Abstract
Background
Currently, drug-resistance is a major challenge in the treatment of renal cancer. Although microRNAs (miRNAs) have been reported to contribute to the incidence of drug resistance in renal cancer, the bio-functional roles and underlying regulatory mechanisms of novel miRNAs in cisplatin resistance remain largely unclear.
Methods
In this study, miRNA microarray analysis was applied to evaluate miRNA changes induced by cisplatin on RCC (renal cell carcinoma) cell lines. Then, Caki-1 and 786-0 cells were transfected with miR (miRNA)-124 mimics to observe cisplatin resistance in RCC cell lines after up-regulation of miR-124. TargetScan was used to identify putative protein-coding gene targets of miR-124. Further, the interaction between calpain small subunit 1 (Capn4) and CCR4-NOT transcription complex subunit 3 (CNOT3) was detected by quantitative real-time PCR (qPCR) and western blotting, and confirmed by co-immunoprecipitation. The effect of Capn4 and/or CNOT3 on cell viability and half maximal inhibitory concentration (IC50) value of miR-124 overexpressed Caki-1 and 786-O cells to cisplatin was evaluated using the Cell Counting Kit-8 (CCK-8) assay. And the effect of Capn4 and/or CNOT3 on the level of necroptosis in miR-124 overexpressed Caki-1 and 786-O cells to cisplatin was evaluated by flow cytometric analysis. Then, four groups of 786-0 cells (miR-124, miR-124+ Capn4, miR-124+ CNOT3, miR-124+ Capn4+ CNOT3) were inoculated into nude mice to observe the effect of cisplatin on tumor formation.
Results
miR-124 was found to be markedly elevated in renal cancer cells by cisplatin. Functionally, the overexpression of miR-124 reduced the sensitivity of renal cancer cells to cisplatin and CAPN4 was found to be a direct target of miR-124, which can negatively regulated CAPN4 expression. Moreover, ectopic expression of CAPN4 reversed the impairment of miR-124 on cisplatin-sensitivity and cisplatin-induced necroptosis. Mechanically, the present study revealed that CAPN4 could directly interact with CNOT3 and promote its degradation, and that the cisplatin-resistant phenotype was reversed by up-regulation of CNOT3.
Conclusions
Therefore, miR-124 is an important inhibitor in cisplatin-induced necroptosis, and the miR-124-CAPN4-CNOT3 signaling axis plays a critical role in the emergence of cisplatin-resistance.
Keywords: Renal cell carcinoma (RCC), cisplatin-resistance, miRNA-124, calpain small subunit 1 (Capn4), CCR4-NOT transcription complex subunit 3 (CNOT3)
Introduction
Renal cell carcinoma (RCC) is a common neoplasm in the urinary system, which ranks second in urinary tumor mortality, accounting for nearly 4% of all adult malignancies (1-3). Owing to its simplicity and rapid effect, chemotherapy is recognized as the most ideal treatment method (3-6). However, tumor insensitivity to chemotherapy and the gradual development of drug resistance has presented difficulties, which is a significant challenge for RCC treatment. Therefore, exploring the underlying mechanisms of drug resistance is critical to the development of effective treatment strategies.
MicroRNAs (miRNAs) are a kind of small non-coding RNA of 18–25 nt in length that can induce post-transcriptional inhibition by binding directly to the mRNA (messenger RNA) 3'-untranslational region (UTR) (7,8). It has been well documented that miRNAs play an important role in various important biological processes, including cell growth, differentiation, proliferation, and programmed cell death (9-11). In addition, numerous miRNAs are involved in RCC, including down-regulated miRNAs [miR-141 (12), miR-200c (13), miR-23b (14), miR-27b (15), and miR-135a (16)] and up-regulated miRNAs [miR-221 (17)], which have been found in RCC cells or tissues.
However, RCC resistance is not fully understood in terms of the underlying mechanisms of miRNAs regulation. MiR-124 has been reported to play an important role in the anticancer mechanism of various cancers, including glioma, ovarian cancer, gastric cancer, non-small cell lung cancer, and liver cancer (18-22). It is also thought to play an oncogenic role in RCC (23,24). Furthermore, miR-124 contributes to glioma drug-resistance by targeting R-RAS and N-RAS (25); however, its role and mechanism of action in the development of drug resistance in human RCC remains unclear. Recently, studies on therapy of RCC mainly focused on targeted therapy and immunotherapy, but rarely on chemotherapy. This study was the first to investigate the potential mechanism of miR-124 in cisplatin-resistance in RCC.
In this study, we demonstrated that miR-124 is an essential inhibitor in cisplatin-induced necroptosis, and the miR-124-CAPN4-CNOT3 signaling axis plays a key role in inducing cisplatin-resistance. Therefore, our data elucidates the novel role of miR-124 in inducing drug resistance in RCC and provides an effective therapeutic strategy for the treatment of RCC. We present the following article in accordance with the ARRIVE reporting checklist (available at https://dx.doi.org/10.21037/tau-21-777).
Methods
Cells and reagents
Human RCC cell lines, 786-O and Caki-1, were purchased from the ATCC (American Type Culture Collection) and cultured in RPMI (Roswell Park Memorial Institute) 1640 for 786-O or McCoy’s 5a for Caki-1 (ThermoFisher, USA), supplemented with 10% FBS (Fetal Bovine Serum, ThermoFisher, USA) and 2 mM L-glutamine (ThermoFisher, USA). Cisplatin was obtained from Selleck (Shanghai, China).
MiRNA microarray
A total RNA isolation kit of mirVana miRNA (Ambion, USA) was used to extract total RNA from cultured cells. cRNA (complementary RNA) per sample was synthesized and labeled in the buffer. The labeled cRNA fragments were used in the MicroRNA2.0 array (Affymetrix, USA) and hybridized in a GeneChip hybrid oven 640 (Affymetrix, USA) After cleaning and staining, the array was scanned using GeneChip Scanner 3000 (Affymetrix, USA). Partek GS 6.5 (Partek Inc., USA) was used to normalize and compare the miRNA expression levels of the samples. Average link hierarchy clustering was used for the data and Treeview was used to display the clustering results (http://rana.lbl.gov).
Cell proliferation and drug half maximal inhibitory concentration (IC50) determination
A CCK -8 (Cell Counting Kit-8) assay was used to evaluate cell proliferation. Briefly, RCC cells were seeded in 96-well plates (103 cells/well) and cultured for 24 h (denoted as day 0). The cells were then treated as indicated with four replications. At the specified time point, a 10 mL CCK-8 reagent solution (Donjido, Japan) was added to each well and incubated for 1 h. The optical density and absorbance at 450 nm were determined by a microplate reader (Bio-Tek, USA). The CCK-8 experiment was repeated at least three times. The data were used to generate drug effects or cell proliferation rate curves.
RNA isolation and qPCR
Total RNA from tissue and cell lines was extracted using TriZol (Thermo Fisher, USA). The quantification of miRNA levels was evaluated using a miRNA quantitative real-time PCR (qPCR) assay kit (GeneCopoeia, USA). To quantify mRNA levels of CAPN4 and CNOT3, a reverse transcription (RT) reaction was performed using a RevertAidTM H Minus First Strand cDNA Synthesis Kit (Fermentas, Canada) and performed using the ABI 7900 system (ABI, USA). U6 and GAPDH were used as normal controls for miRNA and mRNA quantification, respectively. The 2−ΔΔCt method to calculate the relative expression levels. The primers were shown in Table 1.
Table 1. The primers for qPCR.
| Primer name | Forward | Reverse |
|---|---|---|
| MiR-124 | 5’-ACACTCCAGCTGGGTAAGGCACGCGGTGA-3’ | 5’-TGGTGTCGTGGAGTCG-3’ |
| U6 | 5’GCTTCGGCAGCACATATACTAAAAT3’ | 5’CGCTTCACGAATTTGCGTGTCAT3’ |
| CAPN4 | 5’-TCCGACGCTACTCAGATGAAAGT-3’ | 5’-GATTTGTCCAGTGCCATCTTTGT-3’ |
| CNOT3 | 5’-AAGAAGAAAGGCGACAAGGA-3’ | 5’-GTCCAGCATTCGCAGGAT-3’ |
| GAPDH | 5’-TGACTTCAACAGCGACACCCA-3’ | 5’-CACCCTGTTGCTGTAGCCAAA-3’ |
MiRNA transfection
Both the negative control (NC) and miR-124 mimics (mimics) were purchased from GenePharma (Shanghai, China). Lipofectamine 2000 transfection reagent (Invitrogen, USA) was used to transfect cells with NC and simulators according to the manufacturer's protocol. Transfection efficiency was detected by qRT-PCR.
Western blotting
Total cell protein was extracted using RIPA (Radio-Immunoprecipitation Assay) buffer solution containing protease inhibitor PMSF (Phenylmethanesulfonyl fluoride), phosphatase inhibitor NaF (sodium fluoride), and Na3VO4 (sodium orthovanadate) (Roche, Switzerland). Western blotting detection was applied as described previously (26). The level of each protein film band was quantified by Quantity One (Bio-Rad, USA). The primary antibodies [CAPN4 (1:1,000) and CNOT3 (1:2,000)] were purchased from Abcam (UK). All protein expression experiments were repeated at least three times.
Luciferase reporter gene assays
The 3'-UTR of CAPN4 (miR-124 binding site) was synthesized and subcloned into a pGL3 reporter plasmid (Promega, USA). The plasmid was co-transfected with miR-124 mimics or miR-NC into 786-O cells for 48 h. 786-O cells were collected and the activity of luciferase was measured using the dual-luciferase reporter assay kit (Promega, USA).
Apoptosis and necroptosis assay
Annexin V/FITC (fluorescein isothiocyanate) Detection Kit (ThermoFisher, USA) was used to detect apoptosis and necroptosis. After transfection, the cells were incubated for 24 h, collected, and washed with phosphate-buffered saline (PBS) twice. FITC-Annexin V (5 mL) and PI (propidium iodide) (10 mL) were added and incubated for 15 min at 25 °C. The stained cells were counted by flow cytometry and FACScalibur (BD, USA) was used to analyze apoptosis (AnnexinV+/PI−) and necroptosis (AnnexinV−/PI+). The experiment was repeated three times.
Calpain activity
As mentioned earlier, calprotease activity was determined by using the fluorescent substrate N-succinyl-LLVD-AMC and casein enzyme spectrum.
Co-immunoprecipitation
RCC cells were collected 48 h after transfection, washed with PBS, and then scraped into the lysis buffer. After freezing, the cells were centrifuged for clarification. Total lysate (500–800 mol/L) was added based on protein G-sepharose (Amersham Pharmacia, Piscataway, NJ) at 10 mol/L and anti-mouse immunoglobulin G (IgG) or anti-CapN4/CNOT3 antibody at 5 mol/L. After tilting incubation at 4 °C for 4 h, the samples were repeatedly washed three times with 1% Nonidet P (NP)-40 buffer solution and then extracted with SDS (sodium dodecyl sulfate) sample buffer solution. The extract was analyzed on SDS-PAGE (polyacrylamide gel electrophoresis) with a gradient of 4–20% and transferred to cellulose nitrate for western blot analysis.
Xenograft model
The 786-O cells (1×106 cells suspended in PBS containing 50% matrix gel) were stably transfected with LV(lentiviral vector)-miR-124, LV-CAPN4, or LV-CNTO3 (purchased from Genelily Biotechnology Company, Shanghai) and subcutaneously injected into the lateral ventral of BALB/C nude mice (6 weeks old, n=5 in each group). Cisplatin was intraperitoneally administered once every 3 days. Tumor volume was recorded at 5–30 days after transplantation according to the following formula Tumor volume = length × width2/2. Tumor weight of transplanted mice was recorded 30 days after injection. This study was approved by the Ethics Committee of the Third Affiliated Hospital of Soochow University. The operation process of all experimental animals was strictly in accordance with the “Guide to the Care and Use of Experimental Animals” [2016].
Statistical analyses
SPSS 21.0 (IBM, Armonk, NY, USA) was used for statistical analysis. All statistical tests were bilateral. One-way analysis of variance (ANOVA) was used for comparison between groups. P<0.05 was considered statistically significant.
Results
MiR-124 contributed to cisplatin resistance in RCC cells
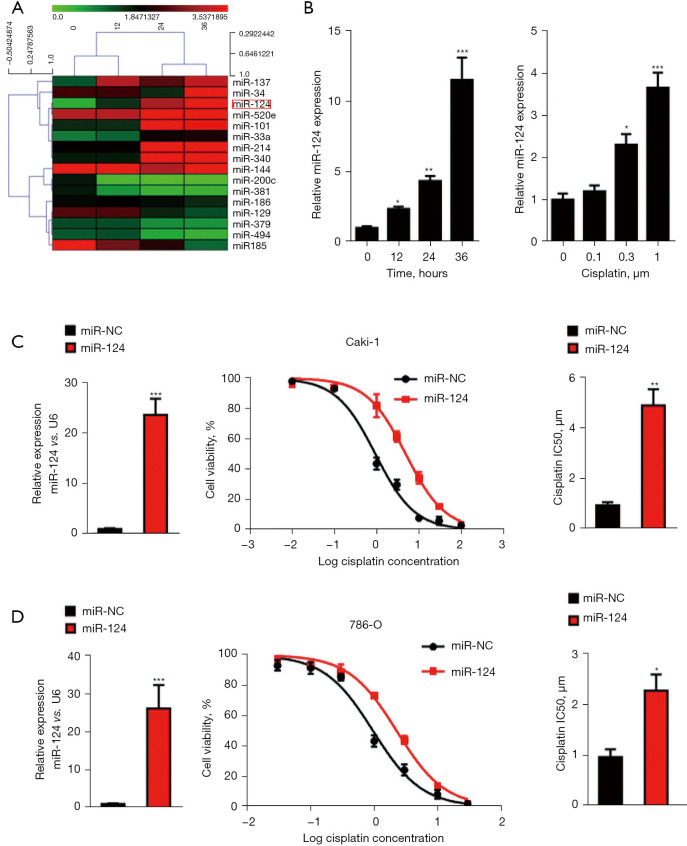
To explore the miRNA expression altered by chemotherapy drugs, the RCC cell line, 786-O, was treated with cisplatin (as shown in Figure 1A). Next, miRNA microarray analysis was performed. A significant increase in miR-124 was observed in cisplatin-induced changes in miRNAs (Figure 1A), which was confirmed by qPCR analysis (Figure 1B). To determine whether the increase in miR-124 was dependent on cisplatin concentration, we treated 786-O cells with different doses of cisplatin for 24 h and found that the expression level of miR-124 increased with the increase of cisplatin dose (Figure 1B).
Figure 1.
MiR-124 contributed to cisplatin resistance in renal cell carcinoma (RCC) cells. (A) 786-O cells were treated with 10 µM cisplatin for 0, 12, 24, and 36 h. The filtered miRNA array data were used for hierarchical clustering analysis. (B) The level of miR-124 was detected by real-time polymerase chain reaction (PCR). (C) The overexpression level of miR-124 in Caki-1 cells was measured by quantitative real-time PCR (qPCR). Cell viability and half maximal inhibitory concentration (IC50) value of Caki-1 cells to cisplatin were evaluated using the Cell Counting Kit-8 (CCK-8) assay. (D) The overexpression level of miR-124 in 786-O cells was measured by qPCR. Cell viability and IC50 value of 786-O cells to cisplatin were evaluated using the CCK-8 assay. *, P<0.05; **, P<0.01; and ***, P<0.001.
To evaluate the biological effect of miR-124 on IC50 value, we cultured regular Caki-1 and 786-O cells in vitro, transfected with miR-124 mimic or scramble, and treated them with several doses of cisplatin (as indicated in Figure 1C,1D) after 24 h of miRNA transfection. The expression of miR-124 was dramatically increased after miR-124 mimic transfection in Caki-1 and 786-O cells. The curves showed that the IC50 value of cisplatin in Caki-1 cells was 0.93±0.12 μM, which increased to 4.89±0.63 μM in the miR-124 mimic-transfected cells. A similar result was observed in 786-O cells. The IC50 value of cisplatin was 0.96±0.15 μM, which increased to 2.27±0.32 μM after miR-124 mimic transfection. These results suggested that miR-124 is contributed to cisplatin resistance in RCC cells.
MiR-124 post-transcriptionally inhibits CAPN4 expression by directly targeting its 3'-UTR
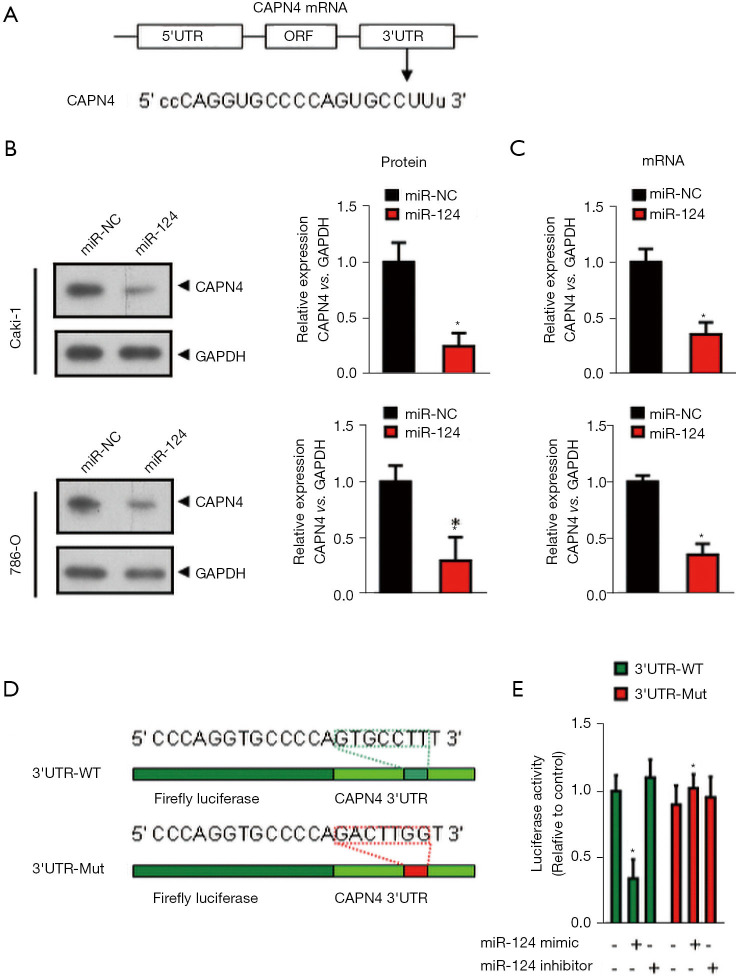
We used the TargetScan bioinformatics analysis program (http://www.targetscan.org) to identify the recognized miR-124 protein-coding gene targets. TargetScan miRNA target prediction revealed sthat the 3'-UTR of CAPN4 mRNA contains a potential binding site for miR-124 at nucleotide 427 (Figure 2A). To determine whether exogenous miR-124 inhibits CANP4 expression, early miR-124 or control miRNA (miRNA-NC) was transfected into Caki-1 and 786-O cells for 24 h. The expression of CAPN4 was analyzed by RT-PCR (Figure 2B) and western blotting (Figure 2C). The ectopic expression of miR-124 significantly inhibited the expression of CAPN4 mRNA (Figure 2B) and protein (Figure 2C).
Figure 2.
MiR-124 post-transcriptionally inhibits calpain small subunit 1 (CAPN4) expression by directly targeting its 3'-UTR. (A) Alignment of the miR-124 sequence with that of the 3'-UTR of CAPN4. (B)Western blot analysis revealed that the protein level of CAPN4 was decreased in the miR-124-overexpressed Caki-1 and 780-O cells. (C) The mRNA level of CAPN4 in the miR-124-overexpressed Caki-1 and 780-O cells was analyzed by real-time polymerase chain reaction (PCR) analysis. (D) CAPN4 3'-UTR constructs mutants at the predicted miR-124 site. (E) Dual-luciferase reporter activities of the pSiCheck2 vectors carrying the luciferase gene and a fragment of the CAPN4 3'-UTR containing the miR-124 binding sites in the control, miR-124 mimic, and miR-124 inhibitor groups. *, P<0.05.
Next, we analyzed whether the 3'-UTR of CAPN4 mRNA is a critical target of miR-124 in RCC. The predicted CAPN4 miRNA binding sequences [including wild-type (wt) and mutant (Mut) sequences] were cloned into a downstream region constructed by a luciferase reporter gene (psiCheck-CAPN4 3 'UT-wt/Mut, Figure 2D). These constructs were transiently transfected into 786-O cells in the presence of miR-124 mimic or miRNA-NC. As shown in Figure 2E, compared with miRNA-NC, miR-124 significantly decreased the luciferase activity of pSiCheck-CAPN4 3'UTR, but miR-124 could not reduce the luciferase activity of pSiCheck-CAPN4 3'UTR-Mut. These results indicate that miR-124 specifically binds CAPN4's 3-UTR nucleotide 427 and reduces the expression of CAPN4.
Overexpression of CAPN4 abolishes the miR-124-induced decrease in cisplatin sensitivity
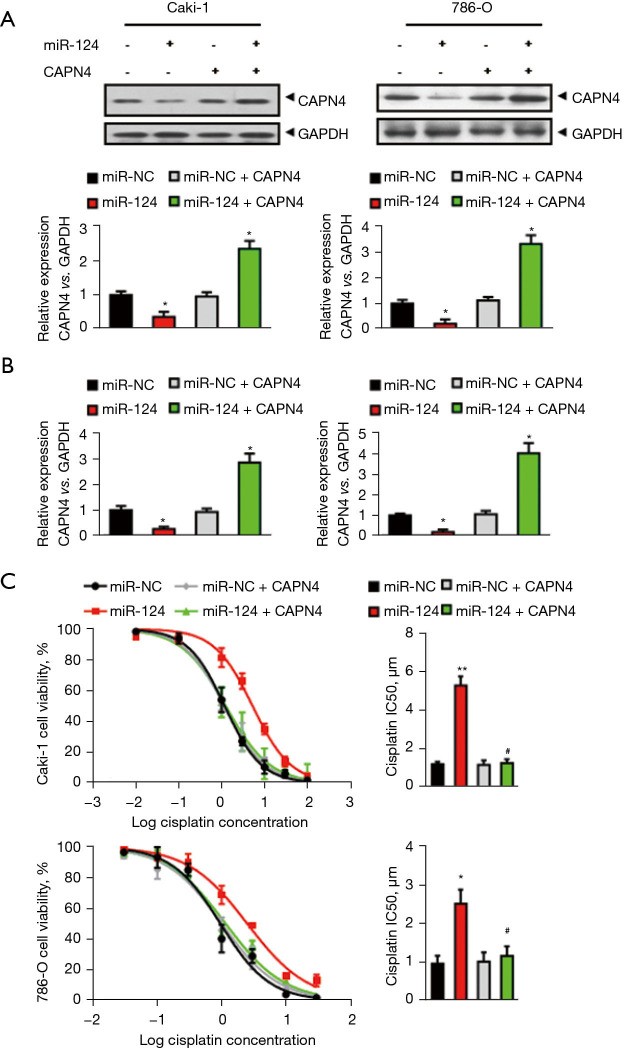
Since miR-124 can inhibit the expression of CAPN4 through direct inhibition of CAPN4 transcripts, we investigated whether increasing the expression of CAPN4 can reduce cisplatin resistance promoted by miR-124. Therefore, miR-124 was expressed in vitro in Caki-1 and 786-O cells and in CAPN4 encoding sequence, but was lacking a 3'-UTR or empty vector. CAPN4 overexpression was confirmed by western blotting (Figure 3A) and RT-PCR (Figure 3B). The curves indicated IC50 values of cisplatin in Caki-1 or 786-O cells (Figure 3C) showed that overexpression of CAPN4 completely abolishes the effect of miR-124 on the decreased cisplatin sensitivity. These results suggested that miR-124 reduced cisplatin sensitivity in RCC by targeting and inhibiting the expression of CAPN4.
Figure 3.
Overexpression of calpain small subunit 1 (CAPN4) abolishes the miR-124-induced decrease in cisplatin sensitivity. (A) The protein levels of CAPN4 in Caki-1 and 786-O cells was analyzed by western blot analysis. (B) The mRNA levels of CAPN4 in Caki-1 and 786-O cells was analyzed by real-time polymerase chain reaction (PCR) analysis. (C) The effect of miR-124 and/or CAPN4 on cell viability and the half maximal inhibitory concentration (IC50) value of Caki-1 and 786-O cells to cisplatin was evaluated using the Cell Counting Kit-8 (CCK-8) assay. *, P<0.05; **, P<0.01 vs. miR-NC group; #, P<0.05 vs. miR-124 group.
MiR-124 decreases the cisplatin-induced necroptosis but not apoptosis by repressing CAPN4
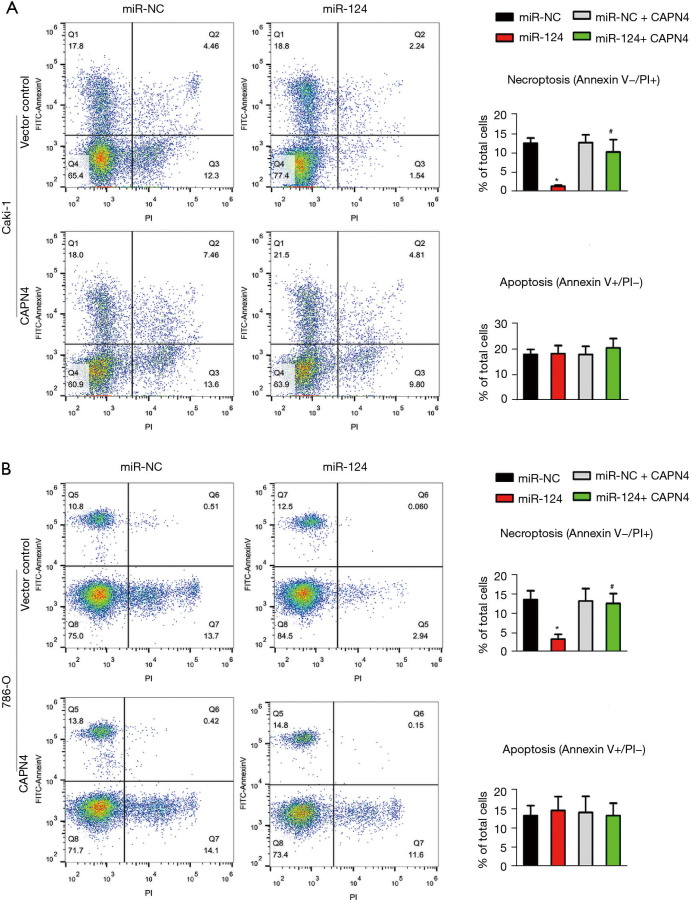
To analyze the effect of miR-124 and CAPN4 on apoptosis and necrosis, Caki-1 and 786-O cells were randomly divided into four groups: miR-NC+vector control group, miR-124+vector control group, miR-NC+CAPN4 group, and miR-124+CPAN4 group. The cells were stimulated by cisplatin for 24 h, and apoptosis and necrosis were detected by flow cytometry after Annexin V and PI staining. The two-dimensional quadrants of apoptosis and necrosis are shown in Figure 4A,4B. The results showed that miR-124 significantly reduced cell necrosis in Caki-1 (Figure 4A) and 786-O (Figure 4B) cells, while inhibition of CAPN4 reversed the cell necrosis induced by miR-124. The apoptotic level changed with the overexpression of miR-124 or CANP4. These results suggested that miR-124 reduces the sensitivity of cisplatin in RCC by inhibiting cisplatin-induced renal cell necrosis rather than apoptosis.
Figure 4.
MiR-124 decreases cisplatin-induced necroptosis but not apoptosis by repressing calpain small subunit 1 (CAPN4). The function of miR-124 and/or on the level of apoptosis and necroptosis in cisplatin-treated Caki-1 (A) and 786-O (B) cells were evaluated by flow cytometric analysis. *, P<0.05 vs. miR-NC group; #, P<0.05 vs. miR-124 group.
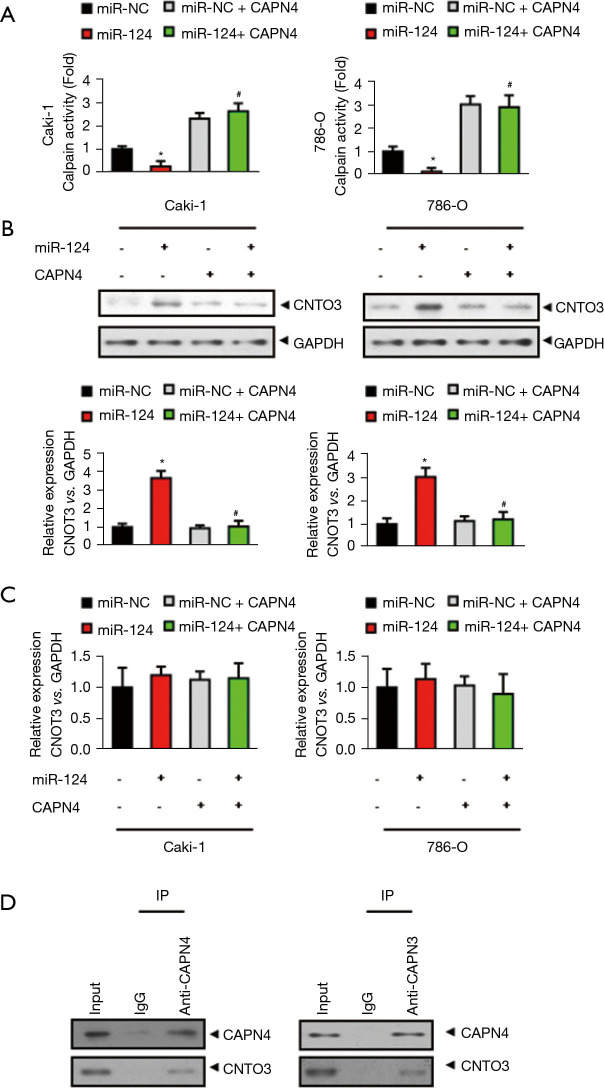
CAPN4 directly interacts with CNOT3 and promotes its degradation
Considering CAPN4 is a subunit of calpain and decreasing CAPN4 abolishes the activity of calpain (27), we hypothesized that the calpain activity and its degraded protein are critical to the miR-124-induced decrease in cisplatin sensitivity in RCC. The results showed that miR-124 markedly decreased the activity of calpain in Caki-1 and 786-O cells, which was significantly reversed after CAPN4 overexpression (Figure 5A).
Figure 5.
Calpain small subunit 1 (CAPN4) directly interacts with CCR4-NOT transcription complex subunit 3 (CNOT3) and promotes its degradation. (A) The calpain activity assay revealed that the decreased calpain activity in Caki-1 and 786-O cells were significantly increased after CAPN4 overexpression. (B) The protein levels of CNOT3 in miR-124-overexpressed Caki-1 and 786-O cells was analyzed by western blot analysis (C) The mRNA levels of CNOT3 in miR-124-overexpressed Caki-1 and 786-O cells was analyzed by real-time PCR(polymerase chain reaction) analysis. (D) Co-immunoprecipitation assays were performed in 786-O cells. Endogenous CAPN4 and CNOT3 were co-immunoprecipitated by anti-CAPN4 and anti CNOT3 antibodies. *, P<0.05 vs. miR-NC group; #, P<0.05 vs. miR-124 group.
As the previous results suggested that CNOT3 could be degraded due to calpain activation (28), we directly examined whether or not CNOT3 was a substrate of calpain in RCC. As shown in Figure 5B, miR-124 significantly increased the protein level of CNOT3 and this increase was completely abolished by CAPN4 overexpression. However, the mRNA level of CNOT3 was unchanged by miR-124 or CAPN4 overexpression (Figure 5C). These results suggest that miR-124/CAPN4 may affect the level of CNOT3 by increasing its degradation, but not inhibition of its expression.
We then analyzed the direct interaction between CANP4 and CNOT4 by co-immunoprecipitation. The result showed that both CANP4 and CNOT4 directly interacted with each other (Figure 5D). From what has been discussed above, these results reveal that CAPN4 could interact with CNOT3 and promote its degradation.
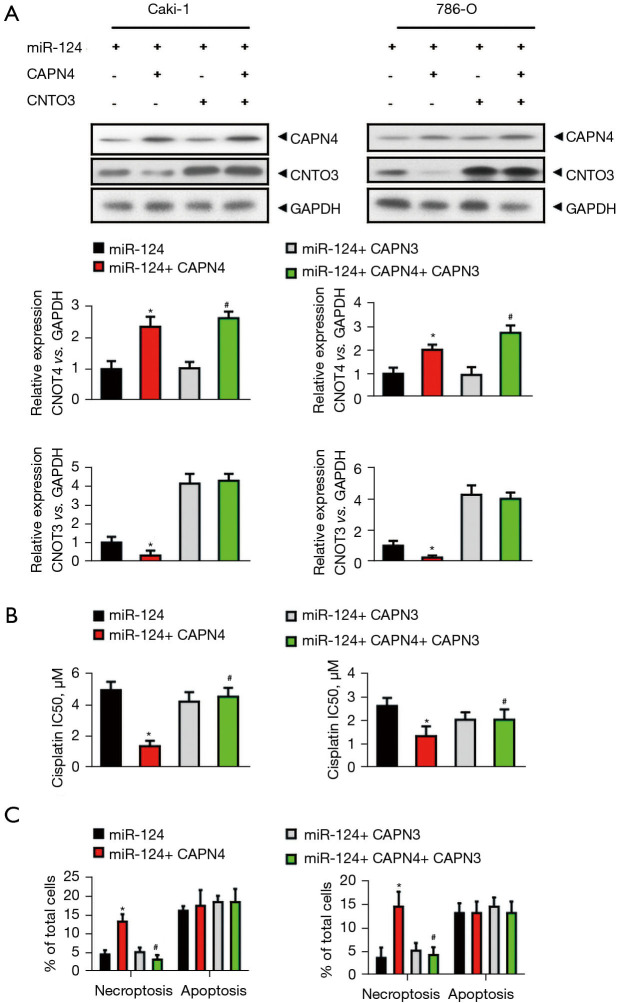
MiR-124 decreases the cisplatin sensitivity and cisplatin-induced necroptosis by inhibiting CAPN4-mediated CNOT3 degradation
To evaluate the specific role of CAPN4-mediated CNOT3 degradation in the miR-124-induced decrease in cisplatin sensitivity and cisplatin-induced necroptosis, Caki-1 and 786-O cells were divided into four groups: miR-124, miR-124+CAPN4, miR-124+CNOT3, and miR-124+CPAN4+CNOT3. The efficiency of CAPN4 and CNOT3 overexpression was confirmed by western blot and qPCR (Figure 6A). The cells were then stimulated by cisplatin for 1 day. After staining with Annexin V and PI, apoptosis and necroptosis were detected by flow cytometry. The results showed that CAPN4 significantly decreased the IC50 of cisplatin in miR-124-overexpressed Caki-1 and 786-O cells, and this decreasing was completely abolished by CNOT3 overexpression (Figure 6B).
Figure 6.
MiR-124 decreases cisplatin sensitivity and cisplatin-induced necroptosis by inhibiting calpain small subunit 1 (CAPN4)-mediated CCR4-NOT transcription complex subunit 3 (CNOT3) degradation. (A) Western blot and quantitative real-time PCR (qPCR) analysis revealed that the protein and mRNA levels of CAPN4 and CNOT3 in different groups in Caki-1 and 786-O cells (miR-124, miR-124+CAPN4, miR-124+CNOT3, miR-124+ CPAN4+ CNOT3). (B) The effect of CAPN4 and/or CNOT3 on cell viability and the IC50 (half maximal inhibitory concentration) value of miR-124-overexpressed Caki-1 and 786-O cells to cisplatin were evaluated using the Cell Counting Kit-8 (CCK-8) assay. (C) The effect of CAPN4 and/or CNOT3 on the level of necroptosis in miR-124-overexpressed Caki-1 and 786-O cells to cisplatin were evaluated by flow cytometric analysis. *, P<0.05 vs. miR-124 group; #, P<0.05 vs. miR-124+ CAPN4 group.
Similarly, CAPN4 markedly increased the level of necroptosis in miR-124-overexpressed Caki-1 and 786-O cells, which was completely abolished by CNOT3 overexpression (Figure 6C). However, CNOT3 alone showed no effect on the IC50 of cisplatin (Figure 6B) and the level of necroptosis (Figure 6C) in miR-124-overexpressed Caki-1 and 786-O cells. Nevertheless, the level of apoptosis was not changed with CAPN4 or CNOT3 overexpression (Figure 6C). In summary, these results reveal that miR-124 decreases the cisplatin sensitivity and cisplatin-induced necroptosis via inhibiting CAPN4-mediated CNOT3 degradation.
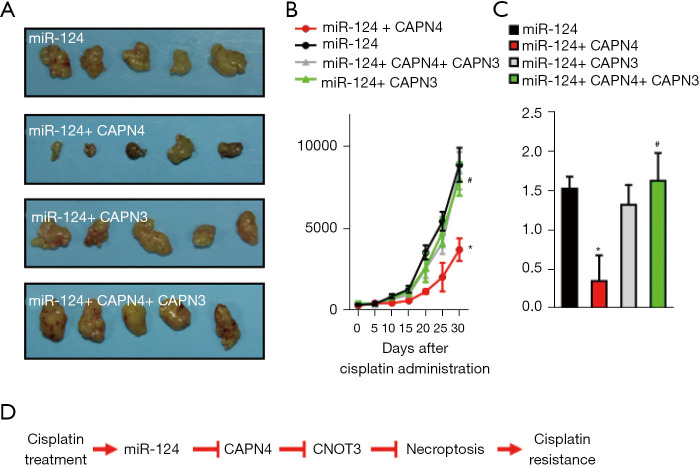
Effect of miR-124-CANP4-CNOT3 on cisplatin-resistance of 786-O cells in vivo
To assess the effects of miR-124-CANP4-CNOT3 on cisplatin resistance of 786-O cells in vivo, we generated xenograft mice with four groups of Caki-1 cells (miR-124, miR-124+CAPN4, miR-124+CNOT3, and miR-124+CPAN4+CNOT3). Consistently, tumors derived from miR-124+CAPN4-transfected Caki-1 cells grew more slowly than those derived from only miR-124-transfected cells after treatment with cisplatin (Figure 7A,7B). However, tumors derived from miR-124+CNOT3- or miR-124+CAPN4+CNOT3-transfected 786-O cells were similar to those derived from only miR-124-transfected cells (Figure 7A,7B). These data were further verified by end-point analysis of explanted tumor weight (Figure 7C). Overall, the in vivo results demonstrate that miR-124 attenuates cisplatin sensitivity via inhibiting CAPN4-mediated CNOT3 degradation (Figure 7D).
Figure 7.
Effect of miR-124-calpain small subunit 1 (CANP4)-CCR4-NOT transcription complex subunit 3 (CNOT3) on the cisplatin-resistance of 786-O cells in vivo. (A) Images of xenograft tumors formation from miR-124-overexpressed 786-O cells with or without CAPN4 and/or CNOT3. (B) The average volume of tumors is shown as mean ± SEM (standard error of mean). (C) Tumors from indicated xenograft mice were weighed immediately after removal and the average tumor weight is indicated as mean ± SEM. (D) A schematic model of cisplatin-induced miR-124 expression and its underlying mechanism in cisplatin resistance in RCC. *, P<0.05 vs. miR-124 group; #, P<0.05 vs. miR-124+CAPN4 group.
Discussion
Although bevacizumab/interferon-α, everolimus and pazopanib are used as first-line agents in advanced RCC, chemotherapy is still widely used in other carcinomas. Cisplatin is one of the most effective chemotherapy drugs, the underlying mechanism of cisplatin resistance has been studied for decades; however, there are no effective pharmacological operations to overcome this form of complex resistance (29-31). It has been widely reported that miRNAs regulate cisplatin resistance in various cancers (31-35), but has rarely been reported in RCC. Recently, miR-126 targeting SERPINE1 (36) and miR-148a targeting Rab14 (37) were identified to regulate cisplatin-resistance in RCC, and it was also reported that miR-124 represses FZD5 to attenuate P-glycoprotein-mediated doxorubicin- and vinblastine-resistance in RCC (38). Therefore, the purpose of this study was to investigate the role of miR-124 in cisplatin resistance in RCC. We used high-throughput miRNA microarray analysis to assess the role of miRNAs in cisplatin resistance in RCC cells. Our data showed that miR-124 was dramatically increased after cisplatin treatment, and that overexpression of miR-124 decreased cisplatin sensitivity. Although numerous descriptive studies have shown that miR-124 inhibits cell proliferation in a variety of tumors, the present study identified the role of miR-124 in regulating cisplatin resistance for the first time.
To understand the mechanism of miR-124 regulation of cisplatin resistance in RCC, we first found that miR-124 targets CAPN4’s 3’-UTR to inhibit its expression. CAPN4 is the regulatory subunit of the catalytic subunits calpain1 and calpain2 (39,40). We previously reported that CAPN4 is a promising biomarker for metastatic RCC (41), which activates and promotes RCC cell proliferation via the focal adhesion kinase (FAK) signaling pathway (26). It has also been reported that CAPN4 is a target of miR-99a and miR-491, and is involved in cisplatin resistance in gastric cancer (42). However, the mechanism of CAPN4 regulating RCC cisplatin-resistance is unknown.
In the current investigation, we discovered a new mechanism: miR-124 decreases the cisplatin sensitivity via inhibition of CAPN4. Furthermore, this study demonstrated that CAPN4 may directly interact with CNOT3 and affect its degradation, but not inhibit its expression. CNOT3 is an integral part of the CCR4 -NOT transcription complex and plays a general transcriptional regulatory role (43). It has been reported that CNOT3 acts as a new biomarker for molecular and prognostic classification of early-stage colon cancer (44). Moreover, CNOT3 targets specific mRNAs to prevent cell necrosis (45). Hence, the connection between necroptosis in cisplatin-resistant cells and miR-124-CAPN4-CNOT3-aixs excited our curiosity. Intriguingly, we identified that cisplatin-elevated miR-124 decreases RCC cells’ necroptosis by preserving CNOT3 from CAPN4 mediated-degradation. Necroptosis, a form cell death regulation independent of apoptotic mechanisms (46), has not received sufficient attention, and may become a novel effective method to eliminate tumor cells (47,48). Thus, its activation may be a promising option for inducing death of drug-resistant cancer cells.
Given that necrosis induces cell death by bypassing apoptosis retardation, efforts are being made to develop necrotic induction compounds for the treatment of drug-resistant cancers (49-52). Two attractive drug resistance strategies are currently being studied that directly address the causes of apoptosis blocking or alternative mechanisms of cell death, including induced necrosis (53). In this study, we discovered that miR-124 decreases cisplatin sensitivity via inhibition of cisplatin-induced necroptosis.
In conclusion, this study showed that miRNA-124 regulates the sensitivity of renal cancer cells to cisplatin-induced necroptosis by inhibiting CAPN4-mediated CNOT3 degradation. Understanding the regulation of cisplatin-resistant necrosis by miR-124 may be useful in investigating new targets for the treatment of RCC, and the miR-124-CAPN4-CNOT3 signaling axis may be a potential alternative therapeutic strategy for overcoming cisplatin-resistant in RCC.
Supplementary
The article’s supplementary files as
Acknowledgments
Funding: None.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study was approved by the Ethics Committee of the Third Affiliated Hospital of Soochow University. The operation process of all experimental animals was strictly in accordance with the “Guide to the Care and Use of Experimental Animals” [2016].
Reporting Checklist: The authors have completed the ARRIVE reporting checklist. Available at https://dx.doi.org/10.21037/tau-21-777
Data Sharing Statement: Available at https://dx.doi.org/10.21037/tau-21-777
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/tau-21-777). The authors have no conflicts of interest to declare.
(English Language Editor: A. Kassem)
References
- 1.Cairns P. Renal cell carcinoma. Cancer Biomark 2010;9:461-73. 10.3233/CBM-2011-0176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mollica V, Di Nunno V, Massari F. Pembrolizumab plus axitinib: a new treatment option for patients with metastatic renal cell carcinoma. Chin Clin Oncol 2019;8:S21. 10.21037/cco.2019.04.05 [DOI] [PubMed] [Google Scholar]
- 3.Adelaiye-Ogala R, Budka J, Damayanti NP, et al. EZH2 Modifies Sunitinib Resistance in Renal Cell Carcinoma by Kinome Reprogramming. Cancer Res 2017;77:6651-66. 10.1158/0008-5472.CAN-17-0899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang X, Cheng Y, Zhu Y, et al. Epigenetic silencing of ASPP1 confers 5-FU resistance in clear cell renal cell carcinoma by preventing p53 activation. Int J Cancer 2017;141:1422-33. 10.1002/ijc.30852 [DOI] [PubMed] [Google Scholar]
- 5.Robinson SP, Boult JKR, Vasudev NS, et al. Monitoring the Vascular Response and Resistance to Sunitinib in Renal Cell Carcinoma In Vivo with Susceptibility Contrast MRI. Cancer Res 2017;77:4127-34. 10.1158/0008-5472.CAN-17-0248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Naito S, Makhov P, Astsaturov I, et al. LDL cholesterol counteracts the antitumour effect of tyrosine kinase inhibitors against renal cell carcinoma. Br J Cancer 2017;116:1203-7. 10.1038/bjc.2017.77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 2005;120:15-20. 10.1016/j.cell.2004.12.035 [DOI] [PubMed] [Google Scholar]
- 8.Wu F, Zhi X, Xu R, et al. Exploration of microRNA profiles in human colostrum. Ann Transl Med 2020;8:1170. 10.21037/atm-20-5709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fan Y, Ma X, Li H, et al. miR-122 promotes metastasis of clear-cell renal cell carcinoma by downregulating Dicer. Int J Cancer 2018;142:547-60. 10.1002/ijc.31050 [DOI] [PubMed] [Google Scholar]
- 10.Li Y, Zhang D, Wang J. MicroRNA-373 promotes tumorigenesis of renal cell carcinoma in vitro and in vivo. Mol Med Rep 2017;16:7048-55. 10.3892/mmr.2017.7443 [DOI] [PubMed] [Google Scholar]
- 11.Zhai W, Sun Y, Guo C, et al. LncRNA-SARCC suppresses renal cell carcinoma (RCC) progression via altering the androgen receptor(AR)/miRNA-143-3p signals. Cell Death Differ 2017;24:1502-17. 10.1038/cdd.2017.74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen X, Wang X, Ruan A, et al. miR-141 is a key regulator of renal cell carcinoma proliferation and metastasis by controlling EphA2 expression. Clin Cancer Res 2014;20:2617-30. 10.1158/1078-0432.CCR-13-3224 [DOI] [PubMed] [Google Scholar]
- 13.Wang X, Chen X, Han W, et al. miR-200c Targets CDK2 and Suppresses Tumorigenesis in Renal Cell Carcinoma. Mol Cancer Res 2015;13:1567-77. 10.1158/1541-7786.MCR-15-0128 [DOI] [PubMed] [Google Scholar]
- 14.Liu W, Zabirnyk O, Wang H, et al. miR-23b targets proline oxidase, a novel tumor suppressor protein in renal cancer. Oncogene 2010;29:4914-24. 10.1038/onc.2010.237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mu W, Hu C, Zhang H, et al. miR-27b synergizes with anticancer drugs via p53 activation and CYP1B1 suppression. Cell Res 2015;25:477-95. 10.1038/cr.2015.23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yamada Y, Hidaka H, Seki N, et al. Tumor-suppressive microRNA-135a inhibits cancer cell proliferation by targeting the c-MYC oncogene in renal cell carcinoma. Cancer Sci 2013;104:304-12. 10.1111/cas.12072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Khella HWZ, Butz H, Ding Q, et al. miR-221/222 Are Involved in Response to Sunitinib Treatment in Metastatic Renal Cell Carcinoma. Mol Ther 2015;23:1748-58. 10.1038/mt.2015.129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xiao HJ, Ji Q, Yang L, et al. In vivo and in vitro effects of microRNA-124 on human gastric cancer by targeting JAG1 through the Notch signaling pathway. J Cell Biochem 2018;119:2520-34. 10.1002/jcb.26413 [DOI] [PubMed] [Google Scholar]
- 19.Shi B, Wang Y, Yin F. MALAT1/miR-124/Capn4 axis regulates proliferation, invasion and EMT in nasopharyngeal carcinoma cells. Cancer Biol Ther 2017;18:792-800. 10.1080/15384047.2017.1373214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li SL, Gao HL, Lv XK, et al. MicroRNA-124 inhibits cell invasion and epithelial-mesenchymal transition by directly repressing Snail2 in gastric cancer. Eur Rev Med Pharmacol Sci 2017;21:3389-96. [PubMed] [Google Scholar]
- 21.Cui M, Wang J, Li Q, et al. Long non-coding RNA HOXA11-AS functions as a competing endogenous RNA to regulate ROCK1 expression by sponging miR-124-3p in osteosarcoma. Biomed Pharmacother 2017;92:437-44. 10.1016/j.biopha.2017.05.081 [DOI] [PubMed] [Google Scholar]
- 22.Liu K, Yao H, Lei S, et al. The miR-124-p63 feedback loop modulates colorectal cancer growth. Oncotarget 2017;8:29101-15. 10.18632/oncotarget.16248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Butz H, Szabó PM, Khella HW, et al. miRNA-target network reveals miR-124as a key miRNA contributing to clear cell renal cell carcinoma aggressive behaviour by targeting CAV1 and FLOT1. Oncotarget 2015;6:12543-57. 10.18632/oncotarget.3815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gebauer K, Peters I, Dubrowinskaja N, et al. Hsa-mir-124-3 CpG island methylation is associated with advanced tumours and disease recurrence of patients with clear cell renal cell carcinoma. Br J Cancer 2013;108:131-8. 10.1038/bjc.2012.537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shi Z, Chen Q, Li C, et al. MiR-124 governs glioma growth and angiogenesis and enhances chemosensitivity by targeting R-Ras and N-Ras. Neuro Oncol 2014;16:1341-53. 10.1093/neuonc/nou084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shen J, Zhuang Q, Chen Z, et al. Capn4 induces human renal cancer cell proliferation by activating NF-κB signaling pathway through FAK phosphorylation. Int J Clin Exp Pathol 2017;10:7466-74. [PMC free article] [PubMed] [Google Scholar]
- 27.Shimada M, Greer PA, McMahon AP, et al. In vivo targeted deletion of calpain small subunit, Capn4, in cells of the osteoblast lineage impairs cell proliferation, differentiation, and bone formation. J Biol Chem 2008;283:21002-10. 10.1074/jbc.M710354200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zheng X, Dumitru R, Lackford BL, et al. Cnot1, Cnot2, and Cnot3 maintain mouse and human ESC identity and inhibit extraembryonic differentiation. Stem Cells 2012;30:910-22. 10.1002/stem.1070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu D, Luo M, Hu J, et al. Osteopontin enhances cisplatin resistance of human A549 lung cancer cells via stimulating the PI3K signaling pathway and upregulating ERCC1 expression. Transl Cancer Res 2020;9:3258-65. 10.21037/tcr.2020.03.60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Endo S, Xia S, Suyama M, et al. Synthesis of Potent and Selective Inhibitors of Aldo-Keto Reductase 1B10 and Their Efficacy against Proliferation, Metastasis, and Cisplatin Resistance of Lung Cancer Cells. J Med Chem 2017;60:8441-55. 10.1021/acs.jmedchem.7b00830 [DOI] [PubMed] [Google Scholar]
- 31.Li B, Wang W, Li Z, et al. MicroRNA-148a-3p enhances cisplatin cytotoxicity in gastric cancer through mitochondrial fission induction and cyto-protective autophagy suppression. Cancer Lett 2017;410:212-27. 10.1016/j.canlet.2017.09.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cheng MW, Shen ZT, Hu GY, et al. Prognostic Significance of microRNA-7 and its Roles in the Regulation of Cisplatin Resistance in Lung Adenocarcinoma. Cell Physiol Biochem 2017;42:660-72. 10.1159/000477884 [DOI] [PubMed] [Google Scholar]
- 33.Li X, Jin Y, Mu Z, et al. MicroRNA-146a-5p enhances cisplatin-induced apoptosis in ovarian cancer cells by targeting multiple anti-apoptotic genes. Int J Oncol 2017;51:327-35. 10.3892/ijo.2017.4023 [DOI] [PubMed] [Google Scholar]
- 34.Tian J, Xu YY, Li L, et al. MiR-490-3p sensitizes ovarian cancer cells to cisplatin by directly targeting ABCC2. Am J Transl Res 2017;9:1127-38. [PMC free article] [PubMed] [Google Scholar]
- 35.Zeng SG, Xie JH, Zeng QY, et al. MicroRNA-497-5p negatively regulates the proliferation and cisplatin resistance of non-small cell lung cancer cells by targeting YAP1 and TEAD1. Transl Cancer Res 2019;8:2470-80. 10.21037/tcr.2019.10.03 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu W, Chen H, Wong N, et al. Pseudohypoxia induced by miR-126 deactivation promotes migration and therapeutic resistance in renal cell carcinoma. Cancer Lett 2017;394:65-75. 10.1016/j.canlet.2017.02.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim EA, Kim TG, Sung EG, et al. miR-148a increases the sensitivity to cisplatin by targeting Rab14 in renal cancer cells. Int J Oncol 2017;50:984-92. 10.3892/ijo.2017.3851 [DOI] [PubMed] [Google Scholar]
- 38.Long QZ, Du YF, Liu XG, et al. miR-124 represses FZD5 to attenuate P-glycoprotein-mediated chemo-resistance in renal cell carcinoma. Tumour Biol 2015;36:7017-26. 10.1007/s13277-015-3369-3 [DOI] [PubMed] [Google Scholar]
- 39.Dai Z, Zhou SL, Zhou ZJ, et al. Capn4 contributes to tumour growth and metastasis of hepatocellular carcinoma by activation of the FAK-Src signalling pathways. J Pathol 2014;234:316-28. 10.1002/path.4395 [DOI] [PubMed] [Google Scholar]
- 40.Arthur JS, Greer PA, Elce JS. Structure of the mouse calpain small subunit gene. Biochim Biophys Acta 1998;1388:247-52. 10.1016/S0167-4838(98)00166-6 [DOI] [PubMed] [Google Scholar]
- 41.Zhuang Q, Qian X, Cao Y, et al. Capn4 mRNA level is correlated with tumour progression and clinical outcome in clear cell renal cell carcinoma. J Int Med Res 2014;42:282-91. 10.1177/0300060513505524 [DOI] [PubMed] [Google Scholar]
- 42.Zhang Y, Xu W, Ni P, et al. MiR-99a and MiR-491 Regulate Cisplatin Resistance in Human Gastric Cancer Cells by Targeting CAPNS1. Int J Biol Sci 2016;12:1437-47. 10.7150/ijbs.16529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Watanabe C, Morita M, Hayata T, et al. Stability of mRNA influences osteoporotic bone mass via CNOT3. Proc Natl Acad Sci U S A 2014;111:2692-7. 10.1073/pnas.1316932111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cejas P, Cavazza A, Yandava CN, et al. Transcriptional Regulator CNOT3 Defines an Aggressive Colorectal Cancer Subtype. Cancer Res 2017;77:766-79. 10.1158/0008-5472.CAN-16-1346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Suzuki T, Kikuguchi C, Sharma S, et al. CNOT3 suppression promotes necroptosis by stabilizing mRNAs for cell death-inducing proteins. Sci Rep 2015;5:14779. 10.1038/srep14779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rosentreter D, Funken D, Reifart J, et al. RIP1-Dependent Programmed Necrosis is Negatively Regulated by Caspases During Hepatic Ischemia-Reperfusion. Shock 2015;44:72-6. 10.1097/SHK.0000000000000371 [DOI] [PubMed] [Google Scholar]
- 47.Yang C, Li J, Yu L, et al. Regulation of RIP3 by the transcription factor Sp1 and the epigenetic regulator UHRF1 modulates cancer cell necroptosis. Cell Death Dis 2017;8:e3084. 10.1038/cddis.2017.483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cekay MJ, Roesler S, Frank T, et al. Smac mimetics and type II interferon synergistically induce necroptosis in various cancer cell lines. Cancer Lett 2017;410:228-37. 10.1016/j.canlet.2017.09.002 [DOI] [PubMed] [Google Scholar]
- 49.Steinhart L, Belz K, Fulda S. Smac mimetic and demethylating agents synergistically trigger cell death in acute myeloid leukemia cells and overcome apoptosis resistance by inducing necroptosis. Cell Death Dis 2013;4:e802. 10.1038/cddis.2013.320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Laukens B, Jennewein C, Schenk B, et al. Smac mimetic bypasses apoptosis resistance in FADD- or caspase-8-deficient cells by priming for tumor necrosis factor α-induced necroptosis. Neoplasia 2011;13:971-9. 10.1593/neo.11610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bonapace L, Bornhauser BC, Schmitz M, et al. Induction of autophagy-dependent necroptosis is required for childhood acute lymphoblastic leukemia cells to overcome glucocorticoid resistance. J Clin Invest 2010;120:1310-23. 10.1172/JCI39987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gerges S, Rohde K, Fulda S. Cotreatment with Smac mimetics and demethylating agents induces both apoptotic and necroptotic cell death pathways in acute lymphoblastic leukemia cells. Cancer Lett 2016;375:127-32. 10.1016/j.canlet.2016.02.040 [DOI] [PubMed] [Google Scholar]
- 53.Xu B, Xu M, Tian Y, et al. Matrine induces RIP3-dependent necroptosis in cholangiocarcinoma cells. Cell Death Discov 2017;3:16096. 10.1038/cddiscovery.2016.96 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The article’s supplementary files as