Abstract
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) can be detected via a nasopharyngeal swab and in sputum, blood, urine, and feces. However, there is only limited data on the real-time reverse transcriptase polymerase chain reaction (RT-PCR) results of coronavirus disease 2019 (COVID-19) patients with pleural fluid. We report a case of COVID-19 with SARS-CoV-2 detected in both sputum and pleural fluid. A 68-year-old male patient came to the hospital with a chief complaint of dyspnea. He was diagnosed with lung cancer. A biopsy was performed, and a pneumothorax was found. As a result, a chest tube was placed into the right pleural space. During his hospital stay, the patient was confirmed as COVID-19 positive. We identified the presence of SARS-CoV-2 through real-time RT-PCR assay from the pleural fluid. Although pleural effusion is an uncommon finding in the COVID-19, care should be taken to avoid exposure when handling the pleural fluid sample.
Keywords: Coronavirus, Pleural effusion, Polymerase chain reaction
Introduction
Although real-time reverse transcriptase polymerase chain reaction (RT-PCR) of nasopharyngeal swabs is currently used for the diagnosis of coronavirus disease 2019 (COVID-19) [1], positive samples have also been reported from sputum, feces, urine, and blood [2]. There is only limited data on the RT-PCR results of COVID-19 patients with pleural fluid. We report a case of COVID-19 with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) detected in both sputum and pleural fluid.
The Institutional Review Board of Seongnam Citizens Medical Center approved the study (approval no. 1-2020-0001-2-001) and waived the requirement for informed consent due to the retrospective nature of the analyses.
Case report
A 68-year-old male patient came to the hospital with a chief complaint of dyspnea. He was a current smoker with a 45-pack-year smoking history. Chest radiograph showed a bulky air-space consolidation in the right upper lung field and a pleural effusion in the right lower lung field. A percutaneous core needle biopsy was performed, and as a result the patient was diagnosed with a malignant tumor, which was suspected to be sarcomatoid carcinoma. The pneumothorax was found after the biopsy was performed, and a chest tube was placed into the right pleural space. The next day, the first case of COVID-19 was diagnosed in the room where the patient stayed. The result for SARS-CoV-2 in this patient was negative. However, on the seventh hospital day, the patient complained of a fever and was confirmed to be COVID-19 positive via RT-PCR using a nasopharyngeal/oropharyngeal swab.
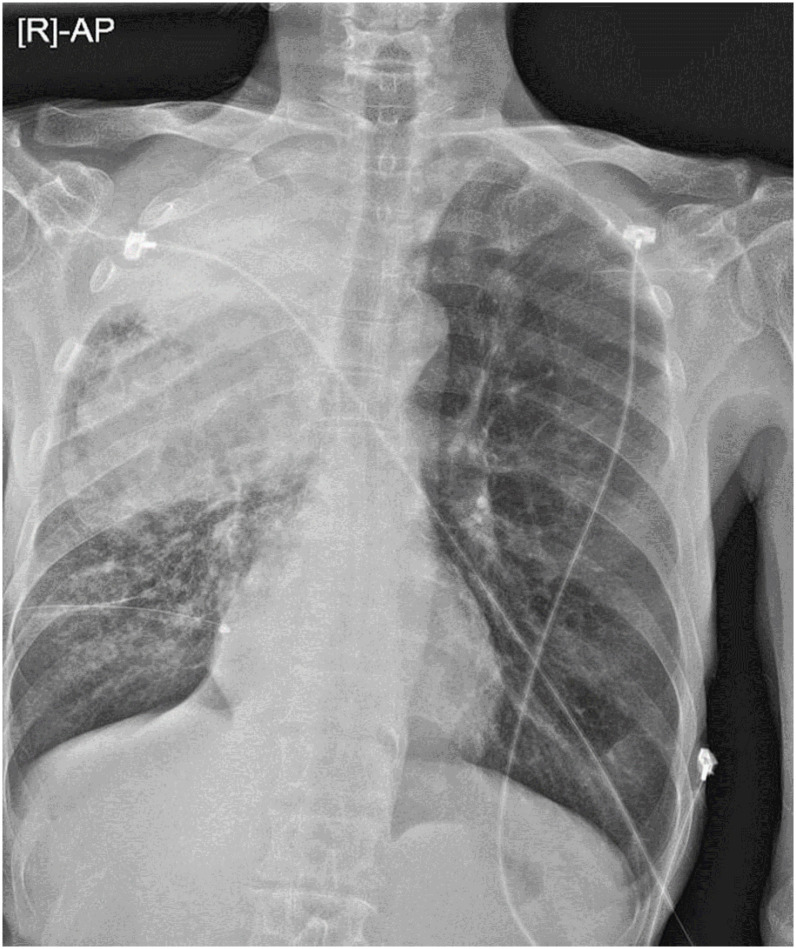
The patient was transferred to Seongnam Citizens Medical Center and was isolated in a negative-pressure room. Upon physical examination, his blood pressure was 113/72 mm Hg, and body temperature was 36.3°C. Oxygen saturation of 100% was measured on 4 L/min via nasal cannula. Laboratory tests showed a normal leukocyte count (5.94 × 103 cells/mm3 with 77.4% neutrophils) and renal function. However, he had an elevated level of C-reactive protein (9.82 mg/dL; normal value, <0.5 mg/dL) and decreased levels of hemoglobin (8.7 g/dL; normal value, 13.0 – 17.0 g/dL) and sodium (126 mmol/L; normal value, 136 – 145 mmol/L). Chest radiograph on admission is shown in Figure 1.
Figure 1. Chest radiograph shows bulky air-space consolidation in the upper lung field and subtle increased interstitial markings in the right basal lung area.
Real-time RT-PCRs for nasopharyngeal swab and sputum were performed at least 48 hours apart. For the treatment of COVID-19 [3], lopinavir 400 mg/ritonavir 100 mg was given twice a day for 8 days. On the third hospital day, cytological examination of the pleural fluid was done due to continuous drainage of approximately 300 mL per day. To evaluate the lung cancer, pleural fluid cell block was performed. Before the procedure, the cytotechnologist put on personal protective equipment; however, the hands were exposed to contaminants when the gloves were removed. For SARS-CoV-2 detection in the pleural fluid, real-time RT-PCR assay and pleural fluid analysis were performed. The pleural fluids have predominantly mononuclear cell exudates (white blood cells of 350/mm3, red blood cells of 117 900/mm3, mononuclear cells of 72%, polymorphonuclear cells of 28%, protein 2.2 g/dL, and lactate dehydrogenase 2017 IU/L). The level of pleural fluid adenosine deaminase was 18.3 IU/L, and there were no malignant cells or bacteria present. We confirmed that RT-PCR results for SARS-CoV-2 were positive in the pleural fluid sample. The cytotechnologist was not infected with COVID-19. On the eighth hospital day, RT-PCR of the nasopharyngeal swab showed negative results, but negative-to-positive RT-PCR conversion was observed (Table 1). On the 10th hospital day, a chest radiograph showed an ill-defined increased opacity in right lower lung zone. An empiric antibiotic with piperacillin/tazobactam (4.5 g every 6 hours for 14 days) was administered intravenously. From the 20th hospital day, nasopharyngeal swab and sputum samples tested negative via RT-PCR. On the 27th hospital day, we identified a negative RT-PCR result for SARS-CoV-2 in the pleural fluid sample. The patient has recovered from COVID-19 pneumonia and is receiving oxygen via nasal cannula.
Table 1. Serial changes in viral load observed for a patient with COVID-19.
| HD 3 | HD 6 | HD 8 | HD 10 | HD 13 | HD 15 | HD 17 | HD 20 | HD 21 | HD 27 | |
|---|---|---|---|---|---|---|---|---|---|---|
| Pharyngeal swab | + | + | − | − | + | − | + | − | − | |
| RdRP assay (Ct cutoff of 40) | 36.98 | 28.91 | UD | UD | 32.85 | UD | 32 | UD | UD | |
| Sputum | + | + | + | + | + | + | + | – | ||
| RdRP assay (Ct cutoff of 35) | 27.23 | 25.42 | 26.44 | 22.36 | 31.2 | 28.66 | 31.79 | UD | ||
| Pleural fluid | + | − | ||||||||
| RdRP assay (Ct cutoff of 35) | 34.51 | UD |
COVID-19 was confirmed by RT-PCR: pharyngeal swab for Allplex 2019-nCoV Assay (Seegene, Inc, Seoul, Korea) and sputum and pleural fluid samples for PowerChek SARS-CoV-2 Real-time PCR Kit (KogeneBiotech, Seoul, Korea), respectively.
COVID-19, coronavirus disease 2019; HD, hospital day; RdRP, RNA-dependent RNA polymerase; Ct, cycle threshold; +, positive; −, negative; UD, undetected; RT-PCR polymerase chain reaction; SARS-CoV-2, syndrome coronavirus 2.
Discussion
In this patient with COVID-19, we identified the presence of SARS-CoV-2 by real-time RT-PCR assay from the pleural fluid. This is the first COVID-19 case of a negative conversion in a pleural fluid PCR test. Pleural effusion in patients with COVID-19 is an infrequent finding and may be observed with disease progression [4]. Recently, Lescure et al. reported that SARS-CoV-2 was detected in the pleural fluid of a COVID-19 patient with a high viral load found on nasopharyngeal swab [5]. They insisted that inhibition of the interferon signaling pathways by SARS-CoV-2 might result in a higher viral load and poor prognosis. In our patient, a positive RT-PCR result in the pleural fluid was observed in the early stage of the disease with a higher viral load. After negative conversion of RT-PCR from sputum and pharyngeal swab samples, a pleural fluid PCR test also showed negative results. Although pleural effusion is an uncommon finding in COVID-19, care should be taken to avoid exposure when handling the pleural fluid sample.
There are some limitations to this study. Serial pleural fluid samples were not obtained on a defined schedule, and pharyngeal and sputum samples were tested using different PCR techniques: pharyngeal swab for Allplex 2019-nCoV Assay (Seegene, Inc, Seoul, Korea) and sputum for PowerChek SARS-CoV-2 Real-time PCR Kit (KogeneBiotech, Seoul, Korea), respectively. Nonetheless, our report can provide clinically relevant data on COVID-19. SARS-CoV-2 can be detected in pleural fluid as well as variety of specimens, such as blood and feces.
Footnotes
Conflict of Interest: No conflicts of interest.
- Conceptualization: CSN, MSB, WYK.
- Data curation: MSB, KJL.
- Writing - original draft: CSN, MSB.
References
- 1.Wang D, Hu B, Hu C, Zhu F, Liu X, Zhang J, Wang B, Xiang H, Cheng Z, Xiong Y, Zhao Y, Li Y, Wang X, Peng Z. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus–infected pneumonia in Wuhan, China. JAMA. 2020;323:1061–1069. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang W, Xu Y, Gao R, Lu R, Han K, Wu G, Tan W. Detection of SARS-CoV-2 in different types of clinical specimens. JAMA. 2020;323:1843–1844. doi: 10.1001/jama.2020.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim JY, Choe PG, Oh Y, Oh KJ, Kim J, Park SJ, Park JH, Na HK, Oh MD. The first case of 2019 novel coronavirus pneumonia imported into Korea from Wuhan, China: Implication for infection prevention and control measures. J Korean Med Sci. 2020;35:e61. doi: 10.3346/jkms.2020.35.e61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Salehi S, Abedi A, Balakrishnan S, Gholamrezanezhad A. Coronavirus disease 2019 (COVID-19): a systematic review of imaging findings in 919 patients. AJR Am J Roentgenol. 2020;215:87–93. doi: 10.2214/AJR.20.23034. [DOI] [PubMed] [Google Scholar]
- 5.Lescure FX, Bouadma L, Nguyen D, Parisey M, Wicky PH, Behillil S, Gaymard A, Bouscambert-Duchamp M, Donati F, Le Hingrat Q, Enouf V, Houhou-Fidouh N, Valette M, Mailles A, Lucet JC, Mentre F, Duval X, Descamps D, Malvy D, Timsit JF, Lina B, van-der-Werf S, Yazdanpanah Y. Clinical and virological data of the first cases of COVID-19 in Europe: a case series. Lancet Infect Dis. 2020;20:697–706. doi: 10.1016/S1473-3099(20)30200-0. [DOI] [PMC free article] [PubMed] [Google Scholar]