Abstract
The transactivator of transcription (Tat) is a key HIV regulatory protein. We aimed to identify the frequency of key polymorphisms in HIV-1C compared with HIV-1B Tat protein, chiefly in the cysteine-, arginine-, and glutamine-rich domains and identify novel point mutations in HIV-1B and C sequences from Southern Brazil. This study was the first to investigate the genetic diversity and point mutations within HIV-1 Tat C in a Brazilian cohort. This was an observational, cross-sectional study, which included sequences of HIV-1B (n = 20) and HIV-1C (n = 21) from Southern Brazil. Additionally, 344 HIV-1C sequences were obtained from the Los Alamos database: 29 from Brazil and 315 from Africa, Asia, and Europe. The frequency of C31S substitution on HIV-1 Tat C in Brazil was 82% vs. 10% in the HIV-1B group (p < 0.0001). The frequency of the R57S substitution among the HIV-1C sequences from Brazil was 74% vs. 20% in HIV-1B (p = 0.004), and that of substitution Q63E in HIV-1C was 80% and 20% in HIV-1B (p < 0.0001). The mutation P60Q was more frequent in HIV-1B than in HIV-1C (55% and 6.12%, respectively, p < 0.0001)). Novel point mutations in the HIV-1C and B Tat functional domains were described. The frequency of C31S and other key point mutations in HIV-1 Tat C in Brazil were similar to those described in Africa, although lower than those in India. The Tat-B and C sequences found in Southern Brazil are consistent with biological differences and have potential implications for HIV-1 subtype pathogenesis.
Keywords: Subtype C, HIV-1, Tat, Dicysteine, Neuropathogenesis, Polymorphism
Introduction
The human immunodeficiency virus type 1 (HIV-1) transactivator of transcription (Tat) is a small basic protein usually with 101 amino acids (aa) and approximately of 14–16 kDa encoded from two separate exons (Williams et al. 2020). Tat is a key HIV regulatory protein that promotes viral replication, pathogenesis, and disease (Darbinian et al. 2008). HIV-1-infected cells, including the central nervous system (CNS) cells, actively secrete Tat protein (Ensoli et al. 1993; Nath 2002; Gurwitz et al. 2017). Tat causes excitotoxicity (Magnuson et al. 1995), which is mediated by direct interactions of the dicysteine region at positions 30 and 31 and crosslinks with cysteine moieties on the N-methyl-d-aspartate receptor (Li et al. 2008). It also plays a pivotal role in monocyte chemotaxis, which is mediated by both direct and indirect processes. Direct chemotaxis has been demonstrated in vitro using recombinant Tat protein and has been mapped to the Tat C30C31 dicysteine motif (Beall et al. 1996; Albini et al. 1998). Tat also induces the secretion of CCL2 from macrophages, thus amplifying the chemokine gradient, which helps recruit additional mononuclear phagocytes across the blood–brain barrier and into the brain (Conant et al. 1998; Weiss et al. 1999).
The aa variation within Tat protein alters its functional properties and, depending on the HIV-1 subtype, may produce Tat phenotypes, differing from virus representatives of each subtype (Spector et al. 2019). Key polymorphisms in Tat include a serine substitution at residue 31 (C31S) and residue 57 (R57S), and a glutamate substitution at residue 63 (Q63E), which have an effect on neurological outcomes and are more frequent in HIV-1C than in B (Williams et al. 2020; Rao et al. 2013; Ruiz et al. 2019). These residue changes indicate that Tat-C has a relatively higher-ordered structure and is less flexible than Tat-B (Siddappa et al. 2006).
HIV-1 is subject to several selective pressures, such as host immune responses, antiretroviral therapy (ART), HIV-1-encoded error-prone reverse transcriptase, and other selection mechanisms, compelling the virus to evolve with optimum replication and adaptation efficiency (Rhee et al. 2005; Konings et al. 2006; Canducci et al. 2009). These pressures cause the accumulation or depletion of specific mutations across the viral genome, leading to the development of large numbers of genetic variants or quasispecies within the patient (Dampier et al. 2016). Tat, encoded by the virus, is also susceptible to mutations and genetic variations within and between patients that can be observed in all HIV-1 subtypes globally (Roy et al. 2015a; Roy et al. 2015b).
Genetic analyses based on DNA polymerase (pol) or envelope (env) sequences of HIV-1 strains from different parts of Brazil show high diversity with a high prevalence of subtype C in Southern Brazil (Raboni et al. 2010), which is lower in the southeastern and northern regions (Guimarães et al. 2015).
This study was the first, to our knowledge, to investigate the genetic diversity and point mutations of HIV-1 Tat C in Brazil. The frequency of Tat polymorphism in the HIV-1C in the cysteine-rich (C30S or C31S) domain in India was 97% and in South Africa was 74% (Rao et al. 2013). We hypothesized that in Brazil, the frequency of the Tat polymorphism in HIV-1C is between this ranges.
The aim of this study was to identify the frequency of point mutations within the HIV-1C Tat protein, chiefly in the cysteine- (C30S or C31S), arginine- (R57S), and glutamine-rich (P60Q and Q63E) domains in Southern Brazil.
Methods
This study, which was a cross-sectional survey of stored cerebrospinal fluid (CSF) samples, was approved by the Hospital de Clínicas-Universidade Federal do Paraná (HC-UFPR, Curitiba, Paraná, Brazil) Institutional Review Board (IRB), the National Commission of Ethics in Research (CONEP, Brasilia, Brazil), and the University of California-San Diego (UCSD, San Diego, CA, USA) IRB. All participants signed consent forms approved by the IRBs in the USA and Brazil. Sequences from blood were obtained using a NIMH-funded protocol (R21 MH076651-01).
Study population
Blood samples from people with HIV (PWH) from Southern Brazil, n = 41, were randomly selected by convenience based on the subtype status from the cohort established in a previous study recruited from the HC-UFPR, Curitiba, state of Paraná, Southern Brazil from 2008 to 2011 (de Almeida et al. 2013). In this cohort, individuals with CNS opportunistic infections were excluded. All volunteers provided blood samples and underwent serological testing to confirm the HIV status before enrollment in accordance with previously published guidelines (Brasil 2018). HIV strains were genotyped by analyzing pol or env sequences. Genotyping indicated that 20 patients were infected with the HIV-1B subtype and 21 with the HIV-1C subtype (HIV-1C BR-study (HIV-1C BR-s)). The demographic, immunological, and virology characteristics of these participants are summarized in Table 1.
Table 1.
Demographic, clinical, and co-morbidities data of people with HIV (PWH) from Curitiba, Southern Brazil by subtype
| HIV-1B (n = 27) |
HIV-1C BR-s (n = 26) |
P | |
|---|---|---|---|
| N | 20 | 21 | |
| Age, years | 44 (36.0; 48.5) | 43 (35.5; 46.5) | 0.676 |
| Gender, n male (%) | 11 (55) | 7 (33.33) | 0.215 |
| Duration of infection, years | 7.26 (3.11; 10.86) | 7.26 (2.25; 10.97) | 0.990 |
| AIDS, n (%) | 17 (85) | 14 (66.67) | 0.277 |
| Current CD4, cell/mm3 | 502 (284; 633) | 372 (193; 518) | 0.235 |
| Nadir CD4, cell/mm3 | 63.5 (10.5; 221.5) | 200 (22; 376) | 0.141 |
| Log Plasma HIV RNA | 1.7 (1.7; 2.81) | 2.24 (1.7; 3.76) | 0.012 |
| Plasma HIV RNA <50 copies/mL, n (%) | 14 (70) | 8 (38) | 0.062 |
| on CART(a), n (%) | 17 (85) | 12 (57.14) | 0.086 |
| CPE(b) | 8 (6; 9) | 7.5 (6; 9) | 0.464 |
Data are presented as median [interquartile range (IQR)] or number of cases (%). HIV-1 subtypes: HIV-1B and HIV-1C (HIV-1C BR-s)
CART, combination anti-retroviral therapy
CPE, anti-retroviral CNS penetration effectiveness (Letendre et al. 2010)
Additionally, 344 HIV-1C sequences from the Los Alamos database, including 29 HIV-1C sequences from Brazil (HIV-1C BR-LA) and 315 from other countries in Africa and Asia where there is a high incidence of HIV-1C (South Africa, ZA, n = 59; Malawi, MW, n = 33; Zambia, ZM, n = 09; India, IN, n = 195; China, CN, n = 19), were studied. Sequences from the Los Alamos database were selected from the same period as that from the HIV-1C BR-s.
Sequence filtering
EDTA blood samples from the participants were collected at the time of enrollment to the study. Proviral DNA was isolated from the peripheral blood mononuclear cells (PBMCs) and separated on a Histopaque® 1077 (Sigma-Aldrich) gradient using the Purelink® Genomic DNA Mini Kit (Invitrogen, Thermo Fisher Scientific, USA) in accordance with the manufacturer’s protocol. We amplified the Tat exon 1 region (HXB2 position 5831–6045) by polymerase chain reaction (PCR) using the Platinum™ SuperFi™ DNA pol (Invitrogen, Thermo Fisher Scientific, USA) according to the manufacturer’s instructions. The primer pair TAT-1_OF (5-AAAGCCACCTYT GCCTAG-3)/TAT-1_OR (5-CTCATTGCCACTGTCTTCTGC-3) and TAT-1_IF (5-GTAGARGATMGATGGAACRA-3)/TAT-1_IR (5-CYCTAATTCTTTYAAYTAACC-3) was used for prenested and nested PCR, respectively (Paul et al. 2014). To purify the PCR products, Nucleosap® was used (Exo + SAP) (Molecular Biotecnologia e Representação Ltda©, Brazil). All PCR products were sequenced on both strands using a BigDye® Terminator 3.1 Cycle Sequencing Kit (Applied Biosystems, Foster City, CA, USA), which were analyzed on an Applied Biosystems® 3130 Genetic Analyzer (Applied Biosystems).
Bioinformatics analysis
Nucleotide sequences were assembled and edited using the DNASTAR Lasergene SeqMan software program (version 7.0; Dnastar, Inc., Madison, WI, USA).
HIV-1 Tat exon 1 reference sequence subtypes B and C datasets from Brazil were obtained from the Los Alamos database (https://www.hiv.lanl.gov/components/sequence/HIV/search/search.html) and aligned with the HIV-1 sequences from the study by ClustalW using MEGA7 software. Nucleotide sequences were translated into aa sequences, and mutations for each patient were identified.
Phylogenetic analysis
Phylogenetic tree construction was performed to characterize relatedness across the HIV sequences downloaded compared with the clinical cohort. Specifically, sequences were imported into the CLC Genomics Workbench v12.0 (https://www.qiagen.com/), and the “Create Alignment” tool was used under default conditions (i.e., alignment mode = very accurate) to generate a multiple sequence alignment. This alignment was then tested using four different procedures supported in the “Model testing” tool (hLRT = hierarchical likelihood ratio test, BIC = Bayesian information criterion, AIC = Akaike Information Criterion, AICc = Akaike Information Criterion corrected) to determine which substitution model (JC = Jukes-Cantor, F81 = Felsenstein 81, K80 = Kimura 80, HKY = Hasegawa-Kishino-Yano, GTR=General Time Reversible) would be the most suitable for the construction of a maximum likelihood tree. The tree was constructed under 100-round bootstrap conditions using the “Maximum Likelihood Phylogeny” tool in conjunction with the optimal substitution model testing indicated (GTR with gamma rate variation and topology variation) and drawn in “circular phylogram” mode. Sequences represented in the tree were grouped by relatedness and separately by type and used to construct multiple sequence alignments. Consensus sequences for each of these alignments were summarized using the WebLogo tool (https://weblogo.berkeley.edu/logo.cgi).
Statistical analysis
The results were presented as the median and interquartile range or the percentage, as appropriate. Comparisons between groups were made using the chi-squared test, Fisher’s exact test, or Mann–Whitney nonparametric test, where appropriate. Results were considered statistically significant at the 5% alpha level.
Results
Participants with HIV-1B and C subtypes were comparable with respect to age, gender, duration of infection, current and nadir CD4, frequency of AIDS cases, and number of participants on combination ART, anti-retroviral CNS penetration effectiveness (Letendre et al. 2010), and the number of participants with controlled HIV. However, the plasma HIV RNA was higher in HIV-1C than in HIV-1B (p = 0.012) (Table 1).
The exon 1 partial Tat sequences from Southern Brazil reported in this study were deposited in GenBank under accession numbers MT624066 to MT624106.
The maximum likelihood phylogenetic tree and WebLog of HIV Tat sequences from PBMC samples of HIV-1B and C from Southern Brazil (HIV-1C BR-s) and HIV-1C sequences from Brazil from Los Alamos (HIV-1C BR-LA) as well as HIV-1B reference sequences are shown in Fig. 1. The majority of HIV-1C BR-s and HIV-1C BR-LA sequences grouped together were distant from HIV-1B sequences. Three sequences from the HIV-1C BR-s group, which did not present the C31S substitution, were grouped among HIV-1C sequences. One sequence genotyped as HIV-1C by pol sequencing did not present the C31S substitution and grouped among HIV-1B, probably because this was a recombinant form. In the HIV-1B group, the C31S substitution on Tat in our sequences was found in two participants (10%). These sequences were genotyped as HIV-1B by env sequencing and were grouped among HIV-1C.
Fig. 1.
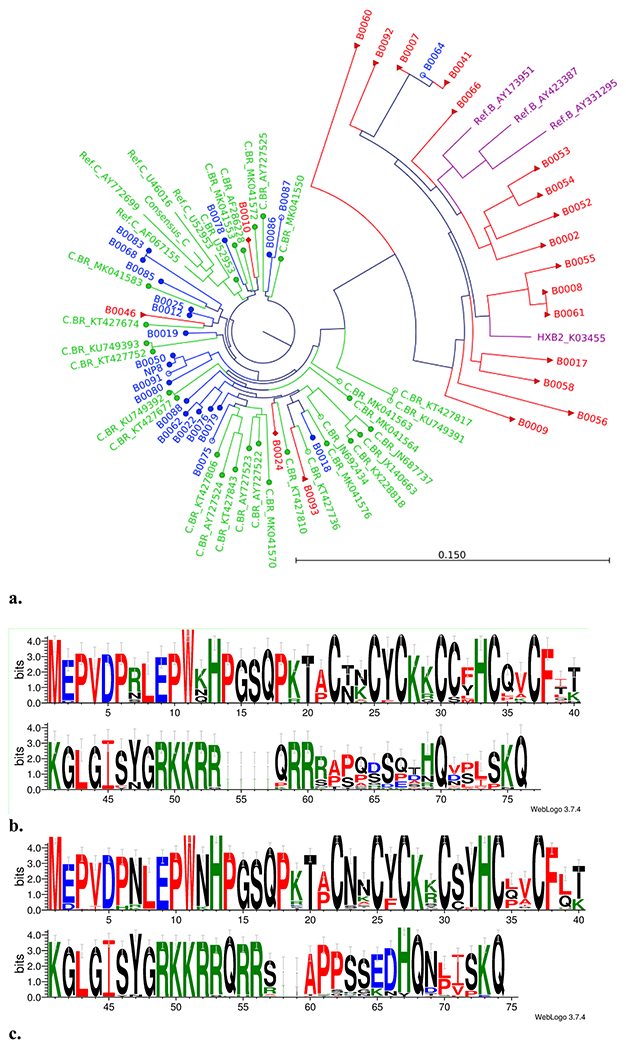
a The maximum likelihood phylogenetic tree analysis representing the similarities on the tat exon-1 gene of HIV-1B and HIV-1C from Southern Brazil. HIV-1B (red), HIV-1C from Curitiba, Southern Brazil (HIV-1C BR-s, blue), HIV-1C from Brazil downloaded from the Los Alamos database (HIV-1C BR-LA, green), and HIV-1B reference sequences (purple). The red full triangle HIV-1B indicates C31C (18); red full diamond HIV-1B indicates C31S mutation (n = 2); full blue circle HIV-1C indicates C31S mutation (n = 17); and empty blue circle HIV-1C indicates C31C (n = 4). Three sequences from the HIV-1C BR-s group that did not present the C31S substitution were grouped among HIV-1C sequences. One sequence initially genotyped as HIV-1C (by pol) did not present the C31S substitution and was grouped among HIV-1B, probably because this was a recombinant form. In the HIV-1B group, the C31S substitution on Tat in our sequences was found in two (10%), initially genotyped as HIV-1B (by env) and grouped among HIV-1C. b Amino acid sequence motif of HIV-1 subtype B represented as Weblogo. c Amino acid sequence motif of HIV-1 subtype C represented as Weblogo
Tat cysteine–rich domain
Overall, for the HIV-1 Tat C sequences studied in Brazil (HIV-1C BR-s and HIV-1C BR-LA), the frequency of C31S substitution was 41/50 (82%). The frequency of the C31S substitution in the HIV-1C BR-s sequences was 17/21 (80.95%) and among HIV-1C BR-LA was 24/29 (82.76%), p = 1.0 (Table 2; Fig. 2). Only two (10%) sequences in the HIV-1B group showed C31S substitution (p < 0.0001). No mutations were found in the Tat C30C residue in HIV-1C or HIV-1B from Brazil (Table 2; Fig. 2).
Table 2.
Frequency of HIV-1 Tat exon 1 variants in HIV-1C and HIV-1B
| Functional domain | Tat residue variant | HIV-1C BR-s |
HIV-1C BR-LA |
HIV-1C Total |
HIV-1B | p |
|---|---|---|---|---|---|---|
| N | 21 | 29 | 50 | 20 | ||
| N-terminal (1–21) | Q2E | 19 (90.48) | 23/28 (82.14) | 42 (84) | 19 (95) | 0.430 |
| Q2D | 2 (9.52) | 5/28 (17.86) | 7 (14) | 1 (5) | 0.422 | |
| P21A | 12 (57.14) | 14/28 (50) | 26 (52) | 14 (70) | 0.193 | |
| Cysteine rich (22–40) | T23N | 19 (90.48) | 27 (93.10) | 46 (92) | 9 (45) | <0.0001 |
| T23S | 1 (4.76) | 2 (6.90) | 3 (6) | 0 | - | |
| K24N | 12 (57.14) | 15 (51.72) | 27 (54) | 10 (50) | 0.796 | |
| K24A | 5 (23.81) | 0 | 5 (10) | 1 (5) | 0.666 | |
| Y26F | 8 (38.10) | 2 (6.90) | 10 (20) | 1 (5) | 0.160 | |
| H29Y | 3 (14.29) | 1(3.45) | 4 (8) | 0 | - | |
| H29K | 10 (47.62) | 11 (37.93) | 21 (42) | 15 (75) | 0.017 | |
| H29R | 8 (38.10) | 9 (31.03) | 17 (34) | 3 (15) | 0.148 | |
| H29S | 0 | 5 (17.24) | 5 (10) | 0 | - | |
| C31S | 17 (80.95) | 24 (82.76) | 41 (82) | 2 (10) | <0.0001 | |
| W32Y | 19 (90.48) | 29 (100) | 48 (96) | 6 (30) | <0.0001 | |
| W32F | 1 (4.76) | 0 | 1 (2) | 10 (50) | <0.0001 | |
| Q35L | 7 (33.33) | 10 (34.48) | 17 (34) | 2 (10) | 0.072 | |
| Q35P | 7 (33.33) | 7 (24.14) | 14 (28) | 2 (10) | 0.127 | |
| Q39T | 0 | 0 | 0 | 6 (30) | - | |
| Q39I | 1 (4.76) | 0 | 1 (2) | 7 (35) | 0.0004 | |
| Q39L | 13 (61.90) | 17 (58.62) | 30 (60) | 3 (15) | 0.0011 | |
| T40K | 5 (23.81) | 7 (24.14) | 12 (24) | 5 (25) | 1.0 | |
| Arginine rich (49–57) | ||||||
| R57S | 14 (66.67) | 23 (79.31) | 37 (74) | 4 (20) | <0.0001 | |
| Glutamine rich (58–72) | P60Q | 2 (9.52) | 1/28 (3.57) | 3 (6.12) | 11 (55) | <0.0001 |
| P60S | 0 | 4 (14.29) | 4 (8.16) | 0 | - | |
| S62D | 1 (4.76) | 0 | 1 (2) | 2 (10) | 0.194 | |
| S62G | 2 (9.52) | 4 (13.79) | 6 (12) | 0 | - | |
| S62N | 0 | 3 (10.34) | 3 (6) | 0 | - | |
| Q63E | 18 (85.71) | 22 (75.86) | 40 (80) | 4 (20) | <0.0001 | |
| T64A | 0 | 0 | 3 (15) | - | ||
| T64D | 17 (80.95) | 27 (93.10) | 44 (88) | 5 (25) | <0.0001 | |
| T64S | 0 | 0 | 3 (15) | - | ||
| T64N | 3 (14.29) | 2 (6.90) | 5 (10) | 1 (5) | 0.666 | |
| S68P | 9 (42.86) | 14 (48.28) | 23 (46) | 8 (40) | 0.791 | |
| S68L | 10 (47.62) | 14 (48.28) | 24 (48) | 3 (15) | 0.014 |
HIV-1C BR-s, sequences from the Brazil study; HIV-1C BR-LA, database sequences from Brazil downloaded from Los Alamos. In italics, mutations described previously
Fig. 2.
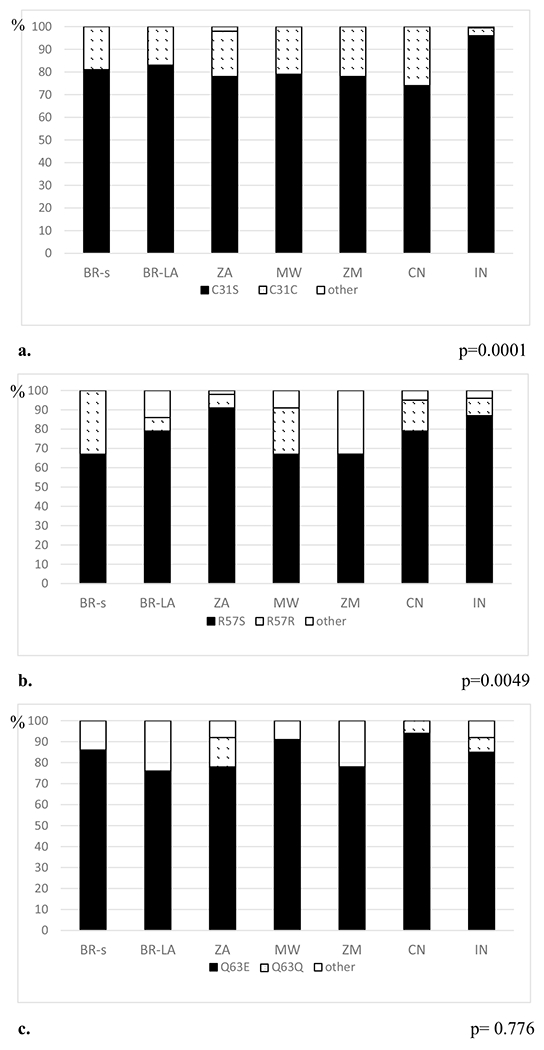
The frequency of C31S, R57S, and Q63E substitutions on HIV-1 Tat C in Brazil and other countries in Africa and Asia. a The frequency of C31S substitution. b The frequency of R57S substitution. c The frequency of Q63E substitution. Brazil sequences from the study (HIV-1C BR-s) and Brazil database sequences from Los Alamos (HIV-1C BR-LA), South Africa (ZA), Malawi (MW), Zambia (ZM), India (IN), and China (CN). Sequences from the Los Alamos database were selected in the same period as that of the cases from the Brazil study (BR-s) from 2008 to 2011
Tat arginine–rich domain
The frequency of the R57S substitution among overall HIV-1C sequences from Brazil was 37 (74%) and among HIV-1B was 4 (20%), p = 0.004 (Table 2; Fig. 2).
For one participant (B0056), there was a mirror-type insertion in the Tat arginine–rich domain between 53 and 54 (53KKRR54). This participant presented with immunological failure despite HIV virological suppression.
Tat glutamine–rich domain
The mutation P60Q was more frequent in HIV-1B (n = 11; 55%) than in HIV-1C (n = 3; 6.12%), p < 0.0001. However, glutamate substitution at residue 63 (Q63E) was more frequent in HIV-1C [40 (80%)] than in HIV-1B [4 (20%)], p < 0.0001 (Table 2), which is in accordance with the results of a previous study (Williams et al. 2020).
The frequencies of C31S, R57S, and Q63E substitutions on HIV-1 Tat C in Brazil and other countries in Africa, Asia, and Europe are shown in Fig. 2. The maximum likelihood phylogenetic tree analysis representing the similarities in the Tat exon-1 gene of HIV-1C from Brazil and other countries in Africa, Asia, and Europe is presented in Supplemental Fig. 1. The nodes of the phylogenetic tree show that most samples from Brazil shared an internal node with most samples from India.
Novel mutations in Tat functional domains
The frequency of HIV-1 Tat exon-1 point mutations by functional domains in HIV-1C and HIV-1B is shown in Table 2. Several novel mutations in the Tat functional domains were identified. Tat mutations were more frequent in HIV-1C than in HIV-1B, T23N: HIV-1C, 46 (92%) vs. HIV-1B, 9 (45%), p < 0.0001; W32Y: HIV-1C, 48 (96%) vs. HIV-1B, 6 (30%), p < 0.0001; Q39L: HIV-1C, 30 (60%) vs. HIV-1B, 3 (15%), p = 0.0011; T64D: HIV-1C, 44 (88%) vs. HIV-1B, 5 (25%), p < 0.0001; S68L: HIV-1C, 24 (48%) vs. HIV-1B, 3 (15%), p = 0.014. Tat mutations were more frequent in HIV-1B than in HIV-1C, H29K: HIV-1C, 21 (42%) vs. HIV-1B, 15 (75%), p = 0.017; W32F: HIV-1C, 1 (2%) vs. HIV-1B, 10 (50%), p < 0.0001; Q39I: HIV-1C, 1 (2%) vs. HIV-1B, 7 (35%), p = 0.0004.
For the dendrogram for the Tat exon-1 gene from Southern Brazil, seven groups of related sequences were selected, and the aa sequence motif is represented as WebLogo for each group (Fig. 3).
Fig. 3.
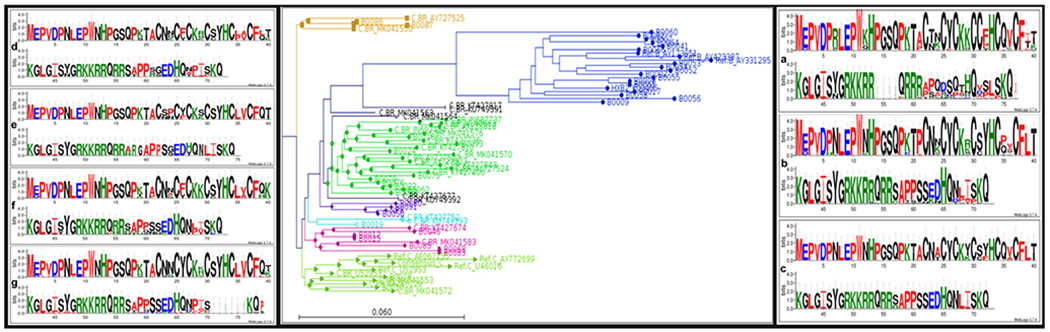
Dendrogram for the Tat exon-1 gene from Brazil. Seven groups of related sequences were selected, and the amino acid sequence motif is represented as WebLogo for each group. Group A (blue) encompasses HIV-1B and only one HIV-1C (B0064), group B (green), C (purple), D (gold), E (cyan), F (pink), and G (lime). The sequences shown in black are sequences which do not group well within any other group of sequences and are not sufficient in number to represent a separate group
Discussion
The frequency of C31S substitution in the cysteine-rich domain in HIV-1C isolates from Brazil in this study was similar to those found previously in Southern African countries, although lower than that in India. In accordance with the features of Tat sequences from South Africa and Zambia, where the occurrence of variants with an intact C30C31 motif is 26% and 20%, respectively, this variant was rare among sequences from India and Bangladesh (3% and 2%, respectively) (Rao et al. 2013). This is in accordance with the origin of HIV-1C in Southern Brazil from a single founder strain closely related to subtype C strains from Burundi (Bello et al. 2008). Previous studies examining HIV-1 env sequences from Northeast India have concluded that Indian HIV-1C has evolved into a distinct sub-clade termed CIN (Shankarappa et al. 2001; Neogi et al. 2012). The Tat gene in the HIV-1C isolates circulating in Southern African countries (South Africa, Zambia, and Botswana) is evolutionarily distinct from those in Southeast Asia (India and Bangladesh), where HIV-1C isolates display a higher proportion (almost 99%) of variants encoding Tat protein with C30S or C31S mutations than that of those in Southern Africa (Rao et al. 2013).
Our results reinforce the findings of previous studies that not all HIV-1C isolates display a C30S31 polymorphism in the Tat protein encoded by them, displaying C30C31 motif, and a small proportion, approximately 1–10%, of HIV-1B displays C30S31 polymorphism (Rao et al. 2013).
In this study, the number of different residues with mutations was higher in HIV-1C than in HIV-1B, which is in accordance with the results of previous studies (Spector et al. 2019; Ruiz et al. 2019; Williams et al. 2020). There are several key protein sequence differences that may account for the altered neuropathogenesis. These include a serine substitution at residue 31 (C31S) and 57 (R57S) as well as a glutamate substitution at residue 63 (Q63E). These polymorphisms in Tat-C account for more potent transactivation capacity and lower levels of monocyte recruitment, neuroinflammation, and neuronal damage both in vitro and in animals (Williams et al. 2020). Tat-C, with an intact dicysteine motif (C30C31), is more neurotoxic to human neuronal cultures than Tat-C lacking this motif (Li et al. 2008; Rao et al. 2013). HIV-1B has a greater neurovirulence but is a less potent transactivator (Williams et al. 2020).
The results of this study support the findings of our previous studies comparing PWH infected by HIV-1 subtypes C and B from the same geographic region (Southern Brazil). These studies showed that HIV-1 subtype C is as neurotropic as subtype B and is able to develop CNS HIV genetic compartmentalization (de Almeida et al. 2017, 2018), despite the findings of the present study showing the occurrence of the C31S mutation in the cysteine-rich domain in the majority of patients with HIV-1C. These findings are contrary to those of previous reports that HIV-1 subtype C is less neuropathogenic than subtype B due to the existence of the C31S mutation (Satishchandra et al. 2000) as well as findings of other experimental studies (Park et al. 2001; Ranga et al. 2004). Moreover, we found similar frequencies of HIV-associated neurocognitive disorder (de Almeida et al. 2013) as well as similar stimulation of ß-chemokines, including CCL2, and inflammatory biomarkers in the CSF of PWH-1C and PWH-1B (de Almeida et al. 2016a, b). HIV-1C and HIV-1B are also comparable in CSF white blood cells (WBC), CSF HIV RNA, and blood–CSF barrier dysfunction (de Almeida et al. 2020). HIV discordance or escape in the CNS occurred at a comparable frequency for PWH-1C and PWH-1B (de Almeida et al. 2020). These findings were corroborated by those of other authors, who also described similar neurovirulence between HIV-1B and HIV-1C (Ortega et al. 2013; Witten et al. 2015; Paul et al. 2017).
Other mutations already described were present in HIV-1 Tat from Southern Brazil in the arginine-rich domain (R57S), more frequently in HIV-1C than in HIV-1B (Spector et al. 2019). This mutation was related to lower inflammation-induced dysfunction in human neuron-astrocyte cultures (Ruiz et al. 2019). The natural polymorphism found in Tat-C (R57S) may be an additional contributor to the lowered inflammatory response. This polymorphism may be responsible for the reduced uptake of Tat by bystander cells and subsequent reduced neuroinflammation in cell cultures. This emphasizes the importance of this polymorphism in Tat uptake and neuroinflammation (Ruiz et al. 2019). We identified a participant with an insertion in the arginine-rich domain (53KKRR54). The arginine-rich domain contains a well-conserved sequence, 49RKKRRQRRR57, which is crucial for the interaction with transactivation response element (TAR) as well as with the secretion and uptake of Tat (Rayne et al. 2010; Hauber et al. 1989; Li et al. 2012). Clinically, this participant showed poor CD4 cell reconstitution despite a good virological response, a phenomenon that has also been correlated with a lower nadir pretreatment CD4 + cell count (Prabhakar et al. 2011). We were not able to find any description of this insertion in the arginine-rich domain in any other previous studies.
In the glutamine-rich domain, the mutation P60Q was more frequent in HIV-1B than in HIV-1C, which corroborates the findings of previous studies (Ronsard et al. 2014). Residues in this position are essential for transactivation through TAR interaction and are also reported to be involved in T cell apoptosis (Ronsard et al. 2014). The transcriptional capacity may be affected by residue polymorphisms present in Tat variants from different HIV-1 subtypes, although there are discordant results among studies (Williams et al. 2020). The Q63E mutation, which is more frequent in Tat-C than in Tat-B, contributes to greater transcriptional activation in human CD4 T cells (Kurosu et al. 2002). The effective binding of Tat to TAR allows HIV to achieve high-level transcription. Sequence variation between Tat-B and Tat-C affects the transactivation capacity, immune modulation, neuronal damage, and pathways related to neuronal damage (Williams et al. 2020).
In addition to the key protein sequence mutations described previously, there are several other Tat functional variants in various functional domains of the Tat protein (Ronsard et al. 2014; Spector et al. 2019). However, the majority of these mutations were not present in Tat-C and Tat-B in the sequences of this study. On the other hand, we identified several other novel Tat mutations not described previously, some of which had significantly different prevalence between HIV-1C and HIV-1B subtypes. The majority of these mutations occurred in the cysteine-rich domain, which is considered highly conserved among different isolates of HIV-1 studies (Williams et al. 2020). Future studies are important to determine whether the mutations are of functional significance.
The Tat first exon contains aa residues 1–72, and the second exon has 73–101 aa residues. HIV Tat has five functional domains as follows: The N-terminal, proline-rich region (aa residues 1–21) has mutations that alter the acidic composition and were originally believed to affect transactivation (Rappaport et al. 1989). The cysteine-rich domain (aa residues 22–40) has seven well-conserved cysteine residues at positions 22, 25, 27, 30, 34, and 37 studies (Williams et al. 2020). Individual mutations in six of the seven cysteine residues abolished Tat function (Koken et al. 1994).
The core domain (aa residues 38–48) contains an RKGLGI motif that is conserved among HIV-1, HIV-2, and SIV Tat. This region, in conjunction with the amino terminus and cysteine domain, has been suggested to circumscribe the minimal activation domain of HIV-1 Tat (Carroll et al. 1991; Derse et al. 1991; Williams et al. 2020). The arginine-rich domain (aa residues 49–57) contains a basic RKKRRQRRR motif. These properties confer TAR RNA-binding properties to Tat (Dingwall et al. 1989; Weeks et al. 1991; Chang et al. 1992) and are important for nuclear localization of the protein (Hauber et al. 1989; Ruben et al. 1989). However, this short basic stretch may be insufficient to determine the entire specificity of Tat-TAR binding, as aa outside of the basic domain also contribute to this interaction Williams et al. 2020). The glutamine-rich domain (aa residues 58–72) is considered the region with the highest rate of sequence variation Williams et al. 2020). The glutamine- and arginine-rich domains, which are referred to as the basic region, are responsible for nuclear localization and mediate binding to CCATT enhancer-binding protein (Siddappa et al. 2006; Spector et al. 2019). The region spanning aa 1–21 is remarkably tolerant of changes. In contrast, changes in aa 22–40 are generally deleterious for transactivation. Although the basic domain (aa residues 49–57) as a unit is necessary for Tat function, individual aa changes do not significantly affect activity (Spector et al. 2019).
We conclude that the frequency of C31S substitution on HIV-1 Tat C in Brazil is high and similar to those found in Africa, although lower than those in India. Novel Tat mutations have been described in all Tat functional domains. The Tat-B and Tat-C sequences found in Southern Brazil are consistent with biological differences and with potential effects on the neuropathophysiology and clinical outcomes. Additional studies are necessary to explore these possibilities.
Supplementary Material
Funding
CFAR (International Pilot Grant P30 AI036214 and CFAR Visiting Researcher Grant PTHMON7) and NIMH (R21 MH076651-01).
Footnotes
Supplementary Information The online version contains supplementary material available at https://doi.org/10.1007/s13365-020-00935-z.
Competing interests The authors declare that they have no competing interests.
References
- Albini A, Benelli R, Giunciuglio D, Cai T, Mariani G, Ferrini S, Noonan DM (1998) Identification of a novel domain of HIV tat involved in monocyte chemotaxis. J Biol Chem 273:15895–15900 [DOI] [PubMed] [Google Scholar]
- Beall CJ, Mahajan S, Kuhn DE, Kolattukudy PE (1996) Site-directed mutagenesis of monocyte chemoattractant protein-1 identifies two regions of the polypeptide essential for biological activity. Biochem J 313:633–640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bello G, Passaes CP, Guimarães ML, Lorete RS, Matos Almeida SE, Medeiros RM, Alencastro PR, Morgado MG (2008) Origin and evolutionary history of HIV-1 subtype C in Brazil. AIDS 22:1993–2000 [DOI] [PubMed] [Google Scholar]
- Brasil, Ministério da Saúde (2018) Programa Nacional de DST/AIDS. http://www.aids.gov.br/assistencia/manualdst/item12.html
- Canducci F, Marinozzi MC, Sampaolo M, Berrè S, Bagnarelli P, Degano M, Gallotta G, Mazzi B, Lemey P, Burioni R, Clementi M (2009) Dynamic features of the selective pressure on the human immunodeficiency virus type 1 (HIV-1) gp120 CD4-binding site in a group of long term non progressor (LTNP) subjects. Retrovirology 6:4. 10.1186/1742-4690-6-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll R, Martarano L, Derse D (1991) Identification of lentivirus Tat functional domains through generation of equine infectious anemia virus/human immunodeficiency virus type 1 tat gene chimera. J Virol 65:3460–3467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang YN, Jeang KT (1992) The basic RNA-binding domain of HIV-2 Tat contributes to preferential trans-activation of a TAR2-containing LTR. Nucleic Acids Res 20:5465–5472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conant K, Garzino-Demo A, Nath A, McArthur JC, Halliday W, Power C, Gallo RC, Major EO (1998) Induction of monocyte chemoattractant protein-1 in HIV-1 Tat-stimulated astrocytes and elevation in AIDS dementia. Proc Natl Acad Sci USA 95:3117–3121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dampier W, Nonnemacher MR, Mell J, Earl J, Ehrlich GD, Pirrone V, Aiamkitsumrit B, Zhong W, Kercher K, Passic S, Williams JW, Jacobson JM, Wigdahl B (2016) HIV-1 genetic variation resulting in the development of new quasispecies continues to be encountered in the peripheral blood of well-suppressed patients. PLoS One 11:e0155382. 10.1371/journal.pone.01553-82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darbinian N, Darbinyan A, Czernik M, Peruzzi F, Khalili K, Reiss K, Gordon J, Amini S (2008) HIV-1 Tat inhibits NGF-induced Egr-1 transcriptional activity and consequent p35 expression in neural cells. J Cell Physiol 216:128–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Almeida SM, Ribeiro CE, de Pereira AP, Badiee J, Cherner M, Smith D, Maich I, Raboni SM, Rotta I, Barbosa FJ, Heaton RK, Umlauf A, Ellis RJ (2013) Neurocognitive impairment in HIV-1 subtype C versus B-infected individuals in Southern Brazil. J Neurovirol 19:550–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Almeida SM, Rotta I, Ribeiro CE, Oliveira MF, Chaillon A, de Pereira AP, Cunha AP, Zonta M, Bents JF, Raboni SM, Smith D, Letendre S, Ellis RJ (2017) Dynamic of CSF and serum biomarkers in HIV-1 subtype C encephalitis with CNS genetic compartmentalization: case study. J Neurovirol 23:460–473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Almeida SM, Oliveira MF, Chaillon A, Rotta I, Ribeiro CE, de Pereira AP, Smith D, Letendre S, Ellis RJ (2018) Transient and asymptomatic meningitis in human immunodeficiency virus-1 subtype C: a case study of genetic compartmentalization and biomarker dynamics. J Neurovirol 24:786–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Almeida SM, Rotta I, Ribeiro CE, Smith D, Wang R, Judicello J, Potter M, Vaida F, Letendre S, Ellis RJ (2016) Blood-CSF barrier and compartmentalization of CNS cellular immune response in HIV infection. J Neuroimmunol 301:41–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Almeida SM, Rotta I, Jiang Y, Li X, Raboni SM, Ribeiro CE, Smith D, Potter M, Vaida F, Letendre S, Ellis RJ (2016) Biomarkers of chemotaxis and inflammation in cerebrospinal fluid and serum in individuals with HIV-1 subtype C versus B. J Neurovirol 22:715–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Almeida SM, Rotta I, de Pereira AP, Tang B, Umlauf A, Ribeiro CEL, Letendre S, Ellis RJ (2020) Cerebrospinal fluid pleocytosis as a predictive factor for CSF and plasma HIV RNA discordance and escape. J Neurovirol 26:241–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derse D, Carvalho M, Carroll R, Peterlin BM (1991) A minimal lentivirus tat. J Virol 65:7012–7015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dingwall C, Ernberg I, Gait MJ, Green SM, Heaphy S, Karn J, Lowe AD, Singh M, Skinner MA, Valerio R (1989) Human immunodeficiency virus 1 tat protein binds trans-activation responsive region (TAR) RNA in vitro. Proc Natl Acad Sci USA 86:6925–6929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ensoli B, Buonaguro L, Barillari G, Fiorelli V, Gendelman R, Morgan RA, Wingfield P (1993) Gallo RC (1993) Release, uptake and effects of extracellular human immunodeficiency virus type 1 Tat protein on cell growth and viral transactivation. J Virol 67:277–287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guimarães ML, Marques BCL, Bertoni N, Teixeira SLM, Morgado MG (2015) Assessing the HIV-1 epidemic in Brazilian drug users: a molecular epidemiology approach. PLoS ONE 10:e0141372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurwitz KT, Burman RJ, Murugan BD, Garnett S, Ganief T, Soares NC, Raimondo JV, Blackburn JM (2017) Time-dependent, HIV-Tat-induced perturbation of human neurons in vitro: towards a model for the molecular pathology of HIV-associated neurocognitive disorders. Front Mol Neurosci 10:163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauber J, Malim MH, Cullen BR (1989) Mutational analysis of the conserved basic domain of human immunodeficiency virus tat protein. J Virol 63:1181–1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koken SE, Greijer AE, Verhoef K, van Wamel J, Bukrinskaya AG, Berkhout B (1994) Intracellular analysis of in vitro modified HIV tat protein. J Biol Chem 269:8366–8375 [PubMed] [Google Scholar]
- Konings FA, Burda ST, Urbanski MM, Zhong P, Nadas A, Nyambi PN (2006) Human immunodeficiency virus type 1 (HIV-1) circulating recombinant form 02_AG (CRF02_AG) has a higher in vitro replicative capacity than its parental subtypes A and G. J Med Virol 78:523–534 [DOI] [PubMed] [Google Scholar]
- Kurosu T, Mukai T, Komoto S, Ibrahim MS, Li YG, Kobayashi T, Tsuji S, Ikuta K (2002) Human immunodeficiency virus type 1 subtype C exhibits higher transactivation activity of tat than subtypes B and E. Microbiol Immunol 46:787–799 [DOI] [PubMed] [Google Scholar]
- Letendre SL, FitzSimons C, Ellis RJ, Clifford D, Collier AC, Gelman B, Marra C, McArthur J, McCutchan JA, Morgello S, Simpson D, Vaida F, Heaton R, Grant I (2010) Correlates of CSF Viral Loads in 1,221 volunteers of the CHARTER cohort. 17th Conference on Retroviruses and Opportunistic Infections [Google Scholar]
- Li W, Huang Y, Reid R, Steiner J, Malpica-Llanos T, Darden TA, Shankar SK, Mahadevan A, Satishchandra P, Nath A (2008) NMDA receptor activation by HIV-Tat protein is clade dependent. J Neurosci 28:12190–12198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li GH, Li W, Mumper RJ, Nath A (2012) Molecular mechanisms in the dramatic enhancement of HIV-1 Tat transduction by cationic liposomes. FASEB J 26:2824–2834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnuson DSK, Knudsen BE, Geiger JD, Brownstone RM, Nath A (1995) Human immunodeficiency virus type 1 tat activates non-N-methyl-D-aspartate excitatory amino acid receptors and causes neurotoxicity. Ann Neurol 37:373–380 [DOI] [PubMed] [Google Scholar]
- Nath A (2002) Human immunodeficiency virus (HIV) proteins in neuropathogenesis of HIV dementia. J Infect Dis 186:S193–S198 [DOI] [PubMed] [Google Scholar]
- Neogi U, Bontell I, Shet A, De Costa A, Gupta S, Diwan V, Laishram RS, Wanchu A, Ranga U, Banerjea AC, Sönnerborg A (2012) Molecular epidemiology of HIV-1 subtypes in India: origin and evolutionary history of the predominant subtype C. PLoS ONE 7:e39819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortega M, Heaps JM, Joska J, Vaida F, Seedat S, Stein DJ, Paul R, Ances BM (2013) HIV clades B and C are associated with reduced brain volumetrics. J Neurovirol 19:479–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park IW, Wang JF, Groopman JE (2001) HIV-1 Tat promotes monocyte chemoattractant protein-1 secretion followed by transmigration of monocytes. Blood 97:352–358 [DOI] [PubMed] [Google Scholar]
- Paul RH, Joska JA, Woods C, Seedat S, Engelbrecht S, Hoare J, Heaps J, Valcour V, Ances B, Baker LM, Salminen LE, Stein DJ (2014) Impact of the HIV Tat C30C31S dicysteine substitution on neuropsychological function in patients with clade C disease. J Neurovirol 20:627–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul RH, Phillips S, Hoare J, Laidlaw DH, Cabeen R, Olbricht GR, Su Y, Stein DJ, Engelbrecht S, Seedat S, Salminen LE (2017) Neuroimaging abnormalities in clade C HIV are independent of Tat genetic diversity. J Neurovirol 23:319–328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prabhakar B, Banu A, Pavithra HB, Chandrashekhara P, Sasthri S (2011) Immunological failure despite virological suppression in HIV seropositive individuals on antiretroviral therapy. Indian J Sex Transm Dis AIDS 32:94–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raboni SM, Almeida SM, Rotta I, Ribeiro CEL, Rosario D, Vidal LR, Nogueira MB, Riedel M, Winhescki MG, Ferreira KA, Ellis R (2010) Molecular epidemiology of HIV-1 clades in Southern Brazil. Mem Inst Oswaldo Cruz 105:1044–1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranga U, Shankarappa R, Siddappa NB, Ramakrishna L, Nagendran R, Mahalingam M, Mahadevan A, Jayasuryan N, Satishchandra P, Shankar SK, Prasad VR (2004) Tat protein of human immunodeficiency virus type 1 subtype C strains is a defective chemokine. J Virol 78:2586–2590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao VR, Neogi U, Talboom JS, Padilla L, Rahman M, Fritz-French C, Gonzalez-Ramirez S, Verma A, Wood C, Ruprecht RM, Ranga U, Azim T, Joska J, Eugenin E, Shet A, Bimonte-Nelson H, Tyor WR, Prasad VR (2013) Clade C HIV-1 isolates circulating in Southern Africa exhibit a greater frequency of dicysteine motif-containing Tat variants than those in Southeast Asia and cause increased neurovirulence. Retrovirology 10:61. 10.1186/1742-4690-10-61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappaport J, Lee SJ, Khalili K, Wong-Staal F (1989) The acidic amino-terminal region of HIV-1 tat protein constitutes an essential activating domain. New Biologist 1:101–110 [PubMed] [Google Scholar]
- Rayne F, Debaisieux S, Yezid H, Lin YL, Mettling C, Konate K, Chazal N, Arold ST, Pugnière M, Sanchez F, Bonhoure A, Briant L, Loret E, Roy C, Beaumelle B (2010) Phosphatidylinositol-(4,5)-bisphosphate enables efficient secretion of HIV-1 Tat by infected T-cells. EMBO J 29:1348–1362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee SY, Fessel WJ, Zolopa AR, Hurley L, Liu T, Taylor J, Nguyen DP, Slome S, Klein D, Horberg M, Flamm J, Follansbee S, Schapiro JM, Shafer RW (2005) HIV-1 Protease and reverse-transcriptase mutations: correlations with antiretroviral therapy in subtype B isolates and implications for drug resistance surveillance. J Infect Dis 192:456–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronsard L, Lata S, Singh J, Ramachandran VG, Das S, Banerjea AC (2014) Molecular and genetic characterization of natural HIV-1 Tat exon-1 variants from North India and their functional implications. PLoS ONE 9:e85452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy CN, Khandaker I, Oshitani H (2015) Intersubtype genetic variation of HIV-1 Tat exon 1. AIDS Res Hum Retroviruses 31:641–648 [DOI] [PubMed] [Google Scholar]
- Roy CN, Khandaker I, Oshitani H (2015b) Evolutionary dynamics of Tat in HIV-1 subtypes B and C. PLoS One 10:e0129896. 10.1371/journal.pone.01298-96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruben S, Perkins A, Purcell R, Joung K, Sia R, Burghoff R, Haseltine WA, Rosen CA (1989) Structural and functional characterization of human immunodeficiency virus tat protein. J Virol 63:1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz AP, Ajasin DO, Ramasamy S, DesMarais V, Eugenin EA, Prasad VR (2019) A naturally occurring polymorphism in the HIV-1 tat basic domain inhibits uptake by bystander cells and leads to reduced neuroinflammation. Sci Rep 9:3308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satishchandra P, Nalini A, Gourie-Devi M, Khanna N, Santosh V, Ravi V, Desai A, Chandramuki A, Jayakumar PN, Shankar SK (2000) Profile of neurologic disorders associated with HIV/AIDS from Bangalore, south India (1989–96). Indian J Med Res 111:14–23 [PubMed] [Google Scholar]
- Shankarappa R, Chatterjee R, Learn GH, Neogi D, Ding M, Roy P, Ghosh A, Kingsley L, Harrison L, Mullins JI, Gupta P (2001) Human immunodeficiency virus type 1 env sequences from Calcutta in eastern India: identification of features that distinguish subtype C sequences in India from other subtype C sequences. J Virol 75:10479–10487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddappa NB, Venkatramanan M, Venkatesh P, Janki MV, Jayasuryan N, Desai A, Ravi V, Ranga U (2006) Transactivation and signaling functions of Tat are not correlated: biological and immunological characterization of HIV-1 subtype-C Tat protein. Retrovirology 3:53. 10.1186/1742-4690-3-53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spector C, Mele AR, Wigdahl B, Nonnemacher MR (2019) Genetic variation and function of the HIV-1 Tat protein. Med Microbiol Immunol 208:131–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weeks KM, Crothers DM (1991) RNA recognition by Tat-derived peptides: interaction in the major groove? Cell 66:577–588 [DOI] [PubMed] [Google Scholar]
- Weiss JM, Nath A, Major EO, Berman JW (1999) HIV-1 Tat induces monocyte chemoattractant protein-1-mediated monocyte transmigration across a model of the human blood-brain barrier and up-regulates CCR5 expression on human monocytes. J Immunol 163:2953–2959 [PubMed] [Google Scholar]
- Williams ME, Zulu SS, Stein DJ, Joska JA, Naudé PJW (2020) Signatures of HIV-1 subtype B and C Tat proteins and their effects in the neuropathogenesis of HIV-associated neurocognitive impairments. Neurobiology of Disease 136:104701. [DOI] [PubMed] [Google Scholar]
- Witten JA, Thomas KG, Westgarth-Taylor J, Joska JA (2015) Executive dyscontrol of learning and memory: findings from a Clade C HIV-positive South African sample. The Clinical Neuropsychologist 29:956–984 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


