Abstract
DNA methylation shows complex correlations with gene expression, and the role of promoter hypermethylation in repressing gene transcription has been well addressed. Emerging evidence indicates that gene body methylation promotes transcription; however, the underlying mechanisms remain to be further investigated. Here, using methylated DNA immunoprecipitation sequencing (MeDIP-seq), bisulfite genomic sequencing, and immunofluorescent labeling, we show that gene body methylation is indeed positively correlated with rRNA gene (rDNA) transcription. Mechanistically, gene body methylation is largely maintained by DNA methyltransferase 1 (DNMT1), deficiency or downregulation of which during myoblast differentiation or nutrient deprivation results in decreased gene body methylation levels, leading to increased gene body occupancy of plant homeodomain (PHD) finger protein 6 (PHF6). PHF6 binds to hypomethylated rDNA gene bodies where it recruits histone methyltransferase SUV4-20H2 to establish the repressive histone modification, H4K20me3, ultimately inhibiting rDNA transcription. These findings demonstrate that DNMT1-mediated gene body methylation safeguards rDNA transcription by preventing enrichment of repressive histone modifications, suggesting that gene body methylation serves to maintain gene expression in response to developmental and/or environmental stresses.
Keywords: gene body methylation, DNMT1, PHF6, H4K20me3, rDNA transcription
Abbreviations: ChIP-qPCR, chromatin immunoprecipitation assays followed by real-time quantitative PCR; DMEM, Dulbecco's modified Eagle medium; DNMT1, DNA methyltransferase 1; FBS, fetal bovine serum; IGS, intergenic spacer; MeDIP-seq, methylated DNA immunoprecipitation sequencing; PHD, plant homeodomain; PHF6, PHD finger protein 6; rRNA, Ribosomal RNA; RT-qPCR, real-time quantitative reverse transcription PCR; UBF, upstream binding factor
DNA methylation exists extensively in eukaryotes and is involved in various physiological processes (1, 2, 3). In mammals, it mainly occurs on the cytosine of CpG dinucleotides and is catalyzed by three well-known DNA methyltransferases: DNMT1, DNMT3A, and DNMT3B (4). DNMT1 is responsible for maintaining existing CpG methylation patterns during DNA replication, while DNMT3A and DNMT3B function as de novo methyltransferases to establish new DNA methylation patterns (5, 6, 7). Extensive studies have demonstrated that hypermethylated promoters suppress gene expression, either by interfering with the binding of transcriptional activators or by recruiting methyl-CpG binding proteins that further interact with chromatin remodelers to establish a repressive chromatin structure (8, 9, 10). In recent years, accumulated evidence has demonstrated that DNA methylation also occurs in coding regions; thus, this type of methylation is called gene body (or intragenic) methylation (11, 12, 13, 14). Furthermore, gene body methylation is positively correlated with gene expression (9, 15, 16, 17). In postnatal neural stem cells, DNMT3A antagonizes the PRC2 complex at gene bodies to facilitate the expression of neurogenic genes (18). In the human colorectal carcinoma cell line, HCT116, many genes implicated in the metabolic activities that are modulated by c-Myc have been reported to be downregulated following treatment with the DNA methyltransferase inhibitor, 5-Aza-CdR (17). In hepatocellular carcinoma (HCC) patient samples, hypermethylated gene bodies are associated with increased expression levels of oncogenes (19). Moreover, locus-specific remethylation of hypomethylated gene body regions of homeobox oncogenes can directly increase their expression (20). Hence, gene body methylation may function as a positive regulator of transcription; nevertheless, the molecular mechanism underlying its regulation remains to be clarified.
Ribosomal RNA (rRNA) is a crucial component of ribosomes and is involved in the control of protein synthesis, of which transcriptional regulation is responsive to alterations in physiological and pathological activities (21, 22, 23). DNA methyltransferases and histone-modifying enzymes have been extensively studied with respect to their regulation of rDNA expression. In short, methylated promoters abolish rDNA transcription by inhibiting the assembly of the transcription initiation complex (24). Moreover, cryptic rDNA transcripts, including long noncoding RNAs and intergenic spacer (IGS) transcripts, promote the establishment of H4K20me3, thereby forming a heterochromatin structure and suppressing rDNA transcription (25, 26). Notably, it was hypothesized that depletion of DNMTs would enhance rDNA transcription; however, knockout of DNMTs actually leads to a reduction in the global methylation status across rDNA repeats, including promoters and gene bodies, and inconsistent changes in pre-rRNA synthesis measured by different methods, prompting us to investigate the relationship of hypomethylation and rDNA transcription and underlying mechanisms (27, 28).
Plant homeodomain (PHD) finger protein 6 (PHF6) is an X-linked gene encoding a highly conserved protein possessing two atypical PHD zinc finger domains, in which a variety of mutations have been identified and shown to be associated with X-linked mental retardation (XLMR), Börjeson–Forssman–Lehmann syndrome (BFLS; OMIM 301900) (29), T-cell acute lymphoblastic leukemia (T-ALL), and acute myeloid leukemia (AML) (30, 31). PHF6 bidirectionally modulates gene expression in leukemic cells, since it maintains chromatin accessibility at B cell-associated genes but forms a repressive conformation at T cell-related genes (32, 33). Additionally, PHF6 is involved in the control of neuronal migration by physically interacting with the PAF1 transcription elongation complex to regulate schizophrenia susceptibility genes (34). Moreover, PHF6 is highly enriched at rDNA gene bodies compared with promoters and inhibits rDNA transcription by reducing the levels of upstream binding factor (UBF) and directly interacting with UBF via its PHD1 domain to impede its release from the rDNA promoter (35, 36). However, as a potential chromatin reader protein, the question of whether PHF6 is able to regulate rDNA transcription by altering chromatin signatures at rDNA gene body regions remains unanswered.
As previously highlighted, gene body methylation appears to be correlated with active rDNA transcription, while H4K20me3 is a well-known histone modification that strongly inhibits rDNA transcription; therefore, it is of great interest to investigate the regulatory mechanism of gene body methylation and its opposite effect to H4K20me3 on rDNA transcription. In the present study, we show that gene body methylation secures rDNA transcription by preventing the recruitment of the epigenetic regulator, PHF6, which guides the histone methyltransferase, SUV4-20H2, to establish H4K20me3 in gene body regions. Loss of gene body DNA methylation induces increased occupancy of PHF6 at gene bodies due to its capability to bind to unmethylated CpG sites. Coimmunoprecipitation and gain-of-function assays reveal that PHF6 can interact with SUV4-20H2 and subsequently establish H4K20me3 marks, which indicate a more compact chromatin structure in gene body regions, thus suppressing rDNA transcription. In conclusion, these findings suggest a gene body methylation-based epigenetic cross talk among PHF6, SUV4-20H2, and the histone modification, H4K20me3, in the regulation of rDNA transcription.
Results
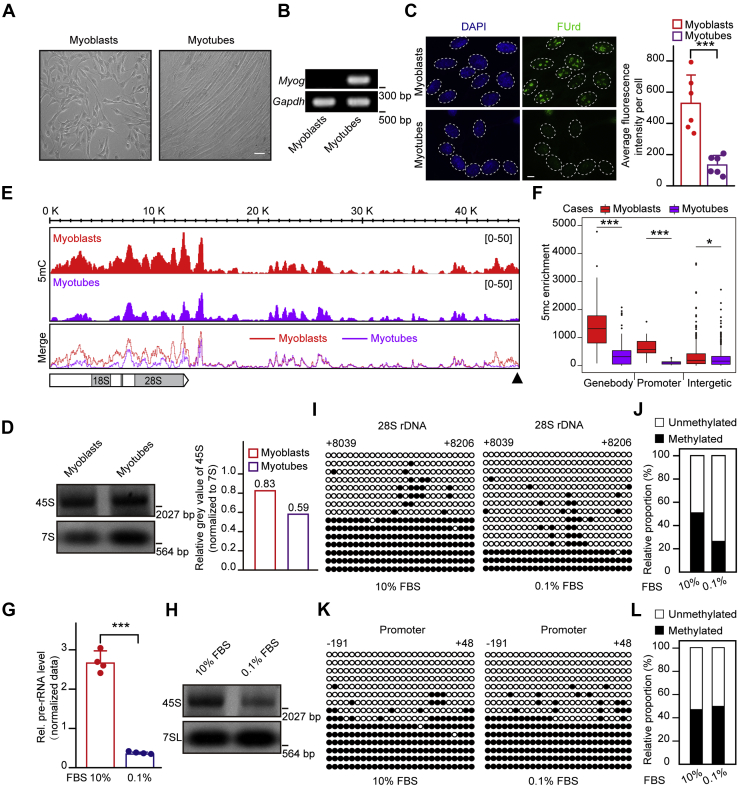
Gene body methylation is positively correlated with rDNA transcription
Previous studies have revealed different effects of hypomethylation caused by DNMTs deficiency on rDNA transcription (27, 28). In view of the fact that gene knockout may lead to side effects by artificial interference with gene expression and nontargets, the functional correlation between hypomethylation and rDNA transcription needs to be verified under physiological conditions. To this end, a C2C12 myoblast differentiation and serum deprivation system was employed, during which rDNA transcription was biologically downregulated (26). Myotubes were formed by incubation of myoblasts with 2% horse serum for 6 days (Fig. 1A), and a high level of Myogenin (Myog) expression was detected correspondingly (Fig. 1B). Fluorouridine (FUrd) was used to pulse-label the nascent rRNA for the detection of pre-rRNA synthesis. We found that pre-rRNA synthesis markedly decreased during the differentiation of C2C12 myoblasts into myotubes (Fig. 1, C and D). Further analysis of the rDNA methylation status by methylated DNA immunoprecipitation followed by sequencing (MeDIP-seq) revealed a reduction in the DNA methylation level in both the rDNA promoter and gene body regions (Fig. 1, E and F). Despite the decreased DNA methylation levels in promoter, the pre-rRNA synthesis was reduced, suggesting a positive correlation between the gene body methylation status and rDNA transcription. Next, we further monitored this correlation in cells with serum deprivation treatment. The pre-rRNA synthesis in serum-deprived HEK293T cells was measured by real-time quantitative reverse transcription PCR (RT-qPCR) and northern blotting, and we found that rDNA transcription was markedly reduced in HEK293T cells following serum deprivation compared with those cultured under normal conditions (Fig. 1, G and H). Intriguingly, a dramatic decrease in the level of gene body methylation was seen in serum-deprived HEK293T cells, comparing with those cultured under normal conditions (Fig. 1, I and J). Notably, the promoter methylation level remained unchanged regardless of the altered nutrient state (Fig. 1, K and L). These findings indicate that maintenance of gene body methylation is positively correlated with rDNA transcription.
Figure 1.
Gene body methylation is positively associated with rDNA transcription.A, morphology of C2C12 myoblasts and myotubes. Scale bar, 50 μm. B, the Myog gene is expressed in myotubes but not in myoblasts. Reverse transcription PCR assays followed by DNA gel electrophoresis showing Myog mRNA expression levels. C, Pre-rRNA synthesis is markedly reduced during differentiation of C2C12 myoblasts to myotubes. Immunostaining showing cell nuclei (blue) and FUrd (green). The pre-rRNA synthesis was measured by FUrd incorporation. Bar graph shows the average FUrd fluorescence intensity per cell of myoblasts or myotubes (n = 6). A total of 82 cells were subjected to statistics. Data are presented as the mean ± SD. Scale bar, 5 μm. D, northern blotting shows the level of 45S pre-rRNA during C2C12 myoblasts differentiation. 7S RNA served as a loading control. Relative gray value of 45S band was measured by the Image J software normalizing to 7S RNA band. E, gene body and promoter methylation levels are markedly decreased upon the differentiation of C2C12 myoblasts. MeDIP-Seq profiles in C2C12 myoblasts and differentiated myotubes in rDNA repeats are displayed. The horizontal bar graph on the bottom left indicates gene body regions for pre-rRNA transcripts, and the black triangle on the bottom right represents the rDNA promoter region. F, statistical analysis for MeDIP-seq results in (E). ∗p < 0.1, ∗∗p < 0.01, ∗∗∗p < 0.001, Wilcoxon test. G, RT-qPCR assays showing levels of pre-rRNA in HEK293T cells cultured in medium containing 10% FBS (normal) and 0.1% FBS (serum-starved). The cycle threshold (Ct) of 45S pre-rRNA was normalized with GAPDH. Data are presented as the mean ± SD. H, northern blotting shows the level of 45S pre-rRNA upon serum starvation. 7SL RNA served as a loading control. HEK293T cells cultured in medium containing 10% FBS (normal) and 0.1% FBS (serum-starved). I, rDNA gene body methylation levels decrease during serum starvation. Bisulfite sequencing showing the rDNA gene body methylation status in HEK293T cells cultured in medium containing 10% FBS (normal) and 0.1% FBS (serum-starved). The black and white circles represent corresponding CpG sites in rDNA gene bodies, denoting methylated and unmethylated CpG sites, respectively. The average ratio of black sites for all colonies represents the relative DNA methylation level. Corresponding sequence is shown in Figure S1G. J, analysis for the results in (I) showing the percentage of methylated and unmethylated gene body CpG sites in HEK293T cells cultured in medium containing 10% FBS (normal) and 0.1% FBS (serum-starved). The relative proportion of unmethylation and methylation was calculated by counting the numbers of black circles (methylated CpG sites) and white circles (unmethylated CpG sites). K, genomic DNA coupled with bisulfite sequencing assays were performed to show promoter methylation levels in control (10% FBS) and serum-starved (0.1% FBS) HEK293T cells. The black and white circles represent corresponding CpG sites in rDNA gene bodies, denoting methylated and unmethylated CpG sites, respectively. The average ratio of black sites for all colonies represents the relative DNA methylation level. Corresponding sequence is shown in Figure S1H. L, analysis for the results in (K) showing the percentage of methylated and unmethylated gene body CpG sites in HEK293T cells cultured in medium containing 10% FBS (normal) and 0.1% FBS (serum-starved). The relative proportion of unmethylation and methylation was calculated by counting the numbers of black circles (methylated CpG sites) and white circles (unmethylated CpG sites). ∗p < 0.1, ∗∗p < 0.01, ∗∗∗p < 0.001, two-tailed unpaired Student's t test.
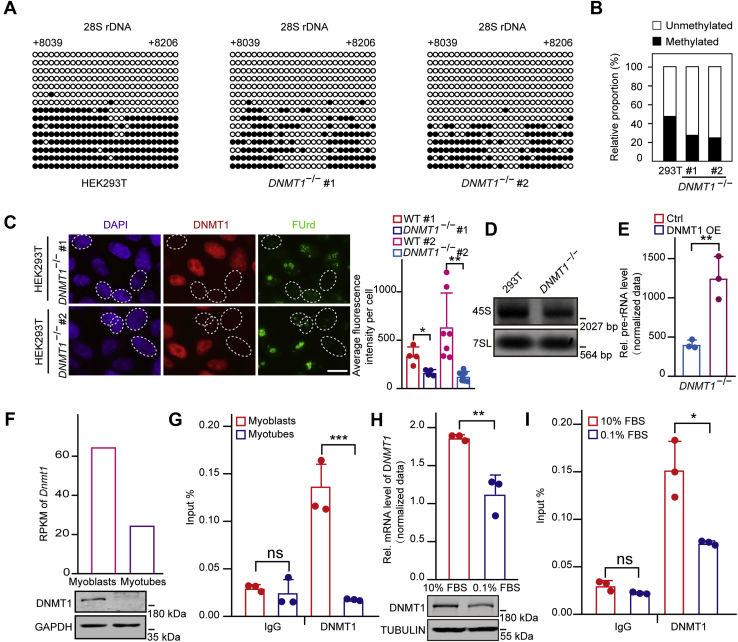
DNMT1 mediates rDNA gene body methylation
To dissect the mechanism underlying the positive correlation between gene body methylation and rDNA transcription, we therefore depleted DNA methyltransferases in HEK293T cells using the CRISPR/Cas9 genome editing technique to determine the methyltransferase responsible for rDNA gene body regions. Previously, the catalytically inactive isoform DNMT3B3 has been shown to be the dominant isoform, with little to no expression of catalytically active DNMT3B1 in somatic cells (37). Loss of DNMT3B does not alter the methylation status in colorectal cancer cells (27). Indeed, we found that DNMT3B was undetectable by western blotting in HEK293T cells (Fig. S1A), and knockout of DNMT3B did not affect rDNA gene body methylations status (Fig. S1, B and C). Furthermore, our previous work has proved that there was almost no change in methylation levels at gene bodies in DNMT3A KO cells compared with wild-type cells (38). Considering that DNMT1 deficiency in colorectal cancer cells causes hypomethylation in rDNA gene bodies, whereas loss of DNMT3B does not alter the methylation status (27), we speculated that DNMT1 may also be responsible for rDNA gene body methylation. Subsequent genome bisulfite sequencing assays showed that the gene body methylation level was reduced in DNMT1 KO cells compared with that in wild-type cells (Figs. S1D and 2, A and B). These results suggest that DNMT1 is the primary methyltransferase for the maintenance of rDNA gene body methylation.
Figure 2.
DNMT1 mediates rDNA gene body methylation.A, knockout of DNMT1 significantly reduces gene body methylation. Genomic DNA coupled with bisulfite sequencing assays were performed to show the gene body methylation levels in control and DNMT1 KO cells. The black and white circles represent corresponding CpG sites in rDNA gene bodies, denoting methylated and unmethylated CpG sites, respectively. The average ratio of black sites for all colonies represents the relative DNA methylation level. Corresponding sequence is shown in Figure S1G. B, analysis for the results in (A) showing the percentage of methylated and unmethylated gene body CpG sites in control and DNMT1 KO cells. The relative proportion of unmethylation and methylation was calculated by counting the numbers of black circles (methylated CpG sites) and white circles (unmethylated CpG sites). C, loss of DNMT1 markedly reduces pre-rRNA synthesis. Immunostaining showing cell nuclei (blue), DNMT1 (red), and FUrd (green). HEK293T and DNMT1 KO cells were cocultured and the pre-rRNA synthesis was measured by FUrd incorporation. Bar graph shows the average FUrd fluorescence intensity per cell of DNMT1 KO or HEK293T cells (n = 4 or n = 7). A total of 155 cells were subjected to statistics. Scale bar, 5 μm. D, northern blotting shows the level of 45S pre-rRNA in DNMT1 KO or HEK293T cells. 7SL RNA served as a loading control. E, RT-qPCR results show the pre-rRNA level in DNMT1 KO and DNMT1-overexpressed DNMT1 KO cells. The cycle threshold (Ct) of 45S pre-rRNA was normalized with GAPDH. Data are presented as the mean ± SD. F, the mRNA and protein levels of Dnmt1 were decreased upon differentiation of C2C12 myoblasts as revealed by RNA-seq and western blotting, respectively. The cycle threshold (Ct) of Dnmt1 was normalized with Gapdh in RT-qPCR. GAPDH served as a loading control in western blotting. G, the occupancy of DNMT1 markedly declines upon differentiation of C2C12 myoblasts. ChIP experiments were performed using primers coated on the 28S rRNA coding region in C2C12 myoblasts and myotubes. Data are presented as the mean ± SD. H, the mRNA and protein levels of DNMT1 were reduced upon serum starvation of HEK293T cells as detected by RT-qPCR and western blotting, respectively. The cycle threshold (Ct) of DNMT1 was normalized with B2M in RT-qPCR. TUBULIN served as a loading control in western blotting. Data are presented as the mean ± SD. I, the occupancy of DNMT1 reduces in serum-deprived HEK293T cells. ChIP experiments were performed using primers coated on the 28S rRNA coding region in serum-deprived and control cells. Data are presented as the mean ± SD. ∗p < 0.1, ∗∗p < 0.01, ∗∗∗p < 0.001, two-tailed unpaired Student's t test.
To elucidate whether DNMT1 is involved in the regulation of rDNA transcription, we pulse-labeled cocultured DNMT1 KO and wild-type HEK293T cells with FUrd. As expected, despite decreases in DNA methylation levels in promoters (Fig. S1, E and F), DNMT1 deletion led to a marked reduction in pre-rRNA synthesis (Fig. 2, C and D). Moreover, the decreased pre-rRNA level in DNMT1 KO cells was rescued through DNMT1 overexpression (Fig. 2E). These results suggest that DNMT1 is required for rDNA transcription. In accordance with this result, both the mRNA and protein levels of DNMT1 were decreased in differentiated myotubes, comparing with those in myoblasts (Fig. 2F). Subsequently, chromatin immunoprecipitation assays followed by real-time quantitative PCR (ChIP-qPCR) showed a decline in the binding of DNMT1 to gene body regions (Fig. 2G). Similarly, we found that serum deprivation led to a decrease in both the mRNA and protein levels of DNMT1 along with its reduced occupancy at gene bodies (Fig. 2, H and I). And ectopic expression of DNMT1 in serum-starved HEK293T cells rescued the previous loss of methylation in rDNA gene body (Fig. S2). Overall, these results indicate that DNMT1-mediated rDNA gene body methylation is positively associated with the regulation of rDNA transcription.
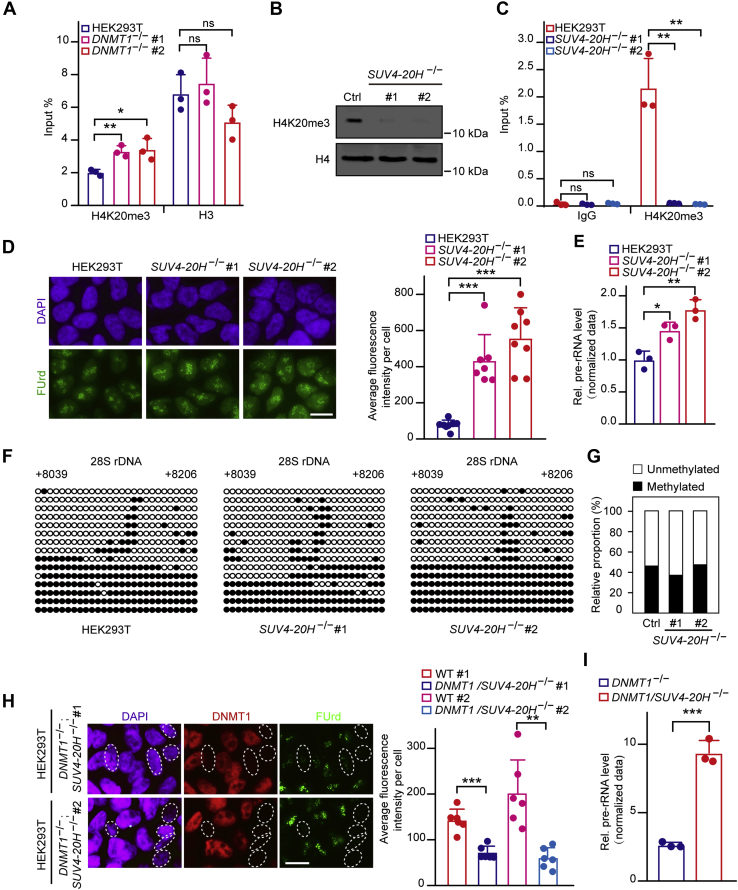
DNMT1 antagonizes H4K20me3 to regulate rDNA transcription
Next, we explored the mechanism of DNMT1 in rDNA transcription regulation. In view of the cross talk between DNA methylation and histone modifications, we assessed the enrichment of several histone modifications (i.e., H3K9me3, H3K27me3, H3K36me3, and H4K20me3) at rDNA gene bodies in DNMT1-depleted cells. The results showed that DNMT1 deficiency results in significant increase in H4K20me3 enrichment in rDNA gene body regions while the occupancy of other histone modifications remains unchanged (Figs. 3A and S3A), which is consistent with the findings that H4K20me3 represses rDNA transcription (26, 39, 40). To further confirm that DNMT1-mediated gene body methylation opposed the enrichment of H4K20me3 to regulate rDNA transcription, we then depleted the major methyltransferases involved with H4K20me3, SUV4-20H1/H2 (designated as SUV4-20H), in HEK293T cells. Since the efficient antibodies for human SUV4-20H1/H2 were not commercially available, H4K20me3 protein levels were used as an indicator for the knockout of SUV4-20H1/H2. Indeed, knockout of SUV4-20H led to a marked decrease in both the abundance of the H4K20me3 protein and its enrichment in rDNA gene body regions (Fig. 3, B and C). FUrd incorporation followed by immunofluorescence microscopy showed that loss of SUV4-20H increased the fluorescence signal, which is indicative of upregulated pre-rRNA synthesis (Fig. 3D). Accordingly, reverse transcription PCR experiments also showed augmented pre-rRNA synthesis in SUV4-20H knockout cells (Fig. 3E).
Figure 3.
DNMT1 antagonizes H4K20me3 to regulate rDNA transcription.A, the enrichment of H4K20me3 is elevated in DNMT1 KO cells. ChIP experiments were performed using primers coated on the 28S rRNA coding region in DNMT1 KO and control cells. Data are presented as the mean ± SD. B, knockout of SUV4-20H1/H2 using the CRISPR/Cas9 system as shown by western blotting. C, loss of SUV4-20H1/H2 leads to a marked reduction in H4K20me3 enrichment at gene bodies as detected by ChIP-qPCR. Data are presented as the mean ± SD. D, depletion of SUV4-20H1/H2 leads to a marked increase in pre-rRNA synthesis. Immunostaining showing cell nuclei (blue) and FUrd (green). The pre-rRNA synthesis was measured by FUrd incorporation. Bar graph shows the average FUrd fluorescence intensity per cell of SUV4-20H KO or HEK293T cells (n = 7 or n = 8). A total of 245 cells were subjected to statistics. Scale bar, 5 μm. E, pre-rRNA synthesis increases significantly upon SUV4-20H depletion compared with that in HEK293T cells. RT-qPCR assays showing levels of pre-rRNA in SUV4-20H DKO and wild-type HEK293T cells. The cycle threshold (Ct) of 45S pre-rRNA was normalized with GAPDH. Data are presented as the mean ± SD. F, knockout (KO) of SUV4-20H does not affect the rDNA gene body methylation level. Genomic DNA coupled with bisulfite sequencing assays were performed to show the gene body methylation level in SUV4-20H DKO and control cells. The black and white circles represent corresponding CpG sites in rDNA gene bodies, denoting methylated and unmethylated CpG sites, respectively. The average ratio of black sites for all colonies represents the relative DNA methylation level. Corresponding sequence is shown in Figure S1G. G, analysis for the results in (F) showing the percentage of methylated and unmethylated gene body CpG sites in SUV4-20H DKO and control cells. The relative proportion of unmethylation and methylation were calculated by counting the numbers of black circles (methylated CpG sites) and white circles (unmethylated CpG sites). H, pre-rRNA synthesis is markedly reduced in DNMT1/SUV4-20H TKO cells. Immunostaining showing cell nuclei (blue), DNMT1 (red), and FUrd (green). The pre-rRNA synthesis was measured by FUrd incorporation. Bar graph shows the average FUrd fluorescence intensity per cell of DNMT1/SUV4-20H TKO or HEK293T cells (n = 6). A total of 227 cells were subjected to statistics. Scale bar, 5 μm. I, pre-rRNA synthesis increases significantly upon triple knockout of DNMT1 and SUV4-20H1/H2 (designated as DNMT1/SUV4-20H−/−) compared with that in DNMT1 KO cells. RT-qPCR assays showing levels of pre-rRNA in DNMT1−/− and DNMT1/SUV4-20H−/− cells. The cycle threshold (Ct) of 45S pre-rRNA was normalized with GAPDH. Data are presented as the mean ± SD. ∗p < 0.1, ∗∗p < 0.01, ∗∗∗p < 0.001, two-tailed unpaired Student's t test.
Moreover, we found that depletion of SUV4-20H neither affected the DNA methylation level (Fig. 3, F and G) nor changed the occupancy of DNMT1 (Fig. S3B) in gene body regions. These results demonstrate that DNMT1 opposes H4K20me3 enrichment in rDNA gene body regions. In addition, triple knockout of DNMT1, SUV4-20H1, and SUV4-20H2 led to a reduction of pre-rRNA synthesis compared with that in HEK293T cells (Figs. S3C and 3H), but a significant increase in rDNA transcription compared with that in DNMT1 KO cells (Fig. 3I), suggesting that hypomethylation of rDNA gene bodies leads to rDNA transcriptional silencing, in part, due to an increase in H4K20me3 enrichment at gene bodies. Taken together, these results suggest that DNMT1-mediated gene body methylation is positively associated with rDNA transcription by antagonizing H4K20me3.
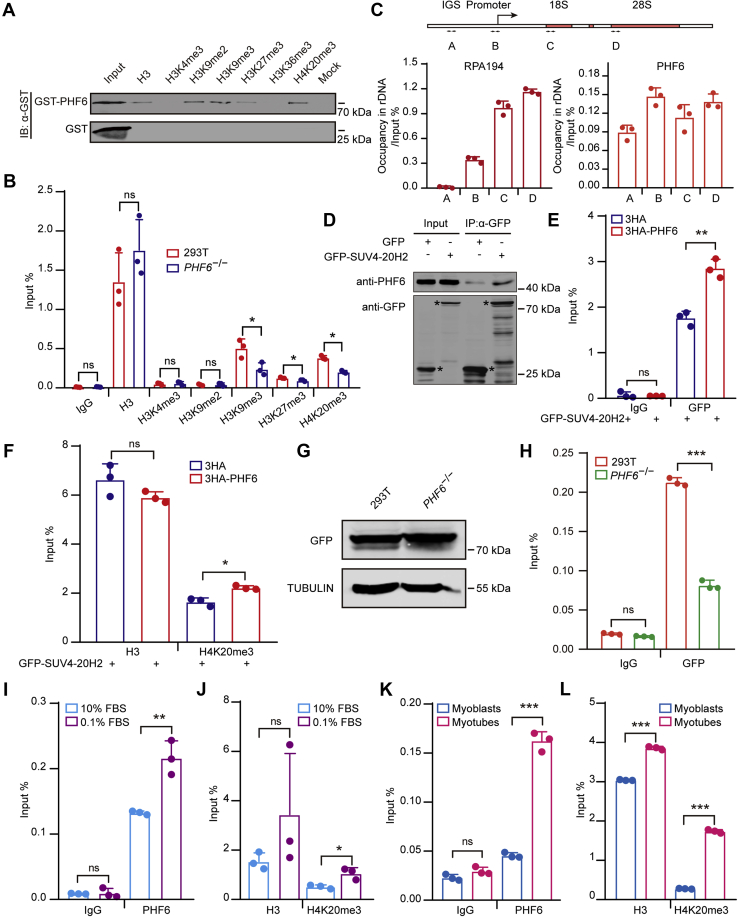
PHF6 recruits SUV4-20H2 to establish H4K20me3 at gene bodies
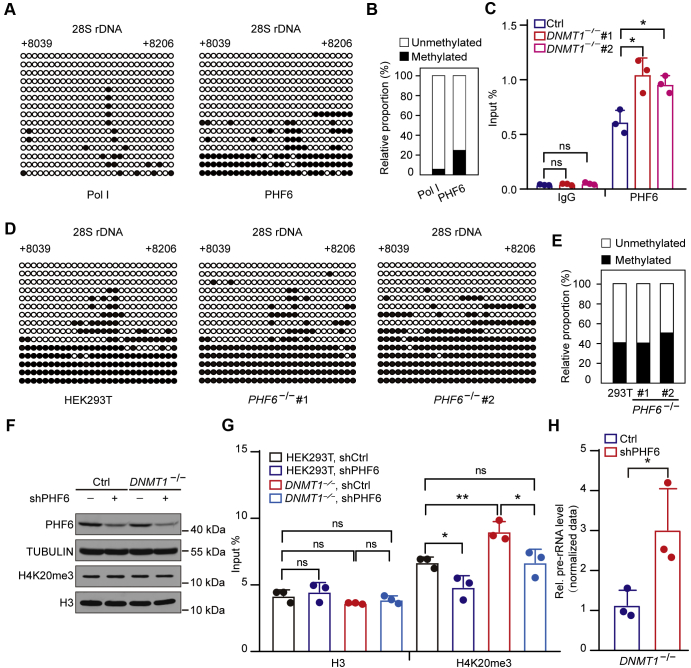
Previous evidence as well as our findings showed that PHF6 was localized in the nucleolus and inhibited rDNA transcription (Fig. S4, A–C) (35, 36). PHF6 has two PHD-like domains, suggesting that it is a potential histone modification reader. To test this hypothesis, we performed peptide binding assays in vitro, the result revealed that PHF6 could bind to repressive histone modifications, such as H3K9me3, H3K27me3, and H4K20me3 (Fig. 4A). In addition, PHF6 has also been shown to physically interact with subunits of the NuRD complex, such as HDAC1 in the nucleoplasm (41, 42, 43), indicating that it may also regulate the occupancy of histone modifications in the nucleolus where rDNA transcription occurs. To explore the regulatory role of PHF6 in histone modifications enrichments, we first assessed the occupancies of different histone modifications (i.e., H3K4me3, H3K9me2, H3K9me3, H3K27me3, and H4K20me3), in PHF6 KO and wild-type cells. The results showed that PHF6 deficiency resulted in declined enrichments of repressive histone modifications, including H4K20me3, at rDNA gene bodies but not in promoter regions, which is of most interest to us (Fig. 4B, Fig. S4D). Meanwhile, lack of PHF6 gave rise to increased occupancy of Pol Ⅰ at gene bodies (Fig. S4E). Next, we explore the distribution of PHF6 in rDNA repeat units using four pairs of primers, each of which was aligned to IGS, promoters, 18S and 28S (Fig. 4C, upper panel), respectively. In accordance with a previous report (36), PHF6 was widely distributed throughout rDNA repeat units, including IGS, promoters, 18S and 28S (Fig. 4C, lower right panel), indicating that PHF6 could function at gene bodies. To further confirm that PHF6 regulates the enrichment of H4K20me3, we depleted SUV-420H and found that loss of SUV4-20H did not alter PHF6 binding to gene bodies (Fig. S4F). These results suggest that PHF6 is involved in establishing H4K20me3 in gene bodies.
Figure 4.
PHF6 recruits SUV4-20H2 to establish H4K20me3 at gene bodies.A, PHF6 interacts with H3K9me2, H3K9me3, H3K27me3, H4K20me3 in vitro. GST-tag and GST-fused PHF6 were incubated with biotin-labeled peptides including H3, H3K4me3, H3K9me2, H3K9me3, H3K27me3, H3K36me3, H4K20me3 in vitro. Bound proteins were analyzed by the anti-GST antibody. B, the enrichment of H3K4me3, H3K9me2, H3K9me3, H3K27me3, and H4K20me3 was shown in PHF6 KO and wild-type HEK293T cells. ChIP experiments were performed using primers coated on the 28S rRNA coding region in PHF6 KO and control cells. Data are presented as the mean ± SD. C, a schematic of the rDNA repeat unit showing the intergenic spacer (IGS), promoter, and coding regions (upper). The arrows indicate the positions of the primers used for the following amplification. Primer sequences are listed in Table S1. PHF6 binds to different regions of the rDNA repeat unit (bottom). ChIP assays were performed in HEK293T cells using the previously annotated primers, showing the occupancies of PHF6 and Pol I (RPA194) on the rDNA repeat unit. Data are presented as the mean ± SD. D, PHF6 directly interacts with SUV4-20H2. GFP-fused full-length SUV4-20H2 plasmids were overexpressed in HEK293T cells, and whole-cell extracts were subsequently incubated with GFP beads. The pulled-down proteins were detected by anti-PHF6 and anti-GFP antibodies. E, PHF6 facilitates the binding of SUV4-20H2 to rDNA gene bodies. ChIP experiments were performed using primers coated on the 28S rDNA coding region in HEK293T cells expressing GFP-tagged SUV4-20H2 and HA-tagged PHF6 or control vector. Data are presented as the mean ± SD. F, PHF6 upregulation leads to the elevated enrichment of H4K20me3 at gene bodies. ChIP experiments were performed using primers coated on the 28S rDNA coding region in HEK293T cells expressing GFP-tagged SUV4-20H2 and HA-tagged PHF6 or control vector. Data are presented as the mean ± SD. G, GFP-SUV4-20H2 plasmids were transfected transiently into wild-type (WT) and PHF6 knockout (PHF6−/−) cells as shown by western blotting. TUBULIN served as a loading control. H, the gene body occupancy of GFP-SUV4-20H2 significantly decreases in PHF6 knockout cells. ChIP assays were performed using primers coated on the 28S rDNA coding region in HEK293T and PHF6 knockout cells. Data are presented as the mean ± SD. I, the gene body occupancy of PHF6 markedly increases upon serum deprivation. ChIP assays were performed using primers coated on the 28S rDNA coding region in HEK293T cells cultured in medium containing 10% FBS (normal) and 0.1% FBS (serum-starved). Data are presented as the mean ± SD. J, the gene body enrichment of H4K20me3 markedly rises following serum deprivation. ChIP assays were performed using primers coated on the 28S rRNA coding region in HEK293T cells cultured in medium containing 10% FBS (normal) and 0.1% FBS (serum-starved). Data are presented as the mean ± SD. K, PHF6 occupancy at gene bodies elevates during C2C12 myoblast differentiation. ChIP assays were performed using primers coated on the 28S rDNA coding region in myoblasts and myotubes. Data are presented as the mean ± SD. L, H4K20me3 enrichment at gene bodies is augmented during C2C12 myoblast differentiation. ChIP assays were performed using primers coated on the 28S rRNA coding region in myoblasts and myotubes. Data are presented as the mean ± SD. ∗p < 0.1, ∗∗p < 0.01, ∗∗∗p < 0.001, two-tailed unpaired Student's t test.
To further elucidate whether PHF6 can directly associate with the main histone methyltransferase for H4K20me3, SUV4-20H2, HEK293T cells were transiently transfected with GFP-tagged SUV4-20H2. Coimmunoprecipitation and western blotting demonstrated that PHF6 interacted with SUV4-20H2 directly (Fig. 4D). Subsequently, we found that ectopic expression of HA-tagged PHF6 gave rise to increased occupancy of SUV4-20H2 in rDNA gene body regions, leading to higher enrichment of H4K20me3 (Fig. 4, E and F). To further verify that PHF6 directly regulates the recruitment of SUV4-20H2 to rDNA gene body, we transfected GFP-SUV4-20H2 plasmids into PHF6 KO cells and found that the occupancy of GFP-SUV4-20H2 was significantly lower in PHF6 KO cells than that in wild-type cells (Fig. 4, G and H). Moreover, increased levels of PHF6 occupancy and H4K20me3 enrichment at gene bodies were exhibited in serum-deprived HEK293T cells (Fig. 4, I and J) as well as in differentiated C2C12 cells (Fig. 4, K and L). Collectively, these results indicate that PHF6 recruits SUV4-20H2 to establish H4K20me3 at rDNA gene bodies for the inhibition of rDNA transcription.
PHF6 mediates the DNMT1-dependent antagonism of H4K20me3
PHF6 has been shown to bind dsDNA via its ePHD2 domain in vitro (42), implying that DNA methylation may affect PHF6 occupancy. To examine whether DNA binding of PHF6 depends on the DNA methylation status at rDNA gene bodies, ChIP assays of PHF6 followed by bisulfite sequencing analysis were carried out. We found that PHF6 was capable of binding to unmethylated and methylated regions with methylated regions accounting for higher proportion (Fig. 5, A and B). These results suggest that the occupancy of PHF6 in rDNA repeat units may be regulated by DNMT1. Furthermore, ChIP-qPCR assays of PHF6 in wild-type and DNMT1 KO cells showed that knockout of DNMT1 led to a marked increase in PHF6 binding to gene body regions (Fig. 5C). However, depletion of PHF6 had no effect on either the rDNA gene body methylation levels (Fig. 5, D and E) or the binding of DNMT1 to gene body regions (Fig. S5). These results suggest that DNMT1 inhibits the occupancy of PHF6 in gene body regions.
Figure 5.
PHF6 mediates the DNMT1-dependent inhibition of H4K20me3.A, PHF6 associates with both methylated and unmethylated DNA, and the latter account for more proportion. Pol I was used as a control, which binds exclusively to unmethylated CpG sites. ChIP assays, followed by bisulfite sequencing, showing the methylation status of DNA bound by specific antibodies in HEK293T cells. The black and white circles represent corresponding CpG sites in rDNA gene bodies, denoting methylated and unmethylated CpG sites, respectively. The average ratio of black sites for all colonies represents the relative DNA methylation level. Corresponding sequence is shown in Figure S1G. B, analysis for the results in (A) showing the percentage of methylated and unmethylated gene body CpG sites bound by PHF6 in HEK293T cells. The relative proportion of unmethylation and methylation was calculated by counting the numbers of black circles (methylated CpG sites) and white circles (unmethylated CpG sites). C, knockout (KO) of DNMT1 facilitates the occupancy of PHF6 at gene bodies. ChIP experiments were performed using primers coated on the 28S rRNA coding region in DNMT1 KO and control cells. Data are presented as the mean ± SD. D, KO of PHF6 does not alter the rDNA gene body methylation status. Genomic DNA coupled with bisulfite sequencing assays were performed to show the gene body methylation level in PHF6 KO and control cells. The black and white circles represent corresponding CpG sites in rDNA gene bodies, denoting methylated and unmethylated CpG sites, respectively. The average ratio of black sites for all colonies represents the relative DNA methylation level. Corresponding sequence is shown in Figure S1G. E, analysis for the results in (D) showing the percentage of methylated and unmethylated gene body CpG sites in PHF6 KO and control cells. The relative proportion of unmethylation and methylation was calculated by counting the numbers of black circles (methylated CpG sites) and white circles (unmethylated CpG sites). F, knockdown of PHF6 in DNMT1 KO and control cells by short hairpin RNAs as revealed by western blotting. TUBULIN and H3 served as a loading control. G, depletion of PHF6 partially reduces the H4K20me3 enrichment regulated by DNMT1 deficiency. ChIP experiments were performed using primers coated on the 28S rRNA coding region in the presence of shCtrl or shRNAs for PHF6 in DNMT1 KO and control cells. Data are presented as the mean ± SD. H, knockdown of PHF6 leads to increased pre-rRNA levels in DNMT1 KO cells compared with that in DNMT1 KO cells with shCtrl. RT-qPCR assays showing levels of pre-rRNA in DNMT1 KO cells with shCtrl or shRNAs for PHF6. The cycle threshold (Ct) of 45S pre-rRNA was normalized with GAPDH. Data are presented as the mean ± SD. ∗p < 0.1, ∗∗p < 0.01, ∗∗∗p < 0.001, two-tailed unpaired Student's t-tests.
Subsequently, to explore whether PHF6 mediates increased H4K20me3 enrichment due to depletion of DNMT1 at gene bodies, we transfected wild-type and DNMT1 KO cells with short hairpin RNAs (shRNAs) targeting PHF6 to downregulate its protein expression level and found that PHF6 depletion did not affect H4K20me3 abundance in control and DNMT1 KO cells (Fig. 5F). ChIP-qPCR analysis showed that loss of PHF6 partially suppressed the increase in H4K20me3 enrichment caused by DNMT1-deficiency (Fig. 5G). Knockdown of PHF6 in DNMT1 KO cells can also rescue pre-rRNA synthesis in DNMT1 KO cells (Fig. 5H). Together, these results suggest that PHF6 functions as a mediator in the regulation of H4K20me3 via DNMT1.
Discussion
DNA methylation in promoters is known to repress rDNA transcription (24). In recent years, genome-wide DNA methylation analyses have shown that DNA methylation is more abundant within gene body regions than in promoters (13, 14). A positive correlation between gene body methylation and gene expression has been revealed (17, 19); however, whether this correlation also exists with rDNA transcription, and the related molecular mechanism, remains to be clarified. Pre-rRNA synthesis has been shown to be downregulated by metabolic ([H3]-uridine) labeling when DNA methylation levels at rDNA loci are reduced by either depletion of DNMT1 and DNMT3B or treatment with aza-dC in the human colorectal carcinoma cell line, HCT116 (28). However, another group showed that DNMT1 deficiency has no significant effect on rDNA transcription rate by northern blot analysis and quantitative RT-qPCR (27). This discrepancy may be explained by the differences among methods: metabolic and FUrd labeling was used to monitor newly synthesized pre-rRNA, which was more appropriate to measure rDNA transcription. While northern blot and quantitative RT-qPCR were used to monitor the pool of pre-rRNA, including newly synthesized pre-rRNA and accumulated pre-rRNA, if any. Likewise, the levels of 47S precursor rRNA measured by northern blot showed an increase in both DNMT1 and DNMT3B KO HCT116 cells compared with that in wild-type cells (44). However, this paper has been retracted by the authors in 2018. In this study, we utilized FUrd labeling to measure new synthesis pre-rRNA in DNMT1 KO cells and found that DNMT1 deficiency led to a significant decrease in pre-rRNA synthesis, which was consistent with the results measured by metabolic labeling (28).
Furthermore, we found that decreased DNA methylation levels in both promoters and gene body regions were accompanied by marked decreases in pre-rRNA synthesis upon growth arrest, i.e., differentiation of C2C12 myoblasts into myotubes and nutrient deprivation in HEK293T cells. These results suggest that, in addition to a hypomethylated promoter, there may be other factors involved in rDNA transcriptional regulation. Indeed, further exploration revealed that gene body methylation secures rDNA transcription by antagonizing H4K20me3 enrichment, which involves interactions between the histone methyltransferase, SUV-420H2, and the histone modification reader, PHF6.
In accordance with previous reports (27, 38), we found that DNMT1 was responsible for rDNA gene body methylation, while depletion of DNMT3A or DNMT3B did not change the DNA methylation status in gene body regions. A lack of DNMT1 has been shown to lead to a reduction in pre-rRNA synthesis (27), which was also observed in the present study. Mechanistically, we revealed that DNMT1 deficiency led to enhanced enrichment of the inhibitory histone modification, H4K20me3, at gene bodies, whereas depletion of SUV420H did not affect gene body methylation levels or alter the occupancy of DNMT1 in gene body regions, verifying that gene body methylation opposes H4K20me3 enrichment. Since H4K20me3 strongly inhibited rDNA transcription, in this way, gene body methylation prevented H4K20me3 enrichment and abrogated its role in transcriptional inhibition, which may partially explain why gene body methylation is beneficial for rDNA transcription. Although gene body methylation is present in actively transcribed regions of both Pol I and Pol II genes, many studies have demonstrated that Pol I and Pol II indeed bind to unmethylated DNA, which raises the question that how Pol I or Pol II overcomes the barrier of gene body methylation to perform transcription. One possible explanation is that there exists a transient methylation/demethylation cycle that forms unmethylated chromatin regions to allow Pol I to bind to actively transcribed gene bodies, as is the case with transcriptionally active promoters of Pol II genes, in which DNMTs play dual roles in methylation and demethylation (45, 46). Moreover, the MERM1/DNMT3A complex maintains the unmethylated window to promote Pol I transcription elongation (38). Nonetheless, more evidence is required to further clarify the speculations regarding the mechanism.
In addition, an elevated acetylation level at lysine 16 of histone H4 (H4K16Ac) in gene bodies has been observed in DNMT1-deficient HCT116 cells (27, 47). H4K16Ac has been shown to inhibit the formation of higher-order chromatin structures in vitro and to be positively correlated with DNA damage repair, indicating an inhibitory role for H4K16Ac and a potential synergistic interplay with H4K20me3 (48, 49). Alternatively, loss of DNA methylation permits cryptic transcription, including antisense RNAs or intergenic spacers, which subsequently interferes with rRNA synthesis (28). In accordance, we observed that the expression levels of antisense RNA coated on 18S, 5.8S, and 28S were elevated in DNMT1 KO cells, which is concomitant with increased occupancy of Pol II (Fig. S6, A and B), suggesting the possibility that antisense RNAs complementary to gene body regions may recruit the histone methyltransferase, SUV4-20H2, to establish H4K20me3 at gene bodies, and implying a versatile role for gene body methylation in rDNA transcriptional regulation. Moreover, it cannot be excluded that the hypomethylated promoter led to the expression of antisense RNA transcribed inversely, which may function as reported in mouse cells (26). Therefore, these findings collectively indicate that removal of DNMT1 represses rDNA transcription by either altering the chromatin structure to a closed state or disrupting the higher-order chromatin structure, establishing a more comprehensive regulatory network for rDNA transcription. Importantly, in mouse postnatal neural cells, DNMT3A binds to and methylates intergenic regions and gene bodies of neurogenic genes to antagonize H3K27me3, promoting gene expression (18). These data are suggestive of a conserved manner in which gene body methylation benefits transcription by antagonizing inhibitory histone modifications. Furthermore, DNMT3B-dependent intragenic DNA methylation ensures gene transcription initiation by preventing spurious transcripts in mouse embryonic stem cells (50). It is conceivable that gene body methylation is catalyzed by different methyltransferases in a cell-type-dependent manner. Further efforts are warranted to clarify the cross talk among these cell- and context-specific scenarios.
In recent years, PHF6 has been reported to reduce the chromatin accessibility of T cell lineage-specific transcriptional factor binding motifs (51). Evidence shows that PHF6 physically interacts with RBBP4, a component of the NuRD complex, in the nucleoplasm, suggesting that it plays a role in the formation of a repressive chromatin structure for PHF6 and extending its regulatory functions in gene expression (41, 42, 43). Our findings reveal that PHF6 recruits SUV4-20H2 to rDNA gene body regions to establish H4K20me3, forming a more compact chromatin structure and inhibiting rDNA transcription, which further implies a potential role for PHF6 in modulating 3D chromatin architecture. Moreover, PHF6 inhibits pre-rRNA synthesis by negatively regulating the protein levels of UBF (35), providing an extra mechanism for rDNA transcriptional regulation by PHF6. Concerning the distribution of PHF6 across rDNA repeat units, albeit at different abundances (36), we cannot exclude the possibility that mechanisms mediated by PHF6 at the promoter may also contribute to rDNA transcription repression. It would be of great interest to dissect the relationships among PHF6-associated gene regulatory networks, either by directly affecting transcription initiation and elongation or by changing the chromatin structure in a H4K20me3-dependent manner. In addition, given that PHF6 deficiency leads to the accumulation of DNA damage at the rDNA locus (35), it is exciting to speculate that the tumor suppressor function of PHF6 may be linked to its roles in the regulation of H4K20me3-dependent maintenance of genomic stability.
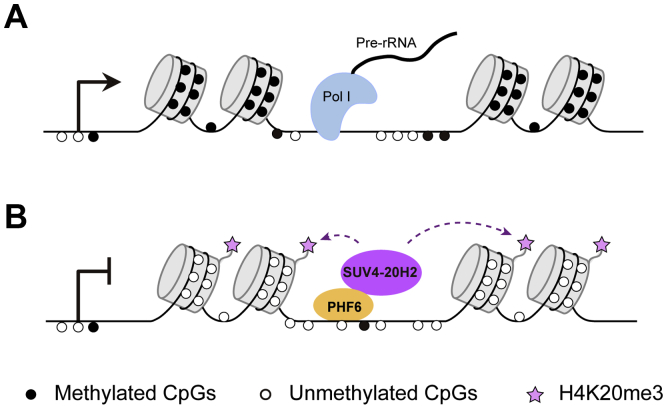
In conclusion, our results may unveil a general mechanism in growth-arrested cells during developmental or environmental alterations that gene body methylation maintained by DNMT1 ensures gene expression by preventing the formation of repressive H4K20me3 marks, which is mediated by PHF6 and SUV4-20H2 (Fig. 6). Further exploration is still warranted to elucidate whether the mechanism is conserved across transcriptional regulation of different genes and in response to different types of stresses.
Figure 6.
Proposed model deciphering the role of gene body methylation in rDNA transcription regulation.A, gene body methylation maintains rDNA transcription. Actively transcribed rDNA contains hypomethylated (represented as white circles) promoter and methylated (denoted as black circles) gene bodies. B, loss of gene body methylation leads to more occupancies of PHF6 at gene bodies, which recruits the histone methyltransferase SUV4-20H2 to establish inhibitory H4K20me3 modification, suppressing rDNA transcription.
Experimental procedures
Cell culture and transfection
HEK293T cells and C2C12 myoblasts were cultured in Dulbecco's Modified Eagle Medium (DMEM, Gibco) supplemented with 10% fetal bovine serum (FBS). Serum-starved cells were maintained in 0.1% FBS for 48 h. C2C12 myoblasts were cultured in the presence of 2% horse serum for differentiation. Plasmid transfection was performed via calcium phosphate coprecipitation.
C2C12 myoblast differentiation
C2C12 myoblasts were passaged at 40−60% confluence and split at a ratio of 1:5. For myoblast differentiation, the medium was replaced at 100% confluence with fresh DMEM supplemented with 2% horse serum and incubated for a further 6 days.
Plasmids and antibodies
The PHF6 and SUV4-20H2 genes were amplified with specific primers using the cDNA of 293T as the template. The amplicons were ligated into pCMV-3× HA and pEGFP-C1, respectively, to generate recombinant expression constructs. pcDNA3/Myc-DNMT1 (Addgene plasmid # 36939) and pcDNA3/Myc-DNMT3A (Addgene plasmid # 35521) were kind gifts from Arthur Riggs.
The anti-DNMT3A (sc-20703), anti-RPA194 (sc-28714), anti-UBF (sc-9131 X), and anti-c Myc (sc-40 X) antibodies were purchased from Santa Cruz. The anti-α-tubulin (T6199), anti-HA (H9658), and anti-bromodeoxyuridine (BrdU) (B2531) antibodies were bought from Sigma. The anti-5-methylcytosine (BI-MECY-0100) antibody was purchased from Eurogentec, and the anti-PHF6 (Bethyl-451A) was purchased from Bethyl Laboratories. The anti-H4K20me3 (ab9053), anti-DNMT1 (ab13537), anti-GFP (ab290), and anti-H3 (ab1791) antibodies were bought from Abcam. The secondary antibodies, IRDye800CW goat anti-mouse IgG (926-32210) and IRDye800CW goat anti-rabbit IgG (926-32211) were purchased from LI-COR.
Immunofluorescence staining and FUrd incorporation
Cells grown on coverslips to 80% confluence were washed twice with ice-cold PBS and fixed in 4% paraformaldehyde at room temperature for 10 min. After rinsing three times with PBS, cells were permeabilized in 0.5% Triton X-100 on ice for 15 min. Subsequently, nonspecific sites were blocked with 1% BSA at room temperature for 30 min, and the cells were incubated with the appropriate primary antibody at 4 °C overnight. After rinsing three times with PBST (0.1% Triton X-100 in PBS), cells were incubated with 150 μl secondary antibody (anti-rabbit, Alexa Fluor 488; anti-mouse, Alexa Fluor 594, Life Technologies) at room temperature for 2 h. After rinsing with 0.1% PBST, cells were stained with DAPI at room temperature for 3 min. Finally, the coverslips were sealed with 10 μl Fluoromount-G (Southern Biotech), incubated for 2 h, and images were subsequently taken using a fluorescence microscope.
For the FUrd incorporation assay, nascent RNA was labeled by incubating cells with 2 mM FUrd at 37 °C, 5% CO2 for 10 min prior to fixation. Cells were subsequently labeled with an anti-BrdU antibody (Sigma).
The ratio of fluorescence intensity to cell numbers in each coverslip was measured by the Image J software, which was used to generate bar graphs and perform statistical test. The cell numbers used in FUrd immunofluorescence analysis are noted in corresponding figure legends.
qPCR analysis and western blotting
At 80% confluence, cells were harvested in TRIzol reagent (Invitrogen, 10296028). RNA and protein were isolated following the manufacturer's protocol. RNA was reverse transcribed by DN6 or a strand-specific primer using the M-MLV Kit (Invitrogen), and qPCR analysis was performed using the Light Cycler 450 (Roche). Primers are listed in Table S1.
For western blotting, proteins were separated by SDS-PAGE, transferred to nitrocellulose membrane using the semidry method (Bio-Rad), and incubated with the appropriate primary antibody overnight at 4 °C. After washing three times in 0.1% PBST, membranes were incubated with secondary antibodies (IRDye800CW, LI-COR) at room temperature for 2 h. Protein bands were detected using the Odyssey infrared imaging system (Odyssey, LI-COR).
Northern blotting
Total RNA was extracted using TRIzol reagent. Fifteen microgram of RNA was subjected to 1% formaldehyde denaturing gel electrophoresis at 25V, 4 °C overnight. RNA in denaturing gel was transferred to positively charged nylon membrane HyBond N+ (Amersham, RPN303B) using upward capillary method, fixing the membrane at 80 °C for 2 h. Interest RNA was hybridized and detected using DIG-High Prime DNA Labeling and Detection Starter Kit II (for chemiluminescent detection with CSPD) (Roche, 11585614910). Probes for detecting interest RNA in northern blotting were prepared using PCR DIG Probe Synthesis Kit (Roche, 11636090910). The primer sequences for amplifying long DNA probes are listed below:
For detecting 45S pre rRNA (391 bp): 5′ TACTATCCAGCGAAACCACAGCC 3′; 5′ CGCCTTAGGACACCTGCGTTA 3′.
For detecting 7S and 7SL RNA (215 bp): 5′ GGGCTGTAGTGCGCTATGCC 3′; 5′ ACGGGGTCTCGCTATGTTGC 3′.
Chromatin immunoprecipitation (ChIP) and quantitative real-time PCR (qPCR)
ChIP assays were performed as previously described (26). Cells were cross-linked with a final concentration of 1% formaldehyde solution (Sigma, F8775) at room temperature for 5−10 min and subsequently quenched by glycine for 5 min at a final concentration of 0.125 M. Cross-linked chromatin was sheared using a Bioruptor Next Gen (Diagenode) to yield fragments of 300−500 base pairs (bp). The supernatant was precleared with salmon sperm DNA/Protein A/G sepharose beads (GE). Subsequently, 10% input was reserved and the remaining supernatant was divided into several aliquots and incubated with the respective antibodies overnight. The next day, the samples were incubated with Protein A/G at 4 °C for 2 h. After washing with a series of wash buffers, the DNA was purified using Chelex-100 resin or phenol-chloroform extraction and analyzed by qPCR using the Light Cycler 450 (Roche). Primers are listed in Table S1. The ChIP enrichment in each sample was calculated by percent of input (input %).
Co-immunoprecipitation assays
Cells grown on 10-cm dishes were washed twice with ice-cold PBS and collected by scraping into 1.5 ml tubes. Following centrifugation at 4 °C, cells were lysed in lysis buffer (10 mM Tris-HCl, pH 7.5, 150 mM NaCl, 0.5 mM EDTA, 0.5% NP-40, 1 mM PMSF, protease inhibitors) on ice for 30 min and recentrifuged at 4 °C, 20,000g for 10 min. The supernatant was then transferred to a clean tube and incubated with GFP-trap_A beads (Chromotek) at 4 °C for 1 h; 10% input was saved for immunoblotting analysis. After incubation, the beads were washed twice with low-salt dilution buffer (10 mM Tris-HCl, pH 7.5, 150 mM NaCl, 0.5 mM EDTA) and once with high-salt dilution buffer (10 mM Tris-HCl, pH 7.5, 500 mM NaCl, 0.5 mM EDTA). The beads were resuspended in 100 μl 2 × SDS loading buffer (80 mM Tris-HCl, pH 6.8, 10% glycerol, 2% SDS, 20% β-mercaptoethanol) and boiled at 95 °C for 10 min. SDS-PAGE was performed for further analysis.
DNA methylation assays
Genomic DNA or DNA enriched by ChIP was treated with the EpiTect Bisulfite Kit (Qiagen). Treated DNA was purified according to the manufacturer's protocol and amplified using primers for methylation analysis. Subsequently, amplicons were purified and ligated into pEasy-T5 Zero (Transgen). Trans T1 competent cells (Transgen) were transformed with the recombinant pEasy-T5 constructs and incubated at 37 °C overnight. At least 20 independent colonies were selected and sequenced, and the results were analyzed using a BiQ analyzer (Max Planck Institute Informatik). Primers are listed Table S1.
Methylated DNA immunoprecipitation and next-generation sequencing (MeDIP-Seq)
MeDIP assays were performed as previously described (38). In brief, DNA libraries were prepared with the NEB Next DNA Library Prep Master Mix Set for Illumina (E6040, NEB), according to the manufacturer's instructions. Firstly, fragmented DNA (200−500 bp) was end-repaired and then purified using AMPure XP beads (A6380, Beckman). dA-tails were added to the DNA, which was purified again using AMPure XP beads. Subsequently, dA-tailed DNA was ligated to adaptors. After purification, treated DNA was denatured at 99 °C for 10 min and then placed immediately on ice. DNA was incubated with a specific antibody against 5 mC (BI-MECY-0100) at 4 °C for 2 h and captured using protein A/G agarose beads at 4 °C for a further 2 h. After washing three times with IP buffer (140 mM NaCl, 0.05% Triton X-100, 3.9 mM NaH2PO4, 6.1 mM Na2HPO4) and digesting with proteinase K, the DNA fragments were extracted twice using phenol-chloroform and amplified by adaptor-mediated PCR. After purification, the DNA fragments were sequenced using the Illumina HiSeq 2000 sequencing platform. The sequencing reads were aligned to the mm9 genome assembly using bowtie2 and the default parameters, following which duplicated reads were removed by Picard (MarkDuplicates). Samtools was then used to select properly mapped reads (samtools view -Sb -h -f 2). Finally, peaks were detected by macs (--nomodel --nolambda -w --space=30).
The raw MeDIP-seq data were deposited in NCBI Gene Expression Omnibus under the accession number, GSE141047 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE141047).
Targeting strategy for the knockout
CRISPR/Cas9 technology was used to knock out genes of interest in HEK293T cells. HEK293T cells were transfected with the plasmids, pcDNA3.1-NLS-hCas9-NLS and pUC19, containing specific guide RNAs, using the calcium phosphate method. A single clone was selected and cultured. Genomic DNA was extracted and analyzed by Sanger sequencing. The guide RNA sequences are provided in Table S1.
Data availability
All data is either provided in this manuscript or will be provided upon request to Dr Wei Tao, Peking University (weitao@pku.edu.cn).
Supporting information
This article contains supporting information.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
We thank Prof Jingwei Xiong for his assistance with the CRISPR/Cas9 systems. We appreciate the staff of the National Center for Protein Sciences at Peking University (Beijing, China), particularly Liying Du, Hongxia Lyu, Guilan Li, and Zhi Dong for their help with biochemical experiments. This work was supported by the National Key Research and Development Project (Grant No. 2020YFC2002903) and the National Natural Science Foundation of China (Grant No. 31871312, 31471205, 31671426).
Author contributions
W. T. funding acquisition; W. T., G. L., X. H., and X. Z. conceptualization; W. T., G. L., X. H., and X. Z project administration; W. T., X. H., and X. Z. data curation; X. H., and X. Z. formal analysis; X. H., X. Z., Le Zong, and Q. G. investigation; X. H., X. Z., Le Zong, Q. G., Y. G., and L. H. methodology; X. H., X. Z., Le Zong, Q. G., Y. G., and L. H. resources; C. Z. and R. W. software; Lijun Zhang supervision; X. H. and X. Z. validation; X. H. and X. Z. visualization; W. T., G. L., X. H., and X. Z. writing–original draft; W. T., G. L., X. H., and X. Z. writing–review and editing.
Edited by John Denu
Contributor Information
Guoliang Lyu, Email: guolianglyu@pku.edu.cn.
Wei Tao, Email: weitao@pku.edu.cn.
Supporting information
References
- 1.Martienssen R.A., Colot V. DNA methylation and epigenetic inheritance in plants and filamentous fungi. Science. 2001;293:1070–1074. doi: 10.1126/science.293.5532.1070. [DOI] [PubMed] [Google Scholar]
- 2.Suzuki M.M., Bird A. DNA methylation landscapes: Provocative insights from epigenomics. Nat. Rev. Genet. 2008;9:465–476. doi: 10.1038/nrg2341. [DOI] [PubMed] [Google Scholar]
- 3.Feng S., Jacobsen S.E., Reik W. Epigenetic reprogramming in plant and animal development. Science. 2010;330:622–627. doi: 10.1126/science.1190614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sinsheimer R.L. The action of pancreatic deoxyribonuclease. II. Isomeric dinucleotides. J. Biol. Chem. 1955;215:579–583. [PubMed] [Google Scholar]
- 5.Stein R., Gruenbaum Y., Pollack Y., Razin A., Cedar H. Clonal inheritance of the pattern of DNA methylation in mouse cells. Proc. Natl. Acad. Sci. U. S. A. 1982;79:61–65. doi: 10.1073/pnas.79.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bestor T.H. Activation of mammalian DNA methyltransferase by cleavage of a Zn binding regulatory domain. EMBO J. 1992;11:2611–2617. doi: 10.1002/j.1460-2075.1992.tb05326.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Okano M., Bell D.W., Haber D.A., Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- 8.Tate P.H., Bird A.P. Effects of DNA methylation on DNA-binding proteins and gene expression. Curr. Opin. Genet. Dev. 1993;3:226–231. doi: 10.1016/0959-437x(93)90027-m. [DOI] [PubMed] [Google Scholar]
- 9.Jones P.A. Functions of DNA methylation: Islands, start sites, gene bodies and beyond. Nat. Rev. Genet. 2012;13:484–492. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- 10.Smith Z.D., Meissner A. DNA methylation: Roles in mammalian development. Nat. Rev. Genet. 2013;14:204–220. doi: 10.1038/nrg3354. [DOI] [PubMed] [Google Scholar]
- 11.Larsen F., Gundersen G., Lopez R., Prydz H. CpG islands as gene markers in the human genome. Genomics. 1992;13:1095–1107. doi: 10.1016/0888-7543(92)90024-m. [DOI] [PubMed] [Google Scholar]
- 12.Hellman A., Chess A. Gene body-specific methylation on the active X chromosome. Science. 2007;315:1141–1143. doi: 10.1126/science.1136352. [DOI] [PubMed] [Google Scholar]
- 13.Ball M.P., Li J.B., Gao Y., Lee J.-H., LeProust E.M., Park I.-H., Xie B., Daley G.Q., Church G.M. Targeted and genome-scale strategies reveal gene-body methylation signatures in human cells. Nat. Biotechnol. 2009;27:361. doi: 10.1038/nbt.1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rauch T.A., Wu X., Zhong X., Riggs A.D., Pfeifer G.P. A human B cell methylome at 100− base pair resolution. Proc. Natl. Acad. Sci. U. S. A. 2009;106:671–678. doi: 10.1073/pnas.0812399106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aran D., Toperoff G., Rosenberg M., Hellman A. Replication timing-related and gene body-specific methylation of active human genes. Hum. Mol. Genet. 2010;20:670–680. doi: 10.1093/hmg/ddq513. [DOI] [PubMed] [Google Scholar]
- 16.Laurent L., Wong E., Li G., Huynh T., Tsirigos A., Ong C.T., Low H.M., Kin Sung K.W., Rigoutsos I., Loring J., Wei C.L. Dynamic changes in the human methylome during differentiation. Genome Res. 2010;20:320–331. doi: 10.1101/gr.101907.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang X., Han H., De Carvalho D.D., Lay F.D., Jones P.A., Liang G. Gene body methylation can alter gene expression and is a therapeutic target in cancer. Cancer Cell. 2014;26:577–590. doi: 10.1016/j.ccr.2014.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu H., Coskun V., Tao J., Xie W., Ge W., Yoshikawa K., Li E., Zhang Y., Sun Y.E. Dnmt3a-dependent nonpromoter DNA methylation facilitates transcription of neurogenic genes. Science. 2010;329:444–448. doi: 10.1126/science.1190485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Arechederra M., Daian F., Yim A., Bazai S.K., Richelme S., Dono R., Saurin A.J., Habermann B.H., Maina F. Hypermethylation of gene body CpG islands predicts high dosage of functional oncogenes in liver cancer. Nat. Commun. 2018;9:3164. doi: 10.1038/s41467-018-05550-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Su J., Huang Y.-H., Cui X., Wang X., Zhang X., Lei Y., Xu J., Lin X., Chen K., Lv J., Goodell M.A., Li W. Homeobox oncogene activation by pan-cancer DNA hypermethylation. Genome Biol. 2018;19:108. doi: 10.1186/s13059-018-1492-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Henderson A., Warburton D., Atwood K. Location of ribosomal DNA in the human chromosome complement. Proc. Natl. Acad. Sci. U. S. A. 1972;69:3394–3398. doi: 10.1073/pnas.69.11.3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Conconi A., Widmer R.M., Koller T., Sogo J. Two different chromatin structures coexist in ribosomal RNA genes throughout the cell cycle. Cell. 1989;57:753–761. doi: 10.1016/0092-8674(89)90790-3. [DOI] [PubMed] [Google Scholar]
- 23.Xie W., Ling T., Zhou Y., Feng W., Zhu Q., Stunnenberg H.G., Grummt I., Tao W. The chromatin remodeling complex NuRD establishes the poised state of rRNA genes characterized by bivalent histone modifications and altered nucleosome positions. Proc. Natl. Acad. Sci. U. S. A. 2012;109:8161–8166. doi: 10.1073/pnas.1201262109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Santoro R., Grummt I. Molecular mechanisms mediating methylation-dependent silencing of ribosomal gene transcription. Mol. Cell. 2001;8:719–725. doi: 10.1016/s1097-2765(01)00317-3. [DOI] [PubMed] [Google Scholar]
- 25.Mayer C., Schmitz K.M., Li J., Grummt I., Santoro R. Intergenic transcripts regulate the epigenetic state of rRNA genes. Mol. Cell. 2006;22:351–361. doi: 10.1016/j.molcel.2006.03.028. [DOI] [PubMed] [Google Scholar]
- 26.Bierhoff H., Dammert M.A., Brocks D., Dambacher S., Schotta G., Grummt I. Quiescence-induced LncRNAs trigger H4K20 trimethylation and transcriptional silencing. Mol. Cell. 2014;54:675–682. doi: 10.1016/j.molcel.2014.03.032. [DOI] [PubMed] [Google Scholar]
- 27.Espada J., Ballestar E., Santoro R., Fraga M.F., Villar-Garea A., Nemeth A., Lopez-Serra L., Ropero S., Aranda A., Orozco H. Epigenetic disruption of ribosomal RNA genes and nucleolar architecture in DNA methyltransferase 1 (Dnmt1) deficient cells. Nucleic Acids Res. 2007;35:2191–2198. doi: 10.1093/nar/gkm118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gagnon-Kugler T., Langlois F., Stefanovsky V., Lessard F., Moss T. Loss of human ribosomal gene CpG methylation enhances cryptic RNA polymerase II transcription and disrupts ribosomal RNA processing. Mol. Cell. 2009;35:414–425. doi: 10.1016/j.molcel.2009.07.008. [DOI] [PubMed] [Google Scholar]
- 29.Lower K.M., Turner G., Kerr B.A., Mathews K.D., Shaw M.A., Gedeon A.K., Schelley S., Hoyme H.E., White S.M., Delatycki M.B., Lampe A.K., Clayton-Smith J., Stewart H., van Ravenswaay C.M., de Vries B.B. Mutations in PHF6 are associated with Borjeson-Forssman-Lehmann syndrome. Nat. Genet. 2002;32:661–665. doi: 10.1038/ng1040. [DOI] [PubMed] [Google Scholar]
- 30.Van Vlierberghe P., Palomero T., Khiabanian H., Van der Meulen J., Castillo M., Van Roy N., De Moerloose B., Philippé J., González-García S., Toribio M.L. PHF6 mutations in T-cell acute lymphoblastic leukemia. Nat. Genet. 2010;42:338. doi: 10.1038/ng.542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Van Vlierberghe P., Patel J., Abdel-Wahab O., Lobry C., Hedvat C.V., Balbin M., Nicolas C., Payer A.R., Fernandez H.F., Tallman M.S. PHF6 mutations in adult acute myeloid leukemia. Leukemia. 2011;25:130. doi: 10.1038/leu.2010.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meacham C.E., Lawton L.N., Soto-Feliciano Y.M., Pritchard J.R., Joughin B.A., Ehrenberger T., Fenouille N., Zuber J., Williams R.T., Young R.A. A genome-scale in vivo loss-of-function screen identifies Phf6 as a lineage-specific regulator of leukemia cell growth. Genes Dev. 2015;29:483–488. doi: 10.1101/gad.254151.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Soto-Feliciano Y.M., Bartlebaugh J.M., Liu Y., Sánchez-Rivera F.J., Bhutkar A., Weintraub A.S., Buenrostro J.D., Cheng C.S., Regev A., Jacks T.E. PHF6 regulates phenotypic plasticity through chromatin organization within lineage-specific genes. Genes Dev. 2017;31:973–989. doi: 10.1101/gad.295857.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang C., Mejia L.A., Huang J., Valnegri P., Bennett E.J., Anckar J., Jahani-Asl A., Gallardo G., Ikeuchi Y., Yamada T., Rudnicki M., Harper J.W., Bonni A. The X-linked intellectual disability protein PHF6 associates with the PAF1 complex and regulates neuronal migration in the mammalian brain. Neuron. 2013;78:986–993. doi: 10.1016/j.neuron.2013.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang J., Leung J.W., Gong Z., Feng L., Shi X., Chen J. PHF6 regulates cell cycle progression by suppressing ribosomal RNA synthesis. J. Biol. Chem. 2013;288:3174–3183. doi: 10.1074/jbc.M112.414839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Todd M.A., Huh M.S., Picketts D.J. The sub-nucleolar localization of PHF6 defines its role in rDNA transcription and early processing events. Eur. J. Hum. Genet. 2016;24:1453–1459. doi: 10.1038/ejhg.2016.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liao J., Karnik R., Gu H., Ziller M.J., Clement K., Tsankov A.M., Akopian V., Gifford C.A., Donaghey J., Galonska C. Targeted disruption of DNMT1, DNMT3A and DNMT3B in human embryonic stem cells. Nat. Genet. 2015;47:469–478. doi: 10.1038/ng.3258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lyu G., Zong L., Zhang C., Huang X., Xie W., Fang J., Guan Y., Zhang L., Ni T., Gu J., Tao W. Metastasis-related methyltransferase 1 (Merm1) represses the methyltransferase activity of Dnmt3a and facilitates RNA polymerase I transcriptional elongation. J. Mol. Cell Biol. U. S. A. 2018;11:78–90. doi: 10.1093/jmcb/mjy023. [DOI] [PubMed] [Google Scholar]
- 39.Khan A., Giri S., Wang Y., Chakraborty A., Ghosh A.K., Anantharaman A., Aggarwal V., Sathyan K.M., Ha T., Prasanth K.V., Prasanth S.G. BEND3 represses rDNA transcription by stabilizing a NoRC component via USP21 deubiquitinase. Proc. Natl. Acad. Sci. 2015;112:8338. doi: 10.1073/pnas.1424705112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Okamoto K., Tanaka Y., Tsuneoka M. SF-KDM2A binds to ribosomal RNA gene promoter, reduces H4K20me3 level, and elevates ribosomal RNA transcription in breast cancer cells. Int. J. Oncol. 2017;50:1372–1382. doi: 10.3892/ijo.2017.3908. [DOI] [PubMed] [Google Scholar]
- 41.Todd M.A.M., Picketts D.J. PHF6 interacts with the nucleosome remodeling and deacetylation (NuRD) complex. J. Proteome Res. 2012;11:4326–4337. doi: 10.1021/pr3004369. [DOI] [PubMed] [Google Scholar]
- 42.Liu Z., Li F., Ruan K., Zhang J., Mei Y., Wu J., Shi Y. Structural and functional insights into the human Borjeson-Forssman-Lehmann syndrome-associated protein PHF6. J. Biol. Chem. 2014;289:10069–10083. doi: 10.1074/jbc.M113.535351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu Z., Li F., Zhang B., Li S., Wu J., Shi Y. Structural basis of plant homeodomain finger 6 (PHF6) recognition by the retinoblastoma binding protein 4 (RBBP4) component of the nucleosome remodeling and deacetylase (NuRD) complex. J. Biol. Chem. 2015;290:6630–6638. doi: 10.1074/jbc.M114.610196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Majumder S., Ghoshal K., Datta J., Smith D.S., Bai S., Jacob S.T. Role of DNA methyltransferases in regulation of human ribosomal RNA gene transcription. J. Biol. Chem. 2018;293:3591. doi: 10.1074/jbc.W118.002433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kangaspeska S., Stride B., Métivier R., Polycarpou-Schwarz M., Ibberson D., Carmouche R.P., Benes V., Gannon F., Reid G. Transient cyclical methylation of promoter DNA. Nature. 2008;452:112. doi: 10.1038/nature06640. [DOI] [PubMed] [Google Scholar]
- 46.Métivier R., Gallais R., Tiffoche C., Le Péron C., Jurkowska R.Z., Carmouche R.P., Ibberson D., Barath P., Demay F., Reid G. Cyclical DNA methylation of a transcriptionally active promoter. Nature. 2008;452:45. doi: 10.1038/nature06544. [DOI] [PubMed] [Google Scholar]
- 47.Espada J., Ballestar E., Fraga M.F., Villar-Garea A., Juarranz A., Stockert J.C., Robertson K.D., Fuks F., Esteller M. Human DNA methyltransferase 1 is required for maintenance of the histone H3 modification pattern. J. Biol. Chem. 2004;279:37175–37184. doi: 10.1074/jbc.M404842200. [DOI] [PubMed] [Google Scholar]
- 48.Shogren-Knaak M., Ishii H., Sun J.-M., Pazin M.J., Davie J.R., Peterson C.L. Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science. 2006;311:844–847. doi: 10.1126/science.1124000. [DOI] [PubMed] [Google Scholar]
- 49.Sharma G.G., So S., Gupta A., Kumar R., Cayrou C., Avvakumov N., Bhadra U., Pandita R.K., Porteus M.H., Chen D.J., Cote J., Pandita T.K. MOF and histone H4 acetylation at lysine 16 are critical for DNA damage response and double-strand break repair. Mol. Cell Biol. 2010;30:3582–3595. doi: 10.1128/MCB.01476-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Neri F., Rapelli S., Krepelova A., Incarnato D., Parlato C., Basile G., Maldotti M., Anselmi F., Oliviero S. Intragenic DNA methylation prevents spurious transcription initiation. Nature. 2017;543:72–77. doi: 10.1038/nature21373. [DOI] [PubMed] [Google Scholar]
- 51.Soto-Feliciano Y.M., Bartlebaugh J.M.E., Liu Y., Sanchez-Rivera F.J., Bhutkar A., Weintraub A.S., Buenrostro J.D., Cheng C.S., Regev A., Jacks T.E., Young R.A., Hemann M.T. PHF6 regulates phenotypic plasticity through chromatin organization within lineage-specific genes. Genes Dev. 2017;31:973–989. doi: 10.1101/gad.295857.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data is either provided in this manuscript or will be provided upon request to Dr Wei Tao, Peking University (weitao@pku.edu.cn).