ABSTRACT
Ralstonia solanacearum is an extremely destructive phytopathogenic bacterium for which there is no effective control method. Though many pathogenic factors have been identified, the survival strategies of R. solanacearum in host plants remain unclear. Transposon insertion sequencing (Tn-seq) is a high-throughput genetic screening technology. This study conducted a Tn-seq analysis using the in planta environment as selective pressure to identify R. solanacearum genes required for survival in tomato plants. One hundred thirty genes were identified as putative genes required for survival in tomato plants. Sixty-three of these genes were classified into four Clusters of Orthologous Groups categories. The absence of genes that encode the outer membrane lipoprotein LolB (RS_RS01965) or the membrane protein RS_RS04475 severely decreased the in planta fitness of R. solanacearum. RS_RS09970 and RS_RS04490 are involved in tryptophan and serine biosynthesis, respectively. Mutants that lack RS_RS09970 or RS_RS04490 did not cause any wilt symptoms in susceptible tomato plants. These results confirmed the importance of genes related to “cell wall/membrane/envelope biogenesis” and “amino acid transport and metabolism” for survival in plants. The gene encoding NADH-quinone oxidoreductase subunit B (RS_RS10340) is one of the 13 identified genes involved in “energy production and conversion,” and the Clp protease gene (RS_RS08645) is one of the 11 identified genes assigned to “posttranslational modification, protein turnover, and chaperones.” Both genes were confirmed to be required for survival in plants. In conclusion, this study globally identified and validated R. solanacearum genes required for survival in tomato plants and provided essential information for a more complete view of the pathogenic mechanism of R. solanacearum.
IMPORTANCE Tomato plant xylem is a nutritionally limiting and dynamically changing habitat. Studies on how R. solanacearum survives in this hostile environment are important for our full understanding of the pathogenic mechanism of this bacterium. Though many omics approaches have been employed to study in planta survival strategies, the direct genome-wide identification of R. solanacearum genes required for survival in plants is still lacking. This study performed a Tn-seq analysis in R. solanacearum and revealed that genes in the categories “cell wall/membrane/envelope biogenesis,” “amino acid transport and metabolism,” “energy production and conversion,” “posttranslational modification, protein turnover, chaperones” and others play important roles in the survival of R. solanacearum in tomato plants.
KEYWORDS: pathogenic bacteria, pathogenicity, Ralstonia solanacearum, transposon sequencing
INTRODUCTION
Ralstonia solanacearum is an aerobic, motile Gram-negative bacterium with a polar flagellar tuft. This soilborne bacterium is probably the most destructive plant-pathogenic bacterium, infecting more than 200 plant species in over 50 families over a broad geographical range (1). The host plants of R. solanacearum include tomato, tobacco, potato, peanut, and many other important commercial crops (2, 3). R. solanacearum is extremely damaging and has caused an estimated US$1 billion loss each year worldwide on potatoes alone because of the lack of an effective control method (4). A better understanding of the pathogenic mechanism is an essential precondition for controlling plant diseases caused by R. solanacearum.
As in most Gram-negative animal- and plant-pathogenic bacteria, the type III secretion system and effectors secreted by this system are the major pathogenicity determinants in R. solanacearum (5). The absence of the structural proteins of type III secretion system apparatus (6) or the key regulators of type III secretion system genes, such as HrpB (7), abolishes the pathogenicity of R. solanacearum. Moreover, type III secretion system effectors are the key host range factors. The gala7 gene extends the host range of R. solanacearum GMI1000 to include the legume Medicago truncatula (8). High-molecular-mass exopolysaccharide (EPS) is another important virulence determinant for R. solanacearum. This EPS contributes to rapid systemic colonization by R. solanacearum and wilt symptoms in susceptible hosts; mutants that lack eps genes cannot cause disease symptoms on host plants (9). Moreover, cell wall-degrading enzymes secreted by the type II secretion system, motility, resistance to stresses within the host plant, nutrient-scavenging systems, and varied regulatory networks play important roles in infection by R. solanacearum (10).
Besides targeted genetic studies, omics approaches have been used to identify pathogenicity genes in R. solanacearum. The most used strategy is to identify genes induced during plant infection. Brown and Allen revealed 153 in planta-expressed genes using in vivo expression technology and suggested that R. solanacearum confronts and overcomes stressful and nutrient-poor environments (11). An in planta transcriptome study revealed that about 12% of R. solanacearum transcriptomes were remarkably altered in planta compared with in rich medium and that the absence of the sucrose uptake and catabolism gene scrA impaired the virulence of R. solanacearum on tomato, potato, and Solanum dulcamara (12). In silico or experimental evolution is another strategy used to screen pathogenicity genes in R. solanacearum. Forty-nine genes in 37 R. solanacearum genomes were identified as nonneutrally evolving and maybe virulence related using Tajima’s D population genetic test (13). The multihost experimental evolution of selected independent mutations in the regulatory gene efpR revealed that it is a determinant conditioning host adaptation of R. solanacearum (14).
Transposon insertion sequencing (Tn-seq) is a high-throughput approach that couples genome-wide transposon mutagenesis with next-generation sequencing (15, 16). Tn-seq can be used to identify genes that contribute to bacterial survival under the selective pressure of interest (17). Tn-seq has been applied to identify genes important for in vivo survival in several plant-pathogenic bacteria using the host environment as the selective pressure. A Tn-seq study inoculated the transposon mutant library of Dickeya dadantii in chicory plant and recovered the mutants from rotten tissue after 2 days; this Tn-seq study revealed that about 100 genes contribute to the survival of D. dadantii in chicory plants (18). A total of 486 genes in Pantoea stewartii subsp. stewartia are essential for survival in corn xylem, as determined through a Tn-seq analysis (19).
A Tn-seq analysis of R. solanacearum in tomato plants was conducted in the present study to acquire a more complete view of the pathogenic mechanism of R. solanacearum. The transposon insertion library was injected into the tomato plant stem and recovered 5 days postinoculation. The transposon interruption of 130 genes reduced the relative fitness of R. solanacearum within tomato plants, providing putative genes required for R. solanacearum survival in tomato plants. Furthermore, targeted gene deletion, pathogenicity assay, in vivo colonization assay, and competition index determination were performed to validate the results of Tn-seq.
RESULTS
Tn-seq analysis to identify putative genes required for survival in tomato plants.
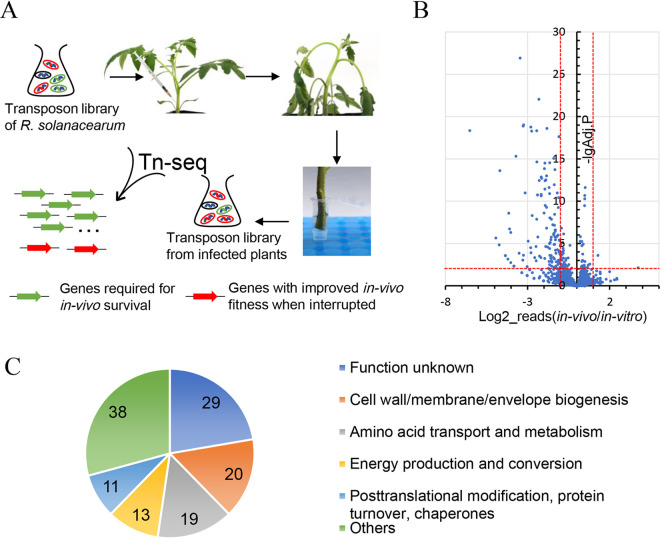
We previously constructed a near-saturated transposon insertion library of R. solanacearum GMI1000 with approximately 240,000 individual insertion mutants, covering 70.44% to 80.96% of all potential insertion sites (20). An in planta Tn-seq analysis was conducted using this near-saturated transposon insertion library to identify R. solanacearum genes essential for in planta survival. As shown in Fig. 1A, the transposon insertion library of R. solanacearum was activated and injected into the tomato plant stem (21). The library was then recovered from the stems of 32 inoculated tomato plants 5 days postinoculation, when the wilting symptom scores of most tomato plants were 3 or 2. The transposon insertion libraries recovered from all tomato plants were then pooled. The total genome DNAs of transposon insertion libraries before (in vitro) and after (in vivo) infection were extracted and split into two groups for technical replicates. The DNA samples were then subjected to Tn-seq to identify the relative abundance of each insertion mutant under the stress of the in planta environment.
FIG 1.
Tn-seq analysis of R. solanacearum genes required for survival in tomato plants. (A) Schematic diagram of Tn-seq analysis in this study. (B) Gene essentiality for in vivo survival. Each dot represents a gene, plotted by the ratio (in vivo/in vitro) of reads mapped within this gene on the horizontal axis and the -lg (adjusted P value) on the vertical. A ratio_reads (in vivo/in vitro) value of <0.5 or >2 with an adjusted P (proportions_reads) value of <0.01 was set as the threshold value to identify genes required for survival in tomato plants or genes resulting in improved in vivo fitness when interrupted. (C) Numbers of genes required for survival in tomato plants classified by COG categories.
The correlation between technical replicates is analyzed and visualized in Fig. S1 in the supplemental material. The correlation coefficients for the in vivo and in vitro replicates were 1.00 and 0.99, respectively, which indicate the reliability and repeatability of this analysis. As shown in Table 1, 6,858,640 reads of in-vivo1 were mapped to the chromosome (NC_003295.1) of R. solanacearum strain GMI1000, and 4,636,990 reads of in-vivo1 were mapped to the megaplasmid (NC_003296.1). These reads hit 135,231 unique locations with 101,663 locations within genes. The output parameters of in-vivo2 were similar to that of in-vivo1. However, more reads were mapped to the chromosome, and fewer reads were mapped to the megaplasmid for the in vitro treatments than for the in vivo treatments. The transposon interruption of a gene essential for survival in plants would reduce the relative fitness of a mutant within tomato plants and result in fewer reads mapped to this gene. As shown in Fig. 1B, 130 genes in R. solanacearum strain GMI1000 were identified as putative genes required for survival in tomato plants when the threshold value was set to a ratio_reads (in vivo/in vitro) value of <0.5 with an adjusted P (proportions_reads) value of <0.01. The interruption of two genes (RS_RS05450 and RS_RS05405) increased the ratio_reads (in vivo/in vitro) value by more than 2-fold (Table S1). Interestingly, 117 of these 132 genes that contribute to the survival in tomato plants are located in the chromosome of R. solanacearum strain GMI1000.
TABLE 1.
Output parameters of the in planta Tn-seq analysis
| Treatment | No. of: |
||
|---|---|---|---|
| Mapped reads (chromosome + megaplasmid) | Unique hits | Unique hits within genes | |
| in-vivo1 | 6,858,640 + 4,636,990 | 135,231 | 101,663 |
| in-vivo2 | 6,835,235 + 4,622,666 | 133,781 | 100,365 |
| in-vitro1 | 7,014,722 + 4,093,910 | 174,518 | 131,536 |
| in-vitro2 | 7,196,108 + 4,206,360 | 175,751 | 132,999 |
Gene essentiality for in vivo survival. Download Table S1, XLSX file, 0.03 MB (34.9KB, xlsx) .
Copyright © 2021 Su et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Coefficient of read coverages for Tn-seq technological replicates (scatter plot) and all Tn-seq samples (heat map). Download FIG S1, TIF file, 0.1 MB (132.4KB, tif) .
Copyright © 2021 Su et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
We classified these genes into Clusters of Orthologous Groups of proteins (COG) categories to obtain an overview of the genes required for survival in tomato plants. As shown in Fig. 1C, 29 of these 130 genes were annotated as “function unknown” or were not mapped to COG categories. Twenty genes involved in “cell wall/membrane/envelope biogenesis” are required for survival in tomato plants. The genes assigned to “amino acid transport and metabolism,” “energy production and conversion,” and “posttranslational modification, protein turnover, chaperones” play important roles in the survival of R. solanacearum in tomato plants.
Cell wall/membrane/envelope biogenesis-related genes required for survival in tomato plants.
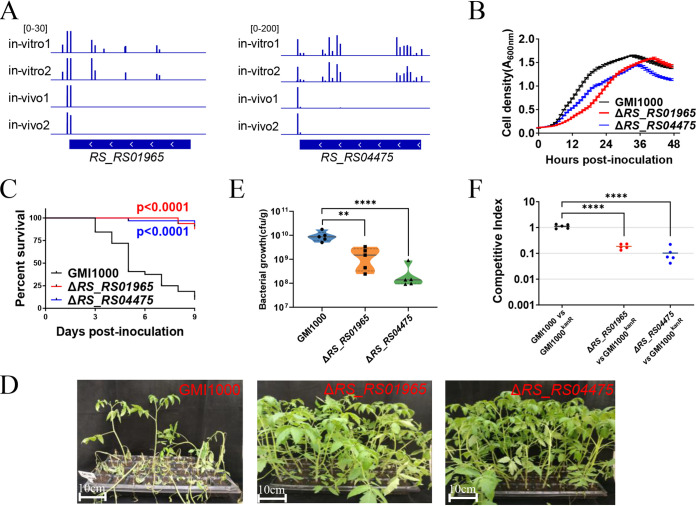
Twenty genes assigned to “cell wall/membrane/envelope biogenesis” were found to be vital for the survival of R. solanacearum in tomato plants by Tn-seq. RS_RS01965 encodes the outer membrane lipoprotein LolB, which is a component of the LolABCDE system, responsible for sorting and localizing lipoprotein. The absence of LolA in the plant pathogen Xanthomonas campestris pv. campestris reduced the pathogen’s attachment, extracellular enzyme production, and virulence (22). As shown in Fig. 2A and Table S1, 6.5 unique hits were found within RS_RS01965 mapped by 26 weighted reads in average before infection, but no transposon insertion mutant of RS_RS01965 was detected from the transposon insertion library recovered from the infected tomato. RS_RS04475 encodes a membrane protein. The relative abundance of the RS_RS04475 mutant was 0.039 times higher in tomato plants than in in vitro (Fig. 2A). RS_RS01965 and RS_RS04475 were deleted in frame individually to validate the contribution of these genes in this category.
FIG 2.
RS_RS01965 and RS_RS04475 are required for survival in tomato plants. (A) Transposon insertion distribution within RS_RS01965 and RS_RS04475 of transposon insertion libraries in vivo and in vitro. (B) Growth of the ΔRS_RS01965 mutant, the ΔRS_RS04475 mutant, and wild-type strain GMI1000 in BG medium. (C) Survival curve of tomato plants inoculated with the ΔRS_RS01965 mutant, the ΔRS_RS04475 mutant, and wild-type strain GMI1000. Kaplan-Meier survival analysis with the Gehan-Breslow-Wilcoxon method was used to compare the pathogenicity between the mutant and wild-type strains. A P value of <0.05 was considered significant. (D) Bacterial wilt symptoms of tomato plants 9 days after inoculation with the ΔRS_RS01965 mutant, the ΔRS_RS04475 mutant, and wild-type strain GMI1000. (E) Colonization of ΔRS_RS01965 and ΔRS_RS04475 mutants. The CFU of R. solanacearum strains in 1 g tomato plant stem were counted 5 days postinoculation. Asterisks indicate significant differences (**, P < 0.01; ****, P < 0.0001; t test). F. In vivo competitive index of ΔRS_RS01965 and ΔRS_RS04475 mutants. R. solanacearum mutants and GMI1000Kanr were coinoculated into the stems of tomato plants. The competitive index was measured 5 days postinoculation. Asterisks indicate significant differences (****, P < 0.0001; t test).
The RS_RS01965 (ΔRS_RS01965) and RS_RS04475 (ΔRS_RS04475) deletion mutants exhibited impaired growth in rich BG medium (Fig. 2B). These two mutants were then injected into the tomato stem to evaluate the pathogenic contribution of RS_RS01965 and RS_RS04475. As shown in Fig. 2C and D, almost all the tested tomato plants were wilted 9 days postinoculation of R. solanacearum wild-type strain GMI1000, whereas almost all tomato plants inoculated with the ΔRS_RS01965 or ΔRS_RS04475 mutant survived. R. solanacearum cells (105 CFU) were injected into the tomato stem to assay the colonization of these mutants. As shown in Fig. 2E, 109.9 CFU R. solanacearum were detected in 1 g tomato plant stem 5 days after inoculation of the wild-type strain. However, this number was 109.0 and 108.2 for the ΔRS_RS01965 and ΔRS_RS04475 mutants, respectively. Moreover, the competitive index was measured to confirm the results of Tn-seq. The ΔRS_RS01965 and ΔRS_RS04475 strains were outcompeted by GMI1000Kanr with competitive index values of 0.19 and 0.10, respectively (Fig. 2F). These results indicated that RS_RS01965 and RS_RS04475 are essential for the survival of R. solanacearum in tomato plants.
Amino acid transport and metabolism-related genes required for survival in tomato plants.
Nineteen amino acid transport and metabolism genes were identified as essential for survival in tomato plants by Tn-seq (Table S1). Ten of these genes were mapped to the pathway of “phenylalanine, tyrosine and tryptophan biosynthesis” in the Kyoto Encyclopedia of Genes and Genomes (KEGG) database (Fig. S2). Four (RS_RS14885, RS_RS07885, RS_RS13320, and RS_RS04510) of these 10 genes are involved in the shikimate pathway, which synthesizes chorismite, an important biochemical intermediate for amino acid biosynthesis. Four genes are responsible for tryptophan biosynthesis, including tryptophan synthase subunit alpha (RS_RS09955; TrpA), tryptophan synthase subunit beta (RS_RS09965; TrpB), anthranilate synthase component I (RS_RS14430; TrpE), and phosphoribosylanthranilate isomerase (RS_RS09970; TrpF). Two genes (RS_RS14785 and RS_RS05000) are involved in the biosynthesis of phenylalanine and tyrosine (Fig. S2). In addition, genes involved in the biosynthesis of serine (RS_RS08265 and RS_RS04490), cysteine (RS_RS05790), methionine (RS_RS00135), and lysine (RS_RS05700) were also identified as essential for the survival of R. solanacearum in tomato plants.
Genes involved in the “phenylalanine, tyrosine, and tryptophan biosynthesis” pathway are required for survival in tomato plants. Download FIG S2, TIF file, 0.7 MB (750KB, tif) .
Copyright © 2021 Su et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
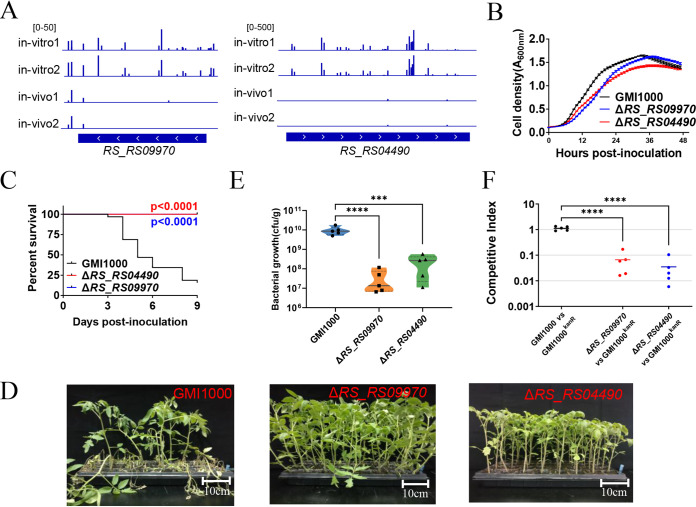
Two genes related to tryptophan and serine biosynthesis were targeted to verify their pathogenic contribution in R. solanacearum. As shown in Fig. 3A and Table S1, 13 unique insertion locations in RS_RS09970 were mapped by 52 weighted reads in average before infection, whereas the transposon insertion mutant of RS_RS09970 was hardly detected from the library after tomato plant infection. The relative abundance of the RS_RS04490 mutant was also sharply reduced after infection (Fig. 3A and Table S1). RS_RS09970 and RS_RS04490 were then deleted individually. As shown in Fig. 3B, the deletion of RS_RS09970 or RS_RS04490 slightly impaired the growth of R. solanacearum in rich BG medium. The functions of RS_RS09970 and RS_RS04490 in amino acid biosynthesis were verified. ΔRS_RS09970 and ΔRS_RS04490 mutants cannot grow in minimal Fahraeus medium. The auxotrophs of the ΔRS_RS09970 and ΔRS_RS04490 mutants could be rescued by supplementation with tryptophan and serine, respectively (Fig. S3). The pathogenicity of these two mutants was then assayed by stem injection. As shown in Fig. 3C and D, ΔRS_RS09970 and ΔRS_RS04490 mutants did not cause any disease symptoms on tomato plants. Moreover, colonization by the ΔRS_RS09970 and ΔRS_RS04490 mutants was remarkably weaker than that of the wild-type strain GMI1000 (Fig. 3E). The ΔRS_RS09970 and ΔRS_RS04490 mutants were outcompeted when coinoculated with GMI1000Kanr into tomato plants with competitive index values of 0.07 and 0.04, respectively (Fig. 3F). These results indicate that RS_RS09970 and RS_RS04490 are essential for fitness in tomato plants.
FIG 3.
RS_RS09970 and RS_RS04490 are required for survival in tomato plants. (A) Transposon insertion distribution within RS_RS09970 and RS_RS04490 of transposon insertion libraries in vivo and in vitro. (B) Growth of the ΔRS_RS09970 mutant, the ΔRS_RS04490 mutant, and wild-type strain GMI1000 in BG medium. (C) Survival curve of tomato plants inoculated with the ΔRS_RS09970 mutant, the ΔRS_RS04490 mutant, and wild-type strain GMI1000. Kaplan-Meier survival analysis with the Gehan-Breslow-Wilcoxon method was used to compare pathogenicity between the mutant and wild-type strains. A P value of <0.05 was considered significant. (D) Bacterial wilt symptoms of tomato plants 9 days after inoculation of the ΔRS_RS09970 mutant, the ΔRS_RS04490 mutant, and wild-type strain GMI1000. (E) Colonization of ΔRS_RS09970 and ΔRS_RS04490 mutants. The CFU of R. solanacearum strains in 1 g tomato plant stem were counted 5 days postinoculation. Asterisks indicate significant differences (***, P < 0.001; ****, P < 0.0001; t test). (F) In vivo competitive index of ΔRS_RS09970 and ΔRS_RS04490 mutants. R. solanacearum mutants and GMI1000Kanr were coinoculated into the stems of tomato plants. The competitive index was measured 5 days postinoculation. Asterisks indicate significant differences (****, P < 0.0001; t test).
The ΔRS_RS09970 and ΔRS_RS04490 mutants are auxotrophs. (A) Growth of the ΔRS_RS09970 mutant, the ΔRS_RS04490 mutant, and wild-type GMI1000 in liquid minimal Fahraeus medium with or without tryptophan (Trp) or serine (Ser). The growth (A600) of each R. solanacearum strain in a given medium was monitored every hour via Bioscreen C Pro. The growth was indicated by the means of three biological replicates. The error bars indicates standard deviations. (B) Growth of the ΔRS_RS09970 mutant, the ΔRS_RS04490 mutant, and wild-type strain GMI1000 on Fahraeus agar medium with or without tryptophan (Trp) or serine (Ser). Gradient-diluted R. solanacearum strains were cultured on Fahraeus medium and photographed 24 h postinoculation. Download FIG S3, TIF file, 0.8 MB (835.2KB, tif) .
Copyright © 2021 Su et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Energy production and conversion-related genes required for survival in tomato plants.
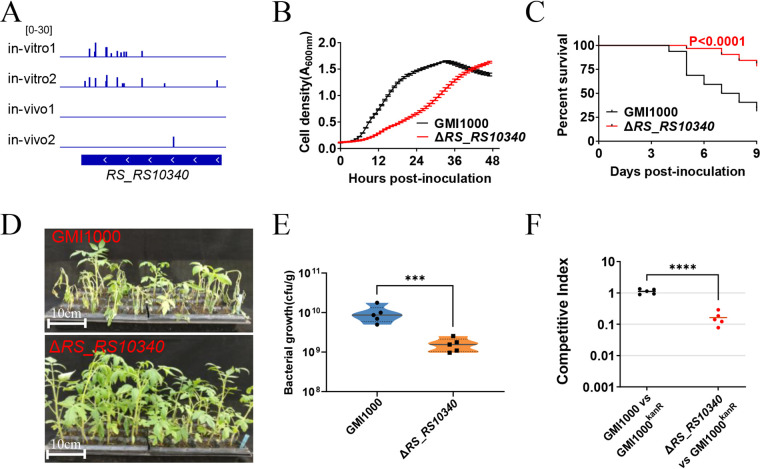
The transposon insertion mutants of 13 energy production and conversion-related genes were less frequently detected in the transposon insertion libraries recovered from the inoculated tomato plants. Thus, energy production- and conversion-related genes are more required for R. solanacearum to survive in tomato plants than in rich medium. Seven of these 13 genes are NADH-quinone oxidoreductase subunit-encoding genes (Table S1). For example, the relative abundance of a NADH-quinone oxidoreductase subunit B (RS_RS10340) mutant in in vivo libraries was 0.06 times higher than that in in vitro libraries (Fig. 4A and Table S1). RS_RS10340 was then deleted in frame to verify the importance of energy production- and conversion-related genes for survival in tomato plants.
FIG 4.
RS_RS10340 is required for survival in tomato plants. (A) Transposon insertion distribution within RS_RS10340 of transposon insertion libraries in vivo and in vitro. (B) Growth of the ΔRS_RS10340 mutant and wild-type strain GMI1000 in BG medium. (C) Survival curve of tomato plants inoculated with the ΔRS_RS10340 mutant and wild-type strain GMI1000. Kaplan-Meier survival analysis with the Gehan-Breslow-Wilcoxon method was used to compare pathogenicity between the mutant and wild-type strains. A P value of <0.05 was considered significant. (D) The bacterial wilt symptom of tomato plants 9 days postinoculation of the ΔRS_RS10340 mutant and wild-type strain GMI1000. (E) Colonization of the ΔRS_RS10340 mutant. The CFU of R. solanacearum strains in 1 g tomato plant stem were counted 5 days postinoculation. Asterisks indicate significant differences (***, P < 0.001; t test). (F) In vivo competitive index of the ΔRS_RS10340 mutant. The ΔRS_RS10340 mutant and GMI1000Kanr were coinoculated into the stems of tomato plants. The competitive index was measured 5 days postinoculation. Asterisks indicate significant differences (****, P < 0.0001; t test).
As shown in Fig. 4B, the final concentration of RS_RS10340 deletion mutant in rich BG medium was similar to that of the wild-type strain GMI1000, though the deletion of RS_RS10340 seriously slowed the growth of R. solanacearum. The ΔRS_RS10340 mutant and wild-type strain GMI1000 were inoculated into tomato plants by injection to assay pathogenicity. As shown in Fig. 4C and D, only 34% of tomato plants survived 9 days after inoculation with wild-type strain, whereas 81% of plants survived 9 days after inoculation with the ΔRS_RS10340 mutant. Moreover, the absence of RS_RS10340 remarkably attenuated the growth of R. solanacearum in tomato plants (Fig. 4E). The competition assay confirmed that the wild-type strain overgrew the ΔRS_RS10340 mutant in tomato plants (Fig. 4F). Thus, RS_RS10340 is required for the survival of R. solanacearum in tomato plants.
Posttranslational modification, protein turnover, and chaperone-related genes required for survival in tomato plants.
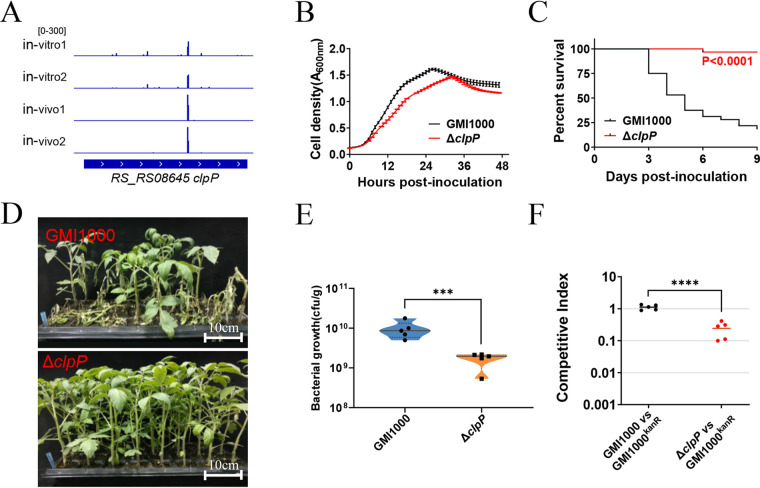
Eleven of the genes required for R. solanacearum survival in tomato plants were categorized as “posttranslational modification, protein turnover, chaperones” (Table S1). Four of these 11 genes, namely, those encoding the FtsH protease activity modulator HflK (RS_RS06120), protease modulator HflC (RS_RS06125), ATP-dependent Clp protease proteolytic subunit (RS_RS08645; clpP), and ATP-dependent Clp protease ATP-binding subunit ClpX (RS_RS08650), are involved in proteolysis. ClpP protease plays an important role in the proteostasis of prokaryotic cells and eukaryotic organelles (23). ClpP was found to be essential for the full virulence of Salmonella enterica serovar Typhimurium, Staphylococcus aureus, and the phytopathogen X. campestris (24–26). Thus, clpP was selected for further functional validation.
As shown in Fig. 5A, three unique insertions in clpP were identified from the in vivo library, and 12 unique hits were identified in clpP for in vitro treatment. The number of weighted reads on average for in vivo treatment is 12, whereas the number for in vitro treatment is 48. As reported in our previous study (27), the targeted deletion of clpP substantially slowed the growth of R. solanacearum in rich BG medium (Fig. 5B). A ΔclpP mutant was inoculated into tomato plant stems to assess the pathogenic role of ClpP. As shown in Fig. 5C and D, 81% of the tomato plants infected with wild-type strain GMI1000 wilted 9 days postinoculation, whereas almost all tomato plants infected with the ΔclpP mutant survived. The result indicates that ClpP protease is essential for the fitness of R. solanacearum. Moreover, colonization assays and competitive index assay confirmed that ClpP protease is required for R. solanacearum survival in tomato plants (Fig. 5E and F).
FIG 5.
clpP is required for survival in tomato plants. (A) Transposon insertion distribution within clpP of transposon insertion libraries in vivo and in vitro. (B) Growth of the ΔclpP mutant and wild-type strain GMI1000 in BG medium. (C) Survival curve of tomato plants inoculated with the ΔclpP mutant and wild-type strain GMI1000. Kaplan-Meier survival analysis with the Gehan-Breslow-Wilcoxon method was used to compare the pathogenicity between the mutant and wild-type strains. P values of <0.05 were considered significant. (D) The bacterial wilt symptom of tomato plants 9 days after inoculation with the ΔclpP mutant and wild-type strain GMI1000. (E) Colonization by the ΔclpP mutant. The CFU of R. solanacearum strains in 1 g tomato plant stem were counted 5 days postinoculation. Asterisks indicate significant differences (***, P < 0.001; t test). (F) In vivo competitive index of the ΔclpP mutant. The ΔclpP mutant and GMI1000Kanr were coinoculated into the stems of tomato plants. The competitive index was measured 5 days postinoculation. Asterisks indicate significant differences (****, P < 0.0001; t test).
DISCUSSION
Besides the genes required for survival in tomato plants, Tn-seq indicated that the interruption of two genes improved relative fitness in vivo compared with in vitro. The read ratio (in vivo/in vitro) of XRE family transcriptional regulator RS_RS05450 was 2.4, which means that the environmental stress within tomato plants improved the relative abundance of the RS_RS05450 mutant by 2.4 times (Fig. S4A and Table S1). Previous experimental evolution and reverse genetic approaches revealed that mutation of RS_RS05450, namely, efpR, is associated with fitness gain on bean, tomato, and other host plants (14, 28). EfpR acts as a global catabolic repressor in R. solanacearum, and the absence of epfR would expand metabolic versatility and improve the relative fitness competing with wild-type R. solanacearum strains (28).
Interruption of efpR (RS_RS05450) and RS_RS05405 resulted in improved relative fitness in vivo compared with in vitro. (A) Transposon insertion distribution within efpR and RS_RS05405 of transposon insertion libraries in vivo and in vitro. (B) Growth of the ΔRS_RS05405 mutant in BG medium. (C) Competitive index of the ΔRS_RS05405 mutant in vitro and in vivo. The ΔRS_RS05405 mutant and GMI1000Kanr containing a kanamycin resistance gene were coinoculated into the stems of tomato plants, and the in vivo competitive index was measured 5 days postinoculation. The ΔRS_RS05405 mutant and GMI1000Kanr were coinoculated into BG medium, and the in vitro competitive index was measured 12 h (log phase) and 36 h (stationary phase) postinoculation. Asterisks indicate significant differences (**, P < 0.01; ***, P < 0.001; t test). Download FIG S4, TIF file, 0.3 MB (316.6KB, tif) .
Copyright © 2021 Su et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Besides efpR, the transposon insertion in RS_RS05405, which encodes nonheme iron oxygenase ferredoxin subunit, improved the relative fitness of R. solanacearum in tomato plants compared with that in vitro. Nonheme-iron-dependent oxygenases catalyze various reactions in the biodegradation of xenobiotics and the biosynthesis of bioactive natural products (29). The relative abundance of the RS_RS05405 mutant in vivo was 13.2 times higher than in vitro (Fig. S4A and Table S1). RS_RS05405 was deleted to validate this finding. As shown in Fig. S4B, RS_RS05405 deletion slowed the growth of R. solanacearum, but the final bacterial concentration of the ΔRS_RS05405 mutant was greater than that of wild-type GMI1000. The pathogenicity of the ΔRS_RS05405 mutant was assayed. However, no statistical difference was found between the pathogenicity of the ΔRS_RS05405 mutant and the wild-type strain. The in vivo and in vitro competitive indexes of the ΔRS_RS05405 mutant were then assayed. Mutant ΔRS_RS05405 and GMI1000Kanr strains containing a kanamycin resistance gene were coinoculated into the stems of tomato plants and rich BG medium. R. solanacearum strains were recovered from tomato plants 5 days postinoculation. The competitive index of the ΔRS_RS05405 mutant in planta was 0.48, whereas the competitive fitness of the ΔRS_RS05405 mutant at the stationary phase in BG was 0.34 (Fig. S4C). The competition assay indicated that RS_RS05405 is required for the survival of R. solanacearum in vivo and in vitro, but this gene may be more critical for in vitro survival than in planta survival.
Lipopolysaccharide (LPS) is a vital component of Gram-negative bacterial outer membrane and protects bacteria from harsh environmental conditions (30). The LPS of pathogenic bacteria has critical roles in biofilm formation, host attachment, and colonization (31). The pathogenic roles of different forms of LPS in R. solanacearum were systematically characterized, and all 13 investigated LPS-defective mutants were unable to cause any disease symptom in tomato plants (32). Consistently, eight of these 13 genes were identified in the present study as essential for R. solanacearum survival in tomato plants. These eight genes were involved in LPS core biogenesis (RS_RS02830, RS_RS04545, and RS_RS04550), LPS O-antigen biogenesis (RS_RS03460, RS_RS03465, and RS_RS11050), LPS O-antigen ligation (RS_RS11060), and mannose metabolism (RS_RS15365). Our targeted deletion also verified that the sorting and localization system of lipoprotein RS_RS01965, as well as the functional known membrane protein RS_RS04475, is required for in vivo survival. These results highlighted the importance of the cell membrane for the pathogenicity and survival of R. solanacearum.
Tomato xylem sap is a nutritionally limiting habitat, and R. solanacearum needs to regulate its metabolism and alter xylem sap biochemistry to adapt to this niche (33–35). Most proteinogenic amino acids are present at micromolar concentrations in xylem sap, and many of them are limiting for the growth of R. solanacearum (34). The biosynthesis of tryptophan is required for the pathogenicity of R. solanacearum on tomato and tobacco. Moreover, the biosyntheses of methionine and leucine are important for survival in tomato but not in tobacco, based on chemical mutagenesis (36). Tn-seq in the present study revealed that 19 genes involved in amino acid transport and metabolism are required for survival in tomato plants. These genes encode enzymes involved in the biosynthesis of chorismite, tryptophan, serine, cysteine, methionine, phenylalanine, tyrosine, and lysine. A Tn-seq study of R. solanacearum cultured in xylem sap extracted from tomato plant was recently reported in a preprint (37). Although the in planta environment is dynamically changing and is different from the environment in extracted xylem sap (38), 12 of the mentioned 19 genes were identified by the Tn-seq study of R. solanacearum cultured in tomato xylem sap (37).
Moreover, RS_RS12160, which encodes phosphoadenylyl-sulfate reductase, and RS_RS06745, which encodes sulfate ABC transporter permease subunit CysT and was here classified in “inorganic ion transport and metabolism (P)” by COG, were associated with cysteine biosynthesis. RS_RS12160 and RS_RS06745 were also previously identified (37). Overall, all the in vivo essential amino acid biosynthesis pathways revealed in the present study were hit by the Tn-seq of R. solanacearum cultured in xylem sap. Although many studies inferred that the biosynthesis of amino acids is important for the pathogenicity and in planta survival of R. solanacearum, targeted verification studies are limited (35). The present study deleted RS_RS01965 and RS_RS04475 in-frame to generate auxotrophic strains and validated that the biosyntheses of tryptophan and serine are essential for the fitness of R. solanacearum in host tomato plants.
Most of the known virulence factors of R. solanacearum are megaplasmid borne, including the type III secretion system, which delivers type III effectors into plant cells and is the key for pathogen and plant interaction (5), type VI protein secretion systems, flagellar motility determinants, chemotaxis genes, and EPS biosynthesis genes (10). However, most (117/132) of the in vivo survival genes identified in our study are chromosome borne, and most of the known virulence factors mentioned were not hit by the present study. Secreted virulence determinants, including type III effectors, type VI proteins, extracellular enzymes, and EPS, serve as public goods for the pathogenic population. Mutants can survive without the secretion of these virulence determinants by benefiting from the public goods secreted by other strains (39, 40). This may explain why these virulence genes were not identified by Tn-seq based on relative fitness. R. solanacearum is virtually nonmotile in plants, and motility and chemotaxis contribute to the early stages of host plant invasion and colonization (41). The transposon insertion library of R. solanacearum was directly injected into the stems of tomato plants in this study. Mutants without motility or chemotaxis could survive inside the stems of tomato plants. Future studies may inoculate the transposon insertion library of R. solanacearum via different methods to identify genes essential for the different stages of R. solanacearum infection.
MATERIALS AND METHODS
Bacterial growth conditions.
R. solanacearum strains were cultured in BG medium (10 g/liter Bacto peptone, 1 g/liter Casamino Acids, 1 g/liter yeast extract, and 5 g/liter glucose) or on BG agar medium at 28°C, except where noted otherwise. Escherichia coli strains were cultured in Luria-Bertani (LB) medium or on LB agar medium at 37°C. A final concentration of 25 μg/ml kanamycin was added to the medium when needed. The growth (A600) of R. solanacearum strains with three biological repeats was monitored every hour via Bioscreen C Pro (Oy Growth Curves Ab Ltd., Turku, Finland) to obtain growth curves. The ΔRS_RS04490 mutant was cultured in Fahraeus medium (21, 42) with or without 17.5 mM serine to assay the auxotroph. The ΔRS_RS09970 mutant was cultured in Fahraeus medium with or without 0.4 mM tryptophan to assay the auxotroph.
Tn-seq of R. solanacearum in tomato plants.
The near-saturated transposon insertion library of R. solanacearum GMI1000 preserved at −80°C was adjusted to 1 × 108 CFU/ml in BG medium and reactivated at 28°C for 1 h. The activated transposon insertion library was injected into the stems of 4-week-old tomato cultivar ‘Zhongshu No. 4’ plants cultured in a classic greenhouse. About 10 μl of transposon insertion library was inoculated per plant. The transposon insertion library before plant inoculation was used as the control group and referred to as “in vitro.” Transposon insertion libraries were recovered from plants 5 days postinoculation, when most plants showed a disease index of 3 or 2. The bacteria were recovered from 32 diseased plants and pooled for further steps to reduce the bottleneck of injected transposon insertion library and enrich bacterial DNA. The recovered transposon insertion library was referred to as “in vivo.” Total DNAs of the transposon insertion libraries before and after infection were extracted and divided into two groups for technical replicates. The total DNA samples were subjected to MmeI digestion, adapter ligation, and PCR amplification to construct Illumina sequencing libraries and subjected to sequencing and raw data preprocessing on the Galaxy web platform as previously described (20, 43). The index sequences of the sequencing manufacturer and transposon sequence in raw reads were trimmed. The reads were then separated based on the barcode for each sample, followed by barcode sequence trimming and read filtering by quality.
The preprocessed reads were mapped to the genome of R. solanacearum GMI1000 (GCA_000009125.1) by using Bowtie tolerating a 0-bp mismatch. The bam files from mapping were subjected to sample correlation coefficient computation and visualization via deepTools (44). We set the transposon insertion library before infection (in vitro) as the control, and gene essentiality in vivo was analyzed by TSAS, a Tn-seq analysis software (45). TSAS was run in two-sample analysis mode, while the other key input parameters were left in default. Ratio_reads (in vivo/in vitro) values of <0.5 with adjusted P (proportions_reads) values of <0.01 were set as the threshold values to identify genes required for survival in tomato plants (Table S1).
The transposon insertion distribution in targeted genes was visualized by Integrative Genomics Viewer using the wig files from TSAS as input (46). The COG categories of genes required for R. solanacearum survival in tomato plants were annotated by eggnog-mapper (47). The pathways of genes classified in amino acid transport and metabolism were analyzed by mapping in the KEGG pathway database and rendered by Pathview (48).
Gene deletion in R. solanacearum.
The gene of interest was deleted in frame as previously described (27). Briefly, two flanking regions of the target gene were amplified using the primers listed in Table S2. These two DNA fragments were ligated via overlapping PCR and cloned into suicide plasmid pK18mobsacB or directly cloned into pK18mobsacB via three appropriate restriction enzyme sites. The recombined plasmid was verified by Sanger sequencing and imported into R. solanacearum wild-type strain GMI1000 by electroporation. The resultant recombinant strain was then cultured in a modified BG medium in which glucose was replaced by 10% sucrose for the second homologous recombination. The mutant that lacks the target gene was selected by PCR using the primers that flank the target gene (1F and 2R in Table S2).
Primers used in this study. Download Table S2, XLSX file, 0.01 MB (11KB, xlsx) .
Copyright © 2021 Su et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Pathogenicity phenotyping of R. solanacearum.
Susceptible tomato cultivar ‘Zhongshu No. 4’ was seeded and transplanted into 32-cell plug trays growing in a greenhouse. The 4-week-old plants were used for pathogenicity assay of R. solanacearum by stem injection following the protocol described elsewhere (21). The tested R. solanacearum mutant and wild-type strains were cultured in BG medium to the log growth phase and adjusted to 1 × 107 CFU/ml with sterilized H2O. About 10 μl of adjusted bacterial suspension was injected into the stem 0.5 cm above the cotyledons using a disposable microsyringe. The wilting symptoms of inoculated plants were scored on a visual scale of 0 (no symptoms) to 4 (complete wilting) once per day. The 32 plants in a tray were inoculated for each R. solanacearum strain. Kaplan-Meier survival analysis with the Gehan-Breslow-Wilcoxon method was used to compare the pathogenicity between the mutant and wild-type strains (49). A P value of <0.05 was considered significant. The pathogenicity was assayed at least three times, and one representative result was presented.
The colonization of R. solanacearum strains was measured according to the previously described protocol (21). We sampled and weighed 1 g of the stem 1 cm above the inoculation point at 5 days postinoculation. The sampled stem was then sterilized in 70% ethanol for 30 s, rinsed in sterile water for 30 s, and ground in sterile mortars. The bacterial concentration was determined by plating on BG agar medium after serial 10× dilutions and expressed as CFU per gram of fresh stem. The bacterial concentrations of five infected tomato plants were measured for each R. solanacearum strain. The colonization differences between mutant and wild-type strains were analyzed with an unpaired t test.
Each R. solanacearum strain for the test was mixed with GMI1000Kanr containing a kanamycin resistance gene at the same bacterial concentration and the same volume to assay the competitive index. The amount of R. solanacearum strains was determined by plating on BG medium with or without 25 μg/ml kanamycin. The amount of GMI1000Kanr was determined by enumerating CFU on BG agar medium with added kanamycin, and the amount of the tested R. solanacearum strain was the number of CFU on BG agar medium without kanamycin minus the amount of GMI1000Kanr. The mixed bacterial suspension was then inoculated into tomato plants by stem injection. The amount of R. solanacearum strains in infected tomato plants was redetermined 5 days postinoculation. Competitive index was calculated as [tested strain CFU/GMI1000Kanr CFU (5 days postinoculation)]/[tested strain CFU/GMI1000Kanr CFU (before inoculation)]. Five biological replicates were performed for each R. solanacearum strain. The competitive index differences between mutant/GMI1000Kanr and GMI1000/GMI1000Kanr were analyzed with an unpaired t test.
Data availability.
The processed reads and raw reads are available in the SRA database of NCBI (PRJNA766096). The wig files from TSAS were deposited in Figshare (https://doi.org/10.6084/m9.figshare.14220053).
ACKNOWLEDGMENTS
This work was supported by the National Natural Science Foundation of China (project 31901847) and the Natural Science Foundation of Guangxi (projects 2019GXNSFBA185010 and GuiKeAD20238056).
Contributor Information
Dehong Zheng, Email: dehong@gxu.edu.cn.
Davide Bulgarelli, University of Dundee.
REFERENCES
- 1.Mansfield J, Genin S, Magori S, Citovsky V, Sriariyanum M, Ronald P, Dow M, Verdier V, Beer SV, Machado MA, Toth I, Salmond G, Foster GD. 2012. Top 10 plant pathogenic bacteria in molecular plant pathology. Mol Plant Pathol 13:614–629. doi: 10.1111/j.1364-3703.2012.00804.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lopes CA, Rossato M. 2018. History and status of selected hosts of the Ralstonia solanacearum species complex causing bacterial wilt in Brazil. Front Microbiol 9:1228. doi: 10.3389/fmicb.2018.01228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jiang G, Wei Z, Xu J, Chen H, Zhang Y, She X, Macho AP, Ding W, Liao B. 2017. Bacterial wilt in China: history, current status, and future perspectives. Front Plant Sci 8:1549. doi: 10.3389/fpls.2017.01549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Elphinstone J, Allen C, Prior P, Hayward A. 2005. The current bacterial wilt situation: a global overview, p 9–28. In Allen C, Prior P, Hayward AC (ed), Bacterial wilt disease and the Ralstonia solanacearum species complex. American Phytopathological Society, St. Paul, MN. [Google Scholar]
- 5.Landry D, González-Fuente M, Deslandes L, Peeters N. 2020. The large, diverse, and robust arsenal of Ralstonia solanacearum type III effectors and their in planta functions. Mol Plant Pathol 21:1377–1388. doi: 10.1111/mpp.12977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lonjon F, Lohou D, Cazalé A-C, Büttner D, Ribeiro BG, Péanne C, Genin S, Vailleau F. 2017. HpaB-dependent secretion of Type III effectors in the plant pathogens Ralstonia solanacearum and Xanthomonas campestris pv. vesicatoria. Sci Rep 7:4879. doi: 10.1038/s41598-017-04853-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Genin S, Gough CL, Zischek C, Boucher CA. 1992. Evidence that the hrpB gene encodes a positive regulator of pathogenicity genes from Pseudomonas solanacearum. Mol Microbiol 6:3065–3076. doi: 10.1111/j.1365-2958.1992.tb01764.x. [DOI] [PubMed] [Google Scholar]
- 8.Angot A, Peeters N, Lechner E, Vailleau F, Baud C, Gentzbittel L, Sartorel E, Genschik P, Boucher C, Genin S. 2006. Ralstonia solanacearum requires F-box-like domain-containing type III effectors to promote disease on several host plants. Proc Natl Acad Sci USA 103:14620–14625. doi: 10.1073/pnas.0509393103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Saile E, McGarvey JA, Schell MA, Denny TP. 1997. Role of extracellular polysaccharide and endoglucanase in root invasion and colonization of tomato plants by Ralstonia solanacearum. Phytopathology 87:1264–1271. doi: 10.1094/PHYTO.1997.87.12.1264. [DOI] [PubMed] [Google Scholar]
- 10.Genin S, Denny TP. 2012. Pathogenomics of the Ralstonia solanacearum species complex. Annu Rev Phytopathol 50:67–89. doi: 10.1146/annurev-phyto-081211-173000. [DOI] [PubMed] [Google Scholar]
- 11.Brown DG, Allen C. 2004. Ralstonia solanacearum genes induced during growth in tomato: an inside view of bacterial wilt. Mol Microbiol 53:1641–1660. doi: 10.1111/j.1365-2958.2004.04237.x. [DOI] [PubMed] [Google Scholar]
- 12.Jacobs JM, Babujee L, Meng F, Milling A, Allen C. 2012. The in planta transcriptome of Ralstonia solanacearum: conserved physiological and virulence strategies during bacterial wilt of tomato. mBio 3:e00114-12. doi: 10.1128/mBio.00114-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eckshtain-Levi N, Weisberg AJ, Vinatzer BA. 2018. The population genetic test Tajima's D identifies genes encoding pathogen-associated molecular patterns and other virulence-related genes in Ralstonia solanacearum. Mol Plant Pathol 19:2187–2192. doi: 10.1111/mpp.12688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guidot A, Jiang W, Ferdy JB, Thebaud C, Barberis P, Gouzy J, Genin S. 2014. Multihost experimental evolution of the pathogen Ralstonia solanacearum unveils genes involved in adaptation to plants. Mol Biol Evol 31:2913–2928. doi: 10.1093/molbev/msu229. [DOI] [PubMed] [Google Scholar]
- 15.van Opijnen T, Bodi KL, Camilli A. 2009. Tn-seq: high-throughput parallel sequencing for fitness and genetic interaction studies in microorganisms. Nat Methods 6:767–772. doi: 10.1038/nmeth.1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cain AK, Barquist L, Goodman AL, Paulsen IT, Parkhill J, van Opijnen T. 2020. A decade of advances in transposon-insertion sequencing. Nat Rev Genet 21:526–540. doi: 10.1038/s41576-020-0244-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fabian BK, Tetu SG, Paulsen IT. 2020. Application of transposon insertion sequencing to agricultural science. Front Plant Sci 11:291. doi: 10.3389/fpls.2020.00291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Royet K, Parisot N, Rodrigue A, Gueguen E, Condemine G. 2019. Identification by Tn-seq of Dickeya dadantii genes required for survival in chicory plants. Mol Plant Pathol 20:287–306. doi: 10.1111/mpp.12754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Duong DA, Jensen RV, Stevens AM. 2018. Discovery of Pantoea stewartii ssp. stewartii genes important for survival in corn xylem through a Tn-Seq analysis. Mol Plant Pathol 19:1929–1941. doi: 10.1111/mpp.12669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Su Y, Xu Y, Li Q, Yuan G, Zheng D. 2020. The essential genome of Ralstonia solanacearum. Microbiol Res 238:126500. doi: 10.1016/j.micres.2020.126500. [DOI] [PubMed] [Google Scholar]
- 21.Morel A, Peeters N, Vailleau F, Barberis P, Jiang G, Berthomé R, Guidot A. 2018. Plant pathogenicity phenotyping of Ralstonia solanacearum strains. Methods Mol Biol 1734:223–239. doi: 10.1007/978-1-4939-7604-1_18. [DOI] [PubMed] [Google Scholar]
- 22.Liao C-T, Chiang Y-C, Hsiao Y-M. 2019. Functional characterization and proteomic analysis of lolA in Xanthomonas campestris pv. campestris. BMC Microbiol 19:20. doi: 10.1186/s12866-019-1387-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bhandari V, Wong KS, Zhou JL, Mabanglo MF, Batey RA, Houry WA. 2018. The role of ClpP protease in bacterial pathogenesis and human diseases. ACS Chem Biol 13:1413–1425. doi: 10.1021/acschembio.8b00124. [DOI] [PubMed] [Google Scholar]
- 24.Knudsen GM, Olsen JE, Aabo S, Barrow P, Rychlik I, Thomsen LE. 2013. ClpP deletion causes attenuation of Salmonella typhimurium virulence through mis-regulation of RpoS and indirect control of CsrA and the SPI genes. Microbiology (Reading) 159:1497–1509. doi: 10.1099/mic.0.065797-0. [DOI] [PubMed] [Google Scholar]
- 25.Farrand AJ, Reniere ML, Ingmer H, Frees D, Skaar EP. 2013. Regulation of host hemoglobin binding by the Staphylococcus aureus Clp proteolytic system. J Bacteriol 195:5041–5050. doi: 10.1128/JB.00505-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li CE, Liao CT, Lo HH, Hsiao YM. 2020. Functional characterization and transcriptional analysis of clpP of Xanthomonas campestris pv. campestris. Curr Microbiol 77:2876–2885. doi: 10.1007/s00284-020-02093-1. [DOI] [PubMed] [Google Scholar]
- 27.Zheng D, Xu Y, Yuan G, Wu X, Li Q. 2020. Bacterial ClpP protease is a potential target for methyl gallate. Front Microbiol 11:598692. doi: 10.3389/fmicb.2020.598692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Perrier A, Peyraud R, Rengel D, Barlet X, Lucasson E, Gouzy J, Peeters N, Genin S, Guidot A. 2016. Enhanced in planta fitness through adaptive mutations in EfpR, a dual regulator of virulence and metabolic functions in the plant pathogen Ralstonia solanacearum. PLoS Pathog 12:e1006044. doi: 10.1371/journal.ppat.1006044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barry SM, Challis GL. 2013. Mechanism and catalytic diversity of rieske non-heme iron-dependent oxygenases. ACS Catal 3:2362–2370. doi: 10.1021/cs400087p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Owens TW, Taylor RJ, Pahil KS, Bertani BR, Ruiz N, Kruse AC, Kahne D. 2019. Structural basis of unidirectional export of lipopolysaccharide to the cell surface. Nature 567:550–553. doi: 10.1038/s41586-019-1039-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Newman MA, Dow JM, Molinaro A, Parrilli M. 2007. Priming, induction and modulation of plant defence responses by bacterial lipopolysaccharides. J Endotoxin Res 13:69–84. doi: 10.1177/0968051907079399. [DOI] [PubMed] [Google Scholar]
- 32.Li CH, Wang KC, Hong YH, Chu TH, Chu YJ, Chou IC, Lu DK, Chen CY, Yang WC, Lin YM, Cheng CP. 2014. Roles of different forms of lipopolysaccharides in Ralstonia solanacearum pathogenesis. Mol Plant Microbe Interact 27:471–478. doi: 10.1094/MPMI-08-13-0248-R. [DOI] [PubMed] [Google Scholar]
- 33.Hamilton CD, Steidl OR, MacIntyre AM, Hendrich CG, Allen C. 2021. Ralstonia solanacearum depends on catabolism of myo-inositol, sucrose, and trehalose for virulence in an infection stage-dependent manner. Mol Plant Microbe Interact 2021:MPMI10200298. doi: 10.1094/MPMI-10-20-0298-R. [DOI] [PubMed] [Google Scholar]
- 34.Zuluaga AP, Puigvert M, Valls M. 2013. Novel plant inputs influencing Ralstonia solanacearum during infection. Front Microbiol 4:349. doi: 10.3389/fmicb.2013.00349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lowe-Power TM, Khokhani D, Allen C. 2018. How Ralstonia solanacearum exploits and thrives in the flowing plant xylem environment. Trends Microbiol 26:929–942. doi: 10.1016/j.tim.2018.06.002. [DOI] [PubMed] [Google Scholar]
- 36.Coplin DL, Sequeira L, Hanson RS. 1974. Pseudomonas solanacearum: virulence of biochemical mutants. Can J Microbiol 20:519–529. doi: 10.1139/m74-080. [DOI] [PubMed] [Google Scholar]
- 37.Georgoulis S, Shalvarjian KE, Helmann TC, Hamilton CD, Carlson HK, Deutschbauer AM, Lowe-Power TM. 2020. Genome-wide identification of tomato xylem sap fitness factors for Ralstonia pseudosolanacearum and Ralstonia syzygii. bioRxiv doi: 10.1101/2020.08.31. [DOI] [PMC free article] [PubMed]
- 38.Lowe-Power TM, Hendrich CG, von Roepenack-Lahaye E, Li B, Wu D, Mitra R, Dalsing BL, Ricca P, Naidoo J, Cook D, Jancewicz A, Masson P, Thomma B, Lahaye T, Michael AJ, Allen C. 2018. Metabolomics of tomato xylem sap during bacterial wilt reveals Ralstonia solanacearum produces abundant putrescine, a metabolite that accelerates wilt disease. Environ Microbiol 20:1330–1349. doi: 10.1111/1462-2920.14020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Czechowska K, McKeithen-Mead S, Al Moussawi K, Kazmierczak BI. 2014. Cheating by type 3 secretion system-negative Pseudomonas aeruginosa during pulmonary infection. Proc Natl Acad Sci USA 111:7801–7806. doi: 10.1073/pnas.1400782111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Friesen ML. 2020. Social evolution and cheating in plant pathogens. Annu Rev Phytopathol 58:55–75. doi: 10.1146/annurev-phyto-010820-012740. [DOI] [PubMed] [Google Scholar]
- 41.Tans-Kersten J, Huang H, Allen C. 2001. Ralstonia solanacearum needs motility for invasive virulence on tomato. J Bacteriol 183:3597–3605. doi: 10.1128/JB.183.12.3597-3605.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fahraeus G. 1957. The infection of clover root hairs by nodule bacteria studied by a simple glass slide technique. J Gen Microbiol 16:374–381. doi: 10.1099/00221287-16-2-374. [DOI] [PubMed] [Google Scholar]
- 43.McCoy KM, Antonio ML, van Opijnen T. 2017. MAGenTA: a Galaxy implemented tool for complete Tn-Seq analysis and data visualization. Bioinformatics 33:2781–2783. doi: 10.1093/bioinformatics/btx320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ramirez F, Ryan DP, Gruning B, Bhardwaj V, Kilpert F, Richter AS, Heyne S, Dundar F, Manke T. 2016. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res 44:W160–W165. doi: 10.1093/nar/gkw257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Burger BT, Imam S, Scarborough MJ, Noguera DR, Donohue TJ. 2017. Combining genome-scale experimental and computational methods to identify essential genes in Rhodobacter sphaeroides. mSystems 2:e00015-17. doi: 10.1128/mSystems.00015-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thorvaldsdóttir H, Robinson JT, Mesirov JP. 2013. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform 14:178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huerta-Cepas J, Szklarczyk D, Forslund K, Cook H, Heller D, Walter MC, Rattei T, Mende DR, Sunagawa S, Kuhn M, Jensen LJ, von Mering C, Bork P. 2016. eggNOG 4.5: a hierarchical orthology framework with improved functional annotations for eukaryotic, prokaryotic and viral sequences. Nucleic Acids Res 44:D286–D293. doi: 10.1093/nar/gkv1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Luo W, Pant G, Bhavnasi YK, Blanchard SG, Jr, Brouwer C. 2017. Pathview Web: user friendly pathway visualization and data integration. Nucleic Acids Res 45:W501–W508. doi: 10.1093/nar/gkx372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Remigi P, Anisimova M, Guidot A, Genin S, Peeters N. 2011. Functional diversification of the GALA type III effector family contributes to Ralstonia solanacearum adaptation on different plant hosts. New Phytol 192:976–987. doi: 10.1111/j.1469-8137.2011.03854.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Gene essentiality for in vivo survival. Download Table S1, XLSX file, 0.03 MB (34.9KB, xlsx) .
Copyright © 2021 Su et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Coefficient of read coverages for Tn-seq technological replicates (scatter plot) and all Tn-seq samples (heat map). Download FIG S1, TIF file, 0.1 MB (132.4KB, tif) .
Copyright © 2021 Su et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Genes involved in the “phenylalanine, tyrosine, and tryptophan biosynthesis” pathway are required for survival in tomato plants. Download FIG S2, TIF file, 0.7 MB (750KB, tif) .
Copyright © 2021 Su et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The ΔRS_RS09970 and ΔRS_RS04490 mutants are auxotrophs. (A) Growth of the ΔRS_RS09970 mutant, the ΔRS_RS04490 mutant, and wild-type GMI1000 in liquid minimal Fahraeus medium with or without tryptophan (Trp) or serine (Ser). The growth (A600) of each R. solanacearum strain in a given medium was monitored every hour via Bioscreen C Pro. The growth was indicated by the means of three biological replicates. The error bars indicates standard deviations. (B) Growth of the ΔRS_RS09970 mutant, the ΔRS_RS04490 mutant, and wild-type strain GMI1000 on Fahraeus agar medium with or without tryptophan (Trp) or serine (Ser). Gradient-diluted R. solanacearum strains were cultured on Fahraeus medium and photographed 24 h postinoculation. Download FIG S3, TIF file, 0.8 MB (835.2KB, tif) .
Copyright © 2021 Su et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Interruption of efpR (RS_RS05450) and RS_RS05405 resulted in improved relative fitness in vivo compared with in vitro. (A) Transposon insertion distribution within efpR and RS_RS05405 of transposon insertion libraries in vivo and in vitro. (B) Growth of the ΔRS_RS05405 mutant in BG medium. (C) Competitive index of the ΔRS_RS05405 mutant in vitro and in vivo. The ΔRS_RS05405 mutant and GMI1000Kanr containing a kanamycin resistance gene were coinoculated into the stems of tomato plants, and the in vivo competitive index was measured 5 days postinoculation. The ΔRS_RS05405 mutant and GMI1000Kanr were coinoculated into BG medium, and the in vitro competitive index was measured 12 h (log phase) and 36 h (stationary phase) postinoculation. Asterisks indicate significant differences (**, P < 0.01; ***, P < 0.001; t test). Download FIG S4, TIF file, 0.3 MB (316.6KB, tif) .
Copyright © 2021 Su et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Primers used in this study. Download Table S2, XLSX file, 0.01 MB (11KB, xlsx) .
Copyright © 2021 Su et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Data Availability Statement
The processed reads and raw reads are available in the SRA database of NCBI (PRJNA766096). The wig files from TSAS were deposited in Figshare (https://doi.org/10.6084/m9.figshare.14220053).