Abstract
The sirtuin (SIRT) protein family has been of major research interest over the last decades because of their involvement in aging, cancer, and cell death. SIRTs have been implicated in gene and metabolic regulation through their capacity to remove acyl groups from lysine residues in proteins in an NAD+-dependent manner, which may alter individual protein properties as well as the histone–DNA interaction. Since SIRTs regulate a wide range of different signaling cascades, a fine-tuned homeostasis of these proteins is imperative to guarantee the function and survival of the cell. So far, however, how exactly this homeostasis is established has remained unknown. Here, we provide evidence that neuronal SIRT degradation in Parkinson’s disease (PD) models is executed by autophagy rather than the proteasome. In neuronal Lund human mesencephalic (LUHMES) cells, all seven SIRTs were substrates for autophagy and showed an accelerated autophagy-dependent degradation upon 1-methyl-4-phenylpyridinium (MPP+) mediated oxidative insults in vitro, whereas the proteasome did not contribute to the removal of oxidized SIRTs. Through blockade of endogenous H2O2 generation and supplementation with the selective radical scavenger phenothiazine (PHT), we could identify H2O2-derived species as the responsible SIRT-oxidizing agents. Analysis of all human SIRTs suggested a conserved regulatory motif based on cysteine oxidation, which may have triggered their degradation via autophagy. High amounts of H2O2, however, rapidly carbonylated selectively SIRT2, SIRT6, and SIRT7, which were found to accumulate carbonylation-prone amino acids. Our data may help in finding new strategies to maintain and modify SIRT bioavailability in neurodegenerative disorders.
Subject terms: Proteolysis, Neurodegeneration
Introduction
The sirtuin (SIRT) protein family and their influence on epigenetics and metabolism have been studied extensively. As such, several SIRTs were correlated to longevity, even across species, and their decline is considered a hallmark of senescence [1]. Indeed, SIRT-activating small molecules are actively pursued in human anti-aging trials [2]. In cancer, however, SIRTs show an astonishing duality, as they can act as either tumor suppressors or oncogenes [3].
SIRTs may be considered peculiar as they require NAD+ as a cofactor but do not reduce it the way other proteins usually do. Instead, SIRTs cleave the nicotinamide group of the ADP-ribose unit and use the latter as an acceptor for the acyl-group they remove from lysine residues [4]. This, however, puts SIRTs in immediate competition with other enzymes that require NAD+. If, for example, poly ADP-ribose polymerases consume high levels of NAD+, mitophagy is compromised through SIRT inhibition [5, 6].
Thus, proper homeostasis of SIRTs is essential to maintain cellular prosperity. Homeostasis of SIRTs, like the homeostasis of the entire proteome, is maintained by their assembly and disassembly, folding, refolding, and degradation [7]. The mechanisms by which dysfunctional SIRTs or excess SIRTs are degraded, however, are only marginally characterized [8].
Aging cells frequently accumulate oxidized biomolecules, which is especially critical in neurons representing a postmitotic, none-replenishable cell type. Indeed, the accumulation of oxidative damage is a hallmark of different neurodegenerative disorders like Parkinson’s disease (PD), Alzheimer’s disease, or amyotrophic lateral sclerosis [9–11]. Autophagy has been implied as a major contributor to remove oxidized and cross-linked proteins [12]. Indeed, previous studies revealed that aging cells might preferentially adopt autophagy to maintain protein homeostasis [13].
SIRTs indeed do contribute to autophagy by directly regulating different components of the autophagic machinery through deacylation. For example, SIRT1 reportedly regulates cytosolic availability of microtubule-associated protein 1 light chain 3 beta (MAP1LC3B, also known as LC3 or LC3B), an autophagosomal component of major importance [14, 15]. Similar observations were published in vivo concerning the acetylation status of ATG5 and ATG7 [16].
In this study, we aimed to investigate systematically how and to what extent increased endogenous reactive oxygen species (ROS) affect human SIRTs, and by which means oxidized or damaged SIRTs are removed or salvaged.
Results
Protein levels of most human SIRTs decrease under oxidative stress
First, we investigated how SIRT protein levels would respond to increased endogenous ROS, induced by the PD models 1-methyl-4-phenylpyridinium (MPP+), rotenone, and paraquat in vitro, using LUHMES cells. MPP+ and rotenone share a similar pathomechanism, as both inhibit the complex I of the mitochondrial respiratory chain. Paraquat mostly acts as a cytosolic redox-cycler [17]. To verify whether SIRT dynamics were related to increased ROS, we included three control treatments. First, the uncoupler FCCP induces mitochondrial stress without immediately increasing ROS formation. Second, we applied the antioxidant phenothiazine (PHT), an established, selective radical scavenger in PD models [18–20]. Finally, a PHT control group was monitored.
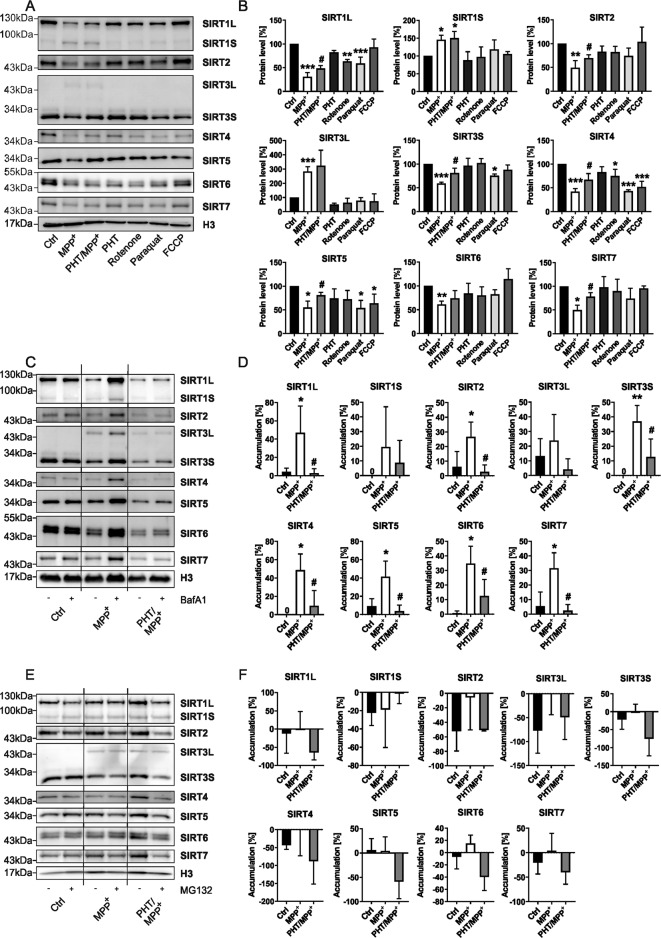
MPP+ at sublethal concentration caused a significant decrease of the main isoforms of all SIRTs (SIRT1–7) in LUHMES cells (Fig. 1A, B). With the exception of SIRT6, these decreases were significantly attenuated by concomitant PHT administration. Two SIRTs presented as long and short isoforms, SIRT1 as SIRT1L at ~130 kDa (main) and SIRT1S at ~75 kDa (minor), and SIRT3 as SIRT3L at ~43 kDa (minor) and SIRT3S at ~28 kDa (main). Interestingly, both minor isoforms, SIRT1S and SIRT3L, were significantly increased in MPP+-treated cells and unaffected by PHT co-treatment. Rotenone treatment caused a significant decline in SIRT1L and SIRT4, while paraquat additionally affected SIRT3S and SIRT5 levels significantly. Interestingly, the non-oxidative uncoupler FCCP induced a selective loss of SIRT4 and SIRT5, possibly through the induction of mitophagy [21]. Sole PHT treatment never affected SIRT protein levels significantly.
Fig. 1. Induction of endogenous ROS provokes a disruption of SIRT protein homeostasis promoted by their autophagic degradation.
A Representative immunoblots of all SIRTs were obtained from differentiated LUHMES cells after 48 h treatments with MPP+ (10 µM), PHT (20 nM), rotenone (10 nM), paraquat (100 µM), or FCCP (1 mM). H3 was used as a loading control. B Bar graphs showing the total SIRT protein levels of the blots presented in a. “*” indicates significant differences compared to the control group, while “#” indicates significant differences compared to the MPP+-treated group. Symbol number indicates the grade of significance with *p < 0.05, ** p < 0.01, and ***p < 0.001. C Representative immunoblots of all SIRTs obtained from differentiated LUHMES cells after 48 h treatments with MPP+ (10 µM) and PHT (20 nM) with and without BafA1 (500 nM) supplementation for 4 h. H3 was used as a loading control. D Bar graphs showing the SIRT accumulation after BafA1 mediated block of autophagic degradation seen in the blots presented in C. “*” indicates significant differences compared to the control group, while “#” indicates significant differences compared to the MPP+-treated group. Symbol number indicates the grade of significance with *p < 0.05 and **p < 0.01. E Representative immunoblots of all SIRTs were obtained from differentiated LUHMES cells after 48 h treatments with MPP+ (10 µM) and PHT (20 nM) with and without MG132 (10 µM) supplementation for 24 h. H3 was used as a loading control. F Bar graphs showing the SIRT accumulation after MG132 mediated block of proteasomal degradation seen in the blots presented in E.
SIRTs are subject to autophagic degradation upon induction of oxidative stress
To clarify the origin of ROS-mediated SIRT protein loss, we focused on the MPP+ and MPP+/PHT treatment groups since MPP+ generally showed the most pronounced effects on SIRT levels (Fig. 1A, B). To inhibit lysosomal acidification and thus degradation of autophagosomal cargo, we added Bafilomycin A1 (BafA1) for the last 4 h of the experiment. Under basal conditions, SIRTs showed no significant accumulation in response to BafA1. However, under MPP+ treatment, all previously downregulated SIRTs significantly accumulated in response to BafA1. Again, the minor isoforms SIRT1S and SIRT3L did not recapitulate this pattern. PHT/MPP+-treated cells demonstrated substantially lowered SIRT accumulation throughout (Fig. 1C, D), indicating that ROS were the general cause of the MPP+-triggered degradation of SIRTs.
Subsequently, we tested whether SIRTs would accumulate when proteasomal degradation was inhibited by MG132. However, no significant effects could be observed (Fig. 1E, F), suggesting that the proteasome does not play a relevant role in SIRT turnover in LUHMES cells. Effective proteasomal inhibition by MG132 was confirmed by measuring the general accumulation of poly-ubiquitinated proteins (Supplementary Fig. 1a, b).
Autophagic SIRT degradation requires oxidative stress
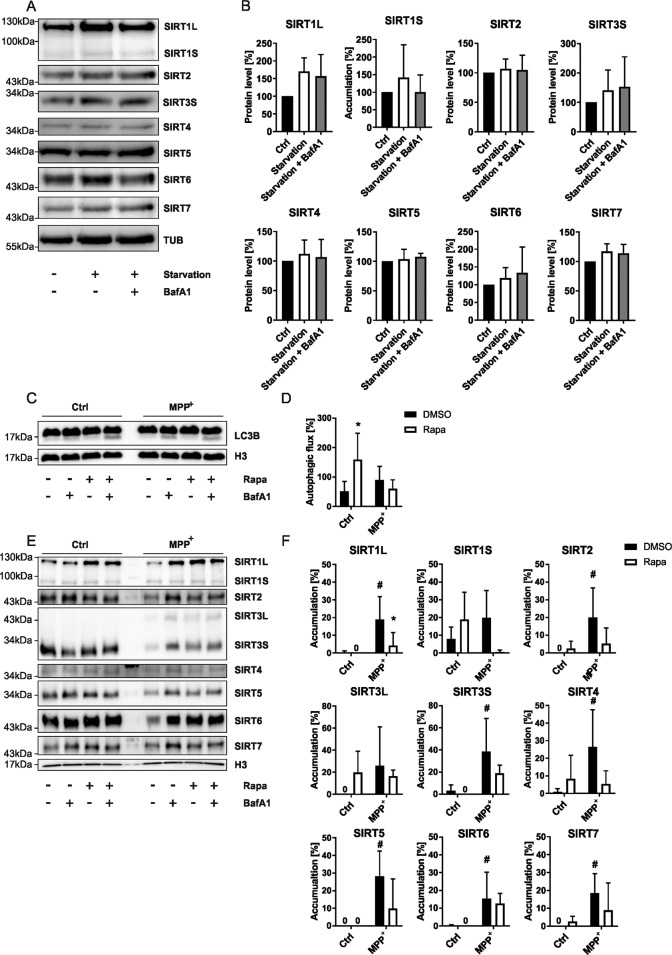
Next, we investigated whether SIRTs become substrates for autophagic-lysosomal degradation under general autophagy-inducing conditions. We subjected LUHMES cells to starvation with and without BafA1. However, we could not observe significant changes regarding any SIRT (Fig. 2A, B).
Fig. 2. SIRT degradation is dependent on an increase in oxidative stress.
A Representative immunoblots of all SIRTs obtained from differentiated LUHMES cells after 4 h of starvation in EBSS with and without BafA1 (500 nM) supplementation for 4 h. TUB was used as a loading control. B Bar graphs showing the total SIRT protein levels of the blots presented in A. C Representative immunoblots of LC3B obtained from differentiated LUHMES cells after 48 h treatments with MPP+ (10 µM) with and without BafA1 (500 nM) and/or rapamycin (500 nM) supplementation for 4 h. H3 was used as a loading control. D Bar graphs showing the autophagic flux derived from the blots presented in C. “*” indicates significant differences compared to the DMSO control group, symbol number indicates the grade of significance with *p < 0.05. E Representative immunoblots of all SIRTs were obtained from differentiated LUHMES cells after 48 h treatments with MPP+ (10 µM) with and without BafA1 (500 nM) and/or rapamycin (500 nM) supplementation for 4 h. H3 was used as a loading control. F Bar graphs showing the SIRT accumulation after BafA1 mediated block of autophagic degradation seen in the blots presented in E. “*” indicates significant differences compared to the control group, while “#” indicates significant differences compared to the DMSO control-treated group. Symbol number indicates the grade of significance with *p < 0.05.
Consequently, we wanted to examine whether SIRT degradation could be further stimulated when the cells were subjected to increased ROS. However, starvation in combination with MPP+ treatment caused a critical decline in LUHMES cell viability. Thus, we chose to induce autophagy pharmacologically by rapamycin (Fig. 2C–F). As expected, rapamycin increased autophagic activity as quantified [22] per the ATG8 paralogue LC3B (Fig. 2C, D) in our control samples. Interestingly, the induction was lost in cells co-treated with MPP+. Rapamycin may have been unable to increase the autophagic flux in MPP+-treated cells, since MPP+ already inhibits mTOR [23]. Thus, the pathway could already be pushed to its limit in this treatment paradigm. Indeed, results in SH-SY5Y cells confirmed that the autophagic flux could no longer be induced after exposure to mild levels (10 µM) of MPP+ [24].
Rapamycin had no immediate effects on autophagic SIRT degradation at baseline (Fig. 2E, F). On the other hand, the manifest induction of SIRT degradation upon MPP+ treatment was prevented by rapamycin treatment (Fig. 2C, D). However, antagonistic effects of rapamycin towards MPP+ mediated toxicity are known [25].
SIRTs co-localize with autophagosomal structures
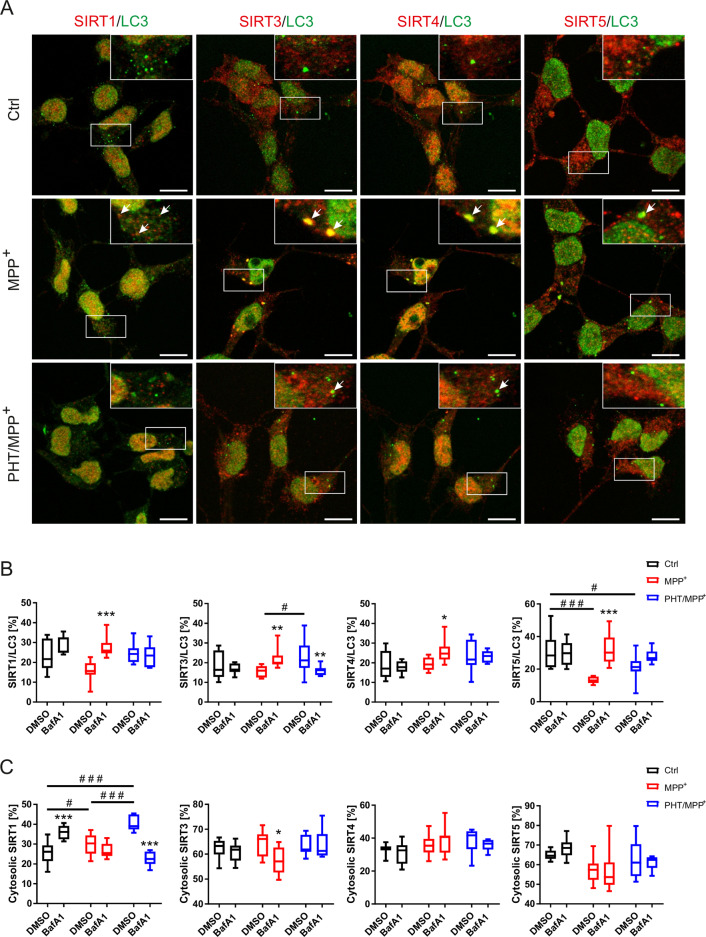
To verify whether SIRTs actually co-localize with autophagosomal structures, we performed immunocytochemistry for SIRT1, SIRT3, SIRT4, SIRT5, and LC3B. Analysis was restricted to cytosolic co-localization to exclude nuclear co-localization not related to autophagosomes [14]. Co-localization between cytosolic SIRTs and LC3B was significantly induced in MPP+- and BafA1-treated cells (versus MPP+-alone treated cells), suggesting higher incorporation of SIRTs into autophagosomal structures (Fig. 3A, B and Supplementary Fig. 2). This effect was reverted by co-treatment with PHT, indicating again an oxidation-related phenomenon. Although recent advances in the field demonstrated the occurrence of LC3B positive, vesicular structures in autophagy-deficient cells [26], their relative absence in our samples not treated with BafA1 would rather suggest a formation of autophagosomes than mere p62 aggregation (Fig. 3A, B and Supplementary Fig. 2).
Fig. 3. SIRTs accumulate in autophagosomal structures.
A Representative LSM images of differentiated LUHMES cells treated after 48 h treatment with MPP+ (10 µM) and/or PHT (20 nM) with BafA1 (500 nM) supplementation for 4 h with 63x magnification. The upper right corner per image highlights the area indicated by the white rectangle. Stained SIRTs are shown in red and LC3B in green. Scale bars represent 10 µm. Arrows indicate SIRT/LC3B co-localized vesicles. B Boxplots describing the cytosolic co-localization of the indicated SIRT with LC3B of images shown in A. “*” indicates significant differences caused by BafA1 treatment, while “#” indicates significant differences between the DMSO treated samples. Symbol number indicates the grade of significance with *p < 0.05, **p < 0.01, and ***p < 0.001. C Boxplots describing the cytosolic localization of the indicated SIRT of images shown in A. “*” indicates significant differences caused by BafA1 treatment, while “#” indicates significant differences between the DMSO treated samples. Symbol number indicates the grade of significance with *p < 0.05 and ***p < 0.001.
As expected, the difference between MPP+-alone treated cells and MPP+/BafA1-treated cells was in part evoked by a loss (of cytosolic) co-localization in MPP+-alone treated cells; this effect was significant for SIRT5 and SIRT1. Hence, MPP+ would have induced the disappearance of SIRTs from the cytosol by the successful execution of autophagy. In order to probe whether this interpretation was correct and unaffected by nuclear shifts, we also analyzed the relative cytosolic fraction of each SIRT (Fig. 3A, C). None of the SIRTs exhibited a smaller cytosolic fraction under MPP+ or PHT/MPP+ treatment at baseline, and except for the borderline significant SIRT3, BafA1 treatment caused no shift in MPP+-treated cells.
SIRT1 was unique in being redistributed into the cytosol by BafA1 alone, and in being redistributed into the nucleus by PHT. Tentatively, we assign this behavior to an unusually rapid turnover of SIRT1, including a rapidly adjusting equilibrium between nucleus and cytosol. This idea is supported by the fact the SIRT1 was the most heavily affected protein by MPP+ treatment (Fig. 1A–D). The rather high levels of the canonically mitochondrial SIRT4 in the nucleus are not unprecedented [27]. Similarly, about 30% of LUHMES cell SIRT5, which has been shown to desuccinylate nuclear proteins [28], was detected in the nucleus (Fig. 3C).
SIRTs are damaged by H2O2 and its products
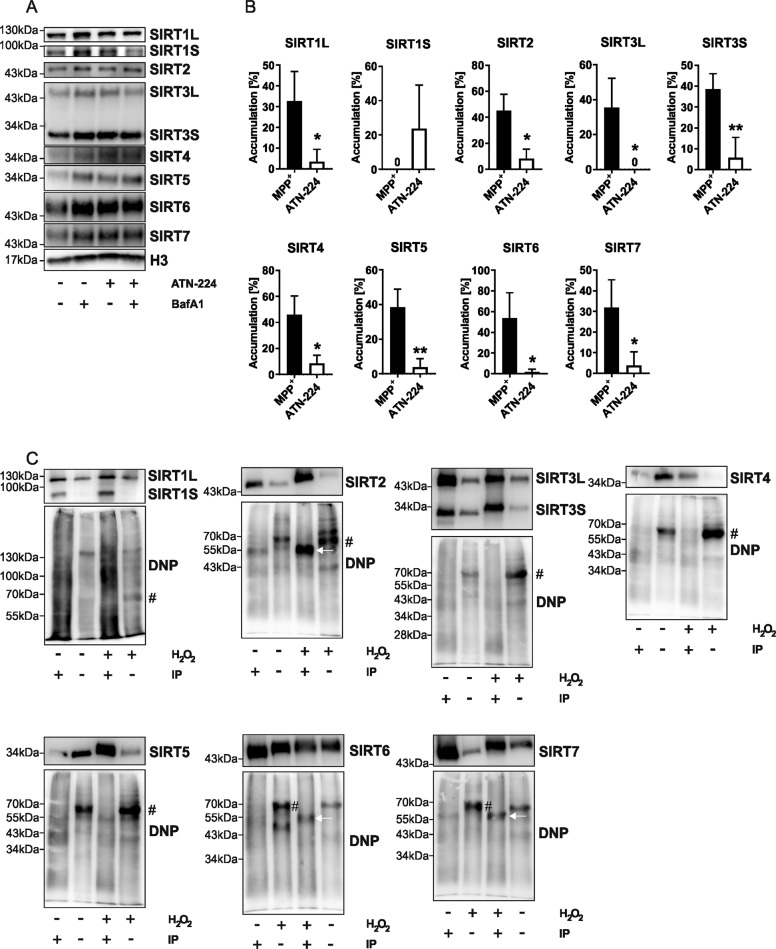
Which ROS could be the mediators of the observed, increased degradation of SIRTs in response to MPP+ treatment? In a first experiment, we administered MPP+-treated LUHMES cells with BafA1 and ATN-224, a pleiotropic SOD inhibitor (Fig. 4A, B). ATN-224 alone had no significant effects on SIRT levels or turnover (Supplementary Fig. 3). Interestingly, we obtained similar results for all SIRTs (except SIRT1S), in that ATN-224 caused a reduction of the MPP+-mediated degradative accumulation (Fig. 4A, B). This observation seems to rule out any direct oxidizing effect of MPP+-induced superoxide on SIRTs. The reported ATN-224-evoked loss of H2O2 generation may well account for our findings [29].
Fig. 4. H2O2, but not superoxide, damages some SIRTs.
A Representative immunoblots of all SIRTs obtained from differentiated LUHMES cells after 48 h treatment with MPP+ (10 µM) with and without BafA1 (500 nM) supplementation for 4 h. ATN-224 (10 µM) was administered over 6 h. H3 was used as a loading control. B Bar graphs showing the SIRT accumulation after BafA1 mediated block of autophagic degradation seen in the blots presented in A. “*” indicates significant differences compared to the control group, while “#” indicates significant differences compared to the DMSO control-treated group. Symbol number indicates the grade of significance with *p < 0.05. C Immunoblots of immunoprecipitated ectopically expressed SIRTs and total protein lysate from HEK-T cells showing carbonylated proteins derivatized with DNP compared to total proteins after 1 h of 20 mM H2O2 treatment. The upper blot shows the signal generated using a SIRT-specific antibody without prior DNP derivatization, lower blot shows the signal generated using the DNP-specific antibody after DNP derivatization. H2O2 indicates which samples were submitted to an oxidative environment, IP indicates which lanes were loaded with the IP purified protein post oxidation. A “#” indicates carbonylated proteins at ~70 kDa, while the white arrows point at carbonylated SIRTs.
Endogenous H2O2 cannot easily be enriched in a similar manner to superoxide in vitro. Thus, we decided to take a different approach and analyzed SIRT protein oxidation by H2O2 in terms of protein carbonylation of ectopically expressed SIRTs from HEK-T cells (Fig. 4C). SIRT1, SIRT3, SIRT4, and SIRT5 showed no increase in protein carbonylation after high-dose H2O2 treatment in vitro, whereas SIRT2, SIRT6, and SIRT7 did. Strikingly, three of the four none-carbonylated SIRTs are mitochondrial. This prompted us to look at possible footprints of natural selection towards or away from oxidative resistance in the amino acid composition of SIRTs across the Bilateria clade.
Natural selection favored none-mitochondrial SIRTs that can be regulated through oxidation
Briefly revisiting the phylogeny of the SIRTs (Supplementary Fig. 4) confirmed the described classification of human SIRTs: class I (SIRT1, SIRT2, and SIRT3), class II (SIRT4), class III (SIRT5), and class IV (SIRT6 and SIRT7) [30]. Eukaryotic SIRT5 appeared distant from other eukaryotic SIRTs, but in proximity to those of the archaea. Contrarily, SIRT4 appeared close to SIRTs from Wolbachia, which are related to proto-mitochondria [31].
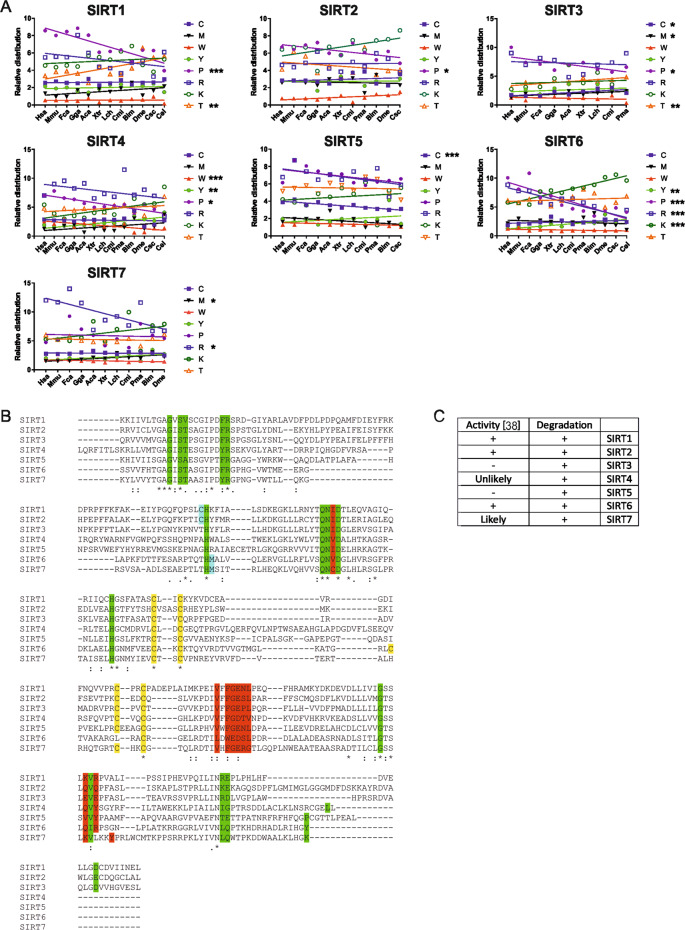
Cysteine is arguably the most rapidly oxidized amino acid under standard conditions. Therefore, previous studies have suggested that along with the evolution of aerobic organisms, cysteine (C) residues were selectively depleted [32, 33]. The observed C frequency in human proteins averages ~2.2% [34]. With the exception of SIRT3, which showed a significant decline, human SIRTs generally harbored a (modestly) higher amount of evolutionarily stable C residues (Fig. 5A). Methionine (M) also decreased in SIRT3; a progression only shared by SIRT7. The mitochondrial nature of SIRT3 could have required it to attain a more oxidation-resilient structure. SIRT5, in turn, behaved completely differently, as its generally high C content progressively increased. Next, we looked at the usually antioxidative amino acids tyrosine (Y) and tryptophan (W) [33, 35]. Again, most SIRTs demonstrated no significant changes, except for SIRT4, in which W and Y appeared to be mutually replaced over time (Fig. 5A).
Fig. 5. SIRT vulnerability to oxidative stress is conserved.
A Shift in indicated amino acid compositions of SIRTs in single letter code throughout bilateria evolution. Plotted species included Homo sapiens (Hsa), Mus musculus (Mmu), Felis catus (Fca), Gallus gallus (Gga), Anolis carolinensis (Aca), Xenopus tropicalis (Xtr), Latimeria chalumane (Lch), Callorhinchus milii (Cmi), Petromyzon marinus, Bombus impatiens, Drosophila melanogaster, Centruroides sculpturatus (Csc), and Caenorhabditis elegans (Cel). For each amino acid, a linear regression was generated and calculated whether the regression showed a significant progression. Symbol number indicates the grade of significance with *p < 0.05, **p < 0.01, and ***p < 0.001. B Alignment of the enzymatic centers of human SIRTs. Green color indicates NAD+ binding, red substrate binding, yellow the Zn2+-tetrathiolate, and blue the possible activity regulating sulfurous amino acids. * indicates identical amino acids, conserved and semi-conserved sites. C Table comparing which SIRT’s activity is regulated by oxidation, based on [38], and which SIRT is degraded by autophagy upon oxidation.
A carbonylation is a terminal form of protein damage that leads to proteasomal degradation [36]. Accordingly, protein carbonyls predominantly arise from relatively stable amino acids such as arginine (R), lysine (K), proline (P), and, to some degree, threonine (T) [37]. Notably, the carbonylated SIRTs all showed net accumulation of these amino acids (higher P in SIRT2; higher P and R, but lower K in SIRT6; higher R in SIRT7) (Fig. 5A).
Hence, the evenly distributed susceptibility of SIRTs to oxidant-induced autophagic degradation most likely arises from the oxidation of their C residues. Indeed, a conserved Zn2+-tetrathiolate was reported highly oxidation-prone (Fig. 5B) [38]. Interestingly, an alignment of the catalytic domain of human SIRTs revealed sulfurous amino acids next to an NAD+-binding histidine exclusively in none-mitochondrial SIRTs (Fig. 5B). A comparison with recently published data [38] indicates that the reported activity pattern would fit oxidation of these sulfurous amino acids, while the autophagic degradation pattern reported here may originate from plain oxidation of the tetrathiolate (Fig. 5C).
Discussion
The homeostasis of SIRT expression is of high biomedical interest since essentially all SIRTs appear to antagonize aspects of aging [39–45] or neurodegeneration [46]. These observations are, however, still discordant in invertebrates [47, 48]. Furthermore, similar results were obtained where a general overexpression of SIRT1 did not lead to increased longevity in mice. Yet, overexpression limited to the brain (BRASTO) caused a significantly later onset of senescence [39]. So far, the knowledge about the general regulation of SIRT activity in mammalian cells has remained incomplete. Here, we provide data indicating that endogenous ROS in form of H2O2 or its decomposition products broadly oxidize SIRTs and induce their autophagic degradation in dopaminergic neuronal cells in models of PD. This is also supported by the absence of degradation through SOD inhibition or its presence upon paraquat administration.
Previous studies have revealed that SIRT1 becomes a target of autophagy in aged tissues, causing a decline in protein level [8]. We were able to confirm this not only for SIRT1 but for all SIRTs with the exception of the minor isoforms SIRT1S and SIRT3L (Fig. 6). The surprising necessity of autophagy to degrade SIRTs may be related to a conserved tetrathiolate. Indeed, high levels of free Zn2+ have been observed in autolysosomes, and cells starved for Zn2+ demonstrate increased autophagy of Zn2+ binding proteins [49, 50]. Since different SIRTs occur more frequently in different cellular compartments, a future approach, which may allow distinguishing between mitophagy and nucleophagy may help gain an even better understanding of these processes. Meanwhile, future projects may also look at different knockout systems (e.g. ATG5, ATG7, or LC3B) to further understand how and when the SIRTs are transferred into the autophagosomes.
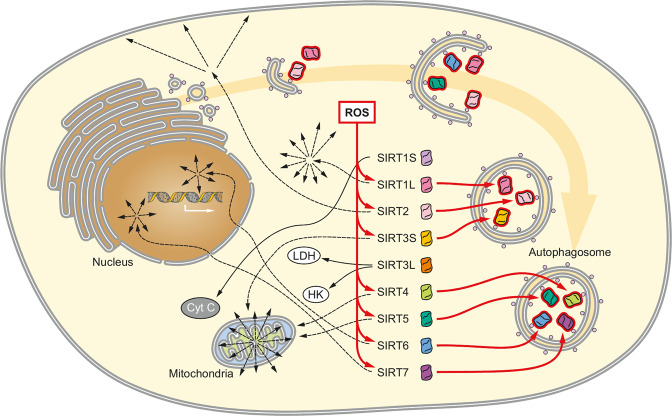
Fig. 6. Graphical model of the SIRT status in LUHMES cells subjected to chronically increased oxidative stress.
Most SIRTs become substrates for autophagic degradation when cells are subjected to chronic oxidative stress. SIRTs are shown in the cellular compartment where they are most commonly encountered. SIRT1S and SIRT3L are not targeted to autophagic degradation, but rather act antiapoptotic (SIRT1S) by pinning cytochrome C (CytC), or enhance compensatory mechanism to satisfy bioenergetics demands.
Interestingly, not all SIRTs have been shown to exhibit decreased activity after being exposed to oxidation [38], yet we have found all of their major forms to be subjected to autophagic degradation. Thus, their increased degradation may be decoupled from protein activity. Their persisting activity and a lack of carbonylation in most SIRTs would berate a clean damage-related response. Interestingly, this persisting activity was restricted to mostly mitochondrial SIRTs, SIRT3 and SIRT5, despite their Zn2+-tetrathiolate being oxidized [38]. The exact function of this thiolate is still not well understood. However, since all previously tested SIRTs were oxidized, while we observed an increased turnover of all SIRTs, this tetrathiolate may mediate the autophagic degradation of the proteins. The previously observed activity pattern would rather fit the conserved H327 (human SIRT1) having obtained sulfurous neighbors. Indeed, it was confirmed that mutation of the C adjacent to H327 resulted in a functional, ROS-resistant SIRT1 [51]. This further strengthens our hypothesis that activity and degradation of SIRT are regulated independently.
SIRT evolution reveals that these sulfurous neighbors must have developed independently, since class IV SIRTs (SIRT6 and SIRT7) evolved an M downstream, while class I SIRTs (SIRT1 and SIRT2) evolved a C upstream. SIRT3 has lost this C again since it became mitochondrial. Thus, we conclude that SIRT7’s activity may also be susceptible toward oxidation, while SIRT4 would not.
The question remains why cells would deplete their active SIRTs under oxidative stress. Induction of apoptosis appears unlikely since the upregulated cytosolic SIRT1S acts antiapoptotically through the scavenging of free cytochrome C [52]. Additionally, SIRT3L has been shown to regulate transcription in favor of cell survival and activate glycolytic enzymes [53–55]. On the other hand, ROS decreasing the bioavailability of SIRTs would leave the cell’s proteome hyper-acetylated, which could be protective in an oxidative environment in two ways. First, the constant damage to biomolecules would require a rapid replacement. Acetylation-mediated stimulation of transcription may facilitate this. Second, the nucleophilic nature of lysine allows it to cross-link with other amino acids or nucleic acids [56, 57]. Acetyl-lysine, however, is chemically more inert and does not form cross-links.
Oxidative stress propels senescence and is a proven stimulus that leads to neurodegenerative events that may develop into disorders of higher-order like PD. While SIRTs are often considered potential treatment targets to prevent or delay senescence, the question needs to be asked whether this would truly prove beneficial for a cell that suffers from chronic oxidative stress. The conserved mechanism of SIRT degradation independent of their activity we describe here would rather suggest that the cell actively removes SIRTs when facing an oxidative dysbalance. Thus, a possible intervention to prolong SIRT availability through, e.g., ectopic expression, may not be as beneficial as the removal of the oxidation source, which would already reestablish a SIRT homeostasis.
Methods
Chemicals
Cell culture materials were purchased from Invitrogen and chemicals from Sigma-Aldrich unless stated otherwise. Used primary antibodies included: anti-SIRT1 (9475, CST); anti-SIRT2 (12672, CST); anti-SIRT3 (2627, CST); anti-SIRT4 (NB100-1406, Novus Biologicals); anti-SIRT5 (8782, CST); anti-SIRT6 (12486, CST); anti-SIRT7 (5360, CST); anti-histone H3 (14269, CST); anti-ubiquitin (3936, CST); anti-tubulin (T9026, Sigma-Aldrich); anti-MAP1LC3B (0260–100, NanoTools) for immunocytochemistry, (L7543, Sigma-Aldrich) for immunoblotting; anti-DNP-KLH (A6430, Invitrogen).
Cell culture
Lund human mesencephalic (LUHMES) cells were grown as published [18]. Four days post differentiation, cells were treated with small molecules in fresh media: MPP+ (10 µM, 48 h), PHT (20 nM, 48 h), rotenone (10 nM, 48 h), carbonyl cyanide-p-trifluoromethoxyphenylhydrazone (FCCP) (1 mM, 48 h, Tocris), paraquat (100 µM, 48 h), BafA1 (500 nM, 4 h, Toronto research chemicals), rapamycin (500 nM, 4 h Enzo Lifesciences), MG132 (10 µM, 24 h, Enzo Lifesciences), ATN-224 (10 µM, 6 h, Cayman chemicals). Starvation occurred with EBSS for 4 h.
Immunoblotting
Immunoblotting was performed according to published protocols [14, 18]. Primary antibodies (1:1000) were detected by horseradish peroxidase-conjugated secondary antibodies (Dianova, 1:10000) through luminescence and quantified using densitometry. PageRulerTM Prestained Protein Ladder (26617, Thermo Fisher Scientific) was used to determine the molecular weight of proteins.
Immunocytochemistry
Immunocytochemistry and co-localization analysis were performed according to published protocols [14, 18]. Primary antibodies (1:200) were labeled with secondary antibodies (Dianova, 1:400). Hoechst 33258 (Invitrogen) was used for nuclear counterstaining. Cells were photographed using a confocal LSM (TCS SP5, Leica Microsystems).
SIRT carbonylation assay
Plasmids encoding human SIRT genes were purchased from GenScript Biotech. HEK-T cells were transfected using calcium phosphate precipitation and harvested in H2O after 48 h. About 500 µg protein homogenate were incubated with 20 mM H2O2 for 1 h at room temperature. Reaction was terminated with 1 mM NaCO3 and 1x IP buffer (50 mM Tris-HCl, pH 7.4; 150 mM NaCl; 2 mM EDTA; 0.5 mM EGTA; 1% Triton X-100; 10% glycerol; protease inhibitor; 1 mM dithiothreitol). SIRTs were extracted via immunoprecipitation (antibody dilution 1:50) following standard procedure [14]. Carbonylation of proteins was analyzed via immunoblot according to published protocols [58].
Phylogenetic analysis
Protein sequences were obtained from UniProt and aligned using Clustal Omega [59, 60]. The phylogenetic tree was constructed using iTOL [61]. Binding sites of NAD+, SIRT substrates, and tetrathiolate location were extrapolated using the InterPro software [62].
Statistical analysis
Depending on data structure, Benjamini and Hochberg adjusted one-way or two-way ANOVA were performed. Post hoc test significances with p values <0.05 are indicated by asterisks or hash signs. Results are presented as mean ± SD. Linear regression was used to evaluate significant amino acid shifts in protein evolution. If not stated otherwise, the number of replicates (n) = 3.
Supplementary information
Poly-ubiquitinated proteins accumulate after exposure to MG132
ATN-224 has no effect on SIRT protein levels
Acknowledgements
This study was supported by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation)—Project-ID 259130777- SFB 1177 and a grant of the Corona Foundation of the Deutsche Stifterverband to C.B. The authors thank Sandra Ritz (Institute of Molecular Biology, Mainz) for her help with the confocal microscopy.
Author contributions
M.W.B., B.M., P.H., A.K. and C.B. conceived and designed the research. M.W.B. and M.S. performed experiments. M.W.B. analyzed data, prepared figures, and drafted the manuscript. M.S., B.M, P.H., A.K. and C.B. edited and revised the paper. All authors approved of the final version.
Funding
Open Access funding enabled and organized by Projekt DEAL.
Data availability
Primary data files and calculations are available upon request from either of the corresponding authors (mbaeken@uni-mainz.de, cbehl@uni-mainz.de).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Marius W. Baeken, Email: mbaeken@uni-mainz.de
Christian Behl, Email: cbehl@uni-mainz.de.
Supplementary information
The online version contains supplementary material available at 10.1038/s41420-021-00683-x.
References
- 1.Tian X, Firsanov D, Zhang Z, Cheng Y, Luo L, Tombline G, et al. SIRT6 is responsible for more efficient DNA double-strand break repair in long-lived species. Cell. 2019;177:622–38. doi: 10.1016/j.cell.2019.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bonkowski MS, Sinclair DA. Slowing ageing by design: the rise of NAD + and sirtuin-activating compounds. Nat Rev Mol Cell Biol. 2016;17:679–90. doi: 10.1038/nrm.2016.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhao E, Hou J, Ke X, Abbas MN, Kausar S, Zhang L, et al. The roles of sirtuin family proteins in cancer progression. Cancers. 2019;11:1949. doi: 10.3390/cancers11121949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Imai S, Guarente L. NAD+ and sirtuins in aging and disease. Trends Cell Biol. 2014;24:464–71. doi: 10.1016/j.tcb.2014.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fang EF, Bohr VA. NAD(+): the convergence of DNA repair and mitophagy. Autophagy. 2017;13:442–3. doi: 10.1080/15548627.2016.1257467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fang EF, Scheibye-Knudsen M, Brace LE, Kassahun H, SenGupta T, Nilsen H, et al. Defective mitophagy in XPA via PARP-1 hyperactivation and NAD(+)/SIRT1 reduction. Cell. 2014;157:882–96. doi: 10.1016/j.cell.2014.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hartl FU, Bracher A, Hayer-Hartl M. Molecular chaperones in protein folding and proteostasis. Nature. 2011;475:324–32. doi: 10.1038/nature10317. [DOI] [PubMed] [Google Scholar]
- 8.Xu C, Wang L, Fozouni P, Evjen G, Chandra V, Jiang J, et al. SIRT1 is downregulated by autophagy in senescence and ageing. Nat Cell Biol. 2020;22:1170–9. doi: 10.1038/s41556-020-00579-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Angelova PR, Choi ML, Berezhnov AV, Horrocks MH, Hughes CD, De S, et al. Alpha synuclein aggregation drives ferroptosis: an interplay of iron, calcium and lipid peroxidation. Cell Death Differ. 2020;27:2781–96. doi: 10.1038/s41418-020-0542-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kadowaki H, Nishitoh H, Urano F, Sadamitsu C, Matsuzawa A, Takeda K, et al. Amyloid beta induces neuronal cell death through ROS-mediated ASK1 activation. Cell Death Differ. 2005;12:19–24. doi: 10.1038/sj.cdd.4401528. [DOI] [PubMed] [Google Scholar]
- 11.Pollari E, Goldsteins G, Bart G, Koistinaho J, Giniatullin R. The role of oxidative stress in degeneration of the neuromuscular junction in amyotrophic lateral sclerosis. Front Cell Neurosci. 2014;8:131. doi: 10.3389/fncel.2014.00131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Filomeni G, De Zio D, Cecconi F. Oxidative stress and autophagy: the clash between damage and metabolic needs. Cell Death Differ. 2015;22:377–88. doi: 10.1038/cdd.2014.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gamerdinger M, Hajieva P, Kaya AM, Wolfrum U, Hartl FU, Behl C. Protein quality control during aging involves recruitment of the macroautophagy pathway by BAG3. EMBO J. 2009;28:889–901. doi: 10.1038/emboj.2009.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baeken MW, Weckmann K, Diefenthäler P, Schulte J, Yusifli K, Moosmann B, et al. Novel insights into the cellular localization and regulation of the autophagosomal proteins LC3A, LC3B and LC3C. Cells. 2020;9:2315. doi: 10.3390/cells9102315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang R, Xu Y, Wan W, Shou X, Qian J, You Z, et al. Deacetylation of nuclear LC3 drives autophagy initiation under starvation. Mol Cell. 2015;57:456–66. doi: 10.1016/j.molcel.2014.12.013. [DOI] [PubMed] [Google Scholar]
- 16.Lee IH, Cao L, Mostoslavsky R, Lombard DB, Liu J, Bruns NE, et al. A role for the NAD-dependent deacetylase Sirt1 in the regulation of autophagy. Proc Natl Acad Sci USA. 2008;105:3374–9. doi: 10.1073/pnas.0712145105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hisahara S, Shimohama S. Toxin-induced and genetic animal models of Parkinson’s disease. Parkinsons Dis. 2010;2011:951709. doi: 10.4061/2011/951709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baeken MW, Moosmann B, Hajieva P. Retrotransposon activation by distressed mitochondria in neurons. Biochem Biophys Res Commun. 2020;525:570–5. doi: 10.1016/j.bbrc.2020.02.106. [DOI] [PubMed] [Google Scholar]
- 19.Tapias V, McCoy JL, Greenamyre JT. Phenothiazine normalizes the NADH/NAD(+) ratio, maintains mitochondrial integrity and protects the nigrostriatal dopamine system in a chronic rotenone model of Parkinson’s disease. Redox Biol. 2019;24:101164. doi: 10.1016/j.redox.2019.101164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hajieva P, Mocko JB, Moosmann B, Behl C. Novel imine antioxidants at low nanomolar concentrations protect dopaminergic cells from oxidative neurotoxicity. J Neurochem. 2009;110:118–32. doi: 10.1111/j.1471-4159.2009.06114.x. [DOI] [PubMed] [Google Scholar]
- 21.Chen G, Han Z, Feng D, Chen Y, Chen L, Wu H, et al. A regulatory signaling loop comprising the PGAM5 phosphatase and CK2 controls receptor-mediated mitophagy. Mol Cell. 2014;54:362–77. doi: 10.1016/j.molcel.2014.02.034. [DOI] [PubMed] [Google Scholar]
- 22.Klionsky DJ, Abdelmohsen K, Abe A, Abedin J, Abeliovich H, Arozena AA. et al. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy. 2016;12:1–222. [DOI] [PMC free article] [PubMed]
- 23.Sakamoto S, Miyara M, Sanoh S, Ohta S, Kotake Y. Mild MPP(+) exposure-induced glucose starvation enhances autophagosome synthesis and impairs its degradation. Sci Rep. 2017;7:46668. doi: 10.1038/srep46668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miyara M, Kotake Y, Tokunaga W, Sanoh S, Ohta S. Mild MPP(+) exposure impairs autophagic degradation through a novel lysosomal acidity-independent mechanism. J Neurochem. 2016;139:294–308. doi: 10.1111/jnc.13700. [DOI] [PubMed] [Google Scholar]
- 25.Malagelada C, Jin ZH, Jackson-Lewis V, Przedborski S, Greene LA. Rapamycin protects against neuron death in in vitro and in vivo models of Parkinson’s disease. J Neurosci. 2010;30:1166–75. doi: 10.1523/JNEUROSCI.3944-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Runwal G, Stamatakou E, Siddiqi FH, Puri C, Zhu Y, Rubinsztein DC. LC3-positive structures are prominent in autophagy-deficient cells. Sci Rep. 2019;9:10147. doi: 10.1038/s41598-019-46657-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ramadani-Muja J, Gottschalk B, Pfeil K, Burgstaller S, Rauter T, Bischof H, et al. Visualization of sirtuin 4 distribution between mitochondria and the nucleus, based on bimolecular fluorescence self-complementation. Cells. 2019;8:1583. doi: 10.3390/cells8121583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Park J, Chen Y, Tishkoff DX, Peng C, Tan M, Dai L, et al. SIRT5-mediated lysine desuccinylation impacts diverse metabolic pathways. Mol Cell. 2013;50:919–30. doi: 10.1016/j.molcel.2013.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Juarez JC, Manuia M, Burnett ME, Betancourt O, Boivin B, Shaw DE, et al. Superoxide dismutase 1 (SOD1) is essential for H2O2-mediated oxidation and inactivation of phosphatases in growth factor signaling. Proc Natl Acad Sci USA. 2008;105:7147–52. doi: 10.1073/pnas.0709451105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Frye RA. Phylogenetic classification of prokaryotic and eukaryotic Sir2-like proteins. Biochem Biophys Res Commun. 2000;273:793–8. doi: 10.1006/bbrc.2000.3000. [DOI] [PubMed] [Google Scholar]
- 31.Roger AJ, Muñoz-Gómez SA, Kamikawa R. The origin and diversification of mitochondria. Curr Biol. 2017;27:R1177–92. doi: 10.1016/j.cub.2017.09.015. [DOI] [PubMed] [Google Scholar]
- 32.Schindeldecker M, Stark M, Behl C, Moosmann B. Differential cysteine depletion in respiratory chain complexes enables the distinction of longevity from aerobicity. Mech Ageing Dev. 2011;132:171–9. doi: 10.1016/j.mad.2011.03.002. [DOI] [PubMed] [Google Scholar]
- 33.Moosmann B, Schindeldecker M, Hajieva P. Cysteine, glutathione and a new genetic code: biochemical adaptations of the primordial cells that spread into open water and survived biospheric oxygenation. Biol Chem. 2020;401:213–31. doi: 10.1515/hsz-2019-0232. [DOI] [PubMed] [Google Scholar]
- 34.Gardini S, Cheli S, Baroni S, Di Lascio G, Mangiavacchi G, Micheletti N, et al. On nature’s strategy for assigning genetic code multiplicity. PLoS ONE. 2016;11:e0148174. doi: 10.1371/journal.pone.0148174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moosmann B, Behl C. Cytoprotective antioxidant function of tyrosine and tryptophan residues in transmembrane proteins. Eur J Biochem. 2000;267:5687–92. doi: 10.1046/j.1432-1327.2000.01658.x. [DOI] [PubMed] [Google Scholar]
- 36.Raynes R, Pomatto LC, Davies KJ. Degradation of oxidized proteins by the proteasome: Distinguishing between the 20S, 26S, and immunoproteasome proteolytic pathways. Mol Asp Med. 2016;50:41–55. doi: 10.1016/j.mam.2016.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stadtman ER, Levine RL. Free radical-mediated oxidation of free amino acids and amino acid residues in proteins. Amino Acids. 2003;25:207–18. doi: 10.1007/s00726-003-0011-2. [DOI] [PubMed] [Google Scholar]
- 38.Kalous KS, Wynia-Smith SL, Summers SB, Smith BC. Human sirtuins are differentially sensitive to inhibition by nitrosating agents and other cysteine oxidants. J Biol Chem. 2020;295:8524–36. doi: 10.1074/jbc.RA119.011988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Satoh A, Brace CS, Rensing N, Cliften P, Wozniak DF, Herzog ED, et al. Sirt1 extends life span and delays aging in mice through the regulation of Nk2 homeobox 1 in the DMH and LH. Cell Metab. 2013;18:416–30. doi: 10.1016/j.cmet.2013.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.North BJ, Rosenberg MA, Jeganathan KB, Hafner AV, Michan S, Dai J, et al. SIRT2 induces the checkpoint kinase BubR1 to increase lifespan. EMBO J. 2014;33:1438–53. doi: 10.15252/embj.201386907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bellizzi D, Rose G, Cavalcante P, Covello G, Dato S, De Rango F, et al. A novel VNTR enhancer within the SIRT3 gene, a human homologue of SIR2, is associated with survival at oldest ages. Genomics. 2005;85:258–63. doi: 10.1016/j.ygeno.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 42.Wood JG, Schwer B, Wickremesinghe PC, Hartnett DA, Burhenn L, Garcia M, et al. Sirt4 is a mitochondrial regulator of metabolism and lifespan in Drosophila melanogaster. Proc Natl Acad Sci USA. 2018;115:1564–9. doi: 10.1073/pnas.1720673115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kumar S, Lombard DB. Functions of the sirtuin deacylase SIRT5 in normal physiology and pathobiology. Crit Rev Biochem Mol Biol. 2018;53:311–34. doi: 10.1080/10409238.2018.1458071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kanfi Y, Naiman S, Amir G, Peshti V, Zinman G, Nahum L, et al. The sirtuin SIRT6 regulates lifespan in male mice. Nature. 2012;483:218–21. doi: 10.1038/nature10815. [DOI] [PubMed] [Google Scholar]
- 45.Bi S, Liu Z, Wu Z, Wang Z, Liu X, Wang S, et al. SIRT7 antagonizes human stem cell aging as a heterochromatin stabilizer. Protein Cell. 2020;11:483–504. doi: 10.1007/s13238-020-00728-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hor JH, Santosa MM, Lim VJW, Ho BX, Taylor A, Khong ZJ, et al. ALS motor neurons exhibit hallmark metabolic defects that are rescued by SIRT3 activation. Cell Death Differ. 2021;28:1379–97. doi: 10.1038/s41418-020-00664-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Burnett C, Valentini S, Cabreiro F, Goss M, Somogyvári M, Piper MD, et al. Absence of effects of Sir2 overexpression on lifespan in C. elegans and Drosophila. Nature. 2011;477:482–5. doi: 10.1038/nature10296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhao Y, Wang H, Poole RJ, Gems D. A fln-2 mutation affects lethal pathology and lifespan in C. elegans. Nat Commun. 2019;10:5087. doi: 10.1038/s41467-019-13062-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fang H, Geng S, Hao M, Chen Q, Liu M, Liu C, et al. Simultaneous Zn(2+) tracking in multiple organelles using super-resolution morphology-correlated organelle identification in living cells. Nat Commun. 2021;12:109. doi: 10.1038/s41467-020-20309-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ding B, Zhong Q. Zinc deficiency: an unexpected trigger for autophagy. J Biol. Chem. 2017;292:8531–2. doi: 10.1074/jbc.H116.762948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shao D, Fry JL, Han J, Hou X, Pimentel DR, Matsui R, et al. A redox-resistant sirtuin-1 mutant protects against hepatic metabolic and oxidant stress. J Biol Chem. 2014;289:7293–306. doi: 10.1074/jbc.M113.520403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Oppenheimer H, Gabay O, Meir H, Haze A, Kandel L, Liebergall M, et al. 75-kd sirtuin 1 blocks tumor necrosis factor alpha-mediated apoptosis in human osteoarthritic chondrocytes. Arthritis Rheum. 2012;64:718–28. doi: 10.1002/art.33407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schwer B, North BJ, Frye RA, Ott M, Verdin E. The human silent information regulator (Sir)2 homologue hSIRT3 is a mitochondrial nicotinamide adenine dinucleotide-dependent deacetylase. J Cell Biol. 2002;158:647–57. doi: 10.1083/jcb.200205057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Scher MB, Vaquero A, Reinberg D. SirT3 is a nuclear NAD+-dependent histone deacetylase that translocates to the mitochondria upon cellular stress. Genes Dev. 2007;21:920–8. doi: 10.1101/gad.1527307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Iwahara T, Bonasio R, Narendra V, Reinberg D. SIRT3 functions in the nucleus in the control of stress-related gene expression. Mol Cell Biol. 2012;32:5022–34. doi: 10.1128/MCB.00822-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xu X, Muller JG, Ye Y, Burrows CJ. DNA-protein cross-links between guanine and lysine depend on the mechanism of oxidation for formation of C5 vs C8 guanosine adducts. J Am Chem Soc. 2008;130:703–9. doi: 10.1021/ja077102a. [DOI] [PubMed] [Google Scholar]
- 57.Duff AP, Cohen AE, Ellis PJ, Hilmer K, Langley DB, Dooley DM, et al. The 1.23 Angstrom structure of Pichia pastoris lysyl oxidase reveals a lysine-lysine cross-link. Acta Crystallogr D Biol Crystallogr. 2006;62:1073–84. doi: 10.1107/S0907444906026333. [DOI] [PubMed] [Google Scholar]
- 58.Kunath S, Schindeldecker M, De Giacomo A, Meyer T, Sohre S, Hajieva P, et al. Prooxidative chain transfer activity by thiol groups in biological systems. Redox Biol. 2020;36:101628. doi: 10.1016/j.redox.2020.101628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Uniprot Consortium. UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res. 2019;47:D506–15. doi: 10.1093/nar/gky1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Madeira F, Park YM, Lee J, Buso N, Gur T, Madhusoodanan N, et al. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 2019;47:W636–41. doi: 10.1093/nar/gkz268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Letunic I, Bork P. Interactive tree of life (iTOL) v3: An online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016;44:W242–45. doi: 10.1093/nar/gkw290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hunter S, Apweiler R, Attwood TK, Bairoch A, Bateman A, Binns D, et al. InterPro: the integrative protein signature database. Nucleic Acids Res. 2009;37:D211–5. doi: 10.1093/nar/gkn785. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Poly-ubiquitinated proteins accumulate after exposure to MG132
ATN-224 has no effect on SIRT protein levels
Data Availability Statement
Primary data files and calculations are available upon request from either of the corresponding authors (mbaeken@uni-mainz.de, cbehl@uni-mainz.de).