Abstract
There is limited information on how eukaryotic RNA polymerases (Pol) recognize their cognate preinitiation complex. We have characterized a polypeptide copurifying with yeast Pol III. This protein, C17, was found to be homologous to a mammalian protein described as a hormone receptor. Deletion of the corresponding gene, RPC17, was lethal and its regulated extinction caused a selective defect in transcription of class III genes in vivo. Two-hybrid and coimmunoprecipitation experiments indicated that C17 interacts with two Pol III subunits, one of which, C31, is important for the initiation reaction. C17 also interacted with TFIIIB70, the TFIIB-related component of TFIIIB. The interaction domain was found to be in the N-terminal, TFIIB-like half of TFIIIB70, downstream of the zinc ribbon and first imperfect repeat. Although Pol II similarly interacts with TFIIB, it is notable that C17 has no similarity to any Pol II subunit. The data indicate that C17 is a novel specific subunit of Pol III which participates together with C34 in the recruitment of Pol III by the preinitiation complex.
Transcription of small genes by RNA polymerase III (Pol III) involves a multistep assembly of transcription factors into a preinitiation complex which recruits RNA Pol III (for a review, see reference 55). The multisubunit factor TFIIIC functions in promoter recognition and acts as an assembly factor to direct the binding of the initiation factor TFIIIB to an upstream gene position. Once assembled into a highly stable protein-DNA complex at Pol III promoters, TFIIIB can direct multiple rounds of transcription by Pol III in vitro in the absence of TFIIIC (28, 29). TFIIIB is composed of three loosely associated polypeptides, the TATA-binding protein (TBP) (26, 30), a general transcription factor utilized for transcription by all eukaryotic and archeal RNA polymerases (21, 44), B" or TFIIIB90, which has no equivalent among the other general transcription factors (33, 45, 47), and Brf1 or TFIIIB70, which is related to the Pol II factor TFIIB (9, 13, 36). TFIIIB70 appears as a central bridging factor between the basal components of the class III transcription machinery, in a way that is much similar to the role proposed for TFIIB in the case of class II genes. TFIIIB70 is the first target of TFIIIC-DNA complex in the TFIIIB assembly process; it interacts with TBP and with TFIIIC and Pol III via their τ131 and C34 subunits, respectively (10, 23, 34, 52). Similarly, TFIIB is a target of various transcriptional activators that facilitate its incorporation into the preinitiation complex; there it binds TBP and is involved directly in Pol II-TFIIF complex recruitment through interaction with both Pol II and TFIIF (40, 46). TFIIB has further been implicated in selecting the Pol II transcription start site (19, 41, 43). Interestingly, the N-terminal half of TFIIIB70 is structurally homologous to the full length of TFIIB. Both proteins contain at the extreme amino terminus a cysteine-rich, putative zinc-binding domain that, in the case of TFIIB, is implicated in recruiting the Pol II-TFIIF complex, and both contain a core of two imperfect direct repeats that, in TFIIB, interact with TBP-DNA complex (35, 39). The role of these protein domains is apparently different in TFIIIB70 as it is the C-terminal half of the protein that interacts strongly with TBP (2, 10, 14, 32), while the N-terminal half interacts with τ131, a subunit of TFIIIC (10).
There is only limited information on how Pol III recognizes the preinitiation TFIIIB-TFIIIC-DNA complex. Pol III is the most complex of all three forms of nuclear RNA polymerases. Saccharomyces cerevisiae Pol III comprises 17 polypeptides with sizes ranging from 10 to 160 kDa. The genes for 15 subunits have been characterized. All of them proved to be essential for cell viability (11). Two large subunits, related to β′ and β subunits of Escherichia coli RNA polymerase, harbor the active site and form the enzyme structural platform; five subunits are shared with Pol I and Pol II, and two additional ones are shared with Pol I only. The complexity of Pol III is due to the presence of a set of specific polypeptides that have no counterpart in Pol I or Pol II. Three such specific subunits (C82, C34, and C31) interact with each other and spontaneously dissociate from an enzyme form bearing a mutation in the N-terminal zinc-binding domain of the largest subunit C160 (53). These subunits probably play a major role in preinitiation complex recognition, since a small deletion of the C-terminal end of C31 impaired RNA chain initiation (50) and mutations in C34 that affect its interaction with TFIIIB70 also impaired the efficiency of Pol III recruitment and open complex formation (8). Protein-DNA cross-linking of the binary or ternary Pol III transcription complexes has allowed the mapping of eight Pol III subunits over the SUP4 tRNA gene (5, 6, 42). In the binary open complex, the C34 subunit was seen to extend the farthest upstream, a placement in favor of its role in TFIIIB recognition. Recently, human RNA polymerase has been shown to contain a set of three subunits homologous to yeast C82, C34, and C31 subunits (51). These three human polypeptides formed a subcomplex required for transcription initiation, and the human polypeptide homologous to C34 was shown to interact independently with human TBP and with hTFIIIB90, the human homolog of yeast TFIIIB70. These data strongly suggest that this particular set of subunits directs enzyme recruitment by the preinitiation complex and triggers open complex formation. Although the importance of C34-TFIIIB70 interaction has well been established in vivo and in vitro (8), it seems unlikely that Pol III recruitment relies on a unique binary protein-protein contact whereas the whole transcription complex comprises 26 polypeptides amounting to about 1,500 kDa (55).
In the present work we have extended our characterization of yeast Pol III to the C17 polypeptide by cloning the corresponding gene and showing that C17 is a specific and essential component of the enzyme. Using the two-hybrid system, we have sought to identify the elements of the Pol III transcription machinery which interact with C17. We could demonstrate that C17 interacts with TFIIIB70 and could map the protein domains involved in this interaction. Interestingly, it was the TFIIB-like region that was found to interact with C17.
MATERIALS AND METHODS
DNA constructions and yeast strains.
Two oligonucleotides harboring BamHI and XhoI restriction sites, respectively, were used to amplify the open reading frame (ORF) and surrounding sequences of the S. cerevisiae RPC17 gene by PCR. The resulting 1,025-bp genomic DNA fragment was cloned into a centromeric yeast vector, pRS316 (48), to produce the centromeric plasmid pYc17 (CEN6 URA3 RPC17). Two oligonucleotides were used to insert by PCR-mediated mutagenesis the BamHI and NcoI restriction sites at the initiation codon of RPC17 and the SalI and BamHI restriction sites just after the stop codon of RPC17. After sequencing, the BamHI-BamHI amplified DNA fragment was cloned into the centromeric vector pCM185 (CEN TRP1) (17) to produce pCMc17. The BamHI-SalI DNA fragment was cloned into pET28b (Novagen), giving pET-C17. The NcoI-BamHI DNA fragment was inserted into the corresponding sites of pACTII for fusion with GAL4 amino acids 768 to 881 [GAL4(768-881)] and pAS2 for fusion with GAL4(1-147), giving pACT-C17 and pAS-C17, respectively. The yeast strains used in this study were constructed by genetic techniques based on transformation of lithium acetate-treated cells, sexual mating, and tetrad analysis using standard media and growth conditions (4).
Disruption of RPC17 gene.
The whole RPC17 ORF was disrupted by the direct deletion method described by Baudin et al. (7). Two 59-mer oligonucleotides were used to amplify by PCR a DNA fragment containing the HIS3 gene and stop modules flanked by RPC17 promoter and terminator sequences. The 1,090-bp PCR-amplified DNA fragment was directly used to transform strain YPH501 (48). Correct integration of the HIS3 cassette in the resulting diploid disruptants (YOL14) was verified by PCR analysis. These diploid cells were then transformed with plasmid pYc17. The haploid strain YMLF1, deleted at the RPC17 locus and complemented by centromeric plasmid pYc17, was obtained after sporulation and tetrad dissection of the diploid strain.
In vivo labeling of RNA.
Cells expressing pCMc17 or pYc17 were grown to an A600 of 0.4 in Casamino Acids medium supplemented with adenine (20 μg/ml) and, when indicated, doxycycline (5 μg/ml). Labeling and preparation of RNAs have been described previously (20). Small RNA species were analyzed by loading and separating equal amounts of RNA (6 μg per lane) by electrophoresis on a 7 M urea gel (6% polyacrylamide). RNA species revealed by ethidium bromide staining of the gel were quantified using a ImageQuant software (Molecular Dynamics).
Purification of RNA Pol III and recombinant proteins.
Micropurification of RNA polymerase III was performed as described by Huet et al. (24) from a wild-type strain or from strain YGVS003 complemented with the human gene encoding the hABC23 subunit (provided by M. Vigneron, Strasbourg, France). The subunit composition of Pol III was analyzed by electrophoresis on a 12% polyacrylamide gel in denaturing conditions. Recombinant Rpc17 protein (rC17) fused at its N terminus to six histidines and to the T7 epitope was obtained from E. coli BL21(DE3)pLysS transformed with plasmid pET-C17. Recombinant histidine-tagged TFIIIB70 (rTFIIIB70) was expressed in E. coli cells from plasmid pSH360 (a gift from Steve Hahn). Cell culture, protein induction, and crude extract preparation were performed essentially as indicated elsewhere (10) except that buffer A (20 mM Tris-HCl [pH 8.0], 10% glycerol, 0.1% NP-40, 50 mM KCl, 10 mM β-mercaptoethanol) was used as the lysing buffer. Crude cell extracts containing rC17 or rTFIIIB70 were recovered after centrifugation at 40,000 rpm for 45 min at 4°C.
Immunoprecipitation experiments.
For C17-TFIIIB70 interaction, 3 μg of mouse monoclonal anti-T7 antibody (Novagen) was incubated for 30 min at 10°C with 40 μl of magnetic beads (8 × 108 beads/ml in phosphate-buffered saline containing 0.1% bovine serum albumin coated with rat monoclonal antibodies directed against mouse immunoglobulin G2b (Dynal M450). For TBP-TFIIIB70 interaction, 1.2 μg of rabbit polyclonal anti-TBP antibody was incubated for 30 min at 10°C with 40 μl of magnetic beads coated with sheep monoclonal antibodies directed against rabbit immunoglobulin G (Dynal M280). Crude extracts containing or not containing rC17 (60 μg) were incubated with the same amount of crude extracts with or without rTFIIIB70 in the transcription buffer (20 mM HEPES-KOH pH 7.9, 0.1 mM EDTA, 1 mM dithiothreitol, 5 mM MgCl2, 100 mM KCl, 10% glycerol) for 45 min at 25°C. Similarly, purified recombinant TBP (240 ng) was mixed with crude extracts (60 μg) containing or not containing rTFIIIB70. After extensive washing first in phosphate-buffered saline containing 0.1% bovine serum albumin then in transcription buffer, beads were incubated overnight with gentle shaking at 10°C with the crude extract mixtures and then washed three times with 200 μl of transcription buffer. After incubation for 3 min at 95°C, immunoprecipitated proteins were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and revealed by Western blotting with anti-T7, antipentahistidine (Qiagen), or anti-TFIIIB70 antibodies, using an Amersham enhanced chemiluminescence kit. Immune complexes were quantitated with ImageQuant software (Molecular Dynamics).
Two-hybrid assays.
Expression of GAL4(1-147)-C17 and GAL4(768-881)-C17 fusion proteins from plasmids pAS17 and pACT17, respectively, was verified by Western blot analysis on yeast crude extracts, using polyclonal antibodies directed against GAL4. The expression of fusion proteins with TFIIIB70 or truncated forms of TFIIIB70 has been previously assayed (10). GAL1-lacZ activation assays were then performed as described previously (10) after transformation of the yeast strain Y526 with combinations of plasmids. β-Galactosidase activity was measured in yeast extracts exactly as described previously (52).
RESULTS
RPC17 is an essential gene conserved from yeast to human.
Yeast Pol III copurifies with 16 to 17 polypeptides through different purification procedures, including a selective immunoadsorption step using antibodies directed to the two largest subunits (25). In our earlier work, we noted the presence of a protein band with a stoichiometry of about 2 that migrated slightly behind AC19 subunit (shared by Pol I and Pol III) and that coimmunopurified with the other subunits (25). The identification of that component as another Pol III subunit has been delayed by its erratic migration rate, relative to AC19, upon SDS-PAGE. All Pol III preparations contain a polypeptide that migrates either slightly faster or slower than AC19 or show one AC19 band with an abnormally high stoichiometry. This polypeptide was also seen in Pol III preparations purified by a newly devised procedure (Fig. 1, lane 1). Based on Coomassie blue staining, a stoichiometry of 2 was found for this polypeptide, the same as that of ABC27 (25). It was also present, with the same stoichiometry, in a mutant form of Pol III that had the human subunit hABC23 substituted for yeast ABC23 (Fig. 1, lane 2). As the wild-type and mutant Pol III showed different chromatography patterns on heparin Hyper D and on Mono Q columns, this observation prompted us to characterize this polypeptide, named C17, which copurifies with Pol III and to clone its gene.
FIG. 1.
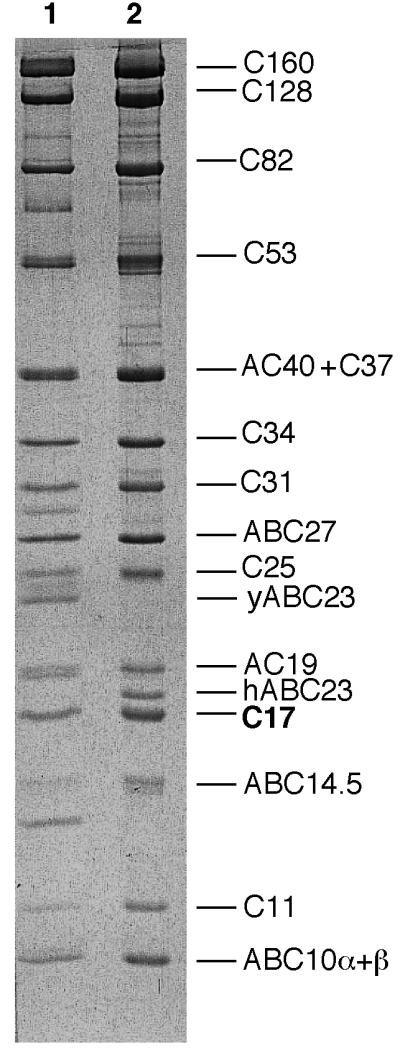
A 17-kDa polypeptide in RNA polymerase III from S. cerevisiae. Pol III purified as described in Materials and Methods was analyzed by electrophoresis under denaturing conditions in an SDS–12% polyacrylamide gel. Proteins were then stained by Coomassie blue. Lane 1, Pol III from a wild-type strain; lane 2, Pol III from a strain in which yABC23 is replaced by its human counterpart hABC23.
Microsequence data were obtained for three tryptic peptides of electrophoretically purified C17 subunit. Three peptide sequences were obtained: VHLY, LQIV, and FLTDLEK. Comparison to the NCBI (National Center for Biotechnology Information) nonredundant database showed that all of these peptide sequences were contained in a unique 161-amino-acid protein encoded by a 486-bp ORF located on chromosome X of S. cerevisiae. This gene, identified as YJL011C, is unique in the yeast genome and was named RPC17. The protein encoded by RPC17 has a theoretical pI of 5.5 and a predicted molecular mass of 17.7 kDa. Comparison of the C17 protein sequence with the NCBI database or with the Stanford database, using the BLAST program server (1), revealed a strong and puzzling sequence similarity (Fig. 2) between C17 and mammalian proteins (31% identities and 46% similarities with the human protein) unrelated to transcription and described as a receptor component for the calcitonin gene-related peptide (CGRP) (37, 38). Similarities were also found with proteins of unknown function from Candida albicans (49% similarities), chicken (45% similarities), and Caenorhabditis elegans (38% similarities). The S. cerevisiae protein was longer than its mammalian orthologs because of the presence of two insertions of 13 and 24 amino acids separating two regions of strong sequence similarity at the N- and C-terminal regions (Fig. 2).
FIG. 2.
Sequence analysis of C17. The yeast C17 protein sequence (GenBank accession no. Z49286) was aligned with different orthologs, as indicated. C. al., Candida albicans protein of unknown function, unfinished fragment of complete genome, Stanford database; C. el., Caenorhabditis elegans protein of unknown function (accession no. CAA87050); H. sap., Homo sapiens CGRP receptor component protein (accession no. AF073792); C. porc., Cavia porcellus CGRP receptor component protein (accession no. U50188); M. mus., Mus musculus CGRP receptor component protein (accession no. AF028242); G. gal., Gallus gallus protein of unknown function, partial sequence (accession no. D26313). The complete sequences are shown except for C. elegans and G. gallus proteins, were residues that showed no homology with C17 are not included. Amino acid positions are indicated; residues identical in five sequences are boxed, and conserved substitutions are shaded.
In contrast to Pol I and Pol II, all subunits of the Pol III enzyme characterized so far are essential for cell viability. To test whether C17 was essential for growth, we deleted RPC17 by a PCR method (7). The whole ORF of RPC17 was replaced by a DNA fragment containing the yeast HIS3 selectable marker surrounded by stop codon modules and inserted in the antisense direction with respect to RPC17. The resulting diploid cells had one chromosome with the deleted allele of RPC17 and one chromosome harboring the wild-type RPC17+ gene. Tetrad analysis of the meiotic offspring generated two viable and two nonviable spores per meiosis. All viable segregants were His−, indicating that the RPC17+ allele was required for growth.
To confirm this conclusion, we cloned RPC17 by PCR amplification of genomic DNA, using primers complementary to sequences located about 400 bp upstream and 100 bp downstream of the coding sequence. This PCR-amplified fragment was inserted into a centromeric yeast vector, pRS316 (48), to produce the centromeric plasmid pYc17 (CEN6 URA3 RPC17). The sequences of five clones obtained from two independent PCR runs were determined; in each case, the sequence diverged from the sequence in the NCBI database by a C→G transversion in the second base of codon 159, resulting in the substitution of glycine for alanine (A159G). The heterozygous rpc17-Δ::HIS3/RPC17 strain was transformed with pYc17 and sporulated. His+ haploid segregants, bearing the rpc17-Δ::HIS3 mutation but harboring the plasmid-borne RPC17 gene (strain YMLF1), were viable.
A new subunit of yeast RNA Pol III.
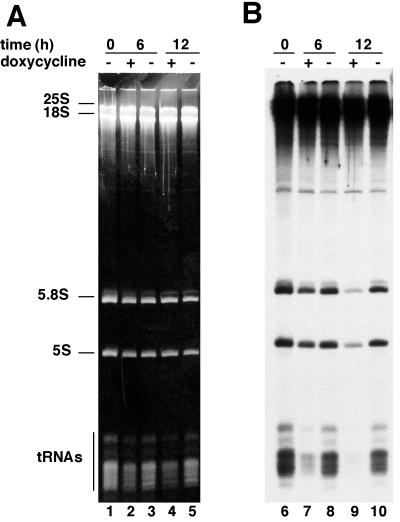
To demonstrate that the 17-kDa polypeptide participates in Pol III transcription system, we cloned the RPC17 ORF into the centromeric vector pCM185 (CEN TRP1) under the control of a tetracycline-regulatable promoter to analyze the effect of RPC17 gene extinction on class III gene expression in vivo (17). The resulting plasmid pCMc17 (CEN TRP1 RPC17) was exchanged for the centromeric plasmid pYc17 (CEN URA3 RPC17) in the strain YMLF1, generating strain YMLF2, which lacks the chromosomal copy of RPC17 but survives by expressing the plasmid-borne RPC17 gene. Plasmid pCMc17 allows modulation of the expression of the essential RPC17 gene by changing doxycycline concentration in the growth medium without imposing any metabolic changes. Analysis of the repression kinetics was carried out with strains YMLF1 and YMLF2. The cell growth rate with or without doxycycline (5 μg/ml) was monitored over 48 h. To study the effect of the repression of RPC17 expression on RNA synthesis, the in vivo labeling of RNA was analyzed after 6 or 12 h of growth in the presence of 5 μg of doxycycline per ml. After a pulse-labeling with tritiated uracil, the RNA species were extracted, analyzed by loading equal amounts of RNA on a 7 M urea gel for electrophoresis, and revealed by ethidium bromide staining (Fig. 3A) or autoradiography (Fig. 3B). The cell growth rate of strain YMLF1 was unaffected by the addition of the antibiotic (90-min generation time). In contrast, YMLF2 cell growth rate began to decline detectably 2 h after the addition of doxycycline to the medium. The doubling time of YMLF2 cells progressively increased from 90 min to 125 and 480 min 6 h and 24 h after the addition of doxycycline, respectively. The presence of the antibiotic did not lead to a complete growth arrest: 48 h after the addition of doxycycline, YMLF2 strain still grew with the same doubling time of 480 min, suggesting that the expression of RPC17 was not completely shut off. Figure 3A shows the steady-state levels of the RNA species synthesized by Pol I (25S RNA, 18S RNA, and 5.8S RNA) or Pol III (5S RNA and tRNAs), revealed by ethidium bromide staining of the gel. A decrease in the steady-state amounts of tRNAs relative to 5.8S RNA was observed when YMLF2 cells were grown in the presence of the antibiotic (compare lanes 2 and 4 to lanes 3 and 5). The tRNA/5.8S RNA ratio showed a 35% decline in tRNA levels 6 h after the addition of doxycycline (lane 2) a 55% decline 12 h after the addition of doxycycline (lane 4), and a 70% decline 24 h after the addition of doxycycline (data not shown). As shown in Fig. 3B, this decrease in the steady-state amounts of tRNA was accounted for a marked inhibition of the de novo synthesis of tRNA. Six hours after the addition of antibiotic (5 μg/ml) in the culture medium, the tRNA synthesis rate was reduced fivefold and became negligible after 12 h (Fig. 3B, lanes 7 and 9). The 5S RNA and 5.8S RNA synthesis started to be affected after 12 h of doxycycline treatment (Fig. 3B, lane 9). The coregulation of these Pol III and Pol I transcripts derives from the fact that the 5S RNA is required for the processing of the 27SB precursor of the mature 25S RNA and 5.8S RNA (15). Thus, the effect of the switch off of RPC17 expression on the synthesis of tRNA and 5S RNA in vivo was as expected for the depletion of an important subunit of Pol III (18, 49).
FIG. 3.
In vivo synthesis of small RNA species after repression of RPC17 gene expression. Strain YMLF2 harboring plasmid pCMc17, which allows the modulation of RPC17 expression by changing doxycycline concentration in the growth medium, was grown at 30°C with (+) or without (−) the addition of doxycycline (5 μg/ml). Tritiated uracil incorporation was allowed for 10 min after 6 or 12 h of growth, as indicated. RNA species were extracted and analyzed by electrophoresis on a 7 M urea gel (6% polyacrylamide), using equal amounts of RNA (6 μg per lane). (A) Ethidium bromide staining of RNA; (B) autoradiography of the dried gel shown in panel A.
C17 interacts with C11, C31, and TFIIIB70.
To gain some insight into the function of the C17 subunit, we used the two-hybrid system to study the interactions between C17 and all components of the Pol III system cloned so far. This approach has been used successfully to elucidate the function of key components of the Pol III transcription system: the interaction of τ131 with TFIIIB70 (10) as well as with TFIIIB90 (47), the interaction of C34 with TFIIIB70 (52), and other interactions between several TFIIIC or Pol III subunits (16). The RPC17 ORF was fused to the DNA-binding domain (amino acids 1 to 147) or to the transcriptional activation domain (amino acids 768 to 881) of the yeast GAL4 protein, and all combinations between these C17 fusion proteins and the TFIIIB (TFIIIB70, TFIIIB90, and TBP), TFIIIC (τ138, τ131, τ95, τ91, τ60, and τ55), TFIIIA, and Pol III (C160, C128, C82, C53, AC40, C34, C31, ABC27, ABC23, AC19, C17, ABC14.5, C11, ABC10α, and ABC10β) complementary fusion proteins were assayed (3, 10, 52). Activation of the lacZ reporter gene was estimated by β-galactosidase assays of selected transformants. The interaction between TFIIIB70 and τ131 (10) was used as a reference. Background levels of β-galactosidase activity were obtained in most cases. In contrast, high levels of β-galactosidase activity were detected when C17, fused to the transcriptional activation domain, was assayed with the C11 complementary fusion (Table 1). The reciprocal combination gave lower but significant levels of β-galactosidase activity. When C17 was fused to the DNA binding domain of GAL4 and assayed with the C31 reciprocal fusion, high levels of β-galactosidase activity were also measured. Interestingly, a significant interaction (about 20-fold the background level) was also observed between C17 and TFIIIB70. These observations on the interaction of C17 with C11, C31, and TFIIIB70 strengthened the conclusion that C17 belongs to Pol III and suggested a role for C17 in Pol III recruitment by TFIIIB.
TABLE 1.
In vivo interaction of C17 with other proteins of the RNA polymerase III transcription system using the two-hybrid system
| Fusion proteins | β-Galactosidase activitya |
|---|---|
| GAL4(1–147)-C17, GAL4(768–881) | 2.4 |
| GAL4(1–147)-C17, GAL4(768–881)-TFIIIB70 | 47 |
| GAL4(1–147)-C17, GAL4(768–881)-C11 | 19 |
| GAL4(1–147)-C17, GAL4(768–881)-C31 | 111 |
| GAL4(768–881)-C17, GAL4(1–147) | 12.6 |
| GAL4(768–881)-C17, GAL4(1–147)-TFIIIB70 | 11 |
| GAL4(768–881)-C17, GAL4(1–147)-C11 | 420 |
| GAL4(768–881)-C17, GAL4(1–147)-C31 | 18.6 |
| GAL4(768–881)-τ131, GAL4(1–147)-TFIIIB70 | 218 |
Nanomoles of o-nitrophenyl β-d-galactoside hydrolyzed per minute and per milligram of protein.
C17 interacts with the TFIIB-related domain of TFIIIB70.
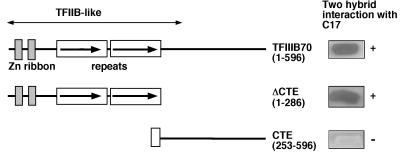
To delineate more precisely the protein domains involved in the interaction between C17 and TFIIIB70, we used the two-hybrid assay to investigate the interaction between C17 and two deleted forms of TFIIIB70 (10). As shown in Fig. 4, the N-terminal half of TFIIIB70 (ΔCTE, residues 1 to 286) was found to be as efficient as the whole protein in activating the reporter gene in combination with the C17 fusion (46 and 47 U of β-galactosidase activity, respectively). In contrast, the C-terminal half of TFIIIB70 (CTE, residues 253 to 596) did not detectably interact with C17. Therefore, the N-terminal part of TFIIIB70 was sufficient to interact with C17 in this assay.
FIG. 4.
Interaction of C17 with wild-type or mutant TFIIIB70 proteins. The two-hybrid system was used to investigate protein-protein interactions between C17 and TFIIIB70. The ORF of C17 was fused in frame with the GAL4 DNA-binding domain (amino acids 1 to 147). Wild-type or mutant TFIIIB70 ORF was fused in frame with the GAL4 activation domain (amino acids 768 to 881). Numbers in parentheses indicate the TFIIIB70 amino acids present in the fusion protein. TFIIIB70 motifs homologous to TFIIB are indicated. Transcription activation of the lacZ reporter gene was assayed by growing the transformed cells on selective medium and overlaying with 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) agar. White (−) and blue (+) coloration of cell patches on X-Gal plates is indicated.
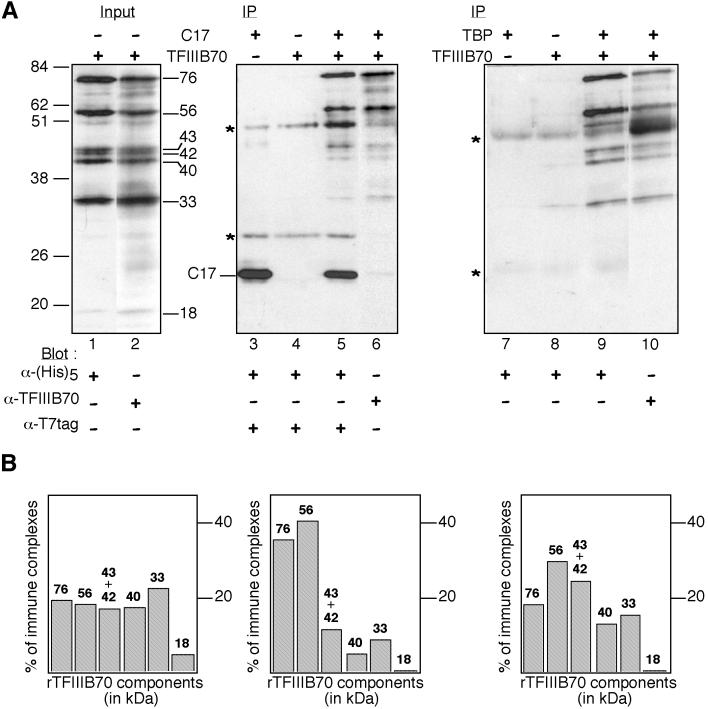
To confirm by an independent method that C17 can directly interact with TFIIIB70, we used a coimmunoprecipitation assay. T7-tagged C17 and TFIIIB70 histidine tagged at its C terminus were expressed separately in E. coli BL21(pLysS). Bacterial crude extracts were mixed and incubated in a transcription buffer at 25°C for 45 min and then subjected to immunoprecipitation using an anti-T7 monoclonal antibody. The protein mixtures were added to magnetic beads precoated with anti-T7 antibodies, the beads were washed, and bound proteins were eluted with SDS. The input and eluted proteins were subsequently analyzed by SDS-PAGE and immunoblotting, using anti-T7, antipentahistidine, or anti-TFIIIB70 antibodies (Fig. 5A). As shown in Figure 5A, the crude protein extracts used for immunoprecipitation contained, in addition to the full-length rTFIIIB70 protein of 76 kDa, several polypeptides that were recognized by anti-TFIIIB70 antibodies (lane 2); these protein bands were characteristic of the proteolyzed band pattern obtained upon bacterial expression of His-tagged TFIIIB70 (22, 23). Most of these polypeptides, ranging from 18 to 56 kDa, were also recognized by antibodies directed to the carboxy-terminal histidine tag (compare lanes 1 and 2), showing that they corresponded to N-terminally deleted forms of rTFIIIB70. Neither the full-length rTFIIIB70 nor the truncated form of rTFIIIB70 was retained on the beads in the absence of rC17 (lane 4). In contrast, the full-length rTFIIIB70 as well as several truncated polypeptides coimmunoprecipitated with T7-tagged C17 (lanes 5 and 6). The coimmunoprecipitation of C17 with rTFIIIB70 polypeptides was also observed when purified recombinant proteins were used instead of bacterial crude extracts, as well as in a reciprocal experiment where anti-TFIIIB70 antibodies were used instead of anti-T7 for immunoprecipitation (data not shown). These results confirmed the interaction between C17 and TFIIIB70 observed with the two-hybrid system (Fig. 4).
FIG. 5.
Coimmunoprecipitation assay of C17 and N-terminally truncated TFIIIB70 fragments. Crude extracts (60 μg) with (+) or without (−) T7-tagged C17 or His6-tagged rTFIIIB70 were preincubated for 45 min at 25°C and then mixed with magnetic beads coated with anti-T7 antibodies. Crude extracts (60 μg) with or without rTFIIIB70 were also preincubated with purified recombinant TBP (240 ng), as indicated, and then mixed with magnetic beads coated with anti-TBP antibodies. The beads were washed, and bound proteins were eluted by boiling the beads in loading buffer. (A) Western blot analysis. The input and bound proteins were analyzed by Western blotting using anti-T7, antipentahistidine, or anti-TFIIIB70 antibodies, as indicated. Asterisks indicate positions of immunoglobulin heavy and light chains; positions and molecular masses (in kilodaltons) of marker bands are indicated at the left. The molecular masses (in kilodaltons) of full-length and truncated rTFIIIB70 polypeptides present in crude extracts are shown. The position of rC17, migrating at 23 kDa, is indicated. IP, immunoprecipitation. (B) Quantification of rTFIIIB70 immune complexes. The immune complexes shown in panel A revealed with anti-TFIIIB70 antibodies (lanes 2, 6, and 10) were quantified with ImageQuant software. The results for each component of rTFIIIB70 are shown as percentage of total immune complexes.
We took advantage of the presence of N-terminally truncated forms of rTFIIIB70 in the bacterial extracts to confirm and map the selective interaction of rC17 with the TFIIB-related domain of rTFIIIB70 that was revealed by the two-hybrid system (Fig. 4). The immune complexes revealed by anti-TFIIIB70 antibodies (Fig. 5A, lanes 2 and 6) were quantified. The results, given as percentage of each rTFIIIB70 polypeptide present in the input or in the immunopurified fraction, are shown in Fig. 5B. The rTFIIIB70 input fraction contained five major polypeptides ranging from 76 to 33 kDa, each corresponding to about 20% of total immune complexes. When C17 was incubated with rTFIIIB70, about 80% of the immunopurified polypeptides corresponded to the 76- and 56-kDa species, which were found in the same ratio as in the input fraction. Therefore, an N-terminal deletion of about 20 kDa (i.e., the Zn-binding domain plus the first imperfect repeat) did not detectably affect C17-TFIIIB70 interaction. On the other hand, C-terminal fragments of ≈40 kDa or less were selectively lost during the immunopurification. As a control experiment, we studied the rTFIIIB70 polypeptides which selectively immunoprecipitated with TBP. Purified recombinant TBP was incubated with the same crude extracts containing the rTFIIIB70 variants, the protein mixtures were subjected to immunoprecipitation using polyclonal anti-TBP antibodies, and the resulting immune complexes were quantified. In previous work, we found by far-western experiments that TBP interacts with the carboxy-terminal extension of TFIIIB70 and, more precisely, with carboxy-terminal fragments of rTFIIIB70 larger than 30 kDa (10, 22). As shown in Fig. 5A (lanes 9 and 10) and B, this result was confirmed by the immunoprecipitation assay. As expected, rTFIIIB70 polypeptides ranging from the full-length product of 76 kDa to 33 kDa (but not the 18-kDa proteolytic component) were all retained to similar extents when incubated with TBP. These results confirmed that the major C17-binding domain of rTFIIIB70 lay in the TFIIB-related domain.
DISCUSSION
Transcription by RNA polymerase III involves the formation of a preinitiation complex by template-bound transcription factors, and the selective recruitment of the enzyme. The modalities of Pol III recruitment are still poorly understood. At least two subunits of Pol III (C34 and C31) are involved in Pol III recruitment and/or RNA chain initiation (8, 50). C34 interacts with TFIIIB70, one component of TFIIIB, and this interaction plays a critical role in enzyme recruitment and open complex formation (8). Here we have characterized a novel subunit of yeast Pol III, C17, and shown by protein-protein interaction assays that it may contribute to the specific recognition of TFIIIB. The essentiality of C17 suggests that its role is not redundant with that of C34.
The possibility that C17 was another subunit of Pol III had not been seriously considered because of its erratic electrophoretic migration (slower or faster than AC19) and of its high stoichiometry, close to 2, based on Coomassie blue staining (Fig. 1). Furthermore, the identification of C17 as a close homolog to a mammalian hormone receptor (37, 38) was not very encouraging. Nevertheless, the data obtained in vivo and in vitro convincingly demonstrate that C17 is a bona fide subunit of Pol III: first, C17 is essential for cell viability, like all the other Pol III subunits; extinction of RPC17 gene expression in vivo causes a strong defect in tRNA synthesis; and finally, C17 was found to interact with at least two Pol III subunits and with a critical component of TFIIIB. In comparison to other specific subunits of Pol III (unrelated to Pol I and Pol II subunits), it is striking that the level of sequence conversion from yeast to human is higher in the case of C17 than in the case of C82 or C31. It is therefore likely that the mammalian proteins homologous to C17 belong to Pol III, which seems at odds with their proposed role as a hormone receptor. The functional replacement of the RPC17 gene by the human gene would deserve to be attempted, although the mammalian homolog of C53 (27), C34 (J. C. Andrau and I. Brun, unpublished results) or C11 (12) subunits could not replace their yeast counterparts.
The in vivo extinction of RPC17 gene expression caused a strong decrease of the de novo synthesis of tRNA that led to a significant fall in the steady-state amounts of tRNA (from 35 to 70% after 6 h or 24 h of antibiotic treatment, respectively). A threefold drop in tRNA levels following C17 gene shutoff was accompanied by a fivefold decline in the cell growth rate. This observation correlates well with a model where the regulation of Pol III transcription would provide a mechanism for regulating the cell growth rate (reference 54 and references therein).
Protein-protein interaction assays showed that C17 binds specifically to at least two components of the Pol III transcription apparatus that play a critical role in chain initiation, namely, C31 on the polymerase side and TFIIIB70. These data support a model in which C17 contributes to the productive positioning of the Pol III by TFIIIB on class III promoters. All evidence garnered to date pointed to C34-TFIIIB70 interaction as the major determinant in Pol III recruitment. Of eight Pol III subunits that could be cross-linked on SUP4 promoter DNA in initiation complexes, C34 extended the furthest upstream, from bp +6 to −17 (5, 6, 42). C34 was found to interact in vitro and in vivo with TFIIIB70 (34, 52), and mutations in C34 decreasing its interaction with TFIIIB70 impaired Pol III recruitment (a defect that could be compensated in vitro by increasing the concentration of the mutant enzyme) or, more unexpectedly, affected the ability of Pol III to form open complexes (8). A proper C34-TFIIIB70 interaction was suggested to be important to trigger the isomerization step required to shift the enzyme into an initiation-competent configuration (8). Interestingly, TFIIIB-DNA complexes assembled with certain truncated versions of TFIIIB70 or B" were found to recruit transcriptionally inactive forms of Pol III unable to initiate transcription (31). These observations underscored the role of TFIIIB-polymerase interactions at postrecruitment steps of transcription initiation (31). C31 is another candidate for participating in the Pol III recruitment/activation step. This polypeptide is part of a subcomplex of three subunits, C82, C34, and C31, conserved in human Pol III, that dissociates from the enzyme under adverse conditions and that is required for initiation (52). A Pol III mutant enzyme containing a truncated form of C31 was shown to be specifically deficient in chain initiation, suggesting a defect in TFIIIB interaction or in open complex formation (50). C82, which belongs to the labile subcomplex, may also be involved in transcription initiation. It was unexpected, therefore, to discover an additional Pol III-specific subunit, C17, that contributes to TFIIIB recognition. The observation that several Pol III-specific subunits appear to be involved in enzyme recruitment and chain initiation strongly suggests that multiple contacts with the preinitiation complex are needed to position the enzyme properly at the start site and to set off the complex process of RNA chain initiation.
C17 being essential for growth, its role cannot be redundant with that of C34 or the other specific subunits. Protein-protein interaction assays indicate that C17 provides another grip on TFIIIB by interacting with the TFIIB-related part of TFIIIB70. GST pull-down experiments showed that C34 also binds to the N-terminal half of TFIIIB70 (34). The interaction of both Pol III-specific subunits, C34 and C17, with the TFIIB-like part of TFIIIB70 accounts well for the major role of this TFIIIB domain in Pol III recruitment. Indeed, the N-terminal half of TFIIIB70 (residues 1 to 282) was shown to retain nearly full transcriptional activity in vitro in a TFIIIC-independent assay using the yeast SNR6 gene (31). The interaction domains on TFIIIB70 for C34 and C17 remain to be mapped precisely. It is known that C34 interacts with the direct repeat region, not with the zinc-binding domain (34). Mutagenesis of TFIIIB70 also revealed C-terminal residues critical for interaction with C34 and TBP (2). Our coimmunoprecipitation experiments showed that a TFIIIB70 fragment lacking ≈20 kDa at its N terminus interacted with C17 as efficiently as the full-length protein. This observation suggested that the C17-binding domain lay C terminal of the first imperfect repeat, within the TFIIB-like half of TFIIIB70.
As mentioned above, the role of C34 and C17 is unlikely to be redundant since both are essential components. Pol III-DNA cross-linking experiments using 4-S-dTMP as the “zero distance” photo-cross-linking agent revealed a direct contact of several Pol III-specific subunits with DNA: C82, C34, and C31, a triad of subunits involved in chain initiation (5). Two unidentified small subunits (18 to 14 kDa) were also found to contact DNA in ternary transcription complexes. Therefore, there is the possibility that C17, like C34, contacts both TFIIIB70 and DNA in the transcription complex. A more precise mapping and mutagenesis of C17-TFIIIB70 interaction domains will hopefully reveal the role of this interaction in Pol III recruitment, start site selection, and/or some subsequent steps leading to RNA chain initiation.
ACKNOWLEDGMENTS
We thank Michel Riva and Françoise Bouet for help in peptide microsequencing, Christian Mark for assistance in sequence analysis, Emmanuel Favry for technical support, and Marc Vigneron for his generous gift of strain YGVS003. G.P. was supported by a fellowship from the French Ministère de l'Enseignement Supérieur et de la Recherche.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Andrau J-C, Sentenac A, Werner M. Mutagenesis of yeast TFIIIB70 reveals C-terminal residues critical for interaction with TBP and C34. J Mol Biol. 1999;288:511–520. doi: 10.1006/jmbi.1999.2724. [DOI] [PubMed] [Google Scholar]
- 3.Arrebola R, Manaud N, Rozenfeld S, Marsolier M C, Lefebvre O, Carles C, Thuriaux P, Conesa C, Sentenac A. τ91, an essential subunit of yeast TFIIIC, cooperates with τ138 in DNA binding. Mol Cell Biol. 1998;18:1–9. doi: 10.1128/mcb.18.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1994. [Google Scholar]
- 5.Bartholomew B, Braun B R, Kassavetis G A, Geiduschek E P. Probing close DNA contacts of RNA polymerase III transcription complexes with the photoactive nucleoside 4-thiodeoxythymidine. J Biol Chem. 1994;269:18090–18095. [PubMed] [Google Scholar]
- 6.Bartholomew B, Durkovich D, Kassavetis G A, Geiduschek E P. Orientation and topography of RNA polymerase III in transcription complexes. Mol Cell Biol. 1993;13:942–952. doi: 10.1128/mcb.13.2.942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baudin A, Ozier-Kalogeropoulos O, Denouel A, Lacroute F, Cullin C. A simple and efficient method for direct gene deletion in Saccharomyces cerevisiae. Nucleic Acids Res. 1993;21:3329–3330. doi: 10.1093/nar/21.14.3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brun I, Sentenac A, Werner M. Dual role of the C34 subunit of RNA polymerase III in transcription initiation. EMBO J. 1997;16:5730–5741. doi: 10.1093/emboj/16.18.5730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Buratowski S, Zhou H. A suppressor of TBP mutations encodes an RNA polymerase III transcription factor with homology to TFIIB. Cell. 1992;71:221–230. doi: 10.1016/0092-8674(92)90351-c. [DOI] [PubMed] [Google Scholar]
- 10.Chaussivert N, Conesa C, Shaaban S, Sentenac A. Complex interactions between yeast TFIIIB and TFIIIC. J Biol Chem. 1995;270:15353–15358. doi: 10.1074/jbc.270.25.15353. [DOI] [PubMed] [Google Scholar]
- 11.Chédin S, Ferri M-L, Peyroche G, Jourdain S, Lefebvre O, Werner M, Carles C, Sentenac A. The yeast RNA polymerase III transcription machinery: a paradigm for eukaryotic gene activation. Cold Spring Harbor Symp Quant Biol. 1998;63:381–389. doi: 10.1101/sqb.1998.63.381. [DOI] [PubMed] [Google Scholar]
- 12.Chédin S, Riva M, Schultz P, Sentenac A, Carles C. The RNA cleavage activity of RNA polymerase III is mediated by an essential TFIIS-like subunit and is important for transcription termination. Genes Dev. 1998;12:3857–3871. doi: 10.1101/gad.12.24.3857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Colbert T, Hahn S. A yeast TFIIB-related factor involved in RNA polymerase III transcription. Genes Dev. 1992;6:1940–1949. doi: 10.1101/gad.6.10.1940. [DOI] [PubMed] [Google Scholar]
- 14.Colbert T, Lee S, Schimmack G, Hahn S. Architecture of protein and DNA contacts within the TFIIIB-DNA complex. Mol Cell Biol. 1998;18:1682–1691. doi: 10.1128/mcb.18.3.1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deschampesme A-M, Koroleva O, Leger-Sylvestre I, Gas N, Camier S. Assembly of 5S ribosomal RNA is required at a specific step of the pre-rRNA processing pathway. J Cell Biol. 1999;145:1369–1380. doi: 10.1083/jcb.145.7.1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Flores A, Briand J-F, Gadal O, Andrau J-C, Rubbi L, Van Mullem V, Boschiero C, Goussot M, Marck C, Carles C, Thuriaux P, Sentenac A, Werner M. A protein-protein interaction map of yeast RNA polymerase III. Proc Natl Acad Sci USA. 1999;96:7815–7820. doi: 10.1073/pnas.96.14.7815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gari E, Piedrafita L, Aldea M, Herrero E. A set of vectors with a tetracycline-regulatable promoter system for modulated gene expression in Saccharomyces cerevisiae. Yeast. 1997;13:837–848. doi: 10.1002/(SICI)1097-0061(199707)13:9<837::AID-YEA145>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 18.Gudenus R, Mariotte S, Moenne A, Ruet A, Mémet S, Buhler J-M, Sentenac A, Thuriaux P. Conditional mutants of RPC160, the gene encoding the largest subunit of RNA polymerase C in Saccharomyces cerevisiae. Genetics. 1988;119:517–526. doi: 10.1093/genetics/119.3.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hawkes N A, Roberts S G. The role of human TFIIB in transcription start site selection in vitro and in vivo and in vivo. J Biol Chem. 1999;14:14337–14343. doi: 10.1074/jbc.274.20.14337. [DOI] [PubMed] [Google Scholar]
- 20.Hermann-Le Denmat S, Werner M, Sentenac A, Thuriaux P. Suppression of yeast RNA polymerase III mutations by FHL1, a gene coding for a fork head protein involved in rRNA processing. Mol Cell Biol. 1994;14:2905–2913. doi: 10.1128/mcb.14.5.2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hernandez N. TBP, a universal eukaryotic transcription factor? Genes Dev. 1993;7:1291–1308. doi: 10.1101/gad.7.7b.1291. [DOI] [PubMed] [Google Scholar]
- 22.Huet J, Conesa C, Carles C, Sentenac A. A cryptic DNA binding domain at the COOH terminus of TFIIIB70 affects formation, stability, and function of preinitiation complexes. J Biol Chem. 1997;272:18341–18349. doi: 10.1074/jbc.272.29.18341. [DOI] [PubMed] [Google Scholar]
- 23.Huet J, Conesa C, Manaud N, Chaussivert N, Sentenac A. Interactions between yeast TFIIIB components. Nucleic Acids Res. 1994;22:3433–3439. doi: 10.1093/nar/22.16.3433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huet J, Manaud N, Dieci G, Peyroche G, Conesa C, Lefebvre O, Ruet A, Riva M, Sentenac A. RNA polymerase III and class III transcription factors from Saccharomyces cerevisiae. Methods Enzymol. 1996;273:249–267. doi: 10.1016/s0076-6879(96)73024-0. [DOI] [PubMed] [Google Scholar]
- 25.Huet J, Riva M, Sentenac A, Fromageot P. Yeast RNA polymerase C and its subunits. Specific antibodies as structural and functional probes. J Biol Chem. 1985;260:15304–15310. [PubMed] [Google Scholar]
- 26.Huet J, Sentenac A. The TATA-binding protein participates in TFIIIB assembly on tRNA genes. Nucleic Acids Res. 1992;20:6451–6454. doi: 10.1093/nar/20.24.6451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ittmann M, Ali J, Greco A, Basilico C. The gene complementing a temperature-sensitive cell cycle mutant of BHK cells is the human homologue of the yeast RPC53 gene, which encodes a subunit of RNA polymerase C (III) Cell Growth Differ. 1993;4:503–511. [PubMed] [Google Scholar]
- 28.Joazeiro C A P, Kassavetis G A, Geiduschek E P. Identical components of yeast transcription factor IIIB are required and sufficient for transcription of TATA box-containing and TATA-less genes. Mol Cell Biol. 1994;14:2798–2808. doi: 10.1128/mcb.14.4.2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kassavetis G A, Braun B R, Nguyen L H, Geiduschek E P. S. cerevisiae TFIIIB is the transcription initiation factor proper of RNA polymerase III, while TFIIIA and TFIIIC are assembly factors. Cell. 1990;60:235–245. doi: 10.1016/0092-8674(90)90739-2. [DOI] [PubMed] [Google Scholar]
- 30.Kassavetis G A, Joazeiro C A P, Pisano M, Geiduschek E P, Colbert T, Hahn S, Blanco J A. The role of the TATA-binding protein in the assembly and function of the multisubunit yeast RNA polymerase III transcription factor, TFIIIB. Cell. 1992;71:1055–1064. doi: 10.1016/0092-8674(92)90399-w. [DOI] [PubMed] [Google Scholar]
- 31.Kassavetis G A, Kumar A, Letts G A, Geiduschek E P. A post-recruitment function for the RNA polymerase III transcription-initiation factor IIIB. Proc Natl Acad Sci USA. 1998;95:9196–9201. doi: 10.1073/pnas.95.16.9196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kassavetis G A, Kumar A, Ramirez E, Geiduschek E P. Functional and structural organization of Brf, the TFIIB-related component of the RNA polymerase III transcription initiation complex. Mol Cell Biol. 1998;18:5587–5599. doi: 10.1128/mcb.18.9.5587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kassavetis G A, Nguyen S T, Kobayashi R, Kumar A, Geiduschek E P, Pisano M. Cloning, expression, and function of TFC5, the gene encoding the B" component of the Saccharomyces cerevisiae RNA polymerase III transcription factor TFIIIB. Proc Natl Acad Sci USA. 1995;92:9786–9790. doi: 10.1073/pnas.92.21.9786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Khoo B, Brophy B, Jackson S P. Conserved functional domains of the RNA polymerase III general transcription factor BRF. Genes Dev. 1994;8:2879–2890. doi: 10.1101/gad.8.23.2879. [DOI] [PubMed] [Google Scholar]
- 35.Lagrange T, Kapanidis A N, Tang H, Reinberg D, Ebright R H. New core promoter element in RNA polymerase II-dependent transcription: sequence-specific DNA binding by transcription factor IIB. Genes Dev. 1998;12:34–44. doi: 10.1101/gad.12.1.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.López-De-León A, Librizzi M, Puglia K, Willis I. PCF4 encodes an RNA polymerase III transcription factor with homology to TFIIB. Cell. 1992;71:211–220. doi: 10.1016/0092-8674(92)90350-l. [DOI] [PubMed] [Google Scholar]
- 37.Luebke A E, Dahl G P, Roos B A, Dickerson I M. Identification of a protein that confers calcitonin gene-related peptide responsiveness to oocytes by using a cystic fibrosis transmembrane conductance regulatory assay. Proc Natl Acad Sci USA. 1996;93:3455–3460. doi: 10.1073/pnas.93.8.3455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Naghashpour M, Rosenblatt M I, Dickerson I M, Dahl G P. Inhibitory effect of calcitonin gene-related peptide on myometrial contractility is diminished at parturition. Endocrinology. 1997;138:4207–4214. doi: 10.1210/endo.138.10.5447. [DOI] [PubMed] [Google Scholar]
- 39.Nikolov D B, Chen H, Halay E D, Usheva A A, Hisatake K, Lee D K, Roeder R G, Burley S K. Crystal structure of a TFIIB-TBP-TATA-element ternary complex. Nature. 1995;377:119–128. doi: 10.1038/377119a0. [DOI] [PubMed] [Google Scholar]
- 40.Orphanides G, Lagrange T, Reinberg D. The general transcription factors of RNA polymerase II. Genes Dev. 1996;10:2657–2683. doi: 10.1101/gad.10.21.2657. [DOI] [PubMed] [Google Scholar]
- 41.Pardee T S, Bangur C S, Ponticelli A S. The N-terminal region of yeast TFIIB contains two adjacent functional domains involved in stable RNA polymerase II binding and transcription start site selection. J Biol Chem. 1998;273:17859–17864. doi: 10.1074/jbc.273.28.17859. [DOI] [PubMed] [Google Scholar]
- 42.Persinger J, Bartholomew B. Mapping the contacts of yeast TFIIIB and RNA polymerase III at various distances from the major groove of DNA by DNA photoaffinity labeling. J Biol Chem. 1996;271:33039–33046. doi: 10.1074/jbc.271.51.33039. [DOI] [PubMed] [Google Scholar]
- 43.Pinto I, Ware D E, Hampsey M. The yeast SUA7 gene encodes a homolog of human transcription factor TFIIB and is required for normal start site selection in vivo. Cell. 1992;68:977–988. doi: 10.1016/0092-8674(92)90040-j. [DOI] [PubMed] [Google Scholar]
- 44.Qureshi S A, Bell S D, Jackson S P. Factor requirements for transcription in the Archaeon Sulfolobus shibatae. EMBO J. 1997;16:2927–2936. doi: 10.1093/emboj/16.10.2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Roberts S, Miller S J, Lane W S, Lee S, Hahn S. Cloning and functional characterization of the gene encoding the TFIIIB90 subunit of RNA polymerase III transcription factor TFIIIB. J Biol Chem. 1996;271:14903–14909. doi: 10.1074/jbc.271.25.14903. [DOI] [PubMed] [Google Scholar]
- 46.Roeder R G. The role of general initiation factors in transcription by RNA polymerase II. Trends Biochem Sci. 1996;21:327–335. [PubMed] [Google Scholar]
- 47.Rüth J, Conesa C, Dieci G, Lefebvre O, Düsterhöft A, Ottonello S, Sentenac A. A suppressor of mutations in the class III transcription system encodes a component of yeast TFIIIB. EMBO J. 1996;15:1941–1949. [PMC free article] [PubMed] [Google Scholar]
- 48.Sikorski R S, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stettler S, Mariotte S, Riva M, Sentenac A, Thuriaux P. An essential and specific subunit of RNA polymerase III (C) is encoded by gene RPC34 in Saccharomyces cerevisiae. J Biol Chem. 1992;267:21390–21395. [PubMed] [Google Scholar]
- 50.Thuillier V, Stettler S, Sentenac A, Thuriaux P, Werner M. A mutation in the C31 subunit of Saccharomyces cerevisiae RNA polymerase III affects transcription initiation. EMBO J. 1995;14:351–359. doi: 10.1002/j.1460-2075.1995.tb07009.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang Z, Roeder R G. Three human RNA polymerase III-specific subunits form a subcomplex with a selective function in specific transcription initiation. Genes Dev. 1997;11:1315–1326. doi: 10.1101/gad.11.10.1315. [DOI] [PubMed] [Google Scholar]
- 52.Werner M, Chaussivert N, Willis I M, Sentenac A. Interaction between a complex of RNA polymerase III subunits and the 70-kDa component of transcription factor IIIB. J Biol Chem. 1993;268:20721–20724. [PubMed] [Google Scholar]
- 53.Werner M, Hermann-Le Denmat S, Treich I, Sentenac A, Thuriaux P. Effect of mutations in a zinc binding domain of yeast RNA polymerase C (III) on enzyme function and subunit association. Mol Cell Biol. 1992;12:1087–1095. doi: 10.1128/mcb.12.3.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.White R J. Regulation of RNA polymerases I and III by the retinoblastoma protein: a mechanism for growth control? Trends Biochem Sci. 1997;22:77–80. doi: 10.1016/s0968-0004(96)10067-0. [DOI] [PubMed] [Google Scholar]
- 55.White R J. RNA polymerase III transcription. Austin, Tex: Molecular Biology Intelligence Unit, R. G. Landes Company; 1998. [Google Scholar]