Abstract
Death-associated protein 5 (DAP5) (also named p97 and NAT1) is a member of the translation initiation factor 4G (eIF4G) family that lacks the eIF4E binding site. It was previously implicated in apoptosis, based on the finding that a dominant negative fragment of the protein protected against cell death. Here we address its function and two distinct levels of regulation during apoptosis that affect the protein both at translational and posttranslational levels. DAP5 protein was found to be cleaved at a single caspase cleavage site at position 790, in response to activated Fas or p53, yielding a C-terminal truncated protein of 86 kDa that is capable of generating complexes with eIF4A and eIF3. Interestingly, while the overall translation rate in apoptotic cells was reduced by 60 to 70%, in accordance with the simultaneous degradation of the two major mediators of cap-dependent translation, eIF4GI and eIF4GII, the translation rate of DAP5 protein was selectively maintained. An internal ribosome entry site (IRES) element capable of directing the translation of a reporter gene when subcloned into a bicistronic vector was identified in the 5′ untranslated region of DAP5 mRNA. While cap-dependent translation from this transfected vector was reduced during Fas-induced apoptosis, the translation via the DAP5 IRES was selectively maintained. Addition of recombinant DAP5/p97 or DAP5/p86 to cell-free systems enhanced preferentially the translation through the DAP5 IRES, whereas neutralization of the endogenous DAP5 in reticulocyte lysates by adding a dominant negative DAP5 fragment interfered with this translation. The DAP5/p86 apoptotic form was more potent than DAP5/p97 in these functional assays. Altogether, the data suggest that DAP5 is a caspase-activated translation factor which mediates cap-independent translation at least from its own IRES, thus generating a positive feedback loop responsible for the continuous translation of DAP5 during apoptosis.
Programmed cell death (PCD) is a fundamental cellular process that provides an intrinsic self-elimination mechanism for the removal of unwanted cells in a wide variety of biological systems. PCD is critical for organ development, tissue remodeling, cellular homeostasis, and elimination of abnormal and damaged cells. However, improper execution of PCD can be quite hazardous and is associated with pathologies including AIDS, neurodegenerative disorders, autoimmune diseases, and others. Altogether, the execution of apoptosis must be tightly regulated. This is achieved by a stringent requirement for an apoptotic trigger on the one hand and protection from inappropriate activation of the cell death program in cells intended to survive on the other hand. The latter mechanism is especially important given that ample proapoptotic proteins are present in normally growing cells, regardless of the presence of an apoptotic trigger.
Death-associated protein 5 (DAP5) (also named p97 and NAT1), a 97-kDa protein homologous to eukaryotic translation initiation factor 4GI (eIF4GI), was isolated independently by several groups (15, 21, 33, 40). In our laboratory, it was rescued as a positive mediator of PCD through a functional approach to gene cloning, which is based on transfections of expression cDNA libraries and selection of cells resistant to apoptosis (10, 21). A death-protective DAP5 cDNA fragment coding for a dominant negative miniprotein was rescued by this method, providing the basis for the isolation of the full-length cDNA. In parallel, Imataka et al. cloned DAP5/p97 in an effort to identify novel genes belonging to the eIF4G family (15). Shaughnessy et al. cloned the mouse DAP5 gene based on its physical linkage to a common retroviral integration site found in myeloid leukemia of BXH2 mice (33). They mapped human DAP5 within a cluster of genes on human chromosome 11p15, which harbors several unidentified tumor suppressor genes. Finally, Yamanaka et al. identified DAP5/NAT1 as a novel target for RNA editing in transgenic mice overexpressing Apobec-1, the catalytic subunit of the editosome complex. In these mice DAP5/NAT1 mRNA was extensively edited, creating multiple stop codons (40). Interestingly, transgenic mice and rabbits overexpressing Apobec-1 developed liver dysplasia and hepatocellular carcinoma, linking oncogenesis with the aberrant hyperediting of target mRNAs. The identification of DAP5 mRNA as a principal editing target in these mice further suggests that DAP5 may be coupled to cell growth control.
Two eIF4G family members, eIF4GI and eIF4GII, serve as a scaffold for the coordinated assembly of the translation initiation complex, leading to the attachment of the template mRNA to the translation machinery at the ribosome, usually through the 5′ cap structure (12, 26). Interestingly, the homology between DAP5 and eIF4GI/GII spans over the central part of the latter proteins, which is responsible for eIF3 and eIF4A binding. In contrast, the N-terminal part of eIF4GI/GII, which binds to the cap binding protein eIF4E, is completely missing from DAP5 protein. Consistent with these structural predictions, it was shown by a few strategies that DAP5/p97/NAT1 binds eIF3 and eIF4A and fails to bind eIF4E, which is essential for mediating cap-dependent translation (15, 16, 40). In this respect, DAP5 resembles the cleaved version of eIF4GI/GII, devoid of its N terminus, which results from infections by several members of the picornavirus family. These cleaved C-terminal fragments of eIF4GI/GII fail to mediate cap-dependent translation (5, 13, 20, 35) but promote cap-independent translation via internal ribosome entry sites (IRES) (5, 20, 29, 30). Despite this structural resemblance, it has been reported that high levels of ectopically expressed DAP5/p97/NAT1 inhibited both cap-dependent and cap-independent translation (from viral IRESes), suggesting that it may function as a general negative regulator of translation (15, 16, 40). The identification of DAP5 as a translation regulator on the one hand and the rescue of the gene as a mediator of apoptosis on the other hand suggest that further studies of this gene should highlight novel mechanisms linking translational control to restriction of cellular outgrowth by PCD (21).
While DAP5 was reported to be ubiquitously and abundantly expressed in normal tissues and growing cell lines (15, 21, 33, 40), the question of its possible regulation during apoptosis has not been investigated yet. In this work, we found that in response to different apoptotic triggers such as activation of Fas receptors or p53 induction, DAP5 was cleaved at a conserved caspase cleavage site to generate a novel p86 form devoid of its C terminus. The cleavage of DAP5 did not interfere with its ability to interact with the translation factors eIF3 and eIF4A. Interestingly, this novel DAP5 form persisted in cells during the apoptotic process at levels comparable to those of the intact p97 form in growing cells. This stood in contrast to the eIF4GI cap-dependent translation factor, the cleavage of which yielded low steady-state levels of products compared to the intact protein in growing cells, thus changing the internal balance within this family of translation factors. Under these apoptotic conditions, total translational activity in the cells was reduced by 60 to 70% and the rate of β-tubulin synthesis was reduced by more than 85%, whereas DAP5 protein itself continued to be selectively translated. We identified in the 5′ untranslated region (5′UTR) of DAP5 mRNA an IRES element which directed cap-independent translation when placed in a bicistronic vector and continued to function during Fas-induced apoptosis, suggesting that it may contribute to the maintained translation of the endogenous DAP5 protein during apoptosis. Finally we show here for the first time that DAP5 IRES-directed translation in rabbit reticulocyte lysate (RRL) is DAP5 dependent and that the cleaved DAP5/p86 form is more potent than the full-length DAP5 in its ability to enhance DAP5 IRES-mediated translation. Altogether, our results suggest that DAP5 is a caspase-activated translation factor responsible at least for the mediation of its own translation during apoptosis.
MATERIALS AND METHODS
Cell cultures and induction of apoptosis.
Human SKW B-lymphoma cells and the murine myeloid leukemic cell line LTR6/M1, carrying a temperature-sensitive p53 mutant, were grown in RPMI 1640 medium supplemented with 10% heat-inactivated fetal calf serum, l-glutamine, penicillin (100 U/ml), and streptomycin (100 μg/ml). The 293 cells were grown as previously described (18). Apoptosis in SKW B-lymphoma cells was induced by addition of soluble protein A (5 μg/ml; Sigma) and anti-Fas agonistic antibodies (100 ng/ml). For the in vivo protease inhibition experiment, SKW B cells were preincubated with the cell-permeable inhibitors BD-Fmk (BD), Z-DEVD-CH2F (DEVD), and Z-YVAD-Fmk (YVAD) (100 μM each; Enzyme Systems Products), GM132 (100 μM), and calpain inhibitor I/ALLN (150 μM; Boehringer) 1 h prior to the addition of anti-Fas antibodies to the cells. p53-induced apoptosis in LTR6/M1 cells was achieved by a temperature shift to 32°C (41). The HeLa-Fas-Bujard (HFB) cells were generated by transfecting a HeLa cell clone (HtTA-1) that expresses a tetracycline-controlled transactivator with a tetracycline-controlled expression vector carrying the Fas receptors (4). The HFB cell line was cultured in Dulbecco modified Eagle medium supplemented with 10% fetal calf serum, l-glutamine, penicillin (100 U/ml), streptomycin (100 μg/ml), and hygromycin B (100 μg/ml; Calbiochem). Apoptosis was induced by addition of soluble protein A (5 μg/ml; Sigma) and anti-Fas agonistic antibodies (100 ng/ml).
Cell death was assessed in SKW B cells and LTR6/M1 cells by using an ApoAlert annexin V-fluorescein isothiocyanate apoptosis kit (Clontech) according to the manufacturer's instructions. Detection of the resulting signal was carried out by flow cytometry according to the manufacturer's instructions.
DNA constructs.
Wild-type and mutant versions of DAP5 were expressed from pECE-Flag vector. pECE-DAP5/p97 was generated by subcloning DAP5 SspI cDNA fragment in frame to the Flag tag in the SmaI site of plasmid pECE-FLAG. pECE-DAP5/p86, pECE-DAP5/DETA, and pECE-DAP5/DHVA were generated by creating point mutations in the pECE-DAP5/p97 plasmid by using a QuikChange site-directed mutagenesis kit (Stratagene), using a set of primers encompassing the inserted point mutations.
The basic luciferase (LUC)-secreted alkaline phosphatase (SeAP) bicistronic (LS) vector, used in this study for insertion of DAP5's 5′UTR element, was provided by QBI Enterprises (Nes-Ziona, Israel) (34). DAP5's 5′UTR (306 nucleotides) was obtained by PCR utilizing DAP5's EcoRI insert as the template with primers encompassing an XhoI restriction site at the 5′ end (5′ GGC GGG CTC GAG CAG CAG TGA GTC GGA GCT CTA TGG AGG TGG CAG CGG GTA) and an NcoI restriction site at the 3′ end (5′ GGA CTC CCA TGG TTG GCG CTT GAC AAC GAA GAA TCT TC). The 5′UTR was inserted between the XhoI (5′) and BsmBI (3′) sites of the intercistronic region of the bicistronic vector, allowing the hybrid NcoI-BsmBI site to recreate the initiator ATG codon of SeAP, giving rise to the LS-DAP5 vector. LS-immunoglobulin heavy chain binding protein, based on the same basic LS bicistronic vector (34), was provided by Eli Keshet. LS-EMCV (encephalomyocarditis virus) was provided by QBI Enterprises (Nes-Ziona, Israel).
pcDNA3-CAT and pcDNA3-hpCAT (provided by Y. Groner, Weizmann Institute) contain CAT (chloramphenicol acetyl transferase)-ORF (open reading frame) and CAT-ORF preceded by a 30-nucleotide hairpin (ΔG = −40 kcal/mol), respectively (1, 3, 37). The bicistronic vectors CAT-LUC-DAP5 and hp-CAT-LUC-DAP5 (CL-DAP5 and hpCL-DAP5, respectively) were generated by placing the DAP5 5′UTR conjugated to LUC (originating from promoter plasmid PGL2 [Promega]) within the polylinker sequence, downstream of the CAT gene, at the BstXI restriction sites.
Cell lysates and immunoprecipitations.
Cells were washed with phosphate-buffered saline and lysed in cold buffer B (100 mM KCl, 0.5 mM EDTA, 20 mM HEPES-KOH [pH 7.6], 0.4% NP-40, 20% glycerol, aprotinin [4 μg/ml], pepstatin [5 μg/ml], leupeptin [5 μg/ml], and 1 mM phenylmethylsulfonyl fluoride) unless indicated otherwise. For immunoprecipitation experiments, 1 mg of protein extract was precleared by protein A-Sepharose CL-4B beads (Pharmacia Biotech) or by protein G-PLUS-agarose beads (Santa Cruz Biotechnology) for 30 min at room temperature. The precleared extracts were incubated with the beads and the relevant antibodies for 6 to 12 h at 4°C. Immunoprecipitates were washed repeatedly with buffer B, eluted with Laemmli buffer, and resolved by polyacrylamide gel electrophoresis (PAGE) on a sodium dodecyl sulfate (SDS)–7.5% polyacrylamide gel unless indicated otherwise.
Antibodies.
Anti-DAP5 rabbit polyclonal antibodies, generated against a fragment of DAP5 corresponding to amino acids 488 to 742 (21), were used at dilutions of 1:350 dilution for Western blotting and 1:50 for immunoprecipitations. Anti-DAP5 monoclonal antibodies generated against a fragment of DAP5 corresponding to amino acids 672 to 830 (Transduction Laboratories) were used for Western blotting at 1:500 dilution. The anti-eIF4A, anti-eIF3/p116, and anti-eIF4GII antibodies (12, 15, 25) were kindly provided by N. Sonenberg. Polyclonal antibodies against bacterially produced N- and C-terminal fragments of eIF4GI expressed from pRSET (Invitrogen) (amino acids 173 to 457 and 934 to 1390, respectively) were prepared in New Zealand White rabbits. Anti-poly (ADP-ribose) polymerase (PARP) polyclonal antibodies (BIOMOL) were used for immunoblotting at 1:5,000 dilution. Anti-β-tubulin antibodies used for immunoprecipitation were purchased from Sigma. Anti-Flag monoclonal antibodies coupled to agarose beads (M2 affinity gel; IBI/Kodak) were used for immunoprecipitation of Flag-tagged proteins.
Metabolic labeling of proteins.
Exponentially growing SKW cells were incubated with methionine-depleted medium for 1 h and then labeled with 80 μCi of [35S]Met per ml for an additional 1.5 h. For assessing total protein translation rate, SKW cells, either nontreated or pretreated for 2 h with cycloheximide (CHX; 10 μg/ml; Sigma) or with anti-Fas agonistic antibodies, were starved and labeled as described above and harvested thereafter (treatment with CHX or with anti-Fas continued during the methionine starvation and the radioactive pulse). Cytoplasmic extracts were prepared, applied to filter paper (Whatman), and boiled in 10% trichloroacetic acid. The level of acid-insoluble radioactivity per microgram of protein extract was calculated. For comparison of DAP5 and β-tubulin translation rates, control and anti-Fas-treated extracts (1.5 × 106 cpm of each) were subjected to immunoprecipitation by the corresponding antibodies. The intensity of the bands was determined with a BAS-2000 phosphorimager (Fuji) or by densitometry.
Enzymatic assays of reporter proteins.
HFB or 293 cells were transiently transfected with bicistronic vectors by the standard calcium phosphate technique. LUC enzyme activity in cell extracts was determined by using a commercial LUC assay system (Promega) as recommended by the supplier. Light emission was quantified with a Lumac/3M BIOCOUNTER M2010 luminometer. The activity of excreted SeAP released into the growth medium was determined following inactivation of the endogenous SeAP by heating the medium to 65°C and clarifying it by centrifugation. Aliquots were diluted in SeAP buffer (1 M diethanolamine [pH 9.8; Sigma], 0.5 mM MgCl2, 10 mM l-homoarginine [Sigma]) and 12 mM p-nitrophenyl phosphate substrate (Sigma). Following incubation at 37°C, the product concentration was determined by measuring absorbency at 405 nm, within the linear range of the assay.
RNA analysis.
Total cellular RNA was isolated by using Tri-Reagent (Molecular Center, Inc.) according to the manufacturer's instructions. Thirty-microgram samples of total cellular RNA were electrophoretically separated on a 1% gel. The RNA was transferred onto a nylon membrane (Amersham) and covalently linked to the membrane by a UV cross-linker (Spectronics Corporation). Prehybridization was performed at 42°C for 4 h in hybridization solution (50% formamide, 5× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate], 4× Denhardt solution, 0.1% sodium pyrophosphate, 100 μg of heat-denatured salmon sperm DNA per ml). Hybridization was carried out overnight under the same conditions and in the presence of 106 cpm of [32P]dCTP-labeled DNA probe. Washes were performed at 50°C in a 0.1% SDS–0.1% SSC–0.1% sodium pyrophosphate washing solution. The intensity of the bands was determined with a BAS-2000 phosphorimager (Fuji).
Translation in cell-free system.
The pcDNA3-based CAT-LUC-DAP5 (CL-DAP5) and hp-CAT-LUC-DAP5 (hpCL-DAP5) vectors were linearized with NotI and used as templates for in vitro transcription from the T7 promoter with T7 RNA polymerase (Promega) for 2 h at 37°C. These RNA transcripts were then translated in RRL (Promega) by conventional procedures, and the products were resolved on a 10% polyacrylamide gel followed by salicylic acid amplification.
For studying the effects of DAP5 forms on translation in vitro, LS-DAP5 and LS-EMCV vectors were linearized with HpaI and served as templates for synthesis of capped transcripts as described above, in the presence of m7GpppG (Biolabs) at a 10-fold molar excess over GTP. The recombinant proteins added to the translation reaction were prepared by the following procedures: glutathione S-transferase (GST) and GST-260 (see Results) were produced in bacteria and affinity purified on GST columns as described previously (21). DAP5/p97 and DAP5/p86 recombinant proteins were immunoprecipitated via a Flag tag from transfected cells. To this end, 293 cells transfected with pECE, pECE-DAP5/p97, or pECE-DAP5/p86 were lysed in buffer B (without EDTA), immunoprecipitated with beads conjugated to anti-Flag antibodies (M2 affinity gel; IBI/Kodak) for 12 h, and then washed twice in lysis buffer and twice in Tris (pH 8) buffer. Each translation reaction mixture consisted of 17.5 μl of RRL (Promega), 0.5 μl of amino acid mixture minus Met (1 mM; Promega), 2.5 μl of [35S]Met (10 μCi/μl), and 2 μl of RNA transcript. When indicated, 4 μl GST or GST-260 protein (∼0.3 μg) was added. Alternatively, the entire reaction mixture was added to the immunoprecipitates and incubated at 30°C for 90 min with continuous stirring. The reaction was terminated by boiling in Laemmli buffer. Half of the reaction mixture was resolved by SDS-PAGE (10%) gel, dried, and analyzed for band intensities with a BAS-2000 phosphorimager (Fuji). The other half was subjected to Western analysis and densitometry.
RESULTS
Induction of a novel form of DAP5 protein during PCD.
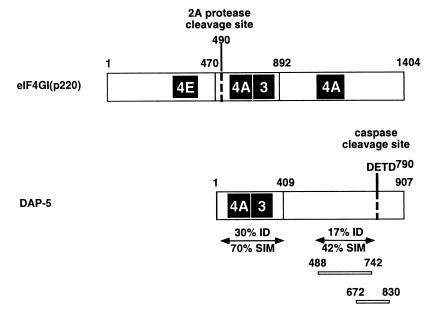
In a search for possible posttranslational regulatory events which may modify the DAP5 protein, several cell lines were induced to undergo apoptosis by different types of stimuli. The fate of DAP5, a 97-kDa protein, was monitored by Western blotting with polyclonal antibodies raised against a fragment of the protein that shows low homology to eIF4G (amino acids 488 to 742; see Fig. 8). The choice of this specific fragment indeed yielded antibodies which did not cross-react with eIF4G proteins (not shown).
FIG. 8.
Amino acid homology alignment of eIF4GI and DAP5. eIF4GI and DAP5 are divided into regions based on the high homology of the central region of eIF4GI with the N-terminal region of DAP5. The shaded boxes indicate binding sites of translation initiation factors: 4E, eIF4E, the cap binding protein; 4A, eIF4A, an ATP-dependent RNA helicase; and 3, eIF3, a ribosome adapter. The site of cleavage of eIF4GI by 2A protease is indicated by a dashed line at position 490. The identified caspase cleavage site of DAP5 is indicated by a dashed line at position 790. DAP5 protein fragments corresponding to amino acids 488 to 742 and 672 to 830 were used for the production of polyclonal and monoclonal antibodies, respectively, against DAP5. Regional homologies at the amino acid level of the conserved N-terminal region and of the less homologous DAP5 miniprotein region between DAP5 and eIF4GI are indicated. Alignment was performed as previously published (21), and homology was determined by using the Dayhoff PAM250 residue weight table. ID, identity; SIM, similarity.
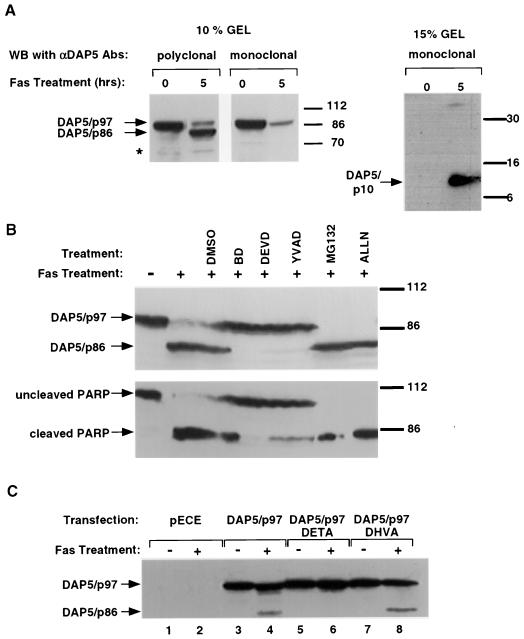
SKW B-lymphoma cells respond to anti-Fas agonistic antibodies in a well-synchronized execution of the cell death program. As early as 5 h after treatment with the anti-Fas antibodies, approximately 70% of the SKW cells exhibit an altered plasma membrane composition, as assessed by flow cytometry analysis with annexin V conjugated to fluorescein isothiocyanate (Fig. 1A). The kinetics of cell death was followed at the level of caspase activation as well. Detection of the caspase cleavage product of PARP was used as a marker for caspase activity. As early as 2 h after activation of the Fas receptors, no intact 112-kDa PARP could be detected, as it was all converted into its cleaved product. This demonstrates both the rapid kinetics of the execution of apoptosis and the synchronized response of SKW cells to Fas activation, making it an ideal system for studying different cell death-associated events. In growing cells, DAP5 appeared as a 97-kDa protein which could be resolved into two bands, depending on the gel fractionation and resolution: a major DAP5 band and a rapidly migrating minor band just beneath, as reported earlier (15). These bands probably reflect some posttranslational modifications of DAP5 protein (Fig. 1A). After Fas stimulation, DAP5 protein displayed a typical pattern of alterations. The level of DAP5/p97 decreased considerably as the apoptotic program progressed, and an 86-kDa protein, recognized by the same anti DAP5 polyclonal antibodies, appeared instead (Fig. 1A). These DAP5 protein alterations could be detected as early as 2 h after activation of the Fas receptors. Again, a minor rapidly migrating band was detected below the major 86-kDa protein as well (Fig. 1A).
FIG. 1.
A novel DAP5/p86 form appears in Fas- and p53-induced apoptosis. (A) Exponentially growing SKW B-lymphoma cells (3 × 106, total cell number) were treated with agonistic anti-Fas antibodies for 0, 2, 5, and 8 h. Treatment was terminated by harvesting all cells and immediately boiling the pellets in Laemmli sample buffer. Immunoblots were reacted with anti-DAP5 polyclonal antibodies (top) and anti-PARP antibodies (bottom). Apoptotic cell death was assessed by detection of changes in the membrane composition by the annexin V fluorescence-activated cell sorting analysis. (B) HFB cells were exposed to anti-Fas agonistic antibodies, for the indicated time periods, in the presence or absence of CHX. The fate of DAP5 was followed by reacting the immunoblots with anti-DAP5 polyclonal antibodies. (C) LTR6/M1 cells, expressing a temperature-sensitive p53 mutant, were induced to undergo apoptosis by a temperature shift to 32°C. Apoptotic cell death was assessed by detection of changes in the membrane composition by annexin V fluorescence-activated cell sorting analysis. The fate of DAP5 was assessed by Western blotting. The sizes of protein markers (in kilodaltons) are shown on the right. The positions of DAP5/p97 and DAP5/p86 are marked by arrows (dashed arrows point to the minor rapidly migrating p97 and p86 bands). The asterisk marks a nonspecific band lightened by the anti-DAP5 antibodies.
The reduction in the level of DAP5/p97 and the concomitant appearance of an 86-kDa protein as apoptosis execution progressed were not confined to a certain cell line or apoptotic trigger. Treatment of HFB cells with anti-Fas agonistic antibodies (Fig. 1B) and activation by temperature shift of a temperature-sensitive p53 mutant expressed in LTR6/M1 cell line (Fig. 1C) resulted in the same alterations of DAP5 protein. However, the extent and time kinetics of these alterations differed from those for the Fas-activated SKW cells, in accordance with the slower kinetics and decreased synchrony of these systems (Fig. 1C).
To confirm that the novel 86-kDa protein appearing during apoptosis is a derivative of DAP5, we attempted to detect it via alternative, nonoverlapping antibodies. Recombinant N-terminally Flag-tagged DAP5 was transfected into HFB cells, and its fate upon death induction was assessed by immunoprecipitation with anti-Flag antibodies. In the normally growing transfectants, only a single tagged protein, approximately 97 kDa in size, was detected, whereas two Flag-tagged proteins of 97 and 86 kDa were recovered upon Fas treatment (Fig. 2C, lanes 3 and 4). The ability of two different epitopes to detect the same novel protein strongly suggests that the 86-kDa protein that appears during cell death is a novel DAP5 form which we have accordingly named DAP5/p86.
FIG. 2.
DAP5/p97 is converted to DAP5/p86 by caspase cleavage. (A) SKW B-lymphoma cells were treated with anti-Fas agonistic antibodies for 5 h. Samples were fractionated on a 10% or 15% polyacrylamide gel, and DAP5 was assessed by reacting the Western blot (WB) with anti-DAP5 polyclonal or monoclonal antibodies (αDAP5 Abs), as indicated (see Fig. 8 for antibody epitopes). A nonspecific band is marked by an asterisk. (B) SKW B-lymphoma cells were treated with agonistic anti-Fas antibodies for 5 h after a 1-h preincubation period with caspase inhibitors (BD, DEVD, and YVAD), proteosome inhibitor (MG132), and calpain I inhibitor (ALLN). As a control, cells were preincubated with dimethyl sulfate solvent alone. Immunoblots were reacted with anti-DAP5 polyclonal antibodies (top) and anti-PARP antibodies (bottom). (C) HFB cells were transiently transfected with the following N-terminal Flag-tagged DAP5 constructs: wild-type DAP5 (DAP5/p97 [lanes 3 and 4]) and two constructs each carrying a potential disrupted caspase cleavage site (DAP5/p97 DETA790 [lanes 5 and 6]; DAP5/p97 DHVA824 [lanes 7 and 8]). An empty pECE vector served as a control (lanes 1 and 2); 24 h posttransfection the cells were exposed to anti-Fas agonistic antibodies for 12 h or to fresh medium alone. The exogenic DAP5 forms were pulled down by immunoprecipitation with anti-Flag antibodies followed by Western blotting with anti-DAP5 polyclonal antibodies.
DAP5/p97 is converted to DAP5/p86 by proteolytic cleavage at the C terminus.
Next, we set out to understand the mechanism underlying the induction of DAP5/p86. Its induction even in the presence of the protein synthesis inhibitor CHX proved that the appearance of DAP5/p86 did not depend on de novo protein synthesis. As shown in Fig. 1B, the DAP5/p86 form was readily detected in HeLa cells stimulated by agonistic anti-Fas antibodies in the presence of CHX. Pulse-chase experiments performed with nontreated and Fas-stimulated SKW cells determined that the half-life of DAP5 protein in growing cells is longer than 5 h and further demonstrated that DAP5/p86 was derived from preexisting labeled DAP5/p97 (not shown).
Wide varieties of posttranslational modifications may affect the migration pattern of a protein on polyacrylamide gels; such modifications include phosphorylation, acetylation, glycosylation, and proteolysis. Since the observed decrease in DAP5's molecular weight was of a relatively great magnitude, and since proteases are well-established regulators and executioners of PCD, we further explored the issue of proteolysis. Because the N-terminal Flag was readily detected by the anti-Flag antibodies in the DAP5/p86 form (Fig. 2C, lanes 4 and 8), the possibility of N-terminal truncations was excluded. To test the possibility that DAP5/p97 is converted to DAP5/p86 by cleavage at its C terminus, monoclonal antibodies directed against DAP5's C-terminal portion (amino acids 672 to 830; see Fig. 8) were used to monitor DAP5 in control and Fas-stimulated SKW cell lysates. The monoclonal antibodies failed to recognize DAP5/p86 in the treated cultures and instead reacted mainly with a protein fragment of approximately 10 kDa (Fig. 2A). These results are consistent with the possibility of a proteolytic cleavage occurring during cell death at the C terminus of DAP5.
DAP5/p97 is cleaved at a conserved caspase cleavage site at position 790.
To assess the contribution of different cellular proteases to the cleavage of DAP5/p97, SKW cells were stimulated with agonistic anti-Fas antibodies in the presence or absence of a panel of protease inhibitors: a proteosome inhibitor (MG132), a calpain I inhibitor (ALLN), and caspase inhibitors (BD, YVAD, and DEVD) (Fig. 2B). Proteosome and calpain I inhibitors did not abrogate the appearance of DAP5/p86 at the expense of DAP5/p97. In contrast, caspase inhibitors prevented the cleavage of DAP5. The ability of the various protease inhibitors to prevent DAP5's cleavage correlated with their ability to abrogate PARP cleavage.
The caspase inhibitors could exert their effects on DAP5 either directly, by blocking a specific caspase responsible for DAP5/p97 cleavage, or indirectly, by interfering with early caspase-dependent events operating upstream to DAP5's conversion. Examination of the amino acid sequence of DAP5 protein revealed the existence of four potential caspase cleavage sites of the motif DXXD (amino acid 185, 592, 790, 824), yet only two of these sites (DETD790 and DHVD824) seemed capable of yielding a cleavage product of the expected molecular weight. To examine directly whether these sites are required for the cleavage of DAP5, we constructed two Flag-tagged DAP5/p97 mutants carrying individual Asp-to-Ala mutations in these potential sites (DXXD→DXXA). We followed the ability of each mutation to abolish the appearance of exogenous DAP5/p86 in Fas-stimulated HFB cells (Fig. 2C). While the p86 form was detected upon transfection with wild-type Flag-DAP5/p97 (lanes 3 and 4) and Flag-DAP5/DHVA824 (lanes 7 and 8), transfection with the DAP5/DETA790 construct failed completely to yield the DAP5/p86 form (lanes 5 and 6). This indicates that DAP5/p97 is converted to DAP5/p86 directly by caspase cleavage and maps the exact cleavage to the DETD790 site.
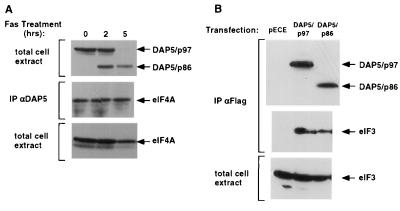
Caspases are known to alter the function of their protein substrates by causing their activation or inactivation or other types of functional modulations (28, 31). We first tested whether the cleavage of DAP5 influenced the binding to translation initiation factors eIF3 and eIF4A. In one line of experiments, endogenous DAP5 was immunoprecipitated with anti-DAP5 polyclonal antibodies from growing or Fas-stimulated SKW cells, and the levels of coimmunoprecipitated eIF4A were subsequently monitored (Fig. 3A). We found that both DAP5/p97 (exclusively appearing in nontreated growing cells) and DAP5/p86 (exclusively appearing in treated cultures at the 5-h point) pulled down very efficiently the endogenous eIF4A. This line of experiments indicated that DAP5/p86 retained its binding capacity to eIF4A and that these complexes exist in cells during apoptosis. In a second line of experiments, we compared eIF3 binding to ectopically expressed p97 and p86 DAP5 forms. The recombinant DAP5/p97 or DAP5/p86 proteins were transiently expressed in 293 cells and immunoprecipitated with anti-Flag antibodies; the levels of the eIF3/p116 subunit in the complex were assessed. Similar to the full-length protein, the truncated p86 form also pulled down the eIF3 protein (Fig. 3B). Hence, the cleavage of DAP5 does not seem to affect its overall ability to bind these critical translation initiation factors.
FIG. 3.
Coimmunoprecipitation of eIF4A and eIF3 with the two DAP5 forms. (A) Growing SKW cells or SKW cells exposed for 2 or 5 h to anti-Fas agonistic antibodies were gently extracted in B buffer. Protein extract (1 mg) was subjected to immunoprecipitation with anti-DAP5 polyclonal antibodies. Coimmunoprecipitation of endogenous eIF4A was assessed by Western blotting the immunoprecipitates with anti-eIF4A antibodies (middle panel). Samples of 100 μg of total cell extracts were assessed for DAP5 and eIF4A levels by direct Western blotting (top and bottom panels, respectively). (B) 293 cells were transiently transfected with Flag-tagged DAP5 constructs in a pECE vector. The constructs included wild-type DAP5 (DAP5/p97), a mutant of DAP5 carrying a stop codon at position 790 (DAP5/p86), and an empty vector (pECE). At 48 h posttransfection the cells were extracted gently in B buffer. The ectopically expressed DAP5 was immunoprecipitated with anti-Flag antibodies and assessed with anti-DAP5 antibodies after resolution of the immunoprecipitates on gels (top); coimmunoprecipitation of endogenous eIF3 was assessed by Western blotting the immunoprecipitates with antibodies against the eIF3/p116 subunit (middle); total endogenous eIF3/p116 was measured by direct Western blotting (bottom).
DAP5 is preferentially translated in apoptotic cells, in the absence of intact eIF4G proteins.
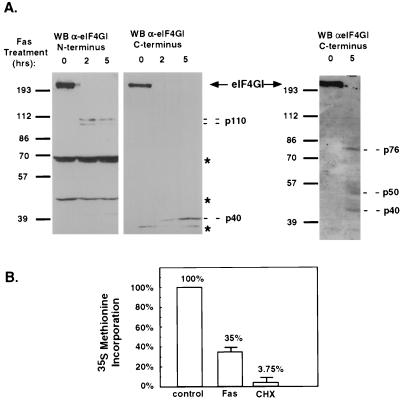
The finding that DAP5 is cleaved during cell death into a novel protein form prompted us to study the fate of the other eIF4G family members during apoptosis. The fate of eIF4GI and eIF4GII, the two main mediators of cap-dependent translation, was assessed in both HFB and SKW cells stimulated by Fas agonistic antibodies. Intact eIF4GI could not be detected at all by polyclonal antibodies raised against its C or N terminus as early as 2 h following treatment (Fig. 4A shows data for SKW cells; similar results were obtained in HFB cells). A more detailed analysis revealed that eIF4GI had already disappeared at 1 h after treatment of SKW cells with anti-Fas agonistic antibodies (not shown). Low levels of truncated eIF4GI forms, about 110-kDa in size (Fig. 4A, left panel) were detected in apoptotic cells with the N-terminal antibodies (an average drop of at least 80% compared to the levels of the intact protein in growing cells, as assessed by densitometry). The C-terminal antibodies could also detect very small amounts of proteins of 40, 50, and 76 kDa in treated cells (Fig. 4A, two right panels). Again, the amounts of the cleaved proteins detected by the anti-eIF4GI antibodies in the treated cells were in the range of 5 to 20% of intact protein levels in control cells. The disappearance of intact eIF4G and the detection of cleaved products is consistent with previous work which showed caspase-mediated cleavage of eIF4GI in different cell systems and in response to various apoptotic triggers (6, 7, 22, 27). Similarly, the eIF4GII protein did not remain in its intact form as well, as assessed by immunoblot analysis of extracts from Fas-induced SKW cells with polyclonal antibodies raised against the N-terminus fragment of the protein (not shown). Thus, the steady-state levels of these two important family members, which are critical for cap-dependent translation, were markedly reduced in the apoptotic cells.
FIG. 4.
Disappearance of intact eIF4GI during cell death. (A) SKW B-lymphoma cells were treated with anti-Fas agonistic antibodies for 0, 2, and 5 h. The fate of eIF4GI was assessed by Western blotting (WB) with polyclonal antibodies generated against eIF4GI N terminus (left panel) or C terminus (two right panels). The position of full-length eIF4GI is indicated by an arrow. Bands exclusively recognized by either anti-eIF4G antibody in the Fas-treated cells are marked by their approximated molecular sizes (in kilodaltons) (p110, p76, p50, and p40). The asterisks mark nonspecific bands lightened by the anti-eIF4GI antibodies. (B) SKW B-lymphoma cells, either nontreated or pretreated for 2 h with CHX or with anti-Fas agonistic antibodies, were incubated in methionine-free medium for 1 h, labeled with [35S]Met for 1.5 h, and harvested thereafter (the treatment with CHX or with anti-Fas continued during the methionine starvation and the radioactive pulse). The level of insoluble radioactivity incorporated per microgram of protein extract was set as 100% in control cells. The results represent the average of four independent experiments.
The well-established fact that cleavage of eIF4GI and eIF4GII during some viral infections leads to a shutdown in cap-dependent cellular translation (13, 20, 35), together with the observation that the apoptotic cells undergo depletion of these key proteins, prompted us to study whether the degradation of these proteins during apoptosis is also correlated with translation shutdown. The overall rate of protein synthesis in SKW cells was measured by incorporation of [35S]methionine into acid-insoluble material. Both control and Fas-pretreated cells were labeled. Fas-treated cells showed a reduced translation rate in the range of 30 to 40% of the control rate (Fig. 4B). Pretreatment with the translation elongation inhibitor CHX, which served as a reference for complete translational shutdown, resulted in a 1 to 10% translation rate compared to control cells. This finding implies that during apoptosis, some translational events still occur.
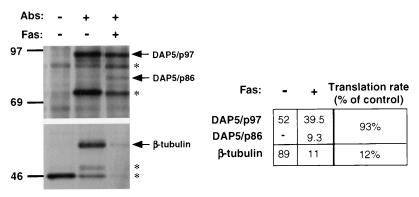
Since overall translation did not cease completely, it was interesting to explore whether the translation of some specific proteins could preferentially continue under these conditions. Being a positive mediator of cell death, DAP5 by itself could belong to this putative group of proteins that continue to be synthesized under this kind of apoptotic stress on translation. Therefore, we compared changes in the translation rate of DAP5 between control and FAS-stimulated SKW cells versus the change in the translation rate of another arbitrarily chosen protein, β-tubulin. Translation rate was determined by metabolically labeling naive or 3-h-Fas-treated cells with [35S]Met for a 1.5-h pulse (a point where intact eIF4GI and eIF4GII were already below detection levels). The labeled cells were lysed, and equal amounts (counts per minute) were immunoprecipitated by the corresponding antibodies. Interestingly, DAP5's synthesis rate was only marginally affected in the apoptotic cells (7% reduction), whereas β-tubulin was reduced by 88% (Fig. 5). Northern blot analysis of DAP5 and β-tubulin RNAs was performed to normalize the translation values. The results clearly showed that the differences in the protein's synthesis rate took place at the translation level, as the levels of both RNA species dropped to the same extent upon Fas activation (not shown).
FIG. 5.
Preferential translation of DAP5 during cell death. SKW cells were treated for 3 h with anti-Fas agonistic antibodies (Abs) or were left untreated. During the last hour, cells were transferred to methionine-depleted medium supplemented with their corresponding treatments; they were then pulse-labeled with 80 μCi [35S]Met per ml for 1.5 h and harvested. Samples of cytoplasmic extracts containing 1.5 × 106 cpm were subjected to immunoprecipitation with anti-DAP5 polyclonal antibodies (top) and with anti-β-tubulin antibodies (bottom). The first lane in each panel represents nonspecific background bound to the Sepharose beads. The asterisks mark protein bands that reacted with the beads or the antibodies nonspecifically. Values from the densitometric analysis of the DAP5 and β-tubulin bands are shown at the right. The translation rate of each protein in the growing cells was set as 100%, and the translation rate of each protein following Fas treatment was calculated accordingly. Similar results were obtained in three additional independent experiments.
An IRES element in DAP5's 5′UTR is activated in apoptotic cells.
One possible explanation for the continuous residual translation during cell death, in the absence of detectable intact eIF4GI/GII, is that mechanisms of cap-independent translation may be preferentially utilized. Along this line, we wondered whether DAP5's preferred translation rate during apoptosis could be mediated by an IRES element in its 5′UTR. The 5′UTR of DAP5 mRNA possesses some characteristics of IRES elements, as it is relatively long (about 300 bp) and encompasses two polypyrimidine-rich tracts (21). The ability of this element to function as an IRES, which directs internal translation initiation, was examined. A stretch of 306 bp from the 5′UTR of DAP5 was inserted between the two cistrons of a bicistronic vector in which the first cistron encodes LUC and the second cistron encodes SeAP. The first cistron is proximal to the mRNA's cap structure and therefore is expected to be translated by the conventional cap-dependent translation mode. The second cistron is distal from the cap site and is separated from the first cistron by multiple stop codons to decrease leaky translation; therefore it is expected to undergo translation only upon insertion of an IRES element between the two cistrons.
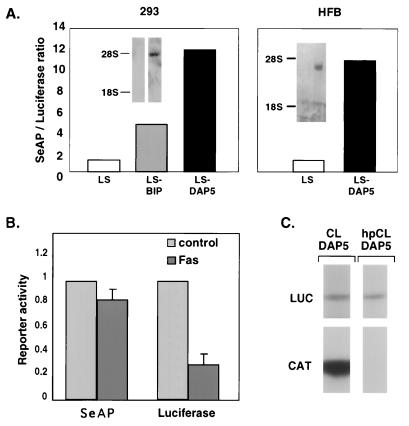
The bicistronic vectors were transiently expressed in 293 or HFB cells, and the resulting SeAP/LUC ratio was determined. A vector lacking an IRES insertion (termed an LS vector) was used as well to estimate the background levels of SeAP. Insertion of DAP5's 5′UTR between the two cistrons (LS-DAP5 vector) enhanced the SeAP/LUC ratio approximately 11-fold both in 293 and HFB cells relative to the LS vector (Fig. 6A; see figure legends for raw data). This value was two- to threefold higher than that for another well-established cellular IRES, BiP, which was examined in parallel. We excluded the presence of a cryptic promoter within the DAP5 5′UTR insert by confirming the existence of a single bicistronic RNA message by Northern blotting (Fig. 6A, inset). To verify that the translation of the second cistron does not result from leaky translation continuing from the first cistron, we switched to a CAT-LUC bicistronic system and interfered with the translation of the first cistron (CAT) by insertion of a stable hairpin at its 5′ end. The effect of the hairpin insertion on the translation of both cistrons was examined by in vitro translation in an RRL. We found that the hairpin insertion led to a strong reduction in the translation of the first cistron (CAT), while the translation from the second cistron (LUC) was significantly less affected (Fig. 6C; eight independent translation experiments yielded average reductions in translation of 70 and 16% from the first and second cistrons, respectively, indicating that the major portion of the translation events from the second cistron were not a consequence of translation initiated at the first cistron).
FIG. 6.
DAP5's 5′UTR possesses an IRES which directs cap-independent translation and is selectively sustained during cell death. (A) 5′UTRs of DAP5 and of BiP were inserted between the two cistrons of a basic bicistronic LS vector, in which the LUC reporter gene is translated in a cap-dependent manner from the first cistron and SeAP is translated in a cap-independent manner from the second cistron, to generate LS-DAP5 and LS-BiP respectively. 293 or HFB cells were transfected with a vector lacking an insert in the intercistronic region (LS), LS-DAP5, or LS-BiP and further assessed 48 h posttransfection. For each experiment, the SeAP/LUC ratio obtained by the LS vector was designated 1, and the relative fold increase in SeAP/LUC ratio in the other vectors was calculated. The results represent the average of three independent experiments. In 293 cells, the average values (in arbitrary units) of the reporter activities for LS, LS-BiP, and LS-DAP5 vectors, respectively, were 995, 784, and 1,167 for LUC and 14, 56, and 189 for SeAP. In HFB cells, the average values (in arbitrary units) of the reporter activities for LS and LS-DAP5 vectors, respectively, were (98 and 85) for LUC and (27 and 286) for SeAP. The inserts correspond to Northern blots of nontransfected (left lane of each) and LS-DAP5 transfected (right lane of each) 293 or HFB cells, respectively, probed by the SeAP cDNA. (B) HFB cells were transfected with LS-DAP5 vector. After 36 h, the medium was replaced by medium containing or lacking anti-Fas agonistic antibodies; 12 h later, the enzymatic activity of each reporter enzyme was determined. For each experiment, the SeAP or LUC value obtained for the control cells was designated 1 and served for normalization of the corresponding reporter activity under Fas-stimulated conditions. These results represent the average of seven independent experiments. The raw data from control and Fas-treated cells, respectively, were 197 and 63, 124 and 24, 767 and 246, 192 and 70, 133 and 35, 606 and 253, and 1290 and 229 for LUC and 138 and 135, 111 and 85, 178 and 148, 147 and 119, 296 and 227, 112 and 105, and 383 and 281 for SeAP. (C) DAP5's 5′UTR was inserted into a bicistronic vector in which CAT is translated from the first cistron and LUC is translated from the second cistron, generating CL-DAP5. Insertion of DAP5's 5′UTR into a bicistronic vector in which CAT, the first cistron, is preceded by a stable hairpin generated hpCL-DAP5. These constructs were transcribed and translated in vitro in the presence of [35S]methionine. The intensity of each band was determined by phosphorimager analysis.
Next, the function of the DAP5 IRES under apoptotic conditions was examined. To this end, the LS and LS-DAP5 bicistronic vectors were transiently expressed in HFB cells, and the effects of anti-Fas agonistic antibodies on SeAP and LUC expression levels were examined by comparing their values to that for control, nontreated cells (Fig. 6B; see the legend for raw data). It was found that as a result of Fas treatment, the extent of DAP5 IRES-mediated translation was not significantly changed and was maintained at approximately 90% of the control rate (n = 14) whereas that of cap-dependent translation was severely impaired, as it dropped to 30% of the control rate (n = 14, P < 0.05). As a consequence, the SeAP/LUC ratio of the LS-DAP5 vector was enhanced almost threefold under apoptotic conditions. In transfections with the LS vector, which lacks a functional IRES element, the SeAP/LUC ratio remained unaltered (not shown). Interestingly, these results matched the data on the preferential in vivo labeling of the endogenous DAP5 protein during cell death (Fig. 5), suggesting that the maintained translation of DAP5 under apoptotic conditions is at least partially attributed to an IRES element in its 5′UTR.
DAP5 protein mediates DAP5-IRES-driven translation.
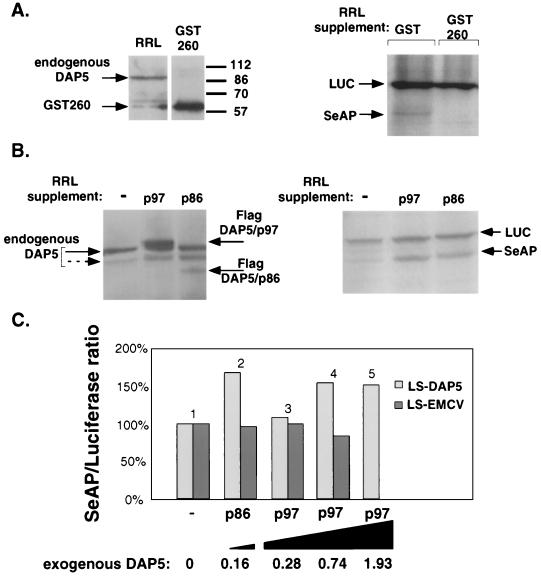
Translation via the DAP5 IRES is preferentially maintained during cell death despite the absence of detectable intact eIF4GI/GII. Under these apoptotic circumstances, DAP5 protein seemed to be the most dominant among the different members of the eIF4G family. It therefore became of interest to test whether DAP5 protein might mediate its own translation in a cap-independent manner. We showed in Fig. 6C that insertion of the DAP5 IRES in a bicistronic vector enables translation of the second cistron in an RRL. Is there DAP5 in the RRL that might contribute to this process? Western analysis of RRL revealed that indeed DAP5 is present in the lysate (Fig. 7A, left panel). To test whether DAP5 IRES-driven translation may be mediated by DAP5 present in the RRL, we attempted to counteract its activity by introducing into the translation reaction a dominant negative fragment of DAP5 (named fragment 260 or DAP5 miniprotein [21]). To this end, we chose to introduce into the translation reaction a bacterially produced GST-fused DAP5 miniprotein (GST-260) purified as previously described (21). The recombinant protein was added to the RRL in excess as shown in Fig. 7A (left panel). Its effect on the translation of both cistrons of the LS-DAP5 bicistronic transcript was measured by comparing the results to the pattern obtained with GST alone (Fig. 7A, right panel). We found that addition of DAP5 miniprotein to RRL significantly interfered with the translation of the second cistron mediated through the DAP5 IRES, causing an average drop to 32% of the translation rates obtained with the GST control reactions (n = 5, P < 0.05). On the other hand, the cap-dependent translation of the first cistron was not significantly affected by the presence of DAP5 miniprotein, as its average value was 93% of its original value (n = 5, P > 0.05). The reduction in the cap-independent translation via the DAP5 IRES and the sustained level of cap-dependent translation, in the presence of the dominant negative DAP5 form, lowered significantly the overall ratio of cap-independent versus cap-dependent translation from the LS-DAP5 vector. Interestingly, when the same procedure was carried out with an LS-EMCV vector, the addition of GST-260 miniprotein to the RRL did not alter the ratio between the two cistrons (not shown), suggesting that the EMCV IRES is not influenced by the miniprotein as is the DAP5 IRES.
FIG. 7.
Translation via the DAP5 IRES in vitro is mediated by DAP5 protein. Capped transcripts of the bicistronic vectors LS-DAP5 or LS-EMCV were translated in vitro in the presence of [35S]methionine. The translation reactions were supplemented with bacterially produced GST-260, using GST supplement as a control (A), or with Flag-DAP5/p97 or Flag-DAP5/p86 immunoprecipitated from transiently transfected 293 cells, using immunoprecipitates from nontransfected 293 cells a control (B and C). Intensities of the LUC and SeAP bands were quantified with a phosphorimager. The SeAP/LUC ratio in the control reaction was set as 1, and that of the DAP5-supplemented reactions was calculated accordingly. (A) LS-DAP5 transcripts were translated as described above, supplemented by equivalent amounts of GST-260 or GST proteins. A typical autoradiogram of the resulting translation products is presented (right). Corresponding amounts of RRL and GST-260, as present in the translation reactions, were separated on a 12% gel and immunoblotted with anti-DAP5 polyclonal antibodies (left). (B) Capped LS-DAP5 transcripts were translated as described above, supplemented with anti-Flag immunoprecipitates from transiently transfected 293 cells overexpressing DAP5 protein forms. The resulting autoradiogram of the translation products is presented at the right. The same translation reactions were separated on 10% gel and immunoblotted with anti-DAP5 polyclonal antibodies (left). The solid and dashed arrows on the left indicate endogenous DAP5 within the RRL; the dashed arrow indicates the minor fast-migrating DAP5 form, present both in cells (Fig. 1) and in RRL. Exogenous DAP5 forms from the immunoprecipitations are marked by the arrows to the right. (C) An experiment similar to that in panel B was performed on both LS-DAP5 and LS-EMCV transcripts in parallel, supplemented by equivalent amounts of exogenous DAP5 proteins (compare within bar pairs) as follows: bar 1, control; bar 2, DAP5/p86; bars 3 to 5, increasing amounts of DAP5/p97. The quantity of supplemented exogenous DAP5 proteins was determined by densitometry of immunoblots of the translation reactions, reacted with anti-DAP5 antibodies. The level of endogenous DAP5 in the RRL was scored as 1, and the calculated relative levels of exogenous DAP5 are presented below each bar pair. The raw band intensity data of the LS-DAP5 translation reactions presented in the graph are detailed in the form of bar no. (luciferase, SeAP, SeAP/LUC ratio): 1 (894, 417, 0.47), 2 (1207, 948, 0.79), 3 (1222, 617, 0.50), 4 (1131, 812, 0.72), and 5 (937, 661, 0.71).
In a reciprocal approach, we tested how DAP5 IRES-mediated translation is affected by addition of recombinant DAP5/p97 or DAP5/p86 on top of the endogenous DAP5 present in the reticulocytes. Exogenous DAP5 proteins were immunoprecipitated from 293 cells overexpressing Flag-DAP5/p97 or Flag-DAP5/p86. Immunoprecipitates from 293 cells transfected with an empty vector were used as a control. The effect of these recombinant proteins on in vitro translation of the LS DAP5 transcript was examined. It was found that addition of either DAP5 form to the cell-free translation system significantly altered the ratio between the two cistrons, resulting in more than a 1.5-fold increase in favor of cap-independent translation from the DAP5 IRES (Fig. 7B, right panel). Western blot analysis of the translation reactions confirmed the presence of each exogenous DAP5 form in the reactions and enabled comparison of their levels to those of the endogenous DAP5 (Fig. 7B, left panel; note that the recombinant p97 form was in large excess over the p86 form). To make more accurate comparisons between DAP5/p97 and DAP5/p86 in these functional assays, we scaled down the amount of DAP5/p97 added to the reactions. Quantitation of exogenous DAP5/p97 and DAP5/p86 protein levels in each reaction mixture of RRL and of their effects on the resulting translation products from LS-DAP5 transcript revealed that DAP5/p86 was more potent than DAP5/p97 in these assays (see the legend to Fig. 7C for the raw data). While low levels of DAP5/p86 (16% of endogenous levels) sufficed to enhance the ratio of DAP5 IRES-mediated translation to cap-dependent translation 1.6-fold, much higher levels of DAP5/p97 (74% of endogenous levels) were required to achieve a similar effect (compare bars 1, 2, and 4 in Fig. 7C). In addition, further dilution of the exogenous DAP5/p97 level to 28% of endogenous levels (still twice the amount of recombinant protein relative to DAP5/p86) hardly affected DAP5 IRES-mediated translation (Fig. 7C, compare bars 1, 2, and 3). These results further suggest that the DAP5/p86 form is more potent in its ability to stimulate DAP5 IRES-mediated translation than the DAP5/p97 form.
Finally, when the same procedure was carried out in parallel with an LS-EMCV transcript, the ratio between cap-dependent and cap-independent translation did not change in any given concentration of the DAP5 recombinant proteins (Fig. 7C). This finding complements previous results showing that EMCV IRES-mediated translation is refractory to GST-260 inhibitory effect. Thus, we conclude that DAP5 IRES-mediated translation, unlike EMCV IRES-mediated translation, is supported by both DAP5 proteins in this system.
DISCUSSION
The requirement for ongoing protein synthesis in PCD differs among various apoptotic systems. Some apoptotic processes are based primarily on activation of preexisting death proteins such as the caspases and do not require any de novo-synthesized proteins. Furthermore, in some instances blockage of protein synthesis per se is a critical event in guiding the cell to undergo apoptosis, as in the case of tumor necrosis factor receptor-mediated apoptosis, where a transcription-translation-dependent death-protective pathway is activated in parallel to the death pathway (2, 38). Conversely, certain scenarios of PCD are translation dependent, as they are abrogated by translation inhibitors. These translation-dependent apoptotic scenarios employ both preexisting and de novo-synthesized death proteins. Some examples are the apoptotic processes in insect and vertebrate embryonic development and the death of tropic factor-deprived sympathetic neurons (9, 23, 24). The relationship between the translation-dependent and translation-independent apoptotic pathways, how they are interconnected, when one is favored over the other, and what determines the dominance of one pathway over the other, are yet unresolved issues.
The functional cloning of a novel eIF4G homolog, DAP5, as a positive mediator of apoptosis provided a molecular tool for understanding translation regulation during apoptosis. DAP5 is one of several genes which were isolated by the technical knockout strategy designed for targeting functionally relevant, rate-limiting death genes (10, 17). The cDNA fragment that served as the basis for DAP5 selection directed the synthesis of a miniprotein (amino acids 488 to 742) that modulated the function of DAP5 and thus conveyed resistance to gamma interferon-induced cell death (21).
In this work we found that in response to two different apoptotic stimuli and in several cell lines, DAP5/p97 was cleaved at a single conserved caspase cleavage site, generating a novel DAP5/p86 form devoid of its C terminus. In addition, we discovered that while there was a general decrease in the translation rate during Fas-induced apoptosis, in accordance with the degradation of eIF4GI and eIF4GII proteins, DAP5 protein was selectively translated. Examination of the 5′UTR sequence of DAP5 revealed the presence of a functional IRES which was preferentially utilized over cap-dependent translation during Fas-induced apoptosis. Such a quality can provide an advantage for a positive mediator of apoptosis under these translation-limiting conditions. However, it should be stressed that unlike the p53-induced death of LTR/M1 cells or the gamma interferon-induced apoptosis, Fas-induced cell death can proceed in the absence of new protein synthesis. Thus, some apoptotic systems clearly utilize dominant protein synthesis-independent pathways although they still display the typical pattern of DAP5 regulation and the described changes in the translation machinery. This further documents the complexity and diversity of apoptotic pathways, which may be differently utilized in various scenarios. Further evidence for this complexity stems from the finding that ectopic expression of DAP5/p86 did not culminate in cell death (not shown), suggesting that DAP5 by itself is not sufficient to induce apoptosis and that additional death signals may be required.
An important aspect highlighted by this work refers to the alterations of the protein translation initiation machinery under apoptotic stress. Early reports noted an eventual shutdown of translation that occurs as apoptosis proceeds (9). Recently the reduction in translation rate in apoptotic cells was correlated with the caspase cleavage of eIF4GI in a wide range of cell types and apoptotic triggers (6, 7, 22, 27). We also correlated the reduction in protein translation rate during Fas-induced apoptosis with the rapid degradation of eIF4GI and eIF4GII. However, we noticed that the protein translation machinery was not completely turned off and that translation continued at one-third of its normal rate. This raised the possibility that the residual translation activity resulted from an alternative translation mechanism which is eIF4GI/GII independent.
Is there a subgroup of death-associated proteins translated under apoptotic stress? Although we failed to detect a unique pattern of metabolically labeled proteins in apoptotic cells by comparing their profile by one-dimensional gel electrophoresis (not shown), a more sensitive comparison might be required to reveal subtle changes in the synthesis of specific proteins. However, by following the translation rate of DAP5 itself, we found this first candidate and further examined the possibility that its advantageous translation in apoptosis may occur via cap-independent mechanisms. Indeed, we found that DAP5's 5′UTR can drive cap-independent translation in reporter studies using bicistronic vectors. Furthermore, transfections with a bicistronic vector containing the DAP5 IRES showed that translation via this IRES was maintained, while cap-dependent translation was strongly reduced, in response to an apoptotic trigger. This observation implies that the DAP5 IRES drives translation under apoptotic stress. Recently, another cellular IRES, the X-linked inhibitor of apoptosis (XIAP) IRES, was also shown to enable continuous translation during apoptosis induced by serum deprivation (14). Identification of additional IRES-containing cellular messages, preferentially translated during cell death, might open a new horizon for identification of major apoptotic genes. It should be mentioned that cap-independent translation has already been associated with stressful cellular situations in which overall protein synthesis (mostly cap-dependent in nature) is significantly inhibited. These situations include heat shock (11) and hypoxia (19, 34).
What is the biochemical activity of DAP5 (in particular DAP5/p86), and how does it contribute to cell death? DAP5 is one of several novel members of the expanding eIF4G protein family which includes, besides eIF4GI and eIF4GII (12), the recently identified poly(A) binding protein-interacting protein (PAIP) (8), all of which share homology in the middle eIF4A and eIF3 binding segment (Fig. 8). According to one working model, DAP5 might function as a translation inhibitor, titrating out initiation factors eIF4A and eIF3. The titrator model is based on the finding that DAP5/p97 overexpression in transient transfection assays led to a twofold reduction in both cap-dependent and cap-independent translation (15, 40). Recently it was found that DAP5, like eIF4GI, interacts with Mnk1 and thus may interfere with the phosphorylation of eIF4E as a second possible titrating mechanism (32, 39). The death-promoting activity of DAP5, according to the titrator model, might be to disrupt the maintenance of cap-dependent synthesis of prosurvival proteins. It should be noted that in transfection-based functional assays, nonphysiological effects due to large excess of the ectopically expressed DAP5 protein may be observed. Moreover, the finding that during apoptosis translation is shut down rapidly and efficiently by cleavage of eIF4GI/GII (6, 7, 22, 27) reduces the need for DAP5 to function as a translation titrator under these circumstances.
An alternative working model proposes that DAP5 itself might be an active functional member of the eIF4G family, capable of supporting cap-independent translation, especially under conditions where cap-dependent translation is unfavorable. According to this model, DAP5 may drive the cell down the apoptotic pathway by selectively translating death genes via “death IRESes.” In this work, we provide new support for this alternative model. The finding that DAP5/p86 becomes the predominant eIF4G form in the dying cells, capable of binding eIF3 and eIF4A, made it an attractive candidate for supporting residual translation, including its own, in the dying cells. Indeed, we found that DAP5 can function as an active translation factor in cell-free systems. We show here that translation from the IRES element found in the DAP5 5′UTR was preferentially stimulated over cap-dependent translation by adding excess of recombinant DAP5 proteins into the RRL. Conversely, it was preferentially suppressed by adding a dominant negative fragment of DAP5. Taking these observations together, we suggest that DAP5 may mediate its own translation under conditions that are unfavorable for cap-dependent translation and thus be responsible for its sustained translation during apoptosis. The next step will be to determine whether it mediates the translation of other proapoptotic genes as well.
Last, we have identified DAP5 as a caspase substrate. Despite the fact that the list of caspase cellular substrates that are cleaved during the execution of the cell death program is expanding rapidly, it is important to keep in mind that caspases are highly selective proteases (28, 31, 36). Cell death is characterized by highly regulated and selective cleavage of discrete, specific proteins rather than by a bulk, nonspecific proteolysis of cellular proteins. We found that the caspase cleavage of DAP5 potentiates its ability to support DAP5 IRES translation in vitro. Further studies will be required to assess more accurately the mechanism by which the C-terminal truncation changes DAP5's properties.
ACKNOWLEDGMENTS
The first two authors contributed equally to this work.
We thank Nahum Sonenberg for kindly providing the anti-eIF4A, eIF3, and eIF4GII antibodies. We thank Eli Keshet, Yoram Groner, and Orna Stein for the different DNA constructs. We thank David Wallach for kindly providing the HFB cell line.
This work was supported by the Israel Foundation, which is administered by the Israel Academy of Sciences and Humanities, and by QBI Enterprises. A.K. is the incumbent of Helena Rubinstein Chair of Cancer Research.
REFERENCES
- 1.Akiri G, Nahari D, Finkelstein Y, Le S Y, Elroy-Stein O, Levi B Z. Regulation of vascular endothelial growth factor (VEGF) expression is mediated by internal initiation of translation and alternative initiation of transcription. Oncogene. 1998;17:227–236. doi: 10.1038/sj.onc.1202019. [DOI] [PubMed] [Google Scholar]
- 2.Ashkenazi A, Dixit V M. Death receptors: signaling and modulation. Science. 1998;281:1305–1308. doi: 10.1126/science.281.5381.1305. [DOI] [PubMed] [Google Scholar]
- 3.Bernstein J, Sella O, Le S Y, Elroy-Stein O. PDGF2/c-sis mRNA leader contains a differentiation-linked internal ribosomal entry site (D-IRES) J Biol Chem. 1997;272:9356–9362. doi: 10.1074/jbc.272.14.9356. [DOI] [PubMed] [Google Scholar]
- 4.Boldin M P, Varfolomeev E E, Pancer Z, Mett I L, Camonis J H, Wallach D. A novel protein that interacts with the death domain of Fas/APO1 contains a sequence motif related to the death domain. J Biol Chem. 1995;270:7795–7798. doi: 10.1074/jbc.270.14.7795. [DOI] [PubMed] [Google Scholar]
- 5.Borman A M, Kirchweger R, Ziegler E, Rhoads R E, Skern T, Kean K M. eIF4G and its proteolytic cleavage products: effect on initiation of protein synthesis from capped, uncapped, and IRES-containing mRNAs. RNA. 1997;3:186–196. [PMC free article] [PubMed] [Google Scholar]
- 6.Bushell M, McKendrick L, Janicke R U, Clemens M J, Morley S J. Caspase-3 is necessary and sufficient for cleavage of protein synthesis eukaryotic initiation factor 4G during apoptosis. FEBS Lett. 1999;451:332–336. doi: 10.1016/s0014-5793(99)00614-6. [DOI] [PubMed] [Google Scholar]
- 7.Clemens M J, Bushell M, Morley S J. Degradation of eukaryotic polypeptide chain initiation factor (eIF) 4G in response to induction of apoptosis in human lymphoma cell lines. Oncogene. 1998;17:2921–2931. doi: 10.1038/sj.onc.1202227. [DOI] [PubMed] [Google Scholar]
- 8.Craig A W, Haghighat A, Yu A T, Sonenberg N. Interaction of polyadenylate-binding protein with the eIF4G homologue PAIP enhances translation. Nature. 1998;392:520–523. doi: 10.1038/33198. [DOI] [PubMed] [Google Scholar]
- 9.Deckwerth T L, Johnson E M., Jr Temporal analysis of events associated with programmed cell death (apoptosis) of sympathetic neurons deprived of nerve growth factor. J Cell Biol. 1993;123:1207–1222. doi: 10.1083/jcb.123.5.1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deiss L P, Kimchi A. A genetic tool used to identify thioredoxin as a mediator of a growth inhibitory signal. Science. 1991;252:117–120. doi: 10.1126/science.1901424. [DOI] [PubMed] [Google Scholar]
- 11.Duncan R F. Translational control during heat shock. In: Hershey J W B, Mathews M B, Sonenberg N, editors. Translational control. Cold Spring Harbor, N.Y: CSHL Press; 1996. pp. 271–293. [Google Scholar]
- 12.Gradi A, Imataka H, Svitkin Y V, Rom E, Raught B, Morino S, Sonenberg N. A novel functional human eukaryotic translation initiation factor 4G. Mol Cell Biol. 1998;18:334–342. doi: 10.1128/mcb.18.1.334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gradi A, Svitkin Y V, Imataka H, Sonenberg N. Proteolysis of human eukaryotic translation initiation factor eIF4GII, but not eIF4GI, coincides with the shutoff of host protein synthesis after poliovirus infection. Proc Natl Acad Sci USA. 1998;95:11089–11094. doi: 10.1073/pnas.95.19.11089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Holcik M, Lefebvre C, Yeh C, Chow T, Korneluk R G. A new internal-ribosome-entry-site motif potentiates XIAP-mediated cytoprotection. Nat Cell Biol. 1999;1:190–192. doi: 10.1038/11109. [DOI] [PubMed] [Google Scholar]
- 15.Imataka H, Olsen H S, Sonenberg N. A new translational regulator with homology to eukaryotic translation initiation factor 4G. EMBO J. 1997;16:817–825. doi: 10.1093/emboj/16.4.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Imataka H, Sonenberg N. Human eukaryotic translation initiation factor 4G (eIF4G) possesses two separate and independent binding sites for eIF4A. Mol Cell Biol. 1997;17:6940–6947. doi: 10.1128/mcb.17.12.6940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kimchi A. DAP genes: novel apoptotic genes isolated by a functional approach to gene cloning. Biochim Biophys Acta. 1998;1377:F13–F33. doi: 10.1016/s0304-419x(98)00002-x. [DOI] [PubMed] [Google Scholar]
- 18.Kissil J L, Cohen O, Raveh T, Kimchi A. Structure-function analysis of an evolutionary conserved protein, DAP3, which mediates TNF-alpha- and Fas-induced cell death. EMBO J. 1999;18:353–362. doi: 10.1093/emboj/18.2.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kraggerud S M, Sandvik J A, Pettersen E O. Regulation of protein synthesis in human cells exposed to extreme hypoxia. Anticancer Res. 1995;15:683–686. [PubMed] [Google Scholar]
- 20.Lamphear B J, Kirchweger R, Skern T, Rhoads R E. Mapping of functional domains in eukaryotic protein synthesis initiation factor 4G (eIF4G) with picornaviral proteases. Implications for cap-dependent and cap-independent translational initiation. J Biol Chem. 1995;270:21975–21983. doi: 10.1074/jbc.270.37.21975. [DOI] [PubMed] [Google Scholar]
- 21.Levy-Strumpf N, Deiss L P, Berissi H, Kimchi A. DAP-5, a novel homolog of eukaryotic translation initiation factor 4G isolated as a putative modulator of gamma interferon-induced programmed cell death. Mol Cell Biol. 1997;17:1615–1625. doi: 10.1128/mcb.17.3.1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marissen W E, Lloyd R E. Eukaryotic translation initiation factor 4G is targeted for proteolytic cleavage by caspase 3 during inhibition of translation in apoptotic cells. Mol Cell Biol. 1998;18:7565–7574. doi: 10.1128/mcb.18.12.7565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martin D P, Schmidt R E, DiStefano P S, Lowry O H, Carter J G, Johnson E M., Jr Inhibitors of protein synthesis and RNA synthesis prevent neuronal death caused by nerve growth factor deprivation. J Cell Biol. 1988;106:829–844. doi: 10.1083/jcb.106.3.829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McCall K, Steller H. Facing death in the fly: genetic analysis of apoptosis in Drosophila. Trends Genet. 1997;13:222–226. doi: 10.1016/S0168-9525(97)01126-8. [DOI] [PubMed] [Google Scholar]
- 25.Methot N, Rom E, Olsen H, Sonenberg N. The human homologue of the yeast Prt1 protein is an integral part of the eukaryotic initiation factor 3 complex and interacts with p170. J Biol Chem. 1997;272:1110–1116. doi: 10.1074/jbc.272.2.1110. [DOI] [PubMed] [Google Scholar]
- 26.Morley S J, Curtis P S, Pain V M. eIF4G: translation's mystery factor begins to yield its secrets. RNA. 1997;3:1085–1104. [PMC free article] [PubMed] [Google Scholar]
- 27.Morley S J, McKendrick L, Bushell M. Cleavage of translation initiation factor 4G (eIF4G) during anti-Fas IgM-induced apoptosis does not require signalling through the p38 mitogen-activated protein (MAP) kinase. FEBS Lett. 1998;438:41–48. doi: 10.1016/s0014-5793(98)01269-1. [DOI] [PubMed] [Google Scholar]
- 28.Nicholson D W, Thornberry N A. Caspases: killer proteases. Trends Biochem Sci. 1997;22:299–306. doi: 10.1016/s0968-0004(97)01085-2. [DOI] [PubMed] [Google Scholar]
- 29.Ohlmann T, Rau M, Pain V M, Morley S J. The C-terminal domain of eukaryotic protein synthesis initiation factor (eIF) 4G is sufficient to support cap-independent translation in the absence of eIF4E. EMBO J. 1996;15:1371–1382. [PMC free article] [PubMed] [Google Scholar]
- 30.Pestova T V, Shatsky I N, Hellen C U. Functional dissection of eukaryotic initiation factor 4F: the 4A subunit and the central domain of the 4G subunit are sufficient to mediate internal entry of 43S preinitiation complexes. Mol Cell Biol. 1996;16:6870–6878. doi: 10.1128/mcb.16.12.6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Porter A G, Ng P, Janicke R U. Death substrates come alive. Bioessays. 1997;19:501–507. doi: 10.1002/bies.950190609. [DOI] [PubMed] [Google Scholar]
- 32.Pyronnet S, Imataka H, Gingras A C, Fukunaga R, Hunter T, Sonenberg N. Human eukaryotic translation initiation factor 4G (eIF4G) recruits mnk1 to phosphorylate eIF4E. EMBO J. 1999;18:270–279. doi: 10.1093/emboj/18.1.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shaughnessy J D, Jr, Jenkins N A, Copeland N G. cDNA cloning, expression analysis, and chromosomal localization of a gene with high homology to wheat eIF-(iso)4F and mammalian eIF-4G. Genomics. 1997;39:192–197. doi: 10.1006/geno.1996.4502. [DOI] [PubMed] [Google Scholar]
- 34.Stein I, Itin A, Einat P, Skaliter R, Grossman Z, Keshet E. Translation of vascular endothelial growth factor mRNA by internal ribosome entry: implications for translation under hypoxia. Mol Cell Biol. 1998;18:3112–3119. doi: 10.1128/mcb.18.6.3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Svitkin Y V, Gradi A, Imataka H, Morino S, Sonenberg N. Eukaryotic initiation factor 4GII (eIF4GII), but not eIF4GI, cleavage correlates with inhibition of host cell protein synthesis after human rhinovirus infection. J Virol. 1999;73:3467–3472. doi: 10.1128/jvi.73.4.3467-3472.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thornberry N A, Lazebnik Y. Caspases: enemies within. Science. 1998;281:1312–1316. doi: 10.1126/science.281.5381.1312. [DOI] [PubMed] [Google Scholar]
- 37.Vagner S, Gensac M C, Maret A, Bayard F, Amalric F, Prats H, Prats A. Alternative translation of human fibroblast growth factor 2 mRNA occurs by internal entry of ribosomes. Mol Cell Biol. 1995;15:35–44. doi: 10.1128/mcb.15.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang C Y, Mayo M W, Korneluk R G, Goeddel D V, Baldwin A S., Jr NF-kappaB antiapoptosis: induction of TRAF1 and TRAF2 and c-IAP1 and c-IAP2 to suppress caspase-8 activation. Science. 1998;281:1680–1683. doi: 10.1126/science.281.5383.1680. [DOI] [PubMed] [Google Scholar]
- 39.Waskiewicz A J, Johnson J C, Penn B, Mahalingam M, Kimball S R, Cooper J A. Phosphorylation of the cap-binding protein eukaryotic translation initiation factor 4E by protein kinase Mnk1 in vivo. Mol Cell Biol. 1999;19:1871–1880. doi: 10.1128/mcb.19.3.1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yamanaka S, Poksay K S, Arnold K S, Innerarity T L. A novel translational repressor mRNA is edited extensively in livers containing tumors caused by the transgene expression of the apoB mRNA-editing enzyme. Genes Dev. 1997;11:321–333. doi: 10.1101/gad.11.3.321. [DOI] [PubMed] [Google Scholar]
- 41.Yonish-Rouach E, Resnitzky D, Lotem J, Sachs L, Kimchi A, Oren M. Wild-type p53 induces apoptosis of myeloid leukaemic cells that is inhibited by interleukin-6. Nature. 1991;352:345–347. doi: 10.1038/352345a0. [DOI] [PubMed] [Google Scholar]