Abstract
Biobanking efforts, to establish and grow the pool of available tissue from which evidence on aetiology, therapeutic susceptibility and prognosis of various diseases, have been underway for decades. This is illustrated nowhere better than in cancer. High incidence cancers such as breast, colorectal and lung have seen massive increases in their requisite formularies that have yielded improved prognoses. These discoveries, on a very fundamental level, were made by scientists who had access to tumour tissue and associated clinical data from patient donors. As the research space for higher incidence malignancies became increasingly crowded, attention has turned towards those malignancies with lower incidence. In the same time span, technology has continued to evolve, allowing the next generation of scientists and clinicians to ask more nuanced questions. Inquiries are no longer limited to the -omics of tumour tissue but also include biomarkers of blood and excretory products, concurrent disease status and composition of the gut microbiome. The impact of these new technologies and the questions now facing researchers in low-incidence cancers will be summarized and discussed. Our experience with pancreatic ductal adenocarcinoma will be used as a model for this review.
Keywords: Tissues, planning, translational medical research, biomarkers, research design
Introduction
Contemporary disease biomarker discovery and validation are direct outputs from tissue and body fluids biobanks. Biomarker assays have evolved from simple immunohistochemical techniques to multi-omic methodologies which have served to stratify cancers of a single organ system into a multitude of diseases.1-3 Through this stratification, novel targeted molecular therapeutics have been discovered that have proven to be effective in improving the prognosis of various biomarker-identified strata.4-6 More recently, there have been a number of therapies developed that are considered to be tumour agnostic which are indicated entirely by biomarker status. 7 These developments have catalysed the erosion of the organ specific paradigm of cancer treatment.
In pancreatic ductal adenocarcinoma (PDAC), we are faced with significant challenges which include: late detection in patients with suboptimal performance status, inherent resistance to therapy and a lack of clinically validated biomarkers which could serve to stratify patients into different therapeutic regimens. 8 These challenges result in a relatively poor prognosis for PDAC patients with greater than 50% of patients presenting with metastatic disease and yielding an aggregate 5-year survival probability of approximately 11%. 9 Recent genomic and transcriptomic studies with tumour tissue have identified subtypes of pancreatic neuroendocrine tumour (PNET) and PDAC but a validated predictive biomarker remains elusive.10-12
In this article, we aim to: 1. Describe the current biobanking strategy employed by our group. 2. Explain how this strategy has served to inform our research priorities. 3. Illustrate the potential pitfalls associated with the constantly evolving landscape of the cancer research environment. 4. Argue that strategies which can accommodate the inevitable changes associated with constant innovation can position teams to succeed in unexpected ways.
Prospective and Retrospective Biobanking in the Context of Pancreatic Cancer
The process for prospective recruitment targets patients undergoing resection for PDAC. They are approached at the time of surgical consult to provide pre-operative informed consent to participate in the biobanking process. Patients may choose to consent during the pre-operative consult or take the form home for consideration. If a patient has not chosen to consent before surgery, their specimen will be excluded from the biobanking process. Our consent success rate is greater than 90%. We attribute this high rate of success to the clinicians who explain the benefits of biobanking to the patients. In the Vancouver Coastal Health Region, we see approximately 100 cases of resected PDAC each year. Of these, approximately 75% have residual tissue not needed for diagnostic purposes and can therefore be utilized for biobanking activities. Tissues from tumour and adjacent grossly uninvolved pancreas are selected, aliquoted, snap-frozen in liquid nitrogen and stored at –80°C until utilized. Corresponding samples are also taken for formalin fixed paraffin embedding (FFPE) and are sectioned for haematoxylin and eosin (H & E) staining. For all consented patients where tissue was acquired for biobanking, blood samples are processed to extract serum, plasma and buffy coat aliquots which are also snap frozen and can be used for proteomics, circulating tumour DNA and as a source of germline DNA respectively.
The process to embed the prospective biobanking process into the clinical workflow required the recruitment of dedicated staff to attend to fresh specimens received in anatomical pathology. These individuals were trained in blood processing, grossing and the non-destructive sampling of resection specimens. On-call pathologists are available to assist with specimens where sampling might compromise the ability for accurate assessment of tumour margins or other elements essential for an accurate diagnosis.
Corresponding patient-level clinico-pathologic and outcome data are also collected and are summarized in Table 1. These data are obtained from the electronic medical record. Where synoptic data are unavailable, narrative reports are coded by individuals who are sufficiently trained in medical terminology. The codes are developed in consultation with a biostatistician who supervises the analyses performed by the research group. The choice of these data elements is derived from our experience with diagnosing and researching of the disease. We acknowledge that there are elements missing from the minimum reporting datasets such as the College of American Pathologists or the Royal College of Pathologists. We have found that elements we have chosen to exclude have similar values across the vast majority of patients and therefore do not add information at the time of multivariable statistical analysis.
Table 1.
Description of patient specific data fields for pancreatic biobank specimens.
| Variable | Description |
|---|---|
| Age (years) | Calculation: (Date of diagnosis - Date of birth) |
| Sex | Sex at birth |
| Date of diagnosis | Date: (dd-mm-yyyy) |
| Date of progression | Date: (dd-mm-yyyy) |
| Date of last follow-up | Date: (dd-mm-yyyy) |
| Status at last follow-up | Vital and disease status at most recent chart review |
| Neoadjuvant chemotherapy agents | List of therapeutic agents or institution specific regimen |
| Adjuvant chemotherapy agents | List of therapeutic agents or institution specific regimen |
| Palliative chemotherapy agents | List of therapeutic agents or institution specific regimen |
| Tumour site a | Location of tumour in the resected specimen |
| Histologic type a | Variant of pancreatic ductal adenocarcinoma |
| Histologic grade a | Extent of glandular differentiation |
| Lymphovascular invasion a | Presence of lympho-vascular invasion |
| Perineural invasion a | Presence of perineurial invasion |
| Primary tumour pT-stage a | Extent of disease in the resected specimen |
| Regional lymph node pN-stage a | Presence of lymph node metastasis |
| Distant metastasis pM-stage a | Presence of peritoneal seeding or positive cytology |
| Resection status a | A summary of tumour involvement for 1 or more margins |
Adopted from the College of American Pathologists Protocol for the Examination of Specimens From Patients With Carcinoma of the Pancreas <https://documents.cap.org/protocols/cp-gihepatobiliary-pancreasexocrine-20-4100.pdf>.
Our strategy for retrospective collection follows a similar pattern to that outlined for prospective collection. Given the relatively poor prognosis of the disease and the fact that the majority of patients were already deceased at the time of cohort assembly, we obtained a waiver of consent from the university ethics board allowing us to proceed without consent of subjects or their next of kin. The cases were chosen based upon sample availability and the inclusion criterion was any patient who had a Whipple’s procedure (pancreatoduodenectomy) whose FFPE samples were available from the archives. There were a multitude of non-PDAC diagnoses such as ampullary carcinoma and neuroendocrine tumours which we elected to include in the retrospective collection. Duplicate 0.6 mm core tissue micro-arrays were constructed for: tumour epithelium, tumour associated stroma and adjacent uninvolved normal appearing pancreas. Pancreatic Intraepithelial Neoplasia (PanIN) TMAs were also constructed using a 2 mm coring needle and a maximum of 3 lesions were taken per case. A limitation of this resource was the lack of high grade genomic DNA as blood samples are not routinely kept by the lab. The retrospective cohort utilizes the same data structure as for the prospective cohort specified in Table 1.
Biobanking Resources as a Research Guide and Facilitator
The approach we used for the establishment of biobank resources has served to guide our research endeavours. Our first overarching goal as a research group was to add knowledge, in the form of biomarkers, to improve our ability to prognosticate PDAC patients. Studies that used a histomorphology approach included: confirmation of the suspicion that, analogous to colorectal cancer, tumour budding was an independent predictor of poor prognosis, 13 and that low gland or neutrophil infiltration was associated with poor prognosis. 14 Subsequent immunohistochemistry studies that followed included: the identification of mismatch repair deficiency as a marker of therapeutic resistance to adjuvant chemotherapy with a pyrimidine analogue, a study confirming human equilibrative nucleoside transporter 1 (hENT1) is a predictive biomarker for sensitivity to gemcitabine and that expression of programmed cell death ligand 1 (PDL1) is a low prevalence phenomenon and an independent marker for poor prognosis in PDAC.15-17
Utilizing our approach to include several pancreas associated malignancies on our TMAs yielded an ability to compare protein expression between variants of ampullary carcinoma and established the rationale to differentiate intestinal from pancreatobilliary variants. 18 Initial findings from our TMA also identified biomarker driven prognostic differences in pancreatic neuroendocrine tumours which formed the basis for a whole genome and transcriptome study. 19 The strategy of not limiting the construction of a TMA to a single diagnosis within an organ site has served to produce information on what are best classified as severely understudied disease entities. It has also served to broaden our ability to collaborate with researchers outside of our core group.
Emerging Research Areas and the Advantage of Being Nimble and Adaptable
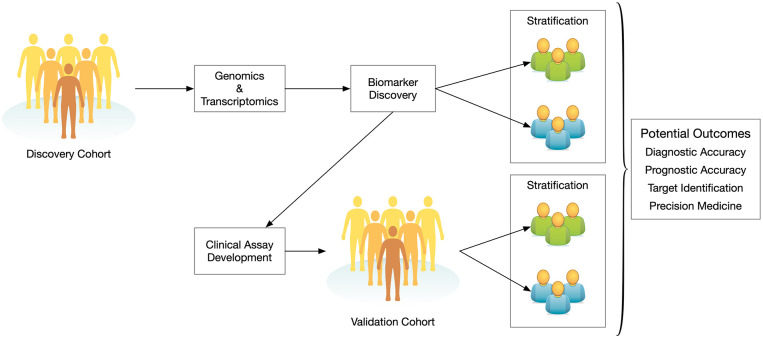
The core prospective and retrospective resources described previously have the ability to serve tumour-based multi-omic approaches to target discovery, identification of biomarker specific strata and validation using clinically practicable assays. 20 This translational medicine cascade is illustrated in Figure 1. It is important to note that the workflow depicted in Figure 1 has multiple entry points allowing for several groups to participate in the translational medicine pipeline for a given biomarker. Additionally, a discovery cohort can yield multiple biomarker leads. Those leads can be further investigated by a different group with a separate validation cohort. This approach serves to enhance external validity thereby lending increased credibility to the research findings. 21 In the cancer literature, an increasing number of studies are using discovery and validation cohorts to substantiate their results and make the argument for clinical translation.22-24
Figure 1.
A representation of the translational medicine pipeline and how cohorts derived from biobanks are the core starting points for discovery and validation.
Recent research has suggested that tumour-tissue specific nucleic acids are not the only component where clinically relevant biomarkers exist. The microbiomes of both the gut and the tumour of PDAC patients have been shown to harbour potentially valuable diagnostic and prognostic information.25,26 The composition of the gut microbiome has also been implicated as a modulating factor affecting the response to immunotherapy in cancer patients. 27 In response to this, our group has added the systematic collection of faecal material to our prospective biobanking activities. It has been asserted that faecal assessment of the gut microbiome is an imperfect method to assess the exact flora present in the gut. 28 However, we hold the view that the benefits of a minimally invasive approach outweigh the increased precision of more invasive methods, specifically in terms of risk to the patient and practicability when viewed through the lens of cost-benefit. By the application of statistical thinking, we are of the opinion that once a sufficient number of faecal samples are investigated, the likelihood of detecting a biologically meaningful signal will shift from possible to probable.
Recent research has also identified that metabolites found in urine detected through field asymmetric waveform ion mobility spectrometry may be used to differentiate PDAC patients from healthy controls. 29 This study demonstrated a fairly high sensitivity of 91% but at the expense of specificity (49%) when chronic and acute pancreatitis samples were combined with PDAC and compared against controls. The study recruited 182 patients but was arguably powered only for comparisons between PDAC and controls which yielded relatively good differentiation between the groups with sensitivity and specificity of 79% respectively.
Based upon these emerging areas of research, we are adding systematic collection of faecal and urine samples to our biobank collection protocol. The ability to expand the scope of our prospective biobanking activities with manageable effort allows us to diversify our focus from tumour tissue derived findings to research questions focussed on early detection, mechanisms of resistance and drug interactions. This will position our group to be a key participant in these emerging research areas in the years to come.
Following the Path of Discovery to Build the Mosaic of Knowledge
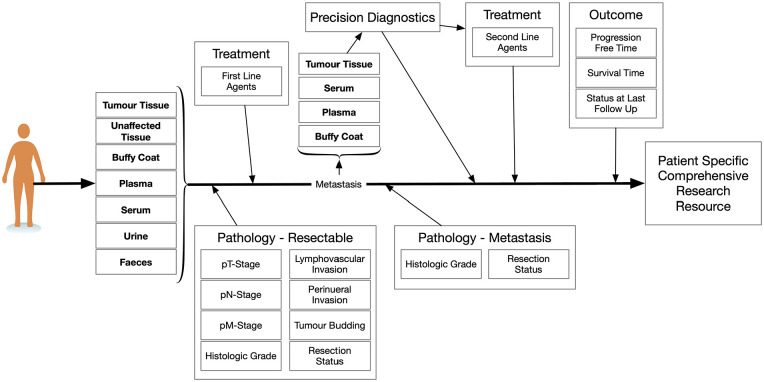
Our current biobanking process is illustrated in Figure 2. The flow from sample collection to patient level information allows for a comprehensive information resource which provides clinical context for scientific discoveries. As for Figure 1, more than 1 entry point exists for biobanking. As most patients with PDAC do not present with resectable disease, metastatic tissue may be collected and utilized for genomic studies as is being done for the clinical trial: Prospectively Defining Metastatic Pancreatic Ductal Adenocarcinoma Subtypes by Comprehensive Genomic Analysis (clinical trial identifier: NCT02869802). 30 In this trial, tissue derived through additional biopsies from metastatic patients is also used to establish organoids. Tight integration between the clinical and research teams ensures that the specimens are transported from the clinic to the research lab in a timely fashion. This has yielded over 10 stable oragnoid cultures so far, with many more in development.
Figure 2.
A biobanking schematic along the disease course of a pancreatic cancer patient. Items in bold represent specimen collection opportunities.
The collection of information is only the first step in the formulation of knowledge. There must be a deliberate strategy to integrate this information in order to discover contingencies, effect modifications and modulations and develop models with solid statistical foundations that have the potential to improve outcomes for patients. To accomplish this, a bi-directional information system must be employed. Merely collecting samples and associated information in a database does not help to generate knowledge. Information should not only be collected but used and dispersed according to ethical and governance guidelines established by a biobank. This is an evolving area of research for which few studies have explicitly examined the biobanking governance processes. 31 Our biobanking governance is administered through a committee composed of lead investigators, researchers and a statistician. We evaluate requests for specimens and data on an individual case basis and judge the merit of requests based upon criteria such as preliminary research and the number of samples requested. We favour studies that have substantial in-vitro mechanistic evidence, a high degree of clinical relevance and subsequent translational potential. While we do not advocate for a 1 size fits all approach to biobanking governance, we believe that there are most likely a set of common principles utilized by biobanks. Increased transparency of those processes is required to provide opportunities to share improvements across the biobanking spectrum. To do so will enhance the possibilities of harmonization across resources.
To react to the ever changing clinical research landscape, the collection and distribution of information through data storage and management solutions must be adaptive in nature. This is especially true in gastrointestinal malignancies where new assays are being implemented such as BRAF testing and other genetic based panels. Databases should focus on user interface designs that are intuitive for end users. The rationale for this approach is to empower the individuals entering the data to be more comfortable using the system. The system should not be imposed upon them but developed with them in a cooperative manner. The goals should be efficiency and accuracy of data input thereby yielding reduced cognitive burden for data entry personnel. For instance, the use of pre-populated value lists for data fields helps ensure that valid data are entered and thereby reduces data cleaning at the time of analysis. Additionally, an efficient data interface is likely to yield higher compliance rates for the timely completion of retrospective data entry from the electronic medical record. 32 This is how our biobanking model operates due to the nature of emerging data through the diagnostic, treatment and follow-up processes. The development of our user interface was derived through a trial and error process and although the process was arduous, it resulted in a product that left users feeling empowered. Querying of the database should be able to be performed by personnel that are not information technology professionals. Research questions should be able to be addressed by researchers without the requirement to have specialist staff devoted to running a biobanking database. The database should also be able to be modified with minimal effort through the ability to add fields, alter value-lists that populate fields and rectify other user interface issues that can detract from usability.
Challenges and Opportunities for Biobanking in Low-Incidence Cancers
Our experience with biobanking in the pancreas has been an evolving process. As with other low-incidence malignancies, the complexity of the questions we can ask is limited by the materials available for study. To compensate for this, we have elected to adopt a comprehensive approach to biobanking that gathers as much as possible from a patient and to ask questions from different perspectives. The possibility of measuring risk of disease progression from an assay based on urine would serve to reduce burden on the patient and would likely reduce healthcare costs. A faecal based test to assess the composition of the gut microbiome may serve to inform if concomitant agents should be administered to enhance the activity of anti-cancer immunotherapy. A topic that is starting to emerge is the creation of organoids derived from metastatic PDAC patient biopsies. The potential for doing compound screening to inform off-label treatment is a tantalizing prospect.
The evolution from high throughput genomics and transcriptomics associated with studies in high-incidence malignancies to the necessity to adopt more nuanced approaches applicable to low-incidence malignancies has spawned new technologies and approaches. Through these innovations, we are slowly starting to move the needle in the right direction for PDAC patients. We encourage other researchers to consider an approach to biobanking that involves nimble, integrated and wide-range biospecimen and associated clinical data collection. Such an approach will best utilize patient donated material, and ultimately capitalize on patients’ contribution towards research and discovery that will help improve outcomes for future patients.
Footnotes
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by unrestricted donor funds from the BC Cancer Foundation and the VGH and UBC Hospital Foundation administered by the Pancreas Centre BC.
Declaration of conflicting interests: The author(s) declared the following potential conflicts of interest with respect to the research, authorship, and/or publication of this article: All authors declare no conflicts of interest related to the submitted work. DJR disclosures include research funding and honoraria from Bayer and Roche, and travel funding and honoraria from Servier, Celgene, Taiho, Ipsen and Astra Zeneca. DFS reports consultant fees from Alimentiv Inc. and honoraria from Merck and Pfizer.
Author Contributions: SEK conceived the and wrote the first draft of the manuscript. JMK, CW, DJR and DFS provided critical feedback and edits.
ORCID iD: Steve E Kalloger  https://orcid.org/0000-0003-1332-7881
https://orcid.org/0000-0003-1332-7881
References
- 1. Köbel M, Kalloger SE, Boyd N, et al. Ovarian carcinoma subtypes are different diseases: implications for biomarker studies. PLoS Med. 2008;5:e232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Curtis C, Shah SP, Chin SF, et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486:346-352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Guinney J, Dienstmann R, Wang X, et al. The consensus molecular subtypes of colorectal cancer. Nat Med. 2015;21:1350-1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lockwood WW, Wilson IM, Coe BP, et al. Divergent genomic and epigenomic landscapes of lung cancer subtypes underscore the selection of different oncogenic pathways during tumor development. PLoS One. 2012;7:e37775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sadanandam A, Lyssiotis CA, Homicsko K, et al. A colorectal cancer classification system that associates cellular phenotype and responses to therapy. Nat Med. 2013;19:619-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Waks AG, Winer EP. Breast cancer treatment: a review. JAMA. 2019;321:288-300. [DOI] [PubMed] [Google Scholar]
- 7. Seligson ND, Knepper TC, Ragg S, Walko CM. Developing drugs for tissue-agnostic indications: a paradigm shift in leveraging cancer biology for prec on medicine. Clin Pharmacol Ther. 2021;109:334-342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Oberstein PE, Olive KP. Pancreatic cancer: why is it so hard to treat? Therap Adv Gastroenterol. 2013;6:321-337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. National Cancer Institute Surveillance, Epidemiology and ERP. Cancer stat facts: pancreatic cancer. 2021. Accessed May 30, 2021. https://seer.cancer.gov/statfacts/html/pancreas.html
- 10. Bailey P, Chang DK, Nones K, et al. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature. 2016;531:47-52. [DOI] [PubMed] [Google Scholar]
- 11. Waddell N, Pajic M, Patch AM, et al. Whole genomes redefine the mutational landscape of pancreatic cancer. Nature. 2015;518:495-501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Scarpa A, Chang DK, Nones K, et al. Whole-genome landscape of pancreatic neuroendocrine tumours. Nature. 2017;543:65-71. [DOI] [PubMed] [Google Scholar]
- 13. O’Connor K, Li-Chang HH, Kalloger SE, et al. Tumor budding is an independent adverse prognostic factor in pancreatic ductal adenocarcinoma. Am J Surg Pathol. 2015;39:472-478. [DOI] [PubMed] [Google Scholar]
- 14. Naso JR, Topham JT, Karasinska JM, et al. Tumor infiltrating neutrophils and gland formation predict overall survival and molecular subgroups in pancreatic ductal adenocarcinoma. Cancer Med. 2021;10:1155-1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Riazy M, Kalloger SE, Sheffield BS, et al. Mismatch repair status may predict response to adjuvant chemotherapy in resectable pancreatic ductal adenocarcinoma. Mod Pathol. 2015;28:1383-1389. [DOI] [PubMed] [Google Scholar]
- 16. Kalloger SE, Riazy M, Tessier-Cloutier B, et al. A predictive analysis of the SP120 and 10D7G2 antibodies for human equilibrative nucleoside transporter 1 (hENT1) in pancreatic ductal adenocarcinoma treated with adjuvant gemcitabine. J Pathol Clin Res. 2017;3:179-190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tessier-Cloutier B, Kalloger SE, Al-Kandari M, et al. Programmed cell death ligand 1 cut-point is associated with reduced disease specific survival in resected pancreatic ductal adenocarcinoma. BMC Cancer. 2017;17:618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Leo JM, Kalloger SE, Peixoto RD, et al. Immunophenotyping of ampullary carcinomata allows for stratification of treatment specific subgroups. J Clin Pathol. 2016;69:431-439. [DOI] [PubMed] [Google Scholar]
- 19. Wong HL, Yang KC, Shen Y, et al. Molecular characterization of metastatic pancreatic neuroendocrine tumors (PNETs) using whole-genome and transcriptome sequencing. Cold Spring Harb Mol Case Stud. 2018;4:1-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zeggini E, Gloyn AL, Barton AC, Wain LV. Translational genomics and precision medicine: moving from the lab to the clinic. Science. 2019;365:1409-1413. [DOI] [PubMed] [Google Scholar]
- 21. Bleeker SE, Moll HA, Steyerberg EW, et al. External validation is necessary in prediction research: a clinical example. J Clin Epidemiol. 2003;56:826-832. [DOI] [PubMed] [Google Scholar]
- 22. Roberts BS, Hardigan AA, Moore DE, et al. Discovery and validation of circulating biomarkers of colorectal adenoma by high-depth small RNA sequencing. Clin Cancer Res. 2018;24:2092-2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Cristescu R, Mogg R, Ayers M, et al. Pan-tumor genomic biomarkers for PD-1 checkpoint blockade-based immunotherapy. Science. 2018;362:80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Croner LJ, Dillon R, Kao A, et al. Discovery and validation of a colorectal cancer classifier in a new blood test with improved performance for high-risk subjects. Clin Proteomics. 2017;14:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Half E, Keren N, Reshef L, et al. Fecal microbiome signatures of pancreatic cancer patients. Sci Rep. 2019;9:16801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Riquelme E, Zhang Y, Zhang L, et al. Tumor microbiome diversity and composition influence pancreatic cancer outcomes. Cell. 2019;178:795-806.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Weersma RK, Zhernakova A, Fu J. Interaction between drugs and the gut microbiome. Gut. 2020;69:1510-1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Tang Q, Jin G, Wang G, et al. Current sampling methods for gut microbiota: a call for more precise devices. Front Cell Infect Microbiol. 2020;10:151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Nissinen SI, Roine A, Hokkinen L, et al. Detection of pancreatic cancer by urine volatile organic compound analysis. Anticancer Res. 2019;39:73-79. [DOI] [PubMed] [Google Scholar]
- 30. Renouf DJ. Prospectively defining metastatic pancreatic ductal adenocarcinoma subtypes by comprehensive genomic analysis (PanGen). ClinicalTrials.gov. 2021. Accessed May 30, 2021. https://clinicaltrials.gov/ct2/show/NCT02869802
- 31. Langhof H, Schwietering J, Strech D. Practice evaluation of biobank ethics and governance: current needs and future perspectives. J Med Genet. 2019;56:176-185. [DOI] [PubMed] [Google Scholar]
- 32. Wilbanks BA, Berner ES, Alexander GL, Azuero A, Patrician PA, Moss JA. The effect of data-entry template design and anesthesia provider workload on documentation accuracy, documentation efficiency, and user-satisfaction. Internet J Med Inform. 2018;118:29-35. [DOI] [PubMed] [Google Scholar]