Abstract
Virus entry, consisting of attachment to and penetration into the host target cell, is the first step of the virus life cycle and is a critical ‘do or die’ event that governs virus emergence in host populations. Most antiviral vaccines induce neutralizing antibodies that prevent virus entry into cells. However, while the prevention of virus invasion by humoral immunity is well appreciated, considerably less is known about the immune defences present within cells (known as intrinsic immunity) that interfere with virus entry. The interferon-induced transmembrane (IFITM) proteins, known for inhibiting fusion between viral and cellular membranes, were once the only factors known to restrict virus entry. However, the progressive development of genetic and pharmacological screening platforms and the onset of the COVID-19 pandemic have galvanized interest in how viruses infiltrate cells and how cells defend against it. Several host factors with antiviral potential are now implicated in the regulation of virus entry, including cholesterol 25-hydroxylase (CH25H), lymphocyte antigen 6E (LY6E), nuclear receptor co-activator protein 7 (NCOA7), interferon-γ-inducible lysosomal thiol reductase (GILT), CD74 and ARFGAP with dual pleckstrin homology domain-containing protein 2 (ADAP2). This Review summarizes what is known and what remains to be understood about the intrinsic factors that form the first line of defence against virus infection.
Subject terms: Innate immunity, Virus-host interactions
Besides neutralizing antibodies, viruses face a range of cell-intrinsic inhibitors that are specialized to limit virus entry into host cells. Majdoul and Compton describe the mechanisms of action of the cellular factors providing this important first line of defence against virus infection, including infection by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2).
Introduction
Among the most significant pathogens throughout human history are enveloped viruses characterized by a host-derived lipid membrane obtained as nascent virus particles (virions) exit the infected cell1. Upon encounter with a target cell, virions adhere to and initiate fusion with host cell membranes at the cell surface or following endocytosis2. Viruses encode fusion proteins, the fusogenic activity of which is activated following receptor binding and, in the case of endocytic viruses, conformational changes triggered by the increasingly acidic environment found within endosomes3 (Box 1). In all cases, membrane fusion is required to deliver the viral core, which contains the viral nucleic acid, into the host cell cytoplasm4 (Fig. 1). As it precedes and determines the extent to which viral genome replication, viral protein synthesis and virion assembly occur, virus entry into cells is a crucial aspect of infection. Factors promoting or limiting virus entry into host cells govern the extent to which viruses spread within and between individuals, while also determining the potential for virus emergence in additional host species.
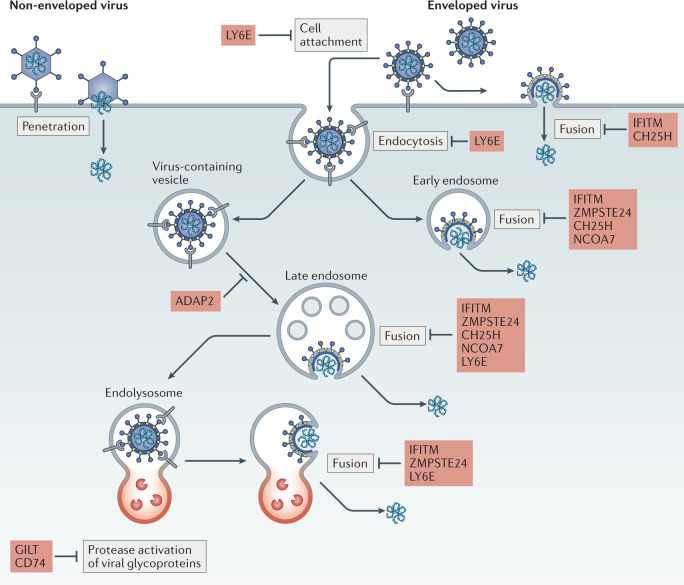
Fig. 1. Pathways for enveloped virus entry into cells and its restriction.
Viral envelope glycoproteins on the surface of the virion mediate cell attachment by interacting directly with a host receptor on the cell surface. Receptor interaction drives conformational changes allowing exposure of the fusion peptide, which inserts itself into host membranes to drive virus–cell fusion. Fusion occurs at the plasma membrane or, in the case of viral glycoproteins that exhibit pH-dependent fusogenic activity and/or activation by cellular proteases, at endosomal or lysosomal (referred to as endolysosomal) membranes. Completion of membrane fusion allows passage of the viral ribonucleoprotein complex, including viral nucleic acid, into the host cell cytoplasm — a prerequisite for subsequent stages of the virus life cycle. Host factors discussed in this Review are listed in red beside the entry step they inhibit. For comparison, entry by non-enveloped viruses usually involves membrane fusion-independent penetration into the host cell, as depicted on the left. ADAP2, ARFGAP with dual pleckstrin homology domain-containing protein 2; CH25H, cholesterol 25-hydroxylase; GILT, interferon-γ-inducible lysosomal thiol reductase; IFITM, interferon-induced transmembrane protein; LY6E, lymphocyte antigen 6E; NCOA7, nuclear receptor co-activator protein 7; ZMPSTE24, zinc metalloproteinase STE24.
Over evolutionary time, animal cells have evolved pathogen defence systems known as cell-autonomous immunity (also known as cell-intrinsic immunity or intrinsic immunity)5–7. Intrinsic immune proteins that are constitutively expressed and exhibit antiviral function are the first line of defence against viruses. Meanwhile, type I interferons (such as interferon-α (IFNα) and IFNβ), type II interferon (IFNγ) and type III interferons (IFNλ1–4) produced following virus sensing by the innate immune system give rise to additional layers of intrinsic protection that halt virus spread between cells8 (Box 2). The collective repertoire of constitutive and induced intrinsic factors targets all steps of the virus life cycle, including entry into cells, to prevent infection before it starts or to contain infection to a limited number of cells. As a result, an effective intrinsic response can prevent or dampen viral disease before the onset of adaptive immunity.
When a novel virus emerges within a naive host population and pre-existing adaptive immunity is lacking, host susceptibility is governed largely by intrinsic immunity. This is evidenced by the fact that many intrinsic defence genes evolve through gene duplication and diversifying (positive) selection9. These telltale patterns of adaptive evolution are believed to result from selective pressures applied by highly pathogenic viruses in the past, those that may have decimated ancestral host populations and selected for genetic variants conferring resistance among the survivors10,11. Prolonged virus–host conflicts extending across generations have given rise to many intrinsic immune defence factors (also known as restriction factors)12. In this Review, we provide an up-to-date overview of the factors providing intrinsic defence against virus entry into cells.
Box 1 Viral glycoproteins in fusion-mediated virus entry.
Following adherence to the host cell surface, enveloped viruses acquire access to the cell cytoplasm by initiating fusion with the plasma membrane or endomembranes (those of early endosomes, late endosomes or endolysosomes). The site at which different viruses undergo fusion with cells depends on the presence of environmental cues needed to activate the fusogenic potential of viral fusion proteins (also known as viral glycoproteins). Three classes of viral glycoprotein exist (class I, class II and class III); these are based on protein structure and the requirements necessary for fusion triggering: low pH, receptor binding and low pH, or receptor binding and cleavage by a host protease153. Some viruses are capable of entering cells via multiple routes, depending on the milieu provided by a particular host cell. Entry of severe acute respiratory syndrome coronavirus 2, for example, requires that the viral glycoprotein (spike) binds to angiotensin-converting enzyme 2 (ACE2) and becomes proteolytically cleaved by one of two host proteases: transmembrane protease serine 2 (TMPRSS2) at the cell surface or cathepsin L/D in endolysosomes. The pH and/or protease triggers provided by the host cell lead to membrane fusion because they drive conformational changes in the viral glycoprotein, allowing exposure of a fusion peptide that inserts itself directly into cell membranes. Enveloped viruses differ in the number of glycoproteins they carry, but only one of them possesses a fusion peptide capable of carrying out membrane merger3.
Much insight into the virus entry process (as well as the host factors that inhibit it) has been gleaned from experimental approaches that focus on the sole and unique properties of specific viral glycoproteins: virus pseudotyping, in which different viral glycoproteins are incorporated into the same viral core (often vectors derived from retroviruses such as murine leukaemia virus and HIV-1 or vesicular stomatitis virus) and their capacity to infect cells is examined; and cell–cell fusion assays, whereby viral glycoproteins are expressed in one set of cells (donor cells) that are co-cultured with cells expressing the cellular receptor (target cells). Viral glycoprotein-dependent fusion of the plasma membranes of donors and targets, which may be triggered by lowering the pH of the cell culture medium and/or by adding protease, leads to the formation of multinucleated syncytia and is used as a proxy for the fusion reaction between the virus particle and the target cell and can be used to identify the cellular cues and requirements that enable it.
Box 2 Types of interferons and induction of the antiviral state in cells.
Interferons are cytokines that are produced by cells following pathogen exposure. There are three distinct interferon families. Type I interferons consist of interferon-α (IFNα; of which there are 13 subtypes in humans), IFNβ and several less characterized cytokines (IFNε, IFNτ, IFNκ, IFNω, IFNδ and IFNζ). Type II interferon is represented by a single member, IFNγ, whereas type III interferons are made up of IFNλ1, IFNλ2, IFNλ3 and IFNλ4 (ref.154). Interferons are produced in response to pathogen recognition by receptors found in the cytosol or membranes of cells, such as RIG-I, cyclic GMP–AMP synthase (cGAS) and Toll-like receptors. All interferon types act in an autocrine and paracrine manner to activate the transcription of antimicrobial genes known as interferon-stimulated genes (ISGs), which regulate innate and adaptive immunity in diverse ways. The potential impact of each interferon type in a given tissue is determined by the expression of interferon genes and the expression of receptors that specifically recognize interferon ligands. Nearly all cell types respond to type I interferons and type II interferon because their receptors (IFNAR1 and IFNAR2, and IFNGR1 and IFNGR2, respectively) are ubiquitously expressed. Type III interferons, meanwhile, bind IFNLR1 and IL-10Rβ, which are preferentially expressed by epithelial cells of mucosal surfaces155. Upon ligation of interferon receptors, Janus kinases phosphorylate signal transducer and activator of transcription 1 (STAT1) and STAT2, which form a complex with interferon regulatory factor 9 (IRF9) and translocate into the nucleus. This complex then binds to interferon-stimulated response elements within the promoters of ISGs. Despite using different receptors, type I and type III interferons induce similar gene expression programmes. However, type I interferon signalling leads to a more rapid and more amplified induction of ISGs. In this manner, type I interferons induce the expression of hundreds of ISGs, including a large number that collectively impose an antiviral state156.
IFITM proteins
The human interferon-induced transmembrane protein (IFITM) family consists of five members, including IFITM1, IFITM2 and IFITM3, which belong to the CD225 superfamily13 and have been studied actively in the context of virus infections. The antiviral properties of IFITM1, IFITM2 and IFITM3 were discovered following a small-interfering RNA-based screen for intrinsic inhibitors of influenza A virus (IAV), West Nile virus (WNV) and dengue virus (DENV) infections14. Their characterization as the first factors known to restrict cellular entry following virus adhesion to the cell surface was demonstrated with use of virus ‘pseudotypes’ and by cell–cell fusion experiments15–17 (Box 1). Together, these techniques have been used to show that IFITM proteins, when expressed in target cells, broadly inhibit the cell entry step of diverse viruses, including orthomyxoviruses (such as IAV), paramyxoviruses (parainfluenza virus, metapneumovirus and respiratory syncytial virus)18–22, rhabdoviruses (vesicular stomatitis virus (VSV)), flaviviruses (WNV, DENV, hepatitis C virus (HCV), Zika virus (ZIKV) and yellow fever virus)14,23–29, filoviruses (Ebola virus (EBOV) and Marburg virus)17,30, poxviruses (vaccinia virus and cowpox virus (CPXV))31, bunyaviruses (Rift Valley fever virus and La Crosse virus)32, alphaviruses (chikungunya virus, Sindbis virus, Semliki Forest virus)33–35, lentiviruses (human and simian immunodeficiency viruses)36–39 and coronaviruses (human coronavirus 229E (hCoV-229E), severe acute respiratory syndrome coronavirus (SARS-CoV), Middle East respiratory syndrome coronavirus (MERS-CoV) and SARS-CoV-2)17,40–44 (Table 1). By contrast, amphotropic murine leukaemia virus, Sendai virus, papillomavirus, cytomegalovirus, adenovirus and the arenaviruses Lassa virus (LASV), Machupo virus and lymphocytic choriomeningitis virus are resistant to inhibition by IFITM1, IFITM2 and IFITM3 (refs45,46). IFITM5 (also known as BRIL) and IFITM10 are not known to participate in human immune functions and, contrary to their name, they are not upregulated by interferon13. Among those that are endowed with antiviral activity, IFITM3 is the best characterized because it exhibits the most potent restriction of important viral pathogens such as IAV and ZIKV26,33,47. Furthermore, single-nucleotide polymorphisms in human IFITM3 have been associated with severe outcomes following infection by IAV48,49 and SARS-CoV-2 (refs50,51). Mice encode IFITM proteins as well, and in addition to the five found in humans, they also possess IFITM6 and IFITM7. Knockout of Ifitm3 alone or the entire IFITM locus was shown to result in elevated morbidity and mortality following experimental infection by IAV48,52,53 (and other viruses as well34,54,55), attesting to the physiological importance of IFITM3 in limiting viral pathogenesis.
Table 1.
Characteristics of intrinsic inhibitors of enveloped virus entry
| Intrinsic inhibitor | Entry step inhibited | Mechanism or mechanisms | Human enveloped viruses targeted | Cell types expressed | Subcellular localization | Induction |
|---|---|---|---|---|---|---|
| IFITM1, IFITM2, IFITM3 | Membrane fusion or endocytic trafficking | Membrane modulation (rigidity, curvature), endosome redirection and virion degradation | VSV, IAV, DENV, ZIKV, HCV, YFV, WNV, EBOV, RVFV, CHIKV, HIV-1, hCoV-229E, SARS-CoV, MERS-CoV, SARS-CoV-2 | Ubiquitous | Plasma membrane, endomembranes (transmembrane) | Constitutive, type I interferons, type II interferon, LPS, IL-6, TGFβ |
| ZMPSTE24 | Membrane fusion | Cooperation with IFITM proteins | VSV, IAV, LCMV, LASV, CPXV, VACV | Ubiquitous | Nuclear envelope, endomembranes (transmembrane) | Constitutive |
| CH25H | Membrane fusion | Membrane modulation via product 25HC (rigidity, curvature, cholesterol depletion) | VSV, EBOV, HIV-1, ZIKV, RVFV, HSV, NiV, SARS-CoV-2 | Immune cells, epithelial cells | Endoplasmic reticulum (transmembrane) | Type I interferons, type II interferon |
| LY6E | Cell attachment or membrane fusion | Receptor downmodulation or endosome redirection | DENV, ZIKV, YFV, WNV, HIV-1, hCoV-229E, hCoV-OC43, SARS-CoV, MERS-CoV, SARS-CoV-2 | Immune cells, epithelial cells | Plasma membrane | Retinoic acid, type I interferons |
| NCOA7 | Viral glycoprotein activation | Enhanced cathepsin activity (virion degradation) | VSV, IAV, HCV | Immune cells, nervous cells, epithelial cells | Nucleus, endomembranes | Type I interferons |
| GILT | Viral glycoprotein activation | Inhibition of cathepsin activity | EBOV, LASV, HIV-1, SARS-CoV | Immune cells | Cytosol, endomembranes (lumen) | Constitutive, type II interferon |
| CD74 | Viral glycoprotein activation | Inhibition of cathepsin activity | EBOV, SARS-CoV-2 | Immune cells | Plasma membrane, endomembranes (transmembrane) | LPS, type II interferon |
| ADAP2 | Endocytic trafficking | Endosome redirection and virion degradation | VSV, DENV | Ubiquitous | Cytosol | Type I interferons |
25HC, 25-hydroxycholesterol; ADAP2, ARFGAP with dual pleckstrin homology domain-containing protein 2; CH25H, cholesterol 25-hydroxylase; CHIKV, chikungunya virus; CPXV, cowpox virus; DENV, dengue virus; EBOV, Ebola virus; GILT, interferon-γ-inducible lysosomal thiol reductase; hCoV-229E, human coronavirus 229E; hCoV-OC43, human coronavirus OC43; HCV, hepatitis C virus; HSV, herpes simplex virus; IAV, influenza A virus; IFITM, interferon-induced transmembrane protein; LASV, Lassa virus; LCMV, lymphocytic choriomeningitis virus; LPS, lipopolysaccharide; LY6E, lymphocyte antigen 6E; MERS-CoV, Middle East respiratory syndrome coronavirus; NCOA7, nuclear receptor co-activator protein 7; NiV, Nipah virus; RVFV, Rift Valley fever virus; SARS-CoV, severe acute respiratory syndrome coronavirus; VACV, vaccinia virus; VSV, vesicular stomatitis virus; WNV, West Nile virus; YFV, yellow fever virus; ZIKV, Zika virus; ZMPSTE24, zinc metalloproteinase STE24.
Inhibition of virus–cell membrane fusion
Mechanistic studies of IFITM1, IFITM2 and IFITM3 suggest that they do not rely on specific recognition of viral components to restrict virus entry. Rather, they alter the mechanical properties of cellular membranes to disfavour the membrane fusion reaction between incoming virus and the target cell. Restriction of virus–cell fusion occurs after hemifusion and before the formation of a dilated fusion pore that allows viral proteins and nucleic acid to access the host cell cytoplasm16,56 (Fig. 2). It was reported that IFITM1, IFITM2 and IFITM3 increase membrane lipid order in cells (increasing rigidity and decreasing fluidity)16,57, and it was subsequently shown that membrane stiffening by IFITM3 is required for its antiviral activity58. The molecular basis by which IFITM3 modifies membranes to render them less fusogenic involves an α-helix that is amphipathic in nature (characterized by both polar and non-polar amino acid residues)59. The helix is found in the amino terminus of IFITM3 and is juxtaposed to the inner, cytoplasmic leaflet of the lipid bilayer, following the consensus that IFITM3 conforms to a type II transmembrane protein topology with its amino terminus in the cytoplasm and its carboxy terminus in the lumen or extracellular space (although alternative topologies are possible)60. When the amphipathic helix is deleted from the context of the IFITM3 polypeptide, the protein loses the capacity to increase membrane rigidity in living cells58. Furthermore, a peptide corresponding to the amphipathic helix of IFITM3 was shown to increase membrane rigidity and curvature in artificial membranes61. These findings align with the notion that amphipathic helices act like wedges and confer membrane-altering properties on the proteins that possess them (Fig. 2). Further evidence demonstrating that IFITM3 remodels membranes to restrict virus entry is the finding that amphotericin B, an antifungal that was shown to counteract the restriction of IAV entry by IFITM3 (ref.62), also prevents the impact of IFITM3 on membrane rigidity58. Amphotericin B is known for its ability to bind sterols in fungal and mammalian lipid bilayers, and it promotes membrane fluidity through an unknown mechanism63. These results collectively suggest that IFITM3 (and likely the other IFITM proteins endowed with antiviral activities) locally modify the membranes in which they reside.
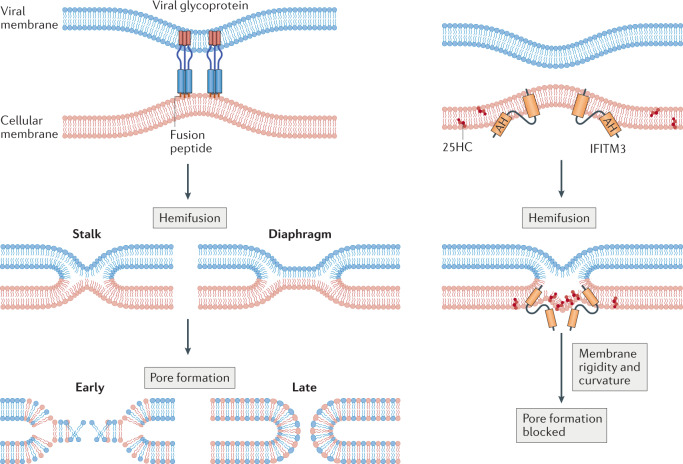
Fig. 2. Stages of virus–cell membrane fusion and the antifusion activities of IFITM3 and 25-hydroxycholesterol.
Close apposition of viral and cellular membranes is induced by viral envelope glycoproteins (shown at the upper left and not drawn thereafter), followed by the formation of a hemifusion stalk. Hemifusion is characterized by lipid mixing between the outer leaflets and alignment of the inner leaflets of each bilayer and may progress from a stalk to a diaphragm-like structure. Finally, further lipid mixing leads to partial opening of the fusion pore, and the pore is further dilated to complete membrane fusion. Interferon-induced transmembrane protein 3 (IFITM3) promotes hemifusion while inhibiting pore formation in a process that requires its amphipathic helix (AH) and protein oligomerization. IFITM3 promotes membrane rigidity and curvature in a manner that may disfavour formation of the fusion pore, and 25-hydroxycholesterol (25HC) produced by cholesterol 25-hydroxylase (CH25H) may function similarly.
Membrane localization
Determinants other than the amphipathic helix are important for IFITM residency in membranes and localization at entry sites preferred by different viruses. While the basis for virus insensitivity to restriction is poorly characterized, it may result from the use of virus entry routes that avoid encounter with IFITM proteins. A tyrosine-based internalization motif recognized by the endocytic adaptor AP-2 and regulated by phosphorylation ensures that IFITM3 and IFITM2 are endocytosed from the cell surface and allowed to accumulate in endosomal and endolysosomal membranes under steady-state conditions64–67. As a result, IFITM2 and IFITM3 reside primarily in early and late endosomes33,68, but phosphorylated pools of protein remain at the plasma membrane. By contrast, IFITM1 is generally observed at or near the plasma membrane, and this distinction may account for the non-overlapping antiviral specificity of the IFITM proteins — IFITM3 and IFITM2 are more potent inhibitors of endocytic viruses such as IAV47 and ZIKV26,27, whereas IFITM1 is more potent against HIV-1 (refs36,69–71), respiratory syncytial virus19 and herpes simplex virus19, viruses that tend to fuse at or near the plasma membrane.
Although the intracellular trafficking patterns may differ among IFITM1, IFITM2 and IFITM3, a conserved post-translational lipid modification known as palmitoylation ensures that they are stably associated with membrane microdomains, such as lipid rafts. Human IFITM1, IFITM2 and IFITM3 contain three cysteine residues to which the palmitoyl lipid is appended, and mutation of these residues results in loss of polypeptide stability, altered subcellular localization and decreased antiviral activity in cells65,72–75.
Endosomal redirection
An early observation was that IFITM protein expression results in virion entrapment within endosomes and their subsequent elimination in endolysosomes15. Later, it was shown that palmitoylation of IFITM3 allows it to travel to virus-containing vesicles and to redirect them towards endolysosomes for degradation76,77. Collectively, these findings suggest that antiviral restriction by IFITM3 may involve a two-part mechanism consisting of, first, prevention of virus entry into the cytoplasm through inhibition of virus–cell membrane fusion and, second, acceleration of endosomal trafficking towards an acidic, degradative endolysosomal compartment15,76. The second step is poorly understood on a molecular level but may be explained by the localization of IFITM3 to late endosomes (also known as multivesicular bodies). IFITM3 is a constituent of intraluminal vesicles, and when they are released into the extracellular space as exosomes, provides antiviral protection to neighbouring cells78,79. Furthermore, it was reported that IFITM3 reduces the ‘back-fusion’ of intraluminal vesicles with the limiting membrane of the late endosome, thereby reducing the release of intraluminal cargo into the cytoplasm80. As a result, IFITM3 and related proteins may promote lysosomal delivery of viral and cellular cargo travelling within late endosomes. Indeed, IFITM3 has been shown to accelerate turnover of epidermal growth factor receptor in endolysosomes76. The full extent to which IFITM3 influences endocytic trafficking remains unknown, but a role in regulating endosome–endosome fusion or endosome–lysosome fusion has not been ruled out.
Cofactors of IFITM restriction
Beyond the sequence determinants present within individual IFITM polypeptides that govern their antiviral activities, the capacity for IFITM protein to bind itself (homomultimerization) or to other proteins (heteromultimerization) has also been implicated in antiviral function57,81. A glycine-based (GxxxG) motif conserved among IFITM1, IFITM2 and IFITM3 from multiple species was shown to promote IFITM3 oligomerization and is required for it to rigidify membranes and to restrict virus infection58. This finding implies that oligomers of IFITM3 presenting multiple amphipathic helices to the inner leaflet of cell membranes are responsible for inhibition of virus–cell fusion. Furthermore, cellular processes that regulate IFITM3 oligomerization are likely to impact its antiviral potential. The same GxxxG motif may also regulate the capacity of IFITM3 to bind IFITM2 or IFITM1, and this heteromultimerization among IFITM proteins may provide an added layer of regulation to their antiviral functions.
Many lines of evidence suggest that IFITM proteins directly alter membrane rigidity and curvature to restrict membrane fusion, but other constituents of the cellular proteome may act as cofactors for IFITM function and play direct or indirect roles in the restriction of virus entry. For example, the E3 ubiquitin ligase NEDD4 binds to the amino termini of IFITM3 and IFITM2 and accelerates their turnover through ubiquitylation and degradation in endolysosomes47,82. As a result, silencing or knockout of NEDD4 results in elevated levels of IFITM2 and IFITM3 and decreased infection by IFITM-sensitive viruses. In addition, the transitional endoplasmic reticulum ATPase valosin-containing protein (VCP; also known as p97) has been identified as interacting with IFITM3 and enables its antiviral functions by guiding its intracellular localization83.
In addition, certain protein interactors have been purported to act as cofactors required for the antiviral effects of IFITM proteins, at least in certain circumstances. IFITM1 was shown to bind to the tetraspanin CD81, and this interaction inhibits HCV, which depends on CD81 for entry23. Another example of a purported cofactor for IFITM function is vesicle-associated membrane protein-associated protein A (VAPA), an important regulator of intracellular cholesterol trafficking. VAPA mediates cholesterol transfer between the endoplasmic reticulum and endosomes by binding to oxysterol-binding protein (OSBP) and OSBP-related proteins84. IFITM3 was shown to promote the accumulation of cholesterol in late endosomes, which may result from its interaction with VAPA and disruption of the VAPA–OSBP interaction85–88. An overabundance of cholesterol in late endosomes is thought to prevent the fusion of intraluminal vesicles with the limiting endosomal membrane89, and it may similarly inhibit fusion between the limiting membrane and enveloped virions. Disruption of proper cholesterol trafficking itself phenocopies the impact of IFITM proteins and results in a block to virus entry90. Nonetheless, the role played by VAPA and cholesterol in the antiviral mechanism of IFITM proteins has been questioned by multiple reports24,40,56,62.
In another example of how IFITM proteins may impact endosomal content, it was reported that they interact with vacuolar ATPase (v-ATPase), an enzyme responsible for the proper acidification of endolysosomes91. Also, IFITM protein overexpression results in expansion of acidic endolysosomes in cells, and v-ATPase recruitment may be central to this observation15. Although it is clear that IFITM3 does not interfere with the pH-dependent triggering of viral fusion proteins15,16,56,92, it remains unclear whether increasing endosomal acidity plays a role in its antiviral functions. Finally, an IFITM3-interacting protein identified as zinc metalloproteinase STE24 (ZMPSTE24; also known as FACE1) was shown to be required for the antiviral activity of IFITM3, and ZMPSTE24 allegedly imposes a block to virus entry itself31. This finding raises the possibility that IFITM3 functions in unison with ZMPSTE24, with the latter serving as the antiviral effector protein (see the next section).
Recent evidence suggests that different models of antiviral function by IFITM proteins are not mutually exclusive. For example, it is possible that the impact of IFITM3 on membrane rigidity and curvature requires the presence of membrane cholesterol, as cholesterol incorporation into lipid bilayers itself decreases membrane fluidity93. Furthermore, the degree to which IFITM3-derived peptides induce membrane curvature in model membranes depends on the presence of cholesterol61. Moreover, different viruses may be inhibited by different effects of IFITM proteins on host cells. Whereas amphotericin B can be used to counteract IFITM3-mediated restriction of IAV, it does not reverse inhibition of EBOV40, suggesting that these two viruses are restricted by distinct consequences of IFITM3 expression in cells. Furthermore, the finding that IFITM proteins can inhibit infection by reovirus, a non-enveloped virus, suggests that restriction may result from their effects on lipid transport or endolysosomal trafficking94.
Coronavirus evasion of and enhancement by IFITM proteins
Other indications suggest that IFITM proteins are multifunctional during virus infections in ways that are not fully understood. Ectopic expression of human IFITM proteins has been shown to inhibit infection mediated by SARS-CoV, MERS-CoV and SARS-CoV-2 spike proteins17,40,41,95. However, cleavage of SARS-CoV-2 spike protein by furin-like proteases and trypsin-like proteases, such as transmembrane protease serine 2 (TMPRSS2), enables partial escape from IFITM-mediated restriction in endosomes41,42,96. Furthermore, mutants of IFITM3 that drive its accumulation on the cell surface do not inhibit but rather promote MERS-CoV and SARS-CoV-2 infection41,43. It has been reported that endogenous IFITM proteins in lung cells enhance SARS-CoV-2 infection by binding to the spike protein and promoting fusion at or near the cell surface97. Similarly, endogenous IFITM3 is a dependency factor required for infection by the seasonal coronavirus hCoV-OC43 (ref.98). Therefore, IFITM proteins exhibit opposing activities during coronavirus infection, and the basis for this duplicity is not understood.
Nonetheless, it is possible that IFITM proteins have shaped the evolution of human coronaviruses in ways that impact their emergence and pathogenicity in humans. Indeed, the single-nucleotide polymorphisms previously identified as determinants for viral disease following IAV infection have also been associated with severe outcomes of COVID-19 (refs50,51). However, the link between genetic variants of human IFITM3 and viral disease may not solely result from the actions of IFITM3 during virus entry. It has been shown that IFITM3 decreases production of the inflammatory cytokine IL-6 in vivo following infection by cytomegalovirus, which is itself insensitive to the entry block imposed by IFITM3 (ref.99). Thus, IFITM proteins can influence the systemic immune response to virus infections both by regulating virus entry into cells and by limiting host-mediated immunopathology, although the mechanisms for the latter have yet to be worked out.
ZMPSTE24
ZMPSTE24 is a constitutively expressed transmembrane protein that promotes the structural integrity of the nuclear membrane by cleaving prelamin A100. Inherited loss-of-function mutations in human ZMPSTE24 are associated with premature ageing and progeria-related diseases101. Additional studies uncovered a role for ZMPSTE24 in removing misfolded proteins from clogged translocons in the endoplasmic reticulum102. ZMPSTE24 had not been implicated in virus infections until affinity purification coupled with mass spectrometry identified it as a high-confidence IFITM1, IFITM2 and IFITM3 interactor31. To address whether the interaction between ZMPSTE24 and IFITM proteins was consequential for virus infection, the study authors showed that while ectopic IFITM3 inhibits IAV infection in normal cells, it fails to do so in cells that are deficient in ZMPSTE24. Further experimentation showed that ZMPSTE24 expression itself results in inhibition of virus–cell fusion in cell culture and inhibition of virus infection in vivo, and that some of the same viruses exhibiting sensitivity to IFITM3 (IAV, VSV, vaccinia virus and CPXV) are also sensitive to ZMPSTE24. Additionally, loss of the Ifitm locus from mouse embryonic fibroblasts results in a marked gain in susceptibility to VSV and vaccinia virus, and further knockdown of ZMPSTE24 results in further gains in susceptibility. These results suggested that IFITM proteins require ZMPSTE24 for their antiviral activities, whereas ZMPSTE24 itself is endowed with independent antiviral function. The study authors proposed that IFITM proteins and ZMPSTE24 act in the same pathway to restrict virus–cell fusion, with ZMPSTE24 acting downstream. Therefore, these findings favour a model whereby IFITM proteins confer antiviral protection on cells via an indirect mechanism. Interestingly, the enzymatic protease activity of ZMPSTE24 is not required for its observed impact on virus entry as loss-of-function mutants still maintained the ability to restrict infection31.
Following this initial report, several questions remain unanswered regarding the antiviral potential of ZMPSTE24 and its relevance to intrinsic control of pathogenic virus infection. For example, it is unclear whether ZMPSTE24 inhibits IFITM-sensitive viruses that undergo fusion at the plasma membrane of cells, such as HIV-1. Also, VSV exhibits greater sensitivity to ZMPSTE24 than it does to IFITM3, whereas the opposite is the case for IAV103. As evidence exists that IFITM proteins restrict virus entry through multiple mechanisms, it is possible that they depend on ZMPSTE24 for the inhibition of some, but not all, viruses. Furthermore, it remains unclear how ZMPSTE24 is capable of restricting virus entry in endosomes when it is associated with a function at the nuclear membrane.
A subsequent report sought to better resolve the interplay between ZMPSTE24 and IFITM proteins by focusing on their individual and collective impact on arenaviruses, which are traditionally viewed as IFITM insensitive. It was found that knockout of human ZMPSTE24 increased cellular susceptibility to pseudovirus infection mediated by arenavirus glycoproteins from Old World and New World arenaviruses, including lymphocytic choriomeningitis virus and LASV, as well as infection by intact Mopeia virus, an arenavirus related to LASV104. Interestingly, while IFITM proteins had little to no effect on arenavirus infections, co-expression of IFITM3 and ZMPSTE24 resulted in enhanced restriction. In addition to confirming that IFITM3 can interact with ZMPSTE24, it was demonstrated that the two proteins affect each other’s subcellular localization. Ectopic co-expression of IFITM3 and ZMPSTE24 resulted in their colocalization in an uncharacterized perinuclear site, which may or may not represent a subset of endolysosomes where arenavirus–cell fusion occurs. To assess whether membrane modulation is associated with the activities of IFITM and ZMPSTE24 on arenavirus infection, the study authors used amphotericin B, an antifungal compound which reduces membrane rigidity, to overcome restriction by IFITM3 (refs58,62). Amphotericin B partially abrogated restriction of arenavirus infection when ZMPSTE24 was expressed alone or in combination with IFITM3, suggesting that ZMPSTE24 and IFITM3 individually and cooperatively increase membrane rigidity within a subset of endosomes to inhibit arenavirus infection104. However, an experimental test of the role played by ZMPSTE24 in the IFITM-mediated enhancement of membrane rigidity is lacking. Nonetheless, the identification of an endosomal compartment that contains ZMPSTE24 as well as IFITM3 may lead to important new insights into the poorly understood arenavirus entry pathway105.
CH25H
Cholesterol 25-hydroxylase (CH25H) is an interferon-induced enzyme that restricts virus–cell membrane fusion during virus entry, and acts broadly to inhibit VSV, HIV-1, herpes simplex virus, EBOV, Rift Valley fever virus, ZIKV, Nipah virus, SARS-CoV, MERS-CoV and SARS-CoV-2 (ref.106). CH25H resides in the endoplasmic reticulum and catalyses the enzymatic oxidation of cholesterol to 25-hydroxycholesterol (25HC), a side chain oxysterol that functions in cholesterol homeostasis. In response to elevated levels of cellular cholesterol, 25HC inhibits the activities of sterol regulatory element-binding protein (SREBP), a transcription factor that drives the expression of genes associated with cholesterol biosynthesis and uptake107. Additionally, 25HC promotes cholesterol trafficking into the endoplasmic reticulum. As a result, CH25H and its product 25HC regulate the cholesterol content of membranes in different subcellular compartments108 (Fig. 3).
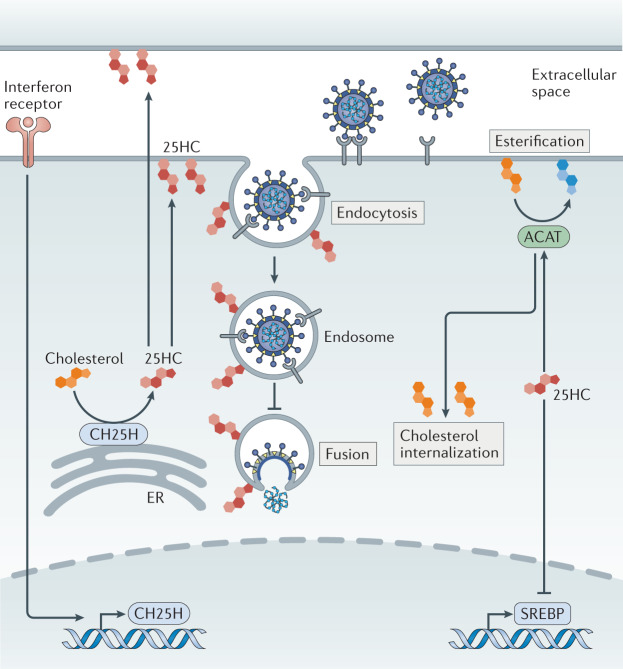
Fig. 3. The impacts of 25-hydroxycholesterol on virus entry and cholesterol homeostasis.
Cholesterol 25-hydroxylase (CH25H) is localized to the endoplasmic reticulum (ER), where it catalyses the oxidation of cholesterol to produce the oxysterol 25-hydroxycholesterol (25HC). 25HC acts in an autocrine and paracrine manner to inhibit virus entry at the level of fusion. 25HC promotes acyl-CoA cholesterol acyltransferase (ACAT) activity to increase cholesterol esterification, which regulates cholesterol availability in membrane compartments. Furthermore, 25HC provides negative feedback in cholesterol metabolism by repressing sterol regulatory element-binding protein (SREBP)-induced genes that promote cholesterol biosynthesis.
Inhibition of virus–cell fusion
The antiviral potential of CH25H was first identified through a systematic screen of genes induced by type I and type II interferons. When 288 interferon-stimulated genes were individually expressed in cells and subsequently challenged with VSV (an RNA virus) or murine gammaherpesvirus 68 (MHV-68; a DNA virus), CH25H was revealed as one of the most potent inhibitors of both109. Further mechanistic work found that resistance to VSV infection is observed in cells expressing CH25H but also in neighbouring cells, indicating that CH25H confers antiviral protection in an autocrine and a paracrine manner by producing a soluble, secreted factor deemed to be 25HC110,111. Although 25HC did not affect adherence (attachment) of VSV to cells, it inhibited fusion mediated by the pH-dependent VSV glycoprotein and the pH-independent HIV-1 envelope protein. Furthermore, 25HC inhibited cell–cell fusion induced by Nipah virus fusion protein, suggesting that this oxysterol produced by CH25H directly modifies cellular membranes, including the plasma membrane and endosomal membranes, to impede the cell entry of diverse viruses. Indeed, addition of cholesterol-containing liposomes caused a dose-dependent reversal of VSV inhibition by 25HC110. As early studies characterizing the cellular impacts of 25HC demonstrated that it could cause membrane perturbations, such as changes in lipid head group spacing and cholesterol accessibility, these results suggest that 25HC inhibits the membrane dynamics required for efficient virus–cell fusion, possibly by modulating membrane rigidity and curvature112 (Fig. 2). While both may alter the mechanical properties of cellular membranes, it seems that IFITM proteins and CH25H act via distinct mechanisms, because MHV-68 entry is sensitive to CH25H/25HC but not to IFITM3 (ref.109).
Regulation of membrane cholesterol
Additional evidence supporting a role for CH25H in regulating membrane cholesterol to impart a broad-spectrum antiviral state was generated from more recent studies involving human coronaviruses. Like IFITM3, CH25H is upregulated in macrophages and epithelial cells from patients with COVID-19 relative to healthy donors. In cell lines permissive to infection, 25HC was found to inhibit entry mediated by spike proteins from SARS-CoV, MERS-CoV and SARS-CoV-2 (refs95,113). 25HC also reduced cell–cell fusion mediated by SARS-CoV-2, suggesting that 25HC inhibits the spike protein-driven membrane fusion reaction113. Similarly to what was shown regarding the effects of 25HC on VSV infection, addition of soluble cholesterol to cells treated with 25HC restored entry driven by MERS-CoV and SARS-CoV-2 spike protein113. This result suggested that the antiviral activities of CH25H and its product 25HC during coronavirus infection may stem from their impact on cholesterol trafficking. In a separate study, it was reported that 25HC activates acyl-CoA cholesterol acyltransferase (ACAT), an enzyme which causes cholesterol esterification and its internalization from the plasma membrane108 (Fig. 3). Accordingly, a specific inhibitor of ACAT counteracted the loss of cell surface cholesterol by 25HC and restored virus spike protein-mediated entry at the plasma membrane of lung cells, providing an important explanation for how CH25H exerts its antiviral activity towards coronaviruses. It is also likely that 25HC affects levels of accessible cholesterol present in endosomal membranes, which can also be used as points of entry for SARS-CoV-2 and other 25HC-sensitive viruses, such as VSV95.
In addition to extensive characterization in vitro and in cell culture experiments, the importance of CH25H in limiting virus entry and the therapeutic potential of its lipid product, 25HC, have been reinforced by in vivo studies. Administration of 25HC to humanized mice decreased their susceptibility to HIV-1 infection and viral disease110. Moreover, mice lacking Ch25h are more susceptible to infection and disease sequelae caused by MHV-68 and ZIKV114. Although 25HC is reported to be non-toxic and tolerable at therapeutic doses, CH25H may contribute to diseases such as atherosclerosis and Alzheimer disease, with evidence accumulating that it intensifies inflammatory signalling115. The fact that CH25H is interferon-inducible likely reflects a requirement to finely regulate its expression to avoid disease.
Cholesterol-independent mechanisms of restriction?
As is likely the case for the IFITM proteins, it is possible that CH25H expression leads to virus inhibition by multiple mechanisms, including some that may not involve the production of 25HC. For example, mutants of CH25H that do not produce 25HC exhibit loss of antiviral activity against MHV-68, but not against HCV, demonstrating that CH25H inhibits the latter through a non-canonical mechanism116. Additionally, CH25H has been shown to inhibit infection by reovirus, papillomavirus, rotavirus and rhinovirus, all non-enveloped viruses that do not enter cells via membrane fusion106,117. Reovirus is sensitive to both CH25H and IFITM3, providing evidence that these two antiviral proteins regulate not only membrane fusion but also endocytic trafficking pathways in cells.
LY6E
Lymphocyte antigen 6E (LY6E) is an interferon-inducible glycosylphosphatidylinositol-anchored protein that was originally known as a marker of T cell activation and has a negative feedback role by inhibiting IL-2 production from T cell receptor-stimulated lymphocytes118. It also acts as an immune modulator in myeloid cells, such as monocytes and macrophages, by downregulating CD14 expression and thereby limiting innate immune signalling in response to lipopolysaccharide119. The antiviral activity of LY6E was discovered through the same screening platform that identified CH25H as an inhibitor of VSV and MHV-68 entry into cells109. Relative to CH25H, which suppresses the membrane merger required for VSV entry into cells110, LY6E exhibited a less potent restriction of VSV. However, later studies using human coronaviruses found that LY6E also restricts the virus–cell membrane fusion step. With use of a cDNA library composed of more than 350 interferon-stimulated genes120, it was observed that LY6E overexpression strongly inhibited infection by hCoV-229E and hCoV-OC43, two seasonal human coronaviruses, as well as SARS-CoV, MERS-CoV and SARS-CoV-2 (ref.121). Although LY6E did not impact coronavirus attachment to host cells, it did inhibit the cell entry step, as pseudovirus infection mediated by coronavirus spike proteins was also inhibited. It also blocked spike protein-mediated cell–cell fusion without affecting the expression or proteolytic cleavage of spike proteins, indicating that LY6E inhibits the fusogenic potential of spike protein required for virus entry121. Its protective role in vivo was determined by the generation of Ly6e-null mice and the demonstration that they were more susceptible to infection and viral disease caused by a mouse coronavirus121. In contrast to IFITM proteins and CH25H, there are few mechanistic details available to explain how LY6E regulates membrane fusion. A separate study reported an antiviral role for LY6E against hCoV-OC43 and SARS-CoV-2, one that cannot be reversed by amphotericin B44. This finding suggests that the mechanism of action of LY6E is distinct from that of IFITM3. Therefore, the antiviral activity that LY6E exhibits against coronaviruses makes it an exciting new addition to the list of intrinsic factors impinging on virus entry, and a surprising one at that.
LY6E was previously shown to promote infections by several important human pathogens, including IAV, yellow fever virus, ZIKV, WNV, DENV and HIV-1 (refs122–124) by enabling the cell entry process at the level of virion internalization or fusion. LY6E is required for a type of endocytosis that is clathrin dependent, microtubule dependent and selective for large cargo such as virions122. Interestingly, a single amino acid substitution at Lys36 that renders LY6E unable to promote entry of yellow fever virus also disrupts its ability to inhibit coronavirus entry, indicating that the opposing functions are controlled by the same determinants121,123. Perhaps the size-dependent endocytosis pathway involving LY6E positively regulates entry for some viruses but not others by funnelling virions towards specific vesicular compartments (early and late endosomes) and away from others (such as endolysosomes, which contain cathepsins that activate coronavirus spike proteins). However, a given virus can be subject to both the enhancing and the inhibiting effects of LY6E, suggesting that it impacts virus infections by multiple, non-redundant mechanisms. Although LY6E can promote HIV-1 infection at the level of fusion, it performs an inhibitory role when levels of CD4, the host cell receptor used to enter cells, are low. CD4 levels on the cell surface were found to be downmodulated by LY6E, inhibiting virus entry and masking its facilitatory role during infection125. This activity of LY6E reflects its capacity to internalize certain cell surface proteins that act in immune signalling, but it does not explain its negative impact on coronavirus infection, as the various receptors used by human coronaviruses were not found to be altered by LY6E121.
Thus, much like the IFITM proteins, LY6E alters the cellular environment to impact virus entry in more than one way, sometimes with opposing effects. As a result, the overall impact of factors such as IFITM proteins and LY6E during virus infection may result from the sum of their individual inhibitory and facilitatory functions, and distinct viruses will respond differently to this balance. The two-faced nature of certain intrinsic immune factors may contribute to some of the deleterious effects observed in humans receiving interferon therapy126. Another possibility that is not mutually exclusive is that certain viruses have evolved to co-opt IFITM proteins and LY6E during the entry process, turning what would normally be a barrier into a springboard to launch infection.
NCOA7
Nuclear receptor co-activator protein 7 (NCOA7) is a member of the oxidation resistance family and is recruited to the nucleus on oestrogen binding to its nuclear receptor127. In complex with the nuclear receptor and oestrogen, NCOA7 acts as a transcription factor and induces the expression of oestrogen-regulated genes128. Interestingly, it was observed that a short isoform of NCOA7 (isoform 4, also known as NCOA7-alternative start) is inducible by type I interferons129.
Through an effort to identify interferon-induced genes that are differentially expressed among cell types permissive to HIV-1 infection, isoform 4 of NCOA7 was revealed as an antiviral factor that, while inactive against HIV-1, was shown to broadly inhibit viruses that enter cells via endocytosis130. HIV-based pseudotypes bearing HIV envelope protein were resistant to NCOA7, whereas those bearing VSV glycoprotein were sensitive, demonstrating that restriction occurs at the entry step. The study authors also showed that NCOA7 inhibited entry at the level of membrane fusion using experiments that assayed the extent of membrane fusion occurring between intact IAV particles and target cell membranes. The viruses sensitive to NCOA7-mediated restriction, including VSV, HCV and IAV, exhibit a shared requirement for pH-dependent virus–cell membrane fusion in endosomal compartments. Mechanistic experiments showed that NCOA7 promotes acidification of endosomes by enhancing v-ATPase activity. Furthermore, NCOA7 expression resulted in elevated cathepsin activity in endolysosomes. These findings suggest that NCOA7 accelerates the turnover of virions by cathepsin-mediated proteolysis, decreasing the likelihood of virus escape into the cytoplasm. Interestingly, this activity has also been cited as a possible mechanism of action of IFITM3 (Fig. 4). Although NCOA7 knockout partially rescued IAV infection in the presence of interferon, the combined knockout of NCOA7 and IFITM3 led to even greater rescue of infection130. Therefore, because they have an additive impact on virus entry, the respective functions of NCOA7 and IFITM3 may be related but are not redundant. This pair of antiviral proteins serves as an example of how factors may perform similar but non-overlapping roles to protect cells from virus invasion.
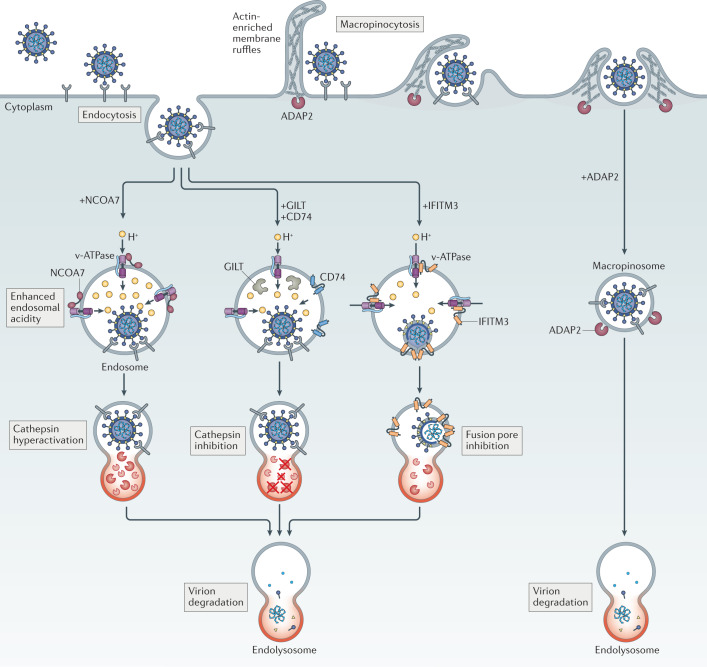
Fig. 4. Intrinsic inhibitors of virus entry promote virion degradation in endolysosomes via distinct mechanisms.
The antiviral activities exhibited by nuclear receptor co-activator protein 7 (NCOA7), interferon-induced transmembrane (IFITM) proteins, interferon-γ-inducible lysosomal thiol reductase (GILT), CD74 and ARFGAP with dual pleckstrin homology domain-containing protein 2 (ADAP2) act against viruses entering cells through pH-dependent fusion in endosomes. Whereas NCOA7 and IFITM3 have been reported to interact with vacuolar ATPase (v-ATPase), only NCOA7 may increase the acidity (lower the pH) of the endosomal lumen. NCOA7-mediated acidification of endosomes is associated with enhanced cathepsin activity in endolysosomes, and this elevated level of proteolytic activity may promote virion degradation before virus–cell fusion occurs. IFITM3, on the other hand, inhibits membrane fusion itself, resulting in endosomal sequestration of virions that are eventually degraded in endolysosomes. This latter effect results from the ability of IFITM3 to promote endolysosomal delivery of viral and cellular cargo, a function that is not yet mechanistically understood. GILT and CD74 are believed to enforce endosomal sequestration of viruses as well, but not by inhibiting membrane fusion — they inhibit the activity of endolysosomal cathepsins, proteases that cleave some viral glycoproteins and render them competent for fusion. ADAP2 promotes internalization of virions by macropinocytosis, bypassing their preferred sites for pH-dependent fusion and resulting in their accelerated disposal in endolysosomes.
GILT
IFNγ-inducible lysosomal thiol reductase (GILT; also known as IFI30) is an enzyme localized to endolysosomes in many cell types and is expressed constitutively or in response to type II interferon131. GILT reduces the disulfide bond of exogenous peptide antigens, including viral glycoproteins, that are internalized into cells through endocytosis132. This function promotes peptide processing and facilitates their presentation by MHC class II and class I molecules133. As a result, GILT plays an important role in the priming of CD4+ T cell and CD8+ T cell responses directed against viral peptides.
Owing to the localization and function of GILT within endolysosomes, it was suspected that GILT may also interfere with the cell entry step of viruses that enter cells via endocytosis. It was shown that GILT expressed in human lung cells inhibited entry mediated by the viral envelope glycoproteins of SARS-CoV, EBOV and LASV, which require endolysosomal delivery to complete cell entry, but not those of IAV or VSV, which complete entry in endosomes134,135. Accordingly, the basis for this restriction required endolysosomal localization of GILT as well as its enzymatic reductase function134. Furthermore, addition of trypsin to cells allowed SARS-CoV pseudovirus to evade inhibition by GILT by facilitating virus entry at the plasma membrane, supporting the notion that restriction occurs in intracellular, endolysosomal membranes134. Unlike NCOA7, GILT was not found to increase endolysosomal cathepsin activity, but rather it inhibited cathepsin L activity. This finding may explain, in part or in whole, its antiviral effect, as SARS-CoV, EBOV and LASV require cathepsin L for proteolytic activation of the viral glycoprotein in endolysosomes. Interestingly, other human coronaviruses, including MERS-CoV and SARS-CoV-2, are insensitive to GILT-mediated restriction, which may reflect differential dependency on endolysosomal cathepsins among the human coronaviruses for cell entry44. Nonetheless, as it was also reported that GILT inhibits entry mediated by HIV-1 envelope protein136, which mediates cell entry in a pH-independent and endolysosome-independent manner137, additional studies are required to understand the antiviral functions of GILT on a molecular level.
CD74
CD74 (also known as invariant chain) is expressed by immune cells and promotes the transport of MHC class II proteins from the endoplasmic reticulum to the cell surface via endosomal intermediates. Consequently, CD74 promotes the presentation of extracellular antigens internalized through endocytic processes. In addition to this role, a portion of the CD74 polypeptide known as the thyroglobulin domain carries out inhibition of endolysosomal cathepsins, the proteolytic enzymes that are co-opted by numerous viruses during the cell entry phase of infection. While multiple isoforms of CD74 exist, two of them possess the thyroglobulin domain138.
By taking advantage of a screening technique whereby transposable elements are inserted in front of and within genes, Bruchez et al. found that MHC class II transactivator (CIITA) inhibited EBOV infection139. CIITA is a transcription factor that controls the expression of MHC class II-related proteins, including CD74, to facilitate antigen presentation to lymphocytes. CIITA blocked pseudovirus infection of particles bearing glycoproteins from EBOV and other filoviruses, whereas VSV glycoprotein and LASV glycoprotein were unaffected, indicating that it selectively blocked some aspect of the filovirus cell entry process. Further experiments revealed that CIITA diminished fusion mediated by EBOV glycoprotein, but not virion uptake into cells, whereas infectious EBOV replication was also reduced in its presence. The suppression of EBOV infection by CIITA was found to require CD74, and CD74 expression itself blocked infection. Both CIITA and CD74 were found to be inducible by type II interferon and lipopolysaccharide. The isoform of CD74 that possessed antiviral activity was p41, one of the isoforms containing the thyroglobulin domain. Accordingly, CD74 p41 inhibited cathepsin-mediated cleavage of EBOV glycoprotein, which is required for membrane fusion in endolysosomes. As one of the cell entry routes used by coronaviruses involves cathepsin-mediated activation of spike protein, the study authors tested whether SARS-CoV-2 was sensitive to CD74-mediated inhibition. In cells lacking the serine protease TMPRSS2, which are permissive to SARS-CoV-2 entry only via the endosomal, cathepsin-mediated entry route, SARS-CoV-2 infection was potently blocked by CD74 (ref.139).
These results suggest that CD74 acts broadly to protect host cells from invasion by a plethora of cathepsin-dependent viruses. This work highlights another example of an intrinsic immune protein that exerts its antiviral function, in whole or in part, by sequestering virions in endolysosomes, which may drive the selection of viruses capable of entering cells elsewhere, such as the plasma membrane.
ADAP2
ARFGAP with dual pleckstrin homology domain-containing protein 2 (ADAP2; also known as CENTA2) is a GTPase-activating protein (GAP) for the GTP-binding protein ARF6 (ref.140), a regulator of actin cytoskeletal rearrangements. ADAP2 activity promotes the GDP-bound form of ARF6, and as a result influences its function. Although ADAP2 is constitutively expressed, it was singled out for studies of antiviral activity when its expression was found to be upregulated by type I interferons141. When ADAP2-expressing cells were challenged with a panel of viruses that enter cells via different routes, it was found that ADAP2 restricted infection by DENV and, to a lesser extent, VSV. By contrast, infection by Sendai virus, which undergoes fusion at the plasma membrane142, was unaffected. These results indicate that ADAP2 selectively blocks virus entry occurring in endosomes, particularly late endosomes. Furthermore, the antiviral activity of ADAP2 does not result from its influence on the expression of other interferon-stimulated genes143, as restriction still occurred in cells in which the gene encoding mitochondrial antiviral signalling protein (MAVS) was knocked out141.
Further investigation revealed that ADAP2 induced macropinocytosis, an actin-dependent membrane protrusion and retraction process that allows the internalization of extracellular fluid and cargo. The antiviral activities of ADAP2 against DENV and induction of macropinocytosis were functionally linked as both effects required its ARF6 GAP activity. This led to a model supported by experimental evidence in which ADAP2 diverts incoming DENV virions away from late endosomes, which provide a favourable environment for virus–cell fusion to occur. Instead, ADAP2 promotes virion trafficking towards a dead-end endolysosomal compartment for disposal. This general mechanism of action implies that other viruses requiring clathrin-mediated endocytosis and pH-dependent fusion with endosomal membranes may be negatively impacted by ADAP2. It was later shown that ADAP2 did not inhibit virus entry mediated by EBOV glycoprotein or by spike proteins from human coronaviruses, including hCoV-OC43, MERS-CoV, SARS-CoV and SARS-CoV-2 (ref.44). The lack of activity reported in this instance may suggest that fusion occurring in endolysosomes (EBOV) or at the plasma membrane (human coronaviruses and Sendai virus) confers resistance to ADAP2. Furthermore, the fact that ADAP2 may not inhibit some of the viruses that are sensitive to IFITM3, CH25H, GILT and LY6E suggests that its respective antiviral function is distinct.
Conclusion
The full network of host factors providing intrinsic defence against virus invasion includes those inhibiting virus–cell membrane fusion from within target cells but also factors that incorporate themselves into virus particles and impair their fusogenic potential144–146. In addition, there are many host proteins that inhibit the earliest steps of virus infection in a more indirect manner. The rise of unbiased screening approaches has helped to identify factors that inhibit steps of the entry process that precede fusion, such as cell attachment and internalization. For example, β-1,4-N-acetylgalactosaminyltransferase 2 (B4GALNT2) modifies the sialic acid moieties used by IAV for cell attachment and, as a result, limits virion binding to the cell surface147. Other factors, including RIN2, TM9SF2 and RAB15, await further characterization but may inhibit virus entry owing to their roles as negative regulators of cellular endocytosis and endosome maturation147–150. When expressed in the same cells, these factors may act in concert with the antiviral effectors highlighted in this Review, resulting in additive inhibition of virus entry.
While cutting-edge genetic screening methods have accelerated the discovery of host factors that regulate virus entry into cells, a molecular understanding of their mechanisms of action often remains elusive. Overcoming this problem will require the development and large-scale adoption of infection models that capture the three-dimensional physiology of human tissues, such as organoids for the study of neurotropic viruses in the brain or respiratory viruses in the lung151. Non-human animal models remain an integral part to our understanding of virus infections. However, conditional (tissue-specific) gene knockouts would provide greater clarity on the contextual roles played by certain host factors, especially those that have been shown to perform multiple and opposing functions in cell culture. Lastly, the utility of traditional monoculture of transformed cells can be improved through combinatorial gene knockouts and hybrid small interfering RNA and CRISPR–Cas9 screening platforms. These latter approaches would help to distinguish between the direct and indirect mechanisms of antiviral activity and would also facilitate the characterization of host restriction modalities that involve two or more factors.
The continued identification of cell-intrinsic factors that block virus entry will provide new targets for novel antiviral therapies against enveloped viruses (and also non-enveloped viruses152). Restriction factors may be amplified or potentiated by gene therapy, whereas dependency factors required by viruses for their entry into cells, such as v-ATPase and cathepsins, may be inactivated by inhibitory drugs. Compared with traditional antivirals targeting viral components, these host-directed therapies may prove less likely to lead to drug resistance among circulating viruses and are more likely to extinguish the threat of virus emergence in the future.
Acknowledgements
This work was supported by the US National Institutes of Health Intramural Research Program, Center for Cancer Research, National Cancer Institute.
Author contributions
The authors contributed equally to all aspects of the article.
Competing interests
The authors declare no competing interests.
Footnotes
Peer review information
Nature Reviews Immunology thanks N. Altan-Bonnet, J. MacMicking and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Parvez MK, Parveen S. Evolution and emergence of pathogenic viruses: past, present, and future. Intervirology. 2017;60:1–7. doi: 10.1159/000478729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dimitrov DS. Virus entry: molecular mechanisms and biomedical applications. Nat. Rev. Microbiol. 2004;2:109–122. doi: 10.1038/nrmicro817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.White JM, Delos SE, Brecher M, Schornberg K. Structures and mechanisms of viral membrane fusion proteins: multiple variations on a common theme. Crit. Rev. Biochem. Mol. Biol. 2008;43:189–219. doi: 10.1080/10409230802058320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marsh M, Helenius A. Virus entry: open sesame. Cell. 2006;124:729–740. doi: 10.1016/j.cell.2006.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bieniasz PD. Intrinsic immunity: a front-line defense against viral attack. Nat. Immunol. 2004;5:1109–1115. doi: 10.1038/ni1125. [DOI] [PubMed] [Google Scholar]
- 6.Yan N, Chen ZJ. Intrinsic antiviral immunity. Nat. Immunol. 2012;13:214–222. doi: 10.1038/ni.2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Randow F, MacMicking JD, James LC. Cellular self-defense: how cell-autonomous immunity protects against pathogens. Science. 2013;340:701–706. doi: 10.1126/science.1233028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Platanias LC. Mechanisms of type-I- and type-II-interferon-mediated signalling. Nat. Rev. Immunol. 2005;5:375–386. doi: 10.1038/nri1604. [DOI] [PubMed] [Google Scholar]
- 9.Emerman M, Malik HS. Paleovirology — modern consequences of ancient viruses. PLoS Biol. 2010;8:e1000301. doi: 10.1371/journal.pbio.1000301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Daugherty MD, Malik HS. Rules of engagement: molecular insights from host-virus arms races. Annu. Rev. Genet. 2012;46:677–700. doi: 10.1146/annurev-genet-110711-155522. [DOI] [PubMed] [Google Scholar]
- 11.Compton AA, Malik HS, Emerman M. Host gene evolution traces the evolutionary history of ancient primate lentiviruses. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2013;368:20120496. doi: 10.1098/rstb.2012.0496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Duggal NK, Emerman M. Evolutionary conflicts between viruses and restriction factors shape immunity. Nat. Rev. Immunol. 2012;12:687–695. doi: 10.1038/nri3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Coomer CA, Rahman K, Compton AA. CD225 proteins: a family portrait of fusion regulators. Trends Genet. 2021;7:1–4. doi: 10.1016/j.tig.2021.01.004. [DOI] [PubMed] [Google Scholar]
- 14.Brass AL, et al. The IFITM proteins mediate cellular resistance to influenza A H1N1 virus, west nile virus, and dengue virus. Cell. 2009;139:1243–1254. doi: 10.1016/j.cell.2009.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Feeley EM, et al. IFITM3 inhibits influenza A virus infection by preventing cytosolic entry. PLoS Pathog. 2011;7:e1002337. doi: 10.1371/journal.ppat.1002337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li K, et al. IFITM proteins restrict viral membrane hemifusion. PLoS Pathog. 2013;9:e1003124. doi: 10.1371/journal.ppat.1003124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang IC, et al. Distinct patterns of IFITM-mediated restriction of filoviruses, SARS coronavirus, and influenza A virus. PLoS Pathog. 2011;7:e1001258. doi: 10.1371/journal.ppat.1001258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McMichael TM, et al. IFITM3 restricts human metapneumovirus infection. J. Infect. Dis. 2018;10:1582–1591. doi: 10.1093/infdis/jiy361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smith SE, et al. Interferon-induced transmembrane protein 1 restricts replication of viruses that enter cells via the plasma membrane. J. Virol. 2019;93:e02003–18. doi: 10.1128/JVI.02003-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rabbani MAG, Ribaudo M, Guo J-T, Barik S. Identification of interferon-stimulated gene proteins that inhibit human parainfluenza virus type 3. J. Virol. 2016;90:11145–11156. doi: 10.1128/JVI.01551-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang W, et al. Human respiratory syncytial virus infection is inhibited by IFN-induced transmembrane proteins. J. Gen. Virol. 2015;96:170–182. doi: 10.1099/vir.0.066555-0. [DOI] [PubMed] [Google Scholar]
- 22.Everitt AR, et al. Defining the range of pathogens susceptible to Ifitm3 restriction using a knockout mouse model. PLoS ONE. 2013;8:e80723. doi: 10.1371/journal.pone.0080723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wilkins C, et al. IFITM1 is a tight junction protein that inhibits hepatitis C virus entry. Hepatology. 2013;57:461–469. doi: 10.1002/hep.26066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Narayana SK, et al. The Interferon-induced transmembrane proteins, IFITM1, IFITM2, and IFITM3 inhibit hepatitis C virus entry. J. Biol. Chem. 2015;290:25946–25959. doi: 10.1074/jbc.M115.657346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Raychoudhuri A, et al. ISG56 and IFITM1 proteins inhibit hepatitis C virus replication. J. Virol. 2011;85:12881–12889. doi: 10.1128/JVI.05633-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Monel B, et al. Zika virus induces massive cytoplasmic vacuolization and paraptosis-like death in infected cells. EMBO J. 2017;36:1653–1668. doi: 10.15252/embj.201695597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Savidis G, et al. The IFITMs inhibit Zika virus replication. Cell Rep. 2016;15:2323–2330. doi: 10.1016/j.celrep.2016.05.074. [DOI] [PubMed] [Google Scholar]
- 28.Wrensch F, et al. IFITMs mediate viral evasion in acute and chronic hepatitis C virus infection. Hepatology. 2019;70:1506–1520. doi: 10.1002/hep.30699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jiang D, et al. Identification of five interferon-induced cellular proteins that inhibit West Nile virus and dengue virus infections. J. Virol. 2010;84:8332–8341. doi: 10.1128/JVI.02199-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wrensch F, et al. Interferon-induced transmembrane protein-mediated inhibition of host cell entry of ebolaviruses. J. Infect. Dis. 2015;212:S210–S218. doi: 10.1093/infdis/jiv255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fu B, Wang L, Li S, Dorf ME. ZMPSTE24 defends against influenza and other pathogenic viruses. J. Exp. Med. 2017;214:919–929. doi: 10.1084/jem.20161270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mudhasani R, et al. IFITM-2 and IFITM-3 but not IFITM-1 restrict Rift Valley fever virus. J. Virol. 2013;87:8451–8464. doi: 10.1128/JVI.03382-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weston S, et al. Alphavirus restriction by IFITM proteins. Traffic. 2016;17:997–1013. doi: 10.1111/tra.12416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Poddar S, Hyde JL, Gorman MJ, Farzan M, Diamond MS. The interferon-stimulated gene IFITM3 restricts infection and pathogenesis of arthritogenic and encephalitic alphaviruses. J. Virol. 2016;90:8780–8794. doi: 10.1128/JVI.00655-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Franz S, et al. Human IFITM3 restricts chikungunya virus and Mayaro virus infection and is susceptible to virus-mediated counteraction. Life Sci. Alliance. 2021;4:e202000909. doi: 10.26508/lsa.202000909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lu J, et al. The IFITM proteins inhibit HIV-1 infection. J. Virol. 2011;85:2126–2137. doi: 10.1128/JVI.01531-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wilkins J, Zheng Y-M, Yu J, Liang C, Liu S-L. Nonhuman primate IFITM proteins are potent inhibitors of HIV and SIV. PLoS ONE. 2016;11:e0156739. doi: 10.1371/journal.pone.0156739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Qian J, et al. Primate lentiviruses are differentially inhibited by interferon-induced transmembrane proteins. Virology. 2015;474:10–18. doi: 10.1016/j.virol.2014.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Foster TL, et al. Resistance of transmitted founder HIV-1 to IFITM-mediated restriction. Cell Host Microbe. 2016;12:429–442. doi: 10.1016/j.chom.2016.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wrensch F, Winkler M, Pöhlmann S. IFITM proteins inhibit entry driven by the MERS-coronavirus spike protein: evidence for cholesterol-independent mechanisms. Viruses. 2014;6:3683–3698. doi: 10.3390/v6093683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shi G, et al. Opposing activities of IFITM proteins in SARS‐CoV‐2 infection. EMBO J. 2020;40:e106501. doi: 10.15252/embj.2020106501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Winstone H, et al. The polybasic cleavage site in the SARS-CoV-2 spike modulates viral sensitivity to type I interferon and IFITM2. J. Virol. 2021;95:e02422–20. doi: 10.1128/JVI.02422-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhao X, et al. Identification of residues controlling restriction versus enhancing activities of IFITM proteins on entry of human coronaviruses. J. Virol. 2018;92:e01535–17. doi: 10.1128/JVI.01535-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhao X, et al. LY6E restricts entry of human coronaviruses, including currently pandemic SARS-CoV-2. J. Virol. 2020 doi: 10.1128/JVI.00562-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Diamond MS, Farzan M. The broad-spectrum antiviral functions of IFIT and IFITM proteins. Nat. Rev. Immunol. 2013;13:46–57. doi: 10.1038/nri3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Warren CJ, et al. The antiviral restriction factors IFITM1, 2 and 3 do not inhibit infection of human papillomavirus, cytomegalovirus and adenovirus. PLoS ONE. 2014;9:e96579. doi: 10.1371/journal.pone.0096579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Compton AA, et al. Natural mutations in IFITM3 modulate post-translational regulation and toggle antiviral specificity. EMBO Rep. 2016;17:1657–1671. doi: 10.15252/embr.201642771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Everitt AR, et al. IFITM3 restricts the morbidity and mortality associated with influenza. Nature. 2012;484:519–523. doi: 10.1038/nature10921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Allen EK, et al. SNP-mediated disruption of CTCF binding at the IFITM3 promoter is associated with risk of severe influenza in humans. Nat. Med. 2017;23:975–983. doi: 10.1038/nm.4370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang Y, et al. Interferon-induced transmembrane protein-3 genetic variant rs12252-C is associated with disease severity in COVID-19. J. Infect. Dis. 2020;222:34–37. doi: 10.1093/infdis/jiaa224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nikoloudis D, Kountouras D, Hiona A. The frequency of combined IFITM3 haplotype involving the reference alleles of both rs12252 and rs34481144 is in line with COVID-19 standardized mortality ratio of ethnic groups in England. PeerJ. 2020;8:e10402. doi: 10.7717/peerj.10402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bailey CC, Huang IC, Kam C, Farzan M. Ifitm3 limits the severity of acute influenza in mice. PLoS Pathog. 2012;8:e1002909. doi: 10.1371/journal.ppat.1002909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kenney AD, et al. IFITM3 protects the heart during influenza virus infection. Proc. Natl Acad. Sci. USA. 2019;116:18607–18612. doi: 10.1073/pnas.1900784116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gorman MJ, Poddar S, Farzan M, Diamond MS. The interferon-stimulated gene ifitm3 restricts west nile virus infection and pathogenesis. J. Virol. 2016;90:8212–8225. doi: 10.1128/JVI.00581-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zani A, Yount JS. Antiviral protection by IFITM3 in vivo. Curr. Clin. Microbiol. Rep. 2018;5:229–237. doi: 10.1007/s40588-018-0103-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Desai TM, et al. IFITM3 restricts influenza A virus entry by blocking the formation of fusion pores following virus-endosome hemifusion. PLoS Pathog. 2014;10:e1004048. doi: 10.1371/journal.ppat.1004048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.John SP, et al. The CD225 domain of IFITM3 is required for both IFITM protein association and inhibition of influenza A virus and dengue virus replication. J. Virol. 2013;87:7837–7852. doi: 10.1128/JVI.00481-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rahman K, et al. Homology-guided identification of a conserved motif linking the antiviral functions of IFITM3 to its oligomeric state. eLife. 2020;9:e58537. doi: 10.7554/eLife.58537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chesarino NM, et al. IFITM3 requires an amphipathic helix for antiviral activity. EMBO Rep. 2017;18:1740–1751. doi: 10.15252/embr.201744100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ling S, et al. Combined approaches of EPR and NMR illustrate only one transmembrane helix in the human IFITM3. Sci. Rep. 2016;6:24029. doi: 10.1038/srep24029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Guo X, et al. Interferon-induced transmembrane protein 3 blocks fusion of diverse enveloped viruses by altering mechanical properties of cell membranes. ACS Nano. 2021;15:8155–8170. doi: 10.1021/acsnano.0c10567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lin T-Y, et al. Amphotericin B increases influenza a virus infection by preventing IFITM3-mediated restriction. Cell Rep. 2013;5:895–908. doi: 10.1016/j.celrep.2013.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kamiński DM. Recent progress in the study of the interactions of amphotericin B with cholesterol and ergosterol in lipid environments. Eur. Biophys. J. 2014;43:453–467. doi: 10.1007/s00249-014-0983-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chesarino NM, McMichael TM, Hach JC, Yount JS. Phosphorylation of the antiviral protein interferon-inducible transmembrane protein 3 (IFITM3) dually regulates its endocytosis and ubiquitination. J. Biol. Chem. 2014;289:11986–11992. doi: 10.1074/jbc.M114.557694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chesarino NM, McMichael TM, Yount JS. Regulation of the trafficking and antiviral activity of IFITM3 by post-translational modifications. Future Microbiol. 2014;9:1151–1163. doi: 10.2217/fmb.14.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jia R, et al. The N-terminal region of IFITM3 modulates its antiviral activity by regulating IFITM3 cellular localization. J. Virol. 2012;86:13697–13707. doi: 10.1128/JVI.01828-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jia R, et al. Identification of an endocytic signal essential for the antiviral action of IFITM3. Cell. Microbiol. 2014;16:1080–1093. doi: 10.1111/cmi.12262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shi G, Ozog S, Torbett BE, Compton AA. mTOR inhibitors lower an intrinsic barrier to virus infection mediated by IFITM3. Proc. Natl Acad. Sci. USA. 2018;115:E10069–E10078. doi: 10.1073/pnas.1811892115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.OhAinle M, et al. A virus-packageable CRISPR screen identifies host factors mediating interferon inhibition of HIV. eLife. 2018;7:e39823. doi: 10.7554/eLife.39823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jia R, et al. The C-terminal sequence of IFITM1 regulates its anti-HIV-1 activity. PLoS ONE. 2015;10:e0118794. doi: 10.1371/journal.pone.0118794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li K, et al. A sorting signal suppresses ifitm1 restriction of viral entry. J. Biol. Chem. 2014;290:4248–4259. doi: 10.1074/jbc.M114.630780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yount JS, et al. Palmitoylome profiling reveals S-palmitoylation-dependent antiviral activity of IFITM3. Nat. Chem. Biol. 2010;6:610–614. doi: 10.1038/nchembio.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yount JS, Karssemeijer RA, Hang HC. S-palmitoylation and ubiquitination differentially regulate interferon-induced transmembrane protein 3 (IFITM3)-mediated resistance to influenza virus. J. Biol. Chem. 2012;287:19631–19641. doi: 10.1074/jbc.M112.362095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Garst EH, et al. Site-Specific lipidation enhances IFITM3 membrane interactions and antiviral activity. ACS Chem. Biol. 2021;16:844–856. doi: 10.1021/acschembio.1c00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Benfield CTO, et al. Bat IFITM3 restriction depends on S-palmitoylation and a polymorphic site within the CD225 domain. Life Sci. Alliance. 2020;3:e201900542. doi: 10.26508/lsa.201900542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Spence JS, et al. IFITM3 directly engages and shuttles incoming virus particles to lysosomes. Nat. Chem. Biol. 2018;15:259–268. doi: 10.1038/s41589-018-0213-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Suddala KC, et al. Interferon-induced transmembrane protein 3 blocks fusion of sensitive but not resistant viruses by partitioning into virus-carrying endosomes. PLoS Pathog. 2019;15:e1007532–e1007535. doi: 10.1371/journal.ppat.1007532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhu X, et al. IFITM3‐containing exosome as a novel mediator for anti‐viral response in dengue virus infection. Cell. Microbiol. 2015;17:105–118. doi: 10.1111/cmi.12339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zou X, Yuan M, Zhang T, Zheng N, Wu Z. EVs containing host restriction factor IFITM3 inhibited ZIKV infection of fetuses in pregnant mice through trans-placenta delivery. Mol. Ther. 2021;29:176–190. doi: 10.1016/j.ymthe.2020.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Perrin P, et al. Retrofusion of intralumenal MVB membranes parallels viral infection and coexists with exosome release. Curr. Biol. 2021 doi: 10.1016/j.cub.2021.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Winkler M, et al. Analysis of IFITM-IFITM interactions by a flow cytometry-based FRET assay. Int. J. Mol. Sci. 2019;20:3859. doi: 10.3390/ijms20163859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chesarino NM, McMichael TM, Yount JS. E3 ubiquitin ligase NEDD4 promotes influenza virus infection by decreasing levels of the antiviral protein IFITM3. PLoS Pathog. 2015;11:e1005095. doi: 10.1371/journal.ppat.1005095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wu X, et al. Site-specific photo-crosslinking proteomics reveal regulation of IFITM3 trafficking and turnover by VCP/p97 ATPase. Cell Chem. Biol. 2020;27:571–585. doi: 10.1016/j.chembiol.2020.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Holthuis JCM, Levine TP. Lipid traffic: floppy drives and a superhighway. Nat. Rev. Mol. Cell Biol. 2005;6:209–220. doi: 10.1038/nrm1591. [DOI] [PubMed] [Google Scholar]
- 85.Amini-Bavil-Olyaee S, et al. The antiviral effector IFITM3 disrupts intracellular cholesterol homeostasis to block viral entry. Cell Host Microbe. 2013;13:452–464. doi: 10.1016/j.chom.2013.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ridgway ND, Zhao K. Cholesterol transfer at endosomal-organelle membrane contact sites. Curr. Op. Lipidol. 2018;29:212–217. doi: 10.1097/MOL.0000000000000506. [DOI] [PubMed] [Google Scholar]
- 87.Zhao K, Ridgway ND. Oxysterol-binding protein-related protein 1L regulates cholesterol egress from the endo- lysosomal system. Cell Rep. 2017;19:1807–1818. doi: 10.1016/j.celrep.2017.05.028. [DOI] [PubMed] [Google Scholar]
- 88.Wyles JP, McMaster CR, Ridgway ND. Vesicle-associated membrane protein-associated protein-A (VAP-A) interacts with the oxysterol-binding protein to modify export from the endoplasmic reticulum. J. Biol. Chem. 2002;277:29908–29918. doi: 10.1074/jbc.M201191200. [DOI] [PubMed] [Google Scholar]
- 89.Sobo K, et al. Late Endosomal cholesterol accumulation leads to impaired intra-endosomal trafficking. PLoS ONE. 2007;2:e851. doi: 10.1371/journal.pone.0000851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kühnl A, et al. Late endosomal/lysosomal cholesterol accumulation is a host cell-protective mechanism inhibiting endosomal escape of influenza A virus. mBio. 2018;9:e01345–18. doi: 10.1128/mBio.01345-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wee YS, Roundy KM, Weis JJ, Weis JH. Interferon-inducible transmembrane proteins of the innate immune response act as membrane organizers by influencing clathrin and v-ATPase localization and function. Innate Immun. 2012;18:834–845. doi: 10.1177/1753425912443392. [DOI] [PubMed] [Google Scholar]
- 92.Desai TM, Marin M, Mason C, Melikyan GB. pH regulation in early endosomes and interferon-inducible transmembrane proteins control avian retrovirus fusion. J. Biol. Chem. 2017;292:7817–7827. doi: 10.1074/jbc.M117.783878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Yang S-T, Kreutzberger AJB, Lee J, Kiessling V, Tamm LK. The role of cholesterol in membrane fusion. Chem. Phys. Lipids. 2016;199:136–143. doi: 10.1016/j.chemphyslip.2016.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Anafu AA, Bowen CH, Chin CR, Brass AL, Holm GH. Interferon-inducible transmembrane protein 3 (IFITM3) restricts reovirus cell entry. J. Biol. Chem. 2013;288:17261–17271. doi: 10.1074/jbc.M112.438515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zang R, et al. Cholesterol 25-hydroxylase suppresses SARS-CoV-2 replication by blocking membrane fusion. Proc. Natl Acad. Sci. USA. 2020;117:32105–32113. doi: 10.1073/pnas.2012197117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Bertram S, et al. TMPRSS2 activates the human coronavirus 229E for cathepsin-independent host cell entry and is expressed in viral target cells in the respiratory epithelium. J. Virol. 2013;87:6150–6160. doi: 10.1128/JVI.03372-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Bozzo CP, et al. IFITM proteins promote SARS-CoV-2 infection and are targets for virus inhibition in vitro. Nat. Commun. 2021;12:4584. doi: 10.1038/s41467-021-24817-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zhao X, et al. Interferon induction of IFITM proteins promotes infection by human coronavirus OC43. Proc. Natl Acad. Sci. USA. 2014;111:6756–6761. doi: 10.1073/pnas.1320856111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Stacey MA, et al. The antiviral restriction factor IFN-induced transmembrane protein 3 prevents cytokine-driven CMV pathogenesis. J. Clin. Invest. 2017;127:1463–1474. doi: 10.1172/JCI84889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Bergo MO, et al. Zmpste24 deficiency in mice causes spontaneous bone fractures, muscle weakness, and a prelamin A processing defect. Proc. Natl Acad. Sci. USA. 2002;99:13049–13054. doi: 10.1073/pnas.192460799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Varela I, et al. Accelerated ageing in mice deficient in Zmpste24 protease is linked to p53 signalling activation. Nature. 2005;437:564–568. doi: 10.1038/nature04019. [DOI] [PubMed] [Google Scholar]
- 102.Ast T, Michaelis S, Schuldiner M. The protease Ste24 clears clogged translocons. Cell. 2016;164:103–114. doi: 10.1016/j.cell.2015.11.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Li S, Fu B, Wang L, Dorf ME. ZMPSTE24 is downstream effector of interferon-induced transmembrane (IFITM) antiviral activity. DNA Cell Biol. 2017;36:513–517. doi: 10.1089/dna.2017.3791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Stott RJ, Foster T. Inhibition of arenavirus entry and replication by the cell-intrinsic restriction factor ZMPSTE24 is enhanced by IFITM antiviral activity. bioRxiv. 2021 doi: 10.1101/2021.04.12.439453v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Rojek JM, Sanchez AB, Nguyen NT, de la Torre JC, Kunz S. Different mechanisms of cell entry by human-pathogenic Old World and New World arenaviruses. J. Virol. 2008;82:7677–7687. doi: 10.1128/JVI.00560-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Zhao J, Chen J, Li M, Chen M, Sun C. Multifaceted functions of CH25H and 25HC to modulate the lipid metabolism, immune responses, and broadly antiviral activities. Viruses. 2020;12:727. doi: 10.3390/v12070727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Schoggins JW, Randall G. Lipids in innate antiviral defense. Cell Host Microbe. 2013;14:379–385. doi: 10.1016/j.chom.2013.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Abrams ME, et al. Oxysterols provide innate immunity to bacterial infection by mobilizing cell surface accessible cholesterol. Nat. Microbiol. 2020;5:929–942. doi: 10.1038/s41564-020-0701-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Liu S-Y, Sanchez DJ, Aliyari R, Lu S, Cheng G. Systematic identification of type I and type II interferon-induced antiviral factors. Proc. Natl Acad. Sci. USA. 2012;109:4239–4244. doi: 10.1073/pnas.1114981109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Liu S-Y, et al. Interferon-inducible cholesterol-25-hydroxylase broadly inhibits viral entry by production of 25-hydroxycholesterol. Immunity. 2013;38:92–105. doi: 10.1016/j.immuni.2012.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Bauman DR, et al. 25-Hydroxycholesterol secreted by macrophages in response to Toll-like receptor activation suppresses immunoglobulin A production. Proc. Natl Acad. Sci. USA. 2009;106:16764–16769. doi: 10.1073/pnas.0909142106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Gale SE, et al. Side chain oxygenated cholesterol regulates cellular cholesterol homeostasis through direct sterol-membrane interactions. J. Biol. Chem. 2009;284:1755–1764. doi: 10.1074/jbc.M807210200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Wang S, et al. Cholesterol 25‐hydroxylase inhibits SARS‐CoV‐2 and other coronaviruses by depleting membrane cholesterol. EMBO J. 2020;39:e106057. doi: 10.15252/embj.2020106057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Li C, et al. 25-Hydroxycholesterol protects host against zika virus infection and its associated microcephaly in a mouse model. Immunity. 2017;46:446–456. doi: 10.1016/j.immuni.2017.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Gold ES, et al. 25-Hydroxycholesterol acts as an amplifier of inflammatory signaling. Proc. Natl Acad. Sci. USA. 2014;111:10666–10671. doi: 10.1073/pnas.1404271111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Chen Y, et al. Interferon-inducible cholesterol-25-hydroxylase inhibits hepatitis C virus replication via distinct mechanisms. Sci. Rep. 2014;4:7242–7248. doi: 10.1038/srep07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Doms A, Sanabria T, Hansen JN, Altan-Bonnet N, Holm GH. 25-Hydroxycholesterol production by the cholesterol-25-hydroxylase interferon-stimulated gene restricts mammalian reovirus infection. J. Virol. 2018;92:e01047–18. doi: 10.1128/JVI.01047-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Kosugi A, Saitoh S, Narumiya S, Miyake K, Hamaoka T. Activation-induced expression of thymic shared antigen-1 on T lymphocytes and its inhibitory role for TCR-mediated IL-2 production. Int. Immunol. 1994;6:1967–1976. doi: 10.1093/intimm/6.12.1967. [DOI] [PubMed] [Google Scholar]
- 119.Xu X, et al. IFN-stimulated gene LY6E in monocytes regulates the CD14/TLR4 pathway but inadequately restrains the hyperactivation of monocytes during chronic HIV-1 infection. J. Immunol. 2014;193:4125–4136. doi: 10.4049/jimmunol.1401249. [DOI] [PubMed] [Google Scholar]
- 120.Schoggins JW, et al. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature. 2012;472:481–485. doi: 10.1038/nature09907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Pfaender S, et al. LY6E impairs coronavirus fusion and confers immune control of viral disease. Nat. Microbiol. 2020;5:1330–1339. doi: 10.1038/s41564-020-0769-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Hackett BA, Cherry S. Flavivirus internalization is regulated by a size-dependent endocytic pathway. Proc. Natl Acad. Sci. USA. 2018;115:4246–4251. doi: 10.1073/pnas.1720032115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Mar KB, et al. LY6E mediates an evolutionarily conserved enhancement of virus infection by targeting a late entry step. Nat. Commun. 2018;9:3603. doi: 10.1038/s41467-018-06000-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Yu J, Liang C, Liu S-L. Interferon-inducible LY6E protein promotes HIV-1 infection. J. Biol. Chem. 2017;292:4674–4685. doi: 10.1074/jbc.M116.755819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Yu J, Liang C, Liu S-L. CD4-dependent modulation of HIV-1 entry by LY6E. J. Virol. 2019;93:e01866–18. doi: 10.1128/JVI.01866-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Lin F, Young HA. Interferons: success in anti-viral immunotherapy. Cytokine Growth Factor. Rev. 2014;25:369–376. doi: 10.1016/j.cytogfr.2014.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Durand M, et al. The OXR domain defines a conserved family of eukaryotic oxidation resistance proteins. BMC Cell Biol. 2007;8:13. doi: 10.1186/1471-2121-8-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Shao W, Halachmi S, Brown M. ERAP140, a conserved tissue-specific nuclear receptor coactivator. Mol. Cell. Biol. 2002;22:3358–3372. doi: 10.1128/MCB.22.10.3358-3372.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Yu L, et al. Induction of a unique isoform of the NCOA7 oxidation resistance gene by interferon β-1b. J. Interf. Cytokine Res. 2015;35:186–199. doi: 10.1089/jir.2014.0115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Doyle T, et al. The interferon-inducible isoform of NCOA7 inhibits endosome-mediated viral entry. Nat. Microbiol. 2018;3:1369–1376. doi: 10.1038/s41564-018-0273-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.West LC, Cresswell P. Expanding roles for GILT in immunity. Cur. Op. Immunol. 2013;25:103–108. doi: 10.1016/j.coi.2012.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Arunachalam B, Phan UT, Geuze HJ, Cresswell P. Enzymatic reduction of disulfide bonds in lysosomes: characterization of a gamma-interferon-inducible lysosomal thiol reductase (GILT) Proc. Natl Acad. Sci. USA. 2000;97:745–750. doi: 10.1073/pnas.97.2.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Singh R, Cresswell P. Defective cross-presentation of viral antigens in GILT-free mice. Science. 2010;328:1394–1398. doi: 10.1126/science.1189176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Chen D, et al. GILT restricts the cellular entry mediated by the envelope glycoproteins of SARS-CoV, Ebola virus and Lassa fever virus. Emerg. Microbes Infect. 2019;8:1511–1523. doi: 10.1080/22221751.2019.1677446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Mingo RM, et al. Ebola virus and severe acute respiratory syndrome coronavirus display late cell entry kinetics: evidence that transport to NPC1+ endolysosomes is a rate-defining step. J. Virol. 2015;89:2931–2943. doi: 10.1128/JVI.03398-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Kubo Y, et al. Gamma-interferon-inducible, lysosome/endosome-localized thiolreductase, GILT, has anti-retroviral activity and its expression is counteracted by HIV-1. Oncotarget. 2016;7:71255–71273. doi: 10.18632/oncotarget.12104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Jakobsdottir GM, et al. On the whereabouts of HIV-1 cellular entry and its fusion ports. Trends Mol. Med. 2017;23:932–944. doi: 10.1016/j.molmed.2017.08.005. [DOI] [PubMed] [Google Scholar]
- 138.Wells AI, Coyne CB. Inhibiting Ebola virus and SARS-CoV-2 entry. Science. 2020;370:167–168. doi: 10.1126/science.abe2977. [DOI] [PubMed] [Google Scholar]
- 139.Bruchez A, et al. MHC class II transactivator CIITA induces cell resistance to Ebola virus and SARS-like coronaviruses. Science. 2020;370:241–247. doi: 10.1126/science.abb3753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Van Acker T, Tavernier J, Peelman F. The small GTPase Arf6: an overview of its mechanisms of action and of its role in host− pathogen interactions and innate immunity. Int. J. Mol. Sci. 2019;20:2209. doi: 10.3390/ijms20092209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Shu Q, Lennemann NJ, Sarkar SN, Sadovsky Y, Coyne CB. ADAP2 is an interferon stimulated gene that restricts RNA virus entry. PLoS Pathog. 2015;11:e1005150. doi: 10.1371/journal.ppat.1005150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Fan DP, Sefton BM. The entry into host cells of Sindbis virus, vesicular stomatitis virus and Sendai virus. Cell. 1978;15:985–992. doi: 10.1016/0092-8674(78)90282-9. [DOI] [PubMed] [Google Scholar]
- 143.Bist P, et al. ArfGAP domain-containing protein 2 (ADAP2) integrates upstream and downstream modules of RIG-I signaling and facilitates type I interferon production. Mol. Cell. Biol. 2017;37:e00537-16. doi: 10.1128/MCB.00537-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Foster TL, Pickering S, Neil SJD. Inhibiting the ins and outs of HIV replication: cell-intrinsic antiretroviral restrictions at the plasma membrane. Front. Immunol. 2018;8:1853. doi: 10.3389/fimmu.2017.01853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Chemudupati M, et al. From APOBEC to ZAP: diverse mechanisms used by cellular restriction factors to inhibit virus infections. Biochim. Biophys. Acta Mol. Cell Res. 2019;1866:382–394. doi: 10.1016/j.bbamcr.2018.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Shi G, Schwartz O, Compton AA. More than meets the I: the diverse antiviral and cellular functions of interferon-induced transmembrane proteins. Retrovirology. 2017;14:53. doi: 10.1186/s12977-017-0377-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Heaton BE, et al. A CRISPR activation screen identifies a pan-avian influenza virus inhibitory host factor. Cell Rep. 2017;20:1503–1512. doi: 10.1016/j.celrep.2017.07.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Silvin A, et al. Constitutive resistance to viral infection in human CD141+ dendritic cells. Sci. Immunol. 2017;2:eaai8071. doi: 10.1126/sciimmunol.aai8071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Strick DJ, Elferink LA. Rab15 effector protein: a novel protein for receptor recycling from the endocytic recycling compartment. Mol. Biol. Cell. 2005;16:5699–5709. doi: 10.1091/mbc.e05-03-0204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Zuk PA, Elferink LA. Rab15 differentially regulates early endocytic trafficking. J. Biol. Chem. 2000;275:26754–26764. doi: 10.1016/S0021-9258(19)61440-7. [DOI] [PubMed] [Google Scholar]
- 151.Katsura H, et al. Human lung stem cell-based alveolospheres provide insights into SARS-CoV-2-mediated interferon responses and pneumocyte dysfunction. Cell Stem Cell. 2020;27:890–904.e8. doi: 10.1016/j.stem.2020.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Lipovsky A, et al. The cellular endosomal protein stannin inhibits intracellular trafficking of human papillomavirus during virus entry. J. Gen. Virol. 2017;98:2821–2836. doi: 10.1099/jgv.0.000954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.White JM, Whittaker GR. Fusion of enveloped viruses in endosomes. Traffic. 2016;17:593–614. doi: 10.1111/tra.12389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.McNab F, Mayer-Barber K, Sher A, Wack A, O’Garra A. Type I interferons in infectious disease. Nat. Rev. Immunol. 2015;15:87–103. doi: 10.1038/nri3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Park A, Iwasaki A. Type I and type III interferons - induction, signaling, evasion, and application to combat COVID-19. Cell Host Microbe. 2020;27:870–878. doi: 10.1016/j.chom.2020.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Schoggins JW. Interferon-stimulated genes: what do they all do? Annu. Rev. Virol. 2019;6:567–584. doi: 10.1146/annurev-virology-092818-015756. [DOI] [PubMed] [Google Scholar]