Abstract
Species of infraorder Gryllidea, or crickets, are useful invertebrate models for studying developmental biology and neuroscience. They have also attracted attention as alternative protein sources for human food and animal feed. Mitochondrial genomic information on related invertebrates, such as katydids, and locusts, has recently become available in attempt to clarify the controversial classification schemes, although robust phylogenetic relationships with emphasis on crickets remain elusive. Here, we report newly sequenced complete mitochondrial genomes of crickets to study their phylogeny, genomic rearrangements, and adaptive evolution. First, we conducted de novo assembly of mitochondrial genomes from eight cricket species and annotated protein-coding genes and transfer and ribosomal RNAs using automatic annotations and manual curation. Next, by combining newly described protein-coding genes with public data of the complete Gryllidea genomes and gene annotations, we performed phylogenetic analysis and found gene order rearrangements in several branches. We further analyzed genetic signatures of selection in ant-loving crickets (Myrmecophilidae), which are small wingless crickets that inhabit ant nests. Three distinct approaches revealed two positively selected sites in the cox1 gene in these crickets. Protein 3D structural analyses suggested that these selected sites could influence the interaction of respiratory complex proteins, conferring benefits to ant-loving crickets with a unique ecological niche and morphology. These findings enhance our understanding of the genetic basis of cricket evolution without relying on estimates based on a limited number of molecular markers.
Keywords: mitochondrial genome, Gryllidea, Myrmecophilidae, adaptation, phylogenetic study
Significance
Crickets are valuable invertebrate models for studies in developmental biology and neuroscience. Their robust phylogenetic relationships and phylogenetic-based genetic characteristics remain to be elucidated. In this study, we conducted phylogenetic analysis and identified gene rearrangements in several branches. Moreover, our analysis of the genetic signatures underlying the selection of ant-loving crickets (Myrmecophilidae) indicated adaptations to their ecological niches and evolution of morphologies characteristic in these crickets. These results expanded our understanding of the genetic basis of cricket evolution without depending on limited number of molecular markers.
Introduction
Gryllidea is one of the most species-rich infraorders in the order Orthoptera, comprising over 6,000 extant species. The biodiversity in Gryllidea has attracted research interest in acoustic communications, neurobiology, regenerative medicine, and alternative animal protein sources of recent date (Bentley and Hoy 1974; Forrest 1982; van Huis et al. 2013; Yoshimasa et al. 2015). The Orthoptera Species File (http://orthoptera.speciesfile.org) (Cigliano et al. 2018) lists true crickets, sword-tailed crickets, ant-loving crickets, and mole crickets under infraorder Gryllidae, representing diverse body sizes and ecological niches. It contains two superfamilies, Grylloidea (Laicharting 1781) and Gryllotalpoidea (Leach 1815). Gryllotalpoidea is further subdivided into two families, Gryllotalpidae (mole crickets) and Myrmecophilidae (ant-loving crickets). Dated phylogenetic analysis demonstrates that the divergence between Grylloidea and Gryllotalpoidea took place approximately 200 Ma in the Early Jurassic (Zhou et al. 2017). Recent phylogenetic studies have presented a highly resolved phylogenetic classification of crickets using a wide taxonomic and character sample set, which relied on a few nuclear and mitochondrial gene segments (Chintauan et al. 2016). Another approach using whole mitochondrial genome sequences (mtgenomes), which generally provide sufficient resolution for reconstructing a robust phylogeny, was toward the phylogenetic analysis of Ensifera, including crickets, katydids, wetas, and related species, although the majority of the studied samples were from katydids, and few cricket samples were included (Zhou et al. 2017).
Mtgenome-based phylogenetic analysis likely better resolves the phylogenetic relationship than that based on a few nuclear and/or mitochondrial genes and provides valuable information for species-tree estimation (Mandal et al. 2014; Meiklejohn et al. 2014). Insect mtgenomes are generally highly conserved and possess 37 genes, namely 13 protein-coding, 22 transfer RNA (tRNA), and two ribosomal RNA (rRNA) genes (Cameron 2014a). The mitochondrial genes are encoded in both strands: the strand with most of the genes is named the “majority” or H (heavy) strand, whereas the other is the “minor” or L (light) strand (Cameron 2014a). According to National Center for Biotechnology Information (NCBI) Taxonomy database, Gryllidea, which occupies a large group of Ensifera, still has a relatively small number of publicly available mtgenomes.
Insects can be found in diverse and harsh ecological niches with different morphological traits. For instance, in a hypoxic environment, insects experience low oxygen supply and, thus adapt their body size and increase the function of their tracheal respiratory system and metabolic rates (Zhang et al. 2013; Li et al. 2018). Grasshoppers and locusts in high-altitude environments with low atmospheric oxygen levels functionally evolved their mtgenomes to confer robustness against hypoxia (Zhang et al. 2013; Li et al. 2018). Body size is also known to be affected by environmental oxygen levels, as in Tibetan locusts (Ding et al. 2018). These findings suggest that mtgenomes may undergo selective pressure in some insects with peculiar ecological and size traits.
Among crickets with diverse ecological niches and morphologies, ant-loving crickets (genus Myrmecophilus Berthold, 1827) demonstrate noticeable traits. They are all small and wingless crickets that live in subterranean ant nests, representing the smallest body size among all crickets. They are kleptoparasitic on their host ants, obtaining food resources from the ant hosts, either by stealing directly from ant nests or via mouth-to-mouth feeding by ants (Wetter and Hugen 2008). In addition, Myrmecophilus crickets can chemically disguise themselves as their host ants by matching their cuticular hydrocarbon profile to that of their ant hosts, thereby enabling them to escape from ant attacks (Akino et al. 1996). In addition, Myrmecophilus crickets scrape cuticular hydrocarbons from their host ants (Akino et al. 1996). Thus, Myrmecophilus crickets are considered to have unique characteristics, such as body size, habitat, nutrient type, and feeding opportunities, compared with other Gryllidea species.
Regardless of these distinctive characteristics in Myrmecophilus crickets, the evolutionary relationships in Gryllidea remain controversial, which might be in part due to paucity of the genomic data of these species. In order to provide a robust evidence for the evolutionary history in Gryllidea, we have newly described mtgenomes of two Myrmecophilus spp., Myrmecophilus kubotai and Myrmecophilus sp., as well as mtgenomes of Teleogryllus emma, Teleogryllus occipitalis, Gryllus bimaculatus, Acheta domesticus, Gryllodes sp., and Tarbinskiellus sp. Using these newly described mtgenomes and those publicly available as of February 2020, we inferred a highly robust mtgenome-based phylogeny in Gryllidea. Based on these data sets, we identified accurate mitochondrial gene annotations, gene order rearrangement, and evolutionary signature in Myrmecophilus crickets.
Results and Discussion
General Characteristics of the Cricket Mtgenomes
We sequenced the genomic DNA of T. emma, T. occipitalis, G. bimaculatus, A. domesticus, Gryllodes sp., Tarbinskiellus sp., M. kubotai, and Myrmecophilus sp., with short-read next generation sequencer and performed de novo assembly of their mtgenomes. The new mtgenomes contained 37 typical insect mitochondrial genes, including 13 protein-coding, 22 tRNA, and two rRNA genes, as previously reported in cricket mtgenomes (Kataoka et al. 2020) (supplementary tables 1 and 2, Supplementary Material online). The length of complete mtgenomes ranges from 15,120 bp in Truljalia hibinonis to 16,589 bp in Ornebius kanetataki (supplementary table 1, Supplementary Material online). The longest mtgenome in O. kanetataki was due to its relatively long control region. The length of the mtgenomes newly reported herein was within this range. The longest intergenic spacer was found to be 322 bp between tRNA-Gln and tRNA-Met in Homoeoxipha nigripes (supplementary table 3, Supplementary Material online). The longest overlap between adjacent genes was 58 bp, found between tRNA-Leu and 16S rRNA in Gryllodes sp. (supplementary tables 2 and 3, Supplementary Material online).
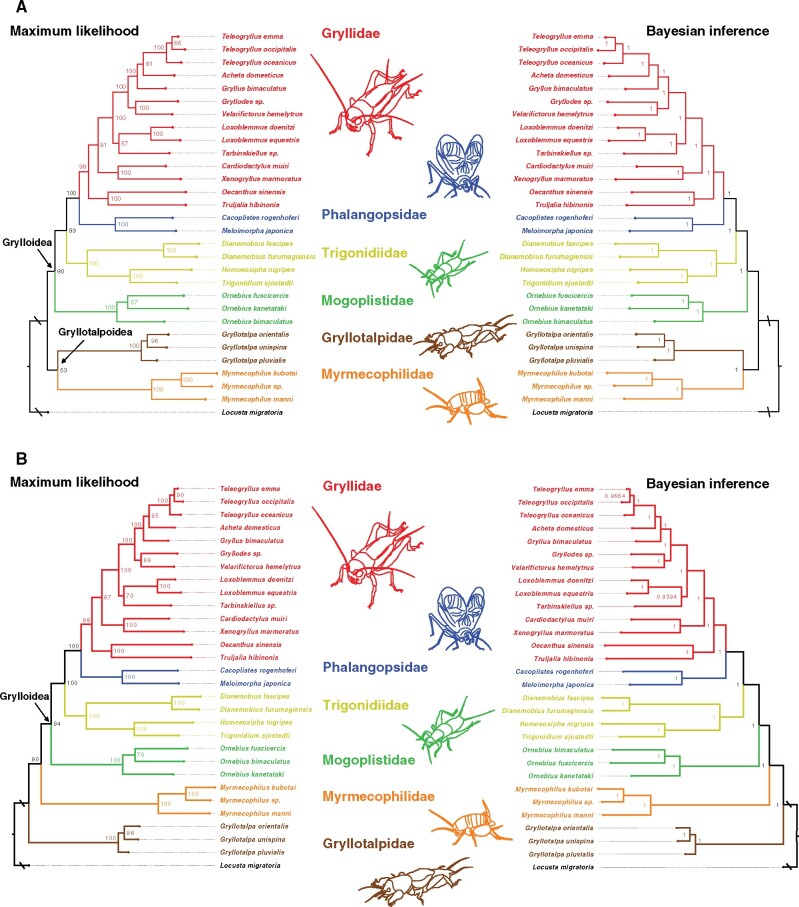
We then analyzed the molecular phylogeny of crickets using complete mtgenomes. To obtain the phylogenetic tree in Gryllidea, we used the concatenated nucleotide sequences of 13 protein-coding genes (PCGs) from 29 crickets, which also included previously published mtgenomes of crickets as well as an outgroup locust, Locusta migratoria. It should be noted that we found mistakes in the annotations of Teleogryllus oceanicus and corrected them using public short-read data (supplementary fig. 1, Supplementary Material online). We analyzed the cricket phylogenies using two independent approaches, maximum likelihood (ML) and Bayesian inference (BI) based on partitioned and nonpartitioned sequence data. In the nonpartitioned data set, both ML and BI methods generated the identical topological trees with the monophyly of Gryllotalpoidea (Gryllotalpidae+Myrmecophilidae) as sister clade of the remaining groups ((Mogoplistidae (Trigonidiidae (Phalangopsidae+Gryllidae)))) (fig. 1A), which is consistent with a previous study that utilized gene segments derived from mitochondrial and nuclear genomes to infer a phylogenetic tree (Fenn et al. 2008; Song et al. 2015; Chintauan‐Marquier et al. 2016). However, the tree topologies with partitioned sequence data were different, which supported the following relationships: (Gryllotalpidae (Myrmecophilidae (Mogoplistidae (Trigonidiidae (Phalangopsidae+Gryllidae))))) (fig. 1B and supplementary table 8, Supplementary Material online). In the ML tree, the partitioning strategy slightly increased the bootstrap values. Just as our analysis inferred two conflicting topological trees, a recent study using large scale transcriptomic data have not inferred a consistent phylogenetic position for Gryllotalpoidea either (Song et al. 2020), in which the authors have pointed out the small sample size in Gryllotalpoidea. Our data set provided highly resolved phylogeny of Gryllidea compared with the study using whole mtgenomes sequences (Zhou et al. 2017), although there is a need for further increase in the sample size with sequence data of nuclear genes.
Fig. 1.
Phylogenetic tree of Gryllidea based on the concatenated nucleotide sequences of 13 protein-coding genes using both maximum likelihood (left) and Bayesian inference (right) analyses based on nonpartitioned (A) and partitioned (B) sequence data. Numbers on branches are Bayesian posterior probabilities and bootstrap values. Locusta migratoria was set as the outgroup in the analysis.
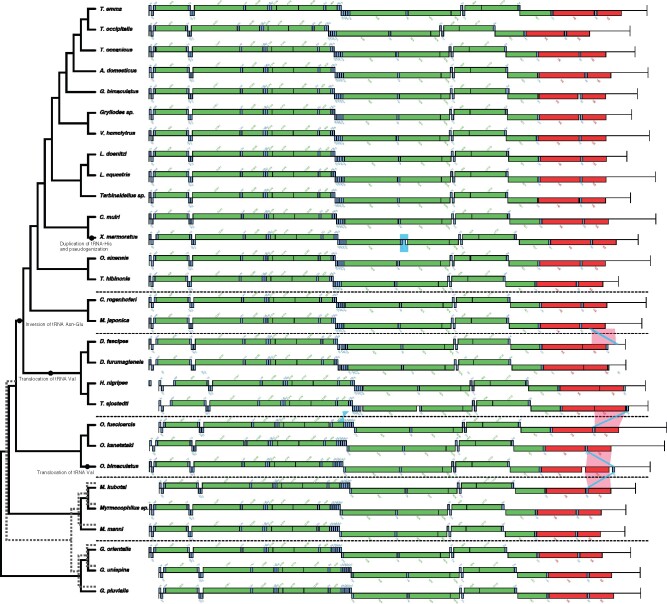
Changes in mitochondrial gene order have been considered to be phylogenetic markers termed “rare genomic changes,” providing valuable insights into phylogenetic relationships that are not obtained from nucleotide substitutions in DNA sequence alone (Boore 1999; Rokas and Holland 2000). We found several gene order rearrangements in the Gryllidea mtgenomes (fig. 2). Mogoplistidae, Gryllotalpidae, and Myrmecophilidae harbored the ARNSEF (Ala, Arg, Asn, Ser, Glu, Phe) tRNA clusters between nad3 and nad5 genes, whereas other subfamilies, including Gryllidae, Phalangopsidae, and Trigonidiidae, showed a transposition in this gene block flanked by tRNA-Glu and tRNA-Asn (ARESNF), indicating that this transposition event occurred in the ancestor of Gryllidae, Phalangopsidae, and Trigonidiidae. Furthermore, translocations between tRNA-Val and 12S rRNA happened twice in Gryllidea; once in the ancestor of Trigonidiidae, and the other before or after the divergence of Ornebius bimaculatus from the ancestor of Ornebiusfuscicercis and O. kanetataki. In addition, Xenogryllus mamoratus probably experienced a duplication of tRNA-His followed by pseudogenization.
Fig. 2.
Mitochondrial gene rearrangements in Gryllidea. Arrangements of mitochondrial genes are shown for protein-coding (green boxes), tRNA (blue boxes), and rRNA (red boxes) genes. tRNA genes are indicated by the three-letter abbreviations for their corresponding amino acids. Genes on the H-strand and the L-strand are shown in the upper and lower positions, respectively. Dotted line indicates the topological tree inferred by nonpartitioning method, whereas solid line is the one inferred by partitioning.
Protein-Coding and Transfer RNA Genes
In many of the mtgenomes, the H-strand encoded 20 genes (9 PCGs and 11 tRNAs), whereas the remaining genes were located on the L-strand (supplementary table 3, Supplementary Material online). The AT content of the 13 PCGs was 73.3% on average, with the highest AT content (78.1%) in Oecanthus sinensis and the lowest (69.6%) in Myrmecophilus manni. Myrmecophilus crickets showed relatively higher GC contents (30.17%, 29.17%, and 30.40%) and lower GC skew (−0.12, −0.13, and −0.14) than all other Gryllidea species, with an average of 26.7% and −0.10, respectively (supplementary table 4, Supplementary Material online). The average AT content of the whole mtgenomes was 73.3%, with the highest AT content (77.6%) in Homoeoxipha nigripes and the lowest (70.0%) in M.kubotai. Myrmecophilus crickets showed relatively high GC contents (30.04%, 29.13%, and 29.82%) (supplementary table 4, Supplementary Material online), similar to those in the PCGs. These results indicated that Myrmecophilidae showed a biased nucleotide composition in their mtgenomes.
We also found that all PCGs had a typical start codon (ATN), except for cox1, which started with TCG. Nonstandard translational start sites for cox1 have also been proposed for number of insects (Sheffield et al. 2008). In a previous study, annotation of the start codon in cox1 was conducted by identifying the most conserved sites downstream of the flanking tRNA with minimized intergenic space and gene overlaps (Cameron 2014a). Thus, by aligning the region encompassing tRNA-Tyr and cox1 from all known Ensifera mtgenomes, conceptual start codons for the cox1 gene were accurately predicted. The first nonoverlapping in-frame codon in cox1 is well conserved throughout Ensifera, and it is possible to choose glutamine (CAA or CAG) as the start codon (supplementary fig. 2, Supplementary Material online). Therefore, it is suggested that glutamine is functionally constrained as the start codon for the cox1 genes in Ensifera, including crickets.
Most of the PCGs finished with the termination codon TAA or TAG, and some genes were terminated with T or TA. The use of such incomplete stop codons (i.e., T or TA) frequently occurred in Gryllidea mtgenomes, which was due to the overlap between the ends of PCGs and the abutting tRNAs. It has been hypothesized that a complete stop codon (TAA) is built through posttranscriptional polyadenylation (Ojala et al. 1981). The presence of partial stop codons in insect mtgenomes has been documented many times (Beard et al. 1993). Complete stop codons were often TAA rather than TAG in the mtgenomes of the current study, which are consistent with the pattern found in mtgenomes published previously (Sheffield et al. 2008).
The Gryllidea mtgenomes analyzed in the present study contained all 22 typical tRNAs. These tRNAs can be folded into typical cloverleaf structures except tRNA-Ser (AGN), which assumed disrupted D-arm structures, as previously described in a broad range of metazoan mtgenomes (Cameron 2014b). Comparative analysis of all tRNA-Ser (AGN) genes in Gryllidea revealed that their secondary structures were similar to others in the same subfamily (supplementary fig. 3, Supplementary Material online). For instance, tRNA-Ser (AGN) structures predicted in Mogoplistidae species showed complete loss of the D-arm, whereas those of the other subfamilies retained partial structures of the D-arm (supplementary fig. 3, Supplementary Material online).
Evolutionary Analysis of PCGs in Ant-Loving Crickets
The morphological and ecological differences between ant-loving crickets and other Gryllidea species may influence their respective metabolic requirements. The characteristic traits of ant-loving crickets, such as their small body size, habitat, nutrient type, and feeding opportunities, could be linked to the molecular evolution of their nuclear and mitochondrial genomes. Our sequencing efforts have provided an increased number of mtgenomes from ant-loving crickets, enabling evolutionary analysis of these insects. Thus, we investigated the potential evolutionary adaptation of 13 PCGs in the mtgenomes of ant-loving crickets based on the phylogenetic tree with partitioning. First, to test the role of selection on the Myrmecophilidae branch, we performed a codeML branch model analysis. The free-ratio model fitted our data significantly better than the null hypothesis (one-ratio model) for all 13 PCGs, suggesting that crickets show different evolutionary rates in mitochondrial genes. The values of dN, dS, and ω for each branch are listed in supplementary table 5, Supplementary Material online. The two-ratio model, used to calculate selective pressures in Myrmecophilidae, significantly fitted for three genes, cox1 and nad1, nad4 when the Myrmecophilidae was labeled as the foreground branch (fig. 3). The nad6 gene showed the highest ω values in Myrmecophilidae compared with other genes. However, the likelihood ratio test (LRT) was unable to indicate the two-ratio model as the best fitting model to explain the differences in ω values for this gene (P = 0.20) (supplementary table 5, Supplementary Material online). To calculate selective pressures acting only on Myrmecophilidae, we next performed an analysis using branch-site models. As a result, cox1 was found to be positively selected in ant-loving crickets, where LRT of the branch-site model (MA vs. MA null) was statistically significant (supplementary table 5, Supplementary Material online). Positively selected sites of cox1 were evaluated using Bayes Empirical Bayes analysis with posterior probabilities of 0.95 or more. As a result, two sites were found to be under selective pressure in cox1 (supplementary table 6, Supplementary Material online). Furthermore, evidence of positive selection was also examined using the fixed-effect likelihood (FEL) approach in the cox1 gene. FEL identified 11 positively selected codons including the two sites in cox1 of ant-loving crickets (FEL significance <0.01). Subsequently, we considered that the two sites in the cox1 gene (positions 484 and 493), which were consistently identified using both methods, underwent positive selection in the mtgenomes of ant-loving crickets (supplementary table 7, Supplementary Material online). We also tested the potential evolutionary adaptation of 13 PCGs using the phylogenetic tree without partitioning (supplementary fig. 4 and tables 5–8, Supplementary Material online). Note that, regardless of the tree with partition or without partition, there was no change in the general interpretation in the evolutionary analysis, except for minor differences in values such as ω.
Fig. 3.
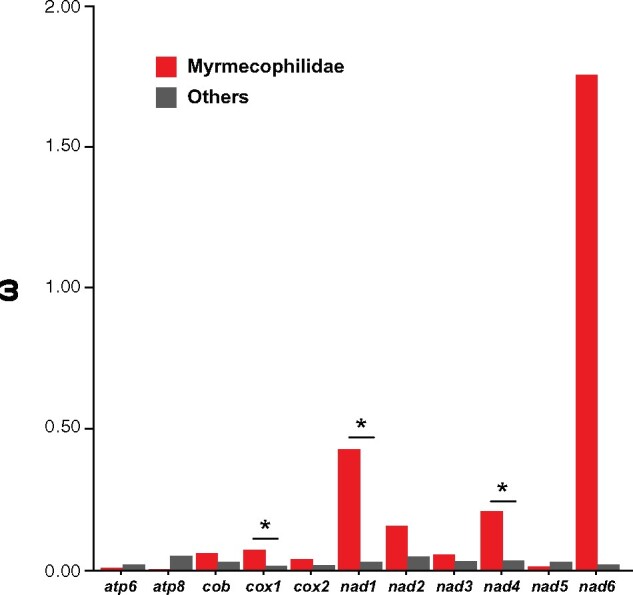
Natural selection strength and the ratio of nonsynonymous to synonymous substitutions (ω) calculated with the two-ratio model in codeML for the 13 protein-coding mitochondrial genes of Myrmecophilidae and other crickets. Genes that significantly fit the two-ratio model are marked with asterisks.
Structural Insights into Positively Selected Sites of Mitochondrial Proteins in Ant-Loving Crickets
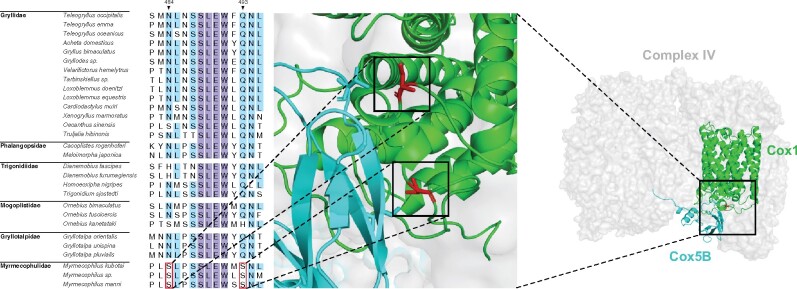
To provide structural insights into the positively selected sites in Myrmecophilus mitochondrial PCGs, we mapped them to the 3D structures of Myrmecophilus cricket cox1 protein predicted using homology modeling. Serine residues under positive selection in ant-loving crickets were in proximity to the neighboring subunit cox5B (fig. 4). Cytochrome c oxidase, or complex IV, has been reported to be functionally modulated as an adaptive response to high-altitude hypoxia in a Tibetan migratory locust (Zhang et al. 2013). Mammalian pika, which also inhabits the Qinghai-Tibetan plateau and is used as an animal model for hypoxia adaptation studies, was found to possess pika-specific amino acid substitutions in cox1, implying possible conformational and functional modifications of cytochrome c oxidase (Yongjun et al. 2008). In the present study, statistically significant changes in Asn or Gln to Ser were detected in Myrmecophilidae cox1 (i.e., N484S and Q493S). The Asn residue has a low pKa of ∼3.9, whereas the Ser residue has a high pKa (∼13), which possibly influence the interaction between Cox1 and Cox5B (fig. 4). Considering that ant-loving crickets inhabit ant nests where oxygen levels can be low (Bollazzi et al. 2012), it would be intriguing to consider that these substitutions in cox1 of ant-loving crickets might alter the interaction between cox1 and cox5B, thereby contributing to hypoxic adaptation.
Fig. 4.
Alignment of partial sequences of cox1 undergoing positive selection in Gryllidea and 3D structures of proteins of respiratory complexes. Conserved or partially conserved residues are indicated by blue and light blue highlights, respectively. Positively selected sites of ant-loving crickets are highlighted by red boxes. Structural model of cox1 (green) with positively selected amino acid residues cox5B (light blue), and complex IV (light gray). In this model, amino acid residues undergoing positive selection are highlighted in red with the side chains by stick models.
Conclusion
Our study represents the first comprehensive comparative analysis of orthopteran mtgenomes with an emphasis on Gryllidea. We found that all species studied here possess 13 PCGs, 22 tRNAs, and two rRNAs, similar to that previously identified from common ancestral insect mtgenomes (Cameron 2014b). We found that the phylogenetic relationship inferred from the concatenated mitochondrial PCGs supported a monophyletic superfamily (Grylloidea and Gryllotalpoidea) with nonpartitioned data set, but not with partitioned dataset. Evolutionary and structural analyses of mtgenomes of ant-loving crickets revealed positive selection in cox1 gene and provided insights into their characteristic biology. Our study provides genetic resources for mitochondrial genomes in crickets and advances the understanding of the evolution of insect mtgenomes.
Materials and Methods
The full Materials and Methods section is in supplementary materials and methods, Supplementary Material online.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
We appreciate the kind support for the collection of giant crickets Tarbinskiellus sp. from Mr. Tan Viet Tran and Gryllodes sp. from Mr. Kosuke Goto. This study was supported by the Cabinet Office, Government of Japan, Cross-ministerial Moonshot Agriculture, Forestry and Fisheries Research and Development Program, “Technologies for Smart Bio-industry and Agriculture” (BRAIN) (JPJ009237).
Data Availability
The mitochondrial genomes for this article have been deposited at GenBank under the accession numbers MZ440652 (Teleogryllus occipitalis, BioSample SAMN19773260), MZ440653 (Teleogryllus emma, BioSample SAMN19773259), MZ440654 (Acheta domesticus, BioSample SAMN19773262), MZ440655 (Tarbinskiellus sp., BioSample SAMN19773264), MZ440656 (Gryllus bimaculatus, BioSample SAMN19773261), MZ440657 (Gryllodes sp., BioSample SAMN19773263), MZ440658 (Myrmecophilus kubotai, BioSample SAMN19773265), and MZ440659 (Myrmecophilus sp., BioSample SAMN19773266) under the BioProject PRJNA739105.
Literature Cited
- Akino T, Mochizuki R, Morimoto M, Yamaoka R.. 1996. Chemical camouflage of myrmecophilous cricket Myrmecophilus sp. to be integrated with several ant species. Jpn J Appl Entomol Z. 40(1):39–46. [Google Scholar]
- Beard C, Hamm DM, Collins F.. 1993. The mitochondrial genome of the mosquito Anopheles gambiae: DNA sequence, genome organization, and comparisons with mitochondrial sequences of other insects. Insect Mol Biol. 2(2):103–124. [DOI] [PubMed] [Google Scholar]
- Bentley D, Hoy R.. 1974. The neurobiology of cricket song. Sci Am. 231(2):34–45. [DOI] [PubMed] [Google Scholar]
- Bollazzi M, Forti L, Roces F.. 2012. Ventilation of the giant nests of Atta leaf-cutting ants: does underground circulating air enter the fungus chambers? Insect Soc. 59(4):487–498. [Google Scholar]
- Boore JL. 1999. Animal mitochondrial genomes. Nucleic Acids Res. 27(8):1767–1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron SL. 2014a. How to sequence and annotate insect mitochondrial genomes for systematic and comparative genomics research. Syst Entomol. 39(3):400–411. [Google Scholar]
- Cameron SL. 2014b. Insect mitochondrial genomics: implications for evolution and phylogeny. Annu Rev Entomol. 59:95–117. [DOI] [PubMed] [Google Scholar]
- Chintauan‐Marquier IC, et al. 2016. Laying the foundations of evolutionary and systematic studies in crickets (Insecta, Orthoptera): a multilocus phylogenetic analysis. Cladistics 32:54–81. [DOI] [PubMed] [Google Scholar]
- Cigliano MM, Braun H, Eades DC, Otte D.. 2018. Orthoptera species file online. Version 5.0/5.0 [WWW document]. Available from: http://orthoptera.speciesfile.org (accessed 2021 Apr 21).
- Ding D, et al. 2018. Genetic variation in PTPN1 contributes to metabolic adaptation to high-altitude hypoxia in Tibetan migratory locusts. Nat Commun. 9(1):1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenn JD, Song H, Cameron SL, Whiting MF.. 2008. A preliminary mitochondrial genome phylogeny of Orthoptera (Insecta) and approaches to maximizing phylogenetic signal found within mitochondrial genome data. Mol Phylogenet Evol. 49(1):59–68. [DOI] [PubMed] [Google Scholar]
- Forrest TG. 1982. Acoustic communication and baffling behaviors of crickets. Fla Entomol. 65(1):33–44. [Google Scholar]
- Kataoka K, et al. 2020. The Draft Genome Dataset of the Asian cricket Teleogryllus occipitalis for molecular research toward entomophagy. Front Genet. 11:470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, et al. 2018. Positive selection drove the adaptation of mitochondrial genes to the demands of flight and high-altitude environments in grasshoppers. Front Genet. 9:605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal DS, Chhakchhuak L, Gurusubramanian G, Kumar NS.. 2014. Mitochondrial markers for identification and phylogenetic studies in insects – a review. DNA Barcodes. 2(1):1–9. [Google Scholar]
- Meiklejohn KA, et al. 2014. Incongruence among different mitochondrial regions: a case study using complete mitogenomes. Mol Phylogenet Evol. 78:314–323. [DOI] [PubMed] [Google Scholar]
- Ojala D, Montoya J, Attardi G.. 1981. tRNA punctuation model of RNA processing in human mitochondria. Nature 290(5806):470–474. [DOI] [PubMed] [Google Scholar]
- Rokas A, Holland PW.. 2000. Rare genomic changes as a tool for phylogenetics. Trends Ecol Evol. 15(11):454–459. [DOI] [PubMed] [Google Scholar]
- Sheffield N, Song H, Cameron S, Whiting M.. 2008. A comparative analysis of mitochondrial genomes in Coleoptera (Arthropoda: Insecta) and genome descriptions of six new beetles. Mol Biol Evol. 25(11):2499–2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song H, et al. 2015. 300 Million years of diversification: elucidating the patterns of orthopteran evolution based on comprehensive taxon and gene sampling. Cladistics 31(6):621–651. [DOI] [PubMed] [Google Scholar]
- Song H, et al. 2020. Phylogenomic analysis sheds light on the evolutionary pathways towards acoustic communication in Orthoptera. Nat Commun. 11(1):4939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Huis A, et al. 2013. Edible insects: future prospects for food and feed security. Rome (Italy): Food and Agriculture Organization of the UN (FAO). [Google Scholar]
- Wetter JK, Hugen S.. 2008. Worldwide spread of the ant cricket Myrmecophilus americanus, a symbiont of the Longhorn crazy ant, Paratrechina longicornis. Sociobiology 52:157–165. [Google Scholar]
- Yongjun L, et al. 2008. Mitochondrial genome analysis of Ochotona curzoniae and implication of cytochrome c oxidase in hypoxic adaptation. Mitochondrion 8:5–6. [DOI] [PubMed] [Google Scholar]
- Yoshimasa H, et al. 2015. Leg regeneration is epigenetically regulated by histone H3K27 methylation in the cricket Gryllus bimaculatus. Development 142:2916–2927. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Chen B, Zhao D, Kang L.. 2013. Functional modulation of mitochondrial cytochrome c oxidase underlies adaptation to high-altitude hypoxia in a Tibetan migratory locust. Proc R Soc Lond B. 280:1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, et al. 2017. Towards a higher-level Ensifera phylogeny inferred from mitogenome sequences. Mol Phylogenet Evol. 108:22–33. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mitochondrial genomes for this article have been deposited at GenBank under the accession numbers MZ440652 (Teleogryllus occipitalis, BioSample SAMN19773260), MZ440653 (Teleogryllus emma, BioSample SAMN19773259), MZ440654 (Acheta domesticus, BioSample SAMN19773262), MZ440655 (Tarbinskiellus sp., BioSample SAMN19773264), MZ440656 (Gryllus bimaculatus, BioSample SAMN19773261), MZ440657 (Gryllodes sp., BioSample SAMN19773263), MZ440658 (Myrmecophilus kubotai, BioSample SAMN19773265), and MZ440659 (Myrmecophilus sp., BioSample SAMN19773266) under the BioProject PRJNA739105.