Abstract
An initiating DNA double strand break (DSB) event precedes the formation of cancer-driven chromosomal abnormalities, such as gene rearrangements. Therefore, measuring DNA breaks at rearrangement-participating regions can provide a unique tool to identify and characterize susceptible individuals. Here, we developed a highly sensitive and low-input DNA break mapping method, the first of its kind for patient samples. We then measured genome-wide DNA breakage in normal cells of acute myeloid leukemia (AML) patients with KMT2A (previously MLL) rearrangements, compared to that of non-fusion AML individuals, as a means to evaluate individual susceptibility to gene rearrangements. DNA breakage at the KMT2A gene region was significantly greater in fusion-driven remission individuals, as compared to non-fusion individuals. Moreover, we identified select topoisomerase II (TOP2)-sensitive and CCCTC-binding factor (CTCF)/cohesin binding sites with preferential DNA breakage in fusion-driven patients. Importantly, measuring DSBs at these sites, in addition to the KMT2A gene region, provided greater predictive power when assessing individual break susceptibility. We also demonstrated that low-dose etoposide exposure further elevated DNA breakage at these regions in fusion-driven AML patients, but not in non-fusion patients, indicating that these sites are preferentially sensitive to TOP2 activity in fusion-driven AML patients. These results support that mapping of DSBs in patients enables discovery of novel break-prone regions and monitoring of individuals susceptible to chromosomal abnormalities, and thus cancer. This will build the foundation for early detection of cancer-susceptible individuals, as well as those preferentially susceptible to therapy-related malignancies caused by treatment with TOP2 poisons.
Keywords: DNA fragility, topoisomerase, acute myeloid leukemia, CTCF, cohesin
1. INTRODUCTION
DNA double-stranded breaks (DSBs) are an incredibly harmful type of DNA damage event that occurs in cells, and the illegitimate repair of these breaks can result in chromosomal abnormalities.1–3 Chromosomal abnormalities are involved in driving oncogenesis of various types of cancer, both solid and hematologic, with 65% of cancer genes being identified in chromosomal abnormality events.4–8 Unfortunately, cancers are often discovered later into the progression of the disease, decreasing the overall prognosis/survival for patients.9,10 Standard cancer screening practices include imaging-based approaches such as mammograms or colonoscopies, as well as genetics-based approaches such as fluorescence in situ hybridization. While these screening/detection methods have improved patient welfare and survival odds in the past,11,12 they cannot be used to preemptively identify healthy patients at-risk for cancer. Most importantly, these methods fail to address the root cause of the mutations, and therefore the cancers, themselves – the DNA breaks.
Multiple factors, both endogenous and exogenous, contribute to the frequency of DNA breaks in cells, such as inter-individual differences in DNA repair capacity, aging, and exposure to environmental or chemotherapy chemicals.13–18 Attempts have been made to assay DNA repair protein function in order to identify individuals with decreased repair potential and therefore increased cancer susceptibility.13,14 Additionally, previous screening methods to determine exposure risks included either anecdotal evidence from patients and/or low-resolution and low-throughput DNA break assays such as comet assays, PCR-based approaches, or the measurement of DNA repair protein activity.19–21 As DNA breaks must precede all chromosomal abnormalities, and measurement of these breaks represents the balance between individual exposure and inherent DNA repair capacity, accurately and sensitively quantifying genome-wide DNA breaks in patients is a key way to identify those most at-risk for developing chromosomal abnormalities and thus cancer.
Recently, many DNA break mapping methods have been developed such as END-seq, BLESS, and DSBCapture.22–24 While all of these methods have shown efficacy with mapping DNA breaks with single-nucleotide resolution genome-wide, two major limitations of these methods exist for use with patient samples – timing/location of break mapping and required input quantity. Most current genome-wide break mapping techniques require capturing DNA breaks within intact cells/nuclei, and as a result, these methods require extremely large quantities of cells in order to capture enough DNA breaks. Quantities of patient samples are frequently scarce due to multiple research groups needing a portion and/or due to standard collection practices in hospitals/clinics. Therefore, the current break mapping techniques are not feasible or sustainable as a screening method with patient samples.
Here we developed a low-input method, based on the previously published DSBCapture method,24 thereby making it applicable to samples with limited sources such as patient samples. We validated the accuracy and sensitivity of this low-input, single-nucleotide, genome-wide DNA break mapping method in human cell culture lines. Importantly, using acute myeloid leukemia (AML) patient samples, we demonstrated that DNA break mapping can differentiate between patients who had fusion-driven AML and patients who did not, as well as discern break susceptibility at different fusion-associated genes. We further identified topoisomerase II (TOP2)-sensitive, CCCTC-binding factor (CTCF), RAD21, and SMC3 binding regions that were preferentially break-prone in fusion-driven AML patients compared to non-fusion patients. Evaluation of DSBs at the sites shared among all four features provided improved assessment when identifying break-prone individuals. Moreover, we demonstrated that DNA break mapping was effective at identifying regions susceptible to chemical-induced DNA breaks in patient cells. This highly sensitive method can be used in future applications such as identifying and monitoring at-risk individuals prior to any cancer formation, screening patients before receiving chemotherapy drugs known to cause secondary malignancies, or identifying novel regions of cancer susceptibility.
2. MATERIALS AND METHODS
2.1. AML patient blood sample procurement
De-identified peripheral blood samples of AML patients were collected by the Oncology Research Information Exchange Network (ORIEN) at the University of Virginia using a protocol approved by the University of Virginia Institutional Review Board (UVA-IRB) for the consenting, specimen procurement and processing, clinical data abstraction, and access to the molecular and clinical data. Peripheral blood mononuclear cells were isolated from fresh patient blood samples using a Ficoll gradient, cultured, and treated with chemicals as described below. The study methodologies were approved by UVA-IRB.
2.2. Cell culture and treatments
HeLa cells (ATCC) and GM13069 cells (ATCC) were grown in DMEM (Gibco) and RPMI 1640 medium (Gibco), respectively, and supplemented with 10% fetal bovine serum (FBS). Patient-derived cells were cultured in RPMI 1640 medium (Gibco) with 10% FBS and treated for 24 hours with either 0.3 μM or 1.5 μM etoposide (Sigma), along with untreated cells.
2.3. Genomic DNA purification
Genomic DNA was carefully extracted from treated and untreated cells. Briefly, genomic DNA was purified by gently lysing cells in 50 mM Tris-HCl (pH 8.0), 100 mM EDTA, 100 mM NaCl, 1% SDS, 1mg/mL Proteinase K for 3 h at 55°C followed by organic extraction purification and ethanol precipitation. Precaution such as gentle pipetting with wide-opening pipette tips to avoid/minimize shearing DNA was taken to avoid introduction of DNA breaks during purification.
2.4. Genome-wide break mapping and sequencing
Detection of DSBs using purified genomic DNA was performed as described.25 Purified genomic DNA was subjected to blunting/A-tailing reactions, Illumina P5 adaptor ligation to capture broken DNA ends. Excess adaptor was removed and then DNA was fragmented by sonication, and subsequently ligated to Illumina P7 adaptor, and the libraries were PCR-amplified for 15 cycles. Prepared libraries were then subjected to whole-genome, 75-bp and 150-bp paired-end sequencing with the Illumina NextSeq 500 and HiSeq X Ten platform, respectively.
2.5. DSB read processing
Sequencing reads were aligned to the human genome (GRCh38/hg38) with bowtie2 (v.2.3.4.1) aligner running in high sensitivity mode (--very-sensitive). Restriction on the fragment length from 100 nt to 2000 nt (-X 2000 -I 100 options) was imposed. Unmapped, non-primary, supplementary and low-quality reads were filtered out with SAMtools (v. 1.7) (-F 2820). Furthermore, PCR duplicates were marked with picard-tools (v. 1.95) MarkDuplicates, and finally, the first mate of non-duplicated pairs (-f 67 -F 1024) were filtered with SAMtools for continued analysis. For each detected break, the most 5’ nucleotide of the first mate defined the DNA break position. Sequencing and alignment statistics for the DSB mapping/sequencing libraries prepared from purified genomic DNA of GM13069, HeLa, and ORIEN samples are listed in Supporting Information Tables S1 and S2, along with data generated in nuclei of GM13069 and HeLa.26 Data of biological duplicates are also included.
2.6. Genome median calculation and normalization
DNA break coverage (RPM) for 1 kb bins was calculated genome-wide for all AML patient samples. The median breakage value was then used to normalize each respective sample’s DSB coverage within a given region. For regions smaller or larger than 1 kb, the median breakage values were appropriately calculated/adjusted to account for size.
2.7. Downstream data analysis
Downstream data analysis following DSB read processing was performed with BEDtools (v. 2.27.1) and standard Linux commands to compute coverages and annotation densities. Results were visualized in Python3 (v. 3.6.5) with matplotlib (v. 2.2.2), numpy (v. 1.15.0), pandas (v. 0.23.3), and seaborn (v. 0.11.0). Statistical tests were performed using Python3 (v. 3.6.5) with scipy stats (v. 0.19.1).
2.8. Processing publicly available data
TOP2-sensitive sites in GM13069 (GRCh38/hg38) can be accessed from Szlachta et al.26 High-throughput sequencing data used in this study were downloaded from Gene Expression Omnibus (GSE numbers), or from ENCODE project27 through the UCSC Genome Browser (GSE and wgENCODE numbers)28 for GM12878 cells, CTCF ChIP-seq (ENCSR000AKB), RAD21 ChIP-seq (ENCSR000EAC), SMC3 ChIP-seq (ENCSR000DZP),27 ATAC-seq (GSE103301),29 and RNA-seq (SRR1153470)30 data; for HeLa cells, CTCF ChIP-seq (ENCSR000AKJ);27 for RPE-1 cells, CC-seq data (SRP187576).31 All data sets used are listed in Supporting Information Table S3.
The publicly available data for GM12878 RNA-seq (SRR1153470) were aligned to the GRCh38/hg38 genome using HISAT232 aligner, and the gene expression (FPKM values) were quantified using StringTie.33 In individual analyses, the expression of the genes was used to define different numbers of bins for further analysis.
The publicly available data for HeLa CTCF (ENCSR000AKJ), GM12878 CTCF (ENCSR000AKB) RAD21 (ENCSR000EAC), SMC3 (ENCSR000DZP), and ATAC-seq (GSE103301), and each associated input data were downloaded and aligned to the GRCh38/hg38 genome using bowtie2 (v 2.3.4.1). Binding peaks were called by macs2 (2.1.1.20160309). In individual analyses the peak strength as defined by macs2 was used to define different numbers of bins for further analyses. For ChIP-seq and ATAC-seq data macs2 was run with default settings with each dataset controlled for with the matching input data. Peak summits were then used to center the regions of interest in all other analyses. Processing of data from Gittens et al. 31 is detailed in “Analysis of CC-seq”.
2.9. Correlation plots
The human genome build GRCh38/hg38 was binned into 10 kb windows using BEDtools makewindows function. Then all bins that intersected the hg38 blacklist34 sites and centromeres were removed. The coverage in all remaining 10 kb bins was calculated for the bam file of each replicate using BEDtools coverage (n = 282,862). The read coverage in each bin is normalized to total read number (reads per million, RPM), then all bins where both samples had zero coverage were removed. Next, the absolute difference in coverage in each bin between replicates is calculated, and the top 0.05% most different (defined as outliers), were removed. Finally, data were read into Python3, read normalized coverage was plotted between the samples, and Pearson correlation was calculated.
2.10. Genomic Regions Annotation
To assign genomic annotations, BEDtools (v. 2.27.1) intersect was used to sequentially assign genomic features with each region only being assigned to one genomic feature. The sequential feature assignment filters out regions as they are assigned to a feature. The order for assigning genomic features was TSS, promoter, TTS, gene body, and those not assigned to any of these features are coded as intergenic. The GRCh38/hg38 build RefSeq genes were downloaded from the UCSC browser. The definitions used for each genomic feature is as follows: promoter region ranging from TSS –1000 nt to –250 nt, TSS region ranging from TSS −250 to +250 nt, gene body region ranging from TSS +250 nt to TTS −250 nt, and TTS regions ranging from TTS −250 nt to +250 nt.
2.11. Single-nucleotide cumulative plots at TOP2-sensitive, CTCF, RAD21, and SMC3 binding sites
To analyze DSBs located at CTCF-binding sites, CTCF ChIP-seq (ENCSR000AKB) was used, because both GM13069 and GM12878 are non-malignant lymphoblastoid cells. The strongest (top 10%) and weakest (bottom 10%) CTCF-binding sites were determined based on macs2 score (n = 4019 each) of CTCF ChIP-seq data. DSB coverage in these regions was determined using BEDtools coverage reporting the depth at each position in the reference regions (-d). Then the merge function was used to compile each region’s coverage into a single line readable to Python3. Using Python3 (v. 3.6.5) with matplotlib (v. 2.2.2), numpy (v. 1.15.0) and pandas (v. 0.23.3), the cumulative single-nucleotide break profiles were plotted over the relative nucleotide position to the CTCF ChIP-seq peak summit, and in the ± 500 bp flanking regions with read normalization (reads per million, RPM). DSBs located at strong CTCF-binding sites (n = 6911) in HeLa cells, using CTCF ChIP-seq (ENCSR000AKJ), were analyzed similarly. For preferential DNA break sites in the KMT2A (previously MLL)-rearranged remission patients at TOP2-sensitive, CTCF, RAD21, and SMC3 binding sites, the same process for coverage and plotting was employed within the regions of interest. Coverage was computed with either each individual patient file or with merged KMT2A-rearranged patient and merged non-fusion patient files.
2.12. Genome track images
Genome track images were made by using igvtools (v. 2.3.68) count with the options to have windows of 5 bp and precompute only 5 bp (-w 5 -z 5) for the GRCh38/hg38 build of the human genome. The resulting tdf files were loaded into the IGV browser, with the track normalization function checked in the track options to read-normalize the data, and break data tracks were set to group auto-scale. Images were then saved out from the current IGV browser view.
2.13. BART analysis on KMT2A patient-specific DNA break-prone regions
BART,35 an unbiased transcriptional regulator prediction method, was used to identify potential transcriptional regulators demonstrating preferential enrichment at KMT2A patient-specific DNA break-prone regions. Read- and size-normalized coverage (RPKM, read per kilobase per million reads) was calculated for all six patient samples at union DNaseI hypersensitive sites (UDHSs), as repertoire of all regulatory regions in the human genome. A differential coverage score on each UDHS was then calculated by subtracting the average coverage of the non-fusion patients from the average coverage of the KMT2A-rearranged patients. BART35 was then applied on the scored UDHS list to infer transcriptional regulators preferentially binding in regions with greater DNA breaks in KMT2A-rearranged patients compared to non-fusion patients.
2.14. Identification of preferential DNA break sites in the KMT2A-rearranged patients at TOP2-sensitive, CTCF, RAD21, and SMC3 binding sites
Read- and median-normalized break coverage was calculated for all six patients for 18791 TOP2-sensitive sites,26 40189 CTCF bindings sites, 33854 RAD21 binding sites, and 16270 SMC3 binding sites.27 We identified regions with a statistically significant difference in normalized DNA break coverage between KMT2A and non-fusion patients (P< 0.05, Student’s t-test): 1114 TOP2-sensitive, 2263 CTCF, 1988 RAD21, and 953 SMC3 binding regions. Among them, we found 695 TOP2-sensitive, 1240 CTCF, 1073 RAD21, and 551 SMC3 binding regions where all of the KMT2A patients have greater DNA breakage than all of the non-fusion patients. Finally, we identified 304 TOP2-sensitive sites, 404 CTCF, 517 RAD21, and 256 SMC3 bindings sites of interest in which the KMT2A to non-fusion breakage ratio was greater than that observed for the KMT2A region.
2.15. Identification and characterization of the 34 shared/common regions
To identify the common regions between the CTCF, TOP2, RAD21, and SMC3 sites, we used the regions for each data set prior to filtering for the KMT2A:NF breakage ratio, as the final stage of filtering (KMT2A:NF ratio) was stringent. We found 59 regions that were shared among the four data sets. Normalized DSB coverage was calculated for the merged common regions, and there were 34 regions with an KMT2A:NF DSB ratio greater than or equal to the KMT2A intron 10/exon 11 region. Genomic coordinates of the 34 common sites (build GRCh38/hg38) are listed in Supporting Information Table S4. The 34 common regions were then further characterized by analyzing for the presence of fragile sites,36 candidate cis-regulatory elements,37 large structural variants,38 and small/single nucleotide variations.39
2.16. Analysis of CC-seq data
The CC-seq data from Gittens et al.31 were downloaded as fastq files from (GSE136943) and then aligned to the human genome (build GRCh38/hg38) following the same processing as break data (as detailed above in ‘DSB read processing’). The matched sets of VP16-treated WT and TOP2B−/− RPE-1 cells in both asynchronous and G1 arrested cells had replicates merged, respectively, and the coverage from each was calculated in the preferential DNA break sites defined above.
2.17. Analysis of gnomAD structural variants
The gnomAD structural variants bed file was downloaded from the project website (https://gnomad.broadinstitute.org/downloads#v2-structural-variants). Start and end coordinates were then separated to single nucleotide coordinates and coded as start or end. Then for each set of preferential DNA break sites, TOP2-sensitive, CTCF, RAD21, and SMC3, the distance to the closest structural variants was determined using the BEDtools closest tools. Then using pybedtools (v. 0.8.0) with the pybedtools.parallel.parallel_apply tool, 1000 shuffled iterations of the DNA preferential break sites, that maintained chromosome identity, were then assessed for their distance to the closest structural variants. Histograms of the regions of preferential DNA break sites and the random shuffled regions were plotted using matplotlib.pyplot.hist with option to normalize between the single set for the regions of interest and the 1000 sets for the random shuffle (density = True). Fold enrichment of regions within 25 kb of gnomAD structural variants was determined by calculating the fold enrichment between the number of regions of preferential DNA break sites that were within 25 kb of structural variants and each iteration of the random shuffle.
2.18. Statistics
Statistical analysis was carried out using scipy stats (v. 0.19.1). Tests are specified in figure legends, and statistical significance is denoted by asterisks; * P-value < 0.05, ** P-value < 0.01, and *** P-value < 0.001; unless stated otherwise.
3. RESULTS
3.1. DNA DSB mapping with low-input purified genomic DNA
Several different DSB mapping methods have been developed,22–24 which map DNA breaks within nuclei and require large quantities of input material (5–70 million, or 30–424 μg DNA) due to reduced ligation efficiency. Patient samples often yield low amounts of genomic DNA (gDNA) due to limited quantities of cells or tissue available for research purposes, making it difficult to sensitively assess DNA breakage levels and subsequent cancer susceptibility in patients. To effectively map and quantify DNA breakage in patient samples, the break mapping method DSBCapture was adapted to accommodate low-input. We propose that mapping DNA breaks with carefully isolated genomic DNA will improve ligation efficiency, allowing for the use of less input. Here, we adapt the DSBCapture method to map DSBs using low amounts of isolated genomic DNA — 2 μg (~330,000 cells) and 500 ng (~83,000 cells) — in comparison to DSBs mapped in nuclei.26
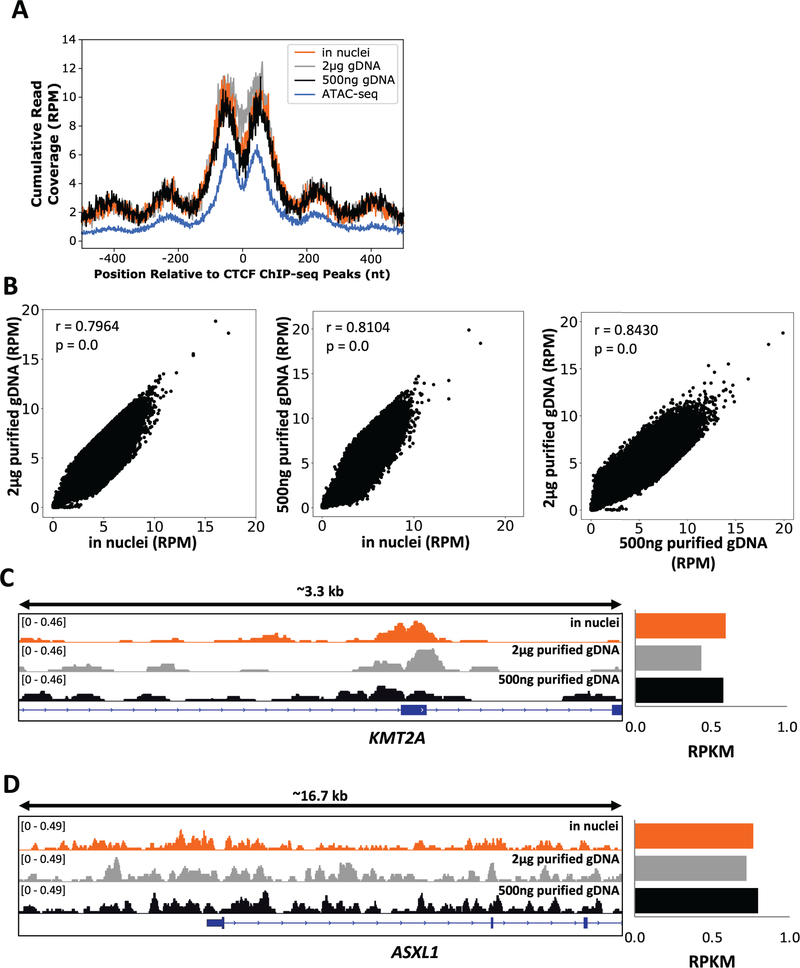
To confirm that the DNA isolation protocol did not introduce a significant amount of artificial breaks, we compared the break signal measured in nuclei and from purified genomic DNA around the chromatin-structuring protein CTCF binding sites. In GM13069 cells (lymphoblastoid cells derived from an apparently normal individual), the DNA break signal for all preparation methods was enriched immediately flanking (+/− 45 nt) the top 10% strongest CTCF binding sites27 (Figure 1A). Importantly, the break pattern also exhibited a periodicity of roughly 200 nt at the regions upstream and downstream of the DNA break peak, indicating that the nucleosome-associated DNA were intact and not sheared during DNA extraction or break mapping processes, as CTCF strongly positions nucleosomes to either side of the binding site.40 Furthermore, the ATAC-seq29 profile at CTCF bindings sites confirmed that the observed periodic DNA break signal was due to nucleosome positioning (Figure 1A). Importantly, no such pattern was observed for both break mapping methods at the weakest (10%) CTCF binding sites (Supporting Information Figure S1). Therefore, the DNA isolation method and subsequent break mapping using isolated genomic DNA captured endogenous DNA breaks and not a significant amount of those that were artificially introduced during processing.
FIGURE 1.
DSBs mapped with purified genomic DNA are strongly correlated with breaks mapped in nuclei. (A) DSBs at CTCF binding sites show nucleosome periodicity as demonstrated by cumulative read-normalized coverage (RPM) of DSBs mapped in GM13069 cells using the in nuclei (orange), the 2 μg purified genomic DNA (grey), and the 500 ng purified genomic DNA (black) methods, as well as GM12878 ATAC-seq data (blue; GSE103301) at the top 10% strongest GM12878 CTCF ChIP-seq peaks (n=4019, ENCSR000AKB). (B) Genome-wide DSB coverage was significantly correlated between the in nuclei and the purified genomic DNA methods in GM13069 cells. Correlation values (Pearson correlation r = 0.7964, P ≈ 0, between the 2 μg genomic DNA and the in nuclei methods; r = 0.8104, P = 0, between the 500 ng genomic DNA and the in nuclei methods; r = 0.8430, P ≈ 0, between the 2 μg and 500 ng genomic DNA method) were calculated genome-wide using 10 kb bins; outliers (0.05% of total bins, 141 bins), bins with zero coverage in both samples, and centromeric/blacklisted regions were removed. (C, D) Gene tracks (left panels) display similar DNA break patterns at two leukemia-related genes: KMT2A (hg38, chr11:118486550–118489829) and ASXL1 (hg38, chr20:32352907–32369562), respectively, for the in nuclei (orange), the 2 μg (grey), and the 500 ng (black) genomic DNA methods in GM13069 cells. DSBs within these gene regions were also quantified and read- and region size-normalized (RPKM, read per kilobase per million reads) (right panels).
To further validate that data generated using either 2 μg or 500 ng of genomic DNA was comparable with the in nuclei method, we compared the DNA break coverage both genome-wide and at specific loci. Genome-wide DNA breakage in purified genomic DNA was significantly correlated with DNA breakage in nuclei in GM13069 cells (Pearson correlation r = 0.7964, P ≈ 0 for 2 μg; r = 0.8104, P ≈ 0 for 500 ng). Additionally, genome-wide DNA breakage was significantly correlated between both purified genomic DNA methods (Pearson correlation r = 0.8430, P ≈ 0) (Figure 1B). At specific gene regions, DNA breakage identified by the break mapping methods showed a similar pattern. A translocation-associated breakpoint cluster region of KMT2A, the intron 10/exon 11 boundary, was enriched for DNA breaks in all three methods. Quantification and read normalization of the data demonstrated that all methods exhibited similar levels of DNA breakage within this region (Figure 1C). Comparable breakage between the three methods was also observed for ASXL1, another leukemia-associated gene (Figure 1D). The reproducibility of the DNA break mapping data at both the gene and genome-wide level between the methods used indicates that low amounts of genomic DNA, both 2 μg and 500 ng, are sufficient to reliably produce DNA break mapping data, and could therefore be used with scarce/precious patient samples to determine DNA break sensitivity and subsequently cancer susceptibility.
To verify that these observations were not specific for lymphoblastoid cells, we mapped DNA breaks in HeLa cells using the in nuclei and the 2 μg genomic DNA methods. When DNA breaks were examined at the top 10% strongest CTCF binding sites in HeLa cells,27 we observed a similar break pattern, with DNA breaks enriched at +/− 45 nt of the CTCF binding peaks and a periodic break signal extending outwards from the peaks (Supporting Information Figure S2A). Genome-wide DNA breaks captured in HeLa cells with both methods were also strongly correlated (Pearson correlation r = 0.8924, P ≈ 0) (Supporting Information Figure S2B), indicating that the 2 μg gDNA method is compatible with multiple cell types. Additionally, DNA breaks at CCAT1 and GUSBP1 were nearly equal in HeLa cells when using the in nuclei or the 2 μg genomic DNA method (Supporting Information Figure S2C and D). Overall, comparisons at both genome-wide and specific loci indicated that data were reliable and reproducible regardless of the starting material or cell type.
3.2. DSBs at rearrangement-specific gene regions are predictive of AML rearrangements
To determine whether DNA fragility at translocation-participating gene regions and other break sensitive regions can serve as a risk assessment for the formation of cancer-associated gene rearrangements, we performed a proof-of-principle experiment in which we compared the DNA break frequencies between break-susceptible (predisposed) individuals and non-susceptible individuals in key regions. The group of predisposed individuals consisted of KMT2A-rearranged remission AML patients, and the group of non-susceptible individuals consisted of AML patients without fusions (Supporting Information Table S5); none of the non-susceptible patients had point mutations in canonical DNA repair proteins. Patients in remission have no detectable leukemia, and therefore no rearrangements present. Absence of a KMT2A rearrangement in these patients was confirmed by examining discordant reads within the translocation breakpoint cluster region of KMT2A (Supporting Information Figure S3, left panels); while a common deletion structural variant on chromosome 8 can be detected, as a positive control for the analysis38 (Supporting Information Figure S3, right panels). We hypothesized that the individuals in the predisposed group would exhibit increased DNA breakage within the AML-driving, translocation-associated region because DNA breaks initiated the formation of the gene translocation event. The non-susceptible group is non-fusion-driven AML patients to serve as a control for the effect of AML on DNA fragility and susceptibility analysis.
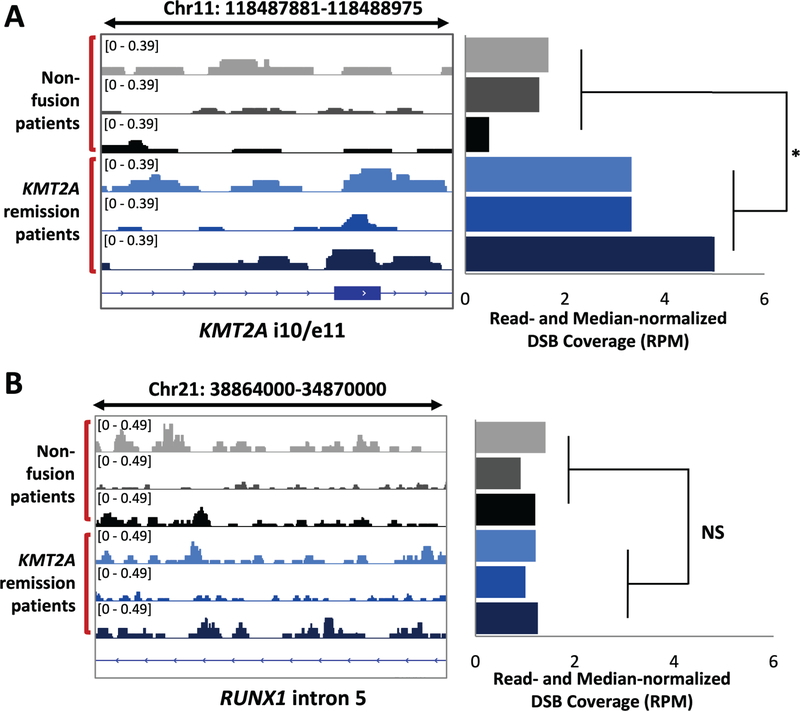
With break mapping in three non-fusion AML and three KMT2A-rearranged remission individuals, DSBs were normalized against each sample’s total reads and each individual’s background breakage level. Each individual’s background breakage level was determined as the median breakage value using genome-wide DSB coverage for 1kb bins. Importantly, there were no significant differences between the median breakage values of all patient samples used here, suggesting the absence of a genome-wide differential instability signature among these patients (data not shown). Quantification of normalized DSBs in the intron 10/exon 11 region of KMT2A (hg38, chr11: 118,488,500–800), the major KMT2A-rearranged region,18 displayed a 2- to 10-fold increase in each KMT2A-rearranged patient as compared to each non-fusion patient, and the group average showed a statistically significant 3.2-fold increase in DNA breakage in the KMT2A-rearranged remission patients (one-tailed Student’s t-test, P = 0.01) (Figure 2A). DSBs were slightly enriched within the respective partner genes for two KMT2A-rearranged patients; however, there was not a significant difference between the non-fusion and KMT2A patients (Supporting Information Figure S4). Previous studies have suggested that spatial proximity rather than high DNA break frequencies within translocation partner genes is important during the initial rearrangement process,41–44 therefore, the lack of significant DSB enrichment within partner genes is plausible. Overall, this indicated that the individuals who had KMT2A rearrangements were indeed more susceptible to DNA breakage at KMT2A region compared to AML patients without rearrangements, supporting the notion that individuals with higher DNA breaks in a rearrangement-participating gene are more susceptible to that particular gene rearrangement, and therefore cancer driven by the rearrangement. Additionally, these results demonstrate that the DNA break mapping method is sensitive to effectively differentiate between non-fusion and rearrangement-driven AML patients.
FIGURE 2.
KMT2A-rearranged AML remission patients exhibit significant KMT2A-specific DNA break sensitivity, as compared to non-fusion AML patients. (A) A read-normalized gene track shows DNA break patterns mapped for three non-fusion AML patients (grey and black) and three KMT2A-rearranged remission AML patients (blue) at the intron 10/exon 11 region of KMT2A (hg38, chr11:118487881–118488975) (left panel), demonstrating patient type specificity of DNA breakage. Read- and genome median-normalized DSB coverage (hg38, chr11:118488500–800) (right panel) indicates that fusion-driven AML remission patients exhibit significant preferential DNA breakage within this key region. (B) A read-normalized gene track showing DNA break patterns mapped at the intron 5 of RUNX1 (hg38, chr21:38864000–34870000) (left panel) suggests that the preferential breakage observed in fusion-driven patients in (A) is specific to the KMT2A gene region; the read- and genome median-normalized DSB coverage (hg38, chr21:34860421–34878550) (right panel) further support this notion. * indicates P = 0.01; one-tailed Student’s t-test.
To establish the feasibility and specificity of DNA fragility as a predictor of rearrangement susceptibility, we assessed another leukemia-associated translocation gene, RUNX1. Within the intron 5 region of RUNX1 (hg38, chr21:34860421–34878550), known as a break cluster region in AML patients, there was no statistically significant difference between the DNA breakage level of the KMT2A-rearranged and the non-fusion individuals (Figure 2B). Furthermore, the DNA breakage level at RUNX1 within the KMT2A-rearranged individuals was approximately two-fold less than that observed within the KMT2A intron10/exon11 region. Therefore, the DNA break mapping can differentiate between non-fusion and rearrangement-driven cancer patients and can also detect differences in DNA breakage between different rearrangement-associated gene regions. These results begin to support the idea that DNA breakage within KMT2A can be used as a reliable and sensitive readout of an individual’s susceptibility for KMT2A-rearranged AML.
3.3. Identifying novel break-prone regions in KMT2A-rearranged patient samples
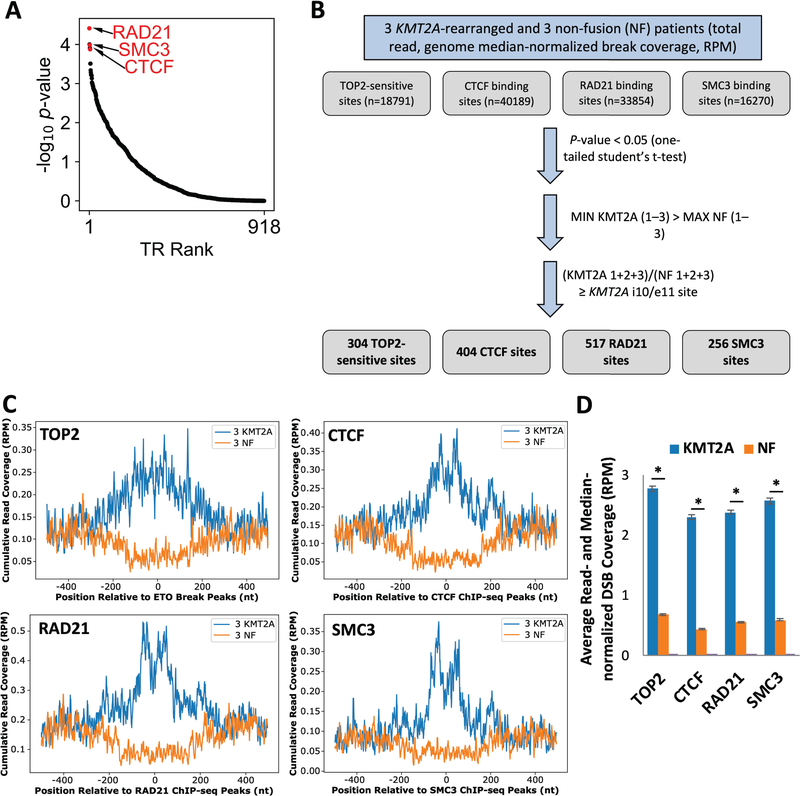
To curate a list of regions that can indicate susceptibility and thus be used in a DNA fragility test, we set out to identify additional regions with preferential DSBs in the KMT2A-rearranged patients compared to non-fusion individuals, and with a break difference greater than that of the intron 10/exon 11 region of KMT2A. A list of such regions will provide a more accurate and representative readout of individual break susceptibility and account for the effects of individual environmental exposure, inherent DNA repair capacity/efficiency, and other factors. First, we used an unbiased analytic tool, BART,35 to infer which transcriptional regulators with binding sites exhibited preferential enrichment at KMT2A patient-specific DNA break-prone regions. This analysis identified CTCF (P = 1.33e-4) and cohesin complex components, RAD21 (P = 3.87e-5) and SMC3 (P = 1.03e-4), with genomic binding sites highly associated with regions of greater DNA breaks in KMT2A-rearranged remission patients than non-fusion individuals (Figure 3A). This suggests that CTCF/cohesin binding sites can be putative genomic features in studying DNA breakage events for cancer susceptibility. Recent studies on the contribution of chromatin higher-order organization to DNA fragility,42,45,46 support this idea. TOP2 activity has also been shown to induce DNA fragility and subsequent formation of chromosomal abnormalities.42,45,47–49 Interestingly, the intron 10/exon 11 region of KMT2A is susceptible to TOP2 cleavage as well as serves as a CTCF/cohesin binding site in lymphoblastoid cells.26,46,50,51 Therefore, we next employed the sensitivity to topoisomerase cleavage and the binding of CTCF/cohesin as two criteria to identify genomic regions with preferential DNA breaks in the KMT2A-rearranged patients, as compared to non-fusion individuals.
FIGURE 3.
TOP2-sensitive, CTCF, RAD21, and SMC3 binding sites significantly exhibit preferential breakage in KMT2A-rearranged remission patients. (A) BART35 analysis identified RAD21, SMC3, and CTCF as top significantly enriched transcriptional regulators in DSBs in KMT2A-rearranged AML patients. (B) The flowchart depicts the three filtering steps used to identify preferential break-prone regions specific to KMT2A-rearranged AML patients from all TOP2-sensitive sites (n=18791),26 CTCF bindings sites (n=40189) (ENCSR000AKB), RAD21 binding sites (n=33854) (ENCSR000EAC), and SMC3 binding sites (n=16270) (ENCSR000DZP). (C) The preferential break-prone regions of TOP2, CTCF, RAD21, and SMC sites displayed higher DSBs in KMT2A-rearranged remission patients than non-fusion patients. Cumulative read-normalized coverage (RPM) of DSBs were mapped in 3 KMT2A-rearranged (blue) and 3 non-fusion (NF, orange) patients at the 304 TOP2-sensitive sites, 404 CTCF binding sites, 517 RAD21 binding sites, and 256 SMC3 binding sites. (D) DSBs at the TOP2, CTCF, RAD21, and SMC sites shown in (C) were quantified within the peak summits ± 150 bp. Error bars represent standard error of the mean. Asterisks indicate significance; P ≈ 0, one-tailed Student’s t-test.
Using four datasets: TOP2-sensitive sites,26 CTCF, RAD21, and SMC327 binding sites, we developed a pipeline (Figure 3B) to first determine regions with significantly higher DNA breaks in KMT2A-rearranged patients than in non-fusion patients (P < 0.05, Student’s t-test). Next, among these sites, we identified 304 TOP2-sensitive sites, 404 CTCF, 517 RAD21, and 256 SMC3 bindings sites, in which all three of the KMT2A patients have greater DNA breakage than all three of the non-fusion patients, and the KMT2A to non-fusion breakage ratio was greater than that observed for the KMT2A region (Figure 3B). These regions demonstrate a striking difference in DNA breaks between KMT2A and non-fusion patient samples, with significantly higher DSBs in the KMT2A-rearranged sample group (P ≈ 0, one-tailed Student’s t-test, Figure 3C and D). The individual patients show the same DSB distribution at these regions as the merged groups (Supporting Information Figure S5A). Furthermore, the differences in DNA breakage were within well-defined sizes (+/− 200 nt surrounding the peak sites), suggesting the specificity of the break sensitivity. Genomic feature analysis found that approximately half of the sites in all four sets were within gene regions (gene body, TSS, promoter, and TTS) (Supporting Information Figure S5B). Interestingly, we found that DNA breakage at the TOP2-sensitive sites that were within genes demonstrated a positive correlation with highly expressed genes; DNA breakage at the CTCF, RAD21, and SMC3 sites that were within gene regions were enriched within the highly expressed genes as well30 (Supporting Information Figure S5C).
Next, we determined common regions shared among the 304 TOP2-sensitive sites, 404 CTCF, 517 RAD21, and 256 SMC3 bindings sites. We found 34 regions with all four features and having an equal or greater DNA break difference than the KMT2A region between KMT2A-rearranged patients and non-fusion patients. None of the partner genes specific to the three patients studied were identified in these common regions, and only the 3’ end, not the breakpoint region, of AFDN contained a break-prone CTCF site. As each KMT2A-rearranged patient had a different rearrangement, and high DNA break frequencies at translocation partner genes have been shown to be less crucial to the formation of a rearrangement,41–44 it is conceivable that we did not detect preferentially break-prone partner gene regions across the three KMT2A-rearranged patients. Two examples of these regions depict the characteristic enrichment of preferential DNA breaks in KMT2A-rearranged patients (Supporting Information Figure S6A). Assessing the extent of DSBs combined at these regions in patient samples could provide more predictive power (Supporting Information Figure S6B, P = 0.0005, compared to Figure 2A, P = 0.01 at the KMT2A region only, one-tailed Student’s t-test), therefore, these regions collectively can be used to determine DNA break susceptibility, and thus risk for KMT2A-rearranged AML.
All 34 common regions are classified as candidate cis-regulatory elements by ENCODE (DNase hypersensitive regions) with 20 regions of distal enhancer-like signatures.37 Based on the Genome Aggregation Database (gnomAD), all 34 regions contained single nucleotide variants (ranging 17–80 different types), 26 regions had indels (−31 to +45 nt changes),39 and 20 regions mapped to large structural variants (gene rearrangements, amplifications and deletions).38 Interestingly, 27 regions are located within known fragile sites.36 The presence of these features suggests that these regions of the genome are more open, and therefore more fragile/prone to DNA breakage, further strengthening the rationale for using the described criteria to identify susceptible regions.
3.4. Exposure to low-dose, non-cytotoxic levels of etoposide enhances DNA breakage sensitivity in KMT2A-rearranged remission patients
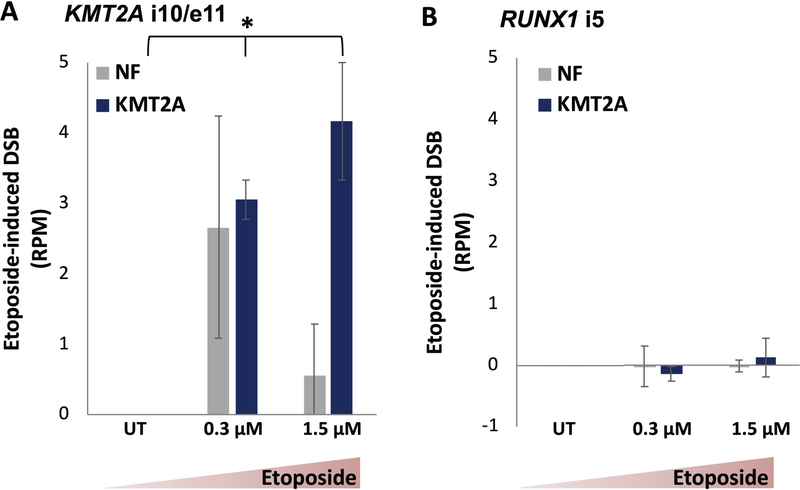
We have shown the potential involvement of TOP2 in these break-sensitive regions, and next, we examine whether etoposide treatment can accentuate susceptibility and increase detection sensitivity. Here, we treated patient-derived peripheral blood mononuclear cells (PBMCs) with low-dose, non-cytotoxic amounts of a chemotherapy chemical, etoposide (0.3 μM and 1.5 μM), for 24 h to model the residual levels the cells would be exposed to in the blood.52 Etoposide, a TOP2 poison, is associated with therapy-related leukemia, specifically those with KMT2A translocation events.53–57 Additionally, we and others have previously shown that etoposide induced DNA breaks at fragile sites and key cancer-associated regions,17,18,58 such as the intron 10/exon 11 region of KMT2A, a known therapy-related breakpoint cluster region.
For each individual and each treatment condition, there were no significant differences in the genome median breakage values between the untreated and etoposide-treated patient samples, indicating that etoposide treatment did not induce higher genome-wide DNA breakage (data not shown). For the intron 10/exon 11 region of KMT2A, the KMT2A-rearranged individuals exhibited a dose-dependent increase of DNA breakage when treated with etoposide (p = 0.02, one sample t-test); the non-fusion patients did not show a dose-dependent increase of DSBs (Figure 4A). Furthermore, when intron 5 of RUNX1 was assessed, there were no significant differences between the KMT2A-rearranged and the non-fusion patients in both the untreated and etoposide-treated conditions (Figure 4B). Importantly, the level of DNA breakage in the KMT2A-rearranged patients within the RUNX1 region was less than that within the KMT2A region, even following etoposide treatment. This demonstrated specifically that the KMT2A-rearranged patients were preferentially susceptible to DNA breaks within the KMT2A region, but not within another fusion-associated region, and that this break susceptibility could be exacerbated with chemical exposure.
FIGURE 4.
A dose-dependent increase of etoposide-induced DSBs presented at the KMT2A region in KMT2A-rearranged AML patients, but not in non-fusion patients. (A) Low-dose etoposide exposure significantly increased DNA breakage in KMT2A-rearranged AML patients at the KMT2A gene region in a dose-dependent manner. (B) This differential phenomenon is not present in the RUNX1 intron 5 region. The read- and genome-median-normalized DSB coverage was calculated for intron 10/exon 11 of KMT2A (hg38, chr11:118488500–118488800) and intron 5 of RUNX1 (hg38, chr21:34860421–34878550) in three non-fusion AML patients (grey) and two KMT2A-rearranged remission AML patients (blue) either untreated or after 24-hour etoposide treatment (0.3 or 1.5 μM). The etoposide-induced DNA breakage was then calculated as the difference between the normalized DSB coverage of each sample and its untreated control. DNA breaks were mapped using 500 ng of purified genomic DNA as input. Dose-dependence at the KMT2A i10/e11 region was determined using one-sample, one-tailed Student’s t-test; P = 0.02 for the KMT2A-rearranged samples; P = 0.42 for the non-fusion samples. Error bars represent standard error of the mean.
To understand how the TOP2-sensitive, CTCF, RAD21, and SMC3 binding sites which we identified as sites of preferential DNA breakage in the KMT2A-rearranged patients, respond to etoposide treatment in patient cells, we examined DSB coverage over these four sets of sites in the treated and untreated samples. We found that the KMT2A-rearranged remission patients had significantly increased DNA breakage in all of the treatments for the TOP2, CTCF, RAD21, and SMC3 sites (P < 0.001, Student’s t-test), as compared to the non-fusion patients (Supporting Information Figure S7A–D). To determine whether DSB frequency was positively correlated with etoposide treatment, the TOP2-sensitive regions were then divided into four equal groups based on the break frequencies upon treatment of 1.5 μM etoposide of MLL-rearranged patients (Group 1, the highest DSB coverage; and Group 4, the lowest coverage) (Supporting Information Figure S8A–D). We found that Group 1 regions exhibited a dose-dependent increase of DNA breaks upon etoposide treatment among the KMT2A-rearranged patients (p < 0.01, Student’s t-test), but not among non-fusion patients. These results indicate that a specific subgroup of TOP2-sensitive regions was preferentially susceptible to etoposide-induced DNA breakage, in a dose-dependent manner, in KMT2A-rearranged remission patients.
Previous studies showed that CTCF, RAD21, and SMC3 binding influence DNA breakage in cultured human and mouse cells.31,42,45,46 Therefore, we next investigated whether the binding strength of CTCF, RAD21, and SMC3 was associated with preferential breakage in KMT2A-rearranged remission patients upon etoposide treatment. Based on the respective ChIP-seq reads of each protein, we divided the 404 CTCF, 517 RAD21, and 256 SMC3 regions into four equal groups (Group 1, the strongest binding; and Group 4, the lowest). We found that DNA breakage decreased as the binding of each protein decreased in response to 1.5 μM and 0.3 μM etoposide treatment (Supporting Information Figure S9), indicating that binding strength was positively associated with more DNA breaks induced by etoposide.
To assess the direct action of TOP2 in the increase of DNA breakage, we analyzed the coverage of TOP2 cleavage complexes (TOP2cc) mapped by Gittens et al.31 in wildtype and TOP2B knockout cells, for the 304 TOP2-senstive, 404 CTCF, 517 RAD21, and 256 SMC3 regions. We found that these regions are indeed sensitive to TOP2 activity as indicated by the significantly decreased TOP2cc coverage observed in the TOP2B knockout condition, as compared to the wild-type cells in both asynchronous and G1 states (P < 0.001, Wilcoxon Rank Sum test) (Supporting Information Figure S10A–D), indicating that TOP2 is involved in generating DNA breaks in these regions. Interestingly, the DSB reduction by knocking out TOP2B is significantly more prominent in G1 versus in asynchronous cells (P < 0.001, Wilcoxon Rank Sum test), indicating TOP2B is a major contributor to generate cleavage complex-associated DSBs (TOP2B, the main TOP2 in G1), and in asynchronous cells TOP2A can compensate the action of TOP2B when it is knocked out.
Finally, we examined the response to etoposide for the 34 common regions identified as TOP2-sensitive and CTCF/cohesin binding sites and having a greater DSB difference than the KMT2A region in KMT2A-rearranged patients as compared to non-fusion patients. We found that 24 of the 34 regions had significantly increased DSBs upon etoposide treatment in the KMT2A-rearranged patients (Supporting Information Figure S11A). Interestingly, the DSB frequency in the other ten regions was significantly higher than the genome-median DSBs in untreated KMT2A-rearranged remission samples (Supporting Information Figure S11B), indicating that these regions were inherently break-prone even in untreated samples. Characterization of these two classes of break-prone sites in the KMT2A-rearranged remission patient samples — those that are sensitive to etoposide treatment and those that are not but have high endogenous DNA breakage — indicates that other factors beyond etoposide treatment, like DNA topology or alternative DNA secondary structure, could contribute to high endogenous TOP2-associated DNA breakage. Altogether, assessing DSBs at many regions will provide a more representative measure of an individual’s sensitivity to DNA breaks and therefore their susceptibility for AML gene rearrangements.
4. DISCUSSION
DNA break mapping methods such as DSBCapture, END-seq, BLESS, and others, while effective, require large quantities of starting material. Here, we have demonstrated the reliability and accuracy of the data generated with the low-input DNA break mapping method which uses 60–850-fold less starting material. With this advancement in the method, we have made it feasible to use DNA break mapping with patient samples, which are often available only in scarce amounts. We demonstrated the utility of the method by distinguishing between different patient populations who had either a cancer-causing gene rearrangement or no fusion events, as well as differentiating between DNA break susceptibility at common fusion breakpoint regions (KMT2A vs. RUNX1). Furthermore, we showed that upon etoposide treatment DNA breakage increased within the breakpoint region of KMT2A, but not in that of RUNX1, in KMT2A-rearranged remission AML patients. This indicated that higher DNA break frequency in rearrangement-participating gene regions could indeed represent a predisposition for rearrangement events in patient samples. Our finding in patient samples is supported by a recent study that utilized FISH to assess DNA break and gene rearrangement frequencies in cultured human cells,42 and we further located the preferential breakage at rearrangement-participating gene regions.
In addition, the genome-wide break mapping allows us to identify previously undiscovered sensitive regions of the genome, and inclusion of these additional regions can increase the power of risk assessment and our understanding of DNA fragility. An unbiased analysis using BART revealed the significant involvement of CTCF, SMC3, and RAD21 proteins in the preferential break sites between KMT2A-rearranged patients and non-fusion patients. We found that predisposed patients, when compared to non-fusion patients, had significantly more DNA breakage at a select group of TOP2-sensitive, CTCF, RAD21, and SMC3 binding sites. A recent study found that CTCF binding was increased in AML patients when compared to normal bone marrow cells, further implicating CTCF in AML pathogenesis.59 More importantly, DNA breakage at these TOP2-sensitive sites specifically increased in a dose-dependent manner, and increased DNA breakage in etoposide-treated samples was also associated with stronger protein binding at CTCF, RAD21, and SMC3 sites. We are also intrigued by the difference in etoposide response among the 34 shared sites (by TOP2, CTCF, RAD21, and SMC3). Within these 34 regions, there were two groups of preferential breakage sites in KMT2A-rearranged remission AML patients — those that respond to etoposide and those that have an intrinsic propensity to break. While 10 regions did not respond to etoposide, the endogenous DNA breakage at these sites was significantly higher than the genome-median breakage value, and 7 of the 10 contained TOP2cc sites.31 This suggests that there is endogenous TOP2 activity at these sites generating high DNA breakage regardless of the etoposide treatment. Hoa et al.60 demonstrated that TOP2 frequently fails to re-ligate the endogenous, transiently-cleaved products even without the presence of inhibitors. These TOP2 cleaved products can be processed into persistent DSBs when a covalently bound pair of TOP2s are both processed by DNA repair machinery to result in free DNA ends. Importantly, the 34 common sites possess the properties of markers for DNA break susceptibility, in parallel with the KMT2A region, as they represent preferential break-prone regions specific to KMT2A-rearranged AML patients.
Our observations strongly suggest that chromatin organization is a contributing factor in predisposition of DNA breakage for gene rearrangements. Previous studies have demonstrated the role of CTCF/cohesin sites in mediating DNA fragility/breakage.42,45,46 Specifically, these studies showed that when a critical CTCF binding site was abrogated or cohesin complex subunits were depleted, DNA breakage at these sites was reduced in either mouse or isogenic human cells.42,45 Furthermore, TOP2 with its known role in DNA fragility, also colocalizes to CTCF binding sites,45,61 and thus could mediate fragility at these sites through regulating DNA topology. Similar to our results in patients, these studies found that etoposide-induced DNA breaks were significantly enriched with CTCF/cohesin binding sites and endogenous break sites,42,46 while many endogenous sites are not overlapped with etoposide-induced DNA breaks. These observations suggest that there are distinct mechanisms among TOP2-related and chromatin organization-related DNA fragility.
Interestingly, we found that among the preferential break sites of the KMT2A-rearranged patients in the four datasets, 33 (TOP2), 37 (CTCF), 51 (RAD21), and 18 (SMC3) unique genes were identified in the BEAT AML study, further supporting the role of these identified regions in leukemia.62 We also investigated if the TOP2-sensitive, CTCF, RAD21, and SMC3 binding regions were associated with chromosomal abnormalities beyond the context of AML. We found that all four sets of regions were closer to publicly-available structural variants (i.e. deletions, insertions, amplifications, etc.),38 as compared to random shuffled control regions (Supporting Information Figure S12A–D). Specifically, TOP2-sensitive regions were approximately 35 times more likely to be within 25 kb of a structural variant; whereas CTCF, RAD21, and SMC3 regions were approximately three times more likely to be close to a structural variant, when compared to shuffled control regions (Supporting Information Figure S12E). This observation indicates the unstable nature of these regions in the general human population.
The current available diagnostic procedures such as examination of blood and bone marrow cells, cytogenetic analysis of chromosomes, and PCR tests are intended to detect the presence of leukemia cells and/or the specific rearrangements. However, many cases are not identified until disease progression has already occurred when treatments are less effective and result in higher mortality. The DNA fragility test, as demonstrated here, to detect DNA breakage – before cancer-causing rearrangements occur – offers an early indication of susceptibility to cancers and facilitates prompt prevention and timely treatments. Similar to stress-induced chromosomal breakage assays used to detect Fanconi anemia,63 we developed a protocol in which we challenged primary, patient-derived PBMCs with chemicals, carefully purified genomic DNA, and performed DNA break mapping. Importantly, many cancers that are driven by this class of mutations are linked to exposure to carcinogenic environmental/chemotherapeutic chemicals.6,64–67 The study here and our previous work examining the effect of chemical exposure on DNA integrity suggests that exposure to such chemicals causes the DNA breaks that promote the generation of chromosomal abnormalities.3 This break mapping approach is also particularly important for patients about to undergo chemotherapy. Therapy-related AML occurs in up to 13% of cancer patients who receive chemotherapy, depending on the agents given.68–70 Identifying those at high risk before chemotherapy begins would be a great advantage, because less leukemogenic agents can be selected for use. To date, there are no DNA tests that can detect cancer susceptibility as it relates to DNA fragility. The method that we developed offers a new diagnostic avenue to these current detection methods; it provides the opportunity to sensitively screen individuals.
Supplementary Material
Acknowledgements:
We acknowledge the contributions of the UVA Cancer Center Support Grant P30CA044579 and the UVA Oncology Research Information Exchange Network (ORIEN) Team and UVA Biorepository and Tissue Research Facility (BTRF) with Partners in Discovery for Total Cancer Care at UVA protocol IRB HSR 18445 for the consenting, specimen procurement and processing, clinical data abstraction, and access to the molecular and clinical data. We would like to thank Sandeep Singh for preliminary data analysis.
Funding information
This work was supported by grants from National Institutes of Health, Grant/Award Numbers: T32CA009109, T32GM008136, and RO1GM101192.
Footnotes
Conflict of interest:
The authors have no conflicts of interest to declare.
Data availability:
DSB mapping datasets generated in this study can be accessed at Sequence Read Archive (SRA) under the accession numbers PRJNA685150. Additional data will be made available upon request to the corresponding author.
REFERENCES
- 1.Ceccaldi R, Rondinelli B, D’Andrea AD. Repair pathway choices and consequences at the double-strand break. Trends Cell Biol. 2016;26(1):52–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arlt MF, Durkin SG, Ragland RL, Glover TW. Common fragile sites as targets for chromosome rearrangements. DNA Repair (Amst). 2006;5(9–10):1126–1135. [DOI] [PubMed] [Google Scholar]
- 3.Gandhi M, Dillon LW, Pramanik S, Nikiforov YE, Wang YH. DNA breaks at fragile sites generate oncogenic RET/PTC rearrangements in human thyroid cells. Oncogene. 2010;29(15):2272–2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kaye FJ. Mutation-associated fusion cancer genes in solid tumors. Mol Cancer Ther. 2009;8(6):1399–1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mitelman F, Johansson B, Mertens F. Fusion genes and rearranged genes as a linear function of chromosome aberrations in cancer. Nat Genet. 2004;36(4):331–334. [DOI] [PubMed] [Google Scholar]
- 6.Mitelman F, Johansson B, Mertens F. The impact of translocations and gene fusions on cancer causation. Nat Rev Cancer. 2007;7(4):233–245. [DOI] [PubMed] [Google Scholar]
- 7.Tate JG, Bamford S, Jubb HC, et al. COSMIC: the Catalogue Of Somatic Mutations In Cancer. Nucleic Acids Res. 2019;47(D1):D941–D947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beroukhim R, Mermel CH, Porter D, et al. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463(7283):899–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cronin KA, Lake AJ, Scott S, et al. Annual Report to the Nation on the Status of Cancer, part I: National cancer statistics. Cancer. 2018;124(13):2785–2800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hayat MJ, Howlader N, Reichman ME, Edwards BK. Cancer statistics, trends, and multiple primary cancer analyses from the Surveillance, Epidemiology, and End Results (SEER) Program. Oncologist. 2007;12(1):20–37. [DOI] [PubMed] [Google Scholar]
- 11.Byers T, Wender RC, Jemal A, Baskies AM, Ward EE, Brawley OW. The American Cancer Society challenge goal to reduce US cancer mortality by 50% between 1990 and 2015: Results and reflections. CA Cancer J Clin. 2016;66(5):359–369. [DOI] [PubMed] [Google Scholar]
- 12.Loud JT, Murphy J. Cancer Screening and Early Detection in the 21(st) Century. Semin Oncol Nurs. 2017;33(2):121–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Berwick M, Vineis P. Markers of DNA repair and susceptibility to cancer in humans: an epidemiologic review. J Natl Cancer Inst. 2000;92(11):874–897. [DOI] [PubMed] [Google Scholar]
- 14.Nagel ZD, Chaim IA, Samson LD. Inter-individual variation in DNA repair capacity: a need for multi-pathway functional assays to promote translational DNA repair research. DNA Repair (Amst) 2014;19:199–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lombard DB, Chua KF, Mostoslavsky R, Franco S, Gostissa M, Alt FW. DNA repair, genome stability, and aging. Cell. 2005;120(4):497–512. [DOI] [PubMed] [Google Scholar]
- 16.Soares JP, Cortinhas A, Bento T, et al. Aging and DNA damage in humans: a meta-analysis study. Aging (Albany NY). 2014;6(6):432–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lehman CE, Dillon LW, Nikiforov YE, Wang YH. DNA fragile site breakage as a measure of chemical exposure and predictor of individual susceptibility to form oncogenic rearrangements. Carcinogenesis. 2017;38(3):293–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thys RG, Lehman CE, Pierce LC, Wang YH. Environmental and chemotherapeutic agents induce breakage at genes involved in leukemia-causing gene rearrangements in human hematopoietic stem/progenitor cells. Mutat Res. 2015;779:86–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Figueroa-Gonzalez G, Perez-Plasencia C. Strategies for the evaluation of DNA damage and repair mechanisms in cancer. Oncol Lett. 2017;13(6):3982–3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gaivao I, Piasek A, Brevik A, Shaposhnikov S, Collins AR. Comet assay-based methods for measuring DNA repair in vitro; estimates of inter- and intra-individual variation. Cell Biol Toxicol. 2009;25(1):45–52. [DOI] [PubMed] [Google Scholar]
- 21.Kianmehr M, Hajavi J, Gazeri J. Assessment of DNA damage in blood lymphocytes of bakery workers by comet assay. Toxicol Ind Health. 2017;33(9):726–735. [DOI] [PubMed] [Google Scholar]
- 22.Canela A, Sridharan S, Sciascia N, et al. DNA breaks and end resection measured genome-wide by end sequencing. Mol Cell. 2016;63(5):898–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Crosetto N, Mitra A, Silva MJ, et al. Nucleotide-resolution DNA double-strand break mapping by next-generation sequencing. Nat Methods. 2013;10(4):361–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lensing SV, Marsico G, Hansel-Hertsch R, Lam EY, Tannahill D, Balasubramanian S. DSBCapture: in situ capture and sequencing of DNA breaks. Nat Methods. 2016;13(10):855–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Szlachta K, Raimer HM, Comeau LD, Wang YH. CNCC: an analysis tool to determine genome-wide DNA break end structure at single-nucleotide resolution. BMC Genomics. 2020;21(1):25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Szlachta K, Manukyan A, Raimer HM, et al. Topoisomerase II contributes to DNA secondary structure-mediated double-stranded breaks. Nucleic Acids Res. 2020;48(12):6654–6671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Consortium EP. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489(7414):57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rosenbloom KR, Sloan CA, Malladi VS, et al. ENCODE data in the UCSC Genome Browser: year 5 update. Nucleic Acids Res. 2013;41(Database issue):D56–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Suzuki M, Liao W, Wos F, et al. Whole-genome bisulfite sequencing with improved accuracy and cost. Genome Res. 2018;28(9):1364–1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tilgner H, Grubert F, Sharon D, Snyder MP. Defining a personal, allele-specific, and single-molecule long-read transcriptome. Proc Natl Acad Sci U S A. 2014;111(27):9869–9874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gittens WH, Johnson DJ, Allison RM, Cooper TJ, Thomas H, Neale MJ. A nucleotide resolution map of Top2-linked DNA breaks in the yeast and human genome. Nat Commun. 2019;10(1):4846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat Methods. 2015;12(4):357–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pertea M, Pertea GM, Antonescu CM, Chang TC, Mendell JT, Salzberg SL. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol. 2015;33(3):290–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Amemiya HM, Kundaje A, Boyle AP. The ENCODE Blacklist: Identification of Problematic Regions of the Genome. Sci Rep. 2019;9(1):9354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang Z, Civelek M, Miller CL, Sheffield NC, Guertin MJ, Zang C. BART: a transcription factor prediction tool with query gene sets or epigenomic profiles. Bioinformatics. 2018;34(16):2867–2869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mrasek K, Schoder C, Teichmann AC, et al. Global screening and extended nomenclature for 230 aphidicolin-inducible fragile sites, including 61 yet unreported ones. Int J Oncol. 2010;36(4):929–940. [DOI] [PubMed] [Google Scholar]
- 37.Consortium EP, Moore JE, Purcaro MJ, et al. Expanded encyclopaedias of DNA elements in the human and mouse genomes. Nature. 2020;583(7818):699–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Collins RL, Brand H, Karczewski KJ, et al. A structural variation reference for medical and population genetics. Nature. 2020;581(7809):444–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Karczewski KJ, Francioli LC, Tiao G, et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581(7809):434–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fu Y, Sinha M, Peterson CL, Weng Z. The insulator binding protein CTCF positions 20 nucleosomes around its binding sites across the human genome. PLoS Genet. 2008;4(7):e1000138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Burrow AA, Williams LE, Pierce LC, Wang YH. Over half of breakpoints in gene pairs involved in cancer-specific recurrent translocations are mapped to human chromosomal fragile sites. BMC Genomics. 2009;10:59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gothe HJ, Bouwman BAM, Gusmao EG, et al. Spatial chromosome folding and active transcription drive DNA fragility and formation of oncogenic MLL translocations. Mol Cell. 2019;75(2):267–283 e212. [DOI] [PubMed] [Google Scholar]
- 43.Gomez-Herreros F DNA Double Strand Breaks and Chromosomal Translocations Induced by DNA Topoisomerase II. Front Mol Biosci. 2019;6:141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Iarovaia OV, Rubtsov M, Ioudinkova E, Tsfasman T, Razin SV, Vassetzky YS. Dynamics of double strand breaks and chromosomal translocations. Mol Cancer. 2014;13:249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Canela A, Maman Y, Huang SN, et al. Topoisomerase II-induced chromosome breakage and translocation Is determined by chromosome architecture and transcriptional activity. Mol Cell. 2019;75(2):252–266 e258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Canela A, Maman Y, Jung S, et al. Genome Organization Drives Chromosome Fragility. Cell. 2017;170(3):507–521 e518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dillon LW, Pierce LC, Lehman CE, Nikiforov YE, Wang YH. DNA topoisomerases participate in fragility of the oncogene RET. PLoS One. 2013;8(9):e75741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Haffner MC, Aryee MJ, Toubaji A, et al. Androgen-induced TOP2B-mediated double-strand breaks and prostate cancer gene rearrangements. Nat Genet. 2010;42(8):668–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schwer B, Wei PC, Chang AN, et al. Transcription-associated processes cause DNA double-strand breaks and translocations in neural stem/progenitor cells. Proc Natl Acad Sci U S A. 2016;113(8):2258–2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cowell IG, Sondka Z, Smith K, et al. Model for MLL translocations in therapy-related leukemia involving topoisomerase IIbeta-mediated DNA strand breaks and gene proximity. Proc Natl Acad Sci U S A. 2012;109(23):8989–8994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang Y, Rowley JD. Chromatin structural elements and chromosomal translocations in leukemia. DNA Repair (Amst). 2006;5(9–10):1282–1297. [DOI] [PubMed] [Google Scholar]
- 52.Kersting G, Willmann S, Wurthwein G, Lippert J, Boos J, Hempel G. Physiologically based pharmacokinetic modelling of high- and low-dose etoposide: from adults to children. Cancer Chemother Pharmacol. 2012;69(2):397–405. [DOI] [PubMed] [Google Scholar]
- 53.Leone G, Mele L, Pulsoni A, Equitani F, Pagano L. The incidence of secondary leukemias. Haematologica. 1999;84(10):937–945. [PubMed] [Google Scholar]
- 54.Broeker PL, Super HG, Thirman MJ, et al. Distribution of 11q23 breakpoints within the MLL breakpoint cluster region in de novo acute leukemia and in treatment-related acute myeloid leukemia: correlation with scaffold attachment regions and topoisomerase II consensus binding sites. Blood. 1996;87(5):1912–1922. [PubMed] [Google Scholar]
- 55.Felix CA. Secondary leukemias induced by topoisomerase-targeted drugs. Biochim Biophys Acta. 1998;1400(1–3):233–255. [DOI] [PubMed] [Google Scholar]
- 56.Pedersen-Bjergaard J, Philip P. Balanced translocations involving chromosome bands 11q23 and 21q22 are highly characteristic of myelodysplasia and leukemia following therapy with cytostatic agents targeting at DNA-topoisomerase II. Blood. 1991;78(4):1147–1148. [PubMed] [Google Scholar]
- 57.McNerney ME, Godley LA, Le Beau MM. Therapy-related myeloid neoplasms: when genetics and environment collide. Nat Rev Cancer. 2017;17(9):513–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Aplan PD, Chervinsky DS, Stanulla M, Burhans WC. Site-specific DNA cleavage within the MLL breakpoint cluster region induced by topoisomerase II inhibitors. Blood. 1996;87(7):2649–2658. [PubMed] [Google Scholar]
- 59.Mujahed H, Miliara S, Neddermeyer A, et al. AML displays increased CTCF occupancy associated with aberrant gene expression and transcription factor binding. Blood. 2020;136(3):339–352. [DOI] [PubMed] [Google Scholar]
- 60.Hoa NN, Shimizu T, Zhou ZW, et al. Mre11 Is essential for the removal of lethal topoisomerase 2 covalent cleavage complexes. Mol Cell. 2016;64(3):580–592. [DOI] [PubMed] [Google Scholar]
- 61.Uuskula-Reimand L, Hou H, Samavarchi-Tehrani P, et al. Topoisomerase II beta interacts with cohesin and CTCF at topological domain borders. Genome Biol. 2016;17(1):182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tyner JW, Tognon CE, Bottomly D, et al. Functional genomic landscape of acute myeloid leukaemia. Nature. 2018;562(7728):526–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cervenka J, Arthur D, Yasis C. Mitomycin C test for diagnostic differentiation of idiopathic aplastic anemia and Fanconi anemia. Pediatrics. 1981;67(1):119–127. [PubMed] [Google Scholar]
- 64.Eastmond DA, Keshava N, Sonawane B. Lymphohematopoietic cancers induced by chemicals and other agents and their implications for risk evaluation: An overview. Mutat Res Rev Mutat Res. 2014;761:40–64. [DOI] [PubMed] [Google Scholar]
- 65.McHale CM, Zhang L, Smith MT. Current understanding of the mechanism of benzene-induced leukemia in humans: implications for risk assessment. Carcinogenesis. 2012;33(2):240–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Poynter JN, Richardson M, Roesler M, et al. Chemical exposures and risk of acute myeloid leukemia and myelodysplastic syndromes in a population-based study. Int J Cancer. 2017;140(1):23–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yunis JJ, Soreng AL, Bowe AE. Fragile sites are targets of diverse mutagens and carcinogens. Oncogene. 1987;1(1):59–69. [PubMed] [Google Scholar]
- 68.Bhatia S Therapy-related myelodysplasia and acute myeloid leukemia. Semin Oncol. 2013;40(6):666–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Krishnan A, Bhatia S, Slovak ML, et al. Predictors of therapy-related leukemia and myelodysplasia following autologous transplantation for lymphoma: an assessment of risk factors. Blood. 2000;95(5):1588–1593. [PubMed] [Google Scholar]
- 70.Zahid MF, Parnes A, Savani BN, Litzow MR, Hashmi SK. Therapy-related myeloid neoplasms - what have we learned so far? World J Stem Cells. 2016;8(8):231–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
DSB mapping datasets generated in this study can be accessed at Sequence Read Archive (SRA) under the accession numbers PRJNA685150. Additional data will be made available upon request to the corresponding author.