Abstract
Purpose
Coronavirus disease 2019 (COVID-19) is caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV2). While the ocular surface is considered one of the major SARS-CoV2 transmission routes, the specific cellular tropism of SARS-CoV2 is not fully understood. In the current study, we evaluated the expression and regulation of two SARS-CoV2 viral entry proteins, TMPRSS2 and ACE2, in human ocular epithelial cells and stem cells.
Methods
TMPRSS2 and ACE2 expression in ABCB5-positive limbal stem cells (LSCs) were assessed by RNAseq, flow cytometry and immunohistochemistry. PAX6, TMPRSS2, and ACE2 mRNA expression values were obtained from the GSE135455 and DRA002960 RNA-seq datasets. siRNA-mediated PAX6 knockdown (KD) was performed in limbal and conjunctival epithelial cells. TMPRSS2 and ACE2 expression in the PAX6 KD cells was analyzed by qRT-PCR and Western blot.
Results
We found that ABCB5-positive LSCs express high levels of TMPRSS2 and ACE2 compared to ABCB5-negative limbal epithelial cells. Mechanistically, gene knockout and overexpression models revealed that the eye transcription factor PAX6 negatively regulates TMPRSS2 expression. Therefore, low levels of PAX6 in ABCB5-positive LSCs promote TMPRSS2 expression, and high levels of TMPRSS2 and ACE2 expression by LSCs indicate enhanced susceptibility to SARS-CoV2 infection in this stem cell population.
Conclusions
Our study points to a need for COVID-19 testing of LSCs derived from donor corneas before transplantation to patients with limbal stem cell deficiency. Furthermore, our findings suggest that expandable human ABCB5+ LSC cultures might represent a relevant novel model system for studying cellular SARS-CoV2 viral entry mechanisms and evaluating related targeting strategies.
Keywords: SARS-CoV2, COVID-19, Limbal stem cells, ACE2, TMPRSS2, PAX6
Abbreviations: COVID19, coronavirus disease 2019; SARS-CoV2, severe acute respiratory syndrome coronavirus 2; LSC, limbal stem cell; ABCB5, ATP-binding cassette subfamily B member 5; LSCD, limbal stem cell deficiency; CK3, Cytokeratin 3; CK12, Cytokeratin 12; CLU, Clusterin; ALDH3A1, Aldehyde dehydrogenase 3 family member A1; AR, androgen receptor; pAb, polyclonal antibody; FSC, forward scatter; SSC, side scatter; A, area; W, width; H, height
Novel coronavirus disease-2019 (COVID-19) is caused by infection with severe acute respiratory syndrome coronavirus 2 (SARS-CoV2), with over 200 million people already affected worldwide. A recent report revealed that 11.2% of COVID-19 patients presented with ocular symptoms such as conjunctivitis, ocular pain, and dry eye, and 16.7% of those patients had RT-PCR-detectable SARS-CoV2 transcripts [1]. Eye protection by goggles or face shields can reduce the risk of SARS-CoV2 infection [2], underlining the significance of the ocular surface as a pathway to viral entry. While being recognized as a route of SARS-CoV2 infectivity, the ocular surface's cellular constituents implicated in SARS-CoV2 tropism, and a potential involvement of the LSC compartment, are currently not fully understood [3].
In the course of human infection, SARS-CoV-2 cellular entry depends on binding of the viral spike (S) protein to the human cellular receptor ACE2 and on subsequent S protein priming by the cellular protease TMPRSS2 [4]. Recent single-cell RNA-sequencing analyses (scRNA-seq) revealed similarly high expression of ACE2 mRNA in all ocular epithelial cells, whereas TMPRSS2 mRNA transcripts were detected at low levels only in the conjunctival epithelium [5]. Similar results were reported by Collin et al. who found, using scRNA-Seq analyses of human adult cornea and conjunctiva, ACE2 and TMPRSS2 co-expression in limbus, peripheral cornea and superficial conjunctival epithelium [6]. In a further study, in situ immunostaining demonstrated ACE2 protein expression localized to superficial ocular epithelial cells, while TMPRSS2 was detected in all epithelial layers, with significantly enhanced expression in cells located at the border of the cornea and conjunctiva, i.e., the limbus [7]. Subsequently, Eriksen et al. showed that the limbus is most susceptible to SARS-CoV-2 infection among various ocular cell types [8]. Despite these observations, it has remained unknown to date whether SARS-CoV-2 entry proteins are expressed homogeneously among all limbal epithelial cell types or whether expression favors a specific cellular sub-population.
The corneal limbus contains stem cells capable of regenerating the entire corneal epithelium, known as limbal stem cells (LSCs) [9]. We recently described ATP-binding cassette subfamily B member 5 (ABCB5) as a first molecular marker suitable for the prospective isolation of LSCs [10]. Transplantation of ABCB5-positive LSCs [[10], [11], [12], [13], [14], [15], [16]] was hereby shown in pre-clinical studies to result in long-term corneal restoration in the setting of limbal stem cell deficiency (LSCD) [10]. As a result of this discovery, an interventional, open-label, phase I/IIa human clinical trial has recently been initiated, which investigates the safety and efficacy of allogeneic ABCB5-positive LSCs for LSCD therapy [17]. Because long-term quiescent ABCB5-positive LSCs in healthy individuals might represent a site of viral entry and a cellular reservoir for clinically observed viral persistence upon SARS-CoV-2 infection, and because ABCB5-positive LSCs represent a stem cell population already in use for experimental transplantation in human clinical trials, we herein sought to examine whether they expressed host viral entry proteins, a prerequisite for potentially rendering this stem cell compartment susceptible to SARS-CoV-2 cellular tropism.
As a master regulator of eye development, the PAX6 transcription factor is essential for the formation of the surface ectoderm-derived ocular epithelial tissues of the cornea and conjunctiva, where it induces the expression of several tissue-specific proteins such as Cytokeratin 3 (CK3), Cytokeratin 12 (CK12), Clusterin (CLU) and Aldehyde dehydrogenase 3 family member A1 (ALDH3A1) [[18], [19], [20], [21]]. The two main PAX6 isoforms, PAX6-a and PAX6-b, are distinguished by an additional exon 5a present in isoform b [[22], [23], [24]]. PAX6-b specifically induces the differentiation marker CK12 in corneal epithelial cells [20]. Because PAX6 induces LSC differentiation, we further hypothesized that it might negatively regulate host viral entry protein expression.
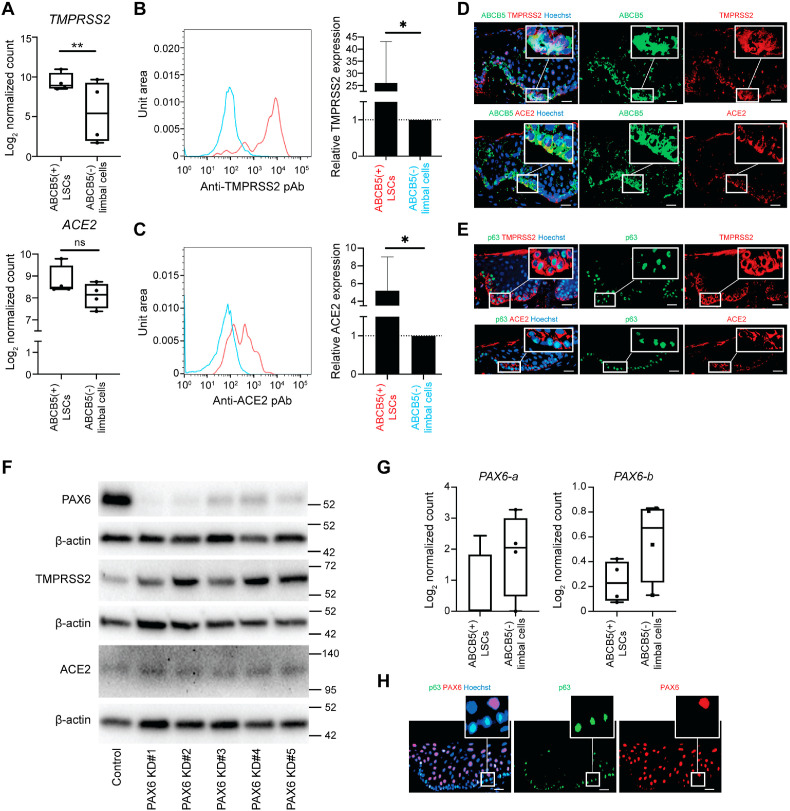
In the current study, we evaluated the expression of two SARS-CoV2 viral entry proteins, TMPRSS2 and ACE2, in human ABCB5-positive LSCs and in more differentiated ocular epithelial cells. First, using RNA sequencing (RNA-seq) analyses of purified ABCB5-positive LSCs and ABCB5-negative limbal epithelial cells isolated as reported previously [10,25] by flow cytometry from n = 4 human donors (Supplementary Fig. 1A), we found that TMPRSS2 transcripts were significantly enriched in ABCB5-positive LSCs (p = 0.0038) (Fig. 1 A). ACE2 mRNA expression was nominally higher in this cell population (Fig. 1A). Consistent with RNA-seq results, flow cytometry analyses revealed that TMPRSS2 (Fig. 1B) and ACE2 (Fig. 1C) proteins were expressed at significantly higher levels by ABCB5-positive LSCs compared to ABCB5-negative limbal epithelial cells (p = 0.0161 and p = 0.0440, respectively). Immunostaining analyses of human corneas showed high expression of TMPRSS2 and ACE2 in ABCB5-positive LSCs (Fig. 1D). TMPRSS2 and ACE2 were also co-expressed with another LSC marker, p63 (Fig. 1E) [26]. These findings demonstrate that amongst limbal epithelial cells, LSCs possess a specific molecular phenotype characterized by TMPRSS2 and ACE2 overexpression, consistent with enhanced susceptibility to SARS-CoV2 cellular tropism.
Fig. 1.
Enhanced TMPRSS2 and ACE2 expression by ABCB5-positive LSCs. (A) Log2 normalized counts of TMPRSS2 and ACE2 expression by ABCB5-positive LSCs and ABCB5-negative limbal epithelial cells (n = 4, ** adjusted p < 0.01). (B) Flow cytometry analysis of TMPRSS2 expression by ABCB5-positive LSCs and ABCB5-negative limbal epithelial cells. Bar graph represents relative TMPRSS2 expression (mean ± SD; n = 6, *p < 0.05). Data were analyzed using the paired-t test. (C) Flow cytometry analysis of ACE2 expression by ABCB5-positive LSCs and ABCB5-negative limbal epithelial cells. Bar graph represents relative ACE2 expression (mean ± SD; n = 6, *p < 0.05). Data were analyzed using the paired-t test. (D) Representative immunostaining analysis of ABCB5 (green) and TMPRSS2 and ACE2 (red) expression in the limbus. Nuclei stained with Hoechst 33,342 (blue). Scale bar: 20 μm. (E) Representative immunostaining analysis of p63 (green) and TMPRSS2 and ACE2 (red) expression in the limbus. Nuclei stained with Hoechst 33,342 (blue). Scale bar: 20 μm. (F) Representative Western blot analyses of PAX6, TMPRSS2 and ACE2 in limbal epithelial cells subjected to PAX6 si-RNA knockdown (KD) (n = 6). (G) Isoforms-specific PAX6 mRNA expression by ABCB5-positive LSCs vs. ABCB5-negative limbal epithelial cells shown as log2 normalized counts. (H) Representative immunostaining analysis of p63 (green) and PAX6 (red) expression in the limbus. Nuclei stained with Hoechst 33,342 (blue). Scale bar: 20 μm.
Given the importance of the eye transcription factor PAX6 in orchestrating ocular gene expression, we hypothesized that PAX6 might play a role in regulating SARS-CoV2 entry protein expression by human limbal epithelial cells. To test this hypothesis, utilizing publicly available RNA-seq results (GSE135455) [27], we first examined the correlation between PAX6, TMPRSS2, and ACE2 mRNA expression in conjunctival swabs collected from 48 human patients. The analyses revealed a moderate negative correlation between PAX6 and TMPRSS2 mRNA expression (r = −0.58, p = 2e-5), while no significant correlation was observed between PAX6 and ACE2 expression (r = −0.26, p = 0.074) (Supplementary Fig. 1B). Of note, a positive correlation was also observed between TMPRSS2 and ACE2 expression (r = 0.5, p = 0.00034) (Supplementary Fig. 1B). Next, utilizing an RNA-seq data set from one of our previously published studies (DRA002960) [20], we examined whether forced overexpression of either PAX6-a or PAX6-b in oral mucosal epithelial cells, which do not normally express these molecules at baseline, regulates TMPRSS2 and ACE2 transcript levels. We found that overexpression of PAX6-b resulted in a significant reduction of TMPRSS2 levels (p < 0.0001), while no significant effect was exerted by overexpression of the PAX6-a isoform (p = 0.5993) (Supplementary Fig. 1C). In contrast, ACE2 mRNA levels were not affected by forced overexpression of either PAX6-a or PAX6-b (p = 0.3102 and p = 0.4804, respectively) (Supplementary Fig. 1C).
To further interrogate a potential role of PAX6 in regulating TMPRSS2 and ACE2 expression in ocular surface epithelial cells, we examined the effects of PAX6 knockdown in cultured primary human limbal epithelial cells and in the immortalized conjunctival epithelial cell line ConjEp-1/p53DD/cdk4R/TERT [28]. Significant knockdown of PAX6 expression was demonstrated in both cell types by five employed distinct siRNAs (Fig. 1F and Supplementary Fig. 1D). Inhibition of PAX6 hereby resulted in the induction of TMPRSS2, indicating that PAX6 serves as a negative regulator of TMPRSS2 expression in ocular surface epithelial cells (Fig. 1F and Supplementary Fig. 1D). In contrast, ACE2 expression was not consistently altered by PAX6 knockdown. Intriguingly, we also found that ABCB5-positive LSCs expressed reduced PAX6-a (ENST00000241001) and PAX6-b (ENST00000419022) mRNA transcripts compared to ABCB5-negative limbal epithelial cells (Fig. 1G). Similarly, lower levels of PAX6 were detected in p63-positive LSCs by immunostaining (Fig. 1H). Using the ChIP sequencing data obtained from limbal epithelial cell cultures (GSE156273) [29], we identified several PAX6 binding sites upstream of TMPRSS2 (Supplementary Fig. 1E). These findings suggest that PAX6 directly suppresses TMPRSS2 expression in more differentiated limbal epithelial cells, and that lower levels of TMPRSS2-inhibitory PAX6 expression by more primitive ABCB5-positive cells or p63-positive cells might be responsible for higher levels of TMPRSS2 expression by these LSCs.
Thus, our study identifies high expression levels of the host SARS-CoV2 viral entry proteins TMPRSS2 and ACE2 by human LSCs, demonstrating a specific molecular phenotype of this cell population suggestive of enhanced susceptibility to SARS-CoV2 cellular tropism. Moreover, we show that PAX6 negatively regulates TMPRSS2 expression in more differentiated ocular epithelial tissues, providing one potential explanation for enhanced TMPRSS2 expression by the PAX6 low-expressing LSC compartment.
While in previous study of oral mucosa [20], both PAX6-a and PAX6-b could negatively regulate TMPRSS2 expression, PAX6-b exerted more pronounced effects. Since we found PAX6-b levels to be significantly reduced in ABCB5-positive LSCs, the relative scarcity of this isoform in this cell population provides a possible explanation for high TMPRSS2 expression by LSCs. While ACE2 was also enriched in LSCs, we did not find any evidence for its direct transcriptional regulation by PAX6, suggesting involvement of additional molecular mechanisms that require further investigation.
The presence of several PAX6 binding sites upstream of the TMPRSS2 gene suggests the possibility of inhibitory PAX6/TMPRSS2 gene interactions. While PAX6 is primarily known as a transcriptional activator of expression of several ocular surface proteins [[18], [19], [20], [21]], the role of PAX6 as a direct transcriptional repressor is less well understood [30]. However, a recent study of murine neural development revealed that Pax6 is capable of functioning as a direct repressor of transcription through co-occupancy with the H3K4 demethylase KDM5C, resulting in decreased H3K4me3 levels [30]. Consistent with this study, we also observed reduced H3K4me3 levels in the majority of PAX6 binding sites upstream of TMPRSS2 (Supplementary Fig. 1E).
Our findings of negative regulation of the SARS-CoV2 viral entry-related protein TMPRSS2 by PAX6, and of preferential expression of viral entry-related proteins by human LSCs, have several important implications: First, to date only androgen receptor (AR)-mediated transcriptional activation had been identified as a molecular mechanism of TMPRSS2 induction, in the context of prostate cancer [31,32]. Therefore, our finding of negative regulation of TMPRSS2 expression by PAX6 identifies a novel mechanism of viral entry protein regulation that now warrants further investigation for potential therapeutic targetability in the development of novel COVID-19 preventive or therapeutic treatment strategies, through drug-activated, PAX6-mediated TMPRSS2 inhibition [33]. For example, the FGF8/FGFR signaling axis represents a key inhibitor of PAX6 activation, and conversely, FGF8/FGFR signaling blockade activates PAX6 expression [34]. Of note, specific FGFR inhibitors with predicted capacity to activate PAX6 and hence, based on the herein presented results, to inhibit TMPRSS2 expression, are already available and clinically approved for the therapy of other disease indications [35].
Second, the presence of several PAX6 binding sites upstream of the TMPRSS2 gene suggests direct inhibitory PAX6/TMPRSS2 gene interactions, a possibility with relevance for potentially cell-protective mechanisms of resistance to SARS-CoV2 tropism of additional developmental and adult PAX6-expressing tissues (such as, for example, the brain [36]), and for potentially enhanced susceptibility to SARS-CoV2 tropism of individuals suffering from genetic syndromes associated with impaired PAX6 function (such as, for example, Aniridia and Gillespie syndrome [37]). Our findings warrant further investigation of such potential vulnerabilities.
Third, because quiescent ABCB5-positive LSCs are long-lived in healthy humans and undergo lower levels of cell turnover than more differentiated corneal cells, their potentially increased susceptibility to SARS-CoV2 infection implies that they might also represent a reservoir for prolonged SARS-CoV2 persistence. This possibility warrants specific attention in the care of patients afflicted by COVID-19-associated eye disease as a possible cause of disease persistence or recurrence. Moreover, because autologous p63-positive LSC-containing donor grafts are in clinical use and because allogeneic ABCB5-positive LSCs are currently also undergoing clinical evaluation in an interventional, open-label, phase I/IIa human clinical trial for safety and efficacy in LSCD therapy [17], our findings highlight a need for specific screening of living limbal graft donors, cadaveric donors, and banked donor tissue for known or occult SARS-CoV2 infection, in order to ensure that only SARS-CoV2 free cells are utilized in the clinical setting to prevent virus spread to graft recipients.
Finally, our finding that human ABCB5-positive LSCs express high levels of SARS-CoV2 viral entry-related proteins suggests that in vitro-expandable human ABCB5-positive LSCs cultures represent an established [17] and relevant novel model system for the study of cellular SARS-CoV2 viral entry mechanisms and the pharmaceutical evaluation of related developmental targeting strategies.
Disclosure/conflict of interest statement
M.H·F., B.R.K. and N·Y.F. are inventors or co-inventors of US and international patents assigned to Brigham and Women's Hospital, Boston Children's Hospital, the Massachusetts Eye and Ear Infirmary and/or the VA Boston Healthcare System, Boston, MA, and licensed to Ticeba GmbH (Heidelberg, Germany) and Rheacell GmbH & Co. KG (Heidelberg, Germany). M.H.F. serves as a scientific advisor to Ticeba GmbH and Rheacell GmbH & Co. KG.
Acknowledgements
We would like to thank the patients for their generous tissue donations that enabled this research. This work was supported by NIH/NEI grants 1K99EY031741 to Y.S., 1R01EY025794 and R24EY028767 to N.Y.F., B.R.K and M.H.F, NIH/NEI Schepens Core grant P30EY003790 to B.R.K., NIH/NIBIB grant 2T32EB016652-06 to C.A.A.L, Alcon Young Investigator Grant to Y.S., Japan Society for the Promotion of Science (JSPS) Overseas Research Fellowships to M.Y., Japan Eye Bank Association Overseas Award to M.Y., and VA R&D Merit Review Award 1I01RX000989 and a Harvard Stem Cell Institute seed grant award to N.Y.F. We thank the Molecular Biology Core Facilities (MBCF) and the Flow Cytometry Core at Dana-Farber Cancer Institute and the Harvard School of Public Health Bioinformatics Core.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jtos.2021.10.002.
Appendix A. Supplementary data
The following is the supplementary data to this article:
References
- 1.Inomata T., Kitazawa K., Kuno T., Sung J., Nakamura M., Iwagami M., et al. Clinical and prodromal ocular symptoms in coronavirus disease: a systematic review and meta-analysis. Invest Ophthalmol Vis Sci. 2020;61:29. doi: 10.1167/iovs.61.10.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chu D.K., Akl E.A., Duda S., Solo K., Yaacoub S., Schunemann H.J., et al. Physical distancing, face masks, and eye protection to prevent person-to-person transmission of SARS-CoV-2 and COVID-19: a systematic review and meta-analysis. Lancet. 2020;395:1973–1987. doi: 10.1016/S0140-6736(20)31142-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Seah I., Agrawal R. Can the coronavirus disease 2019 (COVID-19) affect the eyes? A review of coronaviruses and ocular implications in humans and animals. Ocul Immunol Inflamm. 2020;28:391–395. doi: 10.1080/09273948.2020.1738501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hoffmann M., Kleine-Weber H., Schroeder S., Kruger N., Herrler T., Erichsen S., et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280 e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sungnak W., Huang N., Becavin C., Berg M., Queen R., Litvinukova M., et al. SARS-CoV-2 entry factors are highly expressed in nasal epithelial cells together with innate immune genes. Nat Med. 2020;26:681–687. doi: 10.1038/s41591-020-0868-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Collin J., Queen R., Zerti D., Dorgau B., Georgiou M., Djidrovski I., et al. Co-expression of SARS-CoV-2 entry genes in the superficial adult human conjunctival, limbal and corneal epithelium suggests an additional route of entry via the ocular surface. Ocul Surf. 2021;19:190–200. doi: 10.1016/j.jtos.2020.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou L., Xu Z., Castiglione G.M., Soiberman U.S., Eberhart C.G., Duh E.J. ACE2 and TMPRSS2 are expressed on the human ocular surface, suggesting susceptibility to SARS-CoV-2 infection. Ocul Surf. 2020;18:537–544. doi: 10.1016/j.jtos.2020.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eriksen A.Z., Møller R., Makovoz B., Uhl S.A., Blenkinsop T.A. SARS-CoV-2 infects human adult donor eyes and hESC-derived ocular epithelium. Cell Stem Cell. 2021 doi: 10.1016/j.stem.2021.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davanger M., Evensen A. Role of the pericorneal papillary structure in renewal of corneal epithelium. Nature. 1971;229:560–561. doi: 10.1038/229560a0. [DOI] [PubMed] [Google Scholar]
- 10.Ksander B.R., Kolovou P.E., Wilson B.J., Saab K.R., Guo Q., Ma J., et al. ABCB5 is a limbal stem cell gene required for corneal development and repair. Nature. 2014;511:353–357. doi: 10.1038/nature13426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jongkhajornpong P., Nakamura T., Sotozono C., Nagata M., Inatomi T., Kinoshita S. Elevated expression of ABCB5 in ocular surface squamous neoplasia. Sci Rep. 2016;6:20541. doi: 10.1038/srep20541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kureshi A.K., Dziasko M., Funderburgh J.L., Daniels J.T. Human corneal stromal stem cells support limbal epithelial cells cultured on RAFT tissue equivalents. Sci Rep. 2015;5:16186. doi: 10.1038/srep16186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mathan J.J., Ismail S., McGhee J.J., McGhee C.N.J., Sherwin T. Sphere-forming cells from peripheral cornea demonstrate the ability to repopulate the ocular surface. Stem Cell Res Ther. 2016;7:1–15. doi: 10.1186/s13287-016-0339-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Parfitt G.J., Kavianpour B., Wu K.L., Xie Y., Brown D.J., Jester J.V. Immunofluorescence tomography of mouse ocular surface epithelial stem cells and their niche microenvironment. Investig Ophthalmol Vis Sci. 2015;56:7338–7344. doi: 10.1167/iovs.15-18038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shaharuddin B., Ahmad S., Md Latar N., Ali S., Meeson A. A human corneal epithelial cell line model for limbal stem cell biology and limbal immunobiology. Stem cells translational medicine. 2017;6:761–766. doi: 10.5966/sctm.2016-0175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shaharuddin B., Osei-Bempong C., Ahmad S., Rooney P., Ali S., Oldershaw R., et al. Human limbal mesenchymal stem cells express ABCB5 and can grow on amniotic membrane. Regen Med. 2016;11:273–286. doi: 10.2217/rme-2016-0009. [DOI] [PubMed] [Google Scholar]
- 17.Norrick A., Esterlechner J., Niebergall-Roth E., Dehio U., Sadeghi S., Schroder H.M., et al. Process development and safety evaluation of ABCB5(+) limbal stem cells as advanced-therapy medicinal product to treat limbal stem cell deficiency. Stem Cell Res Ther. 2021;12:194. doi: 10.1186/s13287-021-02272-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Davis J., Duncan M.K., Robison W.G., Jr., Piatigorsky J. Requirement for Pax6 in corneal morphogenesis: a role in adhesion. J Cell Sci. 2003;116:2157–2167. doi: 10.1242/jcs.00441. [DOI] [PubMed] [Google Scholar]
- 19.Kitazawa K., Hikichi T., Nakamura T., Sotozono C., Kinoshita S., Masui S. PAX6 regulates human corneal epithelium cell identity. Exp Eye Res. 2017;154:30–38. doi: 10.1016/j.exer.2016.11.005. [DOI] [PubMed] [Google Scholar]
- 20.Sasamoto Y., Hayashi R., Park S.J., Saito-Adachi M., Suzuki Y., Kawasaki S., et al. PAX6 isoforms, along with reprogramming factors, differentially regulate the induction of cornea-specific genes. Sci Rep. 2016;6:20807. doi: 10.1038/srep20807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shiraishi A., Converse R.L., Liu C.Y., Zhou F., Kao C.W., Kao W.W. Identification of the cornea-specific keratin 12 promoter by in vivo particle-mediated gene transfer. Invest Ophthalmol Vis Sci. 1998;39:2554–2561. [PubMed] [Google Scholar]
- 22.Epstein J.A., Glaser T., Cai J., Jepeal L., Walton D.S., Maas R.L. Two independent and interactive DNA-binding subdomains of the Pax6 paired domain are regulated by alternative splicing. Genes Dev. 1994;8:2022–2034. doi: 10.1101/gad.8.17.2022. [DOI] [PubMed] [Google Scholar]
- 23.Glaser T., Walton D.S., Maas R.L. Genomic structure, evolutionary conservation and aniridia mutations in the human PAX6 gene. Nat Genet. 1992;2:232–239. doi: 10.1038/ng1192-232. [DOI] [PubMed] [Google Scholar]
- 24.Xu H.E., Rould M.A., Xu W., Epstein J.A., Maas R.L., Pabo C.O. Crystal structure of the human Pax6 paired domain-DNA complex reveals specific roles for the linker region and carboxy-terminal subdomain in DNA binding. Genes Dev. 1999;13:1263–1275. doi: 10.1101/gad.13.10.1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sasamoto Y., Sasamoto N., Tran J., Mishra A., Ksander B.R., Frank M.H., et al. Investigation of factors associated with ABCB5-positive limbal stem cell isolation yields from human donors. Ocul Surf. 2020;18:114–120. doi: 10.1016/j.jtos.2019.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pellegrini G., Dellambra E., Golisano O., Martinelli E., Fantozzi I., Bondanza S., et al. p63 identifies keratinocyte stem cells. Proc Natl Acad Sci Unit States Am. 2001;98:3156–3161. doi: 10.1073/pnas.061032098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Derrick T., Habtamu E., Tadesse Z., Callahan E.K., Worku A., Gashaw B., et al. The conjunctival transcriptome in Ethiopians after trichiasis surgery: associations with the development of eyelid contour abnormalities and the effect of oral doxycycline treatment. Wellcome Open Research. 2019;4:130. doi: 10.12688/wellcomeopenres.15419.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rheinwald J.G., Hahn W.C., Ramsey M.R., Wu J.Y., Guo Z., Tsao H., et al. A two-stage, p16(INK4A)- and p53-dependent keratinocyte senescence mechanism that limits replicative potential independent of telomere status. Mol Cell Biol. 2002;22:5157–5172. doi: 10.1128/MCB.22.14.5157-5172.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li M., Huang H., Li L., He C., Zhu L., Guo H., et al. Core transcription regulatory circuitry orchestrates corneal epithelial homeostasis. Nat Commun. 2021;12:420. doi: 10.1038/s41467-020-20713-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gaudenzi G., Dethlefsen O., Walfridsson J., Hermanson O. Pax6 and KDM5C co-occupy a subset of developmentally critical genes including Notch signaling regulators in neural progenitors. bioRxiv. 2019:579219. [Google Scholar]
- 31.Lucas J.M., Heinlein C., Kim T., Hernandez S.A., Malik M.S., True L.D., et al. The androgen-regulated protease TMPRSS2 activates a proteolytic cascade involving components of the tumor microenvironment and promotes prostate cancer metastasis. Cancer Discov. 2014;4:1310–1325. doi: 10.1158/2159-8290.CD-13-1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lin B., Ferguson C., White J.T., Wang S., Vessella R., True L.D., et al. Prostate-localized and androgen-regulated expression of the membrane-bound serine protease TMPRSS2. Cancer Res. 1999;59:4180–4184. [PubMed] [Google Scholar]
- 33.Rabiee B., Anwar K.N., Shen X., Putra I., Liu M., Jung R., et al. Gene dosage manipulation alleviates manifestations of hereditary PAX6 haploinsufficiency in mice. Sci Transl Med. 2020;12 doi: 10.1126/scitranslmed.aaz4894. [DOI] [PubMed] [Google Scholar]
- 34.Bertrand N., Médevielle F., Pituello F. FGF signalling controls the timing of Pax6 activation in the neural tube. Development. 2000;127:4837–4843. doi: 10.1242/dev.127.22.4837. [DOI] [PubMed] [Google Scholar]
- 35.Weaver A., Bossaer J.B. Fibroblast growth factor receptor (FGFR) inhibitors: a review of a novel therapeutic class. J Oncol Pharm Pract. 2020 doi: 10.1177/1078155220983425. [DOI] [PubMed] [Google Scholar]
- 36.Freund C., Horsford D.J., McInnes R.R. Transcription factor genes and the developing eye: a genetic perspective. Hum Mol Genet. 1996;5:1471–1488. doi: 10.1093/hmg/5.supplement_1.1471. [DOI] [PubMed] [Google Scholar]
- 37.Ton C.C., Hirvonen H., Miwa H., Weil M.M., Monaghan P., Jordan T., et al. Positional cloning and characterization of a paired box-and homeobox-containing gene from the aniridia region. Cell. 1991;67:1059–1074. doi: 10.1016/0092-8674(91)90284-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.