Abstract
Background
An optimised standard experimental setup across different hospitals is urgently needed to ensure consistency in nucleic acid test results for SARS-CoV-2 detection. A standard comparison across different nucleic acid tests and their optimal experimental setups is not present. We assessed the performance of three common nucleic acid tests, namely digital PCR (dPCR), quantitative PCR (qPCR), and loop-mediated isothermal amplification (LAMP), to detect SARS-CoV-2 in clinical settings.
Methods
In this systematic review and meta-analysis we compared sensitivity and specificity of qPCR, dPCR, and LAMP and their performances when different experimental setups (namely specimen type used, use of RNA extraction, primer–probe sets, and RNA extraction methods) are applied. We searched PubMed, BioRxiv, MedRxiv, SciFinder, and ScienceDirect for studies and preprints published between Feb 29 and Dec 15, 2020. Included dPCR, qPCR, and LAMP studies using any type of human specimens should report the number of true-positive, true-negative, false-positive, and false-negative cases with Emergency Use Authorization (EUA)-approved PCR assays as the comparator. Studies with a sample size of less than ten, descriptive studies, case studies, reviews, and duplicated studies were excluded. Pooled sensitivity and specificity were computed from the true and false positive and negative cases using Reitsma's bivariate random-effects and bivariate latent class models. Test performance reported in area under the curve (AUC) of the three nucleic acid tests was further compared by pooling studies with similar experimental setups (eg, tests that used RNA extracted pharyngeal swabs but with either the open reading frame 1ab or the N primer). Heterogeneity was assessed and reported in I2 and τ2.
Findings
Our search identified 1277 studies of which we included 66 studies (11 dPCR, 32 qPCR, and 23 LAMP) with 15 017 clinical samples in total in our systematic review and 52 studies in our meta-analysis. dPCR had the highest pooled diagnostic sensitivity (94·1%, 95% CI 88·9–96·6, by Reitsma's model and 95·8%, 54·9–100·0, by latent class model), followed by qPCR (92·7%, 88·3–95·6, and 93·4%, 60·9–99·9) and LAMP (83·3%, 76·9–88·2, and 86·2%, 20·7–99·9), using EUA-approved PCR kits as the reference standard. LAMP was the most specific with a pooled estimate of 96·3% (93·8–97·8) by Reitsma's model and 94·3% (49·1–100·0) by latent class model, followed by qPCR (92·9%, 87·2–96·2, and 93·1%, 47·1–100·0) and dPCR (78·5%, 57·4–90·8, and 73·8%, 0·9–100·0). The overall heterogeneity was I2 0·5% (τ2 2·79) for dPCR studies, 0% (4·60) for qPCR studies, and 0% (3·96) for LAMP studies. AUCs of the three nucleic acid tests were the highest and differed the least between tests (ie, AUC>0·98 for all tests) when performed with RNA extracted pharyngeal swabs using SARS-CoV-2 open reading frame 1ab primer.
Interpretation
All three nucleic acid tests consistently perform better with pharyngeal swabs using SARS-CoV-2 open reading frame 1ab primer with RNA extraction. dPCR was shown to be the most sensitive, followed by qPCR and LAMP. However, their accuracy does not differ significantly. Instead, accuracy depends on specific experimental conditions, implying that more efforts should be directed to optimising the experimental setups for the nucleic acid tests. Hence, our results could be a reference for optimising and establishing a standard nucleic acid test protocol that is applicable in laboratories worldwide.
Funding
University Grants Committee and The Chinese University of Hong Kong.
Introduction
As of Sept 21, 2021, COVID-19 has caused more than 4·6 million deaths.1 Since there is no clinically approved interventional therapy currently available to curb this health crisis, identifying as many infected individuals as possible (both symptomatic and asymptomatic) and isolating them is the most effective way to prevent disease transmission. To this end, nucleic acid and antigen tests are diagnostic methods currently used to screen potential SARS-CoV-2 carriers. Nucleic acid tests are favoured in clinical tests over antigen tests due to their higher sensitivity and specificity.2 Quantitative PCR (qPCR) is the most widely used diagnostic method in humans.3 However, the loop-mediated isothermal amplification (LAMP) assay has emerged as a popular alternative to qPCR in many international airports, hospitals, and testing centres worldwide.4 Digital PCR (dPCR) is a novel technology developed to facilitate the diagnosis of COVID-19 that allows absolute quantification of nucleic acids in hospitals and diagnostic centres. Although less common than qPCR and LAMP, dPCR is growing in popularity because of its high sensitivity.5
Research in context.
Evidence before this study
Quantitative PCR (qPCR), digital PCR (dPCR), and loop-mediated isothermal amplification (LAMP) are nucleic acid amplification tests used for the diagnosis of COVID-19 in hospitals and clinics. qPCR remains the most trusted test type in hospitals. LAMP has been increasingly used in SARS-CoV-2 detection, especially in rapid testing environments such as airports and testing centres worldwide. dPCR has been tested in hospitals in Wuhan, China, and other countries with reported high diagnostic sensitivity and accuracy. The three tests have been evaluated individually in cohort and trial studies using clinical samples. Yet, these studies are not comparable across different assay types, and their experimental setups and performances differ greatly. Studies that compared the three nucleic acid tests but did not account for the experimental variations result in markedly varying conclusions on the accuracy of the nucleic acid tests. Therefore, a comprehensive review that systematically compares all common nucleic acid tests and accounts for different experimental setups is of great importance and urgency.
Added value of this study
The choice of which diagnostic test to use and the optimal conditions to perform the tests to achieve the best diagnostic accuracy and consistency are crucial to effective disease control and prevention. A standardised experimental setup across different hospitals is urgently needed to ensure accuracy and consistency in nucleic acid test results for COVID-19. We performed a systematic review and meta-analysis using bivariate random-effects models on dPCR, qPCR, and LAMP. The results of our analysis, with the largest number of samples (n=15 017) collected from hospitals and clinics and included in a study of this type, showed that dPCR is the most sensitive, followed by qPCR and LAMP. Given its superior sensitivity, dPCR will add value to physicians in diagnosing and reporting COVID-19 cases. We also concluded that the accuracy of nucleic acid-based detection of COVID-19 is similar across different testing types when we controlled for experimental variations. We further showed that assay accuracy is more strongly dependent on the specific experimental setup than on the test type. Our findings, therefore, provide new evidence—to achieve the best performance, optimising the experimental setup is more important than the choice of the test type.
Implications of all the available evidence
As the three nucleic acid tests are similarly accurate under specified conditions, the choice of the gold standard need not be restricted to qPCR, as has been used in almost all studies. LAMP and dPCR could be reliable reference standards for test evaluation and LAMP has similar accuracy to the PCR methods. Considering the scarce medical resources available in low-income and middle-income countries or at a point-of-care settings, LAMP could be a preferable option for the detection of SARS-CoV-2 infection due to its simpler instrumentation. In high-income countries, dPCR should be valued for its exceeding sensitivity and high reproducibility (as it does not require calibration and provides absolute quantification). Our study suggests that the test type can be selected according to the environmental and socioeconomic requirements, but test results will remain consistent if the optimal experimental setup is implemented and standardised.
There are several difficulties in achieving robust and consistent nucleic acid test results from current COVID-19 clinical diagnostic methods. Experimental setups, such as specimen type and primer–probe sets used, can impact test accuracy, which ranges from 47% to 100% in various PCR studies.6, 7, 8, 9, 10, 11, 12, 13, 14, 15 LAMP also differs in reported accuracy (61–100%) depending on the viral content of the sample and reference test used.12, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33 This large variation in reported accuracies of PCR and LAMP is due to the absence of a standardised measure for evaluating diagnostic test accuracy and the experimental setups that vary across studies. These challenges have been reported to hamper diagnostics as a crucial preventive measure.34 A systematic review collating nucleic acid test results could provide a firm conclusion to the most sensitive test while taking different experimental conditions into account.
However, no such literature review has been conducted. Even though many studies have assessed the accuracy of the PCR and LAMP methods separately, their experimental setups differ greatly, leading to test result variations and disagreement in conclusions on which nucleic acid test is the most accurate.6, 7, 8, 9, 10, 11, 13, 14, 15, 17, 20, 21, 24, 25, 26, 28, 29, 35, 36, 37, 38, 39 For example, of the three studies that previously reviewed diagnostic tests, including chest CT, antigen tests, isothermal amplification, and qPCR, a small number of trials and limited subgroups were analysed with large experimental variations.4, 40, 41
We conducted a systematic review and meta-analysis to evaluate the accuracy of dPCR, qPCR, and LAMP by comparing their sensitivities, specificities, diagnostic odd ratios (ORs), and areas under the curve (AUCs). In our meta-analysis we aimed to identify the optimal conditions for these tests in terms of the use of RNA extraction (with extraction or direct), RNA extraction methods (magnetic beads, spin column, or automatic), primer sequence (open reading frame 1ab [ORF1ab] or nucleocapsid [N]), and type of human specimen (pharyngeal, saliva, or sputum). Our study aims to provide a reference to optimise assay conditions to formulate a more standardised protocol across different laboratories while maintaining fairness and confidence in test results for comparisons.
Methods
Search strategy and selection criteria
For this systematic review and meta-analysis, we searched PubMed, BioRxiv, MedRxiv, SciFinder, and ScienceDirect, for studies published between Feb 29 and Dec 15, 2020, and assessed identified diagnostic (observational) studies on dPCR, qPCR, and LAMP by their titles or abstracts under prespecified criteria. Studies should be done in the context of SARS-CoV-2 and use human specimens. They should report either the number of true and false positive and negative cases or sensitivity and specificity (expressed as percentages or decimal numbers). No language restrictions were applied. Studies with a sample size of less than ten, descriptive studies, case studies, and reviews were excluded due to small study size and unquantifiable information. Duplicated studies were removed manually to ensure no overlapping results between studies for meta-analysis. Only studies in which the exact true-positive, false-positive, false-negative, and true-negative cases were provided or could be calculated from the number of patients with or without SARS-CoV-2 infection were selected for the meta-analysis. Selected studies in the meta-analysis should use a uniform reference standard. Keywords used for study repository searches were {[“SARS-CoV-2”OR“COVID-19”OR“covid”] AND [(“quantitative PCR”OR “qPCR”) OR (“LAMP” OR “RT-LAMP”) OR (“digital droplet PCR” OR “dPCR”)] AND “diagnosis”}OR{[“SARS-CoV-2” OR “COVID-19” OR “covid”] AND [(“quantitative PCR” OR “qPCR”) OR (“LAMP”OR“RT-LAMP”) OR (“digital droplet PCR” OR “dPCR”)]}. The same keywords were used in all search tools.
WYA and PPHC screened and selected studies independently based on the criteria described above between Dec 20, 2020 and Jan 9, 2021. WYA extracted and recorded the data from the selected studies. Conflicts were resolved by reaching a consensus between WYA and PPHC. This study followed the Preferred Reporting Items for a Systematic Review and Meta-analysis of Diagnostic Test Accuracy.
Data analysis
Data were extracted from the included studies either for individual samples or summary outcomes for all samples combined. Variables extracted from the studies were the number of true positive, false positive, false negative, and true negative, as well as the target genes, specimen types, RNA extraction methods, patient types (COVID-19 patients, convalescent individuals, or healthy individuals), experimental controls, measurement of the concentration of genetic sequences in the mixture, and choice of the toolkit. We obtained true false positive and negative case numbers from the studies' supplementary documents if they were not reported in the main text. In studies that reported only sensitivity and specificity, case numbers were calculated from the number of patients with or without infection. The main outcomes of our study were pooled sensitivity and specificity. Considering that the reference standard might induce bias in the main outcomes, we first considered two types of reference standard: laboratory-developed tests and Emergency Use Authorization (EUA)-approved PCR kits. We then compared the pooled sensitivity and specificity of the two types of reference standard using Reitsma's bivariate random-effects model42 versus using Bayesian latent class bivariate model.43 Bayesian latent class bivariate model is a model that accounts for imperfect references standard and corrects between-study heterogeneity in sensitivity and specificity. We used as a gold standard for the rest of the meta-analysis the type of reference standard that resulted in more consistent results. Pooled sensitivity and specificity were calculated from true-positive, false-positive, false-negative, and true-negative cases with 0·5 continuity correction for zero events. Suspected or single gene-positive cases were eliminated in the calculation to avoid ambiguous results.
We also did primary and secondary subgroup analyses. In the primary subgroup analysis, we reported sensitivity and specificity of the test of interest using Reitsma's model and the latent class model with studies pooled by specimen type used, use of RNA extraction, primer–probe sets, and RNA extraction methods. Additionally, diagnostic ORs and AUCs were estimated to present sensitivity and specificity as single measures. Computation of diagnostic ORs and heterogeneity indices followed the DerSimonian-Laird method.44 AUCs based on sensitivity and false-positive rate (1–specificity) were computed using the hierarchical summary receiver operator curves method.45 Differences in AUC between two subgroups (studies pooled by specimen type used, use of RNA extraction, primer–probe sets, and RNA extraction methods) were calculated and considered significant if the corresponding bootstrap p value (estimated with the dmetatools package in R) was less than 0·05. Sensitivities, specificities, diagnostic ORs, and AUCs were reported with 95% CIs. As the latent class model assumes the absence of a gold standard, pooled sensitivities and specificities of the test of interest and of the reference test were estimated independently. Our study mainly considered pooled estimates of the tests of interest. For inclusion in the primary subgroup analyses, subgroups must contain at least two studies. For any subgroup consisting of studies fewer than four but more than one, univariate random-effects models were used to estimate accuracy parameters due to non-convergence in bivariate models with a small sample size.
Secondary subgroup analysis pooled only homogeneous studies (ie, studies using the same specimen type and primer–probe set with RNA extraction). Sensitivity, specificity, diagnostic OR, and AUC for at least four homogeneous studies were estimated.
Heterogeneity between studies for each test was assessed by calculating I2 and τ2 according to the Cochrane Handbook for Systematic Reviews.46 Heterogeneity affecting test performance of each test was visualised in the galaxy plots.47 Multivariate small study effect test (MSSET) with p<0·1 indicated a strong small study effect caused by publication bias.48
We consolidated our conclusions on the analytical sensitivities of the three nucleic acid tests by compiling a boxplot that showed the distribution of detection limits as calculated by synthetic viral RNA. Means, medians, and IQR of the distribution of detection limits for each nucleic acid test were calculated. We used R version 4.0.3 (mada, xmeta, rjags, and dmetatool packages) for computation of accuracy parameters and generation of plots.
WYA did a quality assessment following prespecified signalling questions in QUADAS-2 on Review Manager 5.4.1. Studies with uncertain results were re-assessed by PPHC.
Role of the funding source
The funder of the study had no role in study design, data collection, data analysis, data interpretation, or writing of the report.
Results
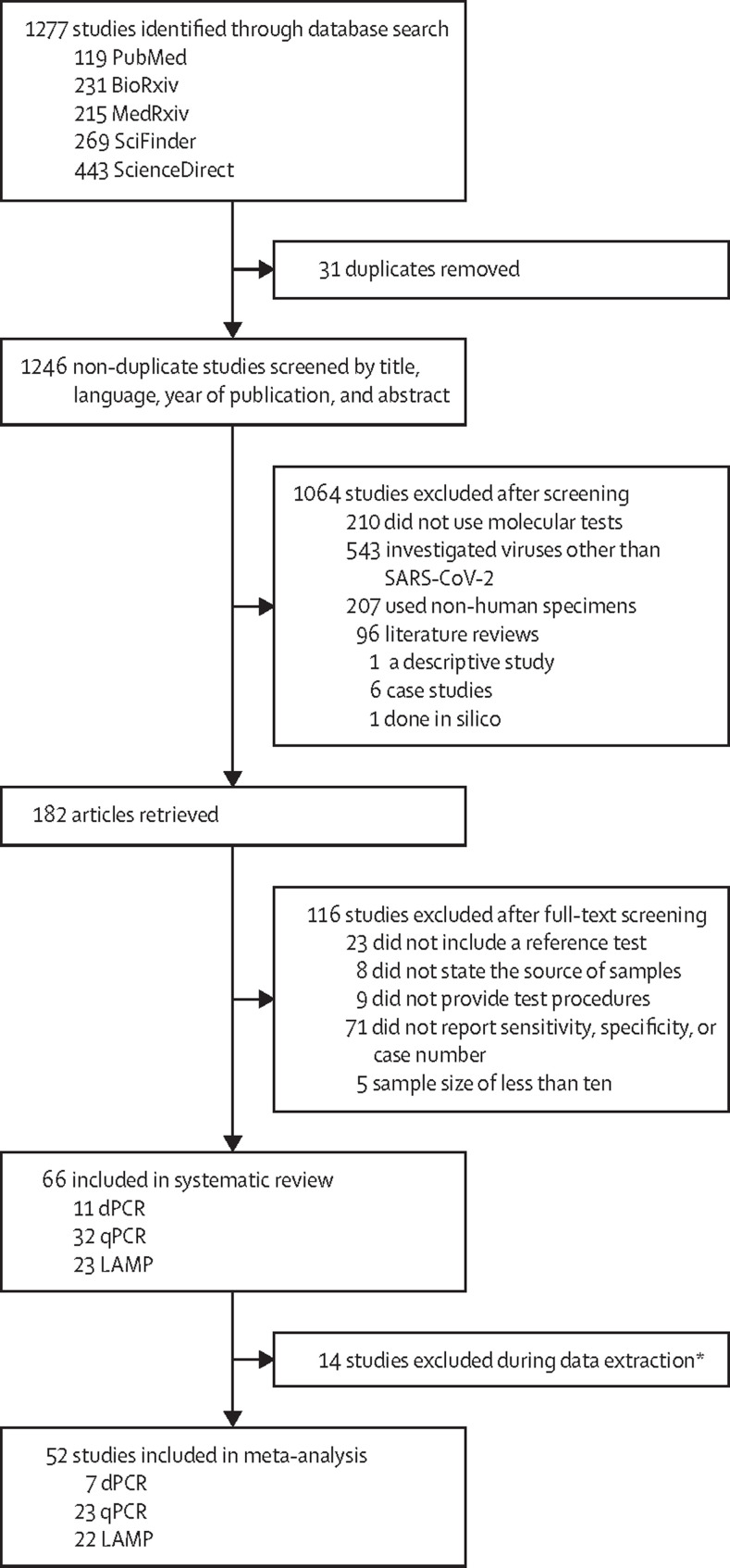
We identified 1277 studies on PubMed, BioRxiv, MedRxiv, SciFinder, and ScienceDirect. 31 duplicate studies were manually removed and 1064 studies were excluded at screening stage because they did not meet the inclusion criteria (figure 1 ). Of the remaining 182 studies, 116 were excluded due to insufficient information on test accuracy, small sample size, or lack of reference methods, sample source, or test procedures. 66 studies were selected for inclusion in our systematic review, of which 11 were on dPCR, 32 on qPCR, and 23 on LAMP (figure 1).5, 6, 7, 8, 9, 10, 11, 13, 14, 15, 17, 20, 21, 26, 28, 29, 35, 36, 37, 38, 39, 49, 50, 51, 52, 53, 54, 55, 56, 57, 58, 59, 60, 61, 62, 63, 64, 65, 66, 67, 68, 69, 70, 71, 72, 73, 74, 75, 76, 77, 78, 79, 80, 81 A total of 15 017 clinical samples were collected from outpatients, hospitalised patients, close contacts, and convalescents. A summary of the study characteristics of the 66 articles is shown in table 1 , with further details in the appendix (pp 2–18). 52 of 66 studies reported true and false positive and negative numbers (dPCR [n=7], qPCR [n=23], and LAMP [n=22]) and were included in the meta-analysis. 37 studies (dPCR [n=6], qPCR [n=19], and LAMP [n=12]) using EUA-approved PCR assays as the reference standard were selected for the computation of pooled overall and subgroup estimates. Experiments within studies in the subgroup analyses were considered individually, yielding 71 experiments (dPCR [n=15], qPCR [n=25], and LAMP [n=31]). We identified four subgroups for each nucleic acid test: specimen, primer–probe set, use of RNA extraction (with or without RNA extraction), and RNA extraction methods (magnetic beads, spin column, or automatic). A list of index tests and reference tests for all individual experiments is provided in the appendix (pp 39–40).
Figure 1.
Preferred Reporting Items for Systematic reviews and Meta-Analyses flow diagram
dPCR=digital PCR. LAMP=loop-mediated isothermal amplification. qPCR=quantitative PCR. *Reported sensitivity and specificity in percentages but did not provide true false-positive and true false-negative case numbers, nor the number of patients with or without infection.
Table 1.
Summary table of study characteristics of the 66 studies
| Total number of studies (%) |
Index test |
||||
|---|---|---|---|---|---|
| dPCR (%) | qPCR (%) | LAMP (%) | |||
| Overall | 66 (100%) | 11 (17%) | 32 (48%) | 23 (37%) | |
| Reference test | |||||
| EUA-approved PCR | 37 (56%) | 6 (9%) | 19 (29%) | 12 (18%) | |
| Laboratory-developed tests | 29 (44%) | 5 (8%) | 13 (20%) | 11 (17%) | |
| Specimen types | |||||
| Swabs (nasopharyngeal and oropharyngeal) | 54 (82%) | 10 (15%) | 27 (41%) | 17 (26%) | |
| Saliva | 16 (24%) | 3 (5%) | 11 (17%) | 2 (3%) | |
| Sputum | 11 (17%) | 4 (6%) | 4 (6%) | 3 (5%) | |
| Other | 7 (11%) | 2 (3%) | 3 (5%) | 2 (3%) | |
| Primer–probe sets | |||||
| ORF1ab | 18 (27%) | 6 (9%) | 4 (6%) | 8 (12%) | |
| N | 47 (71%) | 9 (14%) | 23 (35%) | 15 (23%) | |
| E | 20 (30%) | 2 (3%) | 14 (21%) | 4 (6%) | |
| RdRP | 15 (23%) | 2 (3%) | 9 (14%) | 4 (6%) | |
| Other | 19 (29%) | 0 (0%) | 4 (6%) | 15 (23%) | |
| Use of RNA extraction | |||||
| With RNA extraction | 51 (77%) | 11 (17%) | 22 (33%) | 18 (27%) | |
| Without RNA extraction | 12 (18%) | 0 (0%) | 6 (9%) | 6 (9%) | |
| RNA extraction method | |||||
| Magnetic beads | 16 (24%) | 2 (3%) | 10 (15%) | 4 (6%) | |
| Silica spin column | 12 (18%) | 3 (5%) | 5 (8%) | 4 (6%) | |
| Automatic | 6 (9%) | 2 (3%) | 3 (5%) | 1 (2%) | |
Data are n (%); all percentages were calculated by dividing the number of studies in each category by 66 studies. A given study can straddle two or more study characteristics. A summary of the content of the 66 studies is shown in the appendix (pp 2–18). dPCR=digital PCR. qPCR=quantitative PCR. LAMP=loop-mediated isothermal amplification. EUA=Emergency Use Authorization. ORF1ab=open reading frame 1ab. N=nucleocapsid. E=envelope. RdRP=RNA-dependent RNA polymerase.
Of the 52 studies selected for the meta-analysis, we identified 15 studies that used laboratory-developed tests and 37 that used EUA-approved PCR assays as the comparator in the evaluation of dPCR, qPCR, and LAMP performances. Using the Reitsma's bivariate random-effects model that assumed the presence of a perfect gold standard, we found that laboratory-developed tests studies generally yielded higher average AUCs (95·8% for EUA-approved PCR assays and 97·3% for laboratory-developed tests) and heterogeneity I 2 (0% for EUA-approved PCR assays and 14% for laboratory-developed tests) for the nucleic acid test studies, suggesting a higher variance among the laboratory-developed tests studies and possible overestimation of the accuracy parameters. With the Bayesian latent class bivariate model, which accounted for an imperfect gold standard, we found that studies using EUA-approved PCR assays as reference gave consistent results. Based on this comparison between the two models and estimates of heterogeneity, we decided to use only studies that used EUA-approved PCR assays to compute pool indices.
With EUA-approved PCR assays as the common reference standard, the overall sensitivity and specificity of dPCR, qPCR, and LAMP using Reitsma's bivariate random-effects model were consistent with those using the Bayesian latent class bivariate model. dPCR was shown to be the most sensitive, and LAMP was the most specific by both models (table 2 ). The distribution of sensitivity and specificity of the individual 71 studies were presented in three forest plots for each nucleic acid test (appendix pp 41–43). In general, LAMP was the least diagnostically sensitive, whereas dPCR was the most. The overall heterogeneity was relatively low with I 2 0·5% (τ2 2·79) for dPCR, 0% (4·60) for qPCR, and 0% (3·96) for LAMP studies. This was visualised by the galaxy plots where most studies clustered close to the summary point, suggesting low between-study heterogeneity (appendix p 44). In addition, MSSET showed some degree of small study effect for LAMP studies (p=0·0002) but no effect for dPCR (p=0·14) and qPCR studies (p=0·21).
Table 2.
LAMP, dPCR, and qPCR overall sensitivities and specificities
| LAMP | dPCR | qPCR | |
|---|---|---|---|
| Sensitivity | |||
| Reitsma | 83·3% (76·9–88·2) | 94·1% (88·9–96·9) | 92·7% (88·3–95·6) |
| Latent | 86·2% (20·7–99·9) | 95·8% (54·9–100·0) | 93·4% (60·9–99·9) |
| Specificity | |||
| Reitsma | 96·3% (93·8–97·8) | 78·5% (57·4–90·8) | 92·9% (87·2–96·2) |
| Latent | 94·3% (49·1–100·0) | 73·8% (0·9–100·0) | 93·1% (47·1–100·0) |
Data are % (95% CI). Sensitivities and specificities for each molecular test were estimated using Reitsma's bivariate random-effects model (Reitsma) and Bayesian latent class bivariate model (latent). LAMP=loop-mediated isothermal amplification. dPCR=digital PCR. qPCR=quantitative PCR.
We were able to pool results from at least two studies for each of the four categories: specimen, primer-probe set, use of RNA extraction, and RNA extraction methods. Using Reitsma's bivariate models, we found that both PCR and LAMP assays using pharyngeal swabs were more sensitive compared with the same assays using saliva (table 3 ). We were able to further stratify the LAMP studies by the type of pharyngeal swabs, and both yielded 87% sensitivity. Assays without RNA extraction resulted in lower sensitivity and specificity for qPCR and LAMP. All RNA samples in the dPCR studies were purified, so comparisons were not made. Using the latent class model, all nucleic acid tests using pharyngeal swabs were more sensitive and specific than the same assays using saliva (appendix pp 18–20). LAMP assays with nasopharyngeal and oropharyngeal swabs were similar in sensitivity (84–88%) and specificity (94–95%; appendix pp 18–20). In the significance test for each pair of subgroups (eg, pharyngeal swabs vs saliva), we observed that the AUC of pharyngeal dPCR differed significantly from saliva dPCR (p=0·032) and pharyngeal LAMP differed significantly from saliva LAMP (p=0·017). The AUC of dPCR also differed significantly between ORF1ab and N primers (p=0·045), as did the AUC of qPCR assays using magnetic beads versus spin column as the RNA extraction methods (p=0·031; appendix pp 21–22).
Table 3.
Pooled sensitivity and specificity using Reitsma's random-effects model, pooled diagnostic OR, AUC, and heterogeneity for primary subgroup analysis
| Number of studies (sample size) | Pooled sensitivity (95% CI) | Pooled specificity (95% CI) | Pooled diagnostic OR (95% CI) | AUC | I2(τ2) | ||
|---|---|---|---|---|---|---|---|
| LAMP | |||||||
| Overall | 31 (3453) | 83·3% (76·9–88·2) | 96·3% (93·8–97·8) | 188·37 (84·10–421·91) | 0·963 | 0% (3·96) | |
| Specimen | |||||||
| Nasopharyngeal swabs | 10 (1004) | 87·7% (78·3–93·4) | 92·4% (85·6–96·2) | 107·46 (29·47–391·83) | 0·955 | 19% (3·43) | |
| Oropharyngeal or throat swabs | 11 (1046) | 87·6% (77·9–93·4) | 91·3% (84·9–95·1) | 99·10 (30·17–325·49) | 0·952 | 21% (3·09) | |
| Saliva | 5 (773) | 86·2% (75·5–92·6) | 89·4% (83·0–93·5) | 68·15 (22·631–205·25) | 0·939 | 0·3% (2·72) | |
| Sputum | 7 (513) | 81·7% (76·3–86·1) | 98·6% (96·1–99·5) | 397·60 (127·59–1238·96) | 0·949 | 0% (0·0) | |
| RNA extraction | |||||||
| With RNA extraction | 17 (2445) | 88·0% (82·1–92·2) | 95·1% (90·9–97·4) | 288·67 (88·63–940·17) | 0·961 | 4·14% (4·72) | |
| Without RNA extraction | 14 (1008) | 73·6% (62·4–82·5) | 97·0% (94·1–98·5) | 106·50 (37·60–301·69) | 0·962 | 0% (2·13) | |
| Primer–probe set | |||||||
| ORF1ab primer | 6 (1008) | 86·2% (60·8–96·2) | 98·1% (96·4–99·3) | 362·39 (33·76–3890·00) | 0·984 | 8% (7·14) | |
| N primer | 10 (1136) | 84·4% (72·9–91·6) | 97·5% (93·9–99·0) | 260·98 (115·11–591·70) | 0·975 | 0% (0·0) | |
| E primer | 2 (146)* | 81·6% (71·0–89·5) | 100% (94·9–100) | 300·35 (38·53–2341·37) | 0·901 | 0% (0·0) | |
| RNA extraction method | |||||||
| Magnetic beads | 5 (408) | 81·3% (53·7–94·3) | 97·2% (89·3–99·3) | 180·22 (56·55–574·39) | 0·973 | 0% (0·0) | |
| Silica spin column | 19 (1634) | 83·7% (77·8–88·2) | 95·1% (91·1–97·4) | 198·02 (71·74–546·61) | 0·943 | 0% (3·4) | |
| dPCR | |||||||
| Overall | 15 (783) | 94·1% (88·9–96·9) | 78·5% (57·4–90·8) | 77·47 (24·68–243·19) | 0·946 | 0·5% (2·79) | |
| Specimen | |||||||
| Pharyngeal swabs | 7 (459) | 95·0% (87·5–98·1) | 90·1% (60·3–98·2) | 237·354 (39·35–1431·75) | 0·958 | 7% (3·38) | |
| Saliva | 3 (152)* | 89·7% (75·8–97·1) | 77·0% (68·1–84·4) | 18·76 (3·46–101·62) | 0·865 | 0% (1·16) | |
| Sputum | 2 (98)* | 100% (93·4–100) | 88·6% (75·4–96·2) | 283·29 (32·304–2484·26) | 0·979 | 0% (0·0) | |
| Primer–probe set | |||||||
| ORF1ab primer | 11 (544) | 97·3% (93·8–98·8) | 80·9% (53·7–93·9) | 158·584 (39·77–632·42) | 0·964 | 0·8% (2·46) | |
| N primer | 15 (783) | 94·1% (88·9–96·9) | 78·5% (57·4–90·8) | 77·47 (24·68–243·19) | 0·946 | 0·5% (2·79) | |
| RNA extraction method | |||||||
| Silica spin column | 8 (536) | 92·3% (83·0–96·7) | 81·6% (52·9–94·6) | 76·52 (15·35–381·44) | 0·937 | 10% (3·6) | |
| Automatic | 4 (173) | 96·5% (86·7–99·2) | 91·0% (75·2–97·1) | 310·37 (59·85–1609·41) | 0·972 | 0% (0·0) | |
| qPCR | |||||||
| Overall | 25 (3667) | 92·7% (88·3–95·6) | 92·9% (87·2–96·2) | 231·27 (86·77–616·44) | 0·967 | 0% (4·60) | |
| Specimen | |||||||
| Pharyngeal swabs | 13 (2250) | 96·1% (91·5–98·3) | 94·7% (88·1–97·8) | 638·96 (215·32–1896·09) | 0·982 | 0% (2·07) | |
| Saliva | 10 (1175) | 86·3% (78·6–91·6) | 90·9% (79·3–96·3) | 88·02 (24·89–311·28) | 0·921 | 0% (3·07) | |
| RNA extraction | |||||||
| With RNA extraction | 19 (3230) | 93·7% (88·4–96·6) | 92·4% (85·3–96·3) | 262·47 (81·57–844·57) | 0·971 | 0% (5·63) | |
| Without RNA extraction | 6 (437) | 89·7% (81·9–94·3) | 94·3% (79·2–98·6) | 157·61 (28·96–857·70) | 0·940 | 16% (2·53) | |
| Primer–probe set | |||||||
| ORF1ab primer | 5 (1369) | 97·3% (87·4–99·5) | 88·8% (85·7–91·2) | 527·47 (44·33–6276·02) | 0·892 | 0% (6·16) | |
| N primer | 12 (899) | 92·9% (87·0–96·3) | 91·3% (79·9–96·5) | 192·23 (83·16–444·36) | 0·961 | 4% (0·53) | |
| E primer | 11 (2442) | 92·3% (83·3–96·6) | 94·1% (85·5–97·7) | 208·13 (40·84–1060·61) | 0·971 | 8% (6·3) | |
| RNA extraction method | |||||||
| Magnetic beads | 7 (1170) | 92·9% (86·8–96·3) | 97·0% (88·4–99·3) | 462·91 (83·03–2580·88) | 0·968 | 25% (3·76) | |
| Silica spin column | 10 (1805) | 93·4% (82·9–97·6) | 84·0% (69·4–92·4) | 96·50 (21·89–425·37) | 0·948 | 6% (4·52) | |
| Automatic | 2 (224)* | 96·7% (88·5–99·6) | 91·5% (86·1–95·3) | 350·48 (42·61–2882·13) | 0·961 | 0% (0·0) | |
Data are n (N); sensitivity, specificity, or OR (95% CI); AUC; or I2 (τ2). Summary of subgroup analyses for LAMP, dPCR, and qPCR assays based on the number of true false-positive and false-negative cases reported in the 71 experiments. All dPCR assays required RNA extraction. OR=odds ratio. AUC=area under the curve. LAMP=loop-mediated isothermal amplification. ORF1ab=open reading frame 1 ab. N=nucleocapsid. E=envelope. dPCR=digital PCR. qPCR=quantitative PCR.
A univariate random-effects model was used to estimate sensitivities and specificities for subgroups with less than four included studies, as bivariate models do not converge when the sample size is small.
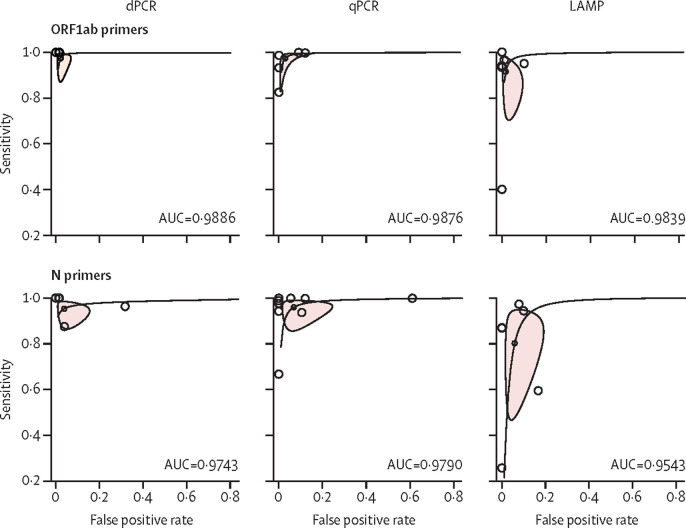
To explore the effect of a specific subgroup on test performance, included studies were stratified into ORF1ab (15 studies, 2896 samples) and N gene clusters (22 studies, 2666 samples), yielding a total of six clusters for three nucleic acid tests. For each cluster, we were able to control for the experimental setup by restricting the studies to those using only pharyngeal swabs with RNA extraction. RNA extraction methods were not specified due to the limited number of studies. The distribution of dPCR studies was the least scattered, followed by qPCR and LAMP, implying that dPCR had the highest accuracy. A descending order in AUC could be seen across the three tests using ORF1ab primers, although the differences in the AUCs were negligible (figure 2 ). We also found that dPCR, qPCR, and LAMP using ORF1ab primers consistently outperformed those using N primers (figure 2). All AUCs were above 0·9, meaning the three nucleic acid tests had excellent test performance. With RNA extraction, diagnostic ORs for all three nucleic acid tests were consistently higher when using pharyngeal swabs and ORF1ab primer sets (diagnostics OR 2082·41 [95% CI 353·44–12 269·35] for dPCR, 2053·37 [680·75–6193·62] for qPCR, 931·31 [94·51–1591·10] for LAMP; appendix p 23). Primer and probe sequences for each study can be found in the appendix (pp 24–38).
Figure 2.
Hierarchical summary receiver operator curves of dPCR, qPCR, and LAMP in the secondary subgroup analysis comparing the use of ORF1ab and N primer–probe sets
All studies from the clusters used pharyngeal swabs and RNA extraction and only clusters consisting of more than three studies are presented. Three clusters were identified for comparing the test performance of dPCR, qPCR, and LAMP using primers targeting the ORF1ab gene, and three were identified using primers targeting the N gene. AUC=area under the curve. dPCR=digital PCR. LAMP=loop-mediated isothermal amplification. N=nucleocapsid. ORF1ab=open reading frame 1ab. qPCR=quantitative PCR.
To consolidate our conclusions on the sensitivities of the three nucleic acid tests in our meta-analysis, we drew a boxplot showing the distribution of detection limit determined by using synthetic viral RNA in the 34 studies (figure 3 ). LAMP had the widest range of detection limits followed by dPCR and qPCR. The small box size of dPCR and qPCR suggests that the reported detection limits were similar between studies and the respective low means of 202·8 (median 140, IQR 103–291) and 1952·4 copies (median 1000, 530–1497) of viral RNA per mL suggested a higher analytical sensitivity for dPCR than for qPCR, in agreement with our results on the overall clinical performance, where dPCR was more sensitive than qPCR. LAMP assay showed a wider spread of detection limits ranging from 80 to 10 000 copies per mL (IQR 400–4000 copies per mL), indicating a less stable sensitivity. Overall a good agreement was observed between the analytical sensitivity (ie, detection limits) and diagnostic sensitivity (ie, pooled estimates) of the three nucleic acid tests.
Figure 3.
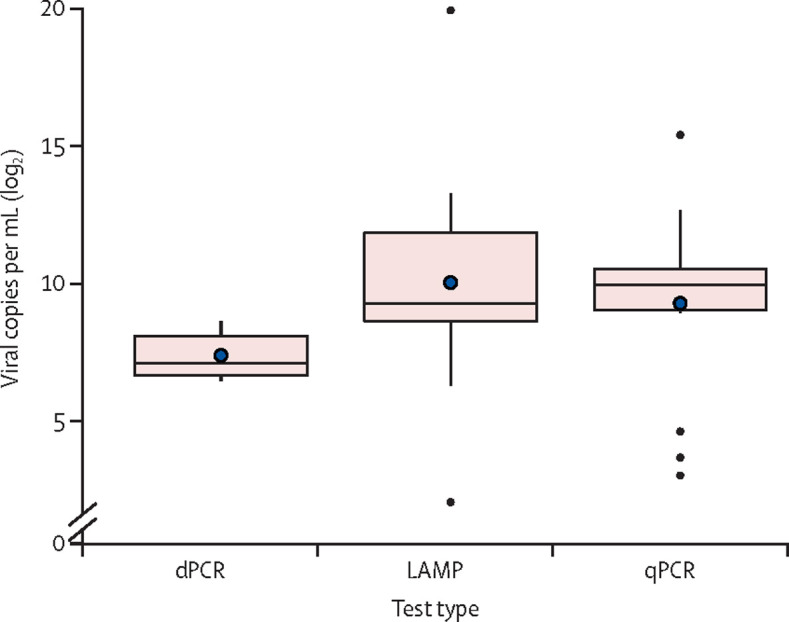
Boxplot of detection limits evaluated using synthetic viral RNA in 34 studies
The blue points represent the mean values of detection limits for each test. dPCR=digital PCR. LAMP=loop-mediated isothermal amplification. qPCR=quantitative PCR.
Quality assessment was done on the 66 original studies according to QUADAS-2. All studies evaluated both the respective molecular tests' analytical performance and clinical application (appendix p 45). However, only 14 (21%) of 66 studies reported details of study design regarding patient selection and the time interval between the reference test and index test. Three (5%) studies had the index test rated as high risk mainly because index tests were assessed using samples with known results by the reference tests, which might increase the potential of bias. 32 (48%) studies only stated the source of clinical samples, and 33 (50%) reported only the technical procedures of the index test, thus receiving an unclear rating for patient selection and index test. Since most studies focused on the experimental details of the PCR and LAMP reactions, only 16 (24%) studies resulted in low risk in the domain of flow and timing of the index and reference tests. The overall methodological quality was fair, and the applicability concerns were deemed acceptable.
Discussion
We performed the first and largest systematic review and meta-analysis to date to compare dPCR, qPCR, and LAMP in the context of SARS-CoV-2 detection. We evaluated various test conditions including primer sets, RNA extraction methods, and types of human specimen, using EUA-approved PCR assays as the reference standard. Overall, dPCR had the highest diagnostic sensitivity but had lower specificity than qPCR and LAMP. This low specificity might result from the threshold effect, in which reduction in specificity was the trade-off for its exceedingly high sensitivity.82
In our primary subgroup analysis, we showed that nucleic acid assays using pharyngeal swabs outperformed saliva assays for dPCR, qPCR, and LAMP with both Reitsma and latent class bivariate models. This finding was consistent with those of studies done in February–April, 2021 (subsequent to the end date of our literature search), which compared saliva specimens with pharyngeal swabs for qPCR and LAMP. Sensitivity of 80–86% and specificity of 98–99% were reported by these saliva studies, which were close to our pooled indices (Reitsma model).83, 84 Our results also agreed with US Centers for Disease Control and Prevention and WHO's recommendations to use pharyngeal swabs over saliva. Overall, tests using pharyngeal swabs should remain the gold standard for diagnosing COVID-19 and a reference standard for rapid tests for detection of SARS-CoV-2 infection in mass screening of the population. Moreover, qPCR and LAMP assays applied to samples without RNA extraction consistently performed poorly in sensitivity and specificity. This finding suggests that without RNA extraction, there is a high probability of failure to identify a patient infected with SARS-CoV-2 compared with tests that include an RNA extraction step. Hence, methods without RNA extraction should only be considered when the importance of time-saving and clinical resources outweigh the need for high test performance.
When testing differences in AUCs between subgroups, we found that the type of specimen was the key factor in determining AUC in dPCR and LAMP, whereas the choice of RNA extraction method strongly influenced qPCR performance. These observations suggest that specimen types and RNA extraction methods should be the main focus in optimising these assays. When we restricted our analysis to studies using only pharyngeal swabs with RNA extraction, the three nucleic acid tests using ORF1ab primers consistently outperformed the tests using N primers. This result is possibly due to less antigenic mutation in the ORF1ab than that in the N region.57 Importantly, we observed that when controlling the experimental setup of each test by specifying the primer–probe set, RNA extraction, and specimen type, the accuracy estimated in AUC did not differ across different nucleic acid tests. However, significant variations in accuracy were observed when the experimental setup was changed. This implies that optimisation efforts should be directed towards designing a robust and consistent experimental setup as the accuracy of SARS-CoV-2 nucleic acid tests is more strongly dependent on the experimental setup than on the type of test used. This conclusion also suggests that LAMP and dPCR could be reliable alternatives to the current qPCR gold standard, as the three nucleic acid tests have similar accuracies when the experimental conditions are similar. Of note, the inclusion of LAMP as a reference standard with similar accuracy to the PCR methods would be advantageous to clinical systems in low-income and middle-income countries or at point-of-care settings where medical resources are scarce. In high-income countries, where dPCR is more readily available, this method has the advantage of providing the absolute number of viral copies in a sample and thus can be used to calibrate other detection methods due to its high sensitivity and reproducibility. Therefore, different test types might be chosen according to environmental and socioeconomic requirements, but test results should remain consistent, provided that the optimal experimental setup is implemented and standardised.
In our analysis of the analytical sensitivity of the nucleic acid tests, the low detection limit of dPCR was consistent with its high pooled sensitivity derived from clinical samples. LAMP studies showed the most variations in the distribution of detection limits, possibly due to inherent differences in detection methods such as colorimetry and turbidity. Although our subgroup analyses were unable to compare the performance of LAMP assays by their detection methods due to the small number of studies per method, future research on LAMP detection methods will be useful in the optimisation of the assay.
Our study has some limitations. Besides the experimental factors used in our study, other factors, such as patient demographics, which were not reported in all diagnostic studies, might affect the test outcome. We also did not evaluate other PCR or isothermal amplification techniques and recombinase polymerase amplification. Moreover, the bias introduced during patient selection, test implementation, and publication might have influenced the experimental outcome and data accessibility, and hence also the results of our analysis.
With rising COVID-19 cases and deaths globally, it is pressing to investigate the utility of current diagnostic tests and the optimal conditions for these tests to achieve the best accuracy and consistency. By synthesising summary estimates for dPCR, qPCR, and LAMP performance, we concluded that all three nucleic acid tests consistently perform better with pharyngeal swabs than saliva samples and with ORF1ab than N primer–probe sets. RNA extraction is a crucial step to safeguard test accuracy in all three tests. dPCR has shown to be the most sensitive, followed by qPCR and LAMP. However, their accuracies do not differ significantly; instead, accuracy depends on specific experimental conditions, implying that more efforts should be directed to optimising the experimental setups for the nucleic acid tests. Our results could be a reference for such optimisation and for the establishment of a standard nucleic acid test protocol that is applicable across laboratories worldwide.
Data sharing
Data in this systematic review with meta-analysis are extracted from published and preprint studies available on the internet. Request for the meta-analysis data can be made by contacting the corresponding author.
Declaration of interests
We declare no competing interests.
Acknowledgments
Acknowledgments
This work was supported by the University Grants Committee General Research Fund (number 16302618) and Chinese University of Hong Kong Department of Chemical Pathology Faculty Startup Fund. We thank the Hong Kong University of Science and Technology, Department of Chemistry, for the opportunity to conduct this study. We thank Chung Heng Au for his assistance with computer programming.
Contributors
PPHC conceived and supervised the study and verified results from study selection and data extraction. WYA screened literature, extracted data from eligible studies, and performed the statistical analyses. Both authors interpreted the data, wrote and finalised the manuscript, had full access to and verified all the data in the study, and had final responsibility for submitting it for publication.
Supplementary Material
References
- 1.WHO Weekly epidemiological update on COVID-19 – 21 September 2021. 2021. https://www.who.int/publications/m/item/weekly-epidemiological-update-on-covid-19-27-july-2021
- 2.Lisboa Bastos M, Tavaziva G, Abidi SK, et al. Diagnostic accuracy of serological tests for COVID-19: systematic review and meta-analysis. BMJ. 2020;370:2516. doi: 10.1136/bmj.m2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jin Y-H, Cai L, Cheng Z-S, et al. A rapid advice guideline for the diagnosis and treatment of 2019 novel coronavirus (2019-nCoV) infected pneumonia (standard version) Mil Med Res. 2020;7:4. doi: 10.1186/s40779-020-0233-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Subsoontorn P, Lohitnavy M, Kongkaew C. The diagnostic accuracy of isothermal nucleic acid point-of-care tests for human coronaviruses: a systematic review and meta-analysis. Sci RepoRtS. 2020;10:22349. doi: 10.1038/s41598-020-79237-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jiang Y, Wang H, Hao S, et al. Digital PCR is a sensitive new technique for SARS-CoV-2 detection in clinical applications. Clin Chim Acta. 2020;511:346–351. doi: 10.1016/j.cca.2020.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fatih Abasiyanik M, Flood B, Lin J, et al. Sensitive detection and quantification of SARS-CoV-2 in saliva. medRxiv. 2020 doi: 10.1101/2020.12.04.20241059. published online Dec 7. (preprint). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alteri C, Cento V, Antonello M, et al. Detection and quantification of SARS-CoV-2 by droplet digital PCR in real-time PCR negative nasopharyngeal swabs from suspected COVID-19 patients. PLoS One. 2020;15:e0236311. doi: 10.1371/journal.pone.0236311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cassinari K, Alessandri-Gradt E, Chambon P, et al. Assessment of multiplex digital droplet RT-PCR as a diagnostic tool for SARS-CoV-2 detection in nasopharyngeal swabs and saliva samples. Clin Chem. 2021;67:736–741. doi: 10.1093/clinchem/hvaa323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dang Y, Liu N, Tan C, et al. Comparison of qualitative and quantitative analyses of COVID-19 clinical samples. Clin Chim Acta. 2020;510:613–616. doi: 10.1016/j.cca.2020.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deiana M, Mori A, Piubelli C, Scarso S, Favarato M, Pomari E. Assessment of the direct quantitation of SARS-CoV-2 by droplet digital PCR. Sci Rep. 2020;10:18764. doi: 10.1038/s41598-020-75958-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dong L, Zhou J, Niu C, et al. Highly accurate and sensitive diagnostic detection of SARS-CoV-2 by digital PCR. Talanta. 2021;224:121726. doi: 10.1016/j.talanta.2020.121726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haq F, Sharif S, Khurshid A, et al. Development optimization and validation of RT-LAMP based COVID-19 facility in Pakistan. bioRxiv. 2020 doi: 10.1101/2020.05.29.124123. ublished online May 29. (preprint). [DOI] [Google Scholar]
- 13.Oberding L, Hu J, Berenger B, Mohon AN, Pillai DR. Quantification of SARS-CoV-2 viral copy number in saliva mouthwash samples using digital droplet PCR. medRxiv. 2020 doi: 10.1101/2020.06.13.20130237. published online June 23. (preprint). [DOI] [Google Scholar]
- 14.Suo T, Liu X, Feng J, et al. ddPCR: a more accurate tool for SARS-CoV-2 detection in low viral load specimens. medRxiv. 2020 doi: 10.1080/22221751.2020.1772678. published online May 5. (preprint). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu L, Wu S, Hao X, et al. Rapid detection of COVID-19 coronavirus using a reverse transcriptional loop-mediated isothermal amplification (RT-LAMP) diagnostic platform. Clin Chem. 2020;66:975–977. doi: 10.1093/clinchem/hvaa102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yvan C, Munderloh UG, Starolis M, et al. Development and validation of a rapid, single-step reverse transcriptase loop-mediated isothermal amplification (RT-LAMP) system potentially to be used for reliable and high-throughput screening of COVID-19. Front Cell Infect Microbiol. 2020;10:331. doi: 10.3389/fcimb.2020.00331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lamb LE, Bartolone SN, Ward E, Chancellor MB. Rapid detection of novel coronavirus/severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) by reverse transcription-loop-mediated isothermal amplification. PLoS One. 2020;15:e0234682. doi: 10.1371/journal.pone.0234682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee JYH, Best N, McAuley J, et al. Validation of a single-step, single-tube reverse transcription loop-mediated isothermal amplification assay for rapid detection of SARS-CoV-2 RNA. J Med Microbiol. 2020;69:1169–1178. doi: 10.1099/jmm.0.001238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mohon AN, Oberding L, Hundt J, et al. Optimization and clinical validation of dual-target RT-LAMP for SARS-CoV-2. J Virol Methods. 2020;286:113972. doi: 10.1016/j.jviromet.2020.113972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Papadakis G, Pantazis AK, Fikas N, et al. Real-time colorimetric LAMP methodology for quantitative nucleic acids detection at the point-of-care. bioRxiv. 2020 doi: 10.1101/2020.07.22.215251. published online July 24. (preprint). [DOI] [Google Scholar]
- 21.Schermer B, Fabretti F, Damagnez M, et al. Rapid SARS-CoV-2 testing in primary material based on a novel multiplex RT-LAMP assay. PLoS One. 2020;15:e0238612. doi: 10.1371/journal.pone.0238612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dao Thi VL, Herbst K, Boerner K, et al. A colorimetric RT-LAMP assay and LAMP-sequencing for detecting SARS-CoV-2 RNA in clinical samples. Sci Transl Med. 2020;12:eabc7075. doi: 10.1126/scitranslmed.abc7075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang Q, Meyerson NR, Clark SK, et al. Saliva TwoStep for rapid detection of asymptomatic SARS-CoV-2 carriers. medRxiv. 2021 doi: 10.1101/2020.07.16.20150250. published online Feb 16. (preprint). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rohaim MA, Clayton E, Sahin I, et al. Artificial intelligence-assisted loop mediated isothermal amplification (AI-LAMP) for rapid detection of SARS-CoV-2. Viruses. 2020;12:972. doi: 10.3390/v12090972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lalli MA, Langmade JS, Chen X, et al. Rapid and extraction-free detection of SARS-CoV-2 from saliva by colorimetric reverse-transcription loop-mediated isothermal amplification. Clin Chem. 2021;67:415–424. doi: 10.1093/clinchem/hvaa267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ali Z, Aman R, Mahas A, et al. iSCAN: an RT-LAMP-coupled CRISPR-Cas12 module for rapid, sensitive detection of SARS-CoV-2. Virus Res. 2020;288:198129. doi: 10.1016/j.virusres.2020.198129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alekseenko A, Barrett D, Pareja-Sanchez Y, et al. Direct detection of SARS-CoV-2 using non-commercial RT-LAMP reagents on heat-inactivated samples. Sci Reports. 2021;11:1820. doi: 10.1038/s41598-020-80352-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ben-Assa N, Naddaf R, Gefen T, et al. Direct on-the-spot detection of SARS-CoV-2 in patients. Exp Biol Med. 2020;245:1187–1193. doi: 10.1177/1535370220941819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang-Ngai Chow F, Tat-Yin Chan T, Raymond Tam A, et al. A rapid, simple, inexpensive, and mobile colorimetric assay COVID-19-LAMP for mass on-site screening of COVID-19. Int J Mol Sci Artic J Mol Sci. 2020;21:5380. doi: 10.3390/ijms21155380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Flynn MJ, Snitser O, Flynn J, et al. A simple direct RT-LAMP SARS-CoV-2 saliva diagnostic. medRxiv. 2020 doi: 10.1101/2020.11.19.20234948. published online Nov 2. (preprint). [DOI] [Google Scholar]
- 31.Fowler VL, Armson B, Gonzales JL, et al. A highly effective reverse-transcription loop-mediated isothermal amplification (RT-LAMP) assay for the rapid detection of SARS-CoV-2 infection. J Infect. 2021;82:117–125. doi: 10.1016/j.jinf.2020.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xuejiao H, Qianyun D, Junmin L, et al. Development and clinical application of a rapid and sensitive loop-mediated isothermal amplification test for SARS-CoV-2 infection. mSphere. 2021;5:e00808–e00820. doi: 10.1128/mSphere.00808-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang WE, Lim B, Hsu C, et al. RT-LAMP for rapid diagnosis of coronavirus SARSCoV-2. Microb Biotechnol. 2020;13:950–961. doi: 10.1111/1751-7915.13586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.West CP, Montori VM, Sampathkumar P. COVID-19 testing. Mayo Clin Proc. 2020;95:1127–1129. doi: 10.1016/j.mayocp.2020.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu C, Shi Q, Peng M, et al. Evaluation of droplet digital PCR for quantification of SARS-CoV-2 virus in discharged COVID-19 patients. Aging (Albany NY) 2020;12:20997–21003. doi: 10.18632/aging.104020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kitagawa Y, Orihara Y, Kawamura R, et al. Evaluation of rapid diagnosis of novel coronavirus disease (COVID-19) using loop-mediated isothermal amplification. J Clin Virol. 2020;129:104446. doi: 10.1016/j.jcv.2020.104446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lu R, Wu X, Wan Z, Li Y, Jin X, Zhang C. A Novel reverse transcription loop-mediated isothermal amplification method for rapid detection of SARS-CoV-2. Int J Mol Sci. 2020;21:2826. doi: 10.3390/ijms21082826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang W, Dang X, Wang Q, et al. Rapid detection of SARS-CoV-2 using reverse transcription RT-LAMP method. medRxiv. 2020 doi: 10.1101/2020.03.02.20030130. published online March 3. (preprint). [DOI] [Google Scholar]
- 39.Rödel J, Egerer R, Suleyman A, et al. Use of the variplex™ SARS-CoV-2 RT-LAMP as a rapid molecular assay to complement RT-PCR for COVID-19 diagnosis. J Clin Virol. 2020;132:104616. doi: 10.1016/j.jcv.2020.104616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Butler-Laporte G, Lawandi A, Schiller I, et al. Comparison of saliva and nasopharyngeal swab nucleic acid amplification testing for detection of SARS-CoV-2: a systematic review and meta-analysis. JAMA Intern Med. 2021;181:353–360. doi: 10.1001/jamainternmed.2020.8876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Böger B, Fachi MM, Vilhena RO, Cobre AF, Tonin FS, Pontarolo R. Systematic review with meta-analysis of the accuracy of diagnostic tests for COVID-19. Am J Infect Control. 2021;49:21–29. doi: 10.1016/j.ajic.2020.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reitsma JB, Glas AS, Rutjes AWS, Scholten RJPM, Bossuyt PM, Zwinderman AH. Bivariate analysis of sensitivity and specificity produces informative summary measures in diagnostic reviews. J Clin Epidemiol. 2005;58:982–990. doi: 10.1016/j.jclinepi.2005.02.022. [DOI] [PubMed] [Google Scholar]
- 43.Ling DI, Pai M, Schiller I, Dendukuri N. A Bayesian framework for estimating the incremental value of a diagnostic test in the absence of a gold standard. BMC Med Res Methodol. 2014;14:67. doi: 10.1186/1471-2288-14-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7:177–188. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 45.Rutter C, Gatsonis C. A hierarchical regression approach to meta-analysis of diagnostic test accuracy evaluations. Stat Med. 2001;2019:2865–2884. doi: 10.1002/sim.942. [DOI] [PubMed] [Google Scholar]
- 46.Macaskill P, Gatsonis C, Deeks J, Harbord R, Takwoingi Y. In: Cochrane handbook for systematic reviews of diagnostic test accuracy version 1.0. Deeks JJ, Bossuyt PM, Gatsonis C, editors. The Cochrane Collaboration; 2010. Analysing and presenting results.https://methods.cochrane.org/sites/methods.cochrane.org.sdt/files/public/uploads/Chapter%2010%20-%20Version%201.0.pdf [Google Scholar]
- 47.Hong C, Duan R, Zeng L, et al. The galaxy plot: a new visualization tool for bivariate meta-analysis studies. Am J Epidemiol. 2020;189:861–869. doi: 10.1093/aje/kwz286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hong C, Salanti G, Morton SC, et al. Testing small study effects in multivariate meta-analysis. Biometrics. 2020;76:1240–1250. doi: 10.1111/biom.13342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Altawalah H, AlHuraish F, Alkandari WA, Ezzikouri S. Saliva specimens for detection of severe acute respiratory syndrome coronavirus 2 in Kuwait: a cross-sectional study. J Clin Virol. 2020;132:104652. doi: 10.1016/j.jcv.2020.104652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Anderson C, Castillo F, Koenig M, Managbanag J. Pooling nasopharyngeal swab specimens to increase testing capacity for SARS-CoV-2. Med J (Ft Sam Houst Tex) 2021;PB 8-21-01/02/03:8–11. [PubMed] [Google Scholar]
- 51.Barra GB, Santa Rita TH, Góes Mesquita P, Jácomo RH, Abdalla Nery LF. Analytical sensibility and specificity of two RT-qPCR protocols for SARS-CoV-2 detection performed in an automated workflow. Genes (Basel) 2020;11:1183. doi: 10.3390/genes11101183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bruce EA, Huang M-L, Perchetti GA, et al. Direct RT-qPCR detection of SARS-CoV-2 RNA from patient nasopharyngeal swabs without an RNA extraction step. PLOS Biol. 2020;18:e3000896. doi: 10.1371/journal.pbio.3000896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chan JF-W, Yip CC-Y, To KK-W, et al. Improved molecular diagnosis of COVID-19 by the novel, highly sensitive and specific COVID-19-RdRp/Hel real-time reverse transcription-PCR assay validated in vitro and with clinical specimens. J Clin Microbiol. 2020;58:e00310–e00320. doi: 10.1128/JCM.00310-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gustavo Dorlass E, Oliveira Monteiro C, Oliveira Viana A, et al. Lower cost alternatives for molecular diagnosis of COVID-19: conventional RT-PCR and SYBR Green-based RT-qPCR. Brazilian J Microbiol. 2020;51:1117–1123. doi: 10.1007/s42770-020-00347-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Freire-Paspuel B, Vega-Mariño P, Velez A, Castillo P, Cruz M, Garcia-Bereguiain MA. Evaluation of nCoV-QS (MiCo BioMed) for RT-qPCR detection of SARS-CoV-2 from nasopharyngeal samples using CDC FDA EUA qPCR kit as a gold standard: an example of the need of validation studies. J Clin Virol. 2020;128:104454. doi: 10.1016/j.jcv.2020.104454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Freire-Paspuel B, Garcia-Bereguiain MA. Analytical sensitivity and clinical performance of a triplex RT-qPCR assay using CDC N1, N2, and RP targets for SARS-CoV-2 diagnosis. Int J Infect Dis. 2021;102:14–16. doi: 10.1016/j.ijid.2020.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hasan MR, Mirza F, Al-Hail H, et al. Detection of SARS-CoV-2 RNA by direct RT-qPCR on nasopharyngeal specimens without extraction of viral RNA. PLoS One. 2020;15:e0236564. doi: 10.1371/journal.pone.0236564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kandel C, Zheng J, McCready J, et al. Detection of SARS-CoV-2 from saliva as compared to nasopharyngeal swabs in outpatients. Viruses. 2020;12:1314. doi: 10.3390/v12111314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Klein S, Müller TG, Khalid D, et al. SARS-CoV-2 RNA extraction using magnetic beads for rapid large-scale testing by RT-qPCR and RT-LAMP. Viruses. 2020;12:863. doi: 10.3390/v12080863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Konrad R, Eberle U, Dangel A, et al. Rapid establishment of laboratory diagnostics for the novel coronavirus SARS-CoV-2 in Bavaria, Germany, February 2020. Eurosurveillance. 2020;25:1. doi: 10.2807/1560-7917.ES.2020.25.9.2000173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lübke N, Senff T, Scherger S, et al. Extraction-free SARS-CoV-2 detection by rapid RT-qPCR universal for all primary respiratory materials. J Clin Virol. 2020;130:104579. doi: 10.1016/j.jcv.2020.104579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.McCormick-Baw C, Morgan K, Gaffney D, et al. Saliva as an alternate specimen source for detection of SARS-CoV-2 in symptomatic patients using Cepheid Xpert Xpress SARS-CoV-2. J Clin Microbiol. 2020;58:e01109–e01120. doi: 10.1128/JCM.01109-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Merindol N, Pépin G, Marchand C, et al. Optimization of SARS-CoV-2 detection by RT-qPCR without RNA extraction. bioRxiv. 2020 doi: 10.1101/2020.04.06.028902. published online April 10. (preprint). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Moreno-Contreras J, Espinoza MA, Sandoval-Jaime C, et al. Saliva sampling and its direct lysis, an excellent option to increase the number of SARS-CoV-2 diagnostic tests in settings with supply shortages. J Clin Microbiol. 2020;58:e01659–e01720. doi: 10.1128/JCM.01659-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pujadas E, Ibeh N, Hernandez MM, et al. Comparison of SARS-CoV-2 detection from nasopharyngeal swab samples by the Roche cobas 6800 SARS-CoV-2 test and a laboratory-developed real-time RT-PCR test. J Med Virol. 2020;92:1695–1698. doi: 10.1002/jmv.25988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ranoa DRE, Holland RL, Alnaji FG, et al. Saliva-based molecular testing for SARS-CoV-2 that bypasses RNA extraction. bioRxiv. 2020 doi: 10.1101/2020.06.18.159434. published online June 18. (preprint). [DOI] [Google Scholar]
- 67.Ratcliff J, Nguyen D, Andersson M, Simmonds P. Evaluation of different PCR assay formats for sensitive and specific detection of SARS-CoV-2 RNA. bioRxiv. 2020 doi: 10.1101/2020.06.24.168013. published online July 1. (preprint). [DOI] [Google Scholar]
- 68.Sun Q, Li J, Ren H, et al. Saliva as a testing specimen with or without pooling for SARS-CoV-2 detection by multiplex RT-PCR test. PLoS One. 2021;16:e0243183. doi: 10.1371/journal.pone.0243183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Vaz SN, Santana DS de. Netto EM, et al. Saliva is a reliable, non-invasive specimen for SARS-CoV-2 detection. Brazilian J Infect Dis. 2020;24:422–427. doi: 10.1016/j.bjid.2020.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Visseaux B, Le Hingrat Q, Collin G, et al. Evaluation of the RealStar SARS-CoV-2 RT-PCR kit RUO performances and limit of detection. J Clin Virol. 2020;129:104520. doi: 10.1016/j.jcv.2020.104520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Vogels CBF, Watkins AE, Harden CA, et al. SalivaDirect: a simplified and flexible platform to enhance SARS-CoV-2 testing capacity. medRxiv. 2020 doi: 10.1101/2020.08.03.20167791. published online Sept 28. (preprint). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang J, Cai K, Zhang R, et al. Novel one-step single-tube nested quantitative real-time PCR assay for highly sensitive detection of SARS-CoV-2. Anal Chem. 2020;92:9399–9404. doi: 10.1021/acs.analchem.0c01884. [DOI] [PubMed] [Google Scholar]
- 73.Wolters F, van de Bovenkamp J, van den Bosch B, et al. Multi-center evaluation of cepheid xpert® xpress SARS-CoV-2 point-of-care test during the SARS-CoV-2 pandemic. J Clin Virol. 2020;128:104426. doi: 10.1016/j.jcv.2020.104426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wozniak A, Cerda A, Ibarra-Henríquez C, et al. A simple RNA preparation method for SARS-CoV-2 detection by RT-qPCR. Sci RepoRtS. 2020;10:16608. doi: 10.1038/s41598-020-73616-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Xiao Y, Li Z, Wang X, et al. Comparison of three TaqMan real-time reverse transcription-PCR assays in detecting SARS-CoV-2. J Virol Methods. 2021;288:114030. doi: 10.1016/j.jviromet.2020.114030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yip CCY, Sridhar S, Leung KH, et al. Development and evaluation of novel and highly sensitive single-tube nested real-time RT-PCR assays for SARS-CoV-2 detection. Int J Mol Sci. 2020;21:1–11. doi: 10.3390/ijms21165674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yu F, Yan L, Wang N, et al. Quantitative detection and viral load analysis of SARS-CoV-2 in infected patients. Clin Infect Dis. 2020;71:793–798. doi: 10.1093/cid/ciaa345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhen W, Berry GJ. Design of a novel multiplex real time RT-PCR assay for SARS-CoV-2 detection. bioRxiv. 2020 doi: 10.1101/2020.06.04.135608. published online June 5. (preprint). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lu X, Wang L, Sakthivel SK, et al. US CDC real-time reverse transcription PCR panel for detection of severe acute respiratory syndrome coronavirus 2. Emerg Infect Dis. 2020;26:1654–1665. doi: 10.3201/eid2608.201246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Perchetti GA, Nalla AK, Huang ML, Jerome KR, Greninger AL. Multiplexing primer/probe sets for detection of SARS-CoV-2 by qRT-PCR. J Clin Virol. 2020;129:104499. doi: 10.1016/j.jcv.2020.104499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yan C, Cui J, Huang L, et al. Rapid and visual detection of 2019 novel coronavirus (SARS-CoV-2) by a reverse transcription loop-mediated isothermal amplification assay. Clin Microbiol Infect. 2020;26:773–779. doi: 10.1016/j.cmi.2020.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Naaktgeboren CA, Ochodo EA, Van Enst WA, et al. Assessing variability in results in systematic reviews of diagnostic studies. BMC Med Res Methodol. 2016;16:6. doi: 10.1186/s12874-016-0108-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Pasomsub E, Watcharananan SP, Boonyawat K, et al. Saliva sample as a non-invasive specimen for the diagnosis of coronavirus disease 2019: a cross-sectional study. Clin Microbiol Infect. 2021;27:285. doi: 10.1016/j.cmi.2020.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Schneider F, Molina L, Picot M, et al. Comparative evaluation of rapid salivary RT-LAMP assay for screening of SARS-CoV-2 infection. SSRN Electron J. 2021 doi: 10.2139/ssrn.3774184. published online Feb 9. (preprint). [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data in this systematic review with meta-analysis are extracted from published and preprint studies available on the internet. Request for the meta-analysis data can be made by contacting the corresponding author.