Abstract
A panel of 18 strains of Neisseria gonorrhoeae, known to be temporally and geographically diverse, was used to evaluate a number of typing systems, including conventional auxotyping and serotyping and the molecular methods of arbitrarily primed PCR (AP-PCR), amplified ribosomal-DNA restriction analysis (ARDRA), opa typing, and pulsed-field gel electrophoresis (PFGE). The discriminatory power of the different typing methods were determined with a collection of 87 clinical isolates from commercial sex workers in Indonesia, and Simpson’s index of diversity was calculated. Of the two traditional techniques, auxotyping and serotyping, the latter gives the highest discriminatory index (DI) (DI, 0.846). The combination of auxotyping and serotyping yields a high DI (DI, 0.928). D11344- and D8635-primed PCR showed low DIs of 0.608 and 0.622, respectively, but a combination of the two primers had a DI of 0.849. The combination of serotyping with D11344-primed or D8635-primed PCR resulted in DIs of 0.936 and 0.937, respectively. ARDRA revealed a low DI of 0.743 alone but a DI of 0.955 in combination with serotyping. PFGE using the restriction enzyme BglII and opa typing produced the highest discrimination (DIs, 0.997 and 0.996, respectively) for isolates of N. gonorrhoeae.
Gonorrhoea is a major sexually transmitted disease prevalent in both developed and developing countries. Precise characterization of Neisseria gonorrhoeae can provide valuable information on gonococcal strain populations in the community, temporal changes, and the emergence and spread of antibiotic-resistant strains. In the absence of a vaccine, better knowledge of the molecular epidemiology of gonococcal infection will be useful for the development of effective prevention and control measures.
Currently, the most widely employed method for differentiation of N. gonorrhoeae strains is based on auxotyping and serological (A/S) characterization. However, the A/S classification system has a number of limitations. Auxotyping is complicated, laborious, and time consuming, and serotyping requires well-characterized and specific polyclonal or monoclonal antigonococcal antibodies (25). Furthermore, there is also skepticism that this system may not provide sufficient discrimination; in most populations only a limited number of A/S classes account for the majority of isolates. It is also possible that unrelated isolates belong to the same A/S class, and isolates that are impossible to auxotype and serotype have been described (8). For strains that carry plasmids, differentiation of strains belonging to the same A/S groups could be achieved. However, plasmid profiling is of limited value when a common plasmid or a common combination of plasmids is present (4).
Isoenzyme typing based on multilocus enzyme electrophoresis is widely applicable for epidemiological studies of diverse groups of pathogens, including N. gonorrhoeae (16, 18, 23). Genetic relatedness of gonococcal isolates has also been assessed by using DNA-based typing techniques, including restriction endonuclease analysis using frequently or rarely cutting enzymes (7, 14, 20, 31, 32), and random and repetitive-motif-based amplification of polymorphic DNA fragments (2, 21). The discriminatory abilities of pulsed-field gel electrophoresis (PFGE) and random and repetitive-motif-based amplification of polymorphic DNA have been shown to be superior to that of traditional A/S typing (2, 14, 21, 32).
In addition, restriction fragment length polymorphism (RFLP) analysis of rRNA genes (ribotyping) has been used to subdivide gonococcal strains of similar auxotypes, but this resulted in only a limited number of ribotypes (13, 15). Furthermore, RFLP analyses of different PCR-amplified genes have been applied. O’Rourke et al. (16) developed a PCR-RFLP method using the opa gene as the target for amplification. The 11 opa genes are amplified with a single pair of primers and digested with frequently cutting restriction enzymes, and the radioactively labeled fragments are separated on polyacrylamide gels to index the strains to particular opa types. This so-called opa typing appears to be highly discriminatory as it was able to establish the close identity of isolates collected from sexual contacts and differentiated isolates from a worldwide collection made over the last 30 years.
Similarly, amplification and RFLP analysis of the por gene have shown a degree of discrimination similar to that of A/S typing (12). Finally, sequencing of the por gene can be highly discriminatory. Cooke et al. (3) demonstrated that the inferred amino acid sequences of the protein I (PI) B molecules of isolates from known sexual contacts were identical, whereas those from unlinked isolates showed significant heterogeneity.
In general, the epidemiology of N. gonorrhoeae has been well delineated by using the A/S classification system. The development and particularly the evaluation of molecular typing techniques are necessary in view of some of the limitations presented by the A/S classification system. Most of the novel molecular typing methods have been developed with reference to serotyping only.
In the present study, a set of 18 temporally and geographically diverse N. gonorrhoeae reference strains was used to evaluate a number of typing methods, including conventional A/S typing and four molecular typing methods, i.e., arbitrarily primed PCR (AP-PCR), amplified ribosomal-DNA restriction analysis (ARDRA), opa typing, and PFGE. The discriminatory power of the different typing methods was determined by using a collection of 87 clinical isolates from Indonesia.
MATERIALS AND METHODS
Bacterial strains.
A set of 105 strains was studied. This set consisted of 18 reference strains selected from a large collection of over 5,000 isolates that differed in their geographic origins and years of isolation (Table 1) and 87 strains isolated in 1996 from female commercial sex workers in Bandung, Indonesia. The latter isolates are referred to hereafter as clinical isolates. All strains were grown on Columbia agar (GIBCO, Life Technologies, Paisley, Scotland) supplemented with 5% defibrinated horse blood at 37°C in 5% CO2 for 24 h.
TABLE 1.
Characteristics of our panel of 18 gonococcal strains
| Strain | Country of isolation | Yr of isolation | Auxotype/serotype | Categorya | RAPDb type
|
ARDRA type | |
|---|---|---|---|---|---|---|---|
| Primer D11344 | Primer D8635 | ||||||
| 1 | United Kingdom | 1993 | NR/IB-6 | — | b | A | III |
| 2 | United Kingdom | 1986 | NR/IB-1 | PPNG | b | B | II |
| 3 | Thailand | 1979 | NR/IA-6 | PPNG | b | C | I |
| 4 | United Kingdom | 1990 | AHU/IA-2 | — | c | A | II |
| 5 | United Kingdom | 1989 | NR/IB-2 | — | b | A | II |
| 6 | United Kingdom | 1989 | PAOU/IB-2 | — | a | A | I |
| 7 | United Kingdom | 1989 | H/IB-5 | PPNG | a | C | IV |
| 8 | United Kingdom | 1989 | NR/IB-3 | — | c | D | I |
| 9 | United Kingdom | 1993 | A/IB-3 | — | b | A | I |
| 10 | United Kingdom | 1993 | P/IB-1 | — | a | C | I |
| 11 | United Kingdom | 1993 | NR/IB-4 | — | a | B | III |
| 12 | United Kingdom | 1993 | P/IB-7 | — | a | C | II |
| 13 | South Africa | 1991 | NR/IB-13 | — | a | A | I |
| 14 | Bahrain | 1988 | NR/IB-19 | — | b | A | II |
| 15 | Tanzania | 1992 | NR/IB-22 | TRNG | a | A | II |
| 16 | South Africa | 1990 | NR/IA-8 | — | b | B | V |
| 17 | South Africa | 1991 | NR/IA-5 | — | a | C | II |
| 18 | Gambia | 1989 | NR/IA-1 | — | b | B | IV |
PPNG, penicillinase producing N. gonorrhoeae; TRNG, tetracycline resistant N. gonorrhoeae; —, neither PPNG or TRNG.
RAPD, randomly amplified polymorphic DNA.
Auxotyping and serotyping.
Auxotyping and determination of the serovar were carried out as described previously (29). Strains were tested for single or multiple requirements for arginine (A), hypoxanthine (H), ornithine (O), proline (P), and uracil (U).
DNA isolation.
Genomic DNA was extracted by the rapid procedure described by Pitcher et al. (17).
PCR-based typing.
Ten arbitrary primers, comprising seven short (10-nucleotide) primers (primers 1247, 1254, 1281, 1283, and 1290 [1] and OPA-O3 and OPA-13 [2]), five long (≥17-nucleotide) primers (primers D8635, D9355, D11344, D14216, and D14307 [1]), and four primers for amplification of repeat motifs (the enterobacterial repetitive intergenic consensus [ERIC] motifs 1R and 2 [28] and the repetitive extragenic palindromic sequences REPIR-Dt and REP-2-Dt [21]), were evaluated for their suitability to differentiate gonococcal isolates. DNA amplification was performed in a DNA thermal cycler (GeneAmp PCR System 9600; Perkin Elmer, Zaventem, Belgium). The 100-μl PCR mixtures consisted of 50 mM KCl, 10 mM Tris-HCl (pH 9.0), 2.5 mM MgCl2, 0.1% Triton X-100, and 0.01% gelatin. Deoxyribonucleotide triphosphates were each used at a final concentration of 0.2 mM. Per reaction mixture, 1.8 U of High Concentration SUPER TAQ (HT Biotechnology Ltd., Cambridge, United Kingdom), 100 pmol of primer (except for ERIC-1R and ERIC-2 [50 pmol]), and 100 ng of extracted DNA were added. PCR conditions were as described previously (2, 21, 27, 30). After amplification, 25-μl aliquots of PCR products were electrophoresed (100 V, 3 h) in 1.5% Pronarose D1 gels (Sphaero Q, Burgos, Spain) and 0.5× TBE (45 mM Tris-HCl, 45 mM boric acid, 1 mM EDTA) running buffer containing 0.5 mg of ethidium bromide/ml. The patterns were visualized under UV light and digitized by the Gel Doc 1000 documentation system (Bio-Rad Laboratories, Nazareth, Belgium). Conversion, normalization, and densitometric analysis of the patterns were done by the GelCompar Software program version 4.0 (Applied Maths, Kortrijk, Belgium). The similarity between all pairs of traces was expressed by the Pearson product-moment correlation coefficient, and clustering was performed by the unweighted pair group method using average linkage (UPGMA) (24). Obtained clusters were always visually confirmed.
ARDRA.
Oligonucleotide primers were derived from conserved regions present in the 16S and 23S rRNA genes (rDNAs). The sequences of the primers were 5′-TTGTACACACCGCCCGTC-3′ and 5′-CCTTTCCCTCACGGTACTG-3′ (Escherichia coli numbering positions 1390 to 1407 [16S rDNA] and 474 to 456 [23S rDNA]) (9). The amplified gene fragment was approximately 1,200 bp and encompassed part of the 16S rDNA gene, the 16S-23S spacer region, and part of the 23S rDNA gene. The composition of the PCR mixtures was the same as described above for PCR-based typing. PCR conditions consisted of an initial denaturation at 95°C for 5 min, followed by 20 cycles of denaturation at 95°C for 30 s, annealing at 55°C for 30 s, extension at 72°C for 1 min, and a final extension at 72°C for 10 min.
PCR products were detected by gel electrophoresis for 1 h at 100 V in 1% Pronarose D1 gels and 0.5× TBE running buffer containing ethidium bromide. Restriction analysis was carried out for 2 h at 37°C in 20-μl volumes of incubation buffer containing 5 U of restriction enzyme (AluI, BamHI, BfaI, BglII, BstNI, DdeI, DraI, EcoRI, EcoRV, HaeIII, HhaI, HindIII, HinfI, HpaI, HpaII, KasI, MaeI, MluI, MnlI, MseI, MspI, NdeI, NheI, NotI, NsiI, PstI, SacI, SalI, Sau3AI, SfiI, SmaI, SpeI, TaqI, XbaI, or XhoI) and 5 to 10 μl of PCR product. Restriction fragment patterns were analyzed by gel electrophoresis of the restriction mixture at 150 V for 1.5 h in 2% agarose gels. Gels were visualized, digitized, and analyzed as described above for PCR. The similarity of ARDRA banding patterns was estimated by using the Dice coefficient (5), and clustering was performed by using UPGMA (24). Strains were considered identical when they showed the same banding pattern.
opa typing.
opa typing was performed by the method of O’Rourke et al. (16). Briefly, isolates were retrieved from storage and subcultured once on nonselective GC agar. Bacterial lysates were prepared by suspending the growth in phosphate-buffered saline, washing once, and then boiling for 5 min before centrifugation. The supernatant (lysate) was stored at −20°C until required. The opa genes were amplified by PCR using the lysate as the DNA source, purified with Nucleiclean (Sigma), and resuspended in water. opa fragments from all isolates were digested with the restriction enzyme TaqI, by the method recommended by the supplier. Isolates that gave identical patterns when digested with TaqI were also digested by using HinPI and HpaII. The ends of the resulting fragments were filled in with [α-32P]dCTP and then separated on a 6% nondenaturing polyacrylamide gel as described previously (16).
The images on the X-ray films were digitized by using a Hewlett-Packard Scanjet IIcx high-resolution scanner. Conversion, normalization, and densitometric analysis of the patterns were done by using GelCompar software. The similarity between all pairs of traces was expressed by the Pearson product-moment correlation coefficient, and clustering was performed by using UPGMA (24). Obtained clusters were always confirmed visually. Gonococci that gave identical fragments with all three enzymes were considered to have the same opa type.
PFGE.
Chromosomal DNA was prepared by the procedure described by Van Looveren et al. (27), except that no lysis step was performed. Four restriction enzymes used in other studies for the typing of gonococci, BglII, NheI, SpeI, and XbaI (13, 14, 19, 22, 32), were tested under various running conditions. All produced comparable results. The endonuclease BglII was chosen for economic reasons. Digestion with BglII (Eurogentec, Seraing, Belgium) was performed at 37°C for 25 h in 250 μl of fresh buffer containing 30 U of restriction endonuclease. The digested plugs were sealed into slots of a 1% agarose gel (Pulsed-Field Certified Agarose; Bio-Rad Laboratories) and subjected to electrophoresis in a contour-clamped homogeneous electric field apparatus with a hexagonal electrode array (CHEF MAPPER; Bio-Rad Laboratories). The electrophoresis was performed in 0.5× TBE buffer equilibrated at 14°C, at a constant voltage of 6 V/cm with pulse times ramping from 1 to 10 s for 18 h and then from 10 to 15 s for 4 h. Gels were stained with ethidium bromide, visualized, digitized, and analyzed as described above for ARDRA. The obtained clusters were always confirmed visually. BglII produced 15 to 20 distinct DNA fragments, ranging from 44 to 674 kb, for all strains. Strains were considered identical if no fragment differences occurred.
Discriminatory power.
The abilities of single or combined typing schemes to discriminate between strains was determined by using Simpson’s index of diversity (6, 11).
RESULTS
Evaluation of the genotypic typing methods with the 18 reference strains.
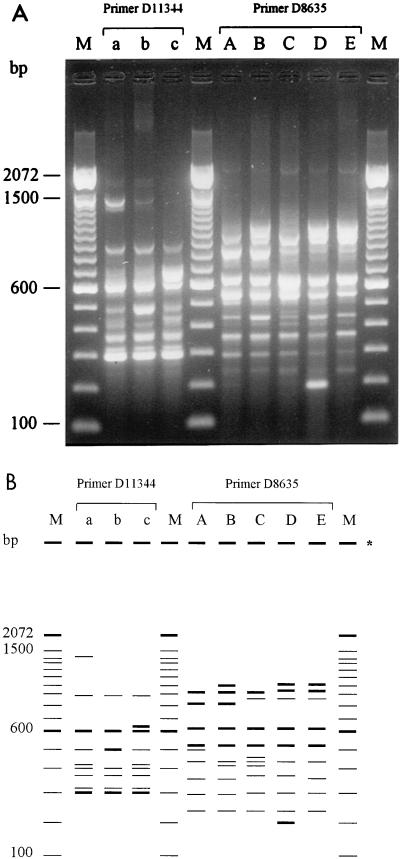
Primers for the PCR-based typing were tested with the panel of 18 reference strains (Table 1) for the production of informative arrays of PCR products. Primers shown to be useful in other typing studies were assessed (1, 2, 21, 28, 30). Primers 1247, 1283, and 1290 (1), ERIC-1R (28), and REP-2-Dt (21) were unable to generate a banding pattern; primers 1254, 1281, D14216, and D14307 (1) resulted in a few amplified bands for each of the 18 strains tested; primers OPA-3 and OPA-13 (2), D9355 (1), and ERIC-2 (28) generated up to 8 bands, most of which were shared by all strains; and primer Repir-Dt (21) generated up to 17 not-well-separated faint bands. In contrast, primers D11344 and D8635 (1) both generated informative arrays of PCR products, with up to 8 clearly distinguishable bands (Fig. 1), and were used in all further studies. In addition, this panel was used to evaluate the ARDRA technique. Thirteen of 35 restriction enzymes (AluI, BfaI, DdeI, HhaI, HindIII, HinfI, HpaII, MaeI, MseI, MspI, Sau3AI, TaqI, and XbaI) tested cut the gonococcal DNA, but only HinfI (5′-GANTC-3′) revealed differences between the strains. As a consequence, HinfI was selected to investigate the 87 clinical isolates. PFGE and opa typing produced distinct patterns with each of the 18 strains.
FIG. 1.
(A) PCR analysis with primers D11344 (lanes a to c) and D8635 (lanes A to E) of strains representative of the various fingerprint groups. Lane M, 100-bp DNA ladder. (B) Schematic representation of the gel shown in panel A. ∗, place where samples were loaded.
Phenotypic methods and validation of the genotypic methods. (i) Auxotyping and serotyping.
The 87 clinical isolates belonged to four different auxotypes (nonrequiring [NR] and proline [P], arginine [A], and proline and arginine [PA] requiring), with the majority of the isolates being nonrequiring (36 isolates) or proline requiring (42 isolates). Serological characterization revealed 12 serovars (IA-4, IA-6, IA-8, IB-1, IB-3, IB-5, IB-6, IB-7, IB-8, IB-10, IB-16, and IB-18), of which serovars IA-8 (22 strains), IB-1 (17 strains), IA-6 (15 strains), and IA-4 (12 strains) were the most abundant. The combination of auxotyping and serotyping resulted in 24 different A/S classes, 13 of which contained one or two strains. P/IB-1 (14 strains) and NR/IA-8 (13 strains) were the most common.
(ii) PCR-based typing.
All strains were examined in one PCR to avoid interrun variability. Based on the differences in DNA banding patterns, strains could be divided into various fingerprint groups. Strains were classified in a different fingerprint group as soon as differences in band position and/or band intensity were observed. Primer D11344 divided the 87 strains into three fingerprint groups (groups a to c), represented by 32, 43, and 12 strains, respectively (Fig. 1). Primer D8635 distinguished four groups (A, B, C, and E), with groups A and C comprising 79% of the strains (48 and 21 isolates, respectively) (Fig. 1) (group D was found among the 18 reference strains). Combination of the data obtained with the two primers produced 11 fingerprint groups each containing 1 to 24 isolates.
(iii) ARDRA.
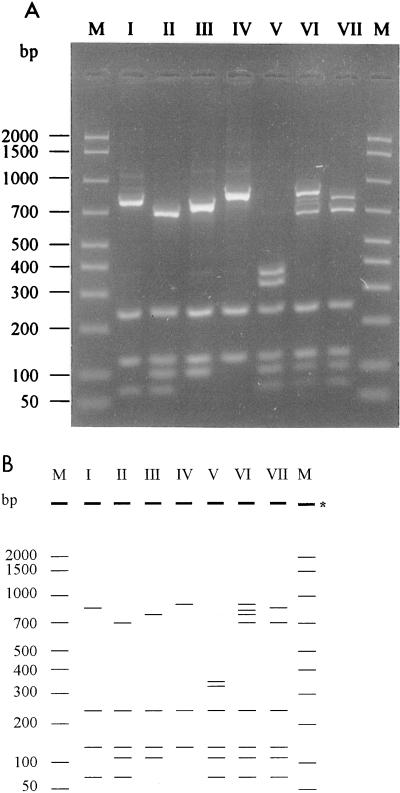
Among the 87 clinical isolates, HinfI distinguished six restriction patterns (patterns I to IV, VI, and VII). They were represented by 34 (pattern I), 9 (pattern II), 13 (pattern III), 24 (pattern IV), 4 (pattern VI), and 3 (pattern VII) isolates (pattern V was found only among the 18 reference isolates). The patterns consisted of three to eight bands, ranging in size from 66 to 854 bp (Fig. 2). They all had 243- and 130-bp bands in common. These were used as an internal standard for the analysis of the DNA patterns. Strains were considered different on the basis of band position; differences in band density were neglected.
FIG. 2.
(A) Representation of the seven different ARDRA patterns encountered amongst the set of 105 strains (lanes I to VII). Lane M, AmpliSize DNA size standard (Bio-Rad). (B) Schematic representation of the gel shown in panel A. ∗, place where samples were loaded.
(iv) PFGE and opa typing.
Cluster analysis and visual inspection of the restriction profiles produced by PFGE and opa typing revealed a large number of distinct profiles, with 7 clusters of identical isolates (five pairs and two triplets) identified by PFGE and 11 clusters (nine pairs, one triplet, and one cluster of four) identified by opa typing. Of these clusters, four were identified by both techniques. Isolates within each cluster mostly belonged to the same ARDRA group, PCR group, and A/S class (Table 2).
TABLE 2.
Phenotypic and genotypic typing data for the 30 gonococcal strains forming the 14 genetic clusters
| Strain | Auxotype/serotype | RAPDa type
|
ARDRA type | opa type | PFGE type | |
|---|---|---|---|---|---|---|
| Primer D11344 | Primer D8635 | |||||
| 1049-1 | P/IA-8 | b | C | I | 1 | 1 |
| 1029-1 | P/IA-8 | b | A | I | 1 | 1 |
| 1242-1 | P/IA-8 | b | C | I | 1 | 1 |
| 1269-1 | NR/IA-8 | b | E | I | 2 | 2 |
| 1267-1 | NR/IA-8 | b | E | I | 2 | 2 |
| 1231-1 | P/IB-1 | c | A | IV | 3 | 3 |
| 1018-2 | P/IB-1 | c | A | IV | 3 | 3 |
| 1167-1 | NR/IA-8 | a | A | VI | 4 | 4 |
| 1184-1 | NR/IA-8 | a | A | VI | 4 | 4 |
| 1031-1 | NR/IB-16 | a | A | VI | 4 | 5 |
| 1312-1 | NR/IA-8 | a | A | VI | 4 | 6 |
| 1042-1 | NR/IA-8 | b | A | III | 5 | 7 |
| 1020-1 | NR/IA-8 | b | A | III | 5 | 8 |
| 1219-1 | P/IB-1 | c | A | IV | 6 | 9 |
| 1249-1 | P/IB-1 | c | A | IV | 6 | 10 |
| 1177-1 | P/IB-7 | b | C | I | 7 | 11 |
| 1069-1 | PA/IB-7 | b | C | I | 7 | 12 |
| 1245-1 | P/IB-6 | b | E | I | 8 | 13 |
| 1274-1 | NR/IB-3 | b | A | I | 8 | 14 |
| 1058-2 | P/IB-1 | a | A | III | 9 | 15 |
| 1171-1 | P/IB-1 | a | A | III | 9 | 16 |
| 1350-1 | NR/IB-3 | b | A | II | 10 | 17 |
| 1199-1 | NR/IB-3 | b | A | II | 10 | 18 |
| 1237-1 | NR/IB-1 | b | A | VII | 11 | 19 |
| 1163-1 | NR/IB-8 | b | A | VII | 11 | 20 |
| 1073-1 | PA/IA-6 | b | C | I | 12 | 21 |
| 1003-2 | PA/IA-6 | b | C | I | 13 | 21 |
| 1165-1 | P/IB-1 | c | A | IV | 14 | 22 |
| 1247-1 | P/IB-1 | c | A | IV | 15 | 22 |
| 1219-1 | P/IB-1 | c | A | IV | 16 | 22 |
RAPD, randomly amplified polymorphic DNA.
(v) Discriminatory power.
The discriminatory power of each method or combination of methods according to Simpson’s index (6, 11) is shown in Table 3. PFGE (DI, 0.997), opa typing (DI, 0.996), and the combination of serotyping and ARDRA (DI, 0.955) produce the highest DIs.
TABLE 3.
DIs of typing methods used to discriminate among 87 N. gonorrhoeae strains
| Typing method | No. of types | Size (% of strains) of largest type | DI |
|---|---|---|---|
| Auxotyping | 4 | 48.3 | 0.597 |
| Serotyping | 12 | 25.3 | 0.846 |
| A/Serovar class | 24 | 16.1 | 0.928 |
| AP-PCR with primer D11344 | 3 | 49.4 | 0.608 |
| AP-PCR with primer D8635 | 4 | 55.2 | 0.622 |
| Combination D11344 and D8635 | 11 | 27.6 | 0.849 |
| Serotyping + AP-PCR with primer D11344 | 22 | 13.8 | 0.936 |
| Serotyping + AP-PCR with primer D8635 | 27 | 16.1 | 0.937 |
| ARDRA | 6 | 39.1 | 0.743 |
| Serotyping + ARDRA | 32 | 10.3 | 0.955 |
| opa typing | 73 | 4.6 | 0.996 |
| PFGE | 78 | 3.5 | 0.997 |
DISCUSSION
The goal of the present study was to identify the strengths and weaknesses of the different typing techniques for N. gonorrhoeae and to determine which method(s) would be best suited for clinical and research purposes. A panel of 18 strains, chosen for their diversity in phenotype, year and country of isolation, and antibiotic susceptibility profiles, was used to evaluate the different genotypic typing methods. The discriminatory power of each method was determined with a collection of clinical isolates which were likely to be heterogeneous.
Each technique tested typed all isolates, but their discriminatory powers differed substantially. The discriminatory power of a typing method is defined as its ability to distinguish between unrelated strains (11). It is determined by the number of types defined by the test method and the relative frequencies of these types. These two facets of discrimination are not generally presented as a single numerical value and therefore cannot be used for a straightforward comparison of different methods. Hunter and Gaston (11) proposed a single numerical index of discrimination, based on the probability that two unrelated isolates would be placed into different typing groups. This probability can be calculated from Simpson’s index of diversity. If typing results are to be interpreted with confidence, a DI of greater than 0.90 is desirable (11).
Since most of the strains had different opa types, even when a single restriction enzyme (TaqI) was used, it is obvious that the opa-typing assay was very discriminatory (DI, 0.996). The discriminatory power of PFGE with the infrequent cutter BglII was comparable to that of opa typing (DI, 0.997). Among the 87 clinical isolates, only 14 clusters (2 to 4 isolates) of strains with identical patterns were found. Whether these isolates were epidemiologically linked is unknown, but as they are indistinguishable by more than one technique and opa typing (16) and PFGE are known to be highly discriminatory and identify linked isolates, these isolates probably originate from patients linked in a transmission chain.
Of the two traditional techniques, auxotyping and serotyping, the latter has the highest DI (DI, 0.846), but this is too low to be satisfactory (11). This discriminatory power is similar to that of PCR-based typing using a single primer. The combination of auxotyping and serotyping yields a high DI (DI, 0.928). Determination of the A/S class is the most widely applied technique for typing gonococci. However, whereas serotyping is a simple and quick technique, auxotyping is more complex, especially for laboratories where it is not routinely performed, and could therefore well be replaced by a technically simpler approach.
Since PCR is being progressively introduced in more clinical laboratories, typing methods based on this technology are becoming more attractive. The combination of serotyping with D11344-primed or D8635-primed PCR resulted in DIs of 0.936 and 0.937, respectively (Table 3). The combination of serotyping and PCR-based typing would thus be a good alternative to A/S classification.
Camarena et al. (2) applied AP-PCR to 70 N. gonorrhoeae isolates and established 40 types with primer OPA-03 (DI, 0.967) and 50 types with primer OPA-13 (DI, 0.978). The discriminatory power based on Simpson’s index was superior to that of either auxotyping (DI, 0.670) or serotyping (DI, 0.934) and comparable to that of a combination of auxotyping and serotyping (DI, 0.968). They concluded that AP-PCR combined with serotyping provided the highest level of discrimination. When we compared auxotyping, serotyping, AP-PCR, determination of A/S class, and the combination of serotyping and PCR typing, we also found that the combination of serotyping and AP-PCR yielded a high DI. However, the discriminatory power of PCR typing was lower with the primers we used. Our evaluation of the primers used by Camarena et al. (2) produced unsatisfactory results, underscoring the problem of interlaboratory reproducibility of AP-PCR. Nevertheless, the results of PCR typing are very appealing and efforts should be undertaken to improve the interlaboratory reproducibility.
ARDRA was previously shown to be useful for typing (10, 26). However, our study revealed a low DI (0.743) when this technique was applied to the 16S-23S rRNA spacer region of gonococci. But this clearly results from the fact that we could find only a sole restriction enzyme to differentiate between our strains, whereas in other studies the combined information from up to five different enzymes was used. The combination of patterns produced by using several enzymes could produce a higher DI but would also make this approach more laborious and complex. Another possibility is to combine ARDRA or AP-PCR with another easy typing method, such as serotyping, both of which gave high DIs.
In conclusion, opa typing as described by O’Rourke et al. (16) and PFGE using the restriction enzyme BglII are the best techniques for typing isolates of N. gonorrhoeae. AP-PCR and ARDRA may also be useful.
ACKNOWLEDGMENTS
This study was performed in the context of the project “Support for STD and HIV/AIDS control and prevention among high risk populations in Jakarta, Surabaya, and Bandung” (contract B7.5046/94/015), financed by the European Commission, DG8-ATF. Work performed at ICSM (C.A.I. and I.M.M.) was supported by the Wellcome Trust. P.V. is indebted to the Fund for Scientific Research-Flanders (Belgium) for a position as a postdoctoral research fellow.
REFERENCES
- 1.Akopyanz N, Bukano N O, Westblom T U, Kresovich S, Berg D E. DNA diversity among clinical isolates of Helicobacter pylori detected by PCR-based RAPD fingerprinting. Nucleic Acids Res. 1992;20:5137–5142. doi: 10.1093/nar/20.19.5137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Camarena J J, Nogueira J M, Dasi M A, Moreno F, Garcia R, Ledesma E, Llorca J, Hernandez J. DNA amplification fingerprinting for subtyping Neisseria gonorrhoeae strains. Sex Transm Dis. 1995;22:128–136. doi: 10.1097/00007435-199503000-00009. [DOI] [PubMed] [Google Scholar]
- 3.Cooke S J, de la Paz H, Poh C L, Ison C A, Heckels J E. Variation within serovars of Neisseria gonorrhoeae detected by structural analysis of outer-membrane protein PIB and by pulsed-field gel electrophoresis. Microbiology. 1997;143:1415–1422. doi: 10.1099/00221287-143-4-1415. [DOI] [PubMed] [Google Scholar]
- 4.Dasi M A, Nogueira J M, Camarena J J, Gil C, Garcia-Verdu R, Barbera J L, Barbera J. Genomic fingerprinting of penicillinase-producing strains of Neisseria gonorrhoeae in Valencia, Spain. Genitourin Med. 1992;68:170–173. doi: 10.1136/sti.68.3.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dice L R. Measure of the amounts of ecological association between species. Ecology. 1945;26:297–302. [Google Scholar]
- 6.Dillon J R, Rahman M, Yeung K. Discriminatory power of typing schemes based on Simpson’s index of diversity for Neisseria gonorrhoeae. J Clin Microbiol. 1993;31:2831–2833. doi: 10.1128/jcm.31.10.2831-2833.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Falk E S, Danielsson D, Bjornvatn B, Melby K, Sorensen B, Kristiansen B-E. Genomic fingerprinting in the epidemiology of gonorrhoea. Acta Dermato-Venereol. 1985;65:235–239. [PubMed] [Google Scholar]
- 8.Gill M J. Serotyping Neisseria gonorrhoeae: a report of the Fourth International Workshop. Genitourin Med. 1991;67:53–57. doi: 10.1136/sti.67.1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gürtler V, Stanisich V A. New approaches to typing and identification of bacteria using the 16S-23S rDNA spacer region. Microbiology. 1996;142:3–16. doi: 10.1099/13500872-142-1-3. [DOI] [PubMed] [Google Scholar]
- 10.Heyndrickx M, Vauterin L, Vandamme P, Kerstens K, De Vos P. Applicability of combined amplified ribosomal DNA restriction analysis (ARDRA) patterns in bacterial phylogeny and taxonomy. J Microbiol Methods. 1996;26:247–259. [Google Scholar]
- 11.Hunter P R, Gaston M A. Numerical index of the discriminatory ability of typing systems: an application of Simpson’s index of diversity. J Clin Microbiol. 1988;26:2465–2466. doi: 10.1128/jcm.26.11.2465-2466.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lau Q C, Chow V T K, Poh C L. Differentation of Neisseria gonorrhoeae strains by polymerase chain reaction and restriction length polymorphism of outer membrane protein IB genes. Genitourin Med. 1995;71:363–366. doi: 10.1136/sti.71.6.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li H, Dillon J-A R. Utility of ribotyping, restriction endonuclease analysis and pulsed-field gel electrophoresis to discriminate between isolates of Neisseria gonorrhoeae of serovar IA-2 which require arginine, hypoxanthine or uracil for growth. J Med Microbiol. 1995;43:208–215. doi: 10.1099/00222615-43-3-208. [DOI] [PubMed] [Google Scholar]
- 14.Ng L-K, Carballo M, Dillon J-A R. Differentiation of Neisseria gonorrhoeae isolates requiring proline, citrulline, and uracil by plasmid content, serotyping, and pulsed-field gel electrophoresis. J Clin Microbiol. 1995;33:1039–1041. doi: 10.1128/jcm.33.4.1039-1041.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ng L-K, Dillon J-A R. Typing by serovar, antibiogram, plasmid content, riboprobing, and isoenzyme typing to determine whether Neisseria gonorrhoeae isolates requiring proline, citrulline, and uracil for growth are clonal. J Clin Microbiol. 1993;31:1555–1561. doi: 10.1128/jcm.31.6.1555-1561.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.O’Rourke M, Ison C A, Renton A M, Spratt B G. opa-typing: a high-resolution tool for studying the epidemiology of gonorrhoeae. Mol Microbiol. 1995;17:865–875. doi: 10.1111/j.1365-2958.1995.mmi_17050865.x. [DOI] [PubMed] [Google Scholar]
- 17.Pitcher D G, Saunders N A, Owen R J. Rapid extraction of bacterial genomic DNA with guanidium thiocyanate. Lett Appl Microbiol. 1989;8:151–156. [Google Scholar]
- 18.Poh C L, Khng H P, Lim C K, Loh G K. Molecular typing of Neisseria gonorrhoeae by restriction fragment length polymorphisms. Genitourin Med. 1992;68:106–110. doi: 10.1136/sti.68.2.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Poh C L, Loh G K, Tapsall J W. Resolution of clonal subgroups among Neisseria gonorrhoeae IB-2 and IB-6 serovars by pulsed-field gel electrophoresis. Genitourin Med. 1995;71:145–149. doi: 10.1136/sti.71.3.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Poh C L, Ocampo J C, Sng E H, Bygdeman S M. Rapid in-situ generation of DNA restriction endonuclease patterns for Neisseria gonorrhoeae. J Clin Microbiol. 1989;27:2784–2788. doi: 10.1128/jcm.27.12.2784-2788.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Poh C L, Ramachandran V, Tapsall J W. Genetic diversity of Neisseria gonorrhoeae IB-2 and IB-6 isolates revealed by whole-cell repetitive element sequence-based PCR. J Clin Microbiol. 1996;34:292–295. doi: 10.1128/jcm.34.2.292-295.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schäfer V, Enzensberger R, Schneider C, Rickmann J, Nitschke-Özbay H, Brade V. Epidemiology of penicillin-resistant Neisseria gonorrhoeae in Frankfurt, Germany. Eur J Clin Microbiol Infect Dis. 1995;14:914–918. doi: 10.1007/BF01691501. [DOI] [PubMed] [Google Scholar]
- 23.Selander R K, Caugant D A, Ochman H, Musser J M, Gilmour M N, Whittam T S. Methods of multilocus enzyme electrophoresis for bacterial population genetics and systematics. Appl Environ Microbiol. 1986;51:873–884. doi: 10.1128/aem.51.5.873-884.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sneath P H A, Sokal R R. Numerical taxonomy: the principles and practice of numerical classification. W. H. San Francisco, Calif: Freeman & Co.; 1973. [Google Scholar]
- 25.Tam M R, Buchanan T M, Sandström E G, Holmes K K, Knapp J S, Siaduk A W, Nowinski R C. Serological classification of Neisseria gonorrhoeae with monoclonal antibodies. Infect Immun. 1982;36:1042–1053. doi: 10.1128/iai.36.3.1042-1053.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vaneechoutte M, De Beenhouwer H, Claeys G, Verschraegen G, De Rouck A, Paepe N, Elaichouni A, Portaels F. Identification of Mycobacterium species by using amplified ribosomal DNA restriction analysis. J Clin Microbiol. 1993;31:2061–2065. doi: 10.1128/jcm.31.8.2061-2065.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Van Looveren M, Vandamme P, Hauchecorne M, Wijdooghe M, Caugant D A, Goossens H. Molecular epidemiology of recent Belgian isolates of Neisseria meningitidis serogroup B. J Clin Microbiol. 1998;36:2828–2834. doi: 10.1128/jcm.36.10.2828-2834.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Versalovic J, Koeuth T, Lupski J R. Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res. 1991;19:6823–6831. doi: 10.1093/nar/19.24.6823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Woodford N, Bindayna K M, Easmon C S F, Ison C A. Associations between serotype and susceptibility to antibiotics of Neisseria gonorrhoeae. Genitourin Med. 1989;65:86–91. doi: 10.1136/sti.65.2.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Woods J P, Kersulyte D, Tolan R W, Jr, Berg C M, Berg D E. Use of arbitrarily primed polymerase chain reaction analysis to type disease and carrier strains of Neisseria meningitidis isolated during a university outbreak. J Infect Dis. 1994;169:1384–1389. doi: 10.1093/infdis/169.6.1384. [DOI] [PubMed] [Google Scholar]
- 31.Xia M, Whittington W L, Holmes K K, Plummer F A, Roberts M C. Pulsed-field gel electrophoresis for genomic analysis of Neisseria gonorrhoeae. J Infect Dis. 1995;171:455–458. doi: 10.1093/infdis/171.2.455. [DOI] [PubMed] [Google Scholar]
- 32.Xia M, Roberts M C, Whittington W L, Holmes K K, Knapp J S, Dillon J-A R, Wi T. Neisseria gonorrhoeae with decreased susceptibility to ciprofloxacin: pulsed-field gel electrophoresis typing of strains from North America, Hawaii, and the Philippines. Antimicrob Agents Chemother. 1996;40:2439–2440. doi: 10.1128/aac.40.10.2439. [DOI] [PMC free article] [PubMed] [Google Scholar]