Abstract
Carex agglomerata C. B. Clarke is a sedge with excellent ornamental characters, it is an important ecosystem stabilizer. Here we report the complete chloroplast genome of C. agglomerata to provide a foundation for further phylogenetic studies on the Cyperaceae. The chloroplast (cp) genome is 184,157 bp in size and consists of a large single-copy (LSC) region 106,654 bp in length, a small single-copy (SSC) region of 36,099 bp, two inverted repeats (IR) regions each 20,702 bp. The total GC content of the cp genome is 33.9% with the LSC, SSC, and IR regions 32, 32.5, and 42.9%, respectively. The cp genome contains 128 genes, including 80 protein-coding, 40 tRNA, and eight rRNA genes. The phylogenetic analysis showed C. agglomerata is in a clade with Carex neurocarpa Maxim and Carex siderosticta Hance. This study provides a basis for further phylogenetic studies of Carex.
Keywords: Carex agglomerata, chloroplast genome phylogenetic analysis sequencing
The genus Carex is an important ecological taxon and is widely distributed in diverse environments ranging from the tundra, dry sand prairies, wetlands, and forests (Gadallah and Jefferies 1995). Species classified in Carex also play important ecological roles and provide food for animals (Gadallah and Jefferies 1995; Shrestha et al. 2012). In addition, some species have medicinal or nutritional properties (Roy et al. 2012). Many sedges have been widely used in gardens and new varieties have been bred due to their excellent ornamental properties (Darke 2007). Species classified to this genus are difficult to distinguish because of the morphological variability. Molecular-level evidence is thus required in order to distinguish species and provide an accurate systematic classification and evolutionary history. One of these species, Carex agglomerate, exhibits excellent ornamental features and is distributed in forests on mountain slopes and wet habitats in valleys at altitudes 1200–3000 m in Gansu, Qinghai, Shaanxi, and Sichuan (Dai et al. 2010). This species is also cultivated as ornamental grass and is used in gardens. Genetic studies of C. agglomerata are limited. SSR markers (Liu et al. 2021) showed a sister relationship between C. agglomerata and Carex dimorpholepis Steud. However, the complete chloroplast genome C. agglomerata has yet to be deciphered. Chloroplast genomes are highly conserved in sequence and structure and are of great importance in species identification and evolutionary studies (Zeng et al. 2017). This study determined the complete genome of C. agglomerata to contribute to the systematics and evolutionary history of the Cyperaceae.
Leaf tissue of C. agglomerata was collected from Ningshan County, Shaanxi province, China (108°29′40″E, 33°28′29.71″N, 2162 m). The voucher specimen was deposited at XBGH [the Herbarium of Xi’an Botanical Garden (HDT022, http://www.xazwy.com/, Bin Li, lbwj@163.com)]. Total genomic DNA was extracted using the CTAB method (Doyle 1987) and sequenced on Illumina Hi-Seq 2500 platform. In total, 3.16 Gb of raw data were obtained (NCBI Sequence Read Archive accession number SRR13414282). The raw reads were filtered using default settings in Trimmomatic v.0.33 (Bolger et al. 2014). The program NOVOPlasty (Dierckxsens et al. 2017) was used to de novo assemble the chloroplast genome. The program PGA (Qu et al. 2019) and Geneious v 11.0.2 (Kearse et al. 2012) performed the annotation followed by manual adjustments. The cp genome sequence was submitted to NCBI GenBank under accession number MT795185.
The cp genome of C. agglomerata is 184,157 bp in length and has a typical quadripartite structure, containing two IR regions 20,702 bp each, separated by an LSC region that is 106,654 bp and an SSC region of 36,099 bp. The total GC content is 33.9%, and the corresponding values of the LSC, SSC, and IR region are 32, 32.5, and 42.9%, respectively. This chloroplast genome has 128 functional genes, including 80 protein-coding (PCGs), 40 tRNA, and eight rRNA. The IR regions have four protein-coding genes (ndhB, rpl2, rps12, and rps7), five tRNA genes, and four rRNA genes. Sixteen genes (atpF, ndhA, ndhB, petB, petD, rpl2, rpl16, rpoC1, rps12, rps16, trnA, trnG, trnI, trnK, trnL, and trnV) contain one intron, and ycf3 has two introns.
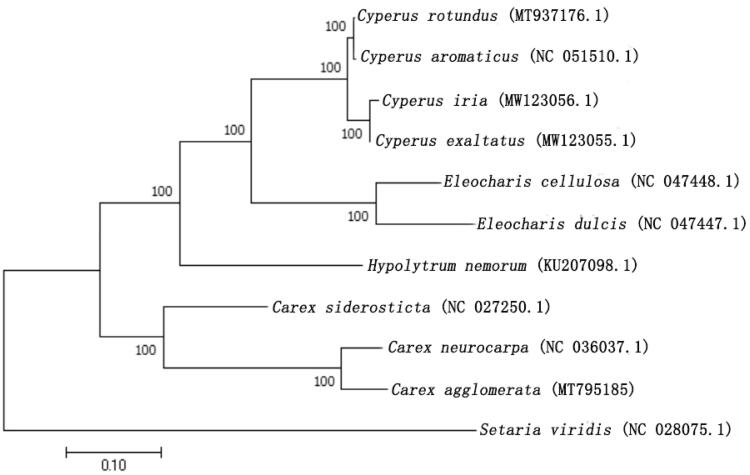
Ten other cp genomes of Cyperaceae and Poaceae from GenBank were used to determine the phylogenetic location of C. agglomerata in Cyperaceae, designating Setaria viridis as the out-group (Figure 1). The complete chloroplast sequences were aligned using MEGA 7.0 (Kumar et al. 2016) and the phylogenetic analysis was performed using the maximum likelihood (ML) method with 1000 bootstrap replicates was built by this program. The data showed that C. agglomerata was in a clade with Carex neurocarpa and Carex siderosticta. These findings are consistent with the results of SSR markers (Liu et al. 2021). The data are useful for future investigations of chloroplast genome evolution and phylogenetic research in the genus Carex.
Figure 1.
The phylogenetic tree was constructed with chloroplast genome sequences of 11 species within the Cyperaceae and Poaceae family and Setaria viridis as an outgroup by MEGA 7.0.
Acknowledgments
Thanks to Dr. Chun Su for her contribution to data analyses.
Funding Statement
This work was supported by Key Research and Development Program of Shaanxi [Program No. 2020NY-084], Fourth National Survey of Chinese Medicine Resources in 2018 [Caishe [2018] No.43], and Agricultural technology research and development project of Xi'an Science and Technology Bureau [20NYYF0056], and Science and Technology Program of Shaanxi Academy of Sciences [2020K-13].
Disclosure statement
The authors report no potential conflict of interest.
Data availability statement
The data that support the findings of this study are available in NCBI GenBank (https://www.ncbi.nlm.nih.gov) with accession number MT795185. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA691435, SRR13414282, and SAMN17255546, respectively.
References
- Bolger AM, Lohse M, Usadel B.. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30(15):2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai L, Songyun L, Shuren Z, Yancheng T, Koyama T, Tucker GC, Simpson DA, Noltie HJ, Strong MT, Bruhl JJ, et al. 2010. Flora of China, Vol. 23. Beijing; St. Louis (MO): Science Press; Botanical Garden Press; p. 379. [Google Scholar]
- Darke R. 2007. The encyclopedia of grasses for livable landscapes. Portland (OR); London: Timber Press. [Google Scholar]
- Dierckxsens N, Mardulyn P, Smits G.. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle JJ. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15. [Google Scholar]
- Gadallah FL, Jefferies RL.. 1995. Comparison of the nutrient contents of the principal forage plants utilized by lesser snow geese on summer breeding grounds. J Appl Ecol. 32(2):263–275. [Google Scholar]
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Tamura K.. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu LY, Fan XF, Tan PH, Wu JY, Zhang H, Han C, Chen C, Xun LL, Guo WE, Chang ZH, et al. 2021. The development of SSR markers based on RNA-sequencing and its validation between and within Carex L. species. BMC Plant Biol. 21(1):17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu XJ, Moore MJ, Li DZ, Yi TS.. 2019. PGA: a software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Meth. 15:50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy B, Giri BR, Mitali C, Swargiary A.. 2012. Ultrastructural and biochemical alterations in rats exposed to crude extract of Carex baccans and Potentilla fulgens. Microsc Microanal. 18(5):1067–1076. [DOI] [PubMed] [Google Scholar]
- Shrestha B, Kindlmann P, Jnawali SR.. 2012. Interactions between the Himalayan Tahr, livestock and snow leopards in the Sagarmantha National Park. In: Kindlmann P, editor. Himalayan biodiversity in the changing world. Dordrecht: Springer; p. 157–176. [Google Scholar]
- Zeng SY, Zhou T, Han K, Yang Y, Zhao JH, Liu ZL.. 2017. The complete chloroplast genome sequences of six Rehmannia species. Genes. 8(3):103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available in NCBI GenBank (https://www.ncbi.nlm.nih.gov) with accession number MT795185. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA691435, SRR13414282, and SAMN17255546, respectively.