Abstract
Although a link between histone acetylation and transcription has been established, it is not clear how acetylases function in the nucleus of the cell and how they access their targets in a chromatin fiber containing H1 and folded into a highly condensed structure. Here we show that the histone acetyltransferase (HAT) p300/CBP-associated factor (PCAF), either alone or in a nuclear complex, can readily acetylate oligonucleosomal substrates. The linker histones, H1 and H5, specifically inhibit the acetylation of mono- and oligonucleosomes and not that of free histones or histone-DNA mixtures. We demonstrate that the inhibition is due mainly to steric hindrance of H3 by the tails of linker histones and not to condensation of the chromatin fiber. Cellular PCAF, which is complexed with accessory proteins in a multiprotein complex, can overcome the linker histone repression. We suggest that linker histones hinder access of PCAF, and perhaps other HATs, to their target acetylation sites and that perturbation of the linker histone organization in chromatin is a prerequisite for efficient acetylation of the histone tails in nucleosomes.
Chromatin, with its associated linker histones, is a highly condensed structure that constrains the genome into the nucleus of the cell and suppresses various DNA-related activities such as transcription and replication. Transcriptional activation has been associated with changes in the structure of both chromatin and nucleosomes (57, 58). These changes are mediated by chromatin remodeling complexes (59) and by reversible modification of histones (46, 56). Indeed, there is a strong correlation between the acetylation state of core histones and the transcriptional competence of specific genes (21, 46, 52). This correlation has been strengthened by the finding that several transcription factors have intrinsic histone acetyltransferase (HAT) activity (28, 46) and that mutants lacking HAT activity fail to activate transcription of their target genes (23, 55). Recent studies suggest that HATs function in the context of multiprotein complexes in vivo and that the acetylase activity of these complexes is more efficient than that of the isolated transcription factors (15, 32, 48). It is conceivable that some of the proteins found in these multiprotein complexes function to facilitate histone acetylation in the context of chromatin.
In chromatin, the N-terminal tails of the core histones are thought to be involved in internucleosomal interactions and have been shown to be required for formation of higher-order, condensed chromatin structure (3, 12, 17). Studies using oligonucleosomes condensed with salt indicate that the HAT GCN5 can efficiently acetylate the N-terminal tail of histone H3 (51), suggesting that at least some of the acetylation targets are available in condensed chromatin. An additional major factor, known to be involved in the formation and stabilization of a higher-order, condensed chromatin structure, is histone H1. Numerous studies have demonstrated that the presence of H1 inhibits transcription and in some cases transcriptional activation is associated with removal of H1 (4, 24, 33). However, some studies have found histone H1 in transcriptionally active genes (11), albeit in an altered chromatin organization (42). The link between histone H1 and core histone acetylation is not clear. It has been suggested that acetylation of H4 during nucleosome assembly regulates the binding of H1 and the ability of chromatin to condense (34, 35). While in some cases active genes are hyperacetylated and contain H1 (10, 31, 37), it has also been reported that while H1 binds to acetylated oligonucleosomes, this binding inhibits transcription (53). In addition, studies have demonstrated that histone acetylation alters the capacity of histone H1 to condense chromatin (36) and that the presence of H1 affects the ability of transcription factors to interact with the DNA (19, 39). Recent studies have also shown that the retinoid receptor, a receptor known to function in part by recruitment of HATs, must also recruit an activity for displacement or remodeling of the linker histone H1 (29). These results argue that displacement of H1 is required prior to acetylation of the target gene and activation of transcription. In addition, studies involving steroid hormone receptors, also known to interact with HATs (14), have shown that activation involves a phosphorylation of H1 that results in a reduced affinity of H1 for chromatin (25). These receptor responsive genes whose activation involves the recruitment of HATs also appear to remodel or remove the linker histone. These data taken together suggest a concerted mechanism for gene activation requiring both histone acetylation and reorganization of H1 on chromatin.
Most studies on the activity of either purified HATs or multiprotein complexes containing HAT activity have been performed with either isolated core histones or purified nucleosome core particles. However, in vivo the true substrate of these HATs is chromatin, which contains histone H1 and is folded into a highly condensed structure. How these various acetylases access their targets in the oligonucleosomal chromatin fiber has not been examined. In this study we examined whether recombinant PCAF (rPCAF) and a multiprotein nuclear complex containing PCAF (cPCAF) could acetylate oligonucleosome arrays in the presence or absence of linker histones. We demonstrate that both rPCAF and cPCAF can acetylate oligonucleosome arrays. Importantly, we demonstrate that saturation of the oligonucleosome with linker histones specifically blocks the ability of both rPCAF and cPCAF to acetylate H3. The H1-induced inhibition of acetylation is due to steric occlusion of the H3 tail by H1 and not to structural changes associated with the formation of a more condensed oligonucleosome array. Furthermore, we demonstrate that in the presence of subsaturating concentrations of H1, the PCAF complex, but not free PCAF, is capable of overcoming the inhibition. The results suggest that H1 hinders access of PCAF and perhaps other acetylases to their target acetylation sites and that perturbation of this steric hindrance is a prerequisite for efficient acetylation of histone tails in chromatin. Our findings raise the possibility that multiprotein complexes that acetylate or remodel chromatin contain components that modify the interaction of H1 with chromatin.
MATERIALS AND METHODS
Materials.
rPCAF (60) and cPCAF (32) were prepared as previously described. A mixture of all isotypes of both the linker histone H1 and core histones were purified from calf thymus and chicken erythrocyte nuclei, respectively (6), all as previously described. The globular domains of H5 (GH5) and H1 (GH1) were prepared from purified H5 and H1, respectively, as previously described (1). [1-14C]acetyl coenzyme A ([1-14C]acetyl-CoA; 55 mCi/mmol) was obtained from Amersham.
HAT assay.
All assays were performed in buffer A (50 mM Tris-HCl [pH 8.0], 10% [vol/vol] glycerol, 1 mM dithiothreitol, 0.1 mM EDTA, 10 mM butyric acid) (5) with addition of 50 mM NaCl (unless otherwise indicated). Oligonucleosome concentrations were 0.1 to 0.25 mg/ml, and the [1-14C]acetyl-CoA concentration was 18 μM. The assay was performed at 37°C and initiated by addition of the enzyme to a mixture containing the substrate and acetyl-CoA in buffer A containing 50 mM NaCl. Since the cPCAF is a more potent HAT (32) than rPCAF, the quantity of rPCAF or cPCAF added to each assay was empirically determined as the amount of preparation required to yield nearly equivalent activities on nucleosome core particles. The amount of PCAF used was empirically determined by using various amounts of the preparation to ensure a linear range for the reaction. All assays were conducted for 20 min at 37°C. The radioactivity incorporated into the protein substrate was detected in a polyacrylamide gel assay (18). In this assay, the reactions were stopped by the addition of an equal volume of a sodium dodecyl (SDS)-gel sample buffer (100 mM Tris-HCl [pH 6.8], 200 mM dithiothreitol, 2% SDS, 0.1% bromophenol blue, 20% glycerol) and boiled for 5 min, and the proteins were resolved on an SDS–15% polyacrylamide gel. Polyacrylamide gel electrophoresis (PAGE) was performed at 15 V/cm and stopped when the bromophenol blue reached the bottom of the gel. The gels were stained with Coomassie blue for estimation of protein quantities and vacuum dried, and the radioactivity incorporated into the protein bands was visualized on a PhosphorImager (Molecular Dynamics) and quantified with ImageQuant software.
Purification of nucleosomal substrates.
Oligonucleosomes, core particles, and chromatosomes were prepared from chicken erythrocyte nuclei (20). Chicken erythrocyte chromatin purified in the absence of histone deacetylase inhibitors represent a pool of histones that are hypoacetylated (27). Purified chicken erythrocyte nuclei were digested with micrococcal nuclease (MNase; at 100 U/mg of DNA) at room temperature for 5 min. The treated nuclei were pelleted by centrifugation (5,000 × g in a Sorvall SS34 rotor) at 4°C. The nuclei were lysed by resuspending the pellet in a buffer containing 0.25 mM EDTA, 10 mM Tris-HCl (pH 7.4), and 1 mM phenylmethylsulfonyl fluoride (PMSF). The resuspended material was rocked gently at 4°C for 3 h, and then the nuclear debris was removed by centrifugation. For preparation of chromatosomes, an additional MNase digestion was performed, and the reaction was stopped by addition of EDTA. The chromatin preparation was then stripped of linker histones and other nonhistone chromosomal proteins. The stripping was accomplished by first gradually bringing the supernatant to 0.45 M NaCl and then adding 200 μl of a slurry of carboxymethyl-Sephadex (in 10 mM Tris-HCl [pH 7.4], 1 mM EDTA, 1 mM PMSF, 0.45 M NaCl) per ml of supernatant. The mixture was gently rocked at 4°C for 1 h, the resin was then removed by centrifugation, and the process was repeated. The resulting supernatant was then concentrated by spin dialysis through a 10-kDa-cutoff membrane. For preparation of core particles, the stripped chromatin was redigested with MNase to yield the core particle, characterized by the 145-bp DNA. The concentrated digested chromatin was then layered onto a 40-ml, 15 to 50% (or 5 to 20% for core particle) sucrose gradient (containing 10 mM Tris-HCl [pH 7.5], 1 mM EDTA, 10 mM NaCl, and 0.1 mM PMSF). The gradients were centrifuged at 28,000 rpm (Beckman SW28 rotor) for 20 h at 4°C. The gradients were fractionated into 0.5-ml fractions, and the DNA content of each fraction was analyzed by agarose gel electrophoresis. The size fractions of interest were pooled, spin dialyzed into a buffer containing 10 mM Tris-HCl (pH 7.4), 10 mM NaCl, and 1 mM EDTA, and concentrated to 1.0 μg/μl. The integrity of the samples was verified by MNase digestion to yield the nucleosomal repeat ladder. Briefly, the oligonucleosomes were digested with various concentrations of MNase for 2 min at room temperature, the reaction was stopped by addition of 2 volumes of MNase (20 mM EDTA, 2% SDS), and each sample was phenol-chloroform extracted twice and ethanol precipitated. The resulting DNA mixtures were then resolved on a 1.0% agarose gel in 0.5× Tris-borate-EDTA (TBE), and the bands were visualized by ethidium bromide staining. The integrity of the protein content of the oligonucleosomes was verified by examination of the samples after SDS-PAGE.
Treatment of oligonucleosomes.
In the oligonucleosome compaction studies, the nucleosome cores or oligonucleosomes were incubated in 2 mM MgCl2 for 30 min at 4°C, and the acetylation assays were performed as described above except that they were performed in buffer A with 2 mM MgCl2 (and 50 mM NaCl was excluded). Oligonucleosomes were reconstituted with linker histones (H1, H5, GH1, or GH5) in buffer A containing 50 mM NaCl and allowed to equilibrate at 20°C for 30 min. MNase digestions were performed as described previously (20) except in the presence of 50 mM NaCl. For MNase digestion prior to acetylation reaction, the digests were performed as described above and stopped with the addition of EDTA and EGTA to 3 and 5 mM, respectively. The digestion reactions were then diluted twofold into 50 mM NaCl with 2× buffer A (containing 36 μM [1-14C]acetyl-CoA), and the acetylation assay was initiated by addition of either rPCAF or cPCAF. Reactions were terminated and analyzed as described above. Chromatosome stop assays were performed on oligonucleosomes reconstituted with linker histones (or globular domains). Briefly, the reconstitutes were digested with MNase (room temperature, 2 min), and the digestion was stopped by addition of 2 volumes of MNase. The mixture was then phenol-chloroform extracted and ethanol precipitated, and the resulting DNA was resolved on a 5% polyacrylamide gel in 0.5× TBE; the bands were then visualized by staining with ethidium bromide.
RESULTS AND DISCUSSION
The linker histones H1 and H5 specifically inhibit acetylation of H3 in oligonucleosomes.
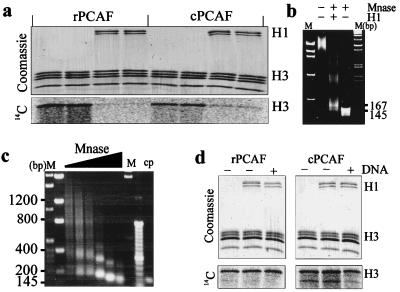
In the nucleus of the cell, the transcription factor PCAF is associated with several proteins in a multiprotein complex, which efficiently acetylates purified chromatin subunits (32). We wished to examine whether either rPCAF (60) or cPCAF (32) could acetylate either H1-depleted or H1-containing chromatin. We purified H1-depleted oligonucleosome arrays (8 to 12-mers) (Fig. 1b) and verified the integrity of the oligonucleosomes by examining the time course of MNase digestion (Fig. 1c). Figure 1c shows that the purified oligonucleosomes exhibit a characteristic (41, 45, 47) nucleosomal repeat of 187 ± 15 bp. We then compared the abilities of these HATs to acetylate the histones in these oligonucleosomes in the presence or absence of added linker histone H1. To ensure proper binding of H1, we examined the reconstituted templates for the appearance of the “chromatosome stop” (44). Figure 1b shows the MNase digestion of the reconstitutes and the appearance of the 167-bp chromatosome stop indicating that H1 was properly bound. As shown in Fig. 1a, addition of 1.4 mol of histone H1 per mol of nucleosome reduced the H3 acetylation by either rPCAF or cPCAF by 90 or 70%, respectively. We specifically tested and found that the reduction was not due to an H1-induced precipitation of the chromatin substrate (not shown). In these assays, histone H1 incorporated no counts, indicating that it is not a substrate and competing for H3 acetylation. The lack of acetylation of H1 is in complete agreement with our previous finding (18), which demonstrated that although histone H1 is an excellent substrate for rPCAF in vitro, it could not function as a substrate when bound to nucleosomes. Significantly, addition of histone H1 to a mixture of free histones, or to a mixture containing free histones and 2,000-bp-long DNA, did not affect the efficiency of H3 acetylation (Fig. 1d). Thus, histone H1 is not a nonspecific inhibitor of HAT activity. We conclude that histone H1 inhibited acetylation specifically, only in the context of chromatin.
FIG. 1.
The linker histone H1 inhibits the acetylation of oligonucleosomes. (a) Oligonucleosomes were acetylated with either rPCAF or cPCAF (top panel, Coomassie blue-stained gel; lower panel, PhosphorImager scan). Note that the addition of H1 inhibits the acetylation of H3. (b) Mnase digestion of the oligonucleosomes devoid of (−) or containing (+) histone H1. The presence of the 167-bp chromatosome stop is indicative of proper H1 placement in chromatin. Lanes: M, molecular weight markers; −, undigested control. (c) MNase digestion of the oligonucleosomes. Lane M, molecular weight standards in base pairs; lane cp, core particles. The nucleosomal repeat length was determined to be 187 ± 15 bp. (d) Mix of individually purified core histones acetylated in the presence or absence of H1 or in the presence of H1 plus DNA (0.2 μg/μl) (top, Coomassie blue-stained gel; bottom, PhosphorImager scan). Note that the presence of H1 did not inhibit the acetylation of free histones or of the histone-DNA mixtures.
rPCAF and cPCAF exhibit different patterns of inhibition as a function of H1.
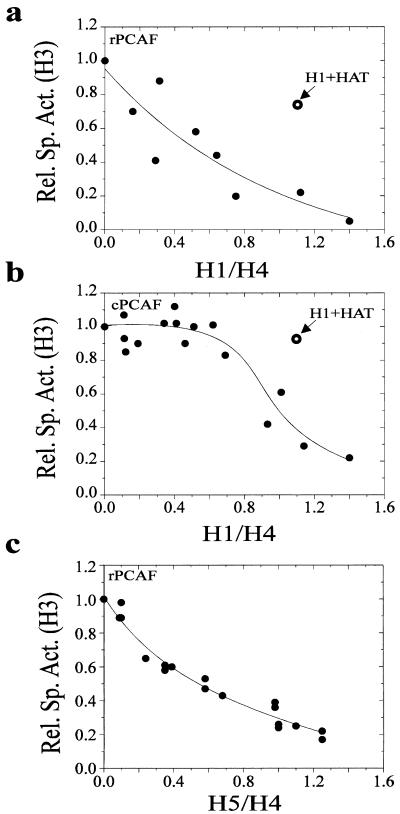
Histone H1 inhibits the activity of both rPCAF and cPCAF in a dose-dependent manner; however, the dose dependency differs significantly between the two types of HATs (Fig. 2). The dose response for rPCAF is linear: an incremental increase in H1 results in a corresponding decrease in acetylation (Fig. 2a). To test whether this was a general effect of linker histone binding, we also tested whether linker histone H5, the avian analog of H1°, could also inhibit rPCAF in a dose-dependent manner (Fig. 2c). Titration with H5 revealed that the pattern of inhibition was indistinguishable from that observed with H1, suggesting that the inhibition of acetylation of the H3 tail in chromatin is a general property of linker histone binding. In contrast, the dose response of H1 inhibition of cPCAF is sigmoidal, with cPCAF inhibited only at relatively high concentrations of H1 (Fig. 2b).
FIG. 2.
The linker histones H1 and H5 inhibit the acetylation of oligonucleosomes in a dose-dependent manner. Oligonucleosomes were reconstituted with varied amounts of H1 or H5 and then acetylated with either rPCAF or cPCAF, as indicated. The relative specific activity (Rel. Sp. Act.) of H3 was determined for each point in the titration and plotted as a function of the H1/H4 ratio (determined from the Coomassie blue-stained gels). Each graph is a composite of at least three independent titrations. The open symbols labeled H1+HAT show the level of acetylation when the acetylases and H1 were added at the same time.
These results suggest differences in the ability of rPCAF and cPCAF to overcome the linker histone-induced repression. Since it has been demonstrated that H1 binds noncooperatively to polynucleosomes (30), we suggest that rPCAF inefficiently acetylates the H3 in a nucleosome containing either H1 or H5 but can still acetylate the neighboring nucleosomes that are devoid of linker histone. In contrast, cPCAF can overcome the presence of H1, so long as the concentration of H1 is not saturating. Indeed, results of competition experiments, in which the acetylases and inhibitory amounts of histone H1 were simultaneously added to the oligonucleosomes, provide additional support for this notion. In these experiments, rPCAF was only slightly (25%) inhibited, and cPCAF was not inhibited at all (Fig. 2a and b). The inhibition of rPCAF is due to competition between H1 and rPCAF for access to the H3 tail. The nucleosomes that bound H1 before they were accessed by rPCAF are refractory to acetylation. In contrast, cPCAF was not inhibited because the enzyme complex was able to bind to oligonucleosomes so long as not all nucleosomes within the array were occupied by H1. We conclude that rPCAF cannot overcome linker histone repression; however, when PCAF is complexed with accessory proteins in a multiprotein complex, it can overcome this repression, provided that a nucleosomal array is not fully saturated. This finding suggests that a function of the accessory proteins may involve overcoming the H1-mediated repression of acetylation.
These data suggest that the H1-mediated inhibition of acetylation is different for rPCAF and cPCAF. rPCAF is simply competing with H1 for access to the individual nucleosome within the array. In contrast, the H1-mediated repression of cPCAF may be mediated by a more global feature of the nucleosomal array, perhaps H1-mediated condensation. Alternatively, it has been demonstrated that more than one H1 can associate per nucleosome within an array (8, 30). It was shown that nucleosomes contain two binding sites for H1, a low-affinity site and a high-affinity site (30). Perhaps the inhibition of cPCAF at high levels of H1 is mediated by the binding of additional H1 molecules per nucleosome.
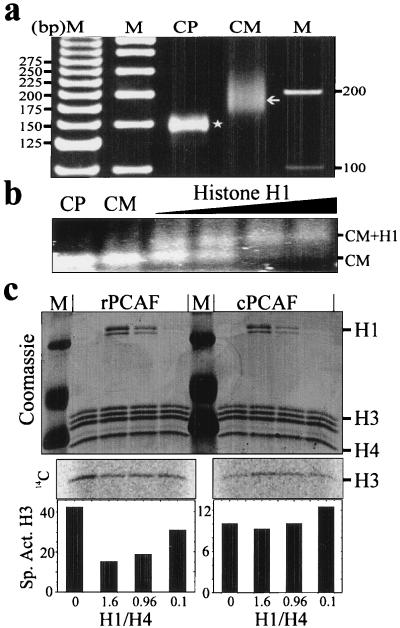
These data show that cPCAF can overcome the H1-mediated inhibition of acetylation, providing that the template is not fully saturated. These results present the interesting possibility that while rPCAF is competing with H1 for the individual nucleosomes within the array; cPCAF is competing for the array. In other word, if cPCAF binds to a nucleosome within the array, it can acetylate and overcome H1 binding in the entire oligonucleosome. To test this quasi-processive mechanism for cPCAF, we examined the ability of H1 to inhibit the acetylation of H3 in chromatin subunits containing linker DNA. We purified these subunits from chicken erythrocyte nuclei and stripped them of endogenous H1. The DNA purified from the H1-stripped chromatin subunit (CM) preparation had an average length of 185 bp of DNA and contained no core particle (Fig. 3a). We then reconstituted the purified CMs with H1 and examined the ability of H1 to inhibit the acetylation by either rPCAF or cPCAF. Figure 3c shows that reconstitution of H1 onto the CMs inhibits the acetylation of H3 by rPCAF, albeit to a reduced extent (60%) compared to that same ratio of H1 on oligonucleosomes (Fig. 2). Interestingly, linker histone H1 did not inhibit the acetylation by cPCAF at any concentration tested. Since the H1-mediated inhibition was either abolished or diminished in assays using CMs, we used a gel mobility shift assay to determine if H1 could bind to the purified CMs. Figure 3b show the results of the gel shift assay performed at ratios of H1 to nucleosome similar to those used in the acetylation experiment. The appearance of the shifted band (CM+H1 in Fig. 3b) indicates that the particles bound H1. These results indicate that cPCAF can overcome the inhibitory effect of H1 and acetylate the template. We conclude that the ability of cPCAF to overcome H1-mediated inhibition does not arise from a processive mechanism. These results suggest that cPCAF can overcome the presence of H1 and that the inhibition observed at saturating concentrations of H1 may arise from a structural feature of the H1 condensed chromatin array.
FIG. 3.
The H3 tails in chromatosomes can be acetylated by cPCAF. Chromatin subunits containing linker DNA were prepared from purified chicken erythrocyte nuclei and stripped of linker histones (designated CM). (a) Length of the DNA obtained from the CM preparation. Lanes: M, molecular weight markers; CP, DNA obtained from purified core particles (145 bp, designated with a star); CM, DNA obtained from the CM preparation (average of 185 bp, designated with an arrow). (b) Gel shift assay performed in the acetylation buffer and resolved on a 0.9% agarose gel in 0.5× TBE. CP, core particle; CM, H1-stripped chromatin subunit preparation; CM+H1, position of the H1-shifted CM. (c) Acetylation assay performed after reconstitution of the CMs with increasing amounts of H1. M, molecular weight markers. Positions of H1 and H3 are designated at the right. The middle section in panel c shows the PhosphorImager scan of the Coomassie-stained gels and represents the incorporation of [14C]acetate into H3. The bottom section shows the calculated specific activity for H3 for each lane in the gel above. Note that rPCAF exhibits a concentration dependence for added H1 whereas the activity of cPCAF is unaffected by the addition of H1.
Linker histone-dependent inhibition of H3 acetylation is not due to chromatin condensation.
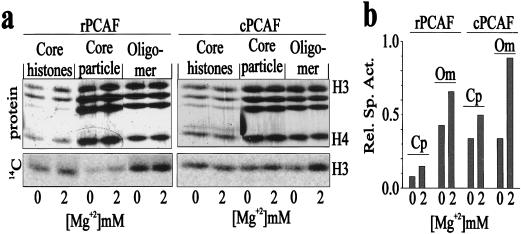
The linker histone H1 binds to nucleosomes and compacts the structure of the chromatin fiber (54). These effects might be especially significant at high concentrations of H1 where the fiber is completely condensed (16) and where more than one H1 molecule can bind per nucleosome (8, 30). Therefore, the inhibition of acetylation by H1 could arise either from a structural change in the histone tails associated with condensation or from steric occlusion arising from either direct interactions of the H3 tail with H1 or an H1-induced conformational change in the H3 tail. To test whether the H1-dependent inhibition of acetylation was due to chromatin compaction, we examined the ability of rPCAF and cPCAF to acetylate oligonucleosomal arrays in the presence of 2 mM Mg2+ ions, conditions which are known to favor condensation of oligonucleosomal arrays (40, 43). Figure 4 shows that Mg2+ ion-dependent condensation of the oligonucleosomes does not inhibit acetylation by either rPCAF or cPCAF. On the contrary, condensation of chromatin with Mg2+ ions results in a stimulation of the activity of both rPCAF and cPCAF for both nucleosome cores and oligonucleosomes but not for a mixture of core histones (Fig. 4). The specific activity of the H3 extracted from the mono- or oligonucleosomes was normalized to that obtained with free histones. In all cases, the specific activity of H3 extracted from oligonucleosomes was significantly higher than that of the H3 extracted from core particles (Fig. 4b). These results indicate that both rPCAF and cPCAF acetylate H3 tails in oligonucleosomes more efficiently than the H3 tails in core particles. Furthermore, while addition of Mg2+ ions did not affect the acetylation of free, uncomplexed histones, the ions did elevate the specific activity of H3 in both core particles and oligonucleosomes. These results indicate that the Mg2+ ion-dependent stimulation is due to changes in the substrate and not to effects on the enzymatic activity of the HATs.
FIG. 4.
Chromatin compaction does not inhibit acetylation of oligonucleosomes by rPCAF or cPCAF. (a) Acetylation of either a mixture of purified core histones, core particles, or oligonucleosome (oligomer) in the presence or absence of 2 mM Mg2+. (b) Relative specific activity (Rel. Sp. Act.), at either 0 or 2 mM Mg2+, of H3 in each of the lanes in panel a, normalized to acetylation of histone H3 in a mix of core histones (core histones). Cp, core particle; Om, oligonucleosome. Note that the condensed oligonucleosomes in 2 mM Mg2+ were very efficiently acetylated by both rPCAF and cPCAF.
These finding are in agreement with a recent report that the ability of GCN5, a close homolog of PCAF, to acetylate histones in chromatin is also stimulated by the addition of Mg2+ ions (51). These results indicate that these HATs prefer condensed chromatin as a substrate. We conclude therefore that both rPCAF and cPCAF readily acetylate condensed chromatin and therefore the inhibitory effects of H1 are not due solely to the induction of a higher-order, more compact chromatin structure.
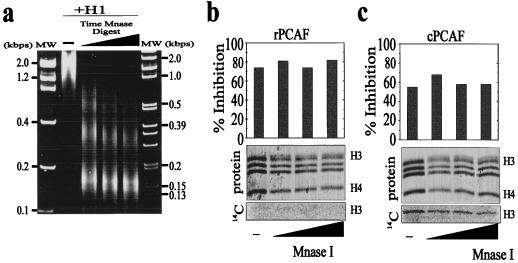
Although Mg2+ ion-induced condensation of chromatin results in a structure that is hydrodynamically similar to an H1-mediated structure (8, 13, 17), the H1-mediated condensed structure must present distinct topological features. We therefore examined the acetylation of H1-reconstituted oligonucleosomes as a function of MNase digestion (Fig. 5). During this digestion, the H1-containing oligonucleosomes are gradually converted to chromatosomes, thereby eliminating any consideration of higher-order chromatin structure. Oligonucleosomes were reconstituted with sufficient linker histone H1 to result in 80 and 60% inhibition of acetylation by rPCAF and cPCAF, respectively. The reconstituted structures were then subjected to a time course of MNase digestion (Fig. 5a represents the products of the MNase digestion prior to acetylation), and each time point was analyzed for the ability of rPCAF and cPCAF to acetylate the mixtures. The inhibitory effects of H1 were not relieved by digestion to chromatosomes, and H3 did not incorporate any additional counts (Fig. 5b and c). Thus, the H1-dependent inhibition of acetylation cannot be due solely to the formation of a higher-order, condensed chromatin structure. Together, these results indicate that H1 inhibits the acetylation of H3 in chromatin by sterically hindering access to the H3 tail. Further, these results combined with those in Fig. 3 indicate that the binding of H1 to oligonucleosomes results in a subunit conformation that is distinct from that of H1 reconstituted onto a purified chromatosomes. In other words, H1 binding to oligonucleosomes forms a stable conformer, and this conformation is maintained when digested to chromatosome, while reconstitution onto chromatosomes previously stripped of H1 results in a conformation that is not repressive to acetylation by the PCAF complex.
FIG. 5.
The H1-mediated inhibition of H3 acetylation is not due to formation of higher-order chromatin structure. MNase digestion of oligonucleosomes to mononucleosomes does not relieve the H1-dependent inhibition of rPCAF or cPCAF. Oligonucleosomes reconstituted with H1 were subjected to time course of digestion with MNase (a; −, no digestion). The MNase concentration and time of digestion was adjusted to yield about 80% monomer at the last point. The digests were stopped, and then the mixture was acetylated with either rPCAF or cPCAF. The bar graphs indicate the percent inhibition relative to the undigested control in the absence of linker histone H1. In the Coomassie blue-stained gels (b and c), the band above H3 is MNase. The lower panel shows the incorporation of [14C]acetate into H3.
The globular domains of H1 and H5 are poor inhibitors of acetylation.
Linker histones are a family of chromatin-associated proteins with evolutionarily conserved sequence and structure (54). They have a tripartite structure composed of highly charged N- and C-terminal tails and a conserved central globular domain (1, 7). The purified globular domains bind to nucleosomes near the dyad axis and interact with two gyres of the nucleosomal DNA in a manner similar to that observed for the full-length protein (1, 50). However, since the globular domain lacks both the C- and N-terminal tails, the binding of this domain does not induce chromatin condensation (2).
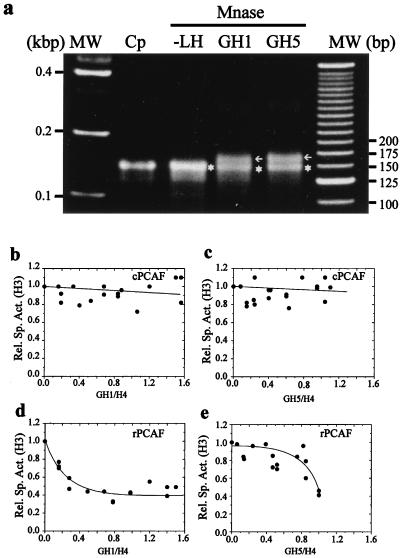
To further examine the nature of the linker histone-induced inhibition of acetylation, we tested the ability of the purified GH1 and GH5 to inhibit the acetylation by either cPCAF or rPCAF. Chromatosome stop assays (Fig. 6a), which are characteristic for proper placement of GH1 and GH5 in nucleosomes (44), confirmed that both GH1 and GH5 were properly bound to the nucleosomes.
FIG. 6.
GH1 and GH5 are poor inhibitors of acetylation. The oligonucleosomes were reconstituted with either GH1 or GH5. Proper binding of the globular domains was verified by the chromatosome stop assay (a) Lanes: MW, DNA molecular weight markers; Cp, DNA purified from chicken core particles; −LH, DNA purified after digestion of the oligonucleosomes in the absence of linker histones; GH1, DNA purified after digestions of oligonucleosomes in the presence of GH1 (GH1:H4 = 1.2:1); GH5, DNA purified after digestion of oligonucleosomes in the presence of GH5 (GH5:H4 = 1.2:1). Arrows indicate positions of the MNase-protected chromatosome stop DNA; stars indicate positions of the core particle DNA. (b to e) Oligonucleosomes reconstituted with different amounts of GH1 or GH5 were acetylated with either cPCAF (b and c) or rPCAF (d and e). The specific activity of H3 normalized to that in the absence of any added globular domain (Rel. Sp. Act.) was plotted as a function of the GH1/H4 or GH5/H4 ratio (determined from the Coomassie blue-stained gel of each reaction). Each graph represents a composite of at least two independent titrations.
Next we tested the ability of the globular domains to sterically block the PCAF-mediated acetylation of the H3 tails in oligonucleosomes. We reconstituted the oligonucleosomes with increasing concentrations of either GH1 or GH5 and examined the ability of either cPCAF or rPCAF to acetylate the H3 tails. The results (Fig. 6b and c) demonstrate that neither GH5 nor GH1 is capable of inhibiting acetylation of cPCAF at any concentration tested. Comparison of these results to those observed for the full-length H1 (Fig. 2b) shows that while H1/H4 ratios of 1.2 resulted in a greater than 80% reduction in H3 acetylation, the same or greater ratio (up to 1.6) of GH1 to H4 had no effect on H3 acetylation. Likewise, the rPCAF-mediated acetylation was inhibited by either GH1 or GH5 (Fig. 6d and e) to a lesser extent than that observed for the full-length proteins (Fig. 2a and c). Thus, while increased ratios of H1 resulted in a gradual decrease in H3 acetylation, leading to complete inhibition of acetylation (Fig. 2a), a GH1/H4 ratio as high as 1.6 resulted in no greater than a 60% inhibition of H3 acetylation (Fig. 6d). Similarly, GH5 was a much poorer inhibitor of rPCAF than the full-length H5 (compare Fig. 2c to Fig. 6e). Taken together, these data indicate that the inhibition of acetylation by the linker histones is steric in nature and largely mediated by the linker histone tails. We note, however, that the globular domains alone partially inhibit the acetylation activity of rPCAF but not that of cPCAF.
The slight differences in the abilities of GH1 and GH5 to inhibit the rPCAF-mediated acetylation may reflect differences in their specific interactions with nucleosomes. Indeed, previous studies of GH1 and GH5 have indicated differences both in their binding to DNA (49) and in their ability to self-associate (26). In addition, prior studies have noted distinct conformations for H1 and H5 (9) that could reflect some differences in their specific contacts with histones in the nucleosome octamer. These differences could account for the observed differences in their ability to inhibit acetylation.
All of our results indicate that the binding of linker histones to nucleosomes sterically hinders access of the H3 tails to rPCAF and that this steric occlusion occurs at the level of the individual nucleosomes within the array. These conclusions are supported by our findings that linker histones inhibit the rPCAF-mediated acetylation on both oligomers and chromatosomes. Furthermore, this steric occlusion is mediated by both the globular domain and the linker histone tails. The partial inhibition observed with the globular domains, in conjunction with the stimulation of acetylation observed by magnesium-induced condensation of the oligonucleosomes, clearly indicates that the inhibition cannot be due solely to the H1-mediated condensation. Taken together, these results strongly indicate that the inhibition of rPCAF is mediated by steric occlusion of the H3 tail by the linker histone. Further studies now under way using truncation mutants of the linker histones will allow for a more detailed understanding of the mechanism by which linker histones inhibit rPCAF.
In contrast, cPCAF is capable of overcoming the steric effect of H1, perhaps by altering the organization of H1 in chromatin. Our results show that acetylation of oligonucleosomes by cPCAF is not inhibited by subsaturating concentrations of H1 or by saturating concentrations of the globular domains. Furthermore, the acetylation of chromatosomes by cPCAF is not inhibited by H1. These results indicate that the complex can overcome the steric effect of linker histones at the level of the individual nucleosome. Thus, it seems that the PCAF complex contains a factor(s) that is capable of reorganizing the H1-containing nucleosomes, thereby allowing access of PCAF to the H3 tail. Like rPCAF, the acetylation activity of cPCAF was not inhibited by magnesium-induced condensation, indicating that the enzyme in complex is not inhibited by condensation of the oligonucleosomes. However, high concentrations of H1 do inhibit the acetylation activity of cPCAF. This inhibition may be due to the binding of more than one H1 molecule per nucleosome in the array (8, 30). Alternatively, the inhibition could result from a structural feature of the fully condensed H1-containing oligonucleosomes that is distinct from that of Mg2+ condensed chromatin. The PCAF complex contains numerous polypeptides (32); purification, identification, and reconstitution of factors within the complex will lead to a more thorough understanding of the mechanism whereby this and other nuclear complexes that target nucleosomes overcome the repressive nature of linker histone H1.
In summary, our findings suggest that efficient acetylation requires changes in the organization of H1 on chromatin and that some members of cPCAF may act to modify the organization of H1. Indeed, others have shown that although H1 is present in actively transcribed regions, it exists in an altered conformation (42). Since GCN5 targets the same acetylation sites as PCAF (22, 38, 51), it is likely that it too will be inhibited by H1. We have recently observed that H1 also inhibits the ability of p300 to acetylate histones in oligonucleosomes (unpublished data). We suggest, therefore, that changes in the chromatin organization of H1 may be a general prerequisite, necessary to allow access to nucleosomes for various regulatory factors that affect the structure and regulate the function of chromatin.
ACKNOWLEDGMENTS
We thank Y. Postinikov, J. Wagner, C. Laufer, M. Bergel, H. Shirakawa, and M. Prymakowska-Bosak for helpful discussions. We also thank J. Allan (Edinburgh University) for providing the globular domains of H1 and H5.
REFERENCES
- 1.Allan J, Hartman P G, Crane-Robinson C, Aviles F X. The structure of histone H1 and its location in chromatin. Nature. 1980;288:675–679. doi: 10.1038/288675a0. [DOI] [PubMed] [Google Scholar]
- 2.Allan J, Mitchel T, Harborne N, Bohm L, Crane-Robinson C. Roles of H1 domains in determining higher order chromatin structure and H1 location. J Mol Biol. 1986;187:591–601. doi: 10.1016/0022-2836(86)90337-2. [DOI] [PubMed] [Google Scholar]
- 3.Ausio J, Dong F, van Holde K E. Use of selectively trypsinized nucleosome core particles to analyze the role of histone “tails” in the stabilization of nucleosomes. J Mol Biol. 1989;206:451–463. doi: 10.1016/0022-2836(89)90493-2. [DOI] [PubMed] [Google Scholar]
- 4.Bresnick E H, Bustin M, Marsaud V, Richard-Foy H, Hager G L. The transcriptionally-active MMTV promoter is depleted of histone H1. Nucleic Acids Res. 1992;20:273–278. doi: 10.1093/nar/20.2.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brownell J E, Allis C D. An activity gel assay detects a single, catalytically active histone acetyltransferase subunit in Tetrahymena macronuclei. Proc Natl Acad Sci USA. 1995;92:6364–6368. doi: 10.1073/pnas.92.14.6364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bustin M. Arrangement of histones in chromatin. Nat New Biol. 1973;245:207–209. doi: 10.1038/newbio245207a0. [DOI] [PubMed] [Google Scholar]
- 7.Bustin M, Cole R D. Bisection of a lysine-rich histone by N-bromosuccinimide. J Biol Chem. 1969;244:5291–5294. [PubMed] [Google Scholar]
- 8.Carruthers L M, Bednar J, Woodcock C L, Hansen J C. Linker histones stabilize the intrinsic salt-dependent folding of nucleosomal arrays: mechanistic ramifications for higher-order chromatin folding. Biochemistry. 1998;37:14776–14787. doi: 10.1021/bi981684e. [DOI] [PubMed] [Google Scholar]
- 9.Cary P D, Hines M L, Bradbury E M, Smith B J, Johns E W. Conformational studies of histone H1° in comparison with histones H1 and H5. Eur J Biochem. 1981;120:371–377. doi: 10.1111/j.1432-1033.1981.tb05714.x. [DOI] [PubMed] [Google Scholar]
- 10.Delcuve G P, Davie J R. Chromatin structure of erythroid-specific genes of immature and mature chicken erythrocytes. Biochem J. 1989;263:179–186. doi: 10.1042/bj2630179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ericsson C, Grossbach U, Bjorkroth B, Daneholt B. Presence of histone H1 on an active Balbiani ring gene. Cell. 1990;60:73–83. doi: 10.1016/0092-8674(90)90717-s. [DOI] [PubMed] [Google Scholar]
- 12.Fletcher T M, Hansen J C. Core histone tail domains mediate oligonucleosome folding and nucleosomal DNA organization through distinct molecular mechanisms. J Biol Chem. 1995;270:25359–25362. doi: 10.1074/jbc.270.43.25359. [DOI] [PubMed] [Google Scholar]
- 13.Fletcher T M, Hansen J C. The nucleosomal array: structure/function relationships. Crit Rev Eukaryotic Gene Expr. 1996;6:149–188. doi: 10.1615/critreveukargeneexpr.v6.i2-3.40. [DOI] [PubMed] [Google Scholar]
- 14.Fryer C J, Archer T K. Chromatin remodelling by the glucocorticoid receptor require the BRG1 complex. Nature. 1998;393:88–91. doi: 10.1038/30032. [DOI] [PubMed] [Google Scholar]
- 15.Grant P A, Duggan L, Cote J, Roberts S M, Brownell J, Candau R, Ohba R, Owen-Hughes T, Allis C D, Winston F, Berger S L, Workman J L. yGCN5 function within multisubunit ADA and SPT/ADA adapter complexes to acetylate nucleosomal histones. Genes Dev. 1997;11:1640–1650. doi: 10.1101/gad.11.13.1640. [DOI] [PubMed] [Google Scholar]
- 16.Graziano V, Gerchman S E, Ramakeichnan V. Reconstitution of chromatin higher-order structure from histone H5 and depleted chromatin. J Mol Biol. 1988;203:997–1007. doi: 10.1016/0022-2836(88)90124-6. [DOI] [PubMed] [Google Scholar]
- 17.Hansen J, Tse C, Wolffe A P. Structure and function of core histone N-termini: more than meets the eye. Biochemistry. 1998;37:17637–17641. doi: 10.1021/bi982409v. [DOI] [PubMed] [Google Scholar]
- 18.Herrera J E, Bergel M, Yang X J, Nakatani Y, Bustin M. The histone acetyltransferase activity of human GCN5 and PCAF is stabilized by coenzymes. J Biol Chem. 1997;272:27253–27258. doi: 10.1074/jbc.272.43.27253. [DOI] [PubMed] [Google Scholar]
- 19.Juan L J, Utley R T, Adams C C, Vetesse-Dadey M, Workman J L. Differential repression of transcription factor binding by histone H1 is regulated by the core histone amino termini. EMBO J. 1994;13:6031–6040. doi: 10.1002/j.1460-2075.1994.tb06949.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kornberg R D, LaPointe J W, Lorch Y. Preparation of nucleosomes and chromatin. Methods Enzymol. 1989;170:3–14. doi: 10.1016/0076-6879(89)70039-2. [DOI] [PubMed] [Google Scholar]
- 21.Kuo M H, Allis C D. Roles of histone acetyltransferases and deacetylases in gene regulation. Bioessays. 1998;20:615–626. doi: 10.1002/(SICI)1521-1878(199808)20:8<615::AID-BIES4>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 22.Kuo M H, Brownell J E, Sobel R E, Ranalli T A, Cook R G, Edmondson D G, Roth S Y, Allis C D. Transcription-linked acetylation by Gcn5p of histones H3 and H4 at specific lysines. Nature. 1996;383:269–272. doi: 10.1038/383269a0. [DOI] [PubMed] [Google Scholar]
- 23.Kuo M H, Zhou J, Jambeck P, Churchill M E, Allis C D. Histone acetyltransferase activity of yeast Gcn5p is required for the activation of target genes in vivo. Genes Dev. 1998;12:627–639. doi: 10.1101/gad.12.5.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Laybourn P J, Kadonaga J T. Role of nucleosomal cores and histone H1 in regulation of transcription by RNA polymerase II. Science. 1991;254:238–245. doi: 10.1126/science.254.5029.238. [DOI] [PubMed] [Google Scholar]
- 25.Lee H L, Archer T K. Prolonged glutocorticoid exposure dephosphorylates histone H1 and inactivates the MMTV promoter. EMBO J. 1998;17:1454–1466. doi: 10.1093/emboj/17.5.1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maman J D, Yager T D, Allan J. Self-association of the globular domain of histone H5. Biochemistry. 1994;33:1300–1310. doi: 10.1021/bi00172a003. [DOI] [PubMed] [Google Scholar]
- 27.Mizzen C A, Brownell J E, Cook R G, Allis C D. Histone acetyltransferase: preparation of substrates and assay procedures. Methods Enzymol. 1999;304:675–696. doi: 10.1016/s0076-6879(99)04041-0. [DOI] [PubMed] [Google Scholar]
- 28.Mizzen C A, Allis C D. Linking histone acetylation to transcriptional regulation. Cell Mol Life Sci. 1998;54:6–20. doi: 10.1007/s000180050121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nagpal S, Ghosn C, Disepio D, Molina Y, Sutter M, Klein E S, Chandraratna R A. Retinoid-dependent recruitment of a histone H1 displacement activity by retinoic acid receptor. J Biol Chem. 1999;274:22563–22568. doi: 10.1074/jbc.274.32.22563. [DOI] [PubMed] [Google Scholar]
- 30.Nelson P P, Albright S C, Wiseman J M, Garrard W T. Reassociation of histone H1 with nucleosomes. J Biol Chem. 1979;254:11751–11760. [PubMed] [Google Scholar]
- 31.Nickel B E, Allis C D, Davie J R. Ubiquitinated histone H2B is preferentially located in transcriptionally active chromatin. Biochemistry. 1989;28:958–963. doi: 10.1021/bi00429a006. [DOI] [PubMed] [Google Scholar]
- 32.Ogryzko V V, Kotani T, Zhang X, Schlitz R L, Howard T, Yang X J, Howard B H, Qin J, Nakatani Y. Histone-like TAFs within the PCAF histone acetylase complex. Cell. 1998;94:35–44. doi: 10.1016/s0092-8674(00)81219-2. [DOI] [PubMed] [Google Scholar]
- 33.Paranjape S M, Kamakaka R T, Kadonaga J T. Role of chromatin structure in the regulation of transcription by RNA polymerase II. Annu Rev Biochem. 1994;63:265–297. doi: 10.1146/annurev.bi.63.070194.001405. [DOI] [PubMed] [Google Scholar]
- 34.Perry C A, Annunziato A T. Histone acetylation reduces H1-mediated nucleosome interactions during chromatin assembly. Exp Cell Res. 1991;196:337–345. doi: 10.1016/0014-4827(91)90269-z. [DOI] [PubMed] [Google Scholar]
- 35.Perry C A, Annunziato A T. Influence of histone acetylation on the solubility, H1 content and DNase I sensitivity of newly assembled chromatin. Nucleic Acids Res. 1989;17:4275–4291. doi: 10.1093/nar/17.11.4275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ridsdale J A, Hendzel M J, Delcuve G P, Davie J R. Histone acetylation alters the capacity of the H1 histones to condense transcriptionally active/competent chromatin. J Biol Chem. 1990;265:5150–5156. [PubMed] [Google Scholar]
- 37.Ridsdale J A, Davie J R. Chicken erythrocyte polynucleosomes which are soluble at physiological ionic strength and contain linker histones are highly enriched in beta-globin gene sequences. Nucleic Acids Res. 1987;15:1081–1096. doi: 10.1093/nar/15.3.1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schiltz R L, Mizzen C A, Vassilev A, Cook R G, Allis C D, Nakatani Y. Overlapping but distinct patterns of histone acetylation by the human coactivators p300 and PCAF within nucleosomal substrates. J Biol Chem. 1999;274:1189–1192. doi: 10.1074/jbc.274.3.1189. [DOI] [PubMed] [Google Scholar]
- 39.Schultz T F, Spiker S, Quatrano R S. Histone H1 enhances the DNA binding activity of the transcription factor EmBP-1. J Biol Chem. 1996;271:25742–25745. doi: 10.1074/jbc.271.42.25742. [DOI] [PubMed] [Google Scholar]
- 40.Schwarz P M, Hansen J C. Formation and stability of higher order chromatin structures. Contributions of the histone octamer. J Biol Chem. 1994;269:16284–16289. [PubMed] [Google Scholar]
- 41.Shaw B R, Herman T M, Kovacic R T, Beaudreau G S, Van Holde K E. Analysis of subunit organization in chicken erythrocyte chromatin. Proc Natl Acad Sci USA. 1976;73:505–509. doi: 10.1073/pnas.73.2.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shick V V, Belyavsky A V, Mirzabekov A D. Primary organization of nucleosomes. J Mol Biol. 1985;185:329–339. doi: 10.1016/0022-2836(85)90407-3. [DOI] [PubMed] [Google Scholar]
- 43.Shwarz P M, Felthauser A, Fletcher T M, Hansen J C. Reversible oligonucleosome self-association: dependence on divalent cations and core histone tail domains. Biochemistry. 1996;35:4009–4015. doi: 10.1021/bi9525684. [DOI] [PubMed] [Google Scholar]
- 44.Simpson R T. Structure of the chromatosome, a chromatin particle containing 160 base pairs of DNA and all the histones. Biochemistry. 1978;17:5524–5531. doi: 10.1021/bi00618a030. [DOI] [PubMed] [Google Scholar]
- 45.Sollner-Web B, Felsenfeld G. A comparison of the digestion of nuclei chromatin by staphlococcal nuclease. Biochemistry. 1975;14:2915–2920. doi: 10.1021/bi00684a019. [DOI] [PubMed] [Google Scholar]
- 46.Struhl K. Histone acetylation and transcriptional regulatory mechanisms. Genes Dev. 1998;12:599–606. doi: 10.1101/gad.12.5.599. [DOI] [PubMed] [Google Scholar]
- 47.Sun X L, Xu Y Z, Bellard M, Chambon P. Digestion of the chicken beta-globin gene chromatin with micrococcal nuclease reveals the presence of an altered nucleosomal array characterized by an atypical ladder of DNA fragments. EMBO J. 1986;5:293–300. doi: 10.1002/j.1460-2075.1986.tb04212.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Syntichaki P, Thireos G. The Gcn5.Ada complex potentiates the histone acetyltransferase activity of Gcn5. J Biol Chem. 1998;273:24414–24419. doi: 10.1074/jbc.273.38.24414. [DOI] [PubMed] [Google Scholar]
- 49.Thomas J O, Rees C, Finch J T. Cooperative binding of the globular domains of histones H1 and H5 to DNA. Nucleic Acids Res. 1992;20:187–194. doi: 10.1093/nar/20.2.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Travers A. The location of the linker histone on the nucleosome. Trends Biochem Sci. 1999;24:4–7. doi: 10.1016/s0968-0004(98)01339-5. [DOI] [PubMed] [Google Scholar]
- 51.Tse C, Georgieva E I, Ruiz-Garcia A B, Sendra R, Hansen J C. Gcn5p, a transcription-related histone acetyltransferase, acetylates nucleosomes and folded nucleosomal arrays in the absence of other protein subunits. J Biol Chem. 1998;273:32388–32392. doi: 10.1074/jbc.273.49.32388. [DOI] [PubMed] [Google Scholar]
- 52.Turner B M, O'Neill L P. Histone acetylation in chromatin and chromosomes. Semin Cell Biol. 1995;6:229–236. doi: 10.1006/scel.1995.0031. [DOI] [PubMed] [Google Scholar]
- 53.Ura K, Kurumizaka H, Dimitrov S, Almouzni G, Wolffe A P. Histone acetylation: influence on transcription, nucleosome mobility and positioning, and linker histone-dependent transcriptional repression. EMBO J. 1997;16:2096–2107. doi: 10.1093/emboj/16.8.2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.van Holde K E. Chromatin. New York, N.Y: Springer-Verlag; 1988. [Google Scholar]
- 55.Wang L, Liu L, Berger S L. Critical residues for histone acetylation by Gcn5, functioning in Ada and SAGA complexes, are also required for transcriptional function in vivo. Genes Dev. 1998;12:640–653. doi: 10.1101/gad.12.5.640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wolffe A P, Hayes J J. Chromatin disruption and modification. Nucleic Acids Res. 1999;27:711–720. doi: 10.1093/nar/27.3.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wolffe A P, Kurumizaka H. The nucleosome: a powerful regulator of transcription. Prog Nucleic Acid Res Mol Biol. 1998;61:379–422. doi: 10.1016/s0079-6603(08)60832-6. [DOI] [PubMed] [Google Scholar]
- 58.Workman J L, Kingston R E. Alteration of nucleosome structure as a mechanism of transcriptional regulation. Annu Rev Biochem. 1998;67:545–579. doi: 10.1146/annurev.biochem.67.1.545. [DOI] [PubMed] [Google Scholar]
- 59.Wu C. Chromatin remodeling and the control of gene expression. J Biol Chem. 1997;272:28171–28174. doi: 10.1074/jbc.272.45.28171. [DOI] [PubMed] [Google Scholar]
- 60.Yang X-J, Ogrryzko V V, Nishikawa J, Howard B H, Nakatani Y. A p300/CBP-associated factor that competes with adenoviral oncoprotein E1A. Nature. 1996;382:319–324. doi: 10.1038/382319a0. [DOI] [PubMed] [Google Scholar]