Abstract
Background
Schizophrenia (SCZ) is a severe psychiatric disorder that affects approximately 0.75% of the global population. Both genetic and environmental factors contribute to development of SCZ. SCZ tends to run in family while both genetic and environmental factor contribute to its etiology. Much evidence suggested that alterations in DNA methylations occurred in SCZ patients.
Methods
To investigate potential inheritable pattern of DNA methylation in SCZ family, we performed a genome-wide analysis of DNA methylation of peripheral blood samples from 106 Chinese SCZ family trios. Genome-wide DNA methylations were quantified by Agilent 1 × 244 k Human Methylation Microarray.
Findings
In this study, we proposed a loci inheritance frequency model that allows characterization of differential methylated regions as SCZ biomarkers. Based on this model, 112 hypermethylated and 125 hypomethylated regions were identified. Additionally, 121 hypermethylated and 139 hypomethylated genes were annotated. The results of functional enrichment analysis indicated that multiple differentially methylated genes (DMGs) involved in Notch/HH/Wnt signaling, MAPK signaling, GPCR signaling, immune response signaling. Notably, a number of hypomethylated genes were significantly enriched in cerebral cortex and functionally enriched in nervous system development.
Interpretation
Our findings not only validated previously discovered risk genes of SCZ but also identified novel candidate DMGs in SCZ. These results may further the understanding of altered DNA methylations in SCZ.
Keywords: Schizophrenia, DNA Methylation, Trios, Chinese
Research in context.
Evidence before this study
Epigenetic dysregulation of the genome can lead to chronic alterations of neurodevelopment, synaptic architecture, and cellular signaling. Therefore, epigenetic dysregulation increases the risk of psychotic disorders. It has been noticed that SCZ frequently ran in families with high heritability. In addition, abnormal DNA methylations have been detected in SCZ patients. However, few reports of genome-wide DNA methylation pattern in SCZ trios have been reported.
Added value of this study
Based on current understanding of DNA methylation, we proposed a novel quantitative method to identify altered DNA methylation regions that are associated with SCZ in large family trio samples, which provided new idea for epigenetic analysis in family-based linkage study. Moreover, this study provided a series of candidate epigenetic markers of SCZ based on information of families of SCZ patients.
Implications of all the available evidence
Our findings demonstrated that familial environment contributed to global DNA methylation pattern. Some identified hypomethylation sites in blood appear to contribute more to SCZ status. Most of our identified epigenetic variations were not genetic in origin. Therefore, we provided new evidence of the important role of DNA methylation in the inheritance and pathogenesis of SCZ.
Alt-text: Unlabelled box
1. Introduction
Schizophrenia (SCZ) is a common psychiatric disorder affecting approximately 0.75% of the worldwide population and often results in lifelong mental disability [1]. SCZ frequently ran in families and its heritability has been estimated to be as high as 70–81% [2]. Mounting evidence have demonstrated the critical role of genetic variants in the emergence of SCZ, including common variants, copy number variants (CNVs), rare and de novo mutations [3], [4], [5], [6], [7], [8], [9]. However, the pathogenesis of SCZ could not be well defined based on existing findings yet. Despite multiple genetic variants have been reported, recent epigenetic analyses of SCZ are providing new insights into our understanding of the complex associations between epidemiological heritability and the phenotypic variation [10,11].
Epigenetic dysregulation of the genome has been shown to lead to chronic alterations of neurodevelopment, synaptic architecture, and cellular signaling. Therefore, specific epigenetic dysregulation may increase the risk of psychotic disorders [12], [13], [14], [15]. In particular, abnormalities in DNA methylation have been detected in the brains of SCZ and bipolar disorder patients. The involvement of alternations in DNA methylation in disease pathophysiology may explain the clinical dynamics observed in these diseases [16,17]. Genome-wide DNA methylation analysis studies using blood and postmortem brain tissues have revealed multiple differentially methylated regions (DMRs) in SCZ patients [14,16,[18], [19], [20], [21], [22]]. Though only a small proportion of identified aberrant DNA methylations in SCZ were in common in these studies, these results still demonstrated that altered DNA methylation occurred in SCZ patients and may serve as candidate biomarker of SCZ.
Omics analysis of SCZ had been widely performed in sporadic cohorts. Compared with case-control design, trios design has several advantages. For example, trio design can be used to identify variants inherited from the parents, detect rare risk variants, avoid population heterogeneity [23]. In 2018, a family-based study reported that inheritance of a translocation linked to major mental illness is associated with differential DNA methylations in a large family [24]. Recently an exome-sequencing study using more than 2000 SCZ trios has identified protein-coding de novo mutations that carried risk of SCZ [25]. However, few DNA methylation analysis in large SCZ trios has been reported.
In this study, we performed a genome wide DNA methylation analysis in 106 SCZ family trios. Genome-wide DNA methylation was profiled on peripheral blood using Agilent 1 × 244 K Methylation microarray with correction for cell type heterogeneity or age-associated loci. We developed a novel scoring model that was able to estimate the loci inheritance frequencies (LIF) in order to rank the DMRs, which may serve as SCZ biomarkers. We expect this study to provide an advance workflow for future family-based analysis of DNA methylation in SCZ trios as well as novel insights into the role of DNA methylations in inheritance and etiology of SCZ.
2. Methods
2.1. Samples
In this study, a total of 200 Chinese Han SCZ family trios (600 participants) were recruited from Shanghai Mental Health Center. SCZ was diagnosed according to DSM (Diagnostic and Statistical Manual of Mental Disorders)-IV criteria by two independent psychiatrists. This study was approved by the Ethical Committee of Bio-X Institutes of Shanghai Jiao Tong University. All subjects gave informed consent for their participation. After quality control of microarray-based methylation analysis, 106 trios were eventually included in further analysis.
2.2. DNA methylation detection
Sample genomic DNAs (gDNAs) were extracted from the peripheral blood of the subjects using QIAmp DNA Blood Kits (Qiagen, USA). Then methylated DNA immunoprecipitation and differentially methylation region (DMR) screening was performed using Agilent 1 × 244 K DNA methylation Microarray (Agilent, USA) according to the manufacturer's instructions. Images of the slides were acquired using Agilent SureScan Microarray Scanner G2505C and transformed to digital features using Agilent Feature Extraction program (version 10.7.1.1). The feature data were analyzed using Agilent GenomicWorkBench (version 7.0) for quality control and DMR calling. Based on BATMAN algorithm [26], the methylation status of all probes in each samples was calculated and shown with three categorized score: “1” (high, DNA methylation > 60%), “0” (moderate, DNA methylation 40 ∼ 60%) and “-1” (low, DNA methylation < 40%) [26]. Probes associated with cellular specificity or aging [27,28] and probes located on sex chromosomes were excluded.
2.3. Loci inheritance frequency (LIF) model designing
2.3.1. Exclusion of the probes based on their combinations among family trios
Among 106 trios enrolled, 86 trios consisted of normal parents, 19 trios consisted of only one affected parent and 1 trio consisted of both affected parents. First, we defined the status of a probe within a family as a combination. For example, for a probe within a family, the status of father, mother and child was high [1], moderate (0), low (-1) respectively and thus the combination of the probe in this family was “10-1”. The distribution of combinations of all probes within all families was summarized (Fig. S1).
As shown in Fig. S1, most combinations were made up of high and low status (approximately more than 80%), while the combinations containing moderate status accounted for less than 20% proportion. The technique we used to scan methylation level was based on methylated DNA immunoprecipitation coupled with CpG island microarray (MeDIP-CGI-arrays) platform. As Rajendram et al. described, MeDIP-CGI array were less reliable in predicting intermediate (moderate) levels of DNA methylation [29]. Hence all the combinations containing moderate status were excluded for further study.
2.3.2. Calculation of the frequencies of the probes fitting the inheritance mode
After excluding moderate methylations, our LIF model hypothesized that methylation levels of sites associated with schizophrenia took place a wide-range alteration (> 20% at least) compared to normal status. For each probe, we assumed a status (hypermethylation or hypomethylation) as a phenotype of schizophrenia. Then we calculated the frequencies of each status of a probe fitting the familial inheritance mode among all family trios except the only one family consisting of both SCZ parents (Fig. S2). The reason why we excluded the family with both SCZ parents were that the combinations of this model would be confounded with the high distribution of “111” and “-1-1-1” combinations.
2.3.3. Identifying high-confident loci
The frequencies of two status of each probe fitting the model were sorted from high to low respectively. And the more one was defined as major type and another one as minor type. The condition that the both were zero was excluded. For major type, top 12.5% loci of high frequencies were extracted as a threshold. And then those of the loci fitting the equation that“Major/Minor > = 3” were extracted. This threshold was determined based on the distribution of the most confident epialleles (Top 100) among all the C/I ratio. As Fig. S3 showed that the distribution of all C/I (conformity/ inconformity) value.
2.3.4. Bioinformatics annotations
We used Venny (https://bioinfogp.cnb.csic.es/tools/venny/, version 2.1) to draw gene venn diagram [30]. We used Cytoscape software with the package “cytohubba” by Maximal Clique Centrality (MCC) algorithm for calculating top 10 hub to construct hub gene network [31]. STRING database was used to conduct gene ontology (GO) analysis, Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis and protein-protein interaction network [32,33]. We used Cis-regulatory Element Annotation System (CEAS) on Cistrome platform (http://cistrome.org/) for enrichment on chromosome and annotation [34,35]. We used Reactome platform (https://reactome.org/) for pathway analysis [36]. We used SZDB database (http://www.szdb.org/) for result evaluation [37,38]. Human protein atlas database [39] and TissueEnrich tools [40] were used to make a tissue-specific gene enrichment of our gene sets (https://tissueenrich.gdcb.iastate.edu/). All the analyses mentioned above were using default parameters. SNPs within identified sites were extracted by UCSC genome browser based on Hg19 common151SNP database (https://genome-asia.ucsc.edu/index.html). Bedtools was used to preprocess datasets for DNA methylation quantitative trait locus (mQTL) enrichment analysis. For the further study, 25 bps were added to each side of each loci. Two mQTL datasets from brain tissue [13] and whole blood [19] respectively was used to estimate their overlapping with identified loci in this study. R package “hlclust” was used for hierarchical clustering. Transcription factor motif enrichment analyses were performed using the MEME suite's AWE software (https://meme-suite.org/meme/index.html). All probes excluded age-associated and cell-type specific sites were set as control sequences. The following command was used: ame –verbose 1 –oc . –scoring avg –method fisher –hit-lo-fraction 0.25 –evalue-report-threshold 10.0 –control –shuffle– –kmer 2 hyper_site_bed_25bp.fasta db/HUMAN/HOCOMOCOv11_core_HUMAN_mono_meme_format.meme
2.3.5. Statistics
As shown in Fig. S2, C/I ratio = Max(Hypermethylation(Frequency(A) + Frequency(B) + Frequency(C)), Hypomethylation(Frequency(A) + Frequency(B) + Frequency(C))) ÷ Min(Hypermethylation(Frequency(A) + Frequency(B) + Frequency(C)), Hypomethylation(Frequency (A) + Frequency(B) + Frequency(C))). Statistical analysis of two parameters was performed using either a two-tailed, unpaired t test, or two-tailed, unpaired Mann-Whitney test based on data distribution.
2.3.6. Role of funding source
The funders of this study had no role in study design, data collection, data analyses, interpretation, or writing of the report.
3. Results
3.1. Data evaluation of all samples
Quality control was performed by filtering the result of microarray-based DNA methylation analysis according to Agilent microarray pipeline. After data preprocessing, a dataset of 106 trios was merged to perform further analyses. Each sample had 252,698 values of probes. Most of the probes were located on CpG islands of human genome. Each probe covered 50 bp genome region.
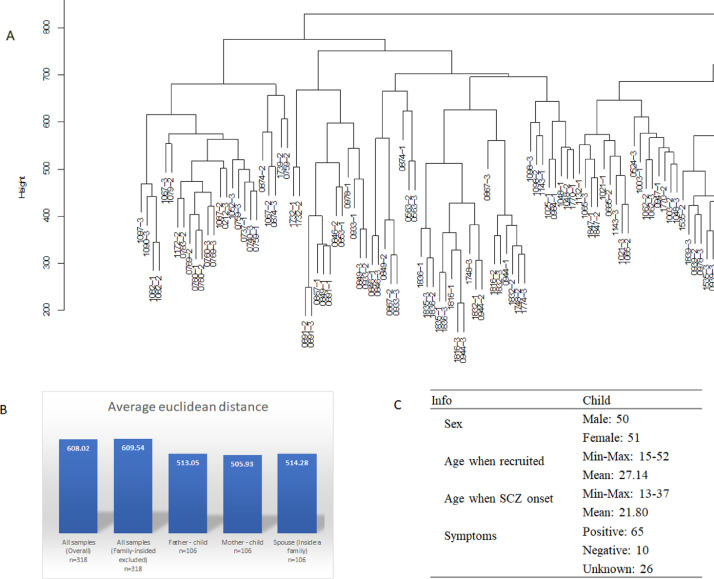
To determine the distributional characteristics of familial DNA methylation data, hierarchical clustering was performed using all 106 families. In addition, the average arithmetic euclidean distances between samples were measured. Interestingly, the family members tended to be clustered together (Part in Fig. 1A, all picture in Fig. S7). The overall average distance between spouses or parent-offspring couple was much lower than that across all samples (pvalue < 2.2 e-16, Fig. 1B). These results suggested that familial factors affected individual DNA methylation pattern significantly and it provided relatively similar epigenetic background within a family, which might be caused by similar life style.
Fig. 1.
Distribution of methylation data among all samples. A. Unsupervised hierarchical cluster of methylation data among 106 family trios (Part). B. Average euclidean distance between samples. The distance was calculated by R function dist() as default parameters. C. Information of the offsprings among 101 family trios.
Among all 106 families, 101 of them had detailed epidemiological data while the rest 5 families with only their family roles and disease status. The summarized epidemiological information of affected-children in the 101 families was presented in Fig. 1C. Among all trios, 86 trios consisted of normal spouses, 19 families consisted of one affected parent and 1 trio consisted of both affected parents.
3.2. Identification of SCZ-related DMRs
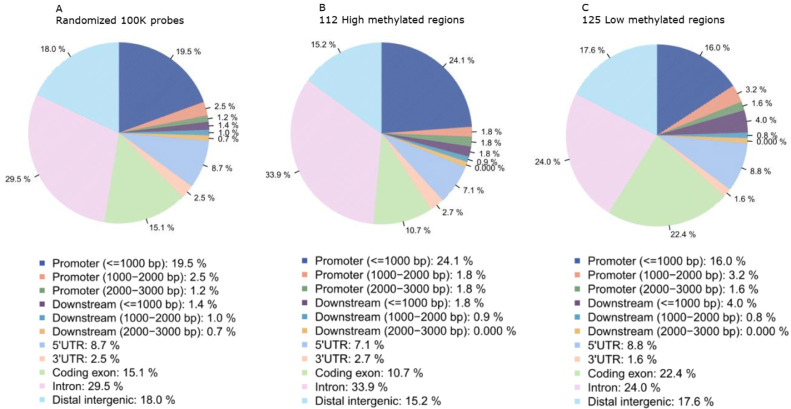
We developed a loci inheritance frequency (LIF) model to identify the top ranked probes that are related with SCZ (Methods). After excluding 51,295 probes associated with cellular specificity or with aging or conserved or located on sex chromosomes, we identified 112 high methylated and 125 low methylated regions using our model with the following criteria: C/I ratio > 3 (Table S1). The hypermethylated regions had no overlap with the hypomethylated regions. The genomic distributions of DMRs from LIF model were presented in Fig. 2 based on reference genome Hg19 [41]. Obviously, most differentially methylated regions (DMRs) located at genes nearby, including proximal promoter (< = 1000 bp), 5’UTR, coding exons, introns and distal intergenic.
Fig. 2.
Genomic distributions of identified DMRs. A. Annotation of randomized 100,000 probes to show the overall distribution of all chromosomes. B. Annotation of identified high methylated regions. C. Annotation of identified low methylated regions.
In addition, we checked if the regions of identified DMRs contain single nucleotide polymorphisms (SNPs). As a result, 21 SNPs were located within the hypermethylated regions and 25 SNPs were located within the hypomethylated regions (Table S6). The result demonstrated that a majority of identified DMRs were SNP-free regions.
3.3. Comprehensive analysis of DMR-related genes
We annotated the DMRs within 5 k bp of the probe based on hg19 reference genome [41]. 121 hypermethylated (High) and 139 hypomethylated (Low) genes were derived from the annotation of top DMRs (Table S2).
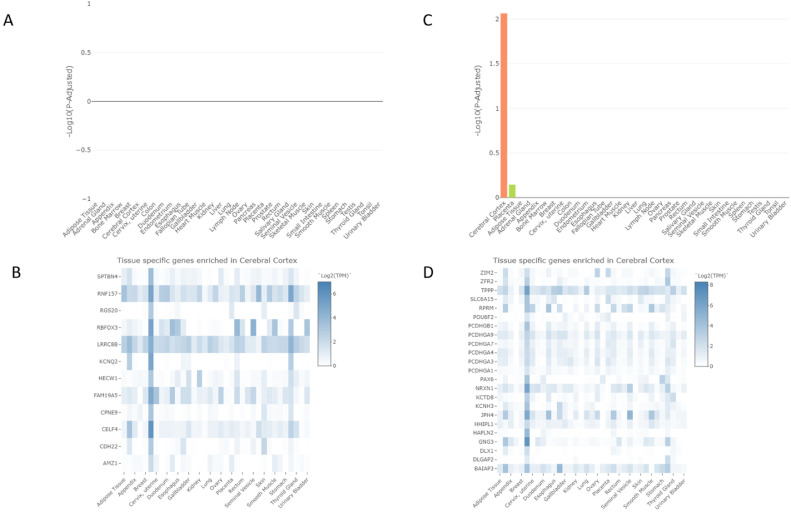
In order to obtain a comprehensive annotation of these genes, we first used Human protein atlas database [39] and TissueEnrich tools [40] to make a tissue-specific gene enrichment of our gene sets (Fig. 3). Hypermethylated genes consisted of 12 tissue specific genes (TSG) in cerebral cortex but showed no significant enrichment (Fig. 3A and B). Hypomethylated genes were significantly enriched in cerebral cortex (23 TSGs, -log10 p-adjusted = 2.06, Fold change = 2.21), Fig. 3C and D) and the second top enriched tissue was placenta (6 TSGs, -log10 p-adjusted = 0.15, Fold change = 2.39, Fig. 3C).
Fig. 3.
Tissue-specific gene enrichment analysis. A. Tissue-specific gene enrichment of hypermethylated genes in 35 tissues. B. Hypermethylated tissue specific genes in cerebral cortex. C. Tissue-specific gene enrichment of hypomethylated genes in 35 tissues. D. Hypomethylated tissue specific genes in cerebral cortex.
Moreover, GO, KEGG and Reactome pathway and protein-protein interaction (PPI) analyses were conducted [36]. GO annotations showed significant enrichment within hypomethylated genes (Table 1, S7), while no significant enrichment within hypermethylated genes. Top 5 enriched pathways of hypermethylated/hypomethylated gene sets from Reactome pathway analysis (Entities p value < 0.05, Tables S3–S4) were summarized in Table 1. Overall, there was Notch/HH/Wnt, GPCR, immune response signaling-related enrichments in hypomethylated group and MAPK, GPCR signaling-related enrichment in hypermethylated group. Moreover, hypomethylated genes showed a significant enrichment in nervous system development. All these pathways such as nervous development, signaling transduction, immune response had been reported to involve with pathogenesis of schizophrenia.
Table 1.
Functional enrichment of DMR genes.
| Hypermethylated genes | Hypomethylated genes | |
|---|---|---|
| BP(GO) | NA | Homophilic cell adhesion via plasma membrane adhesion molecules |
| Cell-cell adhesion via plasma-membrane adhesion molecules | ||
| Cell adhesion | ||
| Nervous system development | ||
| Cell-cell signaling | ||
| MF(GO) | NA | Calcium ion binding |
| Cation binding | ||
| Metal ion binding | ||
| Ion binding | ||
| CC(GO) | NA | Integral component of plasma membrane |
| Plasma membrane | ||
| Membrane | ||
| Local Network Cluster(STRING) |
NA | Calcium/calmodulin-dependent protein kinase II inhibitor and Cadherin, C-terminal catenin-binding domain |
| Cadherin-like, and N-terminal region of Chorein or VPS13 | ||
| mixed, incl. Cadherin cytoplasmic C-terminal, and SHC-transforming protein 4 | ||
| mixed, incl. Cadherin-like, and Transmembrane protein 132 | ||
| integrator complex | ||
| Top 5(Reactome) | Constitutive Signaling by Overexpressed ERBB2 |
FBXW7 Mutants and NOTCH1 in Cancer |
| GRB2 events in ERBB2 signaling | Loss of Function of FBXW7 in Cancer and NOTCH1 Signaling |
|
| TFAP2 (AP-2) family regulates transcription of growth factors and their receptors |
RET signaling | |
| Signaling by ERBB2 ECD mutants | Rap1 signaling | |
| Transcriptional regulation by the AP-2 (TFAP2) family of transcription factors |
Opioid Signaling |
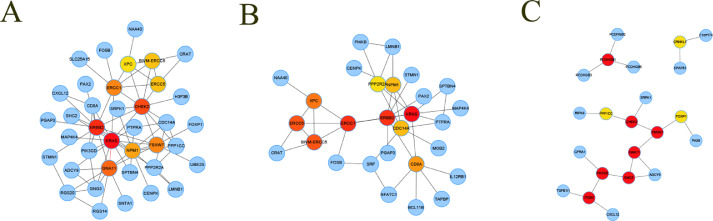
We constructed protein-protein interaction (PPI) network of annotated genes within 5 k bp of DMRs in hypermethylated and hypomethylated group (Figs S4–6). Hub gene networks were extracted from the PPI networks (Fig. 4A–C). Only hypomethylated genes showed significant PPI pathway enrichments (p < 0.05). When Genecards database was used to annotate gene-related diseases, [42] hub gene network of hypomethylated group has two major categories: neurogenesis diseases and immune diseases. The result indicated that hypomethylation of specific sites could be a phenotype of schizophrenia.
Fig. 4.
Protein-protein interaction network of annotated genes. A. Hub gene network of all genes. B. Hub gene network of hypermethylated genes. C. Hub gene network of hypomethylated genes. Each node indicates a gene and each edge denotes an interaction between a pair of genes.
3.4. Estimation of genetic influences on DNA methylation
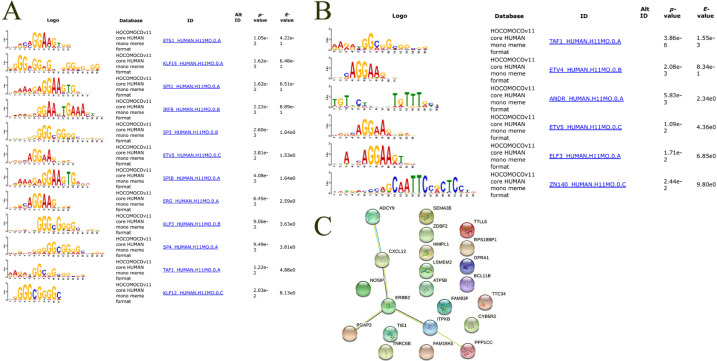
It was clear that differential methylation can occur in a site-specific way [43]. To investigate potential transcriptional factor (TF) binding motif among identified regions, transcription factor motif enrichment analyses were performed using the MEME suite's AWE software. All significant enriched results were shown in Fig. 5A and B. As we expected, some identified motifs in hypermethylated regions recognized GC-rich sequences. For example, SPI1 binds to a purine-rich sequence, Sp1, Sp3, and Sp3 binds to GC-rich motifs. However, the hypomethylated regions lacked this kind of motif.
Fig. 5.
Estimation of genetic influences on DNA methylation. A. Transcription factor motif enrichment analysis of hypermethylated regions (+-25bp). B. Transcription factor motif enrichment analysis of hypomethylated regions (+-25bp). C. Protein-protein interaction network of overlapped mQTL genes. 6 genes were enriched in ERBB-related pathway (ADCY9, CXCL12, ERBB2, PGAP3, ITPKB, PPP1CC).
To investigate the influence of genetics on the identified methylation loci, we first calculated intersections of the identified loci with two published mQTL dataset derived from brain tissue and whole blood, respectively [13,19]. The results showed, that identified hypermethylated sites had 3 overlaps with brain mQTLs and 4 with blood mQTLs and the hypomethylated sites had 10 overlaps with brain mQTLs and 8 with blood mQTLs (Table S5). Overlapped regions were annotated and corresponding gene were enriched in ERBB-related pathway (Fig. 5C). Totally, approximate 10% identified regions overlapped with published mQTL regions.
In sum, these results suggested that most of our identified epigenetic variations were not genetic in origin.
4. Discussion
In this study, we performed methylome-wide analysis of DNA methylation in SCZ family trios with Han Chinese ethnic origins. Through methylated DNA immunoprecipitation (MeDIP) method, we got semi-quantitative methylated status of each regions. Then we proposed an original scoring model to rank SCZ-related DMRs. Our results not only found some reported DMRs in SCZ, but also provided a set of potential SCZ risk genes with altered DNA methylation for further studying.
As far as we know, tools or algorithms taking account the inheritance of DNA methylation were lacking for family-based DNA methylation analysis. The most popular genetic analysis of family-based data is transmission disequilibrium test (TDT) [44]. However, there was not a considerable TDT-like method for epigenetics data because inheritance pattern of epigenetic modification was quite different from that of genetic variations. Considering the binomial and reprogramming pattern of DNA methylation, we established the loci inheritance frequency (LIF) model to rank potential SCZ related DMRs by their C/I ratio among trios. For this model, we assumed (a) some DNA methylation status could be inheritable across generations, (b) DNA methylation could be a biomarker of SCZ, (c) DMRs has a light or moderate but cumulative effect on disease status. In this study, the factor of family disease history was excluded from the LIF model.
It has been found that heritability of DNA methylation elevated at sites which are more variable or with intermediate levels of DNA methylation [43]. The term ‘variable site’ means those where the range of DNA methylation values for the middle 80% of individuals was greater than 5% or with intermediate levels of DNA methylation (i.e. those where the mean level of DNA methylation was between 20% and 80%). These sites were of great value for their study of associations between DNA methylation and diseases. However, due to the different technique platform (MeDIP-CGI-array vs BS-whole_genome-array), sites with intermediate levels of DNA methylation were excluded in our analysis. As Rageen Rajendram et al. described, MeDIP-CGI array were less reliable in predicting intermediate (moderate) levels of DNA methylation [29]. In another word, our study focused on those ‘non-variable sites’ (i.e. those hyper/hypo-methylated sites). These regions might be more conserved and most identified sites in our study seemed to be independent of known tagging genetic variations. In future studies a comprehensive screening including all kinds of epigenetic variations would be necessary.
Among genes with high C/I scores, we found that many of them had been reported to be involved with SCZ. For example, previous studies suggested that NRG1-ERBB signaling was involved in the pathogenesis of SCZ [45,46] and inhibition of ERBB2 could block NRG1-stimulate cell adhesion [47]. ERBB2 also showed peripheral transcription deregulation in treatment resistant SCZ [48]. ERBB2 was a hypermethylated SCZ risk gene in our LIF model. Our result supported deregulation of ERBB2 in SCZ. In additional, FBXW7 was a hypomethylated hub gene in our LIF model. FBXW7 tags the DISC1 protein for destruction via the ubiquitin-proteosome system. The disruption of FBXW7-DISC1 interaction could stabilize DISC1 protein and counteract DISC1 deficiencies observed in neural progenitor cells derived from induced pluripotent stem cells from SCZ patients with a DISC1 frameshift mutation [49]. Also the expression of CDC42EP3, a hypomethylated gene, was reported to be significantly increased in subjects with SCZ [50]. These lines of evidence supported that upregulations of FBXW7 and CDC42EP3 were associated with the pathogenesis of SCZ.
Interestingly, only the hypomethylated genes could be significantly enriched into specific pathways or functions, which indicated an inner connection of these genes. Hub gene-related diseases of hypomethylated group could be divided into two major categories: neurogenesis diseases and immune diseases. This result might contribute to the explanation of SCZ pathogenesis or phenotypes of SCZ patients. Compared with 2 epigenome wide association study of large SCZ cohort published previously [16,22], we replicated the findings of several genes including NFATC1, AGAP1, RAP1GAP2, FOXP1. It is necessary to validate our findings in further studies.
In this study, the limitation of sample size was the main obstacle for statistical analysis. The LIF model was calculated based the inheritance frequencies in all families and thus its confidence should increase with larger sample size. However, we currently do not have enough samples to be used as validation cohort to conduct a validation of the epialleles identified. It is extremely challenging to recruit more qualified patient family samples. We have also searched public databases for any methylation profile data of schizophrenia trios. However, this data type is so rare that we did not find any cohort data from earlier studies that can be used to validate our results. We hope more DNA methylation data of SCZ trios to be published in the future, which could help to validate our findings. Another limitation was whether the DMRs we identified could account for the pathogenesis or the phenotype of SCZ. Accroding to the results, we thought the results might represent the mixture of both. To validate this assumption, more functional experiments were needed. The third limitation is that family-based linkage study for DNA methylation analysis could not reduce the noise caused by various environmental factors such as smoking, drug and many other. However, an advantage of family-based study for DNA methylation analysis was that it provided relatively similar epigenetic background within a family, which could reduce the noise caused by life style for detecting disease risk loci.
In summary, we performed a genome wide DNA methylation analysis using 106 SCZ family trios in this study. We successfully develop a novel method to identify the altered DNA methylation regions associated with SCZ. We not only validated previously characterized risk genes of SCZ, but also identified a number of candidate genes that are involved in the altered methylation in SCZ. Our findings provided new evidence of the role of DNA methylation in the inheritance and pathogenesis of SCZ.
Supplementary material
Fig. S1. The distribution of combinations of all probes within families. A. The distribution of combinations of all probes within 86 families consisted of normal parents. B. The distribution of combinations of all probes within the families consisted of an affected father. C. The distribution of combinations of all probes within the families consisted of an affected mother.
Fig. S2. The combination of probes fitting the disease inheritance mode among all families. C/I ratio = Max(Hypermethylation(Frequency(A) + Frequency(B) + Frequency(C)), Hypomethylation(Frequency(A) + Frequency(B) + Frequency(C))) ÷ Min(Hypermethylation(Frequency(A) + Frequency(B) + Frequency(C)), Hypomethylation(Frequency (A) + Frequency(B) + Frequency(C))).
Fig. S3. Distribution of C/I ratio value of all probes among the fitting model A–C. Frequencies of C/I ratio value among trios consisting of both normal parents (A), affected father (B) and affected mother (C) were shown, respectively.
Fig. S4. PPI network of all identified genes.
Fig. S5. PPI network of all hypermethylated genes.
Fig. S6. PPI network of all hypomethylated genes.
Fig. S7. Unsupervised hierarchical cluster of methylation data among 106 family trios
Table S1. Detail information of identified regions associated with schizophrenia.
Table S2. Genomic annotation of identified regions.
Table S3. Reactome pathway analysis of hypermethylated genes.
Table S4. Reactome pathway analysis of hypomethylated genes.
Table S5. The overlapping between identified regions with reported mQTLs (+-50 bp).
Table S6. SNPs located in identified regions.
Table S7. STRING enrichment of identified genes.
Contributors
Lu Shen, Hailiang Huang and Shengying Qin, conceptualization of the study. Lu Shen, Xiaoying Lv, Mo Li, Cong Huai and Xi Wu, data curation and formal analysis of the study. Hao Wu, Jie Tan, Jingsong Ma, Luan Chen, Ting Wang and Yidan Sun, methodology and validation of the study. Shengying Qin, Chunling Wan and Lin He, funding acquisition. Chao Yang, Lei Cai, Yana Lu, Yan Zhang, Saizheng Weng, Shaobin Tai, clinical investigation and patient recruitment. Na Zhang, project administration. Lu Shen and Xiaoying Lv, writing of original draft.
Data sharing statement
All microarray raw data are available with publication (NGDC database OMIX ID: OMIX530, https://ngdc.cncb.ac.cn/). Analysis scripts can be downloaded from Github (https://github.com/PMC-BioX-SJTU/SczMethylatioS).
Declaration of Competing Interest
The authors declare that no conflict of interest.
Acknowledge
This work was supported by grants from the 863 Program (2012AA02A515, 2012AA021802), the National Nature Science Foundation of China (81421061, 81273596, J1210047, 30900799, 81361120389, 30972823), National key research and development program (2016YFC0905000), the Public Health Key Disciplines in Shanghai-Health Microbiology (12GWZX0801), Shanghai Key Laboratory of Psychotic Disorders 13dz2260500, Public Science and Technology Research Funds (201210056), the Shanghai Jiaotong University Interdisciplinary Research fund, and the Shanghai Leading Academic Discipline Project (B205).
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2021.103609.
Contributor Information
Lin He, Email: helin@sjtu.edu.cn.
Chunling Wan, Email: clwan@sjtu.edu.cn.
Shengying Qin, Email: chinsir@sjtu.edu.cn.
Appendix. Supplementary materials
References
- 1.Costain G., Bassett AS. Clinical applications of schizophrenia genetics: genetic diagnosis, risk, and counseling in the molecular era. Appl Clin Genet. 2012;5:1–18. doi: 10.2147/TACG.S21953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McGue M., Gottesman The genetic epidemiology of schizophrenia and the design of linkage studies. Eur Arch Psychiatry Clin Neurosci. 1991;240(3):174–181. doi: 10.1007/BF02190760. [DOI] [PubMed] [Google Scholar]
- 3.Avramopoulos D. Recent advances in the genetics of schizophrenia. Mol Neuropsychiatry. 2018;4(1):35–51. doi: 10.1159/000488679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pardinas A.F., Holmans P., Pocklington A.J., Escott-Price V., Ripke S., Carrera N. Common schizophrenia alleles are enriched in mutation-intolerant genes and in regions under strong background selection. Nat Genet. 2018;50(3):381–389. doi: 10.1038/s41588-018-0059-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schizophrenia Working Group of the Psychiatric Genomics C Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511(7510):421–427. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lewis C.M., Levinson D.F., Wise L.H., DeLisi L.E., Straub R.E., Hovatta I. Genome scan meta-analysis of schizophrenia and bipolar disorder, part II: schizophrenia. Am J Hum Genet. 2003;73(1):34–48. doi: 10.1086/376549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ng M.Y., Levinson D.F., Faraone S.V., Suarez B.K., DeLisi L.E., Arinami T. Meta-analysis of 32 genome-wide linkage studies of schizophrenia. Mol Psychiatry. 2009;14(8):774–785. doi: 10.1038/mp.2008.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Allen N.C., Bagade S., McQueen M.B., Ioannidis J.P., Kavvoura F.K., Khoury M.J. Systematic meta-analyses and field synopsis of genetic association studies in schizophrenia: the SzGene database. Nat Genet. 2008;40(7):827–834. doi: 10.1038/ng.171. [DOI] [PubMed] [Google Scholar]
- 9.Sun J., Kuo P.H., Riley B.P., Kendler K.S., Zhao Z. Candidate genes for schizophrenia: a survey of association studies and gene ranking. Am J Med Genet B Neuropsychiatr Genet. 2008;147B(7):1173–1181. doi: 10.1002/ajmg.b.30743. [DOI] [PubMed] [Google Scholar]
- 10.Labrie V., Pai S., Petronis A. Epigenetics of major psychosis: progress, problems and perspectives. Trends Genet. 2012;28(9):427–435. doi: 10.1016/j.tig.2012.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cariaga-Martinez A., Saiz-Ruiz J., Alelu-Paz R. From linkage studies to epigenetics: what we know and what we need to know in the neurobiology of schizophrenia. Front Neurosci. 2016;10:202. doi: 10.3389/fnins.2016.00202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gandal M.J., Haney J.R., Parikshak N.N., Leppa V., Ramaswami G., Hartl C. Shared molecular neuropathology across major psychiatric disorders parallels polygenic overlap. Science. 2018;359(6376):693–697. doi: 10.1126/science.aad6469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hannon E., Spiers H., Viana J., Pidsley R., Burrage J., Murphy T.M. Methylation QTLs in the developing brain and their enrichment in schizophrenia risk loci. Nat Neurosci. 2016;19(1):48–54. doi: 10.1038/nn.4182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jaffe A.E., Gao Y., Deep-Soboslay A., Tao R., Hyde T.M., Weinberger D.R. Mapping DNA methylation across development, genotype and schizophrenia in the human frontal cortex. Nat Neurosci. 2016;19(1):40–47. doi: 10.1038/nn.4181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ludwig B., Dwivedi Y. Dissecting bipolar disorder complexity through epigenomic approach. Mol Psychiatry. 2016;21(11):1490–1498. doi: 10.1038/mp.2016.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Montano C., Taub M.A., Jaffe A., Briem E., Feinberg J.I., Trygvadottir R. Association of DNA methylation differences with schizophrenia in an epigenome-wide association study. JAMA Psychiatry. 2016;73(5):506–514. doi: 10.1001/jamapsychiatry.2016.0144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huchegowda R., Shruti A., Amarendra S., Shraddha T., Huchegowda C. Integrative studies to design and validate wearable footwear among neuropathic patients. Diabetes Metab Syndr. 2019;13(3):2075–2079. doi: 10.1016/j.dsx.2019.03.038. [DOI] [PubMed] [Google Scholar]
- 18.Kinoshita M., Numata S., Tajima A., Ohi K., Hashimoto R., Shimodera S. Aberrant DNA methylation of blood in schizophrenia by adjusting for estimated cellular proportions. Neuromol Med. 2014;16(4):697–703. doi: 10.1007/s12017-014-8319-5. [DOI] [PubMed] [Google Scholar]
- 19.Hannon E., Dempster E., Viana J., Burrage J., Smith A.R., Macdonald R. An integrated genetic-epigenetic analysis of schizophrenia: evidence for co-localization of genetic associations and differential DNA methylation. Genome Biol. 2016;17(1):176. doi: 10.1186/s13059-016-1041-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wockner L.F., Noble E.P., Lawford B.R., Young R.M., Morris C.P., Whitehall V.L. Genome-wide DNA methylation analysis of human brain tissue from schizophrenia patients. Transl Psychiatry. 2014;4:e339. doi: 10.1038/tp.2013.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Numata S., Ye T., Herman M., Lipska BK. DNA methylation changes in the postmortem dorsolateral prefrontal cortex of patients with schizophrenia. Front Genet. 2014;5:280. doi: 10.3389/fgene.2014.00280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aberg K.A., McClay J.L., Nerella S., Clark S., Kumar G., Chen W. Methylome-wide association study of schizophrenia: identifying blood biomarker signatures of environmental insults. JAMA Psychiatry. 2014;71(3):255–264. doi: 10.1001/jamapsychiatry.2013.3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ott J., Kamatani Y., Lathrop M. Family-based designs for genome-wide association studies. Nat Rev Genet. 2011;12(7):465–474. doi: 10.1038/nrg2989. [DOI] [PubMed] [Google Scholar]
- 24.McCartney D.L., Walker R.M., Morris S.W., Anderson S.M., Duff B.J., Marioni R.E. Altered DNA methylation associated with a translocation linked to major mental illness. NPJ Schizophr. 2018;4(1):5. doi: 10.1038/s41537-018-0047-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Howrigan D.P., Rose S.A., Samocha K.E., Fromer M., Cerrato F., Chen W.J. Exome sequencing in schizophrenia-affected parent-offspring trios reveals risk conferred by protein-coding de novo mutations. Nat Neurosci. 2020;23(2):185–193. doi: 10.1038/s41593-019-0564-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Down T.A., Rakyan V.K., Turner D.J., Flicek P., Li H., Kulesha E. A Bayesian deconvolution strategy for immunoprecipitation-based DNA methylome analysis. Nat Biotechnol. 2008;26(7):779–785. doi: 10.1038/nbt1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bacos K., Gillberg L., Volkov P., Olsson A.H., Hansen T., Pedersen O. Blood-based biomarkers of age-associated epigenetic changes in human islets associate with insulin secretion and diabetes. Nat Commun. 2016;7:11089. doi: 10.1038/ncomms11089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jaffe A.E., Irizarry R.A. Accounting for cellular heterogeneity is critical in epigenome-wide association studies. Genome Biol. 2014;15(2):R31. doi: 10.1186/gb-2014-15-2-r31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rajendram R., Ferreira J.C., Grafodatskaya D., Choufani S., Chiang T., Pu S. Assessment of methylation level prediction accuracy in methyl-DNA immunoprecipitation and sodium bisulfite based microarray platforms. Epigenetics. 2011;6(4):410–415. doi: 10.4161/epi.6.4.14763. [DOI] [PubMed] [Google Scholar]
- 30.2007. J.C. Oliveros. Venny- an interactive tool for comparing lists with Venn diagrams
- 31.Chin C.H., Chen S.H., Wu H.H., Ho C.W., Ko M.T., Lin C.Y. cytoHubba: identifying hub objects and sub-networks from complex interactome. BMC Syst Biol. 2014;8(Suppl 4):S11. doi: 10.1186/1752-0509-8-S4-S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yu G.C., Wang L.G., Han Y.Y., He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. Omics. 2012;16(5):284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Szklarczyk D., Gable A.L., Lyon D., Junge A., Wyder S., Huerta-Cepas J. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47(D1):D607–DD13. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu T., Ortiz J.A., Taing L., Meyer C.A., Lee B., Zhang Y. Cistrome: an integrative platform for transcriptional regulation studies. Genome Biol. 2011;12(8):R83. doi: 10.1186/gb-2011-12-8-r83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shin H., Liu T., Manrai A.K., Liu X.S. CEAS: cis-regulatory element annotation system. Bioinformatics. 2009;25(19):2605–2606. doi: 10.1093/bioinformatics/btp479. [DOI] [PubMed] [Google Scholar]
- 36.Jassal B., Matthews L., Viteri G., Gong C., Lorente P., Fabregat A. The reactome pathway knowledgebase. Nucleic Acids Res. 2020;48(D1):D498–D503. doi: 10.1093/nar/gkz1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu Y., Li X., Liu J., Luo X.J., Yao Y.G. SZDB2.0: an updated comprehensive resource for schizophrenia research. Hum Genet. 2020;139(10):1285–1297. doi: 10.1007/s00439-020-02171-1. [DOI] [PubMed] [Google Scholar]
- 38.Wu Y., Yao Y.G., Luo X.J. SZDB: a database for schizophrenia genetic research. Schizophr Bull. 2017;43(2):459–471. doi: 10.1093/schbul/sbw102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yu N.Y., Hallstrom B.M., Fagerberg L., Ponten F., Kawaji H., Carninci P. Complementing tissue characterization by integrating transcriptome profiling from the human protein atlas and from the FANTOM5 consortium. Nucleic Acids Res. 2015;43(14):6787–6798. doi: 10.1093/nar/gkv608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jain A., Tuteja G. TissueEnrich: tissue-specific gene enrichment analysis. Bioinformatics. 2019;35(11):1966–1967. doi: 10.1093/bioinformatics/bty890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lander E.S., Linton L.M., Birren B., Nusbaum C., Zody M.C., Baldwin J. Initial sequencing and analysis of the human genome. Nature. 2001;409(6822):860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 42.Stelzer G., Rosen N., Plaschkes I., Zimmerman S., Twik M., Fishilevich S. The GeneCards suite: from gene data mining to disease genome sequence analyses. Curr Protoc Bioinf. 2016;54(1):30 1-1 3. doi: 10.1002/cpbi.5. [DOI] [PubMed] [Google Scholar]
- 43.Hannon E., Knox O., Sugden K., Burrage J., Wong C.C.Y., Belsky D.W. Characterizing genetic and environmental influences on variable DNA methylation using monozygotic and dizygotic twins. PLoS Genet. 2018;14(8) doi: 10.1371/journal.pgen.1007544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Clayton D. A generalization of the transmission/disequilibrium test for uncertain-haplotype transmission. Am J Hum Genet. 1999;65(4):1170–1177. doi: 10.1086/302577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Corfas G., Roy K., Buxbaum J.D. Neuregulin 1-erbB signaling and the molecular/cellular basis of schizophrenia. Nat Neurosci. 2004;7(6):575–580. doi: 10.1038/nn1258. [DOI] [PubMed] [Google Scholar]
- 46.Barros C.S., Calabrese B., Chamero P., Roberts A.J., Korzus E., Lloyd K. Impaired maturation of dendritic spines without disorganization of cortical cell layers in mice lacking NRG1/ErbB signaling in the central nervous system. Proc Natl Acad Sci U S A. 2009;106(11):4507–4512. doi: 10.1073/pnas.0900355106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kanakry C.G., Li Z., Nakai Y., Sei Y., Weinberger D.R. Neuregulin-1 regulates cell adhesion via an ErbB2/phosphoinositide-3 kinase/Akt-dependent pathway: potential implications for schizophrenia and cancer. PLoS One. 2007;2(12):e1369. doi: 10.1371/journal.pone.0001369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mostaid M.S., Lee T.T., Chana G., Sundram S., Shannon W.C., Pantelis C. Peripheral Transcription of NRG-ErbB pathway genes are upregulated in treatment-resistant schizophrenia. Front Psychiatry. 2017;8:225. doi: 10.3389/fpsyt.2017.00225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yalla K., Elliott C., Day J.P., Findlay J., Barratt S., Hughes Z.A. FBXW7 regulates DISC1 stability via the ubiquitin-proteosome system. Mol Psychiatry. 2018;23(5):1278–1286. doi: 10.1038/mp.2017.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ide M., Lewis D.A. Altered cortical CDC42 signaling pathways in schizophrenia: implications for dendritic spine deficits. Biol Psychiatry. 2010;68(1):25–32. doi: 10.1016/j.biopsych.2010.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.