Abstract
From June to November 1994 (period 1) and from February to June 1995 (period 2), multiresistant Acinetobacter baumannii strains were isolated in intensive care units and surgical wards of the Amiens Teaching Hospital Center (Amiens, France). Eighteen isolates were obtained from 17 (1%) of 1,706 patients admitted during both of these periods, giving an incidence rate of nosocomial infection per 1,000 patient days of 0.6%. Of 17 infected patients, 9 had pneumonia, 3 had urinary tract infection, 2 had peritonitis, 1 had septicemia, 1 had a catheter infection, and 1 had pneumonia and urinary tract infection. According to typing results, four antibiotic resistance profiles were detected: a, b, c, and d; seven ribotypes were distinguished by both restriction enzymes EcoRI and SalI (A, B, C, D, E, F, and G). By combining antibiotyping and ribotyping, we obtained eight groups of strains (groups I to VIII). Group I contained five strains (strains 4, 5, 7, 8, and 9) which had antibiogram pattern a and ribopattern A and constituted the outbreak strains. The strains of group II (strains 3, 10, 11, 13, and 14) were closely related to outbreak strain A and appeared to be variants of ribotype A (A2 [strain 3]; A4 [strain 10]; A5 [strains 11, 13, and 14]). Groups III, IV, V, VI, VII, and VIII included strains which were epidemiologically unrelated to the strains of group I and were considered nonoutbreak strains.
Bacteria of the genus Acinetobacter have become increasingly important as nosocomial pathogens. They comprise at least 21 DNA groups identified by DNA-DNA hybridization methods (4, 5, 13, 27). A. baumannii (DNA group 2) (4, 19, 27) seems to be the most prevalent of the Acinetobacter species isolated from clinical specimens in hospitals (6, 7, 15). Most hospital outbreaks have been attributed to this species (14, 15, 24). Various methods have been described for typing A. baumannii strains: biotyping (2, 6, 7, 9), antibiotyping (2, 9), serotyping (28), phage typing (7), plasmid typing (11), cell envelope protein sodium dodecyl sulfate-polyacrylamide gel electrophoresis (7), ribotyping (2, 9, 14, 23), chain reaction fingerprinting (10, 16, 25), and analysis of genomic DNA by pulsed-field gel electrophoresis (15, 23). Using a combination of antibiotyping and ribotyping methods, the aim of this study was to determine whether all A. baumannii isolates identified during the epidemic periods were identical.
MATERIALS AND METHODS
Study population.
The teaching hospital center of Amiens is divided into two main hospitals: the north hospital, which contains a polyvalent intensive care unit (pICU), a neurotraumatological intensive care unit (ntICU), a neurosurgical ward located in the same building, and a maxillofacial-stomatology surgical ward (MFS) located in another building, and the south hospital, which contains two wards located in the same building, namely, a nephrological intensive care unit (npICU) and a pneumological intensive care unit (pnICU). This teaching hospital center is an 1,841-bed medicosurgical hospital comprising 111-bed intensive care units and surgical wards (pICU, 8 beds; ntICU, 6 beds; npICU, 8 beds; pnICU, 10 beds; neurosurgical ward, 60 beds; MFS, 19 beds). The units are very close together, and each one has its staff physician and nurse, with the possibility to transfer a patient from one unit to the other according to his or her pathology.
From June 1994 to November 1994 (period 1) and from February 1995 to June 1995 (period 2), an increase in the number of A. baumannii isolates were observed in some care units, and most cases were associated with severe infection. During these two periods of time, a total of 1,706 patients were hospitalized in the different care units included in this investigation as follows: pICU, 290 patients; ntICU, 228 patients; npICU, 339 patients; pnICU, 297 patients; neurosurgical ward, 420 patients; and MFS, 132 patients. Of these 1,706 patients, 1,276 (74.7%) were male and 430 (25.3%) were female. The subjects had a mean age of 45 years (range, 30 to 63 years). Details of the description of the study population are given in Table 1. The records and clinical conditions of all patients with A. baumannii organisms during the period were studied.
TABLE 1.
Description of the studied population according to the individual periods of the studya
| Care unit | No. of patients
|
||
|---|---|---|---|
| Period 1 (from June to Nov. 1994) | Period 2 (from Feb. to June 1995) | Total | |
| pICU | 109 | 181 | 290 |
| ntICU | 114 | 114 | 228 |
| npICU | 177 | 162 | 339 |
| pnICU | 159 | 138 | 297 |
| Neurosurgical ward | 229 | 191 | 420 |
| MFS ward | 77 | 55 | 132 |
| Total | 865 | 841 | 1706 |
This study concerned a total of 1,706 patients, 865 of whom belonged to period 1 and 841 of whom to period 2.
A suspected index case female patient was identified. She was hospitalized on 23 March 1994, first in a surgical ward because of a left shin bone fracture complicated with leg necrosis and then in the pICU because of acute renal insufficiency and cardiac insufficiency. She was subsequently transferred to another teaching hospital from 29 March 1994 to 15 April 1994 and transferred back to the Amiens surgical ward and then to the pICU on 28 April 1994. A. baumannii was first isolated from peritoneal fluid on 28 April 1994 and was then cultured from vesical tube tips and intubated tube tips. The patient died of a peritoneal nosocomial infection with A. baumannii on 23 June 1994. The antibiogram result showed a strain resistant to multiple antibiotics. During the following months this multiresistant strain was isolated from four other patients (one patient admitted to the pICU, two patients from the ntICU, and one patient from the npICU).
Nosocomial infection and outbreak definitions.
Standard Center for Disease Control and Prevention (CDC) definitions were used (12). Catheter infection was diagnosed when A. baumannii was cultured from a specimen intravascular catheter in significant quantity (≥103 CFU) according to the method of Brun-Buisson (8). In the present study, an outbreak is defined according to the criteria of Tenover (26).
Bacterial-strain origins.
The origins of different A. baumannii strains isolated in the first time period (i.e., in 1994) were as follows: seven (strains 1, 3, 7, 10, 11, 13, and 15) from the pICU, one (strain 14) from the ntICU, and one (strain 17) from the neurosurgical ward. In the second time period (i.e., in 1995), there were two strains (strains 5 and 6) from the pICU, two (strains 8 and 9) from the ntICU, one (strain 2) from the pnICU, one (strain 4) from the npICU, and three (strains 12, 16, and 18) from the MFS. Two of these isolates (strain 5 derived from bronchopulmonary sputum and strain 6 derived from urinary sample) were obtained from the same patient. Strain 7 was isolated from the patient suspected to be the index case (Table 2).
TABLE 2.
Origins and date of isolation of A. baumannii strainsa
| Date of isolation (mo/day/yr) | Strain no. | Care unit | Sample type |
|---|---|---|---|
| 04/28/94 | 7 | pICU | Peritoneal fluid |
| 06/13/94 | 15 | pICU | Urine |
| 06/20/94 | 13 | pICU | Bronchopulmonary sputum |
| 06/30/94 | 14 | ntICU | Bronchopulmonary sputum |
| 07/20/94 | 1 | pICU | Peritonitis fluid |
| 08/10/94 | 3 | pICU | Bronchopulmonary sputum |
| 08/31/94 | 10 | pICU | Blood |
| 08/31/94 | 11 | pICU | Urine |
| 11/04/94 | 17 | Neurosurgical ward | Catheter tip |
| 02/10/95 | 4 | npICU | Urine |
| 03/28/95 | 5 | pICU | Bronchopulmonary sputum |
| 03/28/95 | 6 | pICU | Urine |
| 05/02/95 | 9 | ntICU | Bronchopulmonary sputum |
| 05/03/95 | 8 | ntICU | Bronchopulmonary sputum |
| 05/05/95 | 12 | MFS | Bronchopulmonary sputum |
| 05/11/95 | 18 | MSF | Bronchopulmonary sputum |
| 05/16/95 | 2 | pnICU | Bronchopulmonary sputum |
| 06/16/95 | 16 | MSF | Bronchopulmonary aspirates |
Nine strains were isolated in the first time period (1994), and nine others were isolated in the second time period (1995). Two of these isolates (5b and 6b) were obtained from the same patient. A suspected index case (7a) was identified.
Infection control policy.
During these time periods in an attempt to control the suspected outbreak, extra attention was paid to hand hygiene by using chlorhexidine 4% liquid soap (Zeneca Pharma, Cergy, France) or iodine polyvidone 4% (Betadine Scrub; Asta Medica Laboratory, Merignac, France) for daily hand washing; care of intubated patients was also reviewed. All communal respiratory function equipment had disposable mouthpieces, and all patients had their own respiratory therapy equipment. The patients who were infected with A. baumannii were placed in single rooms and were kept in strict isolation.
Speciation and antimicrobial susceptibility testing.
All strains were identified as belonging to the genus Acinetobacter on the basis of the following properties: the presence of nonmotile coccobacilli that were gram-negative, strictly aerobic, catalase positive, and oxidase negative. All isolates were identified by the API 20NE system (API-bioMérieux, Marcy l’Etoile, France) according to the manufacturer’s instructions (3), and A. baumannii were identified by growth in brain heart infusion broth at 44°C (Difco Laboratories, Detroit, Mich.). Antibiotic susceptibility profiles of the strains were determined by the disc diffusion method (21). Plates of Mueller-Hinton agar (Sanofi Diagnostics Pasteur, Marne-la-Coquette, France) were inoculated with a bacterial suspension equivalent to a 0.5 McFarland standard (22) and incubated aerobically at 37°C for 18 h. Results were expressed as susceptible or resistant according to the criteria adopted by the antibiogram Committee of the French Society for Microbiology (1). The antibiotics tested were ticarcillin (75 μg) and ticarcillin plus clavulanic acid (75 and 10 μg; SmithKline Beecham, Philadelphia, Pa.), piperacillin (75 μg) and piperacillin plus tazobactam (75 and 10 μg; Cyanamid-Lederle, Pearl River, N.Y.), cefsulodin (30 μg); Takeda Laboratories, Puteaux, France), cefoperazone (30 μg; Pfizer Laboratories, Orsay, France), ceftazidime (30 μg; Glaxo, Greenford, United Kingdom), aztreonam (30 μg; Sanofi Winthrop, Gentilly, France), imipenem (10 μg; Merck Sharp & Dohme, West Point, Pa.), colistin (50 μg; Roger Bellon, Neuilly-sur-Seine, France), ofloxacin (5 μg; Roussel, Puteaux, France), ciprofloxacin (5 μg; Bayer-Pharmacia, Puteaux, France); gentamicin (15 μg) and netilmicin (30 μg) Schering-Plough, Levallois-Perret, France), tobramycin (10 μg; Eli Lilly, Indianapolis, Ind.), and amikacin (30 μg; Bristol-Myers Squibb, Paris, France).
Isolation of chromosomal DNA and enzyme digestion.
The procedure followed was as described previously (14) with minor modifications. In brief, strains were grown with shaking in 10 ml of tryptone-casein-soy broth overnight at 37°C and centrifuged at 10,000 × g for 15 min at 4°C. The pellet was resuspended in sucrose buffer (5 g of saccharose in 2 M Tris-HCl [pH 7.5]). The suspension was treated with 0.5 M EDTA and then with lysozyme (0.5 mg of lysozyme in 2 ml of 50 mM Tris-HCl [pH 7.5]) for 10 min at 4°C and finally with 6 M guanidine chloride and 7.5 M ammonium acetate. The cells were lysed by gentle mixing with 10% Sarkosyl and proteinase K (10 mg/ml) at 60°C for 60 min. DNA was precipitated by the addition of 3 volumes of cold 96% ethanol in a freezer for at least 60 min at −20°C. The DNA pellet was resuspended in 2 M Tris-HCl (pH 7.5)–5 M NaCl–0.5 M EDTA solution. Optical densities at 260 and 280 nm were used to estimate the DNA concentration and purity. Purified bacterial DNA (3 μg) was digested by restriction enzymes EcoRI and SalI according to the manufacturer’s instructions (Boehringer Mannheim-GmH Sandhofer, Mannheim, Germany) and separated electrophoretically through a 0.8% horizontal agarose gel (Bioprobe, Montreuil-sous-Bois, France) in TEA buffer (40 mM Tris-acetate, 2 mM EDTA [pH 8.0]) overnight at a constant 25 V. After electrophoresis, the gel was stained with ethidium bromide (1 μg/ml) and visualized under UV light at 302 nm (Macrovue Transilluminator Model; Pharmacia Biotech, Uppsala, Sweden).
Hybridization.
The DNA fragments were vacuum transferred to a nylon membrane (Amersham, les Ulis, France) for 120 min at 37°C. Hybridization was performed at 60°C with a commercially available 16S+23S rRNA from an Escherichia coli probe labelled with acetylaminofluorene (AAF; Eurogentec, Sercung, Belgium) (17). The nylon membranes were hybridized with the AAF-labelled ribosomal RNA probe as described elsewhere (18). Immunoenzymatic detection was carried out according to the manufacturer’s instructions (Boehringer Mannheim). Raoul I (Appligene, Illkirch, France) was used as the size marker in all blots. The hybrids were detected by the presence of purplish blue bands which appeared within a few minutes to 1 h, and the reaction was stopped by washing the film in tap water.
RESULTS
Our study was carried out with a total of 1,706 patients, among whom 865 were hospitalized from June to November 1994 and 841 were hospitalized from February to June 1995; patient days totaled 28,510. The mean length of stay in the care units was 16.7 days (range, 2 to 210 days). There were a total of 18 A. baumannii nosocomial infections in 17 patients (0.98% infected patient), giving an incidence rate of nosocomial infection of 1%, i.e., 0.60 infection per 1,000 patient days (Table 3).
TABLE 3.
Summary of nosocomial infectionsa
| Infections type | No. of infections | Incidence of nosocomial infections (%) | No. of infections/1,000 patient days |
|---|---|---|---|
| Pulmonary infection | 10 | 0.5 | 0.33 |
| Urinary tract infection | 4 | 0.3 | 0.14 |
| Peritonitis | 2 | 0.1 | 0.07 |
| Septicemia | 1 | 0.05 | 0.03 |
| Catheter tip infection | 1 | 0.05 | 0.03 |
| Total | 18 | 1.00 | 0.60 |
A total of 18 nosocomial infections were observed in 17 patients, giving an incidence rate of nosocomial infection of 1%, i.e., 0.60 infections per 1,000 patient days.
These 17 patients were grouped as follows: 8 polytraumatized patients, 5 postoperative patients, and 4 medical patients. A. baumannii was cultured from different sources in these patients. The 18 isolates of A. baumannii were obtained from different sites: bronchopulmonary sputum (n = 10), urine (n = 4), peritoneal fluid (n = 2), blood sample (n = 1), and catheter tips (n = 1). There were 18 cases of nosocomial infection (10 pulmonary infections, 4 urinary tract infections, 2 peritoneal infections, 1 septicemia, and 1 catheter infection). The 17 infected patients presented the following characteristics: 10 patients with pneumonia were mechanically ventilated at the time of A. baumannii isolation for a mean period of 8.5 days (range, 1 to 25 days); 4 patients who presented with urinary tract infections were probed at the time of A. baumannii isolation for a mean period of 11.5 days. We observed two cases of peritonitis; of these, one was spontaneous peritonitis, and the other was postoperative peritonitis. Finally, all of the patients with A. baumannii had one or more catheters. Only one catheter infection and one case of septicemia with positive blood cultures was observed; the origin of this latter infection was unknown. The clinical course was also considered to be affected by A. baumannii in 16 patients, 4 of whom died. The cause of death was known for two patients: one (the index case) succumbed to the aftereffects of A. baumannii peritonitis and the other one died from the aftereffects of pulmonary infection. The cause of death for the two others was not known but was probably due to underlying diseases.
Antibiotyping method.
The outbreak was initially recognized on the basis of the resistance profile of the A. baumannii isolates. Of the 18 isolates, 15 were resistant to ticarcillin, ticarcillin plus clavulanic acid, piperacillin, piperacillin plus tazobactam, cefsulodin, cefoperazone, ceftazidime, and aztreonam for antibiotypes a and c. Regarding the remaining three isolates, two were susceptible to ticarcillin, ticarcillin plus clavulanic acid, piperacillin, and piperacillin plus tazobactam, and were resistant to cefsulodin, cefoperazone, ceftazidime, and aztreonam (antibiotype b), and one was susceptible to ticarcillin, ticarcillin plus clavulanic acid, piperacillin, piperacillin plus tazobactam, cefsulodin, cefoperazone, ceftazidime, and aztreonam (antibiotype d). On the other hand, all of the isolates were susceptible to imipenem and colistin and were resistant to ofloxacin and ciprofloxacin. Resistance to gentamicin, tobramycin, netilmicin, and amikacin was observed in 16 of 18 isolates (antibiotypes a and b); as for the remaining two isolates, one was susceptible to gentamicin and amikacin and was resistant to tobramycin and netilmicin (antibiotype c), and the other was susceptible to gentamicin, tobramycin, netilmicin, and amikacin (antibiotype d). Among the four resistance phenotypes observed, four included isolates of period 1, i.e., (i) for strains 1, 7, 10, 11, 13, and 15 (pICU); (ii) for strain 3 (pICU); (iii) for strain 14 (ntICU); and (iv) for strain 17 (neurosurgical ward); and two included isolates of period 2, i.e., (i) for strains 4 (npICU), 5, and 6 (pICU); strains 8 and 9 (ntICU); and strain 12, 16, and 18 (MFS surgical ward) and (ii) for strain 2 (pnICU). Finally, antibiotyping allowed separation of the 18 isolates into four resistance phenotypes: antibiotype pattern a for strains 1, 4 to 13, 15, 16, and 18 (78.8% of strains); pattern b for strains 2 and 3 (11.2%); pattern c for strain 14 (5.5%); and pattern d for strain 17 (5.5%) (Table 4).
TABLE 4.
Epidemiological markers of A. baumannii isolated from Amiens teaching hospital patientsa
| Strain no. | Antimicrobial resistanceb
|
Antibio-types | Ribotypes
|
Epidemic group | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TIC | TCC | PIP | TAZ | CFS | CFP | CAZ | ATM | IPM | CS | OFX | CIP | GM | TM | NET | AN | EcoRI | SalI | |||
| 1 | R | R | R | R | R | R | R | R | S | S | R | R | R | R | R | R | a | A1 | E | III |
| 4 | R | R | R | R | R | R | R | R | S | S | R | R | R | R | R | R | a | A | F1 | I |
| 5 | R | R | R | R | R | R | R | R | S | S | R | R | R | R | R | R | a | A | F2 | I |
| 6 | R | R | R | R | R | R | R | R | S | S | R | R | R | R | R | R | a | A3 | F1 | IV |
| 7 | R | R | R | R | R | R | R | R | S | S | R | R | R | R | R | R | a | A | F | I |
| 8 | R | R | R | R | R | R | R | R | S | S | R | R | R | R | R | R | a | A | F | I |
| 9 | R | R | R | R | R | R | R | R | S | S | R | R | R | R | R | R | a | A | F | I |
| 11 | R | R | R | R | R | R | R | R | S | S | R | R | R | R | R | R | a | A5 | F1 | II |
| 12 | R | R | R | R | R | R | R | R | S | S | R | R | R | R | R | R | a | B | F3 | V |
| 13 | R | R | R | R | R | R | R | R | S | S | R | R | R | R | R | R | a | A5 | F3 | II |
| 15 | R | R | R | R | R | R | R | R | S | S | R | R | R | R | R | R | a | C | F4 | VI |
| 16 | R | R | R | R | R | R | R | R | S | S | R | R | R | R | R | R | a | A6 | G | VII |
| 18 | R | R | R | R | R | R | R | R | S | S | R | R | R | R | R | R | a | D | F4 | VIII |
| 2 | S | S | S | S | R | R | R | R | S | S | R | R | R | R | R | R | b | A2 | F | IV |
| 3 | S | S | S | S | R | R | R | R | S | S | R | R | R | R | R | R | b | A2 | F1 | II |
| 10 | R | R | R | R | R | R | R | R | S | S | R | R | R | R | R | R | a | A4 | F1 | II |
| 14 | R | R | R | R | R | R | R | R | S | S | R | R | S | R | R | S | c | A5 | F3 | II |
| 17 | S | S | S | S | S | S | S | S | S | S | R | R | S | S | S | S | d | D | F5 | VIII |
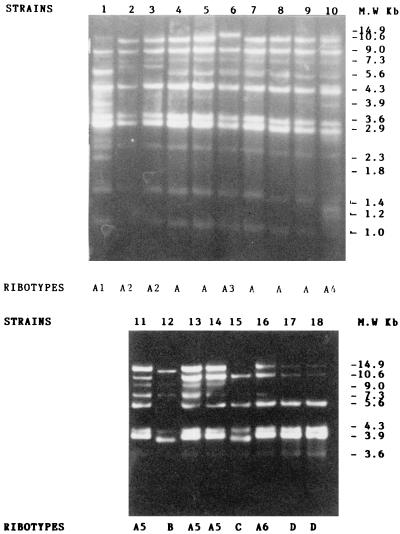
The antibiogram result showed four resistance phenotypes (a, b, c, and d). By using EcoRI, four different ribotypes were found (A, B, C, and D), and five subtypes were found (A1, A2, A3, A4, and A5). With SalI, three ribotypes (E, F, and G) were observed among strains that appeared to be variants of ribotype A (A1, A2, and A6) with EcoRI. Seven ribotypes were distinguished by both restriction enzymes (A, B, C, D, E, F, and G). By combining antibiotyping and ribotyping, eight groups of strains (groups I to VIII) were defined.
Abbreviations: TIC, ticarcillin; TCC, ticarcillin plus clavulanic acid; PIP, piperacillin; TAZ, piperacillin plus tazobactam; CFS, cefsulodin; CFP, cefoperazone; CAZ, ceftazidime; ATM, aztreonam; IPM, imipenem; CS, colistin; OFX, ofloxacin; CIP, ciprofloxacin; GM, gentamicin; TM, tobramycin; NET, netilmicin; AN, amikacin. S, susceptible; R, resistant.
Ribotyping.
Chromosomal DNA restriction patterns were interpreted according to the Tenover criteria for pulsed-field gel electrophoresis (26). Total genomic DNA from the 18 clinical isolates was digested separately with two restriction enzymes, EcoRI and SalI. The ribotypes found are shown in Fig. 1 and 2. With EcoRI, we identified four distinct ribotypes, designated A, B, C, and D, and six subtypes (A1, A2, A3, A4, A5, and A6). Ribotype A included five isolates (strains 4, 5, 7, 8, and 9) genetically indistinguishable regarding the number of bands (i.e., 11) and the size of fragments located in the region between 10.6 and 1 kbp. These isolates were all considered to represent the same strain, and we considered them outbreak strains. The subtype A1 represented by isolate 1 showed 12 bands located in the region between 10.6 and 1 kbp and differed from ribotype A with respect to three fragments which appeared in the 3.9-kbp region between 2.9 and 2.3 kbp and at 1.8 kbp. The subtype A2 corresponded to isolates 2 and 3, included 10 bands, and differed from ribotype A due to the presence of a fragment in the region between 7.3 and 5.6 kbp. The subtype A3 of isolate 6 contained 10 bands located in the region between 14.9 and 1 kbp and differed from ribotype A with respect to a large fragment that appeared in the region of 14.9 kbp. The subtype A4 of isolate 10 included 12 bands (in the region between 10.6 and 1 kbp) and differed from ribotype A with respect to two small fragments located in the regions of 3.9 and 1.2 kbp. All of these subtypes were closely related to the outbreak isolates A and were considered as variants of type A. The subtype A5, represented by isolates 11, 13, and 14, included seven bands and differed from ribotype A due to the presence of fragments in the regions of 14.9 and 3.9 kbp. The subtype A6 (isolate 16) contained seven bands and shared the same bands as subtype A1, except for the lack of a band in the region of 9 kbp and the presence of small bands in the regions of 7.3 and 3.6 kbp. These subtypes 5 and 6 were considered to be possibly related to outbreak isolates A. However, analysis of their ribotype patterns showed that they were not closely related genetically and consequently were less likely to be related epidemiologically, and thus we consider them to be unrelated strains. The ribotype B represented by the isolate 12 showed five bands (in the region between 14.9 and 3.9 kbp) and differed from ribotype A due to the presence of three small fragments located in the regions of 9 and 4.3 kbp. The ribotype C corresponded to isolate 15, included five bands (in the region between 10.6 and 3.6 kbp) and differed from ribotype A in the number of deleted fragments (six total). The ribotype D, corresponding to isolates 17 and 18, included six bands in the region between 14.9 and 3.6 kbp and differed from ribotype A in the presence of two small bands in the regions 14.9 and 10.6 kbp and in the number of deleted fragments (six total). All of these ribotypes were considered to be unrelated to ribotype A.
FIG. 1.
EcoRI ribotype patterns of A. baumannii strains.
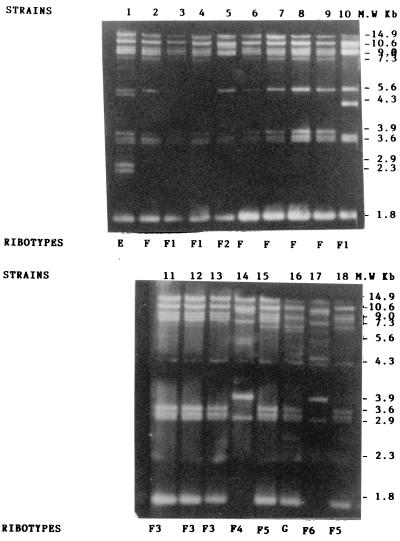
FIG. 2.
SalI ribotype patterns of A. baumannii strains.
Restriction of 18 isolates with SalI gave three different patterns labelled E, F, and G and five subtypes labelled F1, F2, F3, F4, and F5. The three ribotypes included between 8 and 12 bands located in the region between 14.9 and 1.8 kbp. Ribotype F included 5 isolates (strains 2, 6, 7, 8, and 9) genetically indistinguishable due to the size of their different fragments and the presence of the same number of bands (eight total) located in the region between 14.9 and 1.8 kbp. These five isolates were all considered to represent the same strain and were considered to be outbreak strains. The subtype F1 of isolates 3, 4, and 10 differed from outbreak type F in the number of bands (seven total). The subtype of isolate 10 was characterized by the presence of an additional band located in the region of 4.3 kbp, while those of isolates 3 and 4 differed from F due to the absence of a band located in the region of 5.6 kbp. The subtype F2 of isolate 5 differed from outbreak type F due to the number of bands (six total) and the lack of small fragments located in the regions of 7.3 and 3.9 kbp. The subtype F3 concerned three isolates (strains 11, 12, and 13) and included the same number of bands (eight total) as the outbreak type F but differed due to a deletion in the region of 5.6 kbp and the presence of two small fragments in the regions of 4.3 and 2.3 kbp. The subtype 4 of isolate 14 contained six bands and differed from outbreak type F due to the size of a band located in the region of 5.6 kbp and the presence of a fragment in the region of 2.9 kbp. The subtype F5 of isolates 15 and 18 included nine bands and differed from outbreak type F due to the absence of a band in the region of 5.6 kbp and the presence of a fragment in the regions of 4.3 and 2.3 kbp. The subtype F6 of isolate 17 contained nine bands and differed from outbreak type F due to the presence of two small fragments located between the regions 5.6 and 4.3 kbp and to fragments corresponding to regions 3.9 and 2.9 kbp. All of these subtypes were closely related to outbreak pattern F and were considered to be variants of ribotype F. Digestion of isolates 1 and 16 with SalI enabled us to identify two restriction patterns, designated E and G, respectively, that were different from those of outbreak pattern F. Ribotypes E and G comprised 13 and 12 bands, respectively, included in the regions located between 14.9 and 1.8 kbp. These ribotypes differed from F regarding the number of bands: 13 for E and 12 for G. Four additional insertions were found in E; two were located in the regions between 7.3 and 5.6 kbp and between 5.6 and 4.3 kbp, and two others were located between 2.9 and 2.3 kbp. Other differences were also observed in G, including insertions characterized by the presence of three weak bands located in the regions between 14.9 and 10.6 kbp, between 5.6 and 4.3 kbp, and between 2.9 and 2.3 kbp and two deletions near 14.9 and 3.9 kbp. These isolates were not part of the outbreak and were considered to be unrelated to ribotype F.
In total, we found four different ribotypes (A, B, C, and D) and five subtypes (A1, A2, A3, A4, and A5) with EcoRI. Ribotyping yielded between 12 and 5 bands per strain. Digestion of DNA by SalI yielded another three ribotypes (E, F, and G) among strains that appeared to be variants of ribotype A (A1, A2, A3, and A6) with EcoRI. By combining the results obtained with both enzymes, seven ribotypes were distinguished among the strains: A (strains 4, 5, 7, 8, and 9), B (strain 12), C (strain 15), D (strain 17 and 18), E (strain 1), F (strains 2 and 6), and G (strain 16), along with three variants, A2 (strain 3), A4 (strain 10), and A5 (strains 11, 13, and 14), which were closely related to outbreak isolates type A.
Epidemiology.
The combining typing results showed that eight groups of strains (groups I to VIII) were distinguished. The strains within each group were more similar to each other than to other strains (Table 3). Group I included five strains (strains 4, 5, 7, 8, and 9). Strains in this group had antibiogram pattern a and ribotype A, were epidemiologically related, and were outbreak strains; group II comprised five strains (strains 3, 10, 11, 13, and 14) that were characterized by antibiogram type a, except strains 3 and 14, which had antibiogram patterns b and c, respectively. These strains were closely related to outbreak strains A and appeared to be variants of ribotype A (A2 [strain 3]; A4 [strain 10]; A5 [strains 11, 13, and 14]); we considered them to be epidemiologically related isolates. Groups III, V, VI, and VII, comprising four strains (1, 12, 15, and 16, respectively) were distinguished by antibiogram type a and ribotypes E (strain 1), B (strain 12), C (strain 15), and G (strain 16). Group IV included strains 2 and 6. They had antibiogram types b (strain 2) and a (strain 6) and ribotype F. Group VIII contained strains 17 and 18 that had antibiogram types d and a, respectively, and ribotype D. These last eight strains were sporadic isolates from hospitalized patients, seven of whom came from the same geographic area; they were epidemiologically unrelated and represented different strain types. We considered them to be nonoutbreak strains.
Analysis of the strains isolated during the epidemic in different care units showed that of the nine pICU strains, two belonged to group I (strains 5 and 7), four belonged to group II (strains 3, 10, 11, and 13), one belonged to group III (strain 1), one belonged to group IV (strain 6), and one belonged to group VI (strain 15). In this care unit, we isolated both outbreak strains (strains 3, 5, 7, 10, 11, and 13) and nonoutbreak strains (strains 1, 6, and 15). The strains isolated in npICU (strain 4) and ntICU (strains 8, 9, and 14) belonged to group I, except for strain 14 (group II). All of these strains were outbreak strains. The strains isolated in the pnICU (strain 2 [IV]), in the MFS (strains 12 [V], 16 [VII], and 18 [VIII]), and in the neurosurgical unit (strain 17 [VIII]) were nonoutbreak strains.
DISCUSSION
The genus Acinetobacter has been increasingly associated with hospital infection. In our investigation, most nosocomial A. baumannii infections occurred in adult intensive care units and surgical wards, with the respiratory tract and urinary tract being the predominant sites of infection, although other sites have also been observed (blood, peritoneal fluid, and catheter). Using the CDC criteria (12), we found that 16 patients were infected and 1 patient presented with a catheter infection with this microorganism.
A. baumannii was associated with pulmonary infection in 10 cases, with urinary tract infection in 4 cases, peritonitis in 2 cases, septicemia in 1 case, and with catheter infection in 1 case. The data of our study showed a significant increase in the incidence of A. baumannii infection between 1994 and 1995 (during a period of 9 months) of 1% versus 0.4% during a period of 10 months from February to November 1993 and 0.2% in 1996 during the same 10-month period; this suggests an epidemic situation. The rate of nosocomial infections per 1,000 patient days was 0.6, and there was 0.33 pulmonary infection per 1,000 patient days and 0.14 urinary tract infection per 1,000 patient days (Table 3).
Spread of strains and control of measures.
The data concerning the spread of strains showed that the strains of group I, collected in 1994 from patients hospitalized in pICU and in 1995 from patients hospitalized in npICU, pICU, and ntICU, were widely disseminated in the hospital. The strains of group II, recovered in 1994 from patients hospitalized in pICU and ntICU, showed limited epidemic spread. The strains of groups III, IV, V, VI, VII, and VIII were sporadic strains observed during epidemic periods. Due to the absence of a bacteriological study of the medicosurgical material and surface material of patient bedrooms, we were unable to elucidate the reservoir and the mode of spread of the epidemic. It is likely that the infected patients and their environment constituted the source of infection.
This investigation also showed that several A. baumannii outbreaks had coexisted and/or followed one another at this time. Certain strains disappeared as measures of outbreak spread control began to be implemented. This was probably the case for the strains of group II that were observed during the first period of outbreak in 1994. The others (group I strains) were maintained and observed in 1994 as well as in 1995. At the beginning of the outbreak, hand hygiene and contact isolation procedures were implemented to control the acquisition and spread of multiresistant A. baumannii. Despite these control measures, new cases of A. baumannii infection still occurred. It was therefore decided to discharge all patients from the pICU, and the isolation rooms were scrupulously cleaned. During the following months, some new cases were detected despite these infection control measures. Therefore, it was decided to stop new admissions to the pICU. All admitted patients were transferred to other care units. The wards were then thoroughly disinfected. After these operations the incidence of A. baumannii dropped from 1% during the 9 study months (between 1994 and 1995) to 0.2% during the 10 months of 1996 after the interventions.
During the outbreak, one patient was infected in two different sites by two different strains. Strain 5 (group I) was isolated from a respiratory sample, and strain 6 (group IV) was found in a urinary sample from the same patient, thus showing that one patient could be infected by more than one strain of A. baumannii. Such a result was observed by Ling et al. (20). On the basis of the clinical data and the results of the typing methods, we confirmed that the epidemic index case was a patient transferred from a neighborhood teaching hospital to the Amiens teaching hospital. A strain of A. baumannii (strain 7) was isolated from peritoneal fluid as soon as this patient returned to our hospital. The antibioprofile a and ribopattern A of the strain from this patient was the same as that of the epidemic strain (group I).
In conclusion, we investigated various characteristics of 18 clinical isolates of A. baumannii, including the antibiogram and ribotype patterns. Analysis of the antibiogram profiles and band patterns generated by the ribotype method showed that most of the isolates were multiresistant. Among those, five (group I) were outbreak strains, five others (group II) were variants of one ribotype, and the remaining eight were nonoutbreak strains. In addition, the strains of group I, which were involved in the epidemic, had spread through different wards. Therefore, isolation of multiresistant strains of A. baumannii should alert the infection control team and prompt the implementation of stringent infection control measures. Despite the efficacy of the control measures, it is important to emphasize the need to pursue an epidemiological survey of nosocomial infections in the ICU and surgical wards.
REFERENCES
- 1.Antibiogram Committee of the French Society for Microbiology. Statement. Pathol Biol. 1996;44:I–VIII. [PubMed] [Google Scholar]
- 2.Aubert G, Grimont F, Zéni F, Pain P, Michel V P, Vautrin A C, Vedel G, Bouvet P. Acinetobacter baumannii outbreak isolates characterized by three typing methods. Eur J Clin Microbiol Infect Dis. 1995;14:1095–1099. doi: 10.1007/BF01590947. [DOI] [PubMed] [Google Scholar]
- 3.Bernards A T, Van Der Toorn J, Van Boven C P A, Dijkshoorn L. Evaluation of the ability of a commercial system to identify Acinetobacter genomic species. Eur J Clin Microbiol Infect Dis. 1996;15:303–308. doi: 10.1007/BF01695662. [DOI] [PubMed] [Google Scholar]
- 4.Bouvet P J M, Grimont P A D. Taxonomy of the genus Acinetobacter with the recognition of Acinetobacter baumannii sp. nov., Acinetobacter haemolyticus sp. nov., Acinetobacter johnsonii sp. nov., and Acinetobacter junii sp. nov. and emended descriptions of Acinetobacter calcoaceticus and Acinetobacter lwoffii. Int J Syst Bacteriol. 1986;36:228–240. [Google Scholar]
- 5.Bouvet P J M, Jeanjean S. Delineation of new proteolytic genomic species in the genus Acinetobacter. Res Microbiol. 1989;140:291–299. doi: 10.1016/0923-2508(89)90021-1. [DOI] [PubMed] [Google Scholar]
- 6.Bouvet P J M, Grimont P A. Identification and biotyping of clinical isolates of Acinetobacter. Ann Inst Pasteur Microbiol. 1987;138:569–578. doi: 10.1016/0769-2609(87)90042-1. [DOI] [PubMed] [Google Scholar]
- 7.Bouvet P J M, Jeanjean S, Vieu J F, Dijkshoorn L. Species, biotype, and bacteriophage type determinations compared with cell envelope protein profiles for typing Acinetobacter strains. J Clin Microbiol. 1990;28:170–176. doi: 10.1128/jcm.28.2.170-176.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brun-Buisson C. Catheters et infections. Questions actuelles. Lett Infectiol. 1990;V:373–378. [Google Scholar]
- 9.Dijkshoorn L, Aucken H M, Gerner-Smidt P, Kaufmann M E, Ursing J, Pitt T L. Correlation of typing methods for Acinetobacter isolates from hospital outbreaks. J Clin Microbiol. 1993;31:702–705. doi: 10.1128/jcm.31.3.702-705.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ehrenstein B, Bernards A T, Dijkshoorn L, Gerner-Smidt P, Towner K J, Bouvet P J M, Daschner F D, Grundmann H. Acinetobacter species identification by using tRNA spacer fingerprinting. J Clin Microbiol. 1996;34:2414–2420. doi: 10.1128/jcm.34.10.2414-2420.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garcia D C, Nociari M M, Sordelli D O, Di Martino A, Catalano M. The use of plasmid profile analysis and ribotyping for typing Acinetobacter baumannii isolates. J Hosp Infect. 1996;34:139–144. doi: 10.1016/s0195-6701(96)90139-5. [DOI] [PubMed] [Google Scholar]
- 12.Garner J S, Jarvis W R, Emori T G, Horan T C, Hughes J M. CDC definitions for nosocomial infections. Am J Infect Control. 1988;16:128–140. doi: 10.1016/0196-6553(88)90053-3. [DOI] [PubMed] [Google Scholar]
- 13.Gerner-Smidt P. Acinetobacter: epidemiological and taxonomic aspects. AMPIS. 1994;102(Suppl. 47):5–41. [PubMed] [Google Scholar]
- 14.Gerner-Smidt P. Ribotyping of Acinetobacter calcoaceticus-Acinetobacter baumannii complex. J Clin Microbiol. 1992;30:2680–2685. doi: 10.1128/jcm.30.10.2680-2685.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gouby A, Carles-Nurit M J, Bouziges N, Bourg G, Mesnard R, Bouvet P J M. Use of pulsed-field gel electrophoresis for investigation of hospital outbreaks of Acinetobacter baumannii. J Clin Microbiol. 1992;30:1588–1591. doi: 10.1128/jcm.30.6.1588-1591.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Graser Y, Klare I, Halle E, Gantenberg R, Buchholz P, Jacobi H D, Presler W, Schonian G. Epidemiological study of an Acinetobacter baumannii outbreak by using polymerase chain reaction fingerprinting. J Clin Microbiol. 1993;31:2417–2420. doi: 10.1128/jcm.31.9.2417-2420.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grimont F, Chevrier D, Grimont P A D, Lefevre M, Guesdon J L. Acetylaminofluorene-labelled ribosomal RNA for use in molecular epidemiology and taxonomy. Res Microbiol. 1989;140:447–454. doi: 10.1016/0923-2508(89)90065-x. [DOI] [PubMed] [Google Scholar]
- 18.Grimont F, Grimont P A D. Ribosomal nucleic acid gene restriction patterns as potential taxonomic tools. Ann Inst Pasteur Microbiol. 1986;137B:165–175. doi: 10.1016/s0769-2609(86)80105-3. [DOI] [PubMed] [Google Scholar]
- 19.Juni E. Genus III. Acinetobacter, Brisou and Prévot 1954. In: Krieg N R, Holt J G, editors. Bergey’s manual of systematic bacteriology. Vol. 1. Baltimore, Md: The Williams & Wilkins Co.; 1984. pp. 303–307. [Google Scholar]
- 20.Ling J M, Wise R, Woo T H S, Cheng A F. Investigation of the epidemiology of hospital isolates of Acinetobacter anitratus by two molecular methods. J Hosp Infect. 1996;32:29–38. doi: 10.1016/s0195-6701(96)90162-0. [DOI] [PubMed] [Google Scholar]
- 21.National Committee for Clinical Laboratory Standards. Performance standards for antimicrobial disk susceptibility test. Approved standard M2-A4. Villanova, Pa: National Committee for Clinical Laboratory Standards; 1990. [Google Scholar]
- 22.National Committee for Clinical Laboratory Standards. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Approved standard M7-A2. Villanova, Pa: National Committee for Clinical Laboratory Standards; 1990. [Google Scholar]
- 23.Seifert H, Gerner-Smidt P. Comparison of ribotyping and pulsed-field gel electrophoresis for molecular typing of Acinetobacter isolates. J Clin Microbiol. 1995;33:1402–1407. doi: 10.1128/jcm.33.5.1402-1407.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Seifert H, Schulze A, Baginski R, Pulverer G. Comparison of four different methods for epidemiologic typing of Acinetobacter baumannii. J Clin Microbiol. 1994;32:1816–1819. doi: 10.1128/jcm.32.7.1816-1819.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Struelens M J, Carlier E, Macs N, Serruys E, Quint W G V, Van Belkum A. Nosocomial colonization and infection with multiresistant Acinetobacter baumannii outbreak delineation using DNA macrorestriction analysis and PCR-fingerprinting. J Hosp Infect. 1993;25:15–32. doi: 10.1016/0195-6701(93)90005-k. [DOI] [PubMed] [Google Scholar]
- 26.Tenover F C, Arbeit R D, Goering R V, Mickelsen P A, Murray B E, Persing D H, Swaminathan B. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tjernberg I, Ursing J. Clinical strains of Acinetobacter classified by DNA-DNA hybridization. APMIS. 1989;79:595–605. doi: 10.1111/j.1699-0463.1989.tb00449.x. [DOI] [PubMed] [Google Scholar]
- 28.Walter H T. Acinetobacter baumannii serotyping for delineation of outbreaks of nosocomial cross-infection. J Clin Microbiol. 1989;27:2713–2716. doi: 10.1128/jcm.27.12.2713-2716.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]