Abstract
Five sporadic cases of nosocomial Legionnaires’ disease were documented from 1989 to 1997 in a hospital in northern Italy. Two of them, which occurred in a 75-year-old man suffering from ischemic cardiopathy and in an 8-year-old girl suffering from acute leukemia, had fatal outcomes. Legionella pneumophila serogroup 6 was isolated from both patients and from hot-water samples taken at different sites in the hospital. These facts led us to consider the possibility that a single clone of L. pneumophila serogroup 6 had persisted in the hospital environment for 8 years and had caused sporadic infections. Comparison of clinical and environmental strains by monoclonal subtyping, macrorestriction analysis (MRA), and arbitrarily primed PCR (AP-PCR) showed that the strains were clustered into three different epidemiological types, of which only two types caused infection. An excellent correspondence between the MRA and AP-PCR results was observed, with both techniques having high discriminatory powers. However, it was not possible to differentiate the isolates by means of ribotyping and analysis of rrn operon polymorphism. Environmental strains that antigenically and chromosomally matched the infecting organism were present at the time of infection in hot-water samples taken from the ward where the patients had stayed. Interpretation of the temporal sequence of events on the basis of the typing results for clinical and environmental isolates enabled the identification of the ward where the patients became infected and the modes of transmission of Legionella infection. The long-term persistence in the hot-water system of different clones of L. pneumophila serogroup 6 indicates that repeated heat-based control measures were ineffective in eradicating the organism.
Legionella pneumophila is a well-known cause of bacterial pneumonia (22), accounting for up to 30% of cases of nosocomially acquired pneumonia, which most frequently occur in immunologically deficient subjects (10, 12, 24). Fifteen serogroups of L. pneumophila have been described (1, 3). L. pneumophila serogroup 1 is the most frequent among human isolates, and 12 or 15 antigenic subtypes have been recognized with different sets of monoclonal antibodies (MAbs) (references 4 and 11, respectively). L. pneumophila serogroup 6, the second most common serogroup according to the frequency of isolation from clinical samples (17), shows a lower antigenic variability, and up to five antigenic subtypes have been detected in different studies (11, 15, 18).
Epidemiological investigations of legionellosis are complicated by the ubiquity of legionellae in nature. Discriminatory molecular subtyping methods should be applied to clinical and presumptively linked environmental strains in order to detect the source of the infection. MAb subtyping is insufficiently discriminatory when a given serogroup comprises only a few antigenically distinct subtypes, as for L. pneumophila serogroup 6. Furthermore, phenotypic differences have been reported in genotypically similar organisms (9, 23, 25). However, at least in one of these instances (9), it was later shown that macrorestriction analysis (MRA) could differentiate strains showing identical restriction fragment length polymorphism profiles (14). A simultaneous infection with multiple genomic types of a single L. pneumophila serogroup has recently been described (16), leading to discussions as to the number of colonies which should be typed after primary isolation and to the preferable typing method(s). Thus, a combination of antigenic and genomic typing systems has been recommended for the definition of patterns of colonization, and the clone(s) involved in the transmission of the infection (6, 9, 14, 16, 19, 21, 23, 27).
Here we report on an investigation of L. pneumophila serogroup 6 isolates from a hospital in which five sporadic cases of Legionnaires’ disease occurred from 1989 to 1997. In order to determine whether a given clone of L. pneumophila serogroup 6 had found an ecological niche that enabled it to survive over a long period of time in the nosocomial environment and/or whether derivatives of the same organism had infected susceptible people over several years, the human isolates of L. pneumophila serogroup 6 were compared with their environmental counterparts by MAb subtyping, MRA, and arbitrarily primed PCR (AP-PCR).
MATERIALS AND METHODS
Water distribution system and specimen collection and processing.
The hospital consists of a single building with 24 wards and a total of approximately 1,200 beds. The building is 18 years old. It receives water from a single municipal supply. The hot water system consists of four portions that serve three main sections (designated sections A, B, and C) and a minor part (section D) of the hospital. The hot-water temperature is 55 to 56°C. For sections A, B, and C, each section has four 5,000-liter vertical heating tanks. From three of them, vertical pipes deliver hot water at different pressures, depending on the floor served: from the second underground floor to the 1st floor, low pressure, from the 2nd to the 6th floors, medium pressure, and from the 7th to the 12th floors, high pressure. Recirculation of water within each section is accomplished by pumps. The fourth heating tank acts as a reservoir to meet extra demands. Section D of the hospital is served by two 3,000-liter heating tanks with a thermostat-set point to mix warm and cold water and a recirculation pump.
Five-liter hot-water specimens from individual sections of the hospital were collected on the same day in sterile containers from distal outlets after allowing the water to flow for 10 min, and they were collected from the heating tanks through a bottom valve after allowing the water to flow for 2 min. The sampling started on the upper floors and continued to the lower levels.
Total bacterial counts were evaluated as numbers of CFU by the membrane (pore size, 0.45 μm; Millipore, Milan, Italy) filtration method of 10-fold serial dilutions of the samples after transfer of the membranes to the surfaces of two separate tryptone soy agar (Oxoid, Garbagnate Milanese, Italy) plates and incubation at 37°C for 24 h. The concentration of Legionella spp. was determined as the numbers of CFU per liter on buffered charcoal yeast extract (BCYE) agar plates (Oxoid). The water was concentrated by membrane (pore size, 0.2 μm; Millipore) filtration, and then diluted and undiluted specimens were plated and incubated at 37°C in humidified air for at least 10 days. Suspect Legionella colonies, which failed to grow in the absence of cysteine, were further checked by direct immunofluorescence with an L. pneumophila species-specific MAb (Diagnostics Pasteur, Marnes-La-Coquette, France) and then with L. pneumophila serogroup 1 to 6 monovalent fluorescein-labeled antisera (SCIMEDX; Dasit, Cornaredo, Italy, and BIOS; Daltec, San Vittore Olone, Italy).
Strains.
Three groups of L. pneumophila serogroup 6 strains were examined. Their designations and origins are listed in Table 1. The first group consisted of four strains (one from patient 1 and three from two putatively associated hospital environmental sources) isolated in 1989 (26), and the second group consisted of five strains (three from patient 2 and two from the epidemiologically linked source of infection) isolated in 1995. The third group consisted of 15 strains isolated in January 1996 from different sites of the same hospital but not associated in time with human infection. In fact, no isolates were available from three patients with serologically confirmed cases of infection which occurred in 1994, 1995, and 1997. L. pneumophila serogroup 6 reference strains Chicago 2 (ATCC 33215) and Dresden (15) were used as internal controls in molecular typing experiments. Suspensions of strains from cultures derived from a single colony were kept at −80°C in skim milk until they were used.
TABLE 1.
Clinical and environmental L. pneumophila serogroup 6 isolates from the hospital under survey, by time of isolation and origin
| Strain no. | Date (mo/yr) | Source of isolate | Location (block/ward) | Heating tank pressurea | Legionella count (cfu/liter) |
|---|---|---|---|---|---|
| 1 | April/1989 | Patient 1 | C/cardiology, medicine | ||
| 2 | April/1989 | Sink tap water | C/cardiology | MP | 3 × 103 |
| 3 | April/1989 | Sink tap water | C/medicine | MP | NDb |
| 4 | April/1989 | Bath tub water | C/cardiology | MP | 1 × 103 |
| 5 | July/1995 | Shower water | C/pediatric hematology | HP | ND |
| 6 | August/1995 | Shower water | C/pediatric hematology | HP | 8 × 102 |
| 7 | July/1995 | Patient 2 | C/pediatric hematology, room 5 | ||
| 8 | July/1995 | Patient 2 | C/pediatric hematology | ||
| 9 | July/1995 | Patient 2 | C/pediatric hematology | ||
| 10 | January/1996 | Sink tap water | C/pediatric hematology, room 5 | HP | 1 × 102 |
| 11 | January/1996 | Sink tap water | C/pediatric hematology, room 1 | HP | 5 × 102 |
| 12 | January/1996 | Sink tap water | C/pneumology | MP | 2 × 102 |
| 13 | January/1996 | Sink tap water | B/hematology | HP | 1 × 103 |
| 14 | January/1996 | Heating tank | B | MP | >1 × 104 |
| 15 | January/1996 | Heating tank | B | LP | 7 × 102 |
| 16 | January/1996 | Heating tank | A | HP | 4 × 103 |
| 17 | January/1996 | Heating tank | A | MP | 1 × 104 |
| 18 | January/1996 | Sink tap water | A/radiology | LP | 3 × 103 |
| 19 | January/1996 | Sink tap water | D/ICU | LP | 2 × 102 |
HP, high pressure; MP, medium pressure; LP, low pressure.
ND, not determined.
Subtyping with MAbs.
Strains typed as L. pneumophila serogroup 6 by direct immunofluorescence with the monovalent antiserum were further subtyped by indirect immunofluorescence with MAbs 9/2, 4/5, 18/2, and 54/2. These MAbs recognize different epitopes on the lipopolysaccharide of this organism (11, 15). MAb 9/2 specifically recognizes all L. pneumophila serogroup 6 strains. MAb 4/5 also reacts specifically only with serogroup 6 strains, but not with all serogroup 6 strains. MAbs 18/2 and 54/2 recognize antigenic variants of serogroup 6, but they also react with some strains belonging to other serogroups of L. pneumophila (15).
DNA fingerprinting.
MRA, AP-PCR, and ribotyping were performed. For MRA, genomic DNA was prepared by the method described by Lück and coworkers (13). Briefly, the bacteria were grown for 72 h on BCYE agar plates and were then washed twice and suspended in SE buffer (75 mM NaCl, 25 mM EDTA [pH 7.4]). The bacterial suspensions (A600 ≅ 1.5) were mixed with equal volumes of molten 2.0% low-melting-point agarose (Bio-Rad, Milan, Italy) in SE buffer, and the mixture was poured into acrylic casting wells. After the agarose gelled, the blocks were immersed in a digestion solution of 1% sodium lauroylsarcosine, 0.5 M EDTA, and 2 mg of proteinase K (Boehringer Mannheim, Milan, Italy) per ml (pH 9.5) and incubated at 50°C overnight. Agarose blocks were washed four times in TE buffer (10 mM Tris, 1.0 mM EDTA [pH 8.0]) and were stored in the same buffer at 4°C. DNAs were cleaved with NotI, SfiI, and AscI (New England Biolabs, Schwahlbach, Germany) following the manufacturer’s instructions. The blocks were then loaded on 1% agarose (FMC, BIOSPA, Milan, Italy) in 0.25× Tris-borate-EDTA buffer (pH 8.3). Pulsed-field gel electrophoresis (PFGE) was carried out with Rotaphor Type V equipment (Biometra, Gottingen, Germany) at 12°C for 36 h with a voltage decrease from 200 to 180 V and with a constant angle of 135°. Pulse times were 100 to 2, 50 to 2, and 60 to 2 s for DNAs cleaved with NotI, SfiI, and AscI, respectively. Bacteriophage lambda concatemers and Saccharomyces cerevisiae WAY 5-4A (Biometra) were used as DNA size markers. Genomic fragments were stained with ethidium bromide and were photographed under UV illumination.
AP-PCRs were carried out with a set of four oligonucleotides, designated AP5 (5′-TCCCGCTGCG-3′), AP12 (5′-CGGCCCCTGC-3′), CD1 (5′-GGATCCTGAC-3′), and 1247 (5′-AAGAGCCCGT-3′). Amplification reactions were performed in a 50-μl volume containing 10 mM Tris-HCl (pH 8.3), 4.0 mM MgCl2, 0.001% (wt/vol) gelatin, each deoxynucleoside triphosphate at 200 μM, each primer at 2.5 μM, 2 ng of genomic DNA, and 1.25 U of Taq DNA polymerase (Boehringer Mannheim). PCRs were performed in a Perkin-Elmer model 9600 thermal cycler with the fastest available transition times between each temperature. After incubation at 90°C for 60 s and at 95°C for 90 s, the reaction mixtures were cycled 45 times through the following temperature profile: 95°C for 30 s, 37°C for 1 min, and 74°C for 1.5 min. The samples were then incubated at 74°C for 3.5 min and were then held at 4°C. Samples of 10 μl of each amplification mixture were loaded onto a 2.0% (wt/vol) agarose gel with TBE (Tris-borate-EDTA) buffer containing 0.5 mg (wt/vol) of ethidium bromide per ml, and the gel was electrophoresed at 3 V/cm for approximately 5 h. Each strain was tested in three independent experiments performed under identical conditions. Gel photographs were scanned with a Hewlett-Packard Scanjet IIcx scanner. The PFGE and AP-PCR patterns were analyzed by GelCompar, version 4.0, computer software (Applied Maths, Kortrijk, Belgium). Similarity between pairs of strains was calculated as the Dice coefficient, which corresponds to the ratio of twice the number of common fragments to the total number of fragments in the two patterns. Clustering and the linkage level between pairs or groups of strains were calculated by the unweighted pair group method with arithmetic averages and are represented as a dendrogram.
For the analysis of rrn operon polymorphism, the chromosomal DNAs of the strains were digested with HindIII and PstI, and the fragments were separated by electrophoresis through a 0.8% agarose gel in TBE buffer at 40 V for approximately 16 h. Restriction fragments were Southern blotted onto a nylon membrane (Hybond-N; Amersham) and were cross-linked by exposure to UV light. Prehybridization, hybridization with the digoxigenin (DIG)-labelled 7.5-kb BamHI fragment of pKK3535 (5), posthybridization washing, and immunologic detection were performed according to the manufacturer’s instructions (Boehringer Mannheim). DIG-labelled hybrids were detected with an anti-DIG alkaline phosphatase antibody conjugate and the chemiluminescent substrate Lumigen PPD (Boehringer) according to the manufacturer’s specifications. For the detection of the chemiluminescent signal, the membranes were exposed to Kodak XAR film.
Analysis of the 16S-23S rRNA gene spacer regions was performed with primer 2 (5′-TTGTACACACCGCCCGTC-3′), which annealed to the 16S rRNA gene from base pairs 1390 to 1407, and primer 7 (5′-GGTACTTAGATGTTTCAGTTC-3′), which annealed to the 23S rRNA gene from base pairs 188 to 208, according to Gürtler and Stanisich (8). Amplification reactions were carried out by using 10 ng of genomic DNA in a 100-μl reaction mixture containing 1× PCR buffer (Promega), 2.5 mM MgCl2, each deoxynucleoside triphosphate at 50 μM, each primer at 1 μM, and 0.25 U of Taq DNA polymerase (Promega). After incubation at 94°C for 4 min, 30 cycles were performed in a Perkin-Elmer model 9600 thermal cycler, with each cycle comprising 45 s at 94°C, 1 min at 55°C, and 45 s at 72°C. The samples were then incubated at 72°C for 5 min and were then held at 4°C. The amplification products were electrophoresed and visualized as outlined above.
RESULTS
Clinical and environmental investigations.
On 9 March, 1989 a 75-year-old man (patient 1) with a history of nephrosclerosis, hypertension, and ischemic cardiopathy was admitted to the cardiology ward of a hospital in northern Italy. He presented with unstable angina and ulcerative rectocolitis. On 15 March he was transferred to the medicine ward, and then on 2 April he was transferred to the coronary care unit in the cardiology ward due to acute myocardial infarction. Ten days later, after improvement in his clinical condition, he was transferred back to the medicine ward. On 19 April the patient developed acute dyspnea for pulmonary edema, and the chest X ray disclosed a bronchopneumonic picture in the right upper lobe. Ten days later the patient was transferred to the intensive care unit (ICU), where he died 5 h after admission. Antibiotic therapy was not given. L. pneumophila serogroup 6 was isolated from lung tissue obtained at autopsy and from water samples of the cardiology and medicine wards. A semiquantitative evaluation by CFU counts of the samples from the cardiology ward showed 3 × 103 legionellae/liter in the sink tap water and 1 × 103 legionellae/liter in the bathtub water (Table 1).
On 5 June 1995 an 8-year-old girl (patient 2) suffering from acute lymphocytic leukemia was admitted to the pediatric hematology ward of the same hospital to initiate the conditioning regimen prior to bone marrow transplantation. On 14 June she received the transplant, and on the following day she developed a fever (>38°C). Despite antimicrobial therapy, on the 14th posttransplantation day respiratory symptoms appeared and a chest X ray disclosed lower left pulmonary infiltrates. Forced respiration was started, antibiotic therapy was implemented, and the patient was transferred to the ICU. On the 29th posttransplantation day, culture results became available and indicated positivity for L. pneumophila. Despite addition of rifampin to the antibiotic regimen, the patient died from respiratory failure 3 days later. L. pneumophila serogroup 6 was isolated from bronchoalveolar lavage specimens obtained on 7 and 10 July and on the day of death. A microorganism of the same species and serogroup (8 × 102 CFU/liter) as the microorganism isolated from patient 2 was cultured from hot water from the shower of the pediatric hematology ward where the patient had stayed (Table 1).
Three cases of nosocomially acquired Legionnaires’ disease were clinically diagnosed in 1994, 1995, and 1997 and were confirmed by seroconversion of the patients, but cultures of respiratory specimens did not yield Legionella isolates. Since a polyvalent L. pneumophila serogroup 1 to 6 antigen was used for the indirect immunofluorescence test, it was not possible to determine the serogroup that caused the infection.
An environmental investigation was further performed in 1996. L. pneumophila serogroup 6 was isolated from 15 (62.5%) of 24 sites examined. The legionella concentration at the different sites examined did not correlate with the total bacterial counts in the samples (data not shown). No other L. pneumophila serogroups or Legionella species were isolated.
After the first documented case of nosocomial Legionnaires’ disease in 1989, active surveillance was implemented in the hospital. In May 1989 and August 1995 control measures were undertaken by superheating the heating tanks, and water was flushed for 15 min at the distal outlets of the system at a temperature >65°C.
Subtyping with MAbs.
All the strains examined reacted with MAb 9/2, which is specific for L. pneumophila serogroup 6. The other three MAbs were selected because they recognize subgroup-determining epitopes of serogroup 6 strains (11, 15). None of the strains reacted with MAb 18/2. Two strains isolated in 1989 and all those isolated in 1995 were positive with the subgroup-specific MAbs 4/5 and 54/2, as was the Chicago 2 type strain. Two of the strains isolated in 1989 and 11 of the 15 strains isolated in January 1996 were negative with these MAbs (Table 2 and data not shown).
TABLE 2.
MAb, PFGE, and AP-PCR subtyping of clinical and environmental isolates of L. pneumophila serogroup 6
| Strain no.a | Reactivity against MAb:
|
PFGE type after digestion withb:
|
AP-PCR type with primer:
|
Combined type codec | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 9/2 | 4/5d | 18/2d | 54/2d | SfiI | NotI | AscI | AP5 | AP12 | CD1 | 1247 | ||
| 1 | + | + | − | + | A | NC | F | a | e | g | j | I |
| 2 | + | − | − | − | CIII | D | G | c | e | i | l | IIII |
| 3 | + | − | − | − | CI | D | G | c | e | i | l | IIII |
| 4 | + | + | − | + | A | NC | F | a | e | g | j | l |
| 5 | + | + | − | + | B | E | H | b | f | h | k | II |
| 6 | + | + | − | + | B | E | H | b | f | h | k | II |
| 7 | + | + | − | + | B | E | H | b | f | h | k | II |
| 8 | + | + | − | + | B | E | H | b | f | h | k | II |
| 9 | + | + | − | + | B | E | H | b | f | h | k | II |
| 10 | + | − | − | − | CI | D | G | c | e | i | l | IIII |
| 11 | + | − | − | − | CV | D | G | d | e | i | l | IIIII |
| 12 | + | − | − | − | CI | D | G | c | e | i | l | IIII |
| 13 | + | − | − | − | CV | D | G | d | e | i | l | IIIII |
| 14 | + | − | − | − | CII | D | G | c | e | i | l | IIII |
| 15 | + | − | − | − | CIV | D | G | d | e | i | l | IIIII |
| 16 | + | + | − | + | A | NC | F | a | e | g | j | I |
| 17 | + | + | − | + | A | NC | F | a | e | g | j | I |
| 18 | + | − | − | − | CIV | D | G | d | e | i | l | IIIII |
| 19 | + | − | − | − | CII | D | G | c | e | i | l | IIII |
| Chicago 2 | + | + | − | + | ||||||||
| Dresden | + | − | − | − | ||||||||
Strains were numbered as in Table 1.
Indistinguishable patterns are indicated with the same letter code. Minor differences (<20%) within a single PFGE type are indicated by superscript roman numerals; NC, genomic DNA is not cut by the enzyme.
Obtained by combining MAb, PFGE, and AP-PCR types; strains with >80% and 90% similarities at the macrorestriction and AP-PCR levels, respectively, were considered to belong to a single type. Superscript roman numerals indicate minor differences within a single type.
MAbs that recognize monoclonal subgroups of serogroup 6.
Genomic analysis.
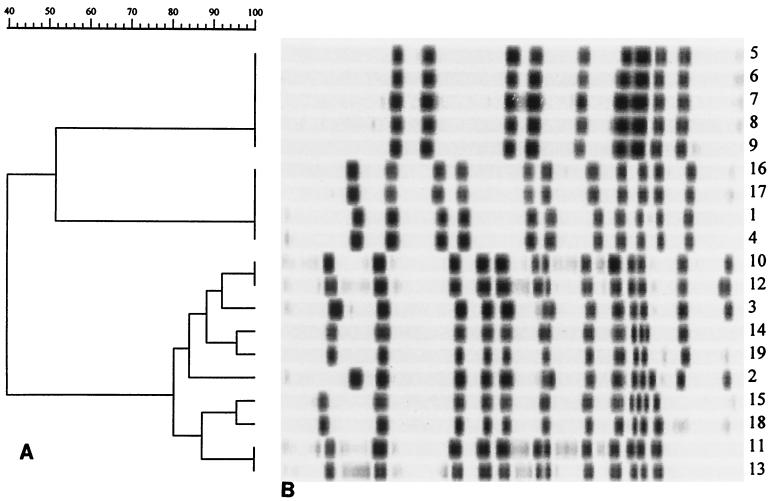
The results obtained by PFGE analysis of SfiI-digested genomic DNAs of 19 representative strains are shown in Fig. 1 and are summarized in Table 2. Among the three enzymes tested, SfiI gave the most complex macrorestriction pattern, allowing the identification of three main clusters, designated clusters A, B, and C. Cluster C included strains characterized by a similarity score of >80%, as deduced by computer-assisted analysis of electropherograms. Within this group, some differences were observed among isolates (at the level of up to four bands), and these differences determined a further subdivision into five subtypes (subtypes CI to CV; Table 2), each of which was composed of strains with >85% similarity. Also, NotI and AscI digestions made it possible to identify three pulsotypes (pulsotypes D, E, and uncut for NotI and pulsotypes F, G, and H for AscI; Table 2). Each pulsotype included strains with identical macrorestriction patterns (data not shown). In addition, the data reported in Table 2 indicate a complete correspondence between groups of strains clustered on the basis of macrorestriction profiles following digestion with the three enzymes.
FIG. 1.
PFGE analysis of SfiI-cleaved genomic DNA of the L. pneumophila serogroup 6 strains listed in Table 1. (A) Dendrogram showing the genetic distance relationships of the 19 isolates designated as indicated in Table 1. (B) Macrorestriction patterns of the isolates.
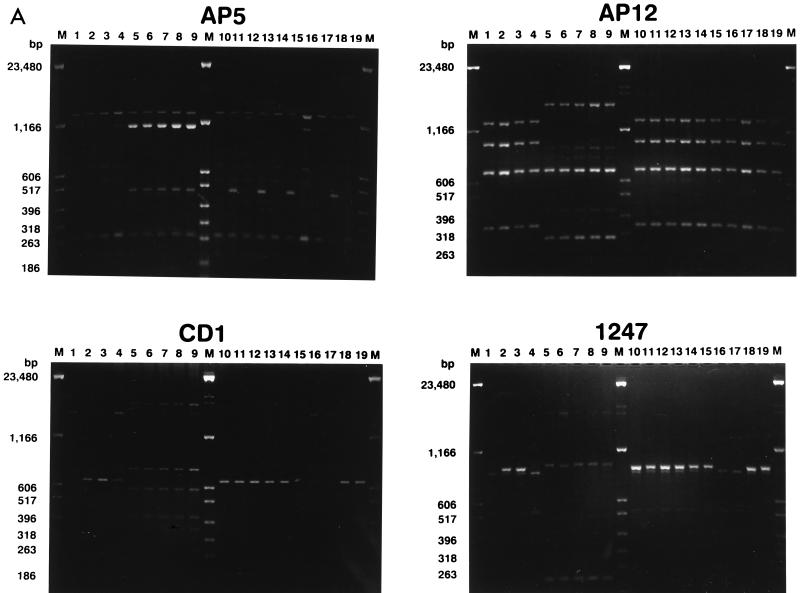
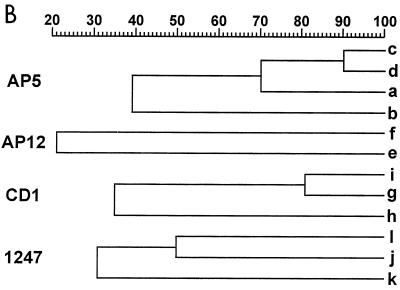
The AP-PCR results obtained by the use of four different oligonucleotide primers are shown in Fig. 2 and are summarized in Table 2. Primers CD1 and 1247 defined three different groups each (groups g, h, and i and groups j, k, and l, respectively), and the groups matched those previously identified by PFGE. Primer AP12 was less discriminatory, since it differentiated the strains into two groups (groups e and f), while AP5 was more discriminatory, allowing the definition of four different groups (groups a, b, c, and d).
FIG. 2.
(A) AP-PCR analysis of genomic DNAs of the L. pneumophila serogroup 6 strains listed by number in Table 1. The primers used for PCR are indicated above each electropherogram. M, molecular weight marker. The numbers on the left of each gel indicate the length (in base pairs) of reference fragments. (B) Dendrogram showing the degree of similarity between the AP-PCR patterns given in Table 2.
There was an excellent correspondence of the results obtained by AP-PCR with primer AP5 and MRA with SfiI. As shown in Table 2, cluster a matched subtype A, cluster b matched subtype B, cluster c matched subtypes CI to CIII, and cluster d matched subtypes CIV and CV. The degree of similarity between subtypes CI, CII, and CIII (corresponding to AP-PCR type c) was nearly 85%. This value is similar to that determined by comparing types CIV and CV, which correspond to AP-PCR type d, and is higher than the value obtained by comparison of all C subtypes (80%; see Fig. 1). Likewise, the degree of similarity between types c and d was 90%, which is the highest value among those determined by analysis of AP-PCR patterns (Fig. 2).
Southern hybridization of HindIII- and PstI-digested genomic DNA with the rrnB gene probe did not differentiate the 19 L. pneumophila isolates listed in Table 2. In addition, PCR of the 16S-23S rrn intergenic region (8) did not reveal any amplicon length polymorphism (data not shown).
By combining the MAb, PFGE, and AP-PCR types presented in Table 2, the strains were subdivided into three different combined type codes, designated types I, II, and III. When the data in Table 1 were examined in light of the typing results (Table 2), it appeared that two unrelated clones of L. pneumophila serogroup 6 were responsible for the infection episodes in 1989 and 1995. The type I strain, which caused the infection in 1989, was found to persist in the water system of the hospital until 1996, when the last environmental sampling was performed. The type II isolate was found to contaminate the high-pressure heating tank of hospital section C only during the summer of 1995, when it was also responsible for the infection of patient 2. Interestingly, from 1989 to 1996 a third clone of L. pneumophila serogroup 6 (type III) was found in the water supply system of the hospital and was found to contaminate different heating tanks and sections but did not cause any documented infection episode.
DISCUSSION
From 1989 to 1997, nosocomially acquired Legionnaires’ disease was documented in five patients in the hospital under study. L. pneumophila serogroup 6 was isolated from clinical specimens from two patients who died, while for the other three patients, all of whom recovered, only seroconversion against a L. pneumophila serogroup 1 to 6 polyvalent antigen was evidenced. Serological typing of presumptively associated environmental strains revealed that L. pneumophila serogroup 6 was responsible for extensive contamination of the hospital hot-water supply system. The legionella concentration at the different sites examined ranged from 102 to >104 CFU per liter, which is an amount considered to be able to cause one or more sporadic cases per year (7). These facts led us to wonder whether a single clone of L. pneumophila serogroup 6 had persisted in the hospital environment for over 7 years and had caused sporadic infections. Therefore, we compared the MAb types, MRA patterns, and AP-PCR types for all the nosocomial L. pneumophila serogroup 6 isolates. Although the discriminatory power of MAb typing is relatively poor for L. pneumophila serogroup 6, it is interesting that strains belonging to the two subtypes Chicago (5 strains) and Dresden (10 strains) contaminated the water system of the hospital over the period of time examined but that only MAb type Chicago had caused infection.
DNA-based typing techniques made it possible to differentiate the isolates into three distinct epidemiological types. The type I and II isolates belonged to MAb type Chicago. Type I was responsible for the infection in 1989 and persisted until 1996. Type II was associated with the infectious episode in 1995 and showed significant differences from type I (<50% similarity at the level of the SfiI MRA and AP-PCR patterns). All the strains included within type III belonged to MAb type Dresden and were indistinguishable when analyzed by PFGE of NotI- and AscI-cleaved DNA or by AP-PCR with primers AP12, CD1, and 1247 but were closely related when analyzed by MRA with SfiI or by AP-PCR with primer AP5 (Fig. 1 and 2).
The results reported here provide useful information. First, the long-term persistence in the water system of the hospital of multiple clones of L. pneumophila serogroup 6, one of which was responsible for a sporadic case of nosocomial legionellosis, is demonstrated. Type I was confined to the hot-water supply system of sections C and A during the years 1989 and 1996, respectively, being responsible for one sporadic human infection, which occurred in 1989, whereas type III was found in the heating tanks of all four hospital sections examined in the 1995 and 1996 period, as well as in the hot water supplied from the tank of section C during the 1989 sampling, but did not cause any documented case of nosocomial legionellosis. It can be speculated that eradication procedures performed after the first case was diagnosed may have altered the relative levels of individual types within the hospital hot-water system. Although the hot water of the hospital was maintained at 5 to 6°C above the thermal threshold for suppression of Legionella multiplication (23) and superheating was performed in May 1989 and August 1995, both control measures failed to eradicate the microorganism. However, while types I and III were able to persist in the hospital water system during the whole period examined, type II was probably eradicated since it was no longer isolated from the water taken during the extensive sampling of 1996. Whether types I and III are more resistant than type II to the thermal shock or whether they are endowed with a greater ecological fitness is still an open question.
Second, this study adds further information on the discriminatory power of DNA-based techniques for the typing of L. pneumophila serogroup 6. Analysis of the 16S rrn operon and of the 16S-23S spacer region did not reveal appreciable genomic polymorphism for the 19 strains examined, suggesting that these two techniques may be inadequate for DNA fingerprinting of L. pneumophila serogroup 6 strains. Digestion of genomic DNAs with either SfiI or AscI gave unique and complex PFGE patterns (nine or more fragments), enabling an accurate discrimination between pulsotypes (Simpson’s index of diversity [D] = 0.37). The complexities of the electropherograms obtained upon NotI digestion were lower (the enzyme either did not cut the DNA or generated two to five fragments), but they were still adequate for differentiation of the isolates (D = 0.37). Interestingly, repeated attempts to obtain NotI digestion of DNA extracted from type I strains were unsuccessful. NotI recognizes and cuts the sequence 5′-GC↓GGCCGC-3′ (the arrow represents the cleavage site) and is sensitive to methylation of the CG residues at positions 4 and 5 of the restriction site. Whether an SssI-like GpC methylase (20) is present in the type I isolates is still unknown, but this activity would certainly block cleavage at all NotI genome sites.
The amplification patterns obtained by AP-PCR with all four primers tested did not differ significantly in terms of complexity, because they produced 3 to 10 major amplicons for each type strain, but the level of discrimination achieved was dependent on the primer used. Thus, primer AP5 gave the best results, in that it generated four distinct patterns (patterns a to d; D = 0.21), while primer AP12 had the lowest discriminatory power and produced only two patterns (patterns e and f; D = 0.59), which, in turn, did not correlate with the observed MAb types. An interesting observation derived from this study is the excellent agreement between MRA with SfiI and AP-PCR with primer AP5. Type III strains show some heterogeneity when analyzed by SfiI digestion and can be considered a single type when an 80% similarity cutoff is imposed on the PFGE analysis, while they are resolved into two clusters at a similarity cutoff of 85%. In the latter case, strains 2, 3, 10, 12, 14, and 19 would be included in one subgroup, while strains 11, 13, 15, and 18 would be split into another subgroup. Of note, these two subgroups perfectly match with the c and d subtypes defined for type III isolates by AP-PCR fingerprinting with primer AP5 (Table 2). From this point of view, AP-PCR proved to be more informative than PFGE analysis. While pairwise comparison of macrorestriction patterns with SfiI and AscI digestion assigned nearly the same extent of similarity to types I, II, and III, AP-PCR revealed significant differences between types, with type I being more closely related to type III than to type II. Moreover, unrelated strains exhibited an overall high degree of polymorphism when tested by both MRA and AP-PCR, indicating comparable discriminatory powers for both techniques. AP-PCR is occasionally reported to suffer from poor reproducibility, but in our study strains tested on more than one occasion with the same primer consistently gave identical results. Thus, a major drawback derived from this observation is that AP-PCR analysis with appropriate primers can provide easily interpretable patterns (consisting of a maximum of 10 major amplicons) and can reach a discriminatory level comparable or even superior to that of MRA.
On the basis of the typing results, we may conclude that the infecting strains were transmitted from the hospital hot-water supply system. High densities of legionellae were found in the hot-water samples and in the heating tanks, which are known to be usual reservoirs of legionellae in hospital settings (28). In addition, legionellae were not isolated from the cold water or from the cooling towers of the hospital air-conditioning system during multiple samplings performed from 1989 to 1996. Strains identical to those isolated from the two patients were present in the central and peripheral hot-water supply system, and there is a close temporal relationship between the isolates from humans and the corresponding isolates from the hot water. Taking into account the temporal sequence of events, it can be assumed that patient 1 became infected in the medicine ward, where he resided during the week preceding the onset of symptoms. Although transmission from the water system (medium-pressure heating tank) of hospital section C to the patient can be hypothesized, we were unable to detect the type I infecting strain from the hot water taken from the medicine ward. However, we have shown that it was present in the hot water of the cardiology ward, where the patient had stayed before being transferred and which is served by the same heating tank as the medicine ward (Table 1). It is therefore possible that the type I strain was also present in the sample taken from the latter ward but that it escaped detection because only one randomly selected colony of L. pneumophila serogroup 6 was sent to the reference laboratory in Rome for typing. The mode of disease acquisition was presumably aspiration. The patient did not shower but was exposed to water by bed bathing, which is a known risk factor (2). For patient 2, who was infected while staying in the pediatric hematology ward, inhalation of aerosol generated from showering appears to be the most likely mode of transmission of the infection. In fact, the causative strain was present in the hot water taken from the shower, and the hospital staff confirmed that the patient took showers on the days preceding the onset of symptoms. Moreover, it was ruled out that she might have drunk tap water and that floor-washing procedures might have constituted a risk factor.
The finding that antigenically similar but genetically different serogroup 6 strains were isolated from the environment associated with the infection has two important consequences. First, serological and MAb typing may be insufficient for discrimination of individual isolates of L. pneumophila serogroup 6. Second, sampling bias can occur, and a large number of environmental isolates should be genotyped to ensure that all types of legionellae present in the sample are recovered and characterized. Despite its undisputed discriminatory power, PFGE typing is time-consuming, relatively expensive, and available only to specialized laboratories. In contrast, AP-PCR is cost-effective, time-saving, and easy to perform. The single-primer reaction can rapidly discriminate large panels of L. pneumophila isolates and can be used for the quick screening of isolates from different sources in local settings.
In conclusion, our results highlight the value of combined MAb typing and genomic analysis in comparing L. pneumophila serogroup 6 strains. In particular, the high discriminatory power and feasibility of AP-PCR make this technique suitable for routine comparison of L. pneumophila serogroup 6 isolates in epidemiological studies aimed at detection of the infection source and validation of the effectiveness of control measures.
ACKNOWLEDGMENTS
We thank Simonetta Ciarrocchi and Sofia Graziani for expert laboratory support.
REFERENCES
- 1.Benson R F, Thacker W L, Wilkinson H W, Fallon R J, Brenner D J. Legionella pneumophila serogroup 14 isolated from patients with fatal pneumonia. J Clin Microbiol. 1988;26:382. doi: 10.1128/jcm.26.2.382-.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blatt S P, Parkinson M D, Pace E, Hoffman P, Dolan D, Lauderdale P, Zajac R A, Melcher G P. Nosocomial Legionnaires’ disease: aspiration as a primary mode of disease acquisition. Am J Med. 1993;95:16–22. doi: 10.1016/0002-9343(93)90227-g. [DOI] [PubMed] [Google Scholar]
- 3.Brenner D J, Steigerwalt A G, Epple P, Bibb W F, McKinney R M, Starnes R W, Colville J M, Selander R K, Edelstein P H, Moss C W. Legionella pneumophila serogroup Lansing 3 isolated from a patient with fatal pneumonia, and description of L. pneumophila subsp. pneumophila subsp. nov., L. pneumophila subsp. fraseri subsp. nov., and L. pneumophila subsp. pascullei subsp. nov. J Clin Microbiol. 1988;26:1695–1703. doi: 10.1128/jcm.26.9.1695-1703.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brindle R J, Bryant T N, Draper P W. Taxonomic investigation of Legionella pneumophila using monoclonal antibodies. J Clin Microbiol. 1989;27:536–539. doi: 10.1128/jcm.27.3.536-539.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brosius J, Ullrich A, Raker M A, Gray A, Dull T J, Gutell R R, Noller H F. Construction and fine mapping of recombinant plasmids containing the rrnB ribosomal RNA operon of Escherichia coli. Plasmid. 1981;6:112–118. doi: 10.1016/0147-619x(81)90058-5. [DOI] [PubMed] [Google Scholar]
- 6.Castellani Pastoris M, Ciceroni L, Lo Monaco R, Goldoni P, Mentore B, Flego G, Cattani L, Ciarrocchi S, Pinto A, Visca P. Molecular epidemiology of an outbreak of Legionnaires’ disease associated with a cooling tower in Genova-Sestri Ponente, Italy. Eur J Clin Microbiol Infect Dis. 1997;16:883–892. doi: 10.1007/BF01700554. [DOI] [PubMed] [Google Scholar]
- 7.Ezzeddine H, Van Ossel C, Delmee M, Wauters C. Legionella spp. in a hospital hot water system: effect of control measures. J Hosp Infect. 1989;13:121–131. doi: 10.1016/0195-6701(89)90018-2. [DOI] [PubMed] [Google Scholar]
- 8.Gürtler V, Stanisich V A. New approaches to typing and identification of bacteria using the 16S-23S rDNA spacer region. Microbiology. 1996;142:3–16. doi: 10.1099/13500872-142-1-3. [DOI] [PubMed] [Google Scholar]
- 9.Harrison T G, Saunders N A, Haththotuwa A, Hallas G, Birtles R J, Taylor A G. Phenotypic variation amongst genotypically homogeneous Legionella pneumophila serogroup 1 isolates: implications for the investigations of outbreaks of Legionnaires’ disease. Epidemiol Infect. 1990;104:171–180. doi: 10.1017/s0950268800059331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hart C A, Makin T. Legionella in hospitals: a review. J Hosp Infect. 1991;18(Suppl. A):481–489. doi: 10.1016/0195-6701(91)90060-l. [DOI] [PubMed] [Google Scholar]
- 11.Helbig J H, Kurtz J B, Castellani Pastoris M, Pelaz C, Lück P C. Antigenic lipopolysaccharide components of Legionella pneumophila recognized by monoclonal antibodies: possibilities and limitations for division of the species and serogroups. J Clin Microbiol. 1997;35:2841–2845. doi: 10.1128/jcm.35.11.2841-2845.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lowry P W, Tompkins L S. Nosocomial legionellosis: a review of pulmonary and extrapulmonary syndromes. Am J Infect Control. 1993;21:21–27. doi: 10.1016/0196-6553(93)90203-g. [DOI] [PubMed] [Google Scholar]
- 13.Lück P C, Bender L, Ott M, Helbig J H, Hacker J. Analysis of Legionella pneumophila serogroup 6 strains isolated from a hospital warm water supply over a three-year period by using genomic long-range mapping techniques and monoclonal antibodies. Appl Environ Microbiol. 1991;57:3226–3231. doi: 10.1128/aem.57.11.3226-3231.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lück P C, Birtles R J, Helbig J H. Correlation of MAb subgroups with genotype in closely related Legionella pneumophila serogroup 1 strains from a cooling tower. J Med Microbiol. 1995;43:50–54. doi: 10.1099/00222615-43-1-50. [DOI] [PubMed] [Google Scholar]
- 15.Lück P C, Helbig J H, Pilz C, Witzleb W. Monoclonal antibodies to Legionella pneumophila serogroup 6: evidence of antigenic diversity. Zenbl Bakteriol Parasitenkd Infektionskr Hyg Abt 1 orig. 1991;274:533–536. doi: 10.1016/s0934-8840(11)80092-7. [DOI] [PubMed] [Google Scholar]
- 16.Lück P C, Wenchel H M, Helbig J H. Nosocomial pneumonia caused by three genetically different strains of Legionella pneumophila and detection of these strains in the hospital water supply. J Clin Microbiol. 1998;36:1160–1163. doi: 10.1128/jcm.36.4.1160-1163.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martson B J, Lipman H B, Breiman R F. Surveillance of Legionnaires’ disease: risk factors for morbidity and mortality. Arch Intern Med. 1994;154:2417–2422. [PubMed] [Google Scholar]
- 18.McKinney R M, Kuffner T A, Bibb W F, Nokkaew C, Wells D E, Arnow P M, Woods I C, Plikaytis B D. Antigenic and genetic variation in Legionella pneumophila serogroup 6. J Clin Microbiol. 1989;27:738–742. doi: 10.1128/jcm.27.4.738-742.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pruckler J M, Mermel L A, Benson R F, Giorgio C, Cassiday P K, Breiman R F, Whitney C G, Fields B S. Comparison of Legionella pneumophila isolates by arbitrarily primed PCR and pulsed-field gel electrophoresis: analysis from seven epidemic investigations. J Clin Microbiol. 1995;33:2872–2875. doi: 10.1128/jcm.33.11.2872-2875.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Renbaum P, Abrahamove D, Fainsold A, Wilson G G, Rottem S, Razin A. Cloning, characterization, and expression in Escherichia coli of the gene coding for the CpG DNA methylase from Spyroplasma sp. strain MQ1 (M.SssI) Nucleic Acids Res. 1990;18:1145–1152. doi: 10.1093/nar/18.5.1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schoonmaker D, Heimberger T, Birkhead G. Comparison of ribotyping and restriction enzyme analysis using pulse-field gel electrophoresis for distinguishing Legionella pneumophila isolates obtained during a nosocomial outbreak. J Clin Microbiol. 1992;30:1491–1498. doi: 10.1128/jcm.30.6.1491-1498.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stout J E, Yu V L. Legionellosis. N Engl J Med. 1997;337:682–687. doi: 10.1056/NEJM199709043371006. [DOI] [PubMed] [Google Scholar]
- 23.Struelens M J, Maes N, Rost F, Deplano A, Jacobs F, Liesnard C, Bornstein N, Grimont F, Lauwers S, McIntyre M P, Serruys E. Genotypic and phenotypic methods for the investigation of a nosocomial Legionella pneumophila outbreak and efficacy of control measures. J Infect Dis. 1992;166:22–30. doi: 10.1093/infdis/166.1.22. [DOI] [PubMed] [Google Scholar]
- 24.Tablan O C, Anderson L J, Arden N H, Breiman R F, Butler J C, McNeil M M the Hospital Infection Control Practices Advisory Committee. Guideline for prevention of nosocomial pneumonia. Part I. Issues on prevention of nosocomial pneumonia. Am J Infect Control. 1994;22:247–292. doi: 10.1016/0196-6553(94)90079-5. [DOI] [PubMed] [Google Scholar]
- 25.VanKetel R J. Similar DNA restriction endonuclease profiles in strains of Legionella pneumophila from different serogroups. J Clin Microbiol. 1988;26:1838–1841. doi: 10.1128/jcm.26.9.1838-1841.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Viganò E F, Giltri G, Cappellini A, Brenna A, Castellani Pastoris M. Polmonite nosocomiale infausta da Legionella pneumophila sierogruppo 6. In: Boemi G, Filadoro F, Mandler F, Mosconi P, editors. Microbiologia delle infezioni ospedaliere. Atti del XVIII Congresso Nazionale Associazione Microbiologi Clinici Italiani 1989. Milan, Italy: Associazione Microbiologi Clinici Italiani; 1990. pp. 197–199. [Google Scholar]
- 27.Willey B M, Low D E, Herbage K, Stout J, McGeer A. Abstracts of the 94th General Meeting of the American Society for Microbiology 1994. Washington, D.C: American Society for Microbiology; 1994. Comparison of restriction enzyme analysis (REA) with pulsed-field gel electrophoresis (PFGE) in the investigation of nosocomial infections caused by Legionella pneumophila serogroup 6, abstr. L23; p. 138. [Google Scholar]
- 28.Yu V L. Nosocomial legionellosis: current epidemiological studies. Curr Clin Top Infect Dis. 1986;7:239–253. [Google Scholar]