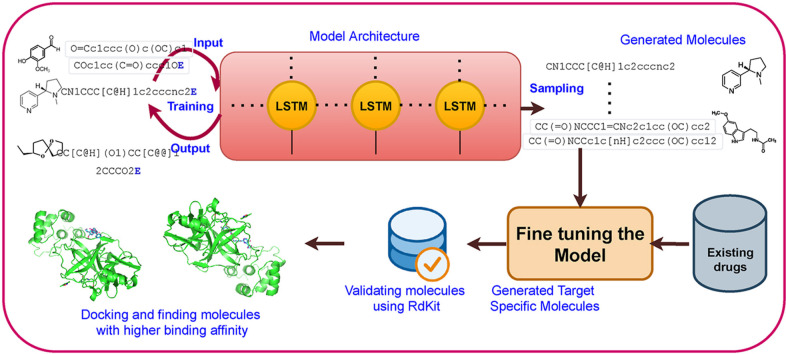
Abstract
The novel Coronavirus outbreak has created a massive economic crisis, and many succumb to death, disturbing the lives of mankind all over the world. Currently, there are no viable treatment for this condition, drug development approaches are being pursued with vigor. The major treatment options are to repurpose existing drugs or to find new ones. Traditional methods for drug discovery take a longer time, so there is an urgent need to develop some alternative techniques that reduces search space for drug candidates. Towards this endeavor, we propose a novel drug discovery method that leverages on long short term memory (LSTM) model to generate novel molecules that are adept at binding with novel Coronavirus protease. Our study demonstrates that the proposed method is able to recreate novel molecules that correlate very much with the properties of trained molecules. Further, we fine-tune the model to generate novel drug-like molecules that are active towards a specific target. We consider 3CLPro, the main protease of novel Coronavirus, as a therapeutic target and demonstrated in silico screening to assess target structural binding affinities with docking simulations. We observed that 80% of generated molecules show docking free energy of less than −5.8 kcal/mol. The top generated drug candidate has the highest binding affinity with a docking score of −8.5 kcal/mol, which is very much lower when compared to approved existing commercial drugs including, Remdesivir. The low binding energy indicates that the generated molecules could be explored as potential drug candidates for Covid-19.
Keywords: Drug discovery, Novel molecules, Deep neural networks, Docking, Covid-19, Generative models
Graphical abstract
1. Introduction
The 2019 novel Coronavirus (2019-nCoV) outbreak has created havoc and caused massive socio-economic shock worldwide resulting in economic crisis and loss of lives. The World Health Organisation (WHO) has declared Covid-19 as a pandemic, and the virus spread rate is worse than previous coronavirus epidemics. According to the WHO, to date (Weekly epidemiological record 96, September 15, 2021) [1] there are 225, 680, 357 Covid-19 cases and more than 4, 644, 740 deaths reported. Since there is no known effective treatment for this disease, has created a sense of urgency towards exploring novel drug discovery approaches for its treatment. However, most of the immediate endeavors were centered towards repurposing of known clinically approved drugs, and virtual screened molecules from large chemical databases have shown minimal effects [2]. Antibody development and small molecular development are two known approaches for drug discovery. In antibody development, antibodies bind to the virus surface protein and stop binding to a host cell receptor. In small molecular development, the novel molecules are designed by employing computational techniques that act as a ligand to inhibit the target proteins.
Besides virtual screening and drug repurposing, de novo molecular design by computational techniques is emerged as a promising field in the branch of drug design and has made outstanding contributions for drug discovery [3]. Over the past few years the deep learning techniques has revolutionized many emerging fields include biomedical data studies [[4], [5], [6]] bioinformatics and computational biology [[7], [8], [9]]. Recently, deep generative models have shown remarkable development in various aspects like musical improvisation [10], creating realistic artworks [11], source-target images translation [12], and facial expression change [13]. In the field of chemo-informatics, there has been increasing demand in developing generative models to generate realistic and valid molecules for de novo drug design. Generally, the molecules are represented in the form of strings known as Simplified Molecular Input Line Entry Specification (SMILES) [14] derived from molecular graphs are popularly used in this area. Recurrent Neural Networks (RNNs) [15] are the most suitable architectures for such representations and consequently, RNN-based generative models with one-hot encoding were commonly adopted [16].
One of the major challenges in drug discovery is the enormous search space for novel molecules. More than 1060 drugs like molecules are estimated to be synthetically accessible [17]. From this corpus of molecules, scientists select and examine molecules that bind to biological targets that inhibits the replication of bacteria or viruses. Moreover, high throughput screening experiments becomes highly expensive. To mitigate these limitations it is preferable to have computational methods to condense the large search space. Generally, virtual screening is used to explore favorable drug-like molecules among billions of existing molecules using similarity based measures that shows how close molecules are related. Whereas in de novo drug design approaches, novel molecules are created which binds towards the biological targets.
Molecular docking and deep learning are two widely investigated computational approaches for drug discovery. Molecular docking examines how well drug molecule binds with biological targets by using 3-dimensional simulation, in which drug molecules (ligands) find its position into targets (proteins) site. However, this approach has two major drawbacks: first, getting the 3D structure of target protein is a difficult task; second, large simulations are expensive and time-consuming. At the same, time deep learning techniques are intriguing as they dramatically reduce the large scale testing of candidate molecules in short time and relatively inexpensive. Computational de novo drug design techniques help to navigate the vast chemical space of entire drugs to screen for candidate molecules that are active against specific targets. Deep learning methods provide automated design and screening of candidate molecules with some desired properties.
This study proposes an entirely data-driven strategy to develop new drug compounds. It employs a generative recurrent neural network based LSTM model for molecular generation that is subjected to rigorous training on large sets of molecules. In our approach, we initially collected nearly 2.9 million molecules from two well known molecular databases namely, Moses and ChemBL. After performing a cleaning operation like removing salts and stereochemical information with the help of chemo-informatics tools like RDKit [18], we retrieved nearly 2.5 million molecules. These molecules are represented in the form of SMILES and they are passed as one-hot encoded input vectors into the proposed generative LSTM model. The model learns the dynamics of SMILE grammar over training data and forms a multinomial probability distribution. The generative model learns how realistic and plausible drug-like molecules are and, upon sampling from the distribution, creates novel drug candidates equivalent to training data. The use of generative models for molecule generation not only saves time and money but also narrows the search field.
We demonstrated that the RNN based LSTM could generate valid novel drug molecules. Furthermore, to generate more focused molecules, i.e., that can bind to novel Coronavirus protein, we fine-tuned our model by training it further on existing commercial antiviral and HIV drugs [19] that are active towards novel Coronavirus protein. Hence, the generator RNN learned knowledge distribution of generalized molecules which can directly produce novel molecules that are biologically active towards target proteins.
2. Related work
Deep learning techniques have the ability to transform the way drugs are discovered [20] and the way diseases are detected [21]. The recent applications of deep learning in drug discovery for Covid-19 are majorly focused on finding repurposed drug candidates [22,23]. Drug repurposing or reposition is a method in which new indictions are given for existing drugs to treat challenging diseases. However, predicting novel molecules for drug discovery to treat Covid–19 is a challenging task. In one of the first efforts at therapeutic possibilities for repurposing drug candidates, Gordon et al. [24] have identified 66 human proteins associated with novel Coronavirus proteins. Li et al. [25] in a study based on genome sequence analyses of three main viral families of SARS-CoV-2, identified 30 repurposed drugs. Kowalewski et al. [26] proposed a machine learning-based method to identify various drug candidates. They collected 65 assay data targeting human proteins, which interacts with novel Coronavirus proteins, and used this data to train their model to predict inhibitory activity. Beck along with his colleagues [19], developed a Molecule Transformer Model for drug-target interaction based on CNN and RNN to predict several existing antiviral drugs that could work for Covid-19. Another prominent way to repurpose the drugs is through construction of medical knowledge graphs. These graphs contain new associations between drugs and diseases. Majorly graph networks use graph embedding techniques to represent nodes and edges in latent dimensional space [27]. Increasing interest in developing graph models leads to different graph representation models. Gysi et al. [23] have developed a graph representation model as a case study on Covid-19 by identifying 81 potential repurposing candidates. On similar grounds, BenevolentAI [28], based on the AI–driven knowledge graph, predicted baricitinib as a potential repurposing drug for SARS-CoV-2. It has targeted Ap2-associated protein AAK1 and found that baricitinib acts as a potential inhibitor. The major limitations of graph models are training the node labels, which costs quadratic complexity, and semantically valid graphs are challenging to generate.
Some deep generative models have also been introduced recently to develop potential drug candidates. Zhavoronkov et al. [2] developed a Generative Auto-encoder and Generative Adversarial Networks (GANs) [29] which uses protein structure and crystallized ligands and homology proteins to identify drug candidates for main protease (3CLPro) of novel Coronavirus. Tang et al. [30] developed techniques based on reinforcement learning to discover drug candidates that inhibit against Covid-19. In an approach towards targeting RNA-dependent RNA polymerase (RdRp) to generate novel molecules that can block viral RNA synthesis. Patankar [31] developed an LSTM model to train IC50 binding data and screened for the molecules that block RdRp. Zhang et al. [32] developed a deep learning method for large scale virtual screening to identify potential protein-ligand binding pairs by using various chemical compound databases.
Machine and deep learning based approaches have explored and developed some methods for drug repurposing against novel Coronavirus. However, there are less to no approaches that explore therapeutic options to find novel drugs. In the present study, we propose a novel data driven approach to create de novo drug molecules. It basically employs a generative recurrent neural network based long short term memory model for molecular generation that is subjected to rigorous training on large sets of molecules. Our major contributions are:
-
1.
We propose a data driven methodology for generating novel molecules based on LSTM, which learns the syntax of molecular representation with great accuracy.
-
2.
This computational model has the capability to learn the probability of molecular patterns and hence can be used to generate novel molecules.
-
3.
Our model successfully exhibits transfer learning when fine-tuned with target specific molecules.
-
4.
The proposed model has ability to generate novel drug candidates with the highest binding affinity when compared to commercially existing antiviral drugs and also Remdesivir.
-
5.
The model identifies a group of novel molecules that are specifically adept at binding with novel Coronavirus main protease structure with higher binding score.
3. Materials and methods
3.1. Molecule representation
To depict molecules in machine readable format it is important to know how the molecules are represented. Usually, the molecules are modeled as Lewis structures or molecular graphs in chemistry [33]. In molecular graphs, each atom is represented as a labeled node, and bonding between the atoms is represented with a connected line known as an edge with the label indicating the bond order (single, double, triple bond). It is therefore better to have a model that reads and outputs a molecular graph. Many scientists have discovered varied ways to store molecules like valence model, connection table, and line notation. Among these representations, line notation is considered the best known way today. We, therefore, employ the Simplified Molecular Input Line Entry System (SMILES) [14] format that depicts the molecular graphs into a compact and human readable format that is simpler to comprehend.
SMILE is a formal grammar, well defined on a set of syntactic rules that is based on alphabet of characters like (B, C, N, O, P, S, F, Cl, Br, I … etc.), special symbols like (-, = , #) and numbers (1, 2, …,9). Usually, SMILE notation consists of a chain of letters, special characters, and numbers that specify the atoms, their connectivity and bond order. It is a single line text representation of a chemical compound of a molecule. As earlier said, SMILES are defined onset of specification rules; for example, atoms are represented by their atomic symbols, metal atoms with symbols in square brackets. Single, double, and triple bonds are represented as -, = , #, respectively. The cyclic structures are represented by aromatic and aliphatic atoms; the former is represented in uppercase and later as lower case. To indicate a closed ring structure, a number is placed at the atoms where the cyclic structure is ended. For example, the SMILE notation of aliphatic Cylcolhexaane is “C1CCCCC1”. Branches are specified by enclosing the atoms in parenthesis and can be nested or arranged. Additionally, some other symbols like/, \, @ are used to represent stereochemistry information. In order to generate realistic molecules, the generative model has to thoroughly learn the SMILE grammar based on production rules to keep track of nested branching and long cyclic structures.
3.2. Datasets
Initially, the molecular SMILE dataset is prepared by combining all the sources of raw SMILES data from two important databanks namely ChEMBL (https://www.ebi.ac.uk/chembl/) and MOSES [34] by retaining them with annotated (k_(d/I)/(B), IC/EC50) nanomolar activities. These two datasets represents about 2.9 million molecules. Then a data pre-processing does a cleanup process which not only removes the salts and stereochemical information but also filter outs long SMILES strings which are out of chemical space to sample. For proper pre-processing, a chemoinformatic tool RDKit [18] is used, which helps in the molecular cleaning process. It applies a series of normalization transforms on the functional group and recombines charges, and also neutralizes ionized acids and bases for cleaning the molecules. Finally, the RNN was trained on nearly 2.5 million canonicalized [35] SMILES retained after data pre-processing with the length between 34 and 128 characters by RDKit.
3.3. Recurrent Neural Networks
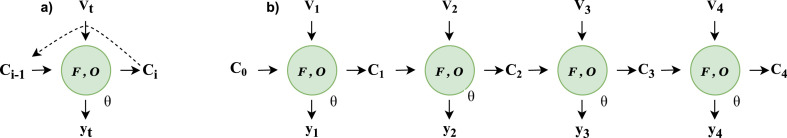
The RNN architecture process data as a sequence of input vectors V 1:n = {v 1, v 2, ….v n} by taking each vector v i in the sequence along with the initial hidden cell state c 0. The RNN takes the input and forwards through a series of gates and returns sequence of hidden cell state vectors C i:n = {c 1, c 2, … … c n} and ouput vectors = {, , ….}. The hidden cell state c i is key to the RNN which is passed over cell loops recursively as shown in Fig. 1 . The RNNs are networks with self loop, has a recursive function F, that takes some input v i and cell state c i and returns a new cell state c i+1. The RNN makes it possible to pass information from one time step to other which allow information to persist. An output function O maps cell state c i to an output vector [36].
| (1) |
| (2) |
| (3) |
Fig. 1.
Schematic diagram showing RNN.(a):Recursively defined RNN (b):Unrolled RNN with weight parameters theta sharing over all time instances.
The cell state vectors allow RNNs to learn the complex representation of data and also possess’ ability to store and persist the information for long term dependencies. If the RNN loop is unrolled, it can be thought of as a deep neural network, each sharing parameter θ to its successor network layers shown Fig. 1.
In the proposed model, a special class of RNN, namely Long Short term memory cell structures, are used, which was introduced by Hochreiter and Schmidhuber [37]. The LSTMs have the ability to regulate the cell states with the help of gates. LSTMs consists of three types of gates, a forget gate: regulates how much of previous information should be passed to next cell. An input gate: manages how much new information the cell stores and finally the output gate outputs the filtered important data. Different gates, which are neural network units decide what information is relevant to keep or forget during training and control the flow of information. Accordingly, LSTMs resolve the problem of vanishing and exploding gradient that normally occurs during backpropagation due to long sequences [38]. At any given time instant, t, LSTM network can be expressed by the following set of Equations.
| (4) |
For each gate there is set of weights and bias associated which are denoted as W f, W i, W c, W o, W v and b f, b i, b c, b o, b v these notations indicates weights and bias of forget, input, candidate cell state, output, and associated softmax layers respectively estimated through back propagation with gradient descent [39] f t, i t, , o t indicates the output of the activation functions sigmoid and tanh and a f, a i, a c, a o represents the input to the activation functions. The cell state c t acts as a memory to LSTM, at each time instance the previous cell state c t−1 multiplies with forget gate f t to decide how much information need to be carry forward then it in turn combines with input gate i t and to form a new cell state.
3.4. Model framework and training
LSTM model can be used to generate molecular sequences as the formal language (SMILES) one token at every time instant t. Generally, an LSTM tends to predict the next symbol of a given input by assigning probability distribution across potential input symbols during every time unit. If the input is of length n symbols, the model predicts (n + 1)th symbol; for example, if the model receives a sequence of characters c1ccccc, then the probability of getting next symbol would be “1” is high as it closes a ring structure benzene. At each time step t, the probability of pth token given previously generated symbols is computed through softmax function as in Equation 5.
| (5) |
Where is output of the model (logits) for the pth token at time instance t and j runs from 1 to set of p tokens. Sampling from probability distribution P(x t+1 = p| x 1,.,x t) of generating next token given already seen tokens would now generate novel molecules. After sampling token at time instance x t+1 then for the next time step t+2 the input vectors are fed into model through ouput vector and softmax to produce next sample x t+2 which again serves as input at next time instance t+3. This token by token sampling is recurring event repeated until some end of line token is encountered. While sampling characters from the model, an additional temperature factor T is employed into softmax which controls the sampling of token. For higher sampling temperature there is lot of structural diversity in generated molecules but decreases the percentage of valid SMILES generated. On other hand lower temperature leads to less structural diversity but increase in validity [40].
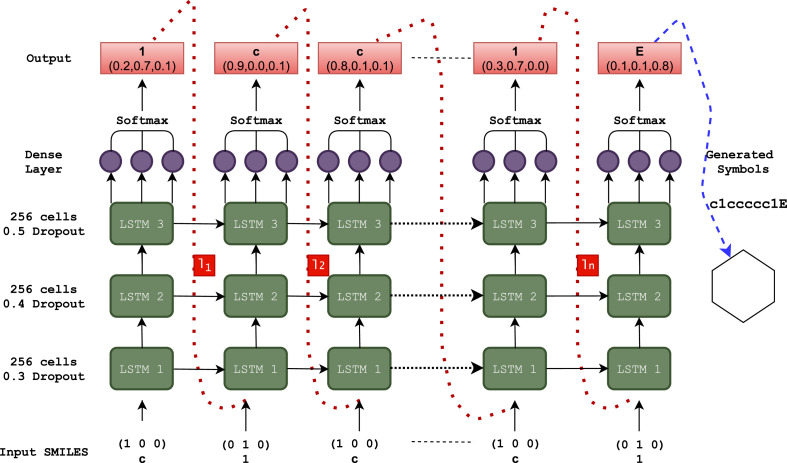
The proposed model architecture is shown in Fig. 2 . It consists of three stacked LSTM layers of 256 hidden cell units with a dropout for regularization [41]. Then it is followed by a dense output layer with softmax activation, which generates the probability of each token as described by Equation 5. The one-hot encoding [42] scheme is employed to convert the SMILES strings into input vectors. Consider a SMILE string S that contains a collection of ‘k’ symbols {s 1, s 2, …,s i, … …,s k } where s i represents a token to passed at time instance t, then a k length vector with its all entries as zeros is constructed as an input vector x t while the ith entry is one. Let {c,1,E} is symbol space of three characters, then c is represented as (1,0,0), 1 as (0,1,0), and finally E as (0,0,1) one-hot vectors respectively. After training the LSTM model, the new molecules are formed by providing a starting symbol as ‘G’ and sampling the next sequence of symbols based on prior generated tokens until an E, end of the string is encountered as shown in Fig. 2.
Fig. 2.
Model Architecture and Molecular generation process.
3.5. Model construction and feature extraction
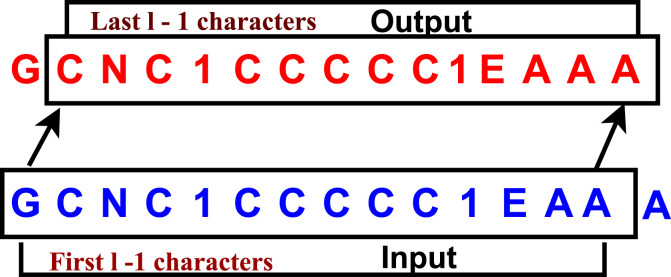
The model constructed for the task of predicting novel molecules is the sequential model with three stacked LSTM layers as shown in Fig. 2. As the inputs are SMILES strings (sequence of tokens), to generate proper molecules, it is highly important to understand the context of given input. LSTMs are best suited for such tasks as it retains sequence information by processing each token of the SMILES string based on the understanding of previous tokens. In the proposed LSTM model, each input molecule was padded to the length l of the longest SMILES string. The first l − 1 characters were taken as the input, and the last l − 1 characters were the target. For each token, the model predicts the next token in the sequence as shown in Fig. 3 . The loss was averaged over all the target tokens in all molecules. This model can capture the syntax of SMILE strings with great accuracy, and the learned probabilities are used to generate novel molecules. The proposed model extracts features by learning the SMILE string's grammar through the employed LSTM units. Every input token processed by the previous LSTM unit is reserved and can be combined with current and future time steps. They extract features by maintaining a pair of long term and short memories. The long term memories are regulated through the forget gate and short memories by input and output gates. In each LSTM unit the model chooses what to be passed from long term and what should be kept from short memory through a series of operations as described in Equation 5. This sequential reservation of features is the way by which the proposed model extracts the features.
Fig. 3.
Training procedure of proposed LSTM model.
3.6. Generator quality metrics
Once the model was trained, we sampled nearly 10,000 molecules and evaluated the quality of molecules generated as it is vital during drug design. To validate the performance of the proposed model we computed the number of valid, novel and unique molecules generated. We summarized these metrics as follows: Consider S m be number of sampled molecules, V m be the set of chemically valid molecules, N is the set molecules from training data.
-
1.
Validity (Q valid) shows percentage of molecules that are chemically valid.
-
2.
Uniqueness(Q Unique) shows percentage that is not duplicates out of total generated valid molecules.
-
3.
Novelty (Q Novel) indicates the molecules that are brand new creation which don't appear in training data.
| (6) |
| (7) |
| (8) |
3.7. Implementation details
All the deep learning models were trained and developed using Keras with Tensorflow as backend in python (v3.6.9) with high end GPUs. All the SMILE strings validation, cleaning, and calculating some physicochemical features were carried out by a chemoinformatic molecular modeling tool kit (RDKit). To minimize the energies of novel generated molecules and to convert them into appropriate ligands (pdbqt format), a virtual screening tool PyRx [43] is used. Further, AutoDock Vina, a molecular docking tool is used to visualize molecules and dock protein ligand compounds to predict binding energies of different conformations. We used OpenBabel [44], a chemical file converter, to convert between file formats and accessed protein data bank for crystal structure of novel Coronavirus protease.
4. Results and discussions
In this study, we address two major aspects; the first is to generate a large set of diverse valid molecules using generative RNNs. Second is to fine-tune the generated model to sample active molecules that specifically inhibits 3CLpro (PDB ID:6LU7) [45] protease of Covid-19. To accomplish the initial task, we have designed a staked LSTM model and trained it on a vast set of molecules to acquire the dynamics of SMILE grammar. Upon sampling, it generated valid molecules from the same training space with similar physicochemical properties. The later and most vital task is to identify novel drug candidates for main protease of novel Coronavirus. In order to achieve this, we perform transfer learning by adding a small set of commercially available known active inhibitors [19], HIV inhibitors [46] to the generated set of valid molecules. Then we fine-tuned the pre-trained model with this small dataset. Further, we sampled new molecules and performed docking studies against the crystal structure of the 3CLpro protease of Covid-19.
4.1. Novel molecule generation
Initially, the proposed model is planned to train for 50 epochs. We employed an early stopping mechanism and it was observed that by 25 epochs, the validation loss flattened, and subsequently, there was no more decrease in loss. After model training, we used the network to sample 10,000 molecules symbol by symbol. Based on the results of our experiments conducted by considering various sample temperatures the proposed model obtained an average of 69.50% validity at T = 0.30, and an average of 55.50% valid SMILES are generated at T = 1.20. The network produced better results at sample temperature T = 0.75 by improving validity to 70.50% and also 99.83% molecules that generated validly are unique, 98.99% molecules are novel. This shows the performance of the network in generating realistic molecules. All these metrics are evaluated after parsing the generated molecules from the RDKit [18] (a chemoinformatic tool for molecular modeling), which removes duplicate and cleans the molecules. It is observed that proportion of valid SMILES increased steadily and become optimal till the temperature factor T reached the value of 0.75. The proportion of valid SMILES decreased steadily with temperature when T > 0.75. This could be because of the increase in randomness of sampling [40], with T = 1.2, the model registered the lowest score which indicates the lower proportion of valid SMILES in higher temperatures. Overall, our results indicate that the model can generate diverse and novel molecules. The percentage of validity, uniqueness, and novelty at various sampling temperatures are shown in Table 1 , a good compromise of validity, uniqueness and novelty was obtained when sampling with T = 0.75.
Table 1.
The proposed model quality metrics at various sampling temperatures.
| Temperature | Validity (%) | Uniqueness (%) | Novelty (%) |
|---|---|---|---|
| 0.30 | 69.50 | 85.26 | 87.41 |
| 0.50 | 70.00 | 90.55 | 87.21 |
| 0.60 | 70.32 | 96.25 | 89.29 |
| 0.75 | 70.50 | 99.83 | 98.99 |
| 0.80 | 65.23 | 97.46 | 96.57 |
| 1.00 | 60.23 | 90.30 | 85.68 |
| 1.20 | 55.50 | 89.25 | 85.39 |
We sampled 10,000 SMILES for each temperature.
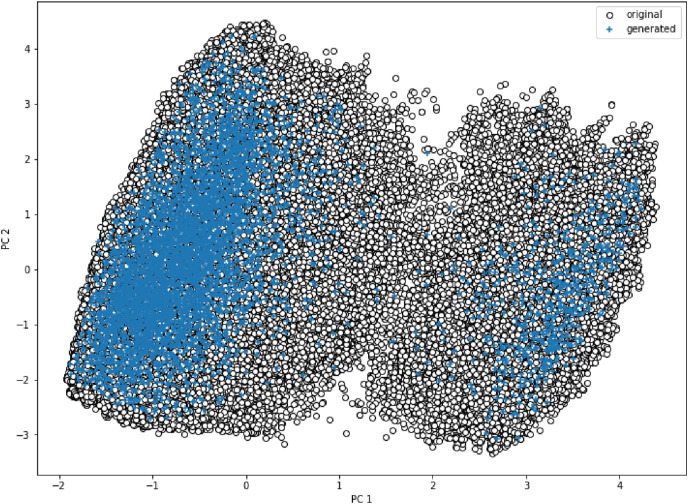
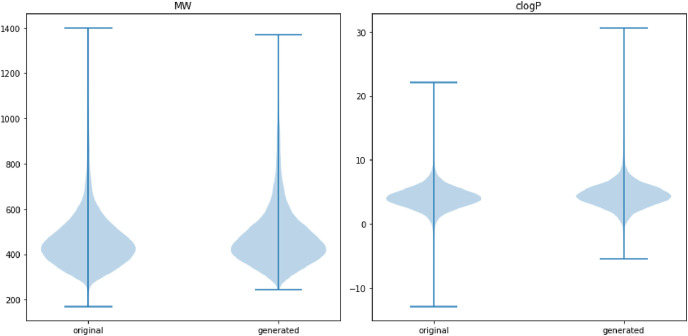
In order to determine the similarity between the molecules generated with the molecules of the training data set, we calculated several physicochemical properties like molecular weight, clogp, H-donors, H-acceptors and more with the RDKit tool. Considering nearly 24 common properties a dimensionality reduction is performed using principle component analysis (PCA) on training datasets and accordingly on newly generated molecules. Fig. 4 shows the distribution of original and newly generated molecules plotted taking the first two principle components. It is apparent that both the training (white circles) and generated molecules (blue symbols) overlap with each other disclosing the fact that our model has very well recreated the molecules from the original training space. Furthermore, from Fig. 5 we can decipher violin plots that show the distribution of molecular weights and clogp values which also infers a similar distribution of generated and original molecules. The wider section of plots represents a higher probability that members of the population take that value and the skinnier sections represent a lower probability. The clogp distribution shows how drug-like molecule is with respect to factors like bioavailability, it is clear from clogp distribution that trained and generated molecules show the same level of drug-likeness.
Fig. 4.
PCA projection of first two principle components performed over various physiochemical properties.
Fig. 5.
Molecular weight (MW) and clogp distribution on original and generated molecules.
4.2. Comparison with other baseline methods in generating realistic molecules
Computational de novo molecular design has become an increasing field of interest in the recent times. To address the issues in the molecular generation, deep generative models have shown promising direction in the design of novel drug molecules. There are several architectures to generate focused set of novel molecules which include, RNNs, autoencoders (AEs), variational autoencoders (VAE), adversarial autoencoders (AAE) [47,48] and generative adversarial networks (GANs) [49]. Here we have summarized all these baseline architectures and made a comparison between the proposed LSTM based generative model and other methods on various parameters as shown in Table 2 . Although, different deep generative models are employed for novel molecular generation, the optimization task on which they applied is different. For instance Segler et al. [33], fine tuned their model for creating molecules against a specific biological target Plasmodium falciparum (parasite). Gupta et al. [16] targeted the peroxisome proliferator-activated receptor (PAPRγ). Likewise, every architecture mentioned in Table 2 has specific biological targets. The proposed LSTM model was fine tuned against the 3CL protease of novel coronavirus. Unlike other models which show generator quality in terms of validity and novelty of molecules, we have also evaluated unique molecules generated. We summarized all results on various parameters as shown in Table 2. Based on the results obtained we strongly believe the proposed model has fairly established a direction in the discovery of novel drug candidates.
Table 2.
Comparison of generator quality metrics and various other parameters of proposed model with other baseline architectures.
| Architecture Type | Model Name | Dataset used | Size of Molecule | No.of Trained Molecules | No.of Generated Molecules | Generator quality metrics | Task |
|---|---|---|---|---|---|---|---|
| RNN and AE based Architecure | Grammar VAE [50] | ZINC | <39 heavy atoms | 250,000 | 100,000 | 7.2% (V) | Penalized logP |
| SD VAE [51] | ZINC | <39 heavy atoms | 250,000 | 100,000 | 43.5% (V) | Penalized logP | |
| AAE [52] | ChEMBL | <121 characters | 1.3 million | no data | 77.4% (V) | Drug analog generation | |
| ECAAE [53] | ZINC | <58 characters | 1.8 million | 10000 | No data | Structural analogs | |
| GAN and RNN based Architecure | ORGAN [54] | QM9 | <52 characters | 5000 | No data | 80.3% (V) | nlogP |
| ORGANIC [55] | QM9 | <10 heavy atoms | 5000 | No data | 0.2–99%(V), 86% (N) | nQED | |
| ATNC [56] | ChemDiv | <91 characters | 15000 | 157986 | 72%(V),77% (N) | No.of unique heterocycles | |
| RANC [57] | ChemDiv | <91 characters | 15000 | 896000 | 58%(V),48% (N) | No.of unique heterocycles | |
| RNN based Architecure with RL | REINVENT [58] | ChEMBL | 10 - 50 heavy atoms | 1.5million | 12800 | 94% (V), 90% (N) | Drug analog generation |
| ReLeaSE [59] | ChEMBL | No data | 1.5 million | 1 million | 95%(V),95.3% (N) | Inhibitor of JAK2 | |
| ChemTS [60] | ZINC | No data | 250,000 | No data | No data | Penalized logP | |
| RNN based Architecure | Segler et al. [33] | ChEMBL | No data | 1.4 million | 976,327 | 97.7% (V), 89.4% (N) | Plasmodium falciparum,5 - HT2A |
| Bjerrum et al. [61] | ZINC | No data | 1,611,889 | 50000 | 98% (V), 63% (N) | Retro-synthetic route of easy/medium/hard group | |
| Gupta et al. [16] | ChEMBL | 34-74 characters | 541,555 | 30107 | 93% (V), 92% (N) | PPARs, Trypsin | |
| Ours | ChEMBL and MOSES | 34-128 characters | 2.9 million | 10000 | 70.50% (V), 99.83% (N),98.99% (U) | Inhibitor of 3CLPro (novel Corona virus main protease) |
Qvalid, QUnique, QNovel are represented as V, U, N respectively. Autoencoders (AE), Adversarial autoencoder (AAE), Generative Adversarial network (GAN).
4.3. Fine tuning to generate active drug molecules to target 3CLpro protease of Covid-19
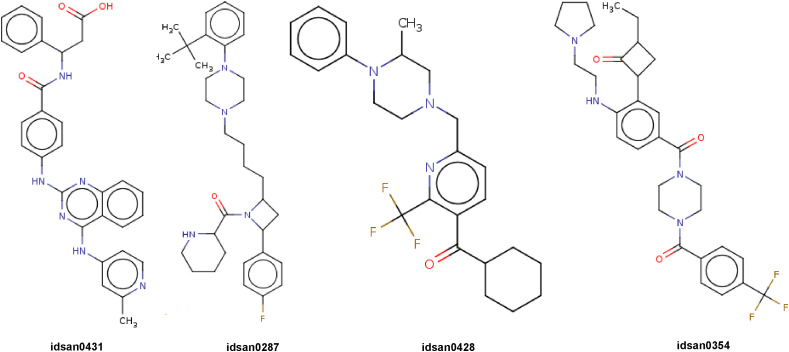
As our model is established to generate valid molecules, we further fine-tuned the model by training it on a smaller list of selected molecules. The goal is to apply unique network expertise of producing realistic molecules to the area of building compounds that are especially proficient at connecting with the major protease (3CLpro) of novel Coronaviruses. In order to generate new ligands, we have collected a small dataset of commercially available antiviral drugs, few HIV inhibitors [19] and added them to the set of validly generated molecules. Then we used this set of molecules to fine-tune with our original model. After 10 epochs of training our fine-tuned model generated a sample set of 1000 molecules of which 65% are valid smiles. Among validly generated molecules 99.68% are unique and 98.36% are novel. Next we have taken validly generated molecules and started docking using Autodock vina [62] from PyRx to find their binding affinity scores. Fig. 6 depicts few generated molecules that binds well with the novel Coronavirus main protease.
Fig. 6.
Few generated molecules that binds well with 3CL-Pro of novel Coronavirus.
4.4. Generated molecule diversity analysis
After fine-tuning the model, a closest neighbor or diversity study is performed using the Tanimoto similarity index to analyze the uniqueness of generated target specific molecules [63]. It is a commonly used measure to represent how closely two molecules are related. The similarity calculation is done based on comparing molecular 2D fingerprints which comprise of structural information about the molecules. For calculating the similarity between two molecules A and B the Tanimoto similarity index is given as in Equation 5. Here N c indicates common attributes in molecule A and B, N a represents individual attributes of A, similarly N b represents individual attributes of B. As a quantitative measure we have calculated Tanimoto similarity between existing commercial antiviral drugs, HIV drugs and Remdesivir. Table 3 shows the molecules generated after fine-tuning the model indicates larger similarity with Remdisivir currently using as therapeutic drug for the treatment of Covid-19 [[64], [65], [66]]. Bulk Tanimoto similarity with the antiviral drugs shows lesser similarity whereas for HIV they are moderately similar.
| (9) |
Table 3.
Tanimoto Similarity Measure of generated molecules with existing antiviral drugs and Remdesivir.
| Ligand Id | Generated SMILE Strings | HIV | AntiViral Drugs | Remdesivir |
|---|---|---|---|---|
| idsan0138 | COc1cc2c(Cc3ccccc3)n(-c3ccc(F)cc3)n(-c3ccc(F)cc3)n2cc1OC | 0.565 | 0.330 | 0.625 |
| idsan0624 | COc1ccc(-c2nc(C(=O)NC3CCC-CC3)cc3s2CCC3(C)C)cc1 | 0.462 | 0.295 | 0.502 |
| idsan0159 | CC(C)(C)C(=O)NC(Cc1ccccc1)C(O)CNCCCCCNC(=O)C(Cc1ccccc1)NC(=O-)CN(Cc1ccc(O)cc1)C(=O)NC(CCC(=O)O)C(=O)C(=O)NC(=CC(N) O)C(=O)O | 0.516 | 0.298 | 0.497 |
| idsan0313 | COc1ccc2c(c1)C(=O)N(CC1CC(C)NC(=O)N2C)C1c1ccc(C(=O)NCCN(C)C)cc1 | 0.468 | 0.298 | 0.494 |
| idsan0344 | CC(C)NC(=O)c1ccc(-n2nc(C(F)(F)F)cc2-c2cccc(C(=O)Nc3ccc(C(F)(F)F)cc3)c2)cc1 | 0.438 | 0.285 | 0.480 |
| idsan0354 | CC1CN(Cc2ccc(C(=O)C3CCCCC3)c(C(F)(F)F)n2)CCN1c1ccccc1 | 0.430 | 0.276 | 0.467 |
| idsan0431 | Cc1cc(Nc2nc(Nc3ccc(C(=O)NC(CC(=O)O)c4ccccc4)cc3)nc3ccccc23)ccn1 | 0.435 | 0.300 | 0.461 |
| idsan0181 | CN(C)C(=O)C1CCN(CC2CCCCC2)CC1Nc1ccc(C(=O)NC(Cc2ccccc2)C(=O)-NC(CC2CCCCC2)C(=O)O)cc1 | 0.440 | 0.274 | 0.449 |
| idsan0615 | Cc1ccccc1C(=O)NC(Cc1ccccc1)C(O)CNC1CCCN1C(=O)C(Cc1ccccc1)-NC(=O)OCc1ccccc1 | 0.480 | 0.275 | 0.446 |
| idsan0410 | Cc1ccc(-c2ccc3nc(NC(=O)N4CC4)cc(NC(C)C)c3n2)cc1 | 0.416 | 0.283 | 0.438 |
| idsan0287 | CC(C)(C)c1ccccc1N1CCN(CCCCC2CC(c3ccc(F)cc3)N2C(=O)C2CCCCN2)CC1 | 0.437 | 0.290 | 0.432 |
| idsan0049 | CC(C)(C)OC(=O)NC(C(=O)NC(Cc1ccccc1)C(O)CNCCCc1ccccc1)C(=O)-NC(CCCC=Cc1cccc2ncccc12)C(N) O | 0.474 | 0.279 | 0.425 |
| idsan0374 | Cc1ccc(C2=NN(C(=O)C(CC(=O)O)NC(=O)C(Cc3ccccc3)NC(=O)–CN3CCNCC3)CC2)cc1 | 0.408 | 0.257 | 0.418 |
| idsan0428 | CCC1CC(c2cc(C(=O)N3CCN(C(=O)c4ccc(C(F)(F)F)cc4)CC3)-ccc2NCCN2CCCC2)C1=O | 0.385 | 0.247 | 0.412 |
| idsan0040 | O=C(NCCCC1CCCCC1)c1cc(-c2cnc(-c3ccccc3)nc2)nc2ccccc12 | 0.412 | 0.271 | 0.387 |
Ligand id indicates novel molecules generated after fine tunning here top 15 molecules are selected which binds well with 3CL Protease of Covid-19.
4.5. Evaluating generated molecules that binds with novel Coronavirus main protease
Various studies performed on structural features of novel Coronavirus have lead to finding drugable targets. Chymotrypsin-like protease (3CLpro) or the main protease of novel Coronavirus is a vital target as it involves in the viral replication step. The main protease is responsible for the formation of non-structural proteins (Nsp) [67] which plays a significant role in replication, therefore we have selected 3CLpro as a drug target for novel Coronavirus and demonstrated docking studies. The molecular docking studies were conducted on PyRx a virtual screening platform used for screen libraries of compounds against potential drug targets. In PyRx we used AutoDock Vina [62] a docking platform that yields binding affinity scores (kcal/mol) between the ligands and target protein. AutoDock vina computationally estimates how well a ligand binds to its receptor of a known 3D structure. As a result, there are numerous ligand conformations obtained along with their associated binding energies. The difference between the energies of the ligand plus protein in the unbound state and the energy created by the protein and ligand complex until it achieves equilibrium is the binding energy of the conformation [68]. This value should be negative for instant binding, lower this energy more stable the ligand will be and highly probable to become a drug candidate.
4.5.1. Obtaining protein structures and ligand preparation
We obtain 3CLpro protease crystal structure (PDB ID:6LU7) from RCSB Protein Data Bank (.pdb format) and loaded it into PyRx and were converted it as macromolecules. All the newly generated SMILES after sampling from fine-tune model were saved as.csv files in pandas data frame then we converted these SMILES to molecules using molecular modeling tool RDKit. Later Open Babel (version 2.3.2) [44] is used to convert molecule by molecule into structured data format (SDF) file. The SDF file options are configured as: generation coordinates are set to 2D and output file format as MDL mol format that holds all information about molecules such as bonds, connectivity atoms. Finally these files were manually loaded into PyRx and converted into pdbqt macromolecules (ligands) as they include hydrogens necessary for binding interactions. Then we perform energy minimization on these ligands to develop a reasonable starting pose before binding. Autodock vina is now used by selecting the 6LU7 protease structure of novel Coronavirus and selected a list of ligands after energy minimization to start binding. After completion of binding it yields binding affinity scores for each ligand with the protease. In order to keep track of generated novel molecules each of them is assigned a unique identifier as ‘idsanXXXX’, when loaded in PyRx. A total of eight binding modes were created; the binding mode with the highest docking score was stored as a.pdb file and used for further examination.
4.5.2. Comparing docking scores of novel generated molecules with other drugs
Molecular docking predicts the drug's interaction with macromolecules. The strength of these interactions is measured in terms of binding affinity. The equilibrium dissociation constant (KD), which is used to assess rank order potency of molecular contacts, is commonly used to estimate and quantify binding affinity. K D is stated as the combination of concentrations of protein and ligands over concentration of protein-ligand complex. We performed blind docking simulations of newly generated molecules and other commercial antiviral drugs along with HIV drugs and Remdesivir. The blind docking expands its search space throughout the surface of the target protein structure for binding site. From the docking results, we observed that nearly 80% of the molecules generated have shown docking free energy of < −6 kcal/mol. We have listed the top 10 generated molecules that have the highest binding affinity and compared them with the binding affinity score of some drugs like Entecavir, Dolutegravir, Efavirenz, Abacavir, Ripivirine. Table 4 shows the docking scores of newly generated molecules when compared with the existing antiviral drugs and Remdesivir which are currently used for Covid-19 treatment. It is apparent that the generated molecules have shown better results and the newly generated molecule with “idsan0431” has yielded the highest binding score. Based upon the docking analysis we recommend the ligands with the highest binding scores to be considered for further investigation.
Table 4.
Docking score comparison.
| Sr.no | Main Protease with PDB ID | Ligand Id | Docking score (kcal/mol) |
|---|---|---|---|
| 1 | 3CLPro (6LU7) | idsan0431 | −8.5 |
| 2 | 3CLPro (6LU7) | idsan0119 | −8.1 |
| 3 | 3CLPro (6LU7) | idsan0539 | −7.8 |
| 4 | 3CLPro (6LU7) | idsan0008 | −7.3 |
| 5 | 3CLPro (6LU7) | idsan0223 | −7.2 |
| 6 | 3CLPro (6LU7) | idsan0029 | −7.1 |
| 7 | 3CLPro (6LU7) | idsan0240 | −7.1 |
| 8 | 3CLPro (6LU7) | idsan0344 | −7.1 |
| 9 | 3CLPro (6LU7) | idsan0146 | −7.0 |
| 10 | 3CLPro (6LU7) | idsan0410 | −6.7 |
| 11 | 3CLPro (6LU7) | Entecavir | −7.4 |
| 12 | 3CLPro (6LU7) | Dolutegravir | −6.9 |
| 13 | 3CLPro (6LU7) | Efavirenz | −6.8 |
| 14 | 3CLPro (6LU7) | Atazanavir* | −6.8 |
| 15 | 3CLPro (6LU7) | Abacavir | −6.6 |
| 16 | 3CLPro (6LU7) | Ripivirine | −6.6 |
| 17 | 3CLPro (6LU7) | ritonavir* | −6.6 |
| 18 | 3CLPro (6LU7) | Remdesivir | −5.5 |
Ligand id indicates novel molecules generated after fine tunning. HIV drugs indicated with *.
4.5.3. Binding affinity analysis of generated top candidates
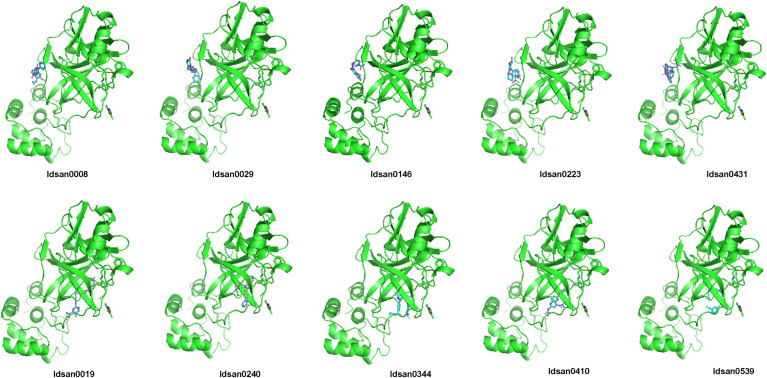
The blind molecular docking analysis of novel generated molecules and Covid-19 main protease have been screened. We used PyMOL [69] a visualization tool to depict protein ligand interactions. Fig. 7 displays the top 10 protein ligand complexes formed and their respective binding affinities are reported in Table 4 for reference. These compounds are ranked according to their binding free energy scores. The top candidate in generated molecules is ‘idsan0431’ with a predicted binding score of −8.5 kcal/mol which is higher than Remdesivir (−5.5 kcal/mol). The presence of multiple hydrogen bond acceptors and donors, as well as the formation of a strong hydrogen bond network with novel Coronavirus protease, account for the high binding affinity. The strong hydrogen bonds formed with the head, body, and tail of top candidates with different residues of SARS-CoV-2 make the interaction with the main protease even more stronger. The hydrogen bond acceptors in the therapeutic candidate compounds are crucial for binding to the novel Coronavirus protease. The hydrogen bond acceptors can establish strong bonds with the Coronavirus protease, inhibit it from replication.
Fig. 7.
Complexes formed between novel Coronavirus main protease and generated top candidates.
5. Conclusions
In this work, we propose a generative long short term memory model which successfully learned molecular grammar and generated novel molecules with the same physicochemical properties as that of training data. We demonstrate transfer learning in our model by using the original network's knowledge of constructing actual molecules. Then transmit this to the area of producing compounds that particularly bind to the novel Coronavirus 3CLpro main protease. We further demonstrated structural screening of novel generated molecules by simulating docking studies to quantitatively measure the best binding score. Based on the binding affinity score we choose the top 10 potential drug candidates and measure similarity score with commercial inhibitors. The novel molecules generated favorable binds well with the main protease with less binding free energy when compared with other existing commercial antiviral drugs including remdesivir. Based upon the generated results we believe our generative model can establish a promising direction in predicting novel molecules. The predicted drug candidates need to be further investigated in vitro and in vivo for more efficacy.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.mondiale de la Santé O., Organization W.H., et al. vol. 96. 2021. p. 36. (Weekly Epidemiological Record). [full issue], Weekly Epidemiological Record= Relevé épidémiologique hebdomadaire 96 (36) (2021) 421–444. [Google Scholar]
- 2.Zhavoronkov A., Aladinskiy V., Zhebrak A., Zagribelnyy B., Terentiev V., Bezrukov D.S., Polykovskiy D., Shayakhmetov R., Filimonov A., Orekhov P., et al. vol. 307. Insilico Medicine Hong Kong Ltd A; 2020. p. E1. (Potential Covid-2019 3c-like Protease Inhibitors Designed Using Generative Deep Learning Approaches). [Google Scholar]
- 3.Hartenfeller M., Schneider G. De novo drug design. Chemoinform. Comput. Chem. Biol. 2010:299–323. doi: 10.1007/978-1-60761-839-3_12. [DOI] [PubMed] [Google Scholar]
- 4.Sua J.N., Lim S.Y., Yulius M.H., Su X., Yapp E.K.Y., Le N.Q.K., Yeh H.-Y., Chua M.C.H. Incorporating convolutional neural networks and sequence graph transform for identifying multilabel protein lysine ptm sites. Chemometr. Intell. Lab. Syst. 2020;206 [Google Scholar]
- 5.Le N.Q.K., Ho Q.-T., Yapp E.K.Y., Ou Y.-Y., Yeh H.-Y. Deep etc: a deep convolutional neural network architecture for investigating and classifying electron transport chain's complexes. Neurocomputing. 2020;375:71–79. [Google Scholar]
- 6.Le N.Q.K., Huynh T.-T. Identifying snares by incorporating deep learning architecture and amino acid embedding representation. Front. Physiol. 2019;10:1501. doi: 10.3389/fphys.2019.01501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Amilpur S., Bhukya R. Edeepssp: explainable deep neural networks for exact splice sites prediction. J. Bioinf. Comput. Biol. 2020;18 doi: 10.1142/S0219720020500249. (04) 2050024. [DOI] [PubMed] [Google Scholar]
- 8.Dasari C.M., Bhukya R. Intersspp: investigating patterns through interpretable deep neural networks for accurate splice signal prediction. Chemometr. Intell. Lab. Syst. 2020;206 [Google Scholar]
- 9.Dasari C.M., Bhukya R. Explainable deep neural networks for novel viral genome prediction. Appl. Intell. 2021:1–16. doi: 10.1007/s10489-021-02572-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jaques N., Gu S., Bahdanau D., Hernández-Lobato J.M., Turner R.E., Eck D. International Conference on Machine Learning. PMLR; 2017. Sequence tutor: conservative fine-tuning of sequence generation models with kl-control; pp. 1645–1654. [Google Scholar]
- 11.Elgammal A., Liu B., Elhoseiny M., Mazzone M. 2017. Can: Creative Adversarial Networks, Generating” Art” by Learning about Styles and Deviating from Style Norms. arXiv preprint arXiv:1706.07068. [Google Scholar]
- 12.Yi Z., Zhang H., Tan P., Gong M. Proceedings of the IEEE International Conference on Computer Vision. 2017. Dualgan: unsupervised dual learning for image-to-image translation; pp. 2849–2857. [Google Scholar]
- 13.Choi Y., Choi M., Kim M., Ha J.-W., Kim S., Choo J. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. 2018. Stargan: unified generative adversarial networks for multi-domain image-to-image translation; pp. 8789–8797. [Google Scholar]
- 14.Weininger D. Smiles, a chemical language and information system. 1. introduction to methodology and encoding rules. J. Chem. Inf. Comput. Sci. 1988;28(1):31–36. [Google Scholar]
- 15.Mikolov T., Karafiát M., Burget L., Černockỳ J., Khudanpur S. Eleventh Annual Conference of the International Speech Communication Association. 2010. Recurrent neural network based language model. [Google Scholar]
- 16.Gupta A., Müller A.T., Huisman B.J., Fuchs J.A., Schneider P., Schneider G. Generative recurrent networks for de novo drug design. Mol. Inform. 2018;37(1–2) doi: 10.1002/minf.201700111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reymond J.-L., Ruddigkeit L., Blum L., van Deursen R. The enumeration of chemical space. Wiley Interdiscipl. Rev.: Comput. Mol. Sci. 2012;2(5):717–733. [Google Scholar]
- 18.Landrum G. 2016. Rdkit: Open-Source Cheminformatics.http://www.rdkit.org Google Scholar There is no corresponding record for this reference. [Google Scholar]
- 19.Beck B.R., Shin B., Choi Y., Park S., Kang K. Predicting commercially available antiviral drugs that may act on the novel coronavirus (sars-cov-2) through a drug-target interaction deep learning model. Comput. Struct. Biotechnol. J. 2020;18:784–790. doi: 10.1016/j.csbj.2020.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ahuja A.S., Reddy V.P., Marques O. 2020. Artificial Intelligence and Covid-19: A Multidisciplinary Approach. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Arslan H., Arslan H. A new covid-19 detection method from human genome sequences using cpg island features and knn classifier, Engineering Science and Technology. Int. J. 2021;24(4):839–847. [Google Scholar]
- 22.Keshavarzi Arshadi A., Webb J., Salem M., Cruz E., Calad-Thomson S., Ghadirian N., Collins J., Diez-Cecilia E., Kelly B., Goodarzi H., et al. Artificial intelligence for covid-19 drug discovery and vaccine development. Front. Artif. Intell. 2020;3:65. doi: 10.3389/frai.2020.00065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou Y., Wang F., Tang J., Nussinov R., Cheng F. Artificial intelligence in covid-19 drug repurposing. The Lancet Digital Health. 2020;2:67–76. doi: 10.1016/S2589-7500(20)30192-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gordon D., Jang G., Bouhaddou M., Xu J., Obernier K., O'Meara M., Guo J., Swaney D., Tummino T., Hüttenhain R., et al. A sars-cov-2-human protein-protein interaction map reveals drug targets and potential drug-repurposing. biorxiv prepr, Serv. Biol. 2020;19(4) [Google Scholar]
- 25.Li X., Yu J., Zhang Z., Ren J., Peluffo A.E., Zhang W., Zhao Y., Wu J., Yan K., Cohen D., et al. Network bioinformatics analysis provides insight into drug repurposing for covid-19. Med. Drug Discov. 2021;10 doi: 10.1016/j.medidd.2021.100090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kowalewski J., Ray A. Predicting novel drugs for sars-cov-2 using machine learning from a> 10 million chemical space. Heliyon. 2020;6(8) doi: 10.1016/j.heliyon.2020.e04639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sun Z., Deng Z.-H., Nie J.-Y., Tang J. 2019. Rotate: Knowledge Graph Embedding by Relational Rotation in Complex Space; p. 10197. arXiv preprint arXiv:1902. [Google Scholar]
- 28.Richardson P., Griffin I., Tucker C., Smith D., Oechsle O., Phelan A., Stebbing J. Baricitinib as potential treatment for 2019-ncov acute respiratory disease. Lancet. 2020;395:E30–E31. doi: 10.1016/S0140-6736(20)30304-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Makhzani A., Shlens J., Jaitly N., Goodfellow I., Frey B. 2015. Adversarial Autoencoders. arXiv preprint arXiv:1511.05644. [Google Scholar]
- 30.Tang B., He F., Liu D., Fang M., Wu Z., Xu D. bioRxiv preprint; 2020. Ai-aided Design of Novel Targeted Covalent Inhibitors against Sars-cov-2. BioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Patankar S. 2020. Deep Learning-Based Computational Drug Discovery to Inhibit the Rna Dependent Rna Polymerase: Application to Sars-Cov and Covid-19. [Google Scholar]
- 32.Zhang H., Saravanan K.M., Yang Y., Hossain M.T., Li J., Ren X., Pan Y., Wei Y. Deep learning based drug screening for novel coronavirus 2019-ncov. Interdiscipl. Sci. Comput. Life Sci. 2020;12:368–376. doi: 10.1007/s12539-020-00376-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Segler M.H., Kogej T., Tyrchan C., Waller M.P. Generating focused molecule libraries for drug discovery with recurrent neural networks. ACS Cent. Sci. 2018;4(1):120–131. doi: 10.1021/acscentsci.7b00512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Polykovskiy D., Zhebrak A., Sanchez-Lengeling B., Golovanov S., Tatanov O., Belyaev S., Kurbanov R., Artamonov A., Aladinskiy V., Veselov M., et al. Molecular sets (moses): a benchmarking platform for molecular generation models. Front. Pharmacol. 2020;11 doi: 10.3389/fphar.2020.565644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schneider N., Sayle R.A., Landrum G.A. Get your atoms in order an open-source implementation of a novel and robust molecular canonicalization algorithm. J. Chem. Inf. Model. 2015;55(10):2111–2120. doi: 10.1021/acs.jcim.5b00543. [DOI] [PubMed] [Google Scholar]
- 36.Goldberg Y. A primer on neural network models for natural language processing. J. Artif. Intell. Res. 2016;57:345–420. [Google Scholar]
- 37.Hochreiter S., Schmidhuber J. Long short-term memory. Neural Comput. 1997;9(8):1735–1780. doi: 10.1162/neco.1997.9.8.1735. [DOI] [PubMed] [Google Scholar]
- 38.Wang J., Zhang L., Guo Q., Yi Z. Recurrent neural networks with auxiliary memory units. IEEE Trans. Neural Networks Learn. Syst. 2017;29(5):1652–1661. doi: 10.1109/TNNLS.2017.2677968. [DOI] [PubMed] [Google Scholar]
- 39.Werbos P.J. Generalization of backpropagation with application to a recurrent gas market model. Neural Network. 1988;1(4):339–356. [Google Scholar]
- 40.Reverdy P., Srivastava V., Leonard N.E. Satisficing in multi-armed bandit problems. IEEE Trans. Automat. Control. 2016;62(8):3788–3803. [Google Scholar]
- 41.Srivastava N., Hinton G., Krizhevsky A., Sutskever I., Salakhutdinov R. Dropout: a simple way to prevent neural networks from overfitting. J. Mach. Learn. Res. 2014;15(1):1929–1958. [Google Scholar]
- 42.Graves A. 2013. Generating Sequences with Recurrent Neural Networks. arXiv preprint arXiv:1308.0850. [Google Scholar]
- 43.Dallakyan S., Olson A.J. Chemical Biology. Springer; 2015. Small-molecule library screening by docking with pyrx; pp. 243–250. [DOI] [PubMed] [Google Scholar]
- 44.O'Boyle N.M., Banck M., James C.A., Morley C., Vandermeersch T., Hutchison G.R. Open babel: an open chemical toolbox. J. Cheminf. 2011;3(1):1–14. doi: 10.1186/1758-2946-3-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mhatre S., Naik S., Patravale V. A molecular docking study of egcg and theaflavin digallate with the druggable targets of sars-cov-2. Comput. Biol. Med. 2021;129 doi: 10.1016/j.compbiomed.2020.104137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gao K., Nguyen D.D., Wang R., Wei G.-W. bioRxiv; 2020. Machine Intelligence Design of 2019-ncov Drugs. bioRxiv. [Google Scholar]
- 47.D. P. Kingma, M. Welling, Auto-encoding Variational Bayes, arXiv preprint arXiv:1312.6114 (2013).
- 48.Rezende D.J., Mohamed S., Wierstra D. International Conference on Machine Learning, PMLR. 2014. Stochastic backpropagation and approximate inference in deep generative models; pp. 1278–1286. [Google Scholar]
- 49.Goodfellow I., Pouget-Abadie J., Mirza M., Xu B., Warde-Farley D., Ozair S., Courville A., Bengio Y. Generative adversarial nets. Adv. Neural Inf. Process. Syst. 2014;27 [Google Scholar]
- 50.Kusner M.J., Paige B., Hernández-Lobato J.M. International Conference on Machine Learning, PMLR. 2017. Grammar variational autoencoder; pp. 1945–1954. [Google Scholar]
- 51.Dai H., Tian Y., Dai B., Skiena S., Song L. 2018. Syntax-directed Variational Autoencoder for Structured Data. arXiv preprint arXiv:1802.08786. [Google Scholar]
- 52.Blaschke T., Olivecrona M., Engkvist O., Bajorath J., Chen H. Application of generative autoencoder in de novo molecular design. Mol. Inform. 2018;37(1–2) doi: 10.1002/minf.201700123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Polykovskiy D., Zhebrak A., Vetrov D., Ivanenkov Y., Aladinskiy V., Mamoshina P., Bozdaganyan M., Aliper A., Zhavoronkov A., Kadurin A. Entangled conditional adversarial autoencoder for de novo drug discovery. Mol. Pharm. 2018;15(10):4398–4405. doi: 10.1021/acs.molpharmaceut.8b00839. [DOI] [PubMed] [Google Scholar]
- 54.Guimaraes G.L., Sanchez-Lengeling B., Outeiral C., Farias P.L.C., Aspuru-Guzik A. 2017. Objective-reinforced Generative Adversarial Networks (Organ) for Sequence Generation Models. arXiv preprint arXiv:1705.10843. [Google Scholar]
- 55.Sanchez-Lengeling B., Outeiral C., Guimaraes G.L., Aspuru-Guzik A. 2017. Optimizing Distributions over Molecular Space. An Objective-Reinforced Generative Adversarial Network for Inverse-Design Chemistry (Organic) [Google Scholar]
- 56.Putin E., Asadulaev A., Vanhaelen Q., Ivanenkov Y., Aladinskaya A.V., Aliper A., Zhavoronkov A. Adversarial threshold neural computer for molecular de novo design. Mol. Pharm. 2018;15(10):4386–4397. doi: 10.1021/acs.molpharmaceut.7b01137. [DOI] [PubMed] [Google Scholar]
- 57.Putin E., Asadulaev A., Ivanenkov Y., Aladinskiy V., Sanchez-Lengeling B., Aspuru-Guzik A., Zhavoronkov A. Reinforced adversarial neural computer for de novo molecular design. J. Chem. Inf. Model. 2018;58(6):1194–1204. doi: 10.1021/acs.jcim.7b00690. [DOI] [PubMed] [Google Scholar]
- 58.Olivecrona M., Blaschke T., Engkvist O., Chen H. Molecular de-novo design through deep reinforcement learning. J. Cheminf. 2017;9(1):1–14. doi: 10.1186/s13321-017-0235-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Popova M., Isayev O., Tropsha A. Deep reinforcement learning for de novo drug design. Sci. Adv. 2018;4(7) doi: 10.1126/sciadv.aap7885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yang X., Zhang J., Yoshizoe K., Terayama K., Tsuda K. Chemts: an efficient python library for de novo molecular generation. Sci. Technol. Adv. Mater. 2017;18(1):972–976. doi: 10.1080/14686996.2017.1401424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bjerrum E.J., Threlfall R. 2017. Molecular Generation with Recurrent Neural Networks (Rnns) arXiv preprint arXiv:1705. [Google Scholar]
- 62.Trott O., Olson A.J. Autodock vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010;31(2):455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chevillard F., Kolb P. Scubidoo: a large yet screenable and easily searchable database of computationally created chemical compounds optimized toward high likelihood of synthetic tractability. J. Chem. Inf. Model. 2015;55(9):1824–1835. doi: 10.1021/acs.jcim.5b00203. [DOI] [PubMed] [Google Scholar]
- 64.Ferner R.E., Aronson J.K. 2020. Remdesivir in Covid-19. [DOI] [PubMed] [Google Scholar]
- 65.Sun D. Remdesivir for treatment of covid-19: combination of pulmonary and iv administration may offer additional benefit. AAPS J. 2020;22:1–6. doi: 10.1208/s12248-020-00459-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liang C., Tian L., Liu Y., Hui N., Qiao G., Li H., Shi Z., Tang Y., Zhang D., Xie X., et al. A promising antiviral candidate drug for the covid-19 pandemic: a mini-review of remdesivir. Eur. J. Med. Chem. 2020 doi: 10.1016/j.ejmech.2020.112527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mody V., Ho J., Wills S., Mawri A., Lawson L., Ebert M.C., Fortin G.M., Rayalam S., Taval S. Identification of 3-chymotrypsin like protease (3clpro) inhibitors as potential anti-sars-cov-2 agents. Commun. Biol. 2021;4(1):1–10. doi: 10.1038/s42003-020-01577-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Raschka S. 2014. Molecular Docking, Estimating Free Energies of Binding, and Autodock's Semi-empirical Force Field.http://sebastianraschka. com/Articles/2014_autodo ck_energycomps. html# table-of-contents can be found under. [Google Scholar]
- 69.DeLano W.L., et al. Pymol: an open-source molecular graphics tool. CCP4 Newsl. Protein Crystallogr. 2002;40(1):82–92. [Google Scholar]