Abstract
The aim of this study was to determine the distribution in France of the Enterobacter aerogenes prevalent clone isolated in the hospitals of the Marseille area (A. Davin-Regli, D. Monnet, P. Saux, C. Bosi, R. Charrel, A. Barthelemy, and C. Bollet, J. Clin. Microbiol. 34:1474–1480, 1996). A total of 123 E. aerogenes isolates were collected from 23 hospital laboratories and analyzed by random amplification of polymorphic DNA and enterobacterial repetitive intergenic consensus-PCR to determine their epidemiological relatedness. Molecular typing revealed that 21 of the 23 laboratories had isolated this prevalent clone harboring the plasmid encoding for extended-spectrum β-lactamase of the TEM-24 type. Most isolates were susceptible only to imipenem and gentamicin. Their dissemination seems to be clonal and was probably the result of the general use of broad-spectrum cephalosporins and quinolones. Four isolates showed an alteration of their outer membrane proteins, causing decrease of susceptibility to third-generation cephalosporins and imipenem and leading to the critical situation of having no alternative therapeutic. The large dissemination of the E. aerogenes prevalent clone probably results from its good adaptation to the antibiotics administered in France and the hospital environment, particularly in intensive care units.
Before 1993, Enterobacter aerogenes was rarely encountered in French hospitals. Since 1995 there have been many reports of its presence both in France (i.e., Bordeaux [2], Dijon [23], Clermont-Ferrand [10, 17], Limoges [26], St. Etienne [14], and Strasbourg [21]) and elsewhere, including Belgium (11, 15), Austria (1), and the United States (13, 25). The first clinical cases described in Marseille hospitals concerned patients in intensive care units (ICUs). In fact, the emergence of this species is associated with medical devices, the widespread administration of antibiotics, and immunodepression of patients (4, 8, 9). Nosocomial infections now concern other medical units (11, 15).
Today, E. aerogenes is the third-most-common pathogen recovered from the respiratory tract in Marseille hospitals and is often isolated in urine and the gastrointestinal tract, as in other countries (16, 29). The dramatic emergence of this pathogen is associated mainly with use of broad-spectrum cephalosporins and quinolones (8, 11, 25). The first outbreaks were caused by isolates presenting an extended-spectrum β-lactamase (ESBL) (9, 13). Today, most isolates involved in nosocomial infections are resistant to multiple antibiotics because they have a chromosomally derepressed cephalosporinase and an ESBL (2, 8, 11, 23). In addition, in some isolates, alteration of the membrane protein composition has led to resistance through impermeability or efflux associated with enzymatic resistance, resulting in multidrug resistance and no availability of an alternative antibiotic (5, 10, 11, 19, 24, 30).
In epidemiology, it is generally agreed that strains indistinguishable by typing scheme and sharing characteristics that can distinguish them from epidemiologically unrelated strains constitute an epidemiological cluster named clone, which arises from a common precursor (27). We had demonstrated by PCR-typing methods that between 1994 and 1995 in the Marseille area hospitals 50% of the E. aerogenes infections or colonizations were due to isolates of the same epidemiological type (8). This prevalent type, herein called clone, was highly resistant to antibiotics except gentamicin, imipenem, and the latest cephalosporins, such as cefepime and cefpirome.
The aim of this study was to establish the prevalence of the clone in France. A representative selection of E. aerogenes isolates sent from 23 French hospital laboratories was analyzed by random amplification of polymorphic DNA (RAPD) and enterobacterial repetitive intergenic consensus-PCR (ERIC-PCR). The relationship between the prevalence of this clone and multidrug resistance was illustrated by determination of the TEM β-lactamase gene sequences and observation of envelope porin expression.
MATERIALS AND METHODS
Characteristics of clinical isolates.
From August 1996 to January 1997, 123 clinical E. aerogenes isolates were collected from infected or colonized patients in 23 hospitals in France (Table 1). These isolates were obtained from bronchial secretions, urine samples, closed cavity drainage fluids, catheters, blood cultures, and wound swabs. Each isolate corresponded to a single patient. All isolates were identified and confirmed to be E. aerogenes by the API 20E identification system (bioMérieux, Marcy l’Etoile, France) according to the manufacturer’s instructions. Most of them were selected by the laboratories for their marked resistance against the usual antibiotics. Susceptibility to 18 to 35 antimicrobial agents and combinations of agents was determined by the standard disk diffusion method (22) in some laboratories or by the Walkaway 40 System (Sanofi-Diagnostics Pasteur, Marnes-la-Coquette, France) in others. These agents and associations were amoxicillin, amoxicillin-clavulanate, ticarcillin, ticarcillin-clavulanate, mezlocillin, piperacillin, cephalothin, cefamandole, cefoxitin, cefotetan, cefotiam, cefsulodin, moxalactam, cefotaxime, ceftazidime, ceftazidime-sulbactam, ceftriaxone, cefoperazone, cefmenoxime, cefepime, cefpirome, imipenem, meropenem, aztreonam, gentamicin, tobramycin, kanamycin, amikacin, chloramphenicol, trimethoprim-sulfamethoxazole, fosfomycin, colistin, pefloxacin, ofloxacin, and ciprofloxacin. The presence of ESBL activity was confirmed with the double-disk diffusion test, in which disks containing cefepime, cefotaxime, ceftazidime, or ceftriaxone were placed near a disk containing a β-lactamase inhibitor (sodium clavulanate) under the conditions described above (9). If an ESBL was present in the organisms, the zones of inhibition around the cephalosporin disks were enhanced between each cephalosporin disk and the clavulanic acid disk. The clinical isolates were divided into two antibiotic resistance phenotypes: the presence of a derepressed cephalosporinase alone or one associated with an ESBL.
TABLE 1.
Characteristics of the 123 E. aerogenes isolates recovered from 23 hospital centers in France
| Medical center location (city) | No. of isolates | No. of strains with:
|
||
|---|---|---|---|---|
| Cefepime-imipenem-gentamicin suscep-tible phenotype | Prevalent RAPD or ERIC type | Porin-deficient phenotype | ||
| Amiens | 5 | 3 | 1 | |
| Angers | 5 | 4 | 4 | |
| Aubagne | 8 | 8 | 7 | 2 |
| Besançon | 5 | 5 | 4 | |
| Bordeaux | 5 | 3 | 3 | |
| Brest | 5 | 5 | 0 | |
| Clermond-Ferrand | 5 | 5 | 4 | |
| La Ciotat | 7 | 7 | 3 | |
| Le Mans | 3 | 2 | 2 | 1 |
| Lille | 5 | 4 | 3 | |
| Limoges | 5 | 5 | 5 | |
| Lyon | 6 | 1 | 1 | |
| Metz | 5 | 2 | 2 | |
| Montpellier | 5 | 3 | 3 | |
| Mulhouse | 5 | 5 | 4 | |
| Nancy | 5 | 5 | 5 | 1 |
| Narbonne | 4 | 0 | 0 | |
| Nîmes | 12 | 10 | 10 | |
| Paris | 5 | 3 | 3 | |
| Quimper | 3 | 3 | 3 | |
| St. Etienne | 5 | 2 | 2 | |
| Strasbourg | 6 | 6 | 6 | |
| Toulouse | 4 | 4 | 4 | |
| Total | 123 | 95 | 79 | 4 |
Epidemiologic typing.
The isolates were investigated by using RAPD with primer AP12H (5′-CGGCCCCTGT-3′) and ERIC-PCR with primer ERIC2 (5′-AAGTAAGTGACTGGGGTGAGCG-3′) as described previously (8, 9, 13, 31).
(i) DNA preparation.
Isolates were grown overnight at 37°C on Mueller-Hinton agar (bioMérieux). Total cellular DNA was extracted by the Chelex technique (12), and DNA concentrations were estimated on agarose gels (28).
(ii) Amplification conditions.
Amplification reactions were performed in a total volume of 47 μl containing 100 μM dATP, 100 μM dCTP, 100 μM dGTP, and 100 μM dTTP, plus 0.2 μM primer, 25 ng of template DNA, and 1.25 U of Taq polymerase (Perkin-Elmer/Cetus, Norwalk, Conn.) in 1× PCR buffer (20 mM Tris-HCl, pH 8.3; 50 mM KCl; 3 mM MgCl2; 0.001% gelatin [wt/vol]). A negative control without template DNA was included in each experiment. The reaction mixtures were overlaid with mineral oil and subjected to amplification in a GenAmp PCR System 9600 (Perkin-Elmer/Cetus) programmed for 45 cycles of 1 min at 94°C, 1 min at 45°C, and 1 min at 74°C. Amplification products (10-μl samples) were electrophoresed in 1.2% agarose gels in Tris-acetate buffer (0.04 M Tris-acetate, 0.001 M EDTA; pH 8.2), stained with ethidium bromide, and photographed on a UV light transilluminator. A molecular weight standard (Marker VI; Boehringer-Mannheim, Mannheim, Germany) was included on each gel. We interpreted and compared the patterns without considering the origin of the isolates. Heterogeneity with respect to the intensity and shape of bands was not considered to be a difference. Interpretation of differences was based on guidelines proposed with fingerprints generated by pulsed-field gel electrophoresis. One band difference is likely to arise as a result of one genetic event, and such isolates can be considered very similar; two to three band differences can arise by two genetic events, and the organism is not similar but may be related, and so on. Four or more band differences are definite evidence of a completely unrelated strain. Thus, according to the total number of bands generated, isolates were considered different if their profiles differed by two or more bands (8, 27, 32).
(iii) Reproducibility.
For the two PCR-based techniques, reproducibility was determined by testing independent DNA preparations extracted from single-colony cultures at different times and amplified separately.
TEM β-lactamase identification. (i) PCR amplification.
PCR amplifications were performed as described previously (18). Amplification was achieved with an initial cycle of 5 min of denaturation at 95°C, followed by 30 cycles of 0.5 min at 94°C, 0.5 min at 55°C, and 0.5 min at 74°C. The primers were 5′-GAC AGT TAC CAA TGC TTA ATC A-3′ and 5′-TTG GGT GCA CGA GTG GGT TA-3′.
(ii) DNA sequencing.
DNA sequencing was carried out by cycle sequencing with fluorescently labeled dideoxynucleotide terminators (Applied Biosystems, Inc., Norwalk, Conn.). The sequencing reactions were analyzed with a 377 automated DNA sequencer (Applied Biosystems, Inc.).
(iii) Sequence analysis.
Amino acid sequences were determined with Translate Tool on the ExPASy worldwide web molecular biology server of the Swiss Institute of Bioinfamatics (12a). Sequences were analyzed by using the table published by G. Jacoby and K. Bush on the worldwide web server of the Lahey Clinic (14a).
SDS-polyacrylamide gel electrophoresis and immunodetection of porins.
Porins were immunodetected on the E. aerogenes isolates presenting a multiresistant phenotype. Exponential bacterial cells grown in Luria-Bertani broth were collected. Bacterial cell pellets were solubilized in loading buffer at 96°C, and samples were loaded onto sodium dodecyl sulfate (SDS)-polyacrylamide gels (10% polyacrylamide, 0.1% SDS) as previously described (3). Electrotransfer to nitrocellulose membranes was performed in the presence of 0.05% SDS to achieve complete transfer of the porins. An initial saturating step with Tris-buffered saline (TBS) (50 mM Tris-HCl, 150 mM NaCl; pH 8) containing 10% bovine serum was carried out overnight at 4°C. The nitrocellulose membranes were then incubated in TBS containing 10% bovine serum and 0.2% Triton X-100 for 2 h at room temperature with polyclonal antibodies directed against denatured Escherichia coli porins (OmpF and OmpC). Polyclonal antibodies directed against the E. coli porins were able to recognize the E. aerogenes porins as reported previously (20). After successive washings in the same buffer, the porins were detected with alkaline phosphatase-conjugated affinitiPure goat anti-rabit immunoglobulin G antibodies (Jackson ImmunoResearch, West Grove, Pa.). The polyclonal antibodies directed against porin monomers OmpF and OmpC have been described elsewhere (19).
RESULTS
Antibiotic susceptibility.
Among the 123 E. aerogenes isolates, 95 were susceptible only to cefepime, imipenem, or gentamicin as observed in clonal isolates (Table 1). In 11 of the 23 hospitals such a phenotype was shown by 100% of the isolates selected (Aubagne, Besançon, Toulouse, etc.). However, in eight other hospitals, 15 to 80% of the isolates were multiresistant. In four hospitals (Lyon, Metz, Narbonne, and St. Etienne), most of the isolates presented a different antibiotic resistance phenotype.
Concerning the 28 other E. aerogenes isolates, their antibiotypes had characteristics suggesting the presence of a derepressed cephalosporinase (i.e., resistance to cephamycins), but they were devoid of ESBL (i.e., they were susceptible to ceftazidime and aminosides).
Epidemiologic typing.
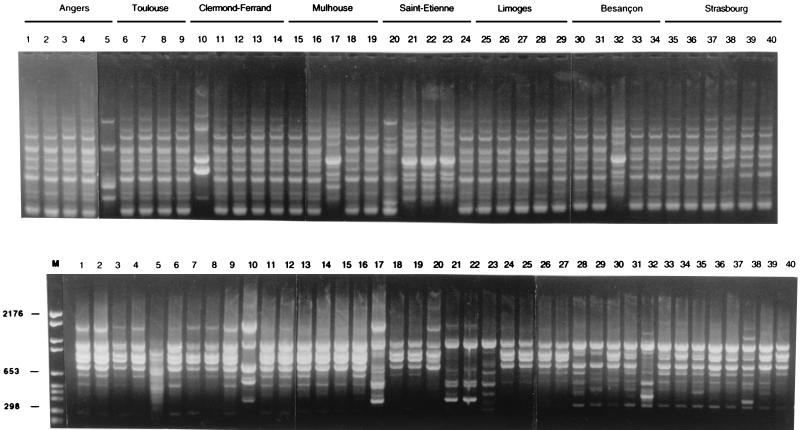
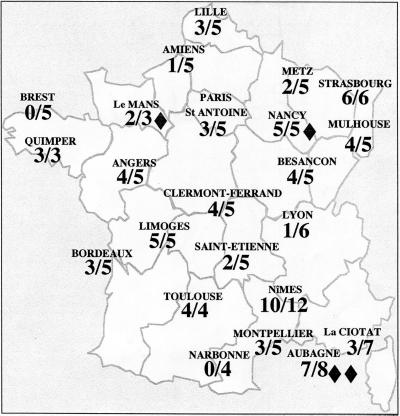
All E. aerogenes isolates were typable by RAPD and ERIC-PCR. The results obtained with the two techniques were concordant. With the RAPD methods, variations of intensity of PCR products were observed with faint bands, but they were not considered to be a difference. Figure 1 shows profiles corresponding to 40 E. aerogenes isolates from eight hospital laboratories. Among the 123 isolates, 79 presented a single type identical to the prevalent clone previously observed in the Marseille area. Figure 2 shows the distribution of the clone in each hospital. The clone was recovered in all hospitals except two (Brest and Narbonne) (Table 1). In most cases, the clone represented the majority of the isolates selected by each laboratory. Among the 79 E. aerogenes isolates belonging to the prevalent clone, 77 presented a multiresistant antibiotype and 2 (Le Mans and Nîmes) presented a phenotype corresponding to a derepressed cephalosporinase alone.
FIG. 1.
ERIC-PCR (upper panel) and RAPD (lower panel) fingerprints of 40 Enterobacter aerogenes isolates from hospital laboratories. Lane M, molecular weight marker (marker VI).
FIG. 2.
Representation of the dissemination of the E. aerogenes prevalent clone among 23 hospital centers in France (prevalent clone/total collected strains); ⧫ represents porin-deficient phenotype.
β-Lactamase identification.
Among the 95 E. aerogenes isolates with a phenotypic resistant profile, analysis of the ESBL by sequencing demonstrated a TEM-24 type.
Immunodetection of outer membrane porins.
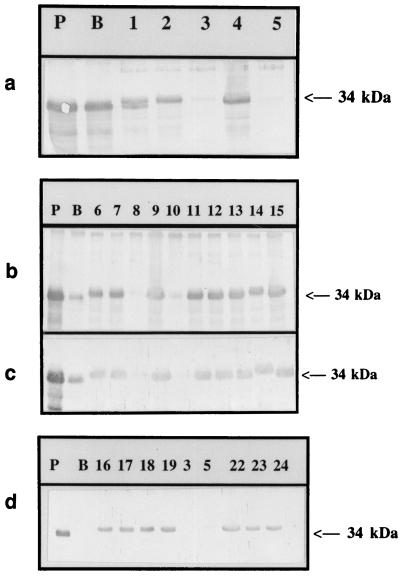
Immunological probes were used to investigate the porin content in clinical isolates and to evaluate the frequency of modification of envelope protein content in a representative population of E. aerogenes isolates. Of the 95 multiresistant isolates, 4 showed a negative response with the immunological probes directed against the unspecific enterobacterial porins, reflecting a porin-deficient phenotype (Fig. 3). Two of these porin-free strains were resistant, and the two others had intermediate resistance to imipenem.
FIG. 3.
Immunodetection of E. aerogenes outer membrane proteins. Immunostaining was done with polyclonal antibodies directed against the OmpF monomer (a and b) or OmpC (c and d). Strains PMY150 (P) and BZB1107 (B) were used as positive and negative samples, respectively. Strains from Aubagne (lanes 3 and 5), Le Mans (lane 8), and Nancy (lane 10) were the four strains that presented a porin-deficient phenotype. Lane numbers do not correspond to those in Fig. 1. Only the relevant part of the blot is shown. The 34-kDa arrow indicates the postulated porin migration.
DISCUSSION
An overview of most recent studies suggest that the epidemiology of E. aerogenes is characterized by the spread of a single clone through the medical units of a hospital (2, 8, 21, 23, 26); in some cases the clone has the imipenem/gentamicin-susceptible phenotype (2, 11, 21). In Belgium, E. aerogenes is responsible for 13% of the nosocomial infections, essentially in the south of the country. In 1995, two epidemiologically distinct studies using ERIC-PCR and PFGE demonstrated the existence of a multidrug resistant clone (11, 15). Isolates presented an ESBL, associated or not with a chromosomally derepressed cephalosporinase and able to resist quinolones in 80 to 100% of cases. Moreover, De Gheldre et al. observed the selection of strains resistant to imipenem and cefepime after the use of imipenem in therapy (11). The same observation was made in 1994 in a Greek hospital (30). In Austria, it has been shown by PFGE, rep-PCR, and RAPD that, during a septicemia outbreak for a 3-month period in an ICU, multidrug-resistant isolates harbored the imipenem/amikacin-sensitive phenotype (1). In the United States, Georgiou et al. studied an outbreak of E. aerogenes which occurred in 1991 in Houston, Tex., and which was due to two clusters of epidemiologically related isolates (13). Most of them (80%) presented an ESBL, and 20% were resistant to fluoroquinolones. However, in 1995 D’Agata et al. observed 11 E. aerogenes isolates in Boston, Mass., that were epidemiologically distinct as shown by PFGE but were resistant to ceftazidime, probably by expression of an ESBL (6). The epidemiology of E. aerogenes in Belgium seems similar to the situation in France. Since molecular typing of such epidemiologically related isolates was not carried out with the same molecular techniques or the same technical conditions as in the present study, the results of these other studies cannot be compared and so relationships between the different clones cannot be determined. However, some E. aerogenes isolates from previous studies may in part correspond to the prevalent clone.
It is difficult to evaluate how patients are colonized by this E. aerogenes prevalent clone. Usually, E. aerogenes is rarely found in the hospital environment. E. aerogenes could be recovered from material in direct contact with patients colonized (e.g., tubings of mechanical ventilators, and water from humidifiers) and occasionally from room surfaces and the hands of health care personnel (8, 11, 15). Also, in many cases patients have no direct contact with other infected patients. Furthermore, patients may already be colonized when they are admitted to hospitals. In France, most patients hospitalized in ICUs are discharged to a general acute care unit and then go to a rest home, a nursing home, or a retirement home. Some of them are still colonized by E. aerogenes, particularly in the urinary tract. Residence of such patients in these care centers, where the nursing personnel are not prepared to face nosocomial bacterial contaminations, favors extra hospital E. aerogenes propagation. This in part accounts for the colonization by multidrug-resistant E. aerogenes in patients coming from these care centers at the time of their first admission at hospital.
The E. aerogenes prevalent clone was multiresistant because it had a plasmid carrying ESBL of the TEM-24 type associated with a chromosomally encoded derepressed cephalosporinase. This multiresistance facilitated the spread of the species in the 21 hospitals. However, two of the prevalent clone isolates were devoid of ESBL, possibly because they had lost their plasmid. In vivo, plasmids can be lost over time, but the phenomenon minimally influence RAPD and ERIC-PCR profiles (27). The prevalent E. aerogenes clone was isolated in all but two hospitals. Selective pressure due to broad-spectrum antibiotic use in hospitals favors colonization of patients by the most resistant nosocomial bacterial species and the spread of such bacteria to all hospital units. However, it is probable that other selective factors are responsible for the exceptional adaptation of the E. aerogenes clone to the hospital environment. Effectively, some multiresistant E. aerogenes strains with different profiles as shown by PCR-typing methods that are isolated in a given hospital were not found elsewhere or showed only limited spread. These isolates apparently did not have the phenotypic or genotypic capacity or the geographic dissemination displayed by the prevalent type.
Moreover, excessive prescription in the community and in the hospital of third-generation cephalosporins or quinolones in monotherapy has made the Enterobacteriaceae strains multidrug resistant in many countries. In multidrug resistant E. aerogenes isolates, resistance to quinolones correlated with the presence of the ESBL of the TEM-24 type even in patients recently hospitalized. For 5 years now, widespread antibiotherapy misuse in France has led to the emergence of multiresistant Enterobacter isolates. This is exemplified by the appearance in 1996 of nosocomial cases of Enterobacter hormaechei infections subsequent to the emergence of a clone that was highly resistant to quinolones. The appearance of the species in patients was always correlated with the use of new quinolones (7).
It is alarming to observe that the emergence of E. aerogenes isolates with a decreased susceptibility to imipenem is now more frequent; they represented 4 to 5% of all of the E. aerogenes studied in our laboratory (3). Arpin et al. reported that 4.6% of their E. aerogenes isolates were resistant to imipenem (2). We found that four E. aerogenes isolates presenting such a phenotype exhibited an alteration of their porin content. This may be another resistance mechanism used by the bacteria. The incidence of such mechanisms in E. aerogenes will probably increase with the use of imipenem in isolates presenting the imipenem/gentamicin-sensitive phenotype. It seems evident that E. aerogenes is capable to perfectly adapt itself to antibiotic pressure. Mallea et al. have recently described clinical E. aerogenes isolates presenting a complex resistance strategy associating β-lactamase production, impermeability, and active efflux (19). The emergence of imipenem resistance is a crucial problem because there are no antibiotherapy possibilities available for such strains expressing a multidrug-resistant phenotype. In every case the survival of patients is seriously impaired.
In conclusion, there is currently in France a hospital pandemic due to a multidrug-resistant E. aerogenes clone particularly well adapted to the hospital environment. Modification of antibiotic use and bacterial monitoring of the hospital ecology and of the patients near the end of their hospitalization seem to be the most important means to stop this development.
ACKNOWLEDGMENTS
We thank the following for providing strains (locations in France are given parenthetically: Y. Assadourian and J. M. Schneider (Aubagne and La Ciotat); F. Barbut (Paris); C. Bebear and A. Allery (Bordeaux); B. Carbonnelle and C. Lemarie (Angers); G. Chabanon and D. Clave (Toulouse); M. Dailloux and A. Lozniewski (Nancy); J. M. Delarbre and M. F. Penner (Mulhouse); F. Denis and M. C. Ploy (Limoges); P. Y. Donnio (Rennes); F. Eb (Amiens); J. Freney, J. Etienne, and P. Girardo (Lyon); F. Geffroy (Quimper); A. Gouby (Nîmes); F. Grattard (St. Etienne); B. Joly (Clermont-Ferrand); D. Lamarca and L. Bertran (Narbonne); A. Marmonier (Le Mans); C. Perez (Montpellier); B. Picard and D. Tande (Brest); Y. Piemont (Strasbourg); C. Plesiat and Y. Michel-Briand (Besançon); P. Roos (Metz); and M. Simonet (Lille). We acknowledge A. Abeille for technical assistance.
This work was supported by Assistance Publique à Marseille (Recherche Clinique) in 1997.
REFERENCES
- 1.Allerberguer F, Koeuth T, Lass-Florl C, Dierich M P, Putensen C, Scmutzhard E, Mohsenipour I, Grundmann H, Hartung D, Bauernfeind A, Eberlein E, Lupski J R. Epidemiology of infections due to multiresistant Enterobacter aerogenes in a university hospital. Eur J Clin Microbiol Infect Dis. 1996;15:517–521. doi: 10.1007/BF01691323. [DOI] [PubMed] [Google Scholar]
- 2.Arpin C, Coze C, Rogues A M, Gachie J P, Bebear C, Quentin C. Epidemiological study of an outbreak due to multidrug-resistant Enterobacter aerogenes in a medical intensive care unit. J Clin Microbiol. 1996;34:2163–2169. doi: 10.1128/jcm.34.9.2163-2169.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Charrel R N, Pagès J-M, De Micco P, Mallea M. Prevalence of outer membrane porin alteration in β-lactam antibiotic-resistant Enterobacter aerogenes. Antimicrob Agents Chemother. 1996;40:2854–2858. doi: 10.1128/aac.40.12.2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chow J W, Yu V L, Shlaes D M. Epidemiologic perspectives on Enterobacter for the infection control professional. Am J Infect Control. 1994;22:195–201. doi: 10.1016/0196-6553(94)90067-1. [DOI] [PubMed] [Google Scholar]
- 5.Chow J W, Shlaes D M. Imipenem resistance associated with the loss of a 40-kDa outer membrane protein of Enterobacter aerogenes. J Antimicrob Chemother. 1991;28:499–504. doi: 10.1093/jac/28.4.499. [DOI] [PubMed] [Google Scholar]
- 6.D’Agata E, Venkataraman L, DeGirolami P, Samore M. Molecular epidemiology of acquisition of ceftazidime-resistant gram-negative bacilli in a nonoutbreak setting. J Clin Microbiol. 1997;35:2602–2605. doi: 10.1128/jcm.35.10.2602-2605.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davin-Regli A, Bosi C, Charrel R N, Ageron E, Papazian L, Grimont P A D, Cremieux A, Bollet C. A nosocomial outbreak due to Enterobacter cloacae strains with the E. hormaechei genotype in patients treated with fluoroquinolones. J Clin Microbiol. 1997;35:1008–1010. doi: 10.1128/jcm.35.4.1008-1010.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davin-Regli A, Monnet D, Saux P, Bosi C, Charrel R, Barthelemy A, Bollet C. Molecular epidemiology of Enterobacter aerogenes acquisition: one-year prospective study in two intensive care units. J Clin Microbiol. 1996;34:1474–1480. doi: 10.1128/jcm.34.6.1474-1480.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davin-Regli A, Saux P, Bollet C, Gouin F, de Micco P. Investigation of a series of outbreaks of Enterobacter aerogenes in an intensive care unit by random amplification polymorphic DNA. J Med Microbiol. 1995;44:89–98. doi: 10.1099/00222615-44-2-89. [DOI] [PubMed] [Google Scholar]
- 10.De Champs C, Henquell C, Guelon D, Sirot D, Gazuy N, Sirot J. Clinical and bacteriological study of nosocomial infections due to Enterobacter aerogenes resistant to imipenem. J Clin Microbiol. 1993;31:123–127. doi: 10.1128/jcm.31.1.123-127.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.De Gheldre Y, Maes N, Rost F, De Ryck R, Clevenbergh P, Vincent J L, Struelens M J. Molecular epidemiology of an outbreak of multidrug-resistant Enterobacter aerogenes infections and in vivo emergence of imipenem resistance. J Clin Microbiol. 1997;35:152–160. doi: 10.1128/jcm.35.1.152-160.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.De Lamballerie X, Zandotti C, Vignoli C, Bollet C, de Micco P. A one-step microbial DNA extraction method using Chelex 100 suitable for gene amplification. Res Microbiol. 1992;143:785–790. doi: 10.1016/0923-2508(92)90107-y. [DOI] [PubMed] [Google Scholar]
- 12a.ExPASy server. 5 January 1999, revision date. Translate tool. [Online.] http://www.expasy.ch/tools/dna.html. [27 April 1999, last date accessed.]
- 13.Georghiou P R, Hamill R J, Wright C E, Versalovic J, Koeuth T, Watson D A, Lupski J R. Molecular epidemiology of infections due to Enterobacter aerogenes: identification of hospital outbreak-associated strains by molecular techniques. Clin Infect Dis. 1995;20:84–94. doi: 10.1093/clinids/20.1.84. [DOI] [PubMed] [Google Scholar]
- 14.Grattard F, Pozetto B, Tabard L, Petit M, Ros A, Gaudin O G. Characterization of nosocomial strains of Enterobacter aerogenes by arbitrarily primed PCR analysis and ribotyping. Infect Control Hosp Epidemiol. 1995;16:224–230. doi: 10.1086/647094. [DOI] [PubMed] [Google Scholar]
- 14a.Jacoby, G., and K. Bush. 8 March 1999, revision date. Table. [Online.] http://www.lahey.org/studies/webt.htm. [27 April 1999, last date accessed.]
- 15.Jalaluddin S, Devaster J-M, Scheen R, Gerard M, Butzler J-P. Molecular epidemiological study of nosocomial Enterobacter aerogenes isolates in a Belgian hospital. J Clin Microbiol. 1998;36:1846–1852. doi: 10.1128/jcm.36.7.1846-1852.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jarvis W R, Martone W J. Predominant pathogens in hospital infections. J Antimicrob Chemother. 1992;29(Suppl. A):19–24. doi: 10.1093/jac/29.suppl_a.19. [DOI] [PubMed] [Google Scholar]
- 17.Livrelli V, De Champs C, Di Martino P, Darfeuille-Michaud A, Forestier C, Joly B. Adhesive properties and antibiotic resistance of Klebsiella, Enterobacter, and Serratia clinical isolates involved in nosocomial infections. J Clin Microbiol. 1996;34:1963–1969. doi: 10.1128/jcm.34.8.1963-1969.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mabilat C, Goussard S. PCR detection and identification of genes for extended-spectrum β-lactamases. In: Persing D H, Smith T F, Tenover F C, White T J, editors. Diagnostic molecular microbiology: principles and applications. Washington, D.C: American Society for Microbiology; 1994. pp. 553–559. [Google Scholar]
- 19.Mallea M, Chevalier J, Bornet C, Eyraud A, Davin-Regli A, Bollet C, Pagès J M. Porin alteration and active efflux: two in vivo drug resistance strategies used by Enterobacter aerogenes. Microbiology. 1998;144:3003–3009. doi: 10.1099/00221287-144-11-3003. [DOI] [PubMed] [Google Scholar]
- 20.Mallea M, Simonet V, Eun-Hee L, Collatz E, Gervier R, Gutmann L, Pagès J M. Biological and immunological comparisons of Enterobacter cloacae and Escherichia coli porins. FEMS Microbiol Lett. 1995;129:273–280. doi: 10.1111/j.1574-6968.1995.tb07592.x. [DOI] [PubMed] [Google Scholar]
- 21.Meunier O, Stoessel P, Saint-Laurent P, Lutun P, Jehl F, Scheftel J M, Monteil H, Tempé J D, Bientz M. Rôle des laboratories d’hygiène et de bactériologie dans la prise en charge d’une épidémie àEnterobacter aerogenes multirésistant aux antibiotiques. Ann Biol Clin. 1997;55:129–137. [PubMed] [Google Scholar]
- 22.National Committee for Clinical Laboratory Standards. Performance standards for antimicrobial disk susceptibility testing, 6th ed. M2A6. Wayne, Pa: National Committee for Clinical Laboratory Standards; 1997. [Google Scholar]
- 23.Neuwirth C, Siebor E, Lopez J, Pechinot A, Kazmierczak A. Outbreak of TEM-24-producing Enterobacter aerogenes in an intensive care unit and dissemination of the extended-spectrum β-lactamase to other members of the family Enterobacteriaceae. J Clin Microbiol. 1996;34:76–79. doi: 10.1128/jcm.34.1.76-79.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nikaido H. Prevention of drug access to bacterial targets: permeability barriers and active efflux. Science. 1994;264:382–388. doi: 10.1126/science.8153625. [DOI] [PubMed] [Google Scholar]
- 25.Pitout J D D, Thomson K S, Hanson N D, Ehrhardt A F, Coudron P, Sanders C C. Plasmid-mediated resistance to expanded-spectrum cephalosporins among Enterobacter aerogenes strains. Antimicrob Agents Chemother. 1998;42:596–600. doi: 10.1128/aac.42.3.596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ploy M C, Grelaud C, Martin C, Denis F. Emergence of Enterobacter aerogenes strains producing derepressed cephalosporinase at CHU in Limoges: molecular epidemiology by the RAPD technique. Pathol Biol (Paris) 1997;45:404–408. [PubMed] [Google Scholar]
- 27.Power E G M. RAPD typing in microbiology. A technical review. J Hosp Infect. 1996;34:247–265. doi: 10.1016/s0195-6701(96)90106-1. [DOI] [PubMed] [Google Scholar]
- 28.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. Quantification of DNA and RNA: appendix E.5. [Google Scholar]
- 29.Schaberg D R, Culver D H, Gaynes R P. Major trends in the microbial etiology of nosocomial infection. Am J Med. 1991;91(Suppl. 3B):72S–75S. doi: 10.1016/0002-9343(91)90346-y. [DOI] [PubMed] [Google Scholar]
- 30.Tzouvelekis L S, Tzelepi E, Kaufmann M E, Mentis A F. Consecutive mutations leading to the emergence in vivo of imipenem resistance in a clinical strain of Enterobacter aerogenes. J Med Microbiol. 1994;40:403–407. doi: 10.1099/00222615-40-6-403. [DOI] [PubMed] [Google Scholar]
- 31.Versalovic J, Koeuth T, Lupski J R. Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res. 1991;19:6823–6831. doi: 10.1093/nar/19.24.6823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Woods C R, Jr, Versalovic J, Koeuth T, Lupski J R. Analysis of relationships among isolates of Citrobacter diversus by using DNA fingerprints generated by repetitive sequence-based primers in the polymerase chain reaction. J Clin Microbiol. 1992;30:2921–2929. doi: 10.1128/jcm.30.11.2921-2929.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]