Abstract
Burkholderia cepacia, which is an important pathogen in cystic fibrosis (CF) owing to the potential severity of the infections and the high transmissibility of some clones, has been recently shown to be a complex of five genomic groups, i.e., genomovars I, II (B. multivorans), III, and IV and B. vietnamiensis. B. gladioli is also involved, though rarely, in CF. Since standard laboratory procedures fail to provide an accurate identification of these organisms, we assessed the ability of restriction fragment length polymorphism (RFLP) analysis of amplified 16S ribosomal DNA (rDNA), with the combination of the patterns obtained with six endonucleases, to differentiate Burkholderia species. This method was applied to 16 type and reference strains of the genus Burkholderia and to 51 presumed B. cepacia clinical isolates, each representative of one clone previously determined by PCR ribotyping. The 12 Burkholderia type strains tested were differentiated, including B. cepacia, B. multivorans, B. vietnamiensis, and B. gladioli, but neither the genomovar I and III reference strains nor the genomovar IV reference strain and B. pyrrociniaT were distinguishable. CF clinical isolates were mainly distributed in RFLP group 2 (which includes B. multivoransT) and RFLP group 1 (which includes B. cepacia genomovar I and III reference strains, as well as nosocomial clinical isolates). Two of the five highly transmissible clones in French CF centers belonged to RFLP group 2, and three belonged to RFLP group 1. The remaining isolates either clustered with other Burkholderia species (B. cepacia genomovar IV or B. pyrrocinia, B. vietnamiensis, and B. gladioli) or harbored unique combinations of patterns. Thus, if further validated by hybridization studies, PCR-RFLP of 16S rDNA could be an interesting identification tool and contribute to a better evaluation of the respective clinical risks associated with each Burkholderia species or genomovar in patients with CF.
Burkholderia cepacia, first described as the agent of the onion soft rot (2), is an opportunistic human pathogen which may be responsible for various nosocomial infections and is especially problematic in patients with cystic fibrosis (CF) due to the potential severity of respiratory infections, the easy patient-to-patient spread of the organism, and its innate resistance to a wide range of antimicrobial agents (8). These data led to the introduction of various preventive guidelines in and outside hospitals (hygiene procedures, segregation of colonized patients, etc.) in order to reduce the risk of acquisition. Nevertheless, the clinical outcome in B. cepacia-infected CF patients is extremely variable, ranging from a fatal necrotizing pneumonia, the “cepacia syndrome” to an unmodified respiratory status (10). In the same way, large epidemics as well as small clusters of infection or unique colonizations have been reported on the basis of the genotypic analysis of isolates (17, 21). Owing to the diversity of clinical situations due to B. cepacia, the identification of pathogenicity and transmissibility markers is essential. The first requirement in achieving this goal is a precise bacteriological knowledge of the responsible organism which, as a matter of fact, proves difficult. First, the accurate identification of B. cepacia with widely used multitest commercial systems has been recently shown to be problematic (12). Second, the genus Burkholderia has undergone multiple changes since its individualization by Yabuuchi et al. in 1992 (28). Some species (formerly B. pickettii and B. solanacearum) have been transferred to another new genus, Ralstonia (29), whereas species either formerly belonging to the genus Pseudomonas (B. cocovenenans [30], B. plantarii and B. glumae [24], and B. pyrrocinia and B. glathei [25, 26]) or newly described (B. vandii [24], B. vietnamiensis [6], and B. thailandensis [1]) have been included in the genus Burkholderia. The involvement of these various species, most of which are phytopathogenic, as opportunistic agents in human pathology has not been assessed so far, whereas infections due to B. gladioli have already been reported in CF (4, 11), chronic granulomatous disease (20), and other immunocompromised states (9). Yet the bacteriological differentiation between B. cepacia and B. gladioli is not always clear-cut (23). Third, organisms presently identified as B. cepacia appear to be very heterogeneous and constitute a “complex” of phenotypically similar species among which five genomic groups, i.e., genomovars I, II, III, and IV and the formerly described B. vietnamiensis, could be differentiated on the basis of whole-cell protein analysis, nucleic acid studies, and cellular fatty acid composition; the individualization of genomovar II as a new species, B. multivorans, has been proposed as well (25). Moreover, the importance of genomovar III among CF isolates, especially epidemic clones, has been emphasized (8, 25).
These differentiation procedures of Burkholderia taxa are not appropriate for routine diagnosis, and the aim of the present study was to develop a molecular identification tool which could be accessible to clinical microbiology laboratories. Restriction fragment length polymorphism (RFLP) analysis of amplified 16S ribosomal DNA (rDNA), previously proven to be discriminant for the differentiation of fluorescent Pseudomonas species (14), was first applied to reference strains belonging to different species of the genus Burkholderia. Next, a large set of clinical isolates, each representative of a given clone previously determined by use of PCR ribotyping (5, 13, 22), was analyzed comparatively to type and reference strains so as to assess the distribution of the various Burkholderia species in CF and nosocomial infections in sporadic and epidemic situations. Furthermore, carbon substrate assimilation tests were performed on strains belonging to the different RFLP types in order to point out differential phenotypic characters.
MATERIALS AND METHODS
Bacterial strains.
All of the strains studied are listed in Tables 1 and 2. Type strains were selected as the gold-standard representatives of each species; if not available, representative strains were chosen among those previously characterized by Vandamme et al. (25) as belonging to the different B. cepacia genomovars. The 18 type and reference strains analyzed (Table 1) were obtained from international culture collections and include species of the B. cepacia complex (25), i.e., genomovars I, II (B. multivorans), III, and IV and B. vietnamiensis; other Burkholderia species (B. pyrrocinia, B. cocovenenans, B. gladioli, B. plantarii, B. caryophilli, B. andropogonis, B. glumae, and B. glathei), and lastly two unrelated species (Ralstonia pickettii and Stenotrophomonas maltophilia). The 51 clinical isolates tested (Table 2), which were recovered from CF or non-CF patients, were selected on the basis of a previously performed genotypic analysis by PCR ribotyping among the French Observatoire cepacia collection and among strains kindly provided by N. Høiby (Copenhagen, Denmark), J. Govan (Edinburg, United Kingdom), K. Poole (Kingston, Ontario, Canada), and D. P. Speert (Vancouver, British Columbia, Canada). Each of these clinical isolates is representative of a different PCR ribotype (designated with capital letters).
TABLE 1.
16S rDNA RFLP profiles of the 18 reference strains analyzed in this study
| Taxona | Strainb | Other designationb | Source of isolation | API 20NE numerical profile, taxon (% identification) | RFLP profiles with:
|
RFLPc
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AluI | CfoI | DdeI | MspI | NciI | BssKI | Group | Subgroup | |||||
| Cepacia complex | ||||||||||||
| B. cepacia genomovar I | ATCC 25416T | LMG 1222T | Allium cepa L. | 1 477 577, B. cepacia (99.9) | A | A | A | A | AH | AH | 1 | 1a |
| B. cepacia genomovar I | ATCC 17759 | LMG 2161 | Forest soil (Trinidad) | 0 067 577, B. cepacia (95.5), P. aureofaciens (4.4) | A | A | A | A | A | A | 1 | 1b |
| B. cepacia genomovar I | ATCC 25609 | LMG 6981 | Human (bronchial washings) | 1 477 576, unidentified | A | A | A | A | A | A | 1 | 1b |
| B. multivorans genomovar II | LMG 13010T | NCTC 13007T | Human (CF patient, Belgium) | 1 477 577, B. cepacia (99.9) | A | A | I | A | A | AI | 2 | 2a |
| B. cepacia genomovar III | LMG 12615 | NCTC 13008 | Human (CF patient, UK) | 0 067 577, B. cepacia (95.5), P. aureofaciens (4.4) | A | A | A | A | A | A | 1 | 1b |
| B. cepacia genomovar IV | LMG 14294 | NCTC 13011 | Human (CF patient, Belgium) | 0 057 577, B. cepacia (50.3), P. aureofaciens (48.9) | A | B | A | B | H | H | 3 | |
| B. vietnamiensis | TVV75T | LMG 10929T | Rice rhizosphere (Vietnam) | 1 067 577, B. cepacia (93.3), P. aureofaciens (6.7) | A | A | J | A | A | A | 4 | |
| Genus Burkholderia (other species) | ||||||||||||
| B. pyrrocinia | LMG 14191T | ATCC 15958T | Soil | 0 067 577, B. cepacia (95.5), P. aureofaciens (4.4) | A | B | A | B | H | H | 3 | |
| B. cocovenenans | LMG 11626T | ATCC 33664T | Poisoned bongkrek (Java) | 1 047 555, unidentified | B | B | B | B | A | F | 5 | 5b |
| B. gladioli pathovar gladioli | CFBP 2427T | ATCC 10248T | Gladiolus sp. | 1 047 577, B. cepacia (68.5), P. aureofaciens (28.7), P. fluorescens (2.8) | B | B | B | B | A | A | 5 | 5a |
| B. gladioli pathovar alliicola | CFBP 2422T | ATCC 19302T | Allium cepa L. (United States) | 1 057 577, B. cepacia (95.5), P. aureofaciens (4.4) | B | B | F | B | A | A | 6 | |
| B. plantarii | LMG 9035T | Oryza sativa L. | 1 077 577, P. aureofaciens (59.9), B. cepacia (39.5) | B | B | G | B | A | A | 7 | ||
| B. caryophilli | LMG 2155T | ATCC 25418T | Dianthus caryophyllus L. | 1 047 544, unidentified | F | B | A | B | B | B | 8 | |
| B. andropogonis | LMG 2129T | ATCC 23061T | Holcus sorghum L. (United States) | 1 047 044, unidentified | C | C | C | C | A | C | 9 | |
| B. glumae | CFBP 2430T | NCPPB 2391T | Oryza sativa (Japan) | 5 077 557, B. cepacia (98.4) | A | B | H | B | A | A | 10 | |
| B. glathei | LMG 14190T | ATCC 29195T | Soil (Germany) | No growth | G | G | L | F | I | J | 11 | |
| Other genera | ||||||||||||
| R. pickettii | ATCC 27511T | Human (tracheotomy) | 1 241 455, R. pickettii (95.2) | D | D | D | D | D | D | 12 | ||
| S. maltophilia | ATCC 13637T | Human (oropharynx) | 1 472 341, S. maltophilia (99.9) | E | E | E | E | E | E | 13 | ||
Genomovars according to Vandamme et al. (25).
T, type strain; ATCC, American Type Culture Collection, Manassas, Va.; CFBP, Collection Française de Bactéries Phytopathogènes, Institut National de la Recherche Agronomique, Station de Pathologie Végétale et Phytobactériologie, Angers, France; LMG, Culture Collection, Laboratorium voor Microbiologie, State University of Ghent, Ghent, Belgium; NCPPB, National Collection of Plant Pathogenic Bacteria, Hatching Green, England; NCTC, National Collection of Type Cultures, London, United Kingdom; TVV, Tran Van Van (6).
The groups are defined on the basis of the RFLP profiles obtained with AluI, CfoI, DdeI, and MspI, and the subgroups are further defined with the NciI and BssKI restriction results.
TABLE 2.
16S rDNA RFLP profiles of 51 presumed B. cepacia clinical isolates
| PCR ribo-type | No. of patients; geographic location; sourcea | Isolate tested (reference) | API 20NE numerical profile, taxon (% identification) | RFLP profiles with:
|
RFLPc
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| AluI | CfoI | DdeI | MspI | NciI | BssKI | Group | Subgroup | ||||
| F | 32 CF patients; France (Giens, 30; Dijon, 1; Pessac 1); sputum, blood culture | 2.31 | 1 447 577, B. cepacia (99.9) | A | A | I | A | A | AI | 2 | 2a |
| X | 22 CF patients; France (Paris, 21; Toulouse, 1); sputum | 1.55 | 5 466 577, B. cepacia (99.9) | A | A | I | A | A | AI | 2 | 2a |
| BN | 1 CF patient; France (Tours); sputum | 3.75 | 5 067 577, B. cepacia (99.9) | A | A | I | A | A | AI | 2 | 2a |
| BF | 1 CF patient; France (St. Nazaire); sputum | 3.6 | 1 067 577, B. cepacia (93.3), P. aureofaciens (6.7) | A | A | I | A | A | AI | 2 | 2a |
| BO | 1 CF patient; France (Lille); sputum | 3.79 | 0 047 577, B. cepacia (75.4), P. aureofaciens (20.3), P. fluorescens (4.3) | A | A | I | A | A | AI | 2 | 2a |
| K | 2 CF siblings; France (Rouen); sputum | 3.67 | 1 047 577, B. cepacia (68.5), P. aureofaciens (28.7), P. fluorescens (2.8) | A | A | I | A | A | AI | 2 | 2a |
| AY | 1 CF patient; France (Rouen); sputum | 2.48 | 5 067 577, B. cepacia (99.9) | A | A | I | A | A | AI | 2 | 2a |
| I | 1 CF patient; France (Giens); sputum | 1.20 | 1 467 577, B. cepacia (99.9) | A | A | I | A | A | AI | 2 | 2a |
| AV | 1 CF patient; Denmark (Copenhagen) | A47 | 1 447 577, B. cepacia (99.9) | A | A | I | A | A | AI | 2 | 2a |
| AO | 1 CF patient; Denmark (Copenhagen) | A40 | 5 467 573, unidentifiedb | A | A | I | A | A | AI | 2 | 2a |
| AQ | 1 CF patient; Denmark (Copenhagen) | A42 | 5 010 004, A. salmonicida (90.2) | A | A | I | A | A | AI | 2 | 2a |
| AI | 1 CF patient; United Kingdom (Edinburgh) | C1911 | 1 067 577, B. cepacia (93.3), P. aureofaciens (6.7) | A | A | I | A | A | AI | 2 | 2a |
| M | 1 CF patient; France (Reims); sputum | 1.45 | 1 447 577, B. cepacia (99.9) | A | A | I | A | A | A | 2 | 2b |
| C | 1 CF patient; France (Nancy); sputum | 1.4 | 5 467 577, B. cepacia (99.9) | A | A | I | A | A | A | 2 | 2b |
| AZ | 1 CF patient; France (Dijon); sputum | 2.60 | 1 047 577, B. cepacia (68.5), P. aureofaciens (28.7), P. fluorescens (2.8) | A | A | I | A | A | A | 2 | 2b |
| BH | 1 CF patient; France (Roscoff); sputum | 3.30 | 1 067 577, B. cepacia (93.3), P. aureofaciens (6.7) | A | A | I | A | A | A | 2 | 2b |
| BL | 1 CF patient; France (Paris); sputum | 3.78 | 1 047 577, B. cepacia (68.5), P. aureofaciens (28.7), P. fluorescens (2.8) | A | A | I | A | A | A | 2 | 2b |
| AT | 1 CF patient; Denmark (Copenhagen) | A45 | 1 467 573, B. cepacia (99.9) | A | A | I | A | A | A | 2 | 2b |
| AE | 1 CF patient; United Kingdom (Edinburgh) | C1409 | 1 467 573, B. cepacia (99.9) | A | A | I | A | A | A | 2 | 2b |
| Q | 1 CF patient; Canada (Vancouver, British Columbia) | FC102 | 5 567 573, unidentified | A | A | I | A | A | A | 2 | 2b |
| Y | 1 CF patient; France (Paris); sputum | 1.78 | 1 047 573, unidentified | A | A | I | A | A | I | 2 | 2c |
| H | 1 CF patient; France (Giens); sputum | 1.15 | 1 467 553, unidentifiedb | A | A | I | A | A | I | 2 | 2c |
| BC | 1 CF patient; France (Suresnes); sputum | 2.65 | 5 467 573, unidentifiedb | A | A | I | A | A | I | 2 | 2c |
| AR | 1 CF patient; Denmark (Copenhagen) | A43 | 5 067 573, unidentifiedb | A | A | I | A | A | I | 2 | 2c |
| AS | 1 CF patient; Denmark (Copenhagen) | A44 | 1 467 573, B. cepacia (99.9) | A | A | I | A | A | I | 2 | 2c |
| AU | 1 CF patient; Denmark (Copenhagen) | A46 | 5 047 577, B. cepacia (99.9) | A | A | I | A | A | I | 2 | 2c |
| L | 5 CF patients; France (Clermont-Ferrand); sputum | 1.36 | 0 467 573, B. cepacia (99.9) | A | A | A | A | AH | AH | 1 | 1a |
| B | 7 CF patients; Canada (Vancouver, British Columbia; ET12 British-Canadian epidemic clone) | C5424 (16) | 0 477 573, B. cepacia (99.9) | A | A | A | A | A | A | 1 | 1b |
| D | 8 CF patients; France (Nancy); sputum | 1.6 | 1 477 577, B. cepacia (99.9) | A | A | A | A | A | A | 1 | 1b |
| A | 26 CF patients; France (Brest, 3; Roscoff, 10; Nancy, 7; Paris, 1; Pessac, 1; Dax, 2; Giens, 1; Dijon, 1); sputum | 1.1 | 1 467 573, B. cepacia (99.9) | A | A | A | A | A | A | 1 | 1b |
| 1 CF patient; Canada (Vancouver, British Columbia) | |||||||||||
| 17 non-CF patients; France (Bordeaux); blood culture, urine, respiratory specimens | |||||||||||
| BP | 12 non-CF patients; Senegal (Dakar); blood culture | DK1 | 0 467 577, B. cepacia (99.9) | A | A | A | A | A | A | 1 | 1b |
| E | 2 CF patients; France (Toulouse); sputum | 1.24 | 0 077 577, B. cepacia (85.7), P. aureofaciens (14.2) | A | A | A | A | A | A | 1 | 1b |
| O | 1 CF patient; France (Roscoff); sputum | 1.51 | 0 456 557, unidentified | A | A | A | A | A | A | 1 | 1b |
| AW | 1 CF patient; Canada (Cleveland); sputum | K128 | 0 477 573, B. cepacia (99.9) | A | A | A | A | A | A | 1 | 1b |
| AH | 1 CF patient; United Kingdom (Edinburgh) | A552 | 0 477 573, B. cepacia (99.9) | A | A | A | A | A | A | 1 | 1b |
| V | 1 CF patient; France (Giens); sputum | 1.58 | 0 077 577, B. cepacia (85.7), P. aureofaciens (14.2) | A | A | A | A | A | A | 1 | 1b |
| BA | 1 CF patient; France (Giens); sputum | 2.64 | 1 067 573, B. cepacia (81.0), P. aureofaciens (19) | A | A | A | A | A | A | 1 | 1b |
| CA | 1 non-CF patient; France (Toulouse); tracheal aspirate, urine | N25 | 0 477 577, B. cepacia (99.9) | A | A | A | A | A | A | 1 | 1b |
| CB | 1 CF patient; France (Dijon); sputum | 3.2 | 1 047 577, B. cepacia (68.5), P. aureofaciens (14.2), P. fluorescens (2.8) | A | A | A | A | A | A | 1 | 1b |
| BK | 1 non-CF patient; France (Toulouse); blood culture | N2 | 5 077 577, B. cepacia (99.9) | A | A | A | A | A | A | 1 | 1b |
| AF | 1 CF patient; United Kingdom (Edinburgh) | A599 | 0 467 573, B. cepacia (99.9) | A | A | A | A | A | AH | 1 | 1c |
| CC | 1 CF patient; France (Giens); sputum | 3.5 | 0 057 577, B. cepacia (50.3), P. aureofaciens (48.9) | A | A | A | A | H | H | 1 | 1d |
| AG | 1 CF patient; United Kingdom (Edinburgh) | A548 | 0 467 573, B. cepacia (99.9) | A | A | A | A | H | H | 1 | 1d |
| R | 1 CF patient; France (Nantes); sputum | 2.44 | 0 456 577, B. cepacia (99.3) | A | B | A | B | H | H | 3 | |
| AC | 1 CF patient; Canada (Calgary, Alberta); sputum | K130 | 4 456 573, unidentifiedb | A | B | A | B | H | H | 3 | |
| U | 2 CF patients; France (Suresnes, 1 patient; Toulouse, 1 patient); sputum | 2.10 | 1 067 577, B. cepacia (93.3), P. aureofaciens (6.7) | A | A | J | A | A | A | 4 | |
| BE | 1 CF patient; France (Dax); sputum | 2.75 | 0 067 553, unidentified | B | B | B | B | A | A | 5 | 5a |
| BG | 1 CF patient; France (Paris); sputum | 2.72 | 1 057 577, P. aureofaciens (59.9), B. cepacia (39.5) | B | B | F | B | A | A | 6 | |
| AP | 1 CF patient; Denmark (Copenhagen) | A41 | 5 067 573, unidentifiedb | A | A | C | A | A | AI | 14 | |
| P | 2 CF patients; Canada (Vancouver, British Columbia) | C5876 | 0 433 077, unidentified | A | A | A | G | A | A | 15 | |
| AX | 1 CF patient; France (Toulouse); sputum | 2.57 | 0 057 577, B. cepacia (50.3), P. aureofaciens (48.9) | B | B | C | B | H | H | 16 | |
If known.
Strain would be identified as B. cepacia (% identification of >99%) if oxidase positive.
The groups are defined on the basis of the RFLP profiles obtained with AluI, CfoI, DdeI, and MspI, and the subgroups are further defined with NciI and BssKI restriction results.
All reference and clinical strains were analyzed with the API 20NE system (bioMérieux, Marcy-l’Etoile, France). Oxidase was checked by using dimethyl-paraphenylenediamine disks (bioMérieux). The results of the API NE biochemical tests and of the oxidase reaction were transformed into a numerical profile, further interpreted by using the API NE analytical profile index, which provided percentages of identification.
RFLP analysis of amplified 16S rDNA.
The 18 reference strains and the 51 clinical isolates were tested. Furthermore, five isolates belonging to PCR ribotype F, four isolates belonging to PCR ribotype X, two isolates belonging to PCR ribotype D, eight isolates belonging to PCR ribotype A, two isolates belonging to PCR ribotype U, and two isolates belonging to PCR ribotype P were analyzed in order to check the homogeneity of the 16S rDNA restriction results within the same PCR ribotype.
DNA was prepared from chocolate agar (bioMérieux) cultures; a bacterial suspension, adjusted to 4 U of McFarland turbidity, was pelleted by centrifugation, washed, resuspended in 500 μl of distilled water, heated for 15 min at 95°C, and then centrifuged; the supernatant was stored at −20°C until used as a template.
Primers fD1 (5′-CCGAATTCGTCGACAACAGAGTTTGATCCTGGCTCAG-3′) and rD1 (5′-CCCGGGATCCAAGCTTAAGGAGGTGATCCAGCC-3′), which are complementary to conserved regions of the 16S rDNA (27), were used in a 100-μl reaction mixture containing 200 μM deoxynucleoside triphosphates (Perkin-Elmer, Saint-Quentin en Yvelines, France), 0.1 μM concentrations of primers (Bioprobe Systems, Montreuil, France), 20 mM Tris-HCl (pH 8.4), 50 mM KCl, 1.5 mM MgCl2, 2.5 U of Taq polymerase (Gibco-BRL Life Technologies, Gaithersburg, Md.), and 10 μl of template DNA or water for the negative control. An initial denaturing step of 95°C for 10 min was followed by 30 cycles of amplification (1 min at 94°C, 1 min at 55°C, and 2 min at 72°C) and a final extension step at 72°C for 10 min. DNA amplification was checked by electrophoresis of 5 μl of PCR product in a 1% agarose gel in Tris-borate-EDTA (TBE) buffer (pH 8.3) and by staining with ethidium bromide. Amplification products were stored at −20°C until digested. Restriction was performed separately with each enzyme by incubating 5 μl of amplified 16S rDNA overnight at the appropriate temperature with 5 U of endonuclease in a final volume of 25 μl. Six of the thirteen enzymes used by Laguerre et al. (14) to differentiate fluorescent Pseudomonas species were selected in our study because of their discriminatory power on Burkholderia species based on published rDNA sequences, i.e., AluI, CfoI, DdeI, MspI, and NciI (Gibco-BRL) and BssKI (i.e., ScrFI) (New England Biolabs, Beverly, Mass.). The restriction fragments were separated by electrophoresis in 3% agarose (Nusieve GTG; FMC, Rockland, Maine) in TBE buffer and stained with ethidium bromide by using marker VIII (Boehringer Mannheim, Meylan, France) as a molecular size ladder. The profiles obtained with each endonuclease were designated with uppercase letters. RFLP groups, designated with arabic numbers, were defined by the combination of the restriction profiles obtained with four of the six enzymes, i.e., AluI, CfoI, DdeI, and MspI, and RFLP subgroups, designated with lowercase letters, were further defined on the basis of NciI and BssKI patterns (Tables 1 and 2).
Carbon substrate assimilation tests.
A complete assimilation study (carbohydrates, amino acids, and organic acids) was performed on the 18 reference strains and on 35 clinical isolates belonging to 35 different PCR ribotypes and 14 16S rDNA genotypes by using Biotype 100 strips inoculated with bacterial suspensions in Biotype medium 1 according to the instructions of the manufacturer (bioMérieux). The strips were incubated at 30°C for 4 days, and growth was recorded at days 2 and 4. Assimilation data were entered in an Apple Macintosh computer with the Taxotron software (Institut Pasteur, Paris, France).
RESULTS
API 20NE identification.
The results obtained with reference strains are given in Table 1. B. cepacia, R. pickettii, and S. maltophilia are the only species among those tested which are included in the API 20NE database. Among the seven strains belonging to the cepacia complex, B. cepacia (ATCC 25416T) and B. cepacia genomovar II (B. multivorans LMG 13010T) were identified as B. cepacia, one strain (ATCC 25609) could not be identified, and the identification was incomplete for the four remaining strains (B. cepacia was not differentiated from P. aureofaciens). The nine type strains belonging to other Burkholderia species were either misidentified (five strains) as B. cepacia or as B. cepacia or P. aureofaciens, not identified (three strains [B. cocovenenans, B. caryophilli, and B. andropogonis]), or not typeable (one strain [B. glathei]). Finally, the type strains of R. pickettii and S. maltophilia were identified with a percentage of identification of >95%.
Among the 51 clinical isolates tested (Table 2), 24 (47%) were identified as B. cepacia with a percentage of identification of >99%, 15 (28%) were incompletely identified (B. cepacia, P. aureofaciens, or P. fluorescens), 1 (2%) was identified as Aeromonas salmonicida because of negative assimilation tests, and 11 (21%) were not identified. However, 6 of the 11 unidentified isolates, which were found to be oxidase negative, would be identified as B. cepacia with a percentage of identification of >99% if considered oxidase positive.
16S rDNA RFLP profiles.
The representatives of the five species or genomovars of the B. cepacia complex were classified in four RFLP groups (Table 1). B. cepacia genomovar I and III strains belonged to the same RFLP group (group 1), whereas B. cepacia genomovar II (B. multivorans), B. cepacia genomovar IV, and B. vietnamiensis belonged to different RFLP groups (groups 2, 3, and 4, respectively).
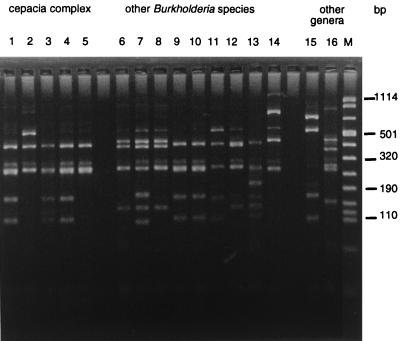
The nine type strains representing other Burkholderia species were classified in eight RFLP groups. B. pyrrocinia harbors the same restriction patterns as B. cepacia genomovar IV, whereas other species are easily differentiated from the B. cepacia complex. Interestingly, B. cocovenenans and B. gladioli pathovar gladioli belong to the same RFLP group, being only differentiated by their BssKI profile, and the two pathovars of B. gladioli could be differentiated on the basis of their DdeI profile. The 16S rDNA genotypes of R. pickettii and S. maltophilia type strains were clearly different from those of Burkholderia strains. Figure 1 illustrates the RFLP profiles obtained with DdeI for the different genomovars and species studied.
FIG. 1.
DdeI restriction patterns (type and reference strains). Lanes: 1 to 5 (cepacia complex strains), B. cepacia genomovar I ATCC 25416T, B. cepacia genomovar II (B. multivorans) LMG 13010T, B. cepacia genomovar III LMG 12615, B. cepacia genomovar IV LMG 14294, and B. vietnamiensis TVV75T, respectively; 6 to 14 (other Burkholderia species), B. gladioli pv. gladioli CFBP 2427T, B. gladioli pv. alliicola CFBP 2422T, B. cocovenenans LMG 11626T, B. pyrrocinia LMG 14191T, B. caryophilli LMG 2155T, B. andropogonis LMG 2129T, B. plantarii LMG 9035T, B. glumae CFBP 2430T, and B. glathei LMG 14190T, respectively; 15 and 16 (other genera), R. pickettii ATCC 27511T and Stenotrophomonas maltophilia ATCC 13637T, respectively; M, molecular weight marker VIII (Boehringer Mannheim).
The analysis of several clinical isolates belonging to the same PCR ribotype confirmed the homogeneity of the 16S rDNA restriction results, thus validating the selection of one isolate per PCR ribotype. The 51 representative CF and non-CF clinical isolates (Table 2) were classified into nine RFLP groups: 1, 2, 3, 4, 5, 6, 14, 15, and 16. The majority of the strains (43 of 51 [83%]) was shown to belong to 2 of the 9 groups defined. (i) Group 2 (26 of 51 strains [51%]) is characterized by an “I” profile with DdeI as for the B. multivorans type strain as shown in Table 1 and is further divided by BssKI into three RFLP subgroups (2a, 2b, and 2c). Subgroups 2b and 2c exhibit a plain BssKI pattern (A and I, respectively), whereas subgroup 2a exhibits the composite AI pattern like the B. multivorans type strain. This group comprises CF strains only, recovered from French, British, Danish, and Canadian patients, and includes two of the five clones proven to be highly transmissible in French CF centers (types F and X). (ii) Group 1 (17 of 51 strains [33%]) is characterized by an “A” profile with DdeI as for B. cepacia genomovar I and III representative strains and is further divided by NciI and BssKI, again according to whether the profile is plain (A and H) or composite (AH), into four RFLP subgroups (1a, 1b, 1c, and 1d), one of which (1b) is largely predominant. This group comprises CF isolates recovered from French, British, and Canadian patients and nosocomial isolates from France and Senegal. Five epidemic clones, involved in CF (the British-Canadian highly transmissible C5424 strain [7, 16] and the French types L, D, and A) and/or in nosocomial infections (types A and BP), belong to this group. Groups 3, 4, 5, and 6 could be respectively related to the B. cepacia genomovar IV reference strain or to B. pyrrociniaT, B. vietnamiensisT, B. gladioli pathovar gladioliT, and B. gladioli pathovar alliicolaT RFLP profiles and comprise a low number of CF strains (one to two strains). The three last groups (14–16) could not be clustered with any of the type or reference strains tested and comprise one CF strain each.
Biotype 100 assimilation tests.
One of the 18 reference strains (B. glathei LMG 14190T) and 5 of the 35 clinical isolates tested did not grow in Biotype 100 strips (typability, 82%). Thus, auxanographic results were available for 17 reference strains and 30 clinical isolates belonging to 8 of the 9 RFLP groups defined as described above. The main discriminating features are presented in Table 3. Strains belonging to RFLP group 2 (17 strains tested) were characterized by the constant absence of growth on sucrose which, in contrast, was assimilated by all the strains belonging to RFLP group 1 (12 strains tested). Sucrose assimilation was found to be generally absent for the type strains of other Burkholderia species, except for the B. vietnamiensis, B. caryophilli, and B. pyrrocinia type strains. Assimilation of dl-lactate was also found to be absent in most of the strains (15 of 17) belonging to RFLP group 2, whereas it did occur with most of the other strains tested (28 of 30). Within RFLP group 1, the results of some auxanographic tests, such as the benzoate, putrescine, and histamine tests, were found to be different for reference strains representative of genomovars I and III and heterogeneous among clinical strains. Numerous auxanographic features observed for the B. vietnamiensis type strain were markedly different from those observed for the RFLP-related clinical strain. In the same way, heterogeneous results were obtained within RFLP group 3.
TABLE 3.
Main discriminant auxanographic features based on the study of 17 reference strains and 30 clinical isolates
| Strain testeda | No. of strains tested | Substrateb
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sucrose | l(−)-Arabitol | d(−)-Tartrate | Ben-zoate | 5-Keto-d-gluconate | Genti-sate | Putre-scine | dl-Lactate | 5-Amino-valerate | Trypt-amine | Histamine | ||
| RFLP group 2 | ||||||||||||
| B. multivoransT | 1 | − | + | − | + | − | + | + | − | + | + | + |
| Clinical strains | 16 | − | 15/16 | 1/16 | 15/16 | 4/16 | + | 10/16 | 2/16 | 13/16 | 11/16 | 12/16 |
| RFLP group 1 | ||||||||||||
| Reference strains of B. cepacia gv I | 3 | + | + | − | + | + | + | + | + | + | 2/3 | + |
| Reference strain of B. cepacia gv III | 1 | + | + | − | − | + | + | − | + | + | + | − |
| Clinical strains | 8 | + | + | − | 5/8 | + | 5/8 | 4/8 | 7/8 | 5/8 | 3/8 | 1/8 |
| RFLP group 3 | ||||||||||||
| Reference strain of B. cepacia gv IV | 1 | − | + | − | + | + | + | + | + | + | − | + |
| B. pyrrociniaT | 1 | + | + | − | + | + | − | + | + | + | + | + |
| Clinical strain | 1 | − | + | − | + | + | + | + | + | + | + | + |
| RFLP group 4 | ||||||||||||
| B. vietnamiensisT | 1 | + | − | − | + | − | − | + | + | + | − | + |
| Clinical strain | 1 | − | + | − | + | − | + | − | − | − | − | − |
| RFLP group 5 | ||||||||||||
| B. gladioli pathovar gladioliT | 1 | − | − | + | + | + | − | − | + | − | − | − |
| B. cocovenenansT | 1 | − | − | + | + | + | − | − | + | − | − | − |
| Clinical strain | 1 | − | − | + | − | − | − | − | + | − | − | − |
| RFLP group 6 | ||||||||||||
| B. gladioli pathovar alliicolaT | 1 | − | − | + | + | + | − | − | + | − | − | − |
| Clinical strain | 1 | − | − | − | − | + | − | − | + | − | − | − |
| RFLP group 15, clinical strain | 1 | + | + | − | + | + | + | + | + | − | − | − |
| RFLP group 16, clinical strain | 1 | + | − | + | − | − | + | − | + | + | − | − |
| RFLP group 7, B. plantariiT | 1 | − | − | + | − | − | − | − | + | − | − | − |
| RFLP group 8, B. caryophilliT | 1 | + | − | − | − | + | − | − | + | − | − | − |
| RFLP group 9, B. andropogonisT | 1 | − | − | − | − | − | − | − | + | − | − | − |
| RFLP group 10, B. glumaeT | 1 | − | − | − | + | − | − | − | + | − | + | − |
| RFLP group 12, R. pickettiiT | 1 | − | − | − | − | − | − | − | + | − | − | + |
| RFLP group 13, S. maltophiliaT | 1 | + | − | − | − | − | − | − | + | − | − | − |
gv, genomovar; T, type strain. The groups are defined on the basis of the RFLP profiles obtained with AluI, CfoI, DdeI, and MspI.
+, all strains tested positive; −, all strains tested negative. Ratios give the number of positive strains over the total number of strains tested.
DISCUSSION
The accurate identification of B. cepacia is critical with regard to the medical and sociological consequences of the diagnosis for patients with CF. Nevertheless, the levels of pathogenicity and transmissibility within colonizing B. cepacia strains have been shown to be highly variable, and biologic prognostic markers are not available yet. On the other hand, there is increasing evidence (8, 25) that B. cepacia is a complex of closely related species rather than a single species. Moreover, another species of the genus Burkholderia, B. gladioli, is also involved in CF, but its pathogenic potential is still being discussed (4, 23). Thus, the unambiguous differentiation of Burkholderia species is undeniably the first obligatory step of any clinicobiological analysis of infections due to these organisms.
Phenotypic identification techniques are generally considered to be insufficiently reliable for this purpose, an observation which was confirmed by the present study. Fifty-one presumed clinical B. cepacia isolates were analyzed with the API 20NE numerical identification system, which is widely used in clinical microbiology laboratories, which was completed by use of the oxidase test. The following difficulties were encountered: (i) the poor detection of oxidase; (ii) the absence of Burkholderia species other than B. cepacia in the database; (iii) the inability to differentiate B. cepacia from P. aureofaciens, as previously reported by Kiska et al. (12) with the API Rapid NFT system; and (iv) the high number of unidentified strains (11 of 51 isolates [21%]). However, misidentifications at the genus level were rare (1 of 51 isolates [2%]).
Genomic analysis might provide more reliable tools for identification. So far, the reference method is the evaluation of DNA-DNA homologies between strains, which requires high technical skill and is time-consuming and thus not appropriate for routine diagnosis. On the other hand, the composition of 16S rDNA sequences, which are generally species specific, can be easily approached by analyzing the endonuclease restriction profiles of the previously amplified 16S rDNA gene. Furthermore, the selection of discriminating enzymes is facilitated by the fact that 16S rDNA sequences are available for many bacterial species. This method has been previously applied by Laguerre et al. (14) to the differentiation of fluorescent Pseudomonas species, and its use in the identification of Burkholderia species was assessed in the present study.
Of the 13 enzymes used by Laguerre et al., 6 (AluI, CfoI, DdeI, MspI, NciI, and BssKI) were used in the present study; four of these (AluI, CfoI, DdeI, and MspI) proved to be the more useful in species classification. However, this method did not allow the differentiation of genomovar I and III representative strains, in contrast to the DNA binding values reported by Vandamme et al. (25) between these strains (maximum, 45%). Further studies are required in order to search for more discriminating enzymes; nevertheless, if the low DNA-binding value is due to the different size of the genome, which was shown to be highly variable among B. cepacia strains (15), this quest might be unsuccessful. On the other hand, the DdeI pattern allowed the differentiation of B. multivoransT (genomovar II) and B. vietnamiensisT from the representative strains of genomovars I and III. Lastly, the 16S rDNA RFLP profile of the B. cepacia genomovar IV reference strain tested was clearly different from those obtained with the other genomovars but was indistinguishable from the genotype of the type strain of B. pyrrocinia (formerly Pseudomonas pyrrocinia). This provides an additional argument for the inclusion of the species pyrrocinia into the genus Burkholderia (25, 26). The relationship between the two species requires further investigation, since DNA hybridization results between these strains were not published. Reference strains belonging to other Burkholderia species, i.e., B. cocovenenans, B. gladioli (pathovar gladioli and pathovar alliicola), B. plantarii, B. caryophilli, B. andropogonis, B. glumae, and B. glathei, were all differentiated. It should be observed that the profiles of B. gladioli pathovar gladioli and of B. cocovenenans are very close, differing only by the BssKI restriction profile; the high level of similarity between these species has been previously reported (26). The easy differentiation of B. gladioli from strains of the cepacia complex is particularly interesting since phenotypic features described as discriminating between B. gladioli and B. cepacia, i.e., negative oxidase and lysine decarboxylase (LDC), the absence of lactose and maltose fermentation, and aminoglycoside susceptibility, may be equivocal (23).
In a second step, the PCR-RFLP method has been applied to a large variety of presumed B. cepacia isolates recovered from CF or nosocomial infections that are either epidemic or sporadic and are from different countries. The previous typing of all the isolates by PCR ribotyping allowed a selection of genetically unrelated strains to assess the diversity within this bacterial group. Nine RFLP groups were delineated among these clinical strains on the basis of the restriction patterns obtained with four of the six restriction enzymes (AluI, CfoI, DdeI, and MspI). However, further variations could be defined within groups 2 and 1, by NciI and/or BssKI analysis, with plain (A, H, and I) and composite (AH and AI) profiles. The existence of composite profiles may be attributed to variations among the different operons, since B. cepacia was shown to harbor at least six rDNA operons (3, 15, 19). Whether the subgroups determined by NciI and/or BssKI reflect intraspecific or interspecific variations is unclear. Since similar variations were observed among the three type and reference strains belonging to genomovar I, the analysis of a large number of reference strains of each species is required to evaluate intraspecific heterogeneity with regard to NciI and BssKI restriction profiles. In the present study, CF isolates were mainly distributed in RFLP groups 2 and 1 which, respectively, include the B. multivorans type strain and B. cepacia reference strains classified by Vandamme et al. in the genomovars I and III, whereas the distribution of the few nosocomial isolates studied herein was restricted to RFLP group 1. The easy differentiation of genomovars I and III would be interesting, since Vandamme et al. showed than genomovar III strains were mainly involved in CF, whereas genomovar I strains were mainly recovered from natural and hospital environments and from non-CF patients (25). None of the phenotypic features described by Vandamme et al. (25) appears to be strictly specific to genomovar I or III, the most constant one being the absence of growth of genomovar I strains at 42°C. Whereas most of the highly transmissible strains and all the isolates associated with acute clinical decline (8, 25) were previously reported to belong to genomovar III, our data show that among the five highly transmissible clones in French CF centers, three (A, L, and D) actually belong to the genomovar I/III-related RFLP group, but two (F and X), of which one (F) has been associated with fatal septicemias, belong to the B. multivorans-related RFLP group. The phenotypic differentiation of B. multivorans proposed by Vandamme et al. (25) is based upon a low number of characters and is not really clear-cut. One of the interesting characters could be LDC determination, reported to be negative in the description of the species B. multivorans but not documented for the other genomovars; LDC has been previously reported to be positive in 80% of B. cepacia strains (18). In our study, phenotypic analysis was restricted to auxanographic data, and the sole distinctive feature was the absence of growth of B. multivorans-related strains on sucrose. The discriminating value of this character requires confirmation on a higher number of strains.
Three other species and/or genomovars are also involved, though rarely, in CF patients: B. gladioli, B. vietnamiensis, and B. cepacia genomovar IV (4, 6, 25). The accurate identification of these species is essential in order to evaluate their pathogenic potential in CF and thus for guiding the segregation and treatment of patients who are infected with these bacteria. In our study, five isolates could be related to these species, though the relation between B. cepacia genomovar IV and B. pyrrocinia, which share the same RFLP pattern, requires further investigation. Finally, three isolates exhibiting unique RFLP profiles (14–16) could not be related to any representative of the Burkholderia species tested, suggesting the involvement of as-yet-uncharacterized species in CF.
In conclusion, the PCR-RFLP method described here appears to be an efficient identification tool for Burkholderia species. Its taxonomic value has to be validated by the analysis of a larger number of reference strains and above all by DNA-DNA hybridization studies. The use of additional endonucleases might also improve its discriminatory power within the B. cepacia complex. If these preliminary results are confirmed, the newly individualized B. multivorans species should be considered to play a significant part in CF and to include transmissible and virulent clones to the same extent as the B. cepacia genomovar III strains.
ACKNOWLEDGMENTS
We are indebted to the following members of the French Observatoire B. cepacia who addressed the CF isolates studied herein: J.-P. Chazalette and J. Carrère, Hôpital Renée Sabran, Giens; F. Huet and A. Pechinot, Hôpital du Bocage, Dijon; P. Domblides and J. Texier-Maugein, Pharmacie du Haut Lévêque, Pessac; J. Navarro and E. Bingen, Hôpital Robert Debré, Paris; A. Clement and H. Vu Thien, Hôpital Trousseau, Paris; D. Hubert and G. Paul, Hôpital Cochin, Paris; B. Sablayrolles, Mucozenne, and J. Lemozy, Hôpital Purpan, Toulouse; F. Varaigne and C. de Gialluly, Hôpital Bretonneau, Tours; G. Picherot and J.-Y. Le Berre, Centre Hospitalier de Saint-Nazaire; T. Perez and M.-O. Husson, Centre Hospitalier, Lille; S. Dominique, O. Mouterde, and M. Nouvellon, Hôpital Charles Nicolle, Rouen; F. Pennaforte and O. Bajolet-Laudinat, American Memorial Hospital, Reims; J. Derelle and M. Weber, Hôpital d’Enfants, Vandœuvre Les Nancy; G. Rault and J. Thubert, Centre Hélio Marin, Roscoff; M. Stern and P. Honderlick, Hôpital Foch, Suresnes; M.-C. Héraud and J. Sirot, Hôtel-Dieu, Clermont-Ferrand; M.-R. Munck and D. Tande, Hôpital A. Morvan, Brest; R. Barbier and J.-P. Lafargue, Centre Hospitalier, Dax; F. Brémont, Hôpital Purpan, and G. Chabanon, Hôpital Rangueil, Toulouse; and V. David and H. Richet, Hôpital Mère Enfant, Nantes. We are grateful to P. Rodriguez, Hôpital Pellegrin, Bordeaux, France, and M. F. Cissé, Hôpital d’Enfants A. Royer, Dakar, Senegal, for providing isolates involved in nosocomial outbreaks. We thank Michèle Cros, Sylvie Daudé, Corinne Fréchou, Aline Prieux, and David Sibrac for technical assistance.
This work was supported by a grant (97-0) from the Association Française de Lutte contre la Mucoviscidose and by the Glaxo Laboratories.
REFERENCES
- 1.Brett P, DeShazer D, Woods D. Burkholderia thailandensis sp. nov., a Burkholderia pseudomallei-like species. Int J Syst Bacteriol. 1998;48:317–320. doi: 10.1099/00207713-48-1-317. [DOI] [PubMed] [Google Scholar]
- 2.Burkholder W H. Sour skin, a bacterial rot of onion bulbs. Phytopathology. 1950;40:115–117. [Google Scholar]
- 3.Cheng H P, Lessie T G. Multiple replicons constituting the genome of Pseudomonas cepacia 17616. J Bacteriol. 1994;176:4034–4042. doi: 10.1128/jb.176.13.4034-4042.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Christenson J C, Welch D F, Mukwaya G, Muszynski M J, Weaver R E, Brenner D J. Recovery of Pseudomonas gladioli from respiratory tract specimens of patients with cystic fibrosis. J Clin Microbiol. 1989;27:270–273. doi: 10.1128/jcm.27.2.270-273.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dasen S E, LiPuma J J, Kostman J R, Stull T L. Characterization of PCR ribotyping for Burkholderia (Pseudomonas) cepacia. J Clin Microbiol. 1994;32:2422–2424. doi: 10.1128/jcm.32.10.2422-2424.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gillis M, Van Van T, Bardin R, Goor M, Hebbar P, Willems A, Segers P, Kersters K, Heulin T, Fernandez M P. Polyphasic taxonomy in the genus Burkholderia leading to an emended description of the genus and proposition of Burkholderia vietnamiensis sp. nov. for N2-fixing isolates from rice in Vietnam. Int J Syst Bacteriol. 1995;45:274–289. [Google Scholar]
- 7.Govan J R, Brown P H, Maddison J, Doherty C J, Nelson J W, Dodd M, Greening A P, Webb A K. Evidence for transmission of Pseudomonas cepacia by social contact in cystic fibrosis. Lancet. 1993;342:15–19. doi: 10.1016/0140-6736(93)91881-l. [DOI] [PubMed] [Google Scholar]
- 8.Govan J R W, Hughes J E, Vandamme P. Burkholderia cepacia: medical, taxonomic and ecological issues. J Med Microbiol. 1996;45:395–407. doi: 10.1099/00222615-45-6-395. [DOI] [PubMed] [Google Scholar]
- 9.Graves M, Robin T, Chipman A M, Wong J, Khashe S, Janda J M. Four additional cases of Burkholderia gladioli infection with microbiological correlates and review. Clin Infect Dis. 1997;25:838–842. doi: 10.1086/515551. [DOI] [PubMed] [Google Scholar]
- 10.Isles A, Maclusky I, Corey M, Gold R, Prober C, Fleming P, Levison H. Pseudomonas cepacia infection in cystic fibrosis: an emerging problem. J Pediatr. 1984;104:206–210. doi: 10.1016/s0022-3476(84)80993-2. [DOI] [PubMed] [Google Scholar]
- 11.Kanj S S, Tapson V, Davis R D, Madden J, Browning I. Infections in patients with cystic fibrosis following lung tranplantation. Chest. 1997;112:924–930. doi: 10.1378/chest.112.4.924. [DOI] [PubMed] [Google Scholar]
- 12.Kiska D L, Kerr A, Jones M C, Caracciolo J A, Eskridge B, Jordan M, Miller S, Hughes D, King N, Gilligan P H. Accuracy of four commercial systems for identification of Burkholderia cepacia and other gram-negative nonfermenting bacilli recovered from patients with cystic fibrosis. J Clin Microbiol. 1996;34:886–891. doi: 10.1128/jcm.34.4.886-891.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kostman J R, Edlind T D, LiPuma J J, Stull T L. Molecular epidemiology of Pseudomonas cepacia determined by polymerase chain reaction ribotyping. J Clin Microbiol. 1992;30:2084–2087. doi: 10.1128/jcm.30.8.2084-2087.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Laguerre G, Rigottier-Gois L, Lemanceau P. Fluorescent Pseudomonas species categorized by using polymerase chain reaction (PCR)/restriction fragment analysis of 16S rDNA. Mol Ecol. 1994;3:479–487. doi: 10.1111/j.1365-294x.1994.tb00126.x. [DOI] [PubMed] [Google Scholar]
- 15.Lessie T, Hendrickson W, Manning B, Devereux R. Genomic complexity and plasticity of Burkholderia cepacia. FEMS Microbiol Lett. 1996;144:117–128. doi: 10.1111/j.1574-6968.1996.tb08517.x. [DOI] [PubMed] [Google Scholar]
- 16.Mahenthiralingam E, Campbell M E, Henry D A, Speert D P. Epidemiology of Burkholderia cepacia infection in patients with cystic fibrosis: analysis by randomly amplified polymorphic DNA fingerprinting. J Clin Microbiol. 1996;34:2914–2920. doi: 10.1128/jcm.34.12.2914-2920.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pitt T L, Kaufman M E, Patel P S, Benge L C A, Gaskin S, Livermore D M. Type characterisation and antibiotic susceptibility of Burkholderia (Pseudomonas) cepacia isolates from patients with cystic fibrosis in the United Kingdom and the Republic of Ireland. J Med Microbiol. 1996;44:203–210. doi: 10.1099/00222615-44-3-203. [DOI] [PubMed] [Google Scholar]
- 18.Richard C, Monteil H, Megraud F, Chatelain R, Laurent B. Caractères phénotypiques de 100 souches de Pseudomonas cepacia. Proposition d’un schéma de biovars. Ann Biol Clin. 1981;39:9–15. [PubMed] [Google Scholar]
- 19.Rodley P D, Römling U, Tümmler B. A physical genome map of the Burkholderia cepacia type strain. Mol Microbiol. 1995;17:57–67. doi: 10.1111/j.1365-2958.1995.mmi_17010057.x. [DOI] [PubMed] [Google Scholar]
- 20.Ross J P, Holland S M, Gill V J, DeCarlo E S, Gallin J I. Severe Burkholderia (Pseudomonas) gladioli infection in chronic granulomatous disease: report of two successfully treated cases. Clin Infect Dis. 1995;21:1291–1293. doi: 10.1093/clinids/21.5.1291. [DOI] [PubMed] [Google Scholar]
- 21.Segonds C, Bingen E, Couetdic G, Mathy S, Brahimi N, Marty N, Plesiat P, Michel-Briand Y, Chabanon G. Genotypic analysis of Burkholderia cepacia isolates from 13 cystic fibrosis centers. J Clin Microbiol. 1997;35:2055–2060. doi: 10.1128/jcm.35.8.2055-2060.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Segonds C, Chabanon G, Bingen E, Michel-Briand Y, Couetdic G. Program and Abstracts of the 31st January 1997 SFM Conference on Pseudomonas et Bactéries Apparentées. Paris, France: Société Française de Microbiologie; 1997. Epidémiologie de la colonisation par Burkholderia cepacia des patients atteints de mucoviscidose en France: approche bioclinique; p. 23. [Google Scholar]
- 23.Simpson I N, Finlay J, Winstaley D J, Dewhurst N, Nelson J W, Butler S L, Govan J R W. Multi-resistance isolates possessing characteristics of both Burkholderia (Pseudomonas) cepacia and Burkholderia gladioli from patients with cystic fibrosis. J Antimicrob Chemother. 1994;34:353–361. doi: 10.1093/jac/34.3.353. [DOI] [PubMed] [Google Scholar]
- 24.Urakami T, Ito-Yoshida C, Araki H, Kijima T, Suzuki K I, Komagata K. Transfer of Pseudomonas plantarii and Pseudomonas glumae to Burkholderia spp. and description of Burkholderia vandii sp. nov. Int J Syst Bacteriol. 1994;44:235–245. [Google Scholar]
- 25.Vandamme P, Holmes B, Vancanneyt M, Coenye T, Hoste B, Coopman R, Revets H, Lauwers S, Gillis M, Kersters K, Govan J R W. Occurrence of multiple genomovars of Burkholderia cepacia in cystic fibrosis patients and proposal of Burkholderia multivorans sp. nov. Int J Syst Bacteriol. 1997;47:1188–1200. doi: 10.1099/00207713-47-4-1188. [DOI] [PubMed] [Google Scholar]
- 26.Viallard V, Poirier I, Cournoyer B, Haurat J, Wiebkin S, Ophel-Keller K, Balandreau J. Burkholderia graminis sp. nov., a rhizospheric Burkholderia species, and reassessment of (Pseudomonas) phenazinium, (Pseudomonas) pyrrocinia, and (Pseudomonas) glathei as Burkholderia. Int J Syst Bacteriol. 1998;48:549–563. doi: 10.1099/00207713-48-2-549. [DOI] [PubMed] [Google Scholar]
- 27.Weisburg W G, Barns S M, Pelletier D A, Lane D J. 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol. 1991;173:697–703. doi: 10.1128/jb.173.2.697-703.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yabuuchi E, Kosako Y, Oyaizu H, Yano I, Hotta H, Hashimoto Y, Ezaki T, Arakawa M. Proposal of Burkholderia gen. nov. and transfer of seven species of the genus Pseudomonas homology group II to the new genus, with the type species Burkholderia cepacia (Palleroni and Holmes 1981) comb. nov. Microbiol Immunol. 1992;36:1251–1275. doi: 10.1111/j.1348-0421.1992.tb02129.x. [DOI] [PubMed] [Google Scholar]
- 29.Yabuuchi E, Kosako Y, Yano I, Hotta H, Nishiuchi Y. Transfer of two Burkholderia and an Alcaligenes species to Ralstonia gen. nov.: proposal of Ralstonia pickettii (Ralston, Palleroni and Doudouroff 1973) comb. nov., Ralstonia solanacearum (Smith 1896) comb. nov. and Ralstonia eutropha (Davis 1969) comb. nov. Microbiol Immunol. 1995;39:897–904. doi: 10.1111/j.1348-0421.1995.tb03275.x. [DOI] [PubMed] [Google Scholar]
- 30.Zhao N, Qu C, Wang E, Chen W. Phylogenetic evidence for the transfer of Pseudomonas cocovenenans (van Damme et al. 1960) to the genus Burkholderia as Burkholderia cocovenenans (van Damme et al. 1960) comb. nov. Int J Syst Bacteriol. 1995;45:600–603. doi: 10.1099/00207713-45-3-600. [DOI] [PubMed] [Google Scholar]