Abstract
Background: Cigarette smoking is a major risk factor for bronchoalveolar epithelial cell (BAEC) injury. Understanding the relevant pathogenesis is important for the treatment of cigarette smoke–related chronic airway diseases such as chronic obstructive pulmonary disease.
Methods: In this study, BAECs were cultured in 5% cigarette smoke extract (CSE) or regular culture medium for 24 h. Differentially expressed genes (DEGs) were detected by next-generation RNA sequencing (RNA-seq) and validated by quantitative reverse transcription polymerase chain reaction (qRT-PCR). Bioinformatic analysis was performed on DEGs. Co-treated BAECs with 5% CSE and the ferroptosis inhibitor, ferrostatin-1 was applied to observe the role of ferroptosis.
Results: In the CSE group, 210 upregulated genes and 159 downregulated genes were identified compared with the control group. Gene Ontology and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses showed that the DEGs were related to oxidative stress and ferroptosis. Ferroptosis-related genes were further verified by qRT-PCR. The mRNA level of GPX4 decreased; the mRNA levels of ACSL4, FTH1 and SLC7A11 increased (p < 0.05). Pretreatment with the ferroptosis inhibitor ferrostatin-1 mitigated CSE-induced ROS accumulation and inflammatory mediator expression in BAECs (p < 0.05).
Conclusion: CSE treatment altered ferroptosis-related gene expression patterns in cultured BAECs. Inhibition of ferroptosis reduced the inflammatory response of CSE-treated BAECs. These data provide a better understanding of the underlying molecular mechanisms of CSE-related lung injury.
Keywords: lung injury, ferroptosis, cigarette smoke extract, RNA sequencing, bronchoalveolar epithelial cell
Introduction
Chronic obstructive pulmonary disease (COPD) is a disease with high morbidity and high mortality (Halpin et al., 2020). COPD is characterized by airway inflammation, and while traditional treatment methods such as bronchodilators and glucocorticoids alleviate symptoms, they cannot prevent disease progression (Nici et al., 2020; Vogelmeier et al., 2020; Baha and Kokturk, 2021). Therefore, it is important to find new targets for COPD prevention and treatment.
Exposure to the harmful components in tobacco is a major risk factor for COPD and directly injures bronchoalveolar epithelial cells. Although previous studies have shown that cigarette smoke extract (CSE) causes airway inflammation and apoptosis of alveolar epithelial cells (Cui et al., 2018; Sun et al., 2019). There are currently no specific treatments to reverse these injuries in clinical practice. In the past decade, advances in next-generation sequencing have provided us with a better understanding of the pathogenesis of many diseases. However, relatively a few studies have focused on gene expression changes induced by CSE in alveolar epithelial cells.
Ferroptosis is an atypical form of regulated cell death, characterized by iron-dependent accumulation of lethal lipid reactive oxygen species (Li et al., 2020). Ferroptosis is distinct from apoptosis, pyroptosis, and necroptosis. It has been reported in a large number of studies in recent years and is associated with multiple pathological processes such as cancer, stroke and obstructive sleep apnea (Chen et al., 2020; Zhou et al., 2020; Xu et al., 2021). In previous study, RNA sequencing (RNA-seq) revealed that several messenger RNAs (mRNAs) related to ferroptosis were enriched in vascular smooth muscle cells pretreated with CSE (Sampilvanjil et al., 2020). However, the role of ferroptosis in CSE-related alveolar epithelial cell injury is not well understood.
The aim of this study was to explore the gene expression profile of CSE-treated alveolar epithelial cells via RNA-seq and perform bioinformatic analysis to clarify the functions of differentially expressed genes (DEGs). A secondary aim was to explore the role of ferroptosis and provide new insights for the treatment of CSE-related airway epithelial lesions.
Materials and Methods
Cigarette Smoke Extract Preparation
Cigarette smoke extract preparation followed a previously reported method with some modifications (Higashi et al., 2014, 2016). One piece of cigarette (Septwolves, Longyan Cigarette Factory, Fujian, China) was burned in a 500 ml bottle with a modified 50-ml syringe-driven apparatus. The smoke was dissolved in 20 ml of RPMI-1640 medium. To remove large particles and bacteria, the resulting solution was filtered through a 0.22-μm pore-size filter and adjusted to a pH of 7.4. The solution was identified as 100% CSE. Concentrations of 5% CSE were obtained by diluting 100% CSE in RPMI-1640 complete medium. Within 30 min of preparation, 5% CSE was used for subsequent experiments.
Establishment of the Cell Model
Bronchoalveolar epithelial cells (BAECs) were obtained from Biyuntian Biological Technology Co., Ltd., (Shanghai, China). Complete medium was made from RPMI-1640 (HyClone Laboratory Inc., United States) supplemented with 10% fetal bovine serum (Gibco Life Technologies Inc., United States). Cells were cultured in the incubator at 37°C (Thermo Fisher Scientific, Waltham, MA, United States) with a humidified atmosphere of a 5% CO2. In CSE group, once BAECs cells reached 70–80% confluency, replaced complete medium with 5% CSE. CSE exposure was applied for 24 h. In CSE + ferrostatin-1 group, co-treated BAECs with 5% CSE and 1 μM ferroptosis inhibitor, ferrostatin-1 was applied for 24 h.
RNA Isolation, Library Preparation, and Illumina Sequencing
After extracting total RNA from BAECs by TRIzol Reagent (Invitrogen, United States), the quality and quantity of the total RNA were quantified via DNA/RNA concentration assay (Pharmathea Inc., China). Poly-T oligo-attached magnetic beads were then used to purify the mRNA. First-strand cDNA and second-strand cDNA was synthesized with random Primers and dUTP respectively. End-repair and ligation with “A-tailing” base adaptors were performed in all synthetic cDNA. After that, polymerase chain reaction (PCR) was used to amplify suitable fragments and construct the cDNA library. The quantity and quality of the sample library were verified by qPCR quantification methods and Agilent 2100 Bioanalyzer (Agilent, United States). The libraries were sequenced by NovaSeq 6000 (Illumina, United States).
Next-Generation RNA Sequencing Data Processing
After quality control and removal of ribosomal RNA, the raw sequence data were detected with adaptor sequences, removed street sequences and short fragments in Read. The trimmed data thus obtained were compared to the reference genome. Fragments per kilobase of exons per million fragments mapped (FRKM) were used to calculate transcript levels. The genes with a FRKM > 0.5 were included for follow-up analysis. Genes with p-value < 0.05 and a CSE vs. control fold change > 1.50 (or <1/1.5) were defined as DEGs.
Bioinformatic Analysis
Volcano graph, scatter plots, and hierarchical clustering were used to visualize the differential gene expression profiles. Functional enrichment of DEGs based on the DAVID database was performed via Gene Ontology (GO) enrichment analysis1. Signal pathways involving DEGs were obtained via Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis2. Significant GO terms and pathways were identified via Fisher’s exact test. A p value < 0.05 was regarded as significant.
Quantitative Reverse Transcription Polymerase Chain Reaction
Total RNA was reverse transcribed into cDNA by PrimeScript RT reagent Kit (Takara Bio Inc., Japan). To determine the mRNA expression levels of Glutathione peroxidase 4 (GPX4), acyl-CoA synthetase long-chain family member 4 (ACSL4), Recombinant Solute Carrier Family 7, Member 11 (SLC7A11), ferritin heavy chain 1 (FTH1), interleukin-6 (IL6) and tumor necrosis factor-alpha (TNF-α), the SYBR Green PCR Master Mix (Takara Bio Inc., Japan) was used and RT-qPCR was performed on an ABI 7500 thermocycler (Applied Biosystems, Foster City, CA, United States). Relevant primers are listed in Table 1. β-Actin was used as an internal control. Fold changes were calculated by the 2–ΔΔCT method.
TABLE 1.
List of primer sequence.
| Genes | Primer sequence |
| FTH1 | F: TCCTACGTTTACCTGTCCATGT |
| R: GTTTGTGCAGTTCCAGTAGTGA | |
| GPX4 | F: CCGCTGTGGAAGTGGATGAAGATC |
| R: CTTGTCGATGAGGAACTGTGGAG | |
| SLC7A11 | F: ATGCAGTGGCAGTGACCTTT |
| R: GGCAACAAAGATCGGAACTG | |
| ACSL4 | F: TCTGCTTCTGCTGCCCAATT |
| R: CGCCTTCTTGCCAGTCTTTT | |
| IL6 | F: GGTGTTGCCTGCTGCCTTCC |
| R: GTTCTGAAGAGGTGAGTGGCTGTC | |
| TNFα | F: TGGCGTGGAGCTGAGAGATAACC |
| R: CGATGCGGCTGATGGTGTGG | |
| Beta-actin | F: TGGCACCCAGCACAATGAA |
| R: CTAAGTCATAGTCCGCCTAGAAGCA |
Cell Viability Test
A Cell Counting Kit-8 assay (Beyotime Bio Inc., China) was used to detect cell viability. Five thousand cells per well were added to a 96-well plate, with five replicate wells in each group. phosphate-buffered saline (PBS) was added to surrounding wells to reduce errors. After 12 h, 5% CSE was added in CSE group. After modeling, 10 μL CCK-8 solution was added to each well and the 96-well plate was incubated at 37°C for 1 h. The absorbance at 450 nm was detected with a microplate reader. Cell viability was calculated as (experimental group-blank control) / (negative control-blank control) × 100%.
Apoptosis Assay
Cells were double stained with Annexin V-fluorescein isothiocyanate (FITC) and propidium iodide (PI) (Beyotime Bio Inc., China). The cell apoptosis rate was then assessed by flow cytometry analysis. In brief, 1 × 105 cells were collected per well, with three replicate wells in each group. After washing in PBS, cells were resuspended in 200 μl binding buffer, mixed with 10 μl of PI and 5 μl of Annexin V-FITC. The suspensions were analyzed by a C6 flow cytometer (Becton Dickinson, United States).
Intracellular Reactive Oxygen Species
Intracellular reactive oxygen species (ROS) were detected by the Reactive Oxygen Species Assay Kit (Beyotime Bio Inc., China). The DCFH-DA fluorescent probe and serum-free culture medium were diluted at a ratio of 1:1,000. One milliliter of diluted DCFH-DA solution was added to one well of the 12-well plates, with three replicate wells in each group. After incubation for 20 min in a 37°C incubator, cells were collected for flow cytometry analysis.
Statistical Analysis
When the data were normally distributed, the mean ± standard deviation was used; otherwise, the interquartile range was used. An independent t test was used to compare the mean of continuous variables in normally distributed data; the Mann–Whitney test was used to compare non-normally distributed data. A p value < 0.05 was considered statistically significant. Statistical analysis was performed by Graphpad Prism 7.0 (GraphPad Software Inc., United States).
Results
Effects of Cigarette Smoke Extract on Cell Viability in Bronchoalveolar Epithelial Cells
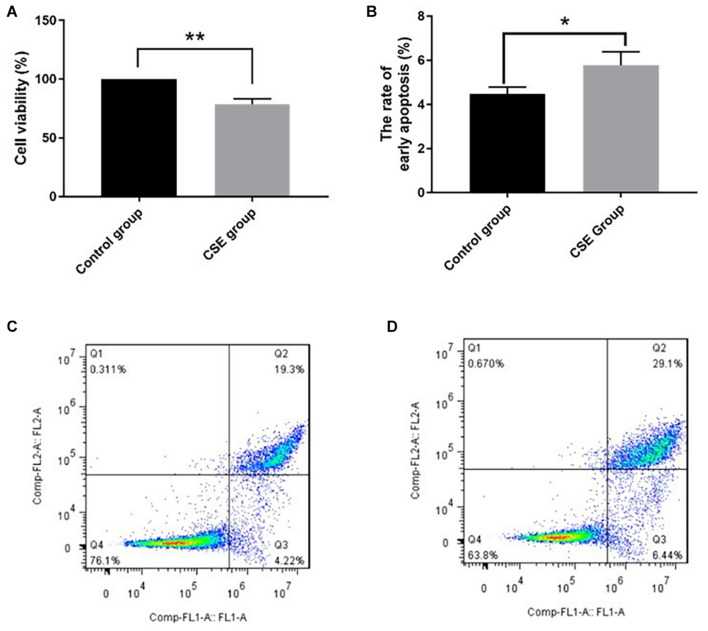
After 24-h exposure to 5% CSE, BAEC viability decreased significantly compared with the control group, (78.7 ± 4.8)% vs. (100.0 ± 0.0)%, p < 0.05. The rate of early apoptotic BAECs increased in the CSE group compared with the control group, (5.78 ± 0.61)% vs. (4.47 ± 0.32)%, p < 0.05. These data support our CSE-induced injury model in BAECs, as shown in Figure 1.
FIGURE 1.
CSE-induced BAEC injury (n = 3). (A) CSE reduced cell viability of BAECs; (B) CSE increased the early apoptosis rate of BAECs; (C) apoptosis rate in the control group; (D) apoptosis rate in the CSE group. CSE, cigarette smoke extract; BAEC, bronchoalveolar epithelial cells. *p < 0.05; **p < 0.01.
Identification of Differentially Expressed Genes
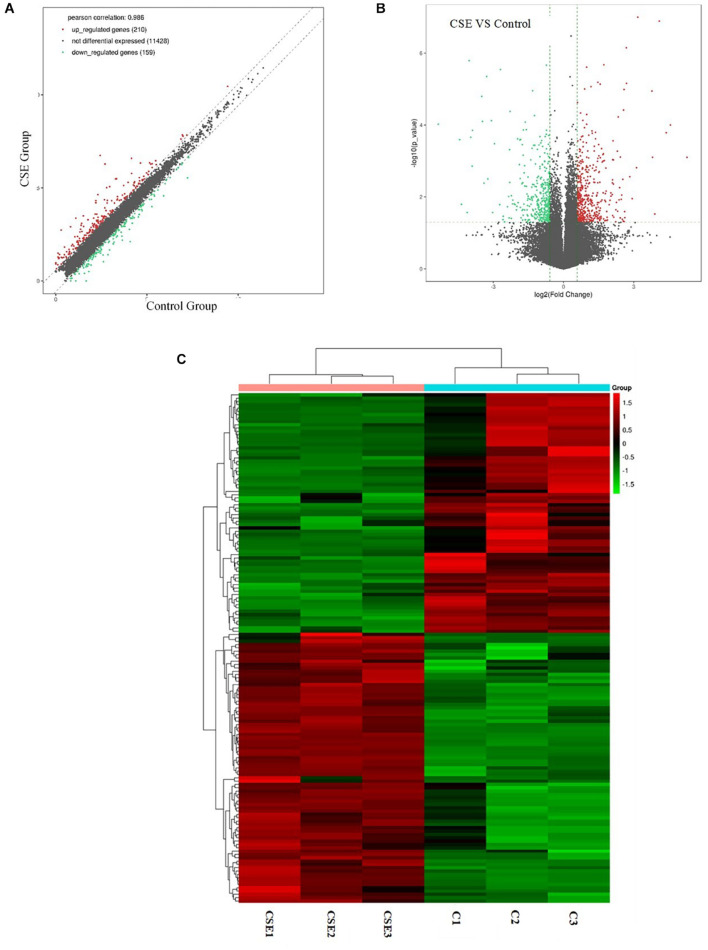
Differentially expressed genes were identified in CSE-treated BAECs by RNA-seq. With a fold change cutoff of 1.5 (or less than 1/1.5) and p < 0.05, 369 dysregulated genes (210 upregulated genes and 159 downregulated genes) were identified in the BAECs exposed to 5% CSE compared with controls. To visualize the DEGs between the CSE group and the control group, volcano plots were performed by fold change values and p values; Gene expression differences are also presented as scatter plots (Figures 2A,B). Hierarchical clustering was conducted to predict the function of DEGs (Figure 2C). SERPINB2, SHISA2, NQO1, PRDM2, CYP1B1, STC2, AHRR, TRIM16L, EREG and SLC7A11 were the top 10 upregulated genes, as shown in Table 2. JAG1, S100A2, NOTCH3, RPL36A-HNRNPH2, MEGF6, SULF1, EPPK1, SLIT3, CD14 and CTGF were the top 10 downregulated genes, as shown in Table 3.
FIGURE 2.
Next-generation RNA sequencing data showing the differentially expressed genes in CSE group and control group of BAECs (n = 3). (A) Differentially expressed genes in the CSE and control groups. (B) Volcano plot showing differentially expressed genes in the CSE and control groups. Genes that were differentially expressed with a fold change > 1.5 and p < 0.05 are marked in color. (C) Heat map of differentially expressed genes in the CSE and control groups. CSE, cigarette smoke extract; BAEC, bronchoalveolar epithelial cells.
TABLE 2.
List of top ten upregulated DEGs in CSE group compared to control group.
| Gene name | Description | Chromosome | Fold change | p value |
| SERPINB2 | Serpin Family B Member 2 | 18 | 5.40 | 0.001 |
| SHISA2 | Shisa Family Member 2 | 13 | 4.28 | 0.001 |
| NQO1 | NAD(P)H Quinone Dehydrogenase 1 | 16 | 3.21 | 0.0002 |
| PRDM2 | PR/SET Domain 2 | 1 | 3.19 | 0.0001 |
| CYP1B1 | Cytochrome P450 Family 1 Subfamily B Member 1 | 2 | 3.06 | 0.0004 |
| STC2 | Stanniocalcin 2 | 5 | 2.84 | 0.023 |
| AHRR | Aryl-Hydrocarbon Receptor Repressor | 5 | 2.68 | 0.011 |
| TRIM16L | Tripartite Motif Containing 16 Like | 17 | 2.63 | 0.001 |
| EREG | Epiregulin | 4 | 2.52 | < 0.001 |
| SLC7A11 | Solute Carrier Family 7 Member 11 | 4 | 2.50 | 0.0004 |
DEP, differentially expressed genes; CSE, Cigarette smoke extract.
TABLE 3.
List of top ten downregulated DEGs in CSE group compared to control group.
| Gene name | Description | Chromosome | Fold change | p value |
| JAG1 | Jagged Canonical Notch Ligand 1 | 20 | 0.51 | 0.003 |
| S100A2 | S100 Calcium Binding Protein A2 | 1 | 0.49 | 0.008 |
| NOTCH3 | Notch Receptor 3 | 19 | 0.49 | 0.031 |
| RPL36A-HNRNPH2 | RPL36A-HNRNPH2 Readthrough | X | 0.47 | 0.042 |
| MEGF6 | Multiple EGF Like Domains 6 | 1 | 0.46 | 0.008 |
| SULF1 | Sulfatase 1 | 8 | 0.46 | 0.002 |
| EPPK1 | Epiplakin 1 | 8 | 0.45 | 0.009 |
| SLIT3 | Slit Guidance Ligand 3 | 5 | 0.44 | 0.001 |
| CD14 | CD14 Molecule | 9 | 0.43 | 0.0002 |
| CTGF | Cellular Communication Network Factor 2 | 6 | 0.42 | 0.005 |
DEP, differentially expressed genes; CSE, Cigarette smoke extract.
Gene Ontology Analysis
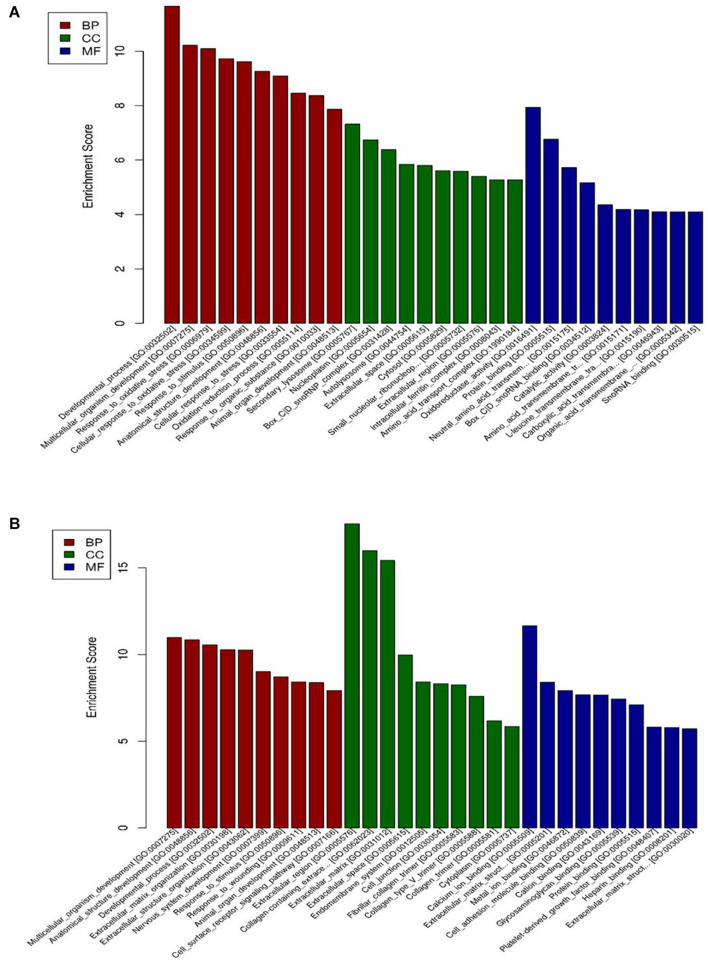
The DEGs were classified by the GO database on the basis of the categories of biological process (BP), cellular component (CC), and molecular function (MF). The top ten GO terms for upregulated genes and downregulated genes are shown in Figure 3. The dysregulated genes were enriched in BP terms including response to oxidative stress and developmental process, CC terms including nucleoplasm and extracellular region, and MF terms including metal ion binding, oxidoreductase activity, and protein binding.
FIGURE 3.
GO enrichment-based clustering analysis of the differentially expressed genes in BAECs (n = 3). (A) The top 10 GO terms associated with upregulated genes in the CSE group compared with the control group; (B) the top 10 GO terms associated with downregulated genes in the CSE group compared with the control group. CSE, cigarette smoke extract; BAEC, bronchoalveolar epithelial cells; CC, Cellular component analysis; BP, Biological process analysis; MF, Molecular function analysis.
Kyoto Encyclopedia of Genes and Genomes Analysis
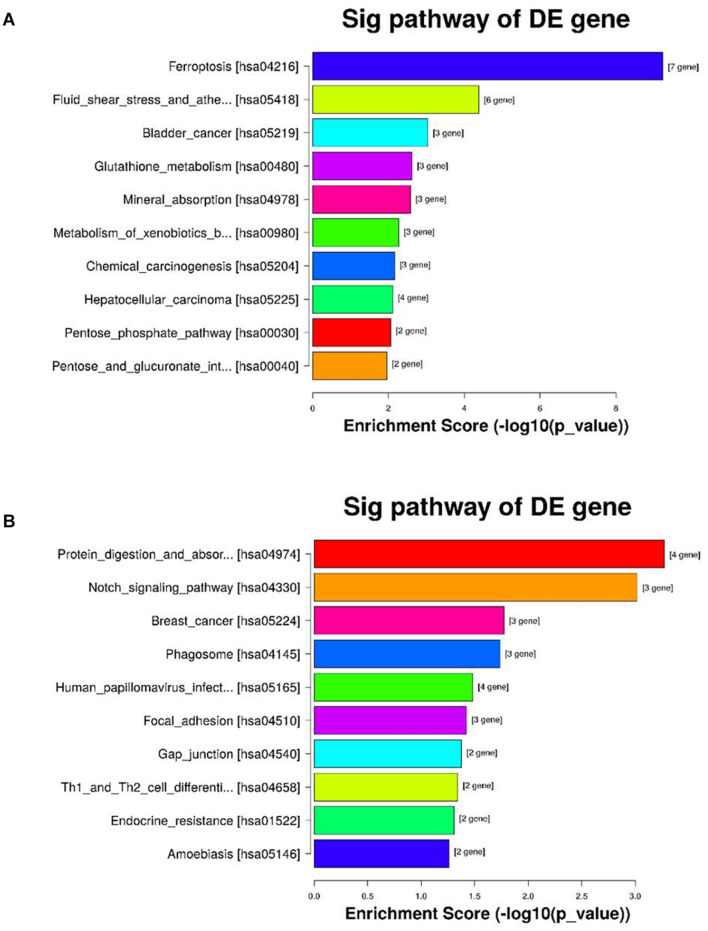
Kyoto encyclopedia of genes and genomes pathway analysis was conducted to find potential biological functions of the significant DEGs. Twenty signaling pathways, including ferroptosis, glutathione metabolism and cancer, were associated with the upregulated DEGs and 12 signaling pathways, including protein digestion and absorption, the Notch signaling pathway, Th1 and Th2 cell differentiation and the Mitogen-activated protein kinase (MAPK) signaling pathway, were associated with the downregulated DEGs. The top 10 enriched pathways sorted by enrichment score are showed in Figure 4.
FIGURE 4.
KEGG pathway enrichment of the differentially expressed genes in the CSE and control groups of BAEC (n = 3). (A) The top 10 KEGG pathways associated with upregulated genes in the CSE group compared with the control group; (B) the top 10 KEGG pathways associated with downregulated genes in the CSE group compared with the control group. KEGG, Kyoto Encyclopedia of Genes and Genomes; CSE, cigarette smoke extract; BAEC, bronchoalveolar epithelial cells.
Validation of Cigarette Smoke Extract-Induced Ferroptosis Gene Expression by Quantitative Reverse Transcription Polymerase Chain Reaction
Quantitative reverse transcription polymerase chain reaction (qRT-PCR) was performed to verify the differential expression of ferroptosis genes in the CSE group compared with the control group. Compared with the control group, ACSL4, FTH1, and SLC7A11 mRNA levels were significantly upregulated in CSE group, while GPX4 mRNA expression was significantly downregulated in CSE group, p < 0.05 (Figure 5A).
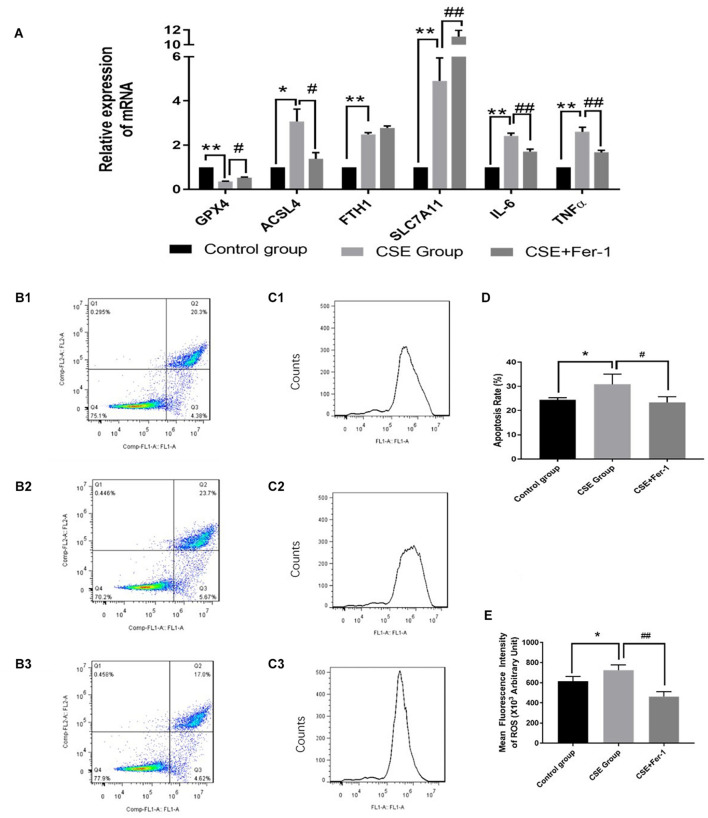
FIGURE 5.
Effects of ferrostatin-1 on mRNA expression levels, apoptosis rates and ROS levels (n = 3). (A) GPX4, ACSL4, FTH1, SLC7A11, IL6 and TNF-α mRNA levels in control group, CSE group and CSE + ferrostatin-1 group of BAECs. (B1–3,D) The apoptosis rate in control group, CSE group and CSE + ferrostatin-1 group of BAECs. (C1–3,E) ROS levels in control group, CSE group and CSE + ferrostatin-1 group of BAECs. CSE, cigarette smoke extract; BAEC, bronchoalveolar epithelial cells. **p < 0.01 compared with the control group; *p < 0.05 compared with the control group; ##p < 0.01 compared with the CSE group; #p < 0.05 compared with the CSE group.
Ferrostatin-1 Alleviated Cigarette Smoke Extract-Induced Inflammation and Ferroptosis in Bronchoalveolar Epithelial Cells
TNF-α and IL6 are biomarkers of inflammation. The qRT-PCR results suggested that CSE treatment increased mRNA levels of TNF-α and IL6, which reflected CSE treatment induced inflammation in BAECs. Therefore, we co-treated BEACs with CSE and the ferroptosis inhibitor, ferrostatin-1. Compared with CSE treatment alone, incubation with CSE + ferrostatin-1 resulted in decreased mRNA expression of TNF-α and IL6, p < 0.05 (Figure 5A). Ferrostatin-1 also changed the mRNA levels of GPX4 and ACSL4 in CSE treated BAECs, p < 0.05 (Figure 5A).
The results of flow cytometry analysis suggested that CSE treatment increased the apoptosis rate of BAECs, while co-treated BEACs with CSE and ferrostatin-1 alleviated apoptosis of BAECs (Figures 5B,D). The DCFH-DA fluorescent probe assay showed increased ROS levels in CSE-treated BAECs. Compared with the CSE group, ROS levels were lower in the CSE + ferrostatin-1-treated group (Figures 5C,E).
Discussion
In this study, we found 210 upregulated genes and 159 downregulated genes in CSE-treated BAECs compared with controls. Gene ontology and KEGG analysis revealed that the DEGs were enriched for the ferroptosis pathway. Moreover, ferrostatin-1, a ferroptosis inhibitor, abrogated part of the CSE-induced gene expression, apoptosis and ROS changes in BAECs.
RNA-seq is an efficient high-throughput technology that has uncovered the potential molecular mechanisms of various diseases (Morrow et al., 2019). However, few RNA-seq studies have focused on CSE-related BAEC injury. In this study, we made a BAECs injury model via CSE and searched for DEPs using RNA-seq technique. In a subsequent step, GO enrichment analysis showed that the upregulated DEGs of the CSE-treated BAECs were involved in oxidative stress pathways and were associated with molecular functions such as protein binding, amino acid, and leucine transport. Oxidative stress plays an important role in cigarette smoke–related COPD (Barnes, 2017, 2020; Zhang et al., 2020). The differential gene expression uncovered in this study may provide new ideas for future studies on the etiology and potential treatment of COPD.
In previous studies, GPX4 expression was downregulated in mice with COPD (Yoshida et al., 2019). GPX4 is a key regulator of ferroptosis (Seibt et al., 2019). In this study, we confirmed downregulation of GPX4 mRNA, and also found that ACSL4, FTH1, and SLC7A11 mRNA expression was significantly increased in CSE-treated BAECs compared with controls. These data further confirm that cigarette smoke likely results in ferroptosis in BAECs. The consumption or inactivation of GPX4 leads to lipid peroxide accumulation in cells and resulting ferroptosis (Imai et al., 2017). The co-culture of ferrostatin-1 may affect the mRNA levels of GPX4 and ACSL4 by inhibiting lipid peroxidation, but did not affect the expression level of FTH1. SLC7A11 is a glutamate-cystine antiporter in the phospholipid bilayer (Okazaki et al., 2019; Koppula et al., 2020). SLC7A11 may indirectly reduce the effect of GPX4 by reducing the transport of glutathione, which leads to intracellular lipid peroxidation and ferroptosis. In this study, the further increased expression of SLC7A11 after ferrostatin-1 co-culture was confusing. Ferrostatin-1 may not affect the transport of glutathione, we will explore the mechanisms in future studies.
In addition to oxidative stress, inflammation also plays an important role in the pathogenesis of COPD (Barnes, 2016; Higham et al., 2019). Ferroptosis induces bronchial epithelial cells to release pro-inflammatory cytokines, which forms an inflammatory cascade that leads to COPD-related airway remodeling and emphysema. Ferrostatin-1 is a highly effective non-apoptotic, erastin-induced ferroptosis inhibitor (Bai et al., 2020; Liu et al., 2020). Previous in vivo and in vitro studies have confirmed that ferrostatin-1 can reduce lung injury caused by lipopolysaccharides (Liu et al., 2020). In our study, CSE increased expression of inflammatory mediators IL-6 and TNF-α in BAECs. In contrast, ferrostatin-1 pretreatment reduced expression of inflammatory mediators, apoptosis rate and ROS in our cultured BAECs. These data suggest that ferroptosis is, at least partially, responsible for CSE-induced BAEC injury in this study.
There are some limitations to this study. First, this is an in vitro cell-based study, and the results have not been validated in animal models or human patients. There may be differences in the results of in vitro and in vivo experiments, which may affect the reproducibility of our conclusions. Secondly, because of funding limitations, the number of samples included in the RNA-seq experiment was relatively small.
In conclusion, our data indicate that CSE exposure alters the gene expression profile of cultured BAECs and that differentially expressed genes are enriched for the ferroptosis pathway. The ferroptosis inhibitor ferrostatin-1 inhibited ROS accumulation and inflammatory mediator expression in CSE-treated BAECs. These results suggest that cigarette smoking may induce ferroptosis in airway epithelium, which could contribute to CSE-related airway dysfunction.
Data Availability Statement
BioProject’s metadata is available at https://www.ncbi.nlm.nih.gov/bioproject/PRJNA761787.
Author Contributions
NL, QZ, and QL: conception and design. NL, QZ, and MC: collection and assembly of data. NL and JC: data analysis and interpretation. NL and JH: manuscript revision. All authors wrote the manuscript and approved the final version of the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Footnotes
Funding
This work was supported by Natural Science Foundation of Fujian Province, China (Grant Number 2019J01440) and Joint Funds for the Innovation of Science and Technology, Fujian Province (Grant Number 2019Y9116).
References
- Baha A., Kokturk N. (2021). Physician’s attitude against COPD guidelines and the choice of first-line treatment for COPD. Respir. Med. 176:106273. 10.1016/j.rmed.2020.106273 [DOI] [PubMed] [Google Scholar]
- Bai T., Li M., Liu Y., Qiao Z., Wang Z. (2020). Inhibition of ferroptosis alleviates atherosclerosis through attenuating lipid peroxidation and endothelial dysfunction in mouse aortic endothelial cell. Free Radic. Biol. Med. 160 92–102. 10.1016/j.freeradbiomed.2020.07.026 [DOI] [PubMed] [Google Scholar]
- Barnes P. J. (2016). Inflammatory mechanisms in patients with chronic obstructive pulmonary disease. J. Allergy Clin. Immunol. 138 16–27. [DOI] [PubMed] [Google Scholar]
- Barnes P. J. (2017). Cellular and molecular mechanisms of asthma and COPD. Clin. Sci. 131 1541–1558. 10.1042/cs20160487 [DOI] [PubMed] [Google Scholar]
- Barnes P. J. (2020). Oxidative stress-based therapeutics in COPD. Redox Biol. 33:101544. 10.1016/j.redox.2020.101544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L.-D., Wu R.-H., Huang Y.-Z., Chen M.-X., Zeng A.-M., Zhuo G.-F., et al. (2020). The role of ferroptosis in chronic intermittent hypoxia-induced liver injury in rats. Sleep Breath. 24 1767–1773. 10.1007/s11325-020-02091-4 [DOI] [PubMed] [Google Scholar]
- Cui W., Zhang Z., Zhang P., Qu J., Zheng C., Mo X., et al. (2018). Nrf2 attenuates inflammatory response in COPD/emphysema: crosstalk with Wnt3a/β-catenin and AMPK pathways. J. Cell. Mol. Med. 22 3514–3525. 10.1111/jcmm.13628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halpin D., Criner G. J., Papi A., Singh D., Vogelmeier C. F. (2020). Global Initiative for the Diagnosis, Management, and Prevention of Chronic Obstructive Lung Disease: the 2020 GOLD Science Committee Report on COVID-19 & COPD. Am. J. Respir. Crit. Care Med. 203 24–36. 10.1164/rccm.202009-3533so [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higashi T., Mai Y., Mazaki Y., Horinouchi T., Miwa S. A. (2016). Standardized Method for the Preparation of a Gas Phase Extract of Cigarette Smoke. Biol. Pharm. Bull. 39 898–902. 10.1248/bpb.b16-00062 [DOI] [PubMed] [Google Scholar]
- Higashi T., Mai Y., Noya Y., Horinouchi T., Terada K., Hoshi A., et al. (2014). A simple and rapid method for standard preparation of gas phase extract of cigarette smoke. PLoS One 9:e107856. 10.1371/journal.pone.0107856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higham A., Quinn A. M., Cançado J. E. D., Singh D. (2019). The pathology of small airways disease in COPD: historical aspects and future directions. Respir. Res. 20:49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai H., Matsuoka M., Kumagai T., Sakamoto T., Koumura T. (2017). Lipid Peroxidation-Dependent Cell Death Regulated by GPx4 and Ferroptosis. Curr. Top. Microbiol. Immunol. 403 143–170. 10.1007/82_2016_508 [DOI] [PubMed] [Google Scholar]
- Koppula P., Zhuang L., Gan B. (2020). Cystine transporter SLC7A11/xCT in cancer: ferroptosis, nutrient dependency, and cancer therapy. Protein Cell 12 599–620. 10.1007/s13238-020-00789-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Cao F., Yin H. L., Huang Z. J., Lin Z. T., Mao N., et al. (2020). Ferroptosis: past, present and future. Cell Death Dis. 11:88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P., Feng Y., Li H., Chen X., Wang G., Xu S., et al. (2020). Ferrostatin-1 alleviates lipopolysaccharide-induced acute lung injury via inhibiting ferroptosis. Cell. Mol. Biol. Lett. 25:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrow J. D., Chase R. P., Parker M. M., Glass K., Seo M., Divo M., et al. (2019). RNA-sequencing across three matched tissues reveals shared and tissue-specific gene expression and pathway signatures of COPD. Respir. Res. 20:65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nici L., Mammen M. J., Charbek E., Alexander P. E., Au D. H., Boyd C. M., et al. (2020). Pharmacologic Management of Chronic Obstructive Pulmonary Disease. An Official American Thoracic Society Clinical Practice Guideline. Am. J. Respir. Crit. Care Med. 201 e56–e69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okazaki S., Umene K., Yamasaki J., Suina K., Otsuki Y., Yoshikawa M., et al. (2019). Glutaminolysis-related genes determine sensitivity to xCT-targeted therapy in head and neck squamous cell carcinoma. Cancer Sci. 110 3453–3463. 10.1111/cas.14182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampilvanjil A., Karasawa T., Yamada N., Komada T., Higashi T., Baatarjav C., et al. (2020). Cigarette smoke extract induces ferroptosis in vascular smooth muscle cells. Am. J. Physiol. Heart Circ. Physiol. 318 H508–H518. [DOI] [PubMed] [Google Scholar]
- Seibt T. M., Proneth B., Conrad M. (2019). Role of GPX4 in ferroptosis and its pharmacological implication. Free Radic. Biol. Med. 133 144–152. 10.1016/j.freeradbiomed.2018.09.014 [DOI] [PubMed] [Google Scholar]
- Sun X., Feng X., Zheng D., Li A., Li C., Li S., et al. (2019). Ergosterol attenuates cigarette smoke extract-induced COPD by modulating inflammation, oxidative stress and apoptosis in vitro and in vivo. Clin. Sci. 133 1523–1536. 10.1042/cs20190331 [DOI] [PubMed] [Google Scholar]
- Vogelmeier C. F., Román-Rodríguez M., Singh D., Han M. K., Rodríguez-Roisin R., Ferguson G. T. (2020). Goals of COPD treatment: focus on symptoms and exacerbations. Respir. Med. 166:105938. 10.1016/j.rmed.2020.105938 [DOI] [PubMed] [Google Scholar]
- Xu W., Deng H., Hu S., Zhang Y., Zheng L., Liu M., et al. (2021). Role of Ferroptosis in Lung Diseases. J. Inflamm. Res. 14 2079–2090. 10.2147/jir.s307081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida M., Minagawa S., Araya J., Sakamoto T., Hara H., Tsubouchi K., et al. (2019). Involvement of cigarette smoke-induced epithelial cell ferroptosis in COPD pathogenesis. Nat. Commun. 10:3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M., Zhang Y., Roth M., Zhang L., Shi R., Yang X., et al. (2020). Sirtuin 3 Inhibits Airway Epithelial Mitochondrial Oxidative Stress in Cigarette Smoke-Induced COPD. Oxid. Med. Cell. Longev. 2020:7582980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Jin Y., Lei Y., Liu T., Wan Z., Meng H., et al. (2020). Ferroptosis Is Regulated by Mitochondria in Neurodegenerative Diseases. Neurodegener. Dis. 20 20–34. 10.1159/000510083 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
BioProject’s metadata is available at https://www.ncbi.nlm.nih.gov/bioproject/PRJNA761787.