and reconciliation after the genome. Beacon Press, Boston, MA. [DOI] [PubMed] [Google Scholar]
- 60.Roberts DE. 2011. Fatal invention: how science, politics, and big business re-create race in the twenty-first century. New Press, New York, NY. [Google Scholar]
- 61.Hiratsuka VY, Beans JA, Reedy J, Yracheta JM, Peercy MT, Saunkeah B, Woodbury RB, O’Leary M, Spicer PG. 2020. Fostering ethical, legal, and social implications research in tribal communities: the Center for the Ethics of Indigenous Genomic Research. J Empir Res Hum Res Ethics 15:271–278. doi: 10.1177/1556264619872640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Campos JS, Wherry EJ, Shin S, Ortiz-Carpena JF. 2021. Challenging systemic barriers to promote the inclusion, recruitment, and retention of URM faculty in STEM. Cell Host Microbe 29:862–866. doi: 10.1016/j.chom.2021.04.001. [DOI] [PubMed] [Google Scholar]
- 63.Barber PH, Hayes TB, Johnson TL, Márquez-Magaña L, 10,234 signatories . 2020. Systemic racism in higher education. Science 369:1440–1441. doi: 10.1126/science.abd7140. [DOI] [PubMed] [Google Scholar]
- 64.Jackson L, Kuhlman C, Jackson F, Fox PK. 2019. Including vulnerable populations in the assessment of data from vulnerable populations. Front Big Data 2:19. doi: 10.3389/fdata.2019.00019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Choi BC, Pak AW. 2006. Multidisciplinarity, interdisciplinarity and transdisciplinarity in health research, services, education and policy: 1. Definitions, objectives, and evidence of effectiveness. Clin Invest Med 29:351–364. [PubMed] [Google Scholar]
- 66.Sankaranarayanan K, Ozga AT, Warinner C, Tito RY, Obregon-Tito AJ, Xu J, Gaffney PM, Jervis LL, Cox D, Stephens L, Foster M, Tallbull G, Spicer P, Lewis CM. 2015. Gut microbiome diversity among Cheyenne and Arapaho individuals from western Oklahoma. Curr Biol 25:3161–3169. doi: 10.1016/j.cub.2015.10.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ma Y, Chen H, Lan C, Ren J. 2018. Help, hope and hype: ethical considerations of human microbiome research and applications. Protein Cell 9:404–415. doi: 10.1007/s13238-018-0537-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gewin V. 2021. How to include Indigenous researchers and their knowledge. Nature 589:315–317. doi: 10.1038/d41586-021-00022-1. [DOI] [PubMed] [Google Scholar]
- 69.R Core Team. 2020. R: a language and environment for statistical computing. Version 3.6.3. R Foundation for Statistical Computing, Vienna, Austria. https://www.R-project.org/.
- 70.Winter DJ. 2017. rentrez: an R package for the NCBI eUtils API. R J 9:520–526. doi: 10.32614/RJ-2017-058. [DOI] [Google Scholar]