Abstract
The basic helix-loop-helix (bHLH) transcription factor genes Hand1 and Mash2 are essential for placental development in mice. Hand1 promotes differentiation of trophoblast giant cells, whereas Mash2 is required for the maintenance of giant cell precursors, and its overexpression prevents giant cell differentiation. We found that Hand1 expression and Mash2 expression overlap in the ectoplacental cone and spongiotrophoblast, layers of the placenta that contain the giant cell precursors, indicating that the antagonistic activities of Hand1 and Mash2 must be coordinated. MASH2 and HAND1 both heterodimerize with E factors, bHLH proteins that are the DNA-binding partners for most class B bHLH factors and which are also expressed in the ectoplacental cone and spongiotrophoblast. In vitro, HAND1 could antagonize MASH2 function by competing for E-factor binding. However, the Hand1 mutant phenotype cannot be solely explained by ectopic activity of MASH2, as the Hand1 mutant phenotype was not altered by further mutation of Mash2. Interestingly, expression of E-factor genes (ITF2 and ALF1) was down-regulated in the trophoblast lineage prior to giant cell differentiation. Therefore, suppression of MASH2 function, required to allow giant cell differentiation, may occur in vivo by loss of its E-factor partner due to loss of its expression and/or competition from HAND1. In giant cells, where E-factor expression was not detected, HAND1 presumably associates with a different bHLH partner. This may account for the distinct functions of HAND1 in giant cells and their precursors. We conclude that development of the trophoblast lineage is regulated by the interacting functions of HAND1, MASH2, and their cofactors.
The placenta is critical for the intrauterine survival of mammalian embryos. In mice, mutations that severely disrupt placentation or establishment of the chorioallantoic circulation result in embryonic lethality by day 10.5 of gestation (E10.5). Defects in placentation also contribute to diseases of human pregnancy, including spontaneous abortion and preeclampsia (11). However, surprisingly little is known regarding the molecular events that regulate development of the trophoblast cell lineage, the epithelial component of the placenta. At the blastocyst stage, trophoblast cells in contact with the inner cell mass (polar trophectoderm) continue to proliferate and later contribute to the chorion and ectoplacental cone (24). In contrast, trophoblast cells distal to the inner cell mass (mural trophectoderm) terminally differentiate to form primary trophoblast giant cells. While mitotically arrested, these cells undergo continued rounds of DNA synthesis (endocycles), thereby acquiring their characteristic giant polyploid nuclei (54). Secondary giant cells subsequently arise due to differentiation of precursor cells present in the ectoplacental cone and, later in gestation, the spongiotrophoblast (17). Trophoblast giant cells participate in a number of processes critical to a successful pregnancy, including blastocyst implantation, remodeling of the maternal decidua, and secretion of hormones that regulate both fetal and maternal development (13).
A limited number of genes have been shown to play direct roles in early trophoblast development (for reviews, see references 39 and 46). Hand1 and Mash2 encode members of the basic helix-loop-helix (bHLH) transcription factor family, which regulate the determination and differentiation of several cell lineages (1). Mash2 mutant mouse conceptuses arrest at E10.5 due to placental defects that include an absence of the spongiotrophoblast layer (derived from trophoblast of the ectoplacental cone), excess trophoblast giant cells, and a poorly developed labyrinthine layer (18, 50). Mash2 function is thus required to maintain spongiotrophoblast at the expense of giant cell differentiation. In contrast, Hand1 mutants arrest at E7.5 due primarily to placental defects that include a block in trophoblast giant cell differentiation and a smaller ectoplacental cone (38). These factors also have opposite activities when overexpressed in the Rcho-1 trophoblast cell line. While Hand1 expression promotes giant cell differentiation, Mash2 inhibits this process (12, 25). As Hand1 and Mash2 have apparently opposing roles in trophoblast development, we wished to determine how their activities are coordinately regulated.
Different mechanisms could in theory ensure that only Hand1 or Mash2 is active in a given cell. One possibility is that the two factors are expressed in nonoverlapping trophoblast subpopulations. Consistent with this model, Mash2 expression is broadened in Hand1 mutants to encompass cells normally fated to differentiate into secondary trophoblast giant cells (38), indicating that Hand1 is essential for repressing Mash2 expression at the onset of giant cell differentiation. While transcripts of Hand1 (12, 15, 19) and Mash2 (18, 32, 41) have been previously localized in the placenta, these separate studies did not resolve if they are coexpressed in individual trophoblast subtypes. Alternatively, HAND1 and MASH2 may be coexpressed, but they could compete for an essential cofactor. Members of the bHLH family dimerize via their HLH domains, allowing the two basic domains to bind DNA (52). Both HAND1 and MASH2 form heterodimers with E factors, the obligate partners of most bHLH factors (12, 23). It is therefore possible that HAND1 and MASH2, if present in the same cell, compete for the same E-factor partners, with the relative abundance and dimerization affinities of HAND1 and MASH2 determining which factor is functional. The different complexes could also compete for DNA-binding sites. MASH2–E-factor dimers bind to and activate transcription from E-box sequences (CANNTG) (23). HAND1–E-factor complexes bind to a different consensus sequence (NNTCTG) (19), which has some overlap with E-box sequences. Thus, it is possible that HAND1 and MASH2 complexes bind to common sequences. If they do, competition for shared sites would ensure that only one of these factors is active at a given time. In this study, we examined each of these possible mechanisms. The results demonstrate that Hand1 and Mash2 are coexpressed in the ectoplacental cone and spongiotrophoblast, intermediate trophoblast subpopulations. In vitro, HAND1 can inhibit MASH2 activity by virtue of competition for E-factor partners. However, analysis of Hand1/Mash2 compound mutants indicates that Hand1 has a distinct role independent of its effects on Mash2. Regulated expression of E-factor genes was also observed, a feature that may further compartmentalize Hand1 and Mash2 functions during trophoblast development.
MATERIALS AND METHODS
Plasmids.
pBS-ALF1B was constructed by ligating a 670-bp N-terminus EcoRI/BamHI fragment from pA-ALF11-706 (36) into pBluescript SK+. pBS-ITF2 was similarly made by ligating a 1,200-bp N-terminal HindIII/XbaI fragment from pCMV-ITF2 into pBluescript SK+. A 600-bp SmaI/HindIII fragment from pALF2 (36) encoding the E2A 3′ untranslated region was ligated into pBluescript SK+ to make pBS-E2A. The vectors pCMV-ITF2 (8), pE-ALF11-706Δ24 (36), pCMV-Mash2 and pβActin-LacZ (12), p2E MCK-CAT (23), pT7-N3 (3), pHis-Mash2 (33), pGAL4-Hand1, pGAL4-Hand1Δb, and pGAL4-Hand1ΔbHLH (19) have been previously described. pCMV-Hand1, pCMV-Hand1Δb (12-amino-acid basic domain deleted), and pCMV-Hand1ΔbHLH (bHLH domain deleted) were constructed by inserting the EcoRI/XbaI fragment from the appropriate pGAL4 fusion vector into pcDNA-1 (Invitrogen). pCMV-VP16Hand1, encoding an N-terminal fusion of the VP16 acidic activation domain to HAND1, was made by ligating the VP16 activation domain (pBS-VP16mSna [33] BamHI/EcoRI fragment) and an EcoRI/XhoI mouse Hand1 cDNA fragment into pcDNA-1. pCMV-FLAGHand1 was constructed by ligating an NdeI (filled-in)/XhoI fragment encoding an N-terminal FLAG epitope-tagged mouse HAND1 (FL-HAND1) into pcDNA3 (Invitrogen). pSV-E47 was made by ligating a HindIII/BamHI human E47 cDNA into pSV2 (Clontech). The glutathione S-transferase (GST)–HAND1 fusion vector pGST-HAND1 was made by inserting an EcoRI/XhoI ovine Hand1 cDNA fragment into pGEX-3X (Pharmacia). The polyhistidine fusion bacterial expression vectors pHis-E47, pHis-Hand1, pHis-ALF1, pHis-ITF2, pHis-c-jun, and pHis-Id-2 were produced by ligating the following fragments into pHK (33): EcoRI from pT7-N3 (3), EcoRI of murine Hand1, KpnI/HindIII of human ALF1, ScaI/HindIII from pCMV-ITF2, EcoRI of c-jun (includes the basic leucine zipper domain), and KpnI/SalI of human Id-2. pT7FL-Hand1ΔN, a bacterial expression vector which encodes a truncated (lacking the sequence N terminal to the bHLH domain) ovine HAND1, was constructed by replacing the E47 cassette in pT7-N3 with a SmaI/BamHI Hand1 fragment. The luciferase reporter constructs pL8G5-Luc and pL8E6-Luc were derived from pL8G5-CAT and pL8E6-CAT (19).
RNA in situ hybridization of histological sections.
E8.5 conceptuses and E10.5 and 12.5 placentas were fixed with 4% paraformaldehyde. Tissues were embedded in paraffin, sectioned, and subjected to RNA in situ hybridization as previously described (30). Antisense 33P-labeled riboprobes were prepared by using an RNA transcription kit (Stratagene). For ALF1, ITF2, and E2A probes, plasmids pBS-ALF1B, pBS-ITF2, and pBS-E2A were used. Probes specific to the placental lactogen 1 gene (Pl1) (21), Tpbp (previously called 4311) (28), Hand1 (12), and Mash2 (18) have been previously described.
Whole-mount in situ hybridization.
Whole-mount in situ hybridization was performed for the Pl1 gene on Hand1 heterozygotes and Hand1, Mash2, and Hand1/Mash2 mutants dissected out of the uterus at E8.5 (E0.5 is defined as noon of the day on which vaginal plugging was detected). Decidual swellings were split longitudinally from the mesometrial to the antimesometrial end. The embryo and visceral yolk sac were removed, leaving the ectoplacental cone, chorion, Reichert's membrane, and trophoblast giant cell layer intact within the decidua. Decidua were then processed as for E10.5 embryos (9). Digoxigenin-labeled Pl1 probe was prepared by using digoxigenin labeling mix (Boehringer Mannheim) and detected by using an anti-digoxigenin-alkaline phosphatase conjugate (Boehringer Mannheim). The resulting Pl1-positive trophoblast giant cells (within the decidua) were photographed. DNA was prepared from the embryo proper and used for genotype analysis with PCR primers specific to Hand1 and Mash2 as previously described (38, 41).
Whole-mount β-galactosidase staining.
E8.5 decidual swellings from Hand1 +/−; Mrj +/6AD1βgeo × Hand1 +/− crosses were dissected as for whole-mount in situ hybridization. Decidua were then processed and subjected to staining for β-galactosidase activity as previously described (20). DNA was prepared from the embryo proper and used for genotype analysis with PCR primers specific to Hand1 as previously described (38).
Coimmunoprecipitations.
In vitro transcription-translation was performed with the Promega TNT rabbit reticulocyte lysate kit and plasmids pT7-N3 (for FL-E47), pT7FL-Hand1ΔN, pHis-Hand1, pHis-Mash2, pHis-E47, and pT7-c-jun according to the manufacturer's instructions. Proteins were labeled via the addition of [35S]methionine to the reaction. Proteins were made in separate reactions and then mixed for 30 min at 4°C in dilution buffer (100 mM NaCl, 50 mM Tris [pH 7.5], 1 mM EDTA, 0.5% Triton X-100) in the presence of protein A/G-agarose (Santa Cruz). Anti-FLAG antibody M2 (IBI) was then added to the supernatant, followed by an overnight incubation at 4°C. Agarose beads were washed five times with dilution buffer, resuspended in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) loading buffer, boiled, and subjected to SDS-PAGE. Gels were washed in Enhance (NEN), allowing the detection of labeled proteins by fluorography.
Cell culture and transfection.
C3H10T1/2 fibroblasts were maintained in Dulbecco modified Eagle medium containing 10% fetal bovine serum (HyClone) plus 50 μM β-mercaptoethanol (Gibco BRL). Rcho-1 cells were cultured in NCTC-135 medium (Sigma) supplemented with 20% serum, 50 μM β-mercaptoethanol, and 1 mM sodium pyruvate as previously described (12, 16). For reporter assays, C3H10T1/2 cells were transiently transfected by using Lipofectamine (Gibco BRL) as previously described (33). Rcho-1 cells were transfected 5 h postplating, using Lipofectamine PLUS (Gibco BRL), with fresh medium added 5 h posttransfection. Plasmid mixtures consisted of 100 ng of pβActin-LacZ, 400 ng of reporter plasmid (p2E MCK-CAT, pL8G5-Luc, or pL8E6-Luc), and expression vector (with empty vector used to a total of 1 μg of DNA/35-mm-diameter well). Cells were harvested 48 h posttransfection. Chloramphenicol acetyltransferase (CAT) levels were measured by enzyme-linked immunosorbent assay using a CAT ELISA kit (Boehringer Mannheim), while luciferase activity was assayed with d(−)-luciferin (Boehringer Mannheim) and an LB 9501 luminometer (Berthold). Values were normalized to β-galactosidase activity (measured as described in reference 45) and are reported as the mean ± standard error (SE) relative to a value of 1.0 for empty expression vector alone. Transfections were performed in duplicate, with each experiment repeated three to six times. Significant differences between values were determined by analysis of variance followed by Student-Newman-Keuls test.
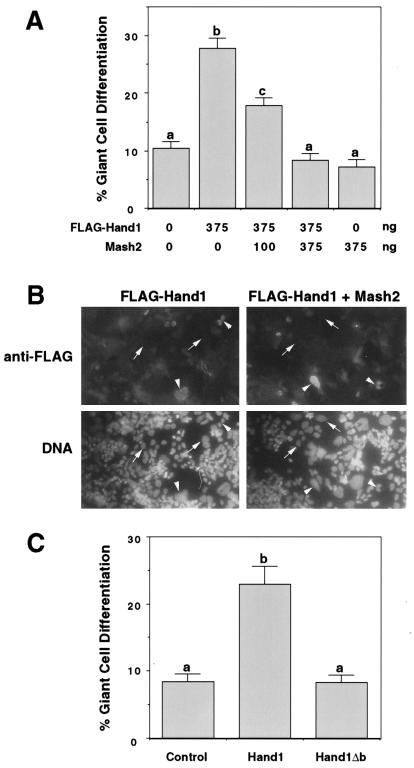
For giant cell differentiation assays (Fig. 6C), Rcho-1 stem cells were transfected by using Lipofectamine PLUS (Gibco BRL) 5 h after plating to coverslips; 100 ng of pβActin-LacZ and 500 ng of expression vector were added per 35-mm-diameter well. Cells were fixed 48 h posttransfection and stained for β-galactosidase activity (45). Giant cell differentiation was scored as the proportion of β-galactosidase-positive cells which had assumed a trophoblast giant cell morphology (12). Percent giant cell differentiation values represent the mean ± SE for approximately 250 cells per treatment group and were similar in two separate experiments. For Mash2 titration experiments (Fig. 6A and B), Rcho-1 cells were transfected as described above, fixed in 4% paraformaldehyde, and permeabilized with methanol. Following incubation with mouse anti-FLAG (1/200 dilution; IBI) and rabbit anti-β-galactosidase (1/400 dilution; Cappell) primary antibodies and anti-mouse-fluorescein isothiocyanate and anti-rabbit-tetramethyl rhodamine isothiocyanate (1/50 dilution; Sigma) secondary antibodies, cells were stained with bisbenzimide and examined by fluorescence microscopy. Giant cell differentiation was scored as the percentage of tetramethyl rhodamine isothiocyanate-positive cells which had the enlarged nuclei characteristic of trophoblast giant cells. Results represent the mean ± SE of 25 fields examined for each treatment group, using a 40× objective.
FIG. 6.
Mutation of the HAND1 basic domain abrogates its ability to promote trophoblast giant cell differentiation. (A and C) Rcho-1 trophoblast cells were transiently cotransfected with a lacZ marker and the indicated expression vector(s). β-Galactosidase-positive cells were scored for giant cell morphology 2 days posttransfection. Different superscripts indicate statistically significant differences (P < 0.05). (B) Immunofluorescent detection of FL-HAND1 expression in transfected cells. Arrows and arrowheads indicate FL-HAND1-negative and -positive cells, respectively.
Electrophoretic mobility shift assays.
In vitro transcription-translation reactions were carried out as described above, with plasmids pHis-ALF1, pHis-ITF2, and pHis-Id-2 used to produce ALF1, ITF2, and ID-2, respectively. GST and GST-HAND1 were produced in Escherichia coli DH5α, using the plasmids pGEX-3X and pGST-HAND1, and batch purified by using glutathione-Sepharose as instructed by the manufacturer (Pharmacia). Control (no DNA) reaction product was added to equalize the amount of reticulocyte lysate present in each lane. Proteins were preincubated for 15 min at 37°C prior to the addition of 10,000 cpm of 32P-labeled probe (labeled via fill-in reactions [45]). Following an additional 20 min at room temperature, reactions were resolved via electrophoresis on 5% nondenaturing acrylamide gels. For competition experiments, a 200-fold excess of unlabeled oligonucleotide was added prior to the first incubation step. Double-stranded oligonucleotides used have all been previously described (33).
RESULTS
Overlapping expression of Hand1 and Mash2 in trophoblast subpopulations.
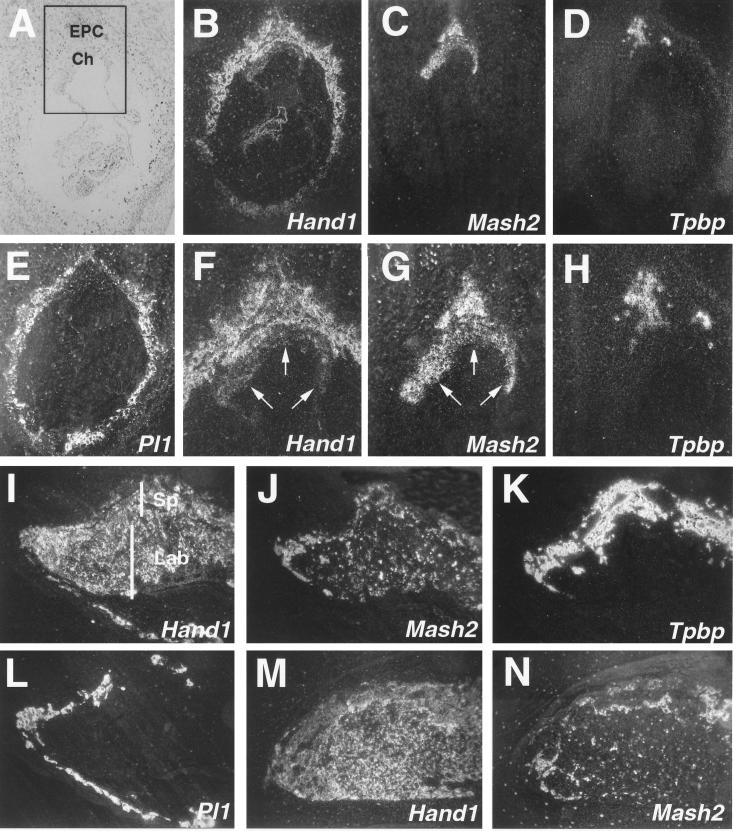
RNA in situ hybridization on serial histological sections from E8.5 murine conceptuses demonstrated that Hand1 and Mash2 expression overlaps in the ectoplacental cone (Fig. 1F and G), a trophoblast subpopulation which also expresses Tpbp (Fig. 1H). Hand1 expression was also detected in both primary and secondary trophoblast giant cells (Fig. 1B, I, and M), which were identified by their expression of Pl1 (Fig. 1E and L). In contrast, Mash2 transcripts were not detectable in giant cells (Fig. 1C, J, and N) but were present at high levels in the chorion, where Hand1 expression was undetectable (compare Fig. 1F and G, arrows). At E10.5 and E12.5, placental expression of Hand1 had expanded to encompass all three trophoblast layers: the outer layer of trophoblast giant cells, the spongiotrophoblast layer, and the inner labyrinthine layer (Fig. 1I and M). However, Hand1 and Mash2 (Fig. 1J and N) hybridization signals were not uniformly detected in the labyrinthine and spongiotrophoblast layers (Fig. 1K), with Mash2 signals limited to an even more restricted portion of the spongiotrophoblast layer by E12.5 (Fig. 1N). As an overlap in Hand1 and Mash2 expression was detected, we concluded that their activities were not segregated based on distinct expression alone. This suggested that their encoded protein products might interact in intermediate trophoblast subpopulations in vivo.
FIG. 1.
Hand1 and Mash2 have intersecting expression domains in trophoblast. (B to E) Serial sections of an E8.5 implantation site following RNA in situ hybridization with antisense probes for Hand1 (B), Mash2 (C), Tpbp (D), and Pl1 (E) shown in dark field. (A) Section B shown in light field. Hand1 and Mash2 expression overlaps in cells of the ectoplacental cone. (F to H) Increased magnification of panels B to D in boxed area depicted in panel A. Arrows indicate the chorion. (I to L) Expression of Hand1 (I), Mash2 (J), Tpbp (K), and Pl1 (L) in E10.5 placenta. (M and N) Expression of Hand1 (M) and Mash2 (N) in E12.5 placenta. (A to E) Panels I to N at ×50 magnification. Ch, chorion; EPC, ectoplacental cone; Lab, labyrinthine layer; Sp, spongiotrophoblast layer.
Restricted expression of E factors in trophoblasts.
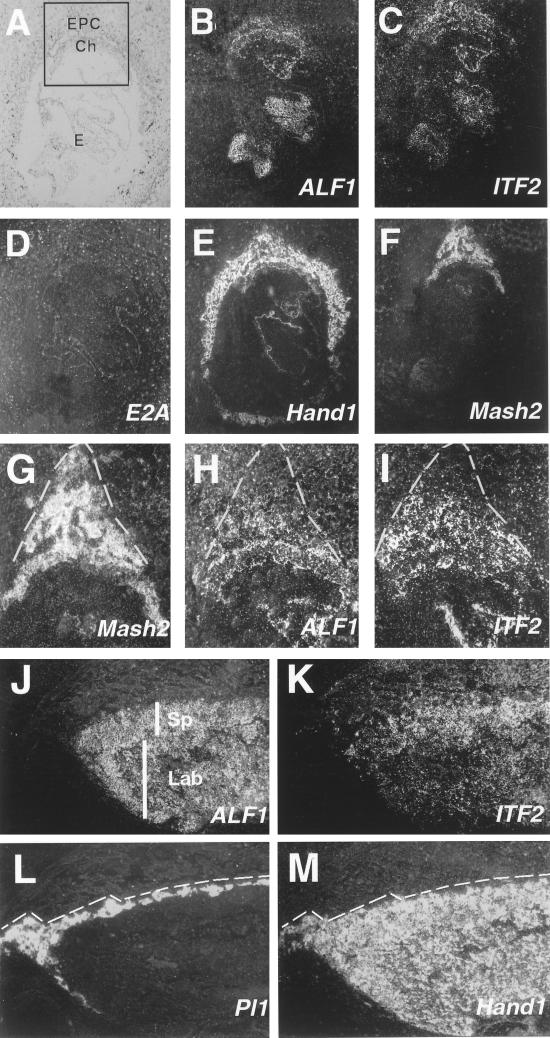
The function of many bHLH factors is dependent on their dimerization with one of the more widely expressed bHLH E factors (6, 26, 31). We examined the trophoblast expression of the three E-factor genes ALF1 (HEB, ITF1 [36]), E2A (E12/E47 [53]), and ITF2 (ME2 [47]), using RNA in situ hybridization. Placental expression of E2A could not be detected between E8.5 and E12.5 except in blood cells (Fig. 2D and data not shown). At E8.5, ALF1 and ITF2 transcripts localized to the embryo proper (not examined further), as well as to trophoblast cells of the chorion and ectoplacental cone (Fig. 2B and C). However, they were not detectable in trophoblast giant cells. In contrast to Mash2, expression of ALF1 and ITF2 did not extend to the limits of the ectoplacental cone (Fig. 2G to I, dotted lines) and was undetectable in the outer region where Tpbp was expressed (compare to Fig. 1H). By E10.5, transcripts of ALF1 and ITF2 remained undetectable in Pl1-positive trophoblast giant cells (Fig. 2J to M). Expression of ALF1 was evident throughout the spongiotrophoblast and labyrinthine layers (Fig. 2J), while that of ITF2 was nonuniform in the spongiotrophoblast layer (Fig. 2K) in a manner similar to that of Mash2 (Fig. 1J). These studies therefore defined a subpopulation of trophoblast cells that coexpress the E-factor genes ALF1 and ITF2, along with Hand1 and Mash2. Intriguingly, E-factor gene expression was undetectable in giant cells (both primary and secondary) at all stages examined.
FIG. 2.
E-factor gene expression during trophoblast development. (A to F) Serial sections of an E8.5 implantation site hybridized with antisense probes specific to ALF1 (B), ITF2 (C), E2A (D), Hand1 (E), and Mash2 (F) shown in dark field. (A) Section B shown in light field. E-factor gene expression is undetectable in trophoblast giant cells. (G to I) Magnification of boxed area shown in panel A for Mash2 (G), ALF1 (H), and ITF2 (I) probes. Note that ALF1 and ITF2 expression does not extend to the periphery of the ectoplacental cone (dotted line). (J to M) Serial sections of E10.5 placenta hybridized with probes for ALF1 (J), ITF2 (K), Pl1 (L), and Hand1 (M). Hand1 is expressed in Pl1-positive giant cells (bounded by the dotted line). (A to G) Panels J to M at ×5 magnification. Ch, chorion; E, embryo; EPC, ectoplacental cone; Lab, labyrinthine layer; Sp, spongiotrophoblast layer.
HAND1 can both homodimerize and heterodimerize with E factors.
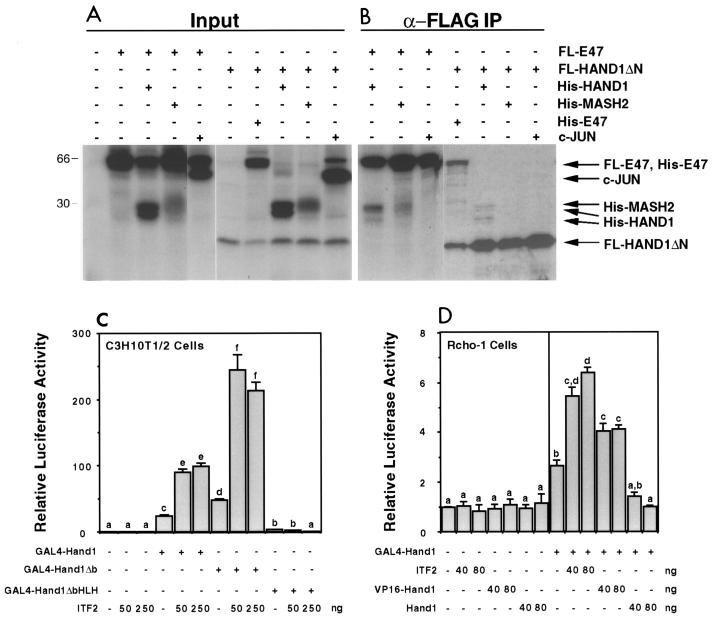
The colocalization of E-factor, Hand1, and Mash2 gene expression in the ectoplacental cone raised the possibility that their protein products could interact in vivo. The dimerization of HAND1 and MASH2 with E factors has been previously analyzed by a number of techniques (12, 19, 23). Coimmunoprecipitation assays were performed with in vitro-translated proteins to determine if other interactions could occur. Both HAND1 and MASH2 were coprecipitated when FL-E47 was used as the bait protein, while the negative control, c-Jun, was not (Fig. 3A). Using FL-HAND1 lacking the N terminus (FL-HAND1ΔN) as the bait, E47, but not MASH2, was coprecipitated (Fig. 3A). As both HAND1 and MASH2 coprecipitated with FL-E47 in the same assay, we conclude that HAND1-MASH2 heterodimers do not readily form. Interestingly, full-length HAND1 was also coprecipitated by FL-HAND1ΔN (Fig. 3A; note that His-HAND1 in vitro translation yields two bands). Compared to E47, a relatively small fraction of the total His-HAND1 was coprecipitated in this assay, suggesting that the affinity of the HAND1-HAND1 interaction in vitro was less than that of HAND1-E47 interaction.
FIG. 3.
HAND1 can both homodimerize and heterodimerize with E factors. (A and B) Coimmunoprecipitation assays using in vitro-translated FL-E47 or FL-HAND1ΔN as bait for untagged HAND1, MASH2, E47, and c-Jun. (A) Proteins were mixed and subjected to SDS-PAGE; (B) mixed proteins were immunoprecipitated (IP) with anti-FLAG antibody M2, washed, and resolved by SDS-PAGE. (C and D) Two-hybrid assays. The pL8G5-Luc reporter, in which luciferase expression is driven by a minimal promoter and five copies of the GAL4 upstream activation sequence DNA-binding site, was used along with 100 ng of the indicated GAL4 fusion construct. Different superscripts indicate statistically significant differences (P < 0.05).
To determine if these interactions could also occur in cells, two-hybrid assays were performed. An expression construct encoding HAND1 fused to the GAL4 DNA-binding domain (GAL4-HAND1) was used to allow recruitment of HAND1 to GAL4 binding sites located upstream of a luciferase reporter. In C3H10T1/2 fibroblasts, transfection of GAL4-HAND1 and GAL4-HAND1Δb (basic domain deleted) expression constructs stimulated reporter activity 25- and 50-fold, respectively. However, transfection of GAL4-HAND1ΔbHLH (bHLH domain deleted) had no significant effect on luciferase activity (Fig. 3C). Therefore, GAL4-HAND1 transcriptional activity was dependent on the HLH dimerization domain, likely due to recruitment of endogenous E factors which contain strong activation domains (37). Consistent with this observation, cotransfection of the E factor ITF2 potentiated GAL4-HAND1 (wild type and Δb)-mediated activation a further fourfold. This effect was likely due to dimerization with GAL4-HAND1, as luciferase activity was not affected by cotransfection of ITF2 when the GAL4-HAND1ΔbHLH mutant was used as bait (Fig. 3B).
In Rcho-1 rat trophoblast cells, transfection of GAL4-HAND1 stimulated reporter activity 2.5-fold (Fig. 3B), a modest level relative to that observed in C3H10T1/2 cells. Transfection of ITF2 potentiated GAL4-HAND1-mediated activation a further 2.5-fold. Wild-type HAND1 abolished GAL4-HAND1 activity, presumably by inhibiting the dimerization of GAL4-HAND1 with E factors. Significantly, transfection of a VP16 activation domain-HAND1 fusion increased GAL4-HAND1 activity 1.5-fold, with this effect being reproducible over a number of experiments. This most likely reflects HAND1 homodimerization, as VP16-HAND1 had no effect when GAL4-HAND1ΔbHLH was used as the bait in this assay (data not shown). The VP16-HAND1 fusion construct activated transcription 100-fold when pGAL4-ITF2 was used as the bait, confirming that VP16-HAND1 was functional (data not shown). Therefore, these results demonstrate that HAND1 homodimerization can occur in trophoblast cells. The ability of HAND1 to homodimerize appeared to vary with cell type, as it was not detected in C3H10T1/2 cells (data not shown).
HAND1 inhibits MASH2 binding to E-box sequences.
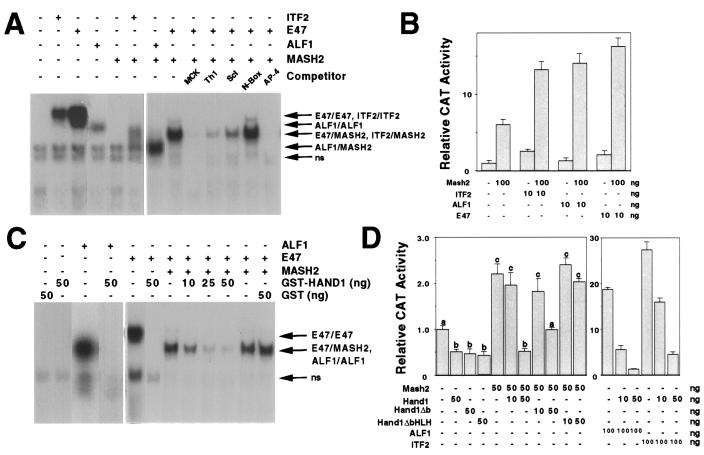
In electrophoretic mobility shift assays, MASH2 binds to canonical E-box sequences (CANNTG), such as the muscle creatine kinase (MCK) E-box (CACCTG), as a heterodimer with the E factors E47, ALF1, and ITF2 (Fig. 4A) (23). Transfection assays were performed in C3H10T1/2 fibroblasts in which a CAT reporter was regulated by two copies of the MCK E box. The MASH2 activity observed was presumably due to binding of heterodimers formed with endogenous E factors present in the cell, as the coaddition of low amounts of ITF2, ALF1, and E47 stimulated transcription with synergistic effects (Fig. 4B). We tested the specificity of this binding in competition assays. An excess of unlabeled oligonucleotides containing MCK and AP-4 (CAGCTG) E-box sequences abolished binding. While no competition was evident when non-E-box sequences were used, partial inhibition was observed when binding sites for Scleraxis (CATGTG [14]) and HAND1 (CATCTG [19]) were added (Fig. 4A).
FIG. 4.
HAND1 inhibits MASH2 binding to MCK E boxes by titrating E factors. (A and C) Electrophoretic mobility shift assay using a labeled MCK E-box probe. For indicated reactions, in vitro-translated FL-E47 (2 μl), His-MASH2 (4 μl), His-ALF1 (2 μl), and His-ITF2 (2 μl) were added. For competition assays, a 200-fold excess of unlabeled oligonucleotide was used. ns, nonspecific complex. (B and D) Transfection assays using C3H10T1/2 cells and the p2E MCK-CAT reporter, in which a CAT gene is driven by a minimal promoter and two copies of an MCK E-box sequence. Different superscripts indicate statistically significant differences (P < 0.05).
Unlike MASH2, GST-HAND1 binding to the MCK E box was not detected, either alone or in conjunction with the E factors E47 and ALF1. However, the addition of GST-HAND1 inhibited E47-MASH2 binding in a concentration-dependent manner (Fig. 4C). Similar results were obtained when E47-MASH2 dimers were formed prior to the addition of GST-HAND1 (data not shown). GST alone had no effect on E47-MASH2 binding (Fig. 4C). As HAND1 forms heterodimers with E factors but not MASH2, this inhibition most likely reflected the formation of E47-HAND1, at the expense of E47-MASH2, dimers. Consistent with this, HAND1 also inhibited binding of E47 and ALF1 homodimers (Fig. 4C). To further study the mechanism of HAND1 inhibition, transfection assays were performed with C3H10T1/2 fibroblasts and the MCK E-box reporter. Basal activity was stimulated 20- to 30-fold by cotransfection with the E factors ALF1 and ITF2 and to a lesser (twofold) extent by MASH2 alone (Fig. 4D). Cotransfection of HAND1 inhibited, in a concentration-dependent manner, the ability of MASH2, ALF1, and ITF2 to stimulate transcription from the reporter. The basic domain, responsible for DNA binding, was not required for this activity, as HAND1Δb similarly inhibited MASH2 and ITF2 (Fig. 4D and data not shown). However, deletion of the HLH domain (HAND1ΔbHLH) abolished HAND1-mediated inhibition. Therefore, while unable to bind to the MCK subclass of E boxes, HAND1 can interfere with E-box-mediated transcription by forming heterodimers with E factors, thereby titrating the pool available for heterodimerization with other bHLH factors such as MASH2.
HAND1 and MASH2 bind competitively to a specific E-box sequence.
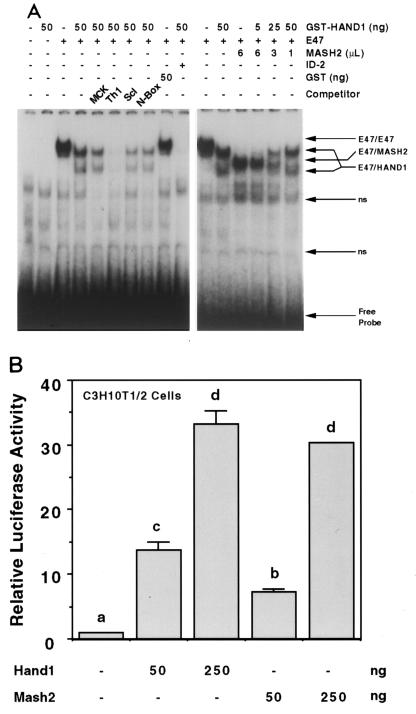
E47-HAND1 complexes bind to DNA with the consensus sequence NNTCTG (Th1 boxes) (19). To further examine this, electrophoretic mobility shift assays were performed with a labeled Th1 E-box oligonucleotide (CATCTG) as a probe. While E47 alone bound this sequence as a homodimer, the further addition of GST-HAND1 resulted in the formation of two new complexes. As partial cleavage of the GST moiety was evident in SDS-PAGE analysis of the purified GST-HAND1 preparation (data not shown), the mobilities of these two complexes were consistent with those of E47–GST-HAND1 and E47-HAND1. Binding of these complexes was abolished by addition of the HLH factor ID-2, which dimerizes with E47 (2). GST-HAND1 alone had no demonstrable binding activity at the concentrations used in these assays. However, binding was evident at 100-fold-higher concentrations (2 to 5 μg of protein), indicating that HAND1 homodimers may also bind DNA (data not shown). To evaluate the DNA-binding specificity of the E47-HAND1 heterodimer, we performed competition assays in which a large excess of unlabeled oligonucleotide was added. Of the sequences used, only the Th1 E box competed for binding (Fig. 5A). E47-HAND1 therefore binds to Th1-box sequences with high specificity, not to related E-box sequences. Interestingly, these latter sequences included sites known to be bound by close bHLH relatives such as Scleraxis (1) and members of the Hairy/E(SPL) family, the latter of which share with HAND1 an atypical proline residue found in the basic domain.
FIG. 5.
HAND1 and MASH2 bind and activate transcription from a Th1 E-box sequence as heterodimers with E factors. (A) Electrophoretic mobility shift assay using a labeled Th1 E-box probe. For indicated reactions, 2 and 4 μl of in vitro-translated FL-E47 and His-MASH2, respectively, were used. For competition assays, a 200-fold excess of unlabeled oligonucleotide was used. ns, nonspecific complex. (B) Transfection assays using C3H10T1/2 fibroblasts. The pL8E6-Luc reporter, in which luciferase expression is driven by a minimal promoter and six copies of the Th1 E box, was used. Different superscripts indicate statistically significant differences (P < 0.05).
To determine if HAND1 could activate transcription from the Th1 E box, transfection assays were performed with C3H10T1/2 fibroblasts and a luciferase reporter regulated by six copies of this sequence. Transfection of a HAND1 expression vector stimulated transcription up to 30-fold in a concentration-dependent manner (Fig. 5B). While these results were consistent within a given experiment, they were observed in only three of six attempts. A variety of culture modifications and different cell lots were tested, with similar results. In negative experiments, a VP16-HAND1 fusion also failed to stimulate transcription (data not shown), whereas HAND1 inhibition of ITF2-mediated activation was always observed (Fig. 4D). This indicates that the ability of HAND1–E-factor complexes to bind to Th1 E-box sequences may be regulated at a step following dimerization.
The competition assays demonstrated that E47-MASH2 also bound to the Th1 E-box sequence with moderate affinity (Fig. 4A). We therefore wished to determine if HAND1 and MASH2 could compete for binding to the same DNA site. In electrophoretic mobility shift assays, E47-MASH2 and E47-HAND1 dimers both bound to the Th1 E box (Fig. 5A). As the amount of GST-HAND1 added was increased and that of MASH2 decreased, E47-MASH2 binding declined in favor of E47-HAND1 complexes (Fig. 5A). At intermediate concentrations, both complexes were evident (Fig. 5A). However, no novel complexes were formed, indicating that E47-HAND1 and E47-MASH2 bind to a single Th1 E box in a mutually exclusive manner. The predominant DNA-binding species therefore mirrors the relative concentrations of MASH2 and HAND1. In transfection experiments, MASH2 stimulated transcription from the Th1 E-box reporter construct to levels comparable to those observed for HAND1 (Fig. 5B). Therefore, MASH2 and HAND1 can bind competitively to a shared subset of target sequences.
Inhibition of Mash2 is not sufficient for Hand1 function in trophoblasts.
To further examine the antagonistic activities of Hand1 and Mash2, giant cell differentiation assays were carried out with transfected Rcho-1 trophoblast cells. Typically, 5 to 10% of Rcho-1 cells in culture differentiate to trophoblast giant cells, a rate which is increased by overexpression of HAND1 (Fig. 6A) (12, 25). We performed transfection experiments in which increasing amounts of an expression vector encoding MASH2 was cotransfected with a fixed amount of a FLAG-HAND1 construct. The ability of FLAG-HAND1 to promote giant cell differentiation was inhibited by the MASH2 expression construct in a concentration-dependent fashion (Fig. 6A). This effect was not due to a decrease in the levels of FLAG-HAND1, as FLAG immunoreactivity was detected in a similar percentage of cells (Fig. 6B and data not shown). In contrast to the effect of MASH2, the giant cell-promoting activity of FLAG-HAND1 was not inhibited by cotransfection of ITF2 (data not shown). Therefore, as suggested by the biochemical data, the relative activities of HAND1 and MASH2 play a role in regulating giant cell differentiation.
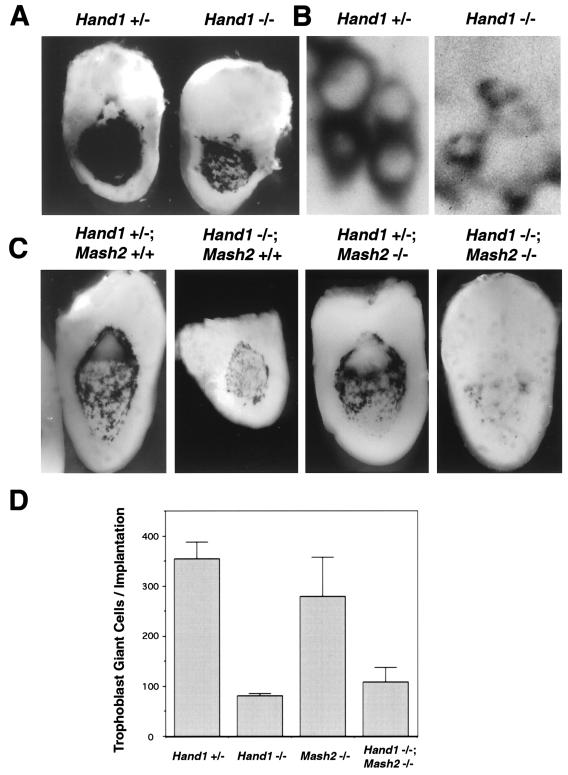
Based on its activities, the function of Hand1 in promoting giant cell differentiation could be mediated, at least in part, by the repression of MASH2-mediated transcription of target genes and/or Mash2 gene expression (38). To determine if the only function of Hand1 is to inhibit Mash2, Hand1/Mash2 double-mutant embryos were generated. If the major function of Hand1 in trophoblast was to inhibit Mash2 expression and/or activity, the Hand1 mutant phenotype might be a consequence of ectopic Mash2 activity, which is known to be sufficient to block giant cell differentiation (12, 25). In this case, the trophoblast defects of Hand1 mutants should be rescued by the further inactivation of Mash2. To examine trophoblast giant cell differentiation, E8.5 implantation sites were bisected and subjected to RNA in situ hybridization with a Pl1 probe. The perinuclear localization of Pl1 mRNA to the rough endoplasmic reticulum of trophoblast giant cells allowed an examination of nuclear size. Strikingly, the nuclei of mural trophectoderm cell derivatives surrounding the implantation site were significantly smaller in Hand1 mutants (Fig. 7B), reflecting a failure for these cells to increase in ploidy and indicating that giant cell transformation was blocked. The number of Pl1-expressing cells was determined (Fig. 7C and D) as a measure of the number of trophoblast cells surrounding each implantation site; the number reflects the derivatives of mural trophectoderm (primary giant cells) plus secondary giant cells that arise after implantation. Because Pl1 expression is reduced in mural trophectodermal derivatives in Hand1 mutants (Fig. 7C) (38), their numbers were independently assessed using the 6AD1βgeo (encoding β-galactosidase) allele of the Mrj gene (Fig. 7A), which is expressed in trophoblast giant cells (20). By either assessment, whereas wild-type implantation sites were lined by about 350 cells, mutants contained only about 80 cells. Strikingly, this low number is not significantly different from the number of mural trophectoderm cells found in the E4.5 blastocyst (10). The lower expression of Pl1, smaller nuclear size, and decreased numbers of trophoblast cells lining the implantation site observed in Hand1 mutants were all unaltered in Hand1/Mash2 compound mutants (Fig. 7C and D). Therefore, Hand1 function in trophoblast is not limited to restricting Mash2 expression and/or activity.
FIG. 7.
Decreased mural trophectoderm cell number and nuclear size in Hand1 mutants is independent of Mash2 function. (A) β-Galactosidase staining for the 6AD1βgeo allele in E8.5 Hand1 +/− and −/− implantation sites. (B) High-magnification view of panel C showing mural trophoblast cells along the lateral side of the implantation site. Pl1 transcript localization is perinuclear. (C) E8.5 implantation sites derived from crosses between Hand1 +/−; Mash2 +/− compound heterozygotes were bisected and subjected to whole-mount in situ hybridization using an antisense Pl1 probe. Shown is a low-magnification view of Pl1 expression in trophoblast giant cells in one-half of the implantation site. (D) Pl1-positive giant cells per conceptus were counted (three implantation sites per genotype).
The ability of Hand1 to promote giant cell differentiation regardless of its effect on Mash2 function could be mediated by two independent mechanisms: (i) indirectly, by inhibiting the dimerization of other bHLH factors, as yet unknown, with their E-factor partners; and (ii) directly, by regulating the transcription of target genes when bound to Th1-box sequences. To discriminate between these activities, we tested the function of the HAND1 basic domain mutant protein in transfected Rcho-1 cells. In contrast to wild-type HAND1, transfection of an expression vector encoding HAND1Δb (DNA-binding basic domain deleted) had no significant effect on giant cell differentiation relative to control (empty expression vector) (Fig. 6C). Importantly, the same HAND1Δb mutant was able to inhibit both E-factor- and MASH2-stimulated transcription (Fig. 4D), indicating that it is expressed and active in transfected cells. These data suggest that in order to promote giant cell differentiation, HAND1 does not solely inhibit the activity of other bHLH proteins but also has a direct (DNA-binding) role in this process.
DISCUSSION
Trophoblast giant cells develop throughout gestation due to the terminal differentiation of precursor cells present first in the ectoplacental cone and later in the spongiotrophoblast layer of the placenta. This process is regulated by bHLH transcription factors encoded by the Hand1 and Mash2 genes, which have stimulatory and repressive roles, respectively. We show here that Hand1 and Mash2 expression patterns overlap in the ectoplacental cone and spongiotrophoblast but that Mash2 expression is down-regulated as these trophoblast cell subpopulations differentiate into giant cells. The expression of the E-factor genes ALF1 and ITF2 is regulated during trophoblast development, as their expression is extinguished in advance of giant cell differentiation. This finding has important implications for understanding both MASH2 and HAND1 function. First, the loss of E-factor partners should restrict MASH2 function, as E factors are essential DNA-binding partners for MASH2. The expression of other bHLH proteins (including HAND1) could also reduce MASH2 function by competing for dimerization with E factors. Second, HAND1 function could change during trophoblast differentiation due to alterations in its DNA-binding specificity. In the ectoplacental cone and spongiotrophoblast, HAND1 can associate with an E-factor bHLH protein, whereas in trophoblast giant cells, where the expression of these factors was not detected, HAND1 presumably associates with a different partner. The switching of dimerization partners could therefore lead to a different DNA-binding specificity, and as a result, the target genes to which HAND1 complexes bind would change. Together, these data suggest that trophoblast differentiation is regulated by interactions between multiple bHLH proteins.
Three distinct expression domains of bHLH factors in the trophoblast lineage.
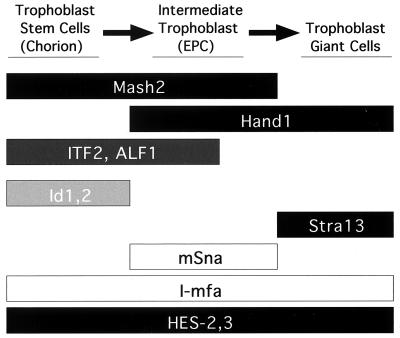
Primary culture experiments have suggested that at least three functionally distinct trophoblast subpopulations exist in the E7.5-8.5 murine placenta (42, 43). Hand1 and Mash2 expression domains fit into these three trophoblast compartments: (i) the chorion, which expresses only Mash2; (ii) the ectoplacental cone, where Mash2 and Hand1 are coexpressed; and (iii) giant cells, where only Hand1 transcripts are present (summarized in Fig. 8). In intermediate trophoblast cells of the ectoplacental cone, the opposing activities of Hand1 and Mash2 clearly must be coordinated. Interestingly, our examination revealed that expression of E factors, the obligate dimerization partners of most bHLH factors, was also subject to regulation. Transcripts of ALF1 and ITF2 were localized to the chorion and ectoplacental cone at E8.5, and to the labyrinthine and spongiotrophoblast layers at later periods, but remained undetectable in trophoblast giant cells at all stages. Strikingly, a more detailed analysis revealed that expression of ALF1 and ITF2 was down-regulated at the periphery of the ectoplacental cone (Fig. 8). As this occurs prior to overt giant cell differentiation, it seems likely that their protein products are not present in giant cells. The absence of an E-factor partner would be expected to effectively abolish MASH2 activity, thereby allowing giant cell differentiation to occur.
FIG. 8.
Distinct bHLH compartments in early trophoblast development. Shown is a summary of expression patterns of bHLH factors and modifiers in trophoblast at E8.5. EPC, ectoplacental cone. Boxes: black, bHLH factors; dark gray, E factors; light gray, HLH factors; white, non-HLH factors.
The E-factor genes are widely considered to be ubiquitously expressed, although this belief primarily stems from whole-tissue Northern blot and low-resolution in situ hybridization analysis (40, 53). Closer examination in the developing central nervous system revealed E-factor gene expression in proliferating neuroblasts and neurons at the initial stages of differentiation yet an absence in more mature, differentiated cells (8, 35, 47). Immunohistochemical evaluation of E12/47 protein in a number of organs demonstrated that expression is restricted primarily to proliferating or relatively undifferentiated cells (44). Additionally, studies of ALF1 expression in Schwann cells revealed a discordance between protein and transcript levels, with protein levels sharply down-regulated in terminally differentiated cells which continue to express abundant levels of the mRNA (48). Therefore, the absence of E-factor gene expression in trophoblast giant cells is generally consistent with findings in other organs, where E-factor expression is down-regulated in advance of terminal differentiation.
Expression of other bHLH factors in trophoblast has been described (Fig. 8). HES-2 and -3 are expressed in all trophoblast subpopulations (32), while expression of Stra13 is restricted to giant cells (4). The dominant-negative HLH genes Id-1 and Id-2 are expressed in the chorion only (22). Overexpression of ID-2 in Rcho-1 trophoblast inhibits giant cell differentiation, demonstrating a role for these factors in trophoblast development (12). In addition, non-bHLH modifiers of bHLH activity are also expressed in trophoblast. The zinc finger transcription factor gene mSna, which inhibits giant cell differentiation in Rcho-1 trophoblast cells, is expressed in the ectoplacental cone and spongiotrophoblast but is down-regulated during giant cell differentiation (33). Interestingly, mSNA binds to E-box sequences identical to those bound by MASH2. I-mfa, first identified as an inhibitor of myogenic bHLH factors (7), is expressed in all three trophoblast layers, and I-mfa mutants have a reduced number of trophoblast giant cells. I-mfa binds to and inhibits MASH2 in vitro and promotes giant cell differentiation in Rcho-1 cells (25). I-mfa can also bind to HAND1 (25), although the functional consequences of this are unknown. The dynamic expression patterns of Hand1, Mash2, E-factor genes, and other regulators therefore define distinct bHLH environments present in the trophoblast subpopulations of the placenta (Fig. 8). Within each of these compartments, the biochemical interplay between the different factors likely allows precise control over the relative actions of HAND1 and MASH2.
Multiple roles of Hand1 in trophoblast development.
Hand1 mutant conceptuses show two distinct phenotypes in the trophoblast lineage. The most striking is that trophoblast giant cell differentiation is arrested at an early stage. Although the trophoblast cell lineage is established in the mutants and the conceptuses implant normally, the derivatives of the mural trophectoderm do not undergo proper primary giant cell transformation, as their nuclei remain relatively small (shown in this study). The enlarged nuclei of giant cells reflect their increased DNA content due to endoreduplication, i.e., continuous rounds of DNA synthesis without intervening mitoses (54). In addition, secondary giant cell differentiation does not occur, based on the failure of peripheral ectoplacental cone cells to activate giant cell-specific genes (e.g., Pl1 and Limk) and to reduce ectoplacental cone-specific genes (e.g., Mash2 and Tpbp) (38). Consistent with the latter data, we demonstrated that the number of trophoblast cells surrounding Hand1 mutant implantation sites at E8.5 (around 80) does not significantly differ from the number of mural trophectoderm cells present at the expanded blastocysts stage (10). In addition to the giant cell phenotype, we have previously observed a phenotype in the ectoplacental cone (38). Trophoblast cells of the ectoplacental cone are precursors of secondary giant cells (43). Therefore, their numbers might be predicted to increase as a consequence of a failure in secondary giant cell differentiation. However, the ectoplacental cones of Hand1 mutants are significantly smaller than wild type (38). Therefore, while Hand1 mutant ectoplacental cone cells are unable to undergo giant cell differentiation, their proliferation and/or maintenance is also directly affected. The precise nature of this latter defect is unknown at present, though the mutant ectoplacental cone cells continue to express the correct cell-specific genes (e.g., Tpbp, mSna, and Mash2) (38). The fact that the number of giant cell precursors is not increased in Hand1 mutants likely indicates that these cells are able to exit the mitotic cell cycle normally but cannot initiate the giant cell differentiation program. It is clear from our analyses, therefore, that Hand1 has distinct functions in two separate trophoblast subpopulations. HAND1 likely regulates different genetic programs in these two trophoblast subpopulations. This suggests additional level(s) of regulation that determine the nature of HAND1 activity in a given cell type.
Regulation of distinct HAND1 activities.
The combination of expression and biochemical data offers insight into how HAND1 could have distinct functions in the trophoblast lineage. In cells at the core of the ectoplacental cone, HAND1 should predominantly form HAND1–E-factor heterodimers, which would presumably regulate target genes involved in the proliferation and/or maintenance of these cells. At the periphery of the ectoplacental cone, prior to secondary trophoblast giant cell differentiation, E-factor expression is down-regulated. Therefore, HAND1 likely promotes giant cell differentiation, in the absence of E factors, as a dimer with a different bHLH partner. We found that HAND1 can homodimerize in both coimmunoprecipitation and mammalian two-hybrid experiments. Alternatively, HAND1 may dimerize with a different partner. Among the bHLH factor genes studied, only Hand1, Stra13 (4), and members of the HES (32) family are known to be expressed in giant cells. The precise nature of the complex functional in giant cells is unknown at present. However, the switch in the HAND1 partner between the different trophoblast subpopulations could alter its DNA-binding site specificity. The DNA target sequences of bHLH factor dimers reflect the half-site binding specificities of the two proteins comprising the dimer complex. HAND1–E-factor heterodimers bind to NNTCTG sequences, representing the half-site sequences bound by the HAND1 (NNT) and E-factor (CAG) molecules (19). Therefore, by switching in giant cells to a partner protein with a half-site specificity different from that of E factors, HAND1 could regulate the transcription of a different set of genes. This paradigm is observed with ARNT, which binds to different DNA sequences in homodimeric versus heterodimeric complexes (49). This may represent a general mechanism through which bHLH factors can have a spectrum of activities in subpopulations of one lineage. The promotion of homodimer formation only in more mature cells may help to ensure in earlier precursor cells that differentiation activities incompatible with their development (e.g., onset of the endocyle) are repressed until appropriate.
It is logical to predict that the lack of E-factor expression in giant cell precursors would facilitate a change in HAND1's dimerization partner. However, it is not clear from our experiments that this down-regulation is actually required. A variety of other mechanisms aside from E-factor availability have been reported to alter bHLH dimerization specificity (27, 29). Indeed we have found that cotransfection of ITF2 does not block the ability of HAND1 to promote giant cell differentiation (I. C. Scott, unpublished results). Further work will be required to scrutinize the predictions of this model for HAND1 functions. Tethered bHLH dimers in which a single polyprotein encodes the two bHLH proteins to be tested separated by a flexible linker can be generated (34). As these complexes are resistant to disruption by other HLH proteins present in the cell, their use would permit dissection of the specific activities of individual complexes (e.g., HAND1–E factor versus HAND1-HAND1) in trophoblast development. Transfection of trophoblast stem cell lines (51) derived from Hand1 mutants would provide the best system for examining these activities.
ACKNOWLEDGMENTS
We thank N. Hattori, who made important contributions to the Rcho-1 differentiation experiments. A. Nagy provided Mash2 mutant mice. S. Hollenberg, T. Neuman, P. Jorgensen, and S. Fisher kindly provided plasmids. We also thank K. Harpal for his aid in histology and tissue sectioning.
This work was supported by a grant from the Medical Research Council of Canada (to J.C.C.). I.C.S. was supported by a studentship from the Natural Science and Engineering Research Council, P.R. was supported by a fellowship from the Wellcome Trust, and J.C.C. is a Scholar of the Medical Research Council of Canada.
REFERENCES
- 1.Atchley W R, Fitch W M. A natural classification of the basic helix-loop-helix class of transcription factors. Proc Natl Acad Sci USA. 1997;94:5172–5176. doi: 10.1073/pnas.94.10.5172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Benezra R, Davis R L, Lockshon D, Turner D L, Weintraub H. The protein Id: a negative regulator of helix-loop-helix DNA binding proteins. Cell. 1990;61:49–59. doi: 10.1016/0092-8674(90)90214-y. [DOI] [PubMed] [Google Scholar]
- 3.Blanar M A, Rutter W J. Interaction cloning: identification of a helix-loop-helix zipper protein that interacts with c-Fos. Science. 1992;256:1014–1018. doi: 10.1126/science.1589769. [DOI] [PubMed] [Google Scholar]
- 4.Boudjelal M, Taneja R, Matsubara S, Bouillet P, Dolle P, Chambon P. Overexpression of Stra13, a novel retinoic acid-inducible gene of the basic helix-loop-helix family, inhibits mesodermal and promotes neuronal differentiation of P19 cells. Genes Dev. 1997;11:2052–2065. doi: 10.1101/gad.11.16.2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Calzonetti T, Stevenson L, Rossant J. A novel regulatory region is required for trophoblast-specific transcription in transgenic mice. Dev Biol. 1995;171:615–626. doi: 10.1006/dbio.1995.1309. [DOI] [PubMed] [Google Scholar]
- 6.Chakraborty T, Brennan T J, Li L, Edmondson D, Olson E N. Inefficient homooligomerization contributes to the dependence of myogenin on E2A products for efficient DNA binding. Mol Cell Biol. 1991;11:3633–3641. doi: 10.1128/mcb.11.7.3633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen C-M A, Kraut N, Groudine M, Weintraub H. I-mf, a novel myogenic repressor, interacts with members of the MyoD family. Cell. 1996;86:731–741. doi: 10.1016/s0092-8674(00)80148-8. [DOI] [PubMed] [Google Scholar]
- 8.Chiaramello A, Soosaar A, Neuman T, Zuber M X. Differential expression and distinct DNA-binding specificity of ME1a and ME2 suggest a unique role during differentiation and neuronal plasticity. Brain Res Mol Brain Res. 1995;29:107–118. doi: 10.1016/0169-328x(94)00236-8. [DOI] [PubMed] [Google Scholar]
- 9.Conlon R A, Reaume A G, Rossant J. Notch1 is required for the coordinate segmentation of somites. Development. 1995;121:1533–1545. doi: 10.1242/dev.121.5.1533. [DOI] [PubMed] [Google Scholar]
- 10.Copp A J. Interaction between inner cell mass and trophectoderm of the mouse blastocyst. I. Study of cellular proliferation. J Embryol Exp Morphol. 1978;48:109–125. [PubMed] [Google Scholar]
- 11.Cross J C. Trophoblast function in normal and preeclamptic pregnancy. Fetal Maternal Med Rev. 1996;8:57–66. [Google Scholar]
- 12.Cross J C, Flannery M L, Blanar M A, Steingrimsson E, Jenkins N A, Copeland N G, Rutter W J, Werb Z. Hxt encodes a basic helix-loop-helix transcription factor that regulates trophoblast cell development. Development. 1995;121:2513–2523. doi: 10.1242/dev.121.8.2513. [DOI] [PubMed] [Google Scholar]
- 13.Cross J C, Werb Z, Fisher S J. Implantation and the placenta: key pieces of the development puzzle. Science. 1994;266:1508–1518. doi: 10.1126/science.7985020. [DOI] [PubMed] [Google Scholar]
- 14.Cserjesi P, Brown D, Ligon K L, Lyons G E, Copeland N G, Gilbert D J, Jenkins N A, Olson E N. Scleraxis: a basic helix-loop-helix protein that prefigures skeletal formation during mouse embryogenesis. Development. 1995;121:1099–1110. doi: 10.1242/dev.121.4.1099. [DOI] [PubMed] [Google Scholar]
- 15.Cserjesi P, Brown D, Lyons G E, Olson E N. Expression of the novel basic helix-loop-helix gene eHAND in neural crest derivatives and extraembryonic membranes during mouse development. Dev Biol. 1995;170:664–678. doi: 10.1006/dbio.1995.1245. [DOI] [PubMed] [Google Scholar]
- 16.Faria T N, Soares M J. Trophoblast cell differentiation: establishment, characterization, and modulation of a rat trophoblast cell line expression members of the placental prolactin family. Endocrinology. 1991;129:2895–2906. doi: 10.1210/endo-129-6-2895. [DOI] [PubMed] [Google Scholar]
- 17.Gardner R L, Papaioannou V E, Barton S C. Origin of the ectoplacental cone and secondary giant cells in mouse blastocysts reconstituted from isolated trophoblast and inner cell mass. J Embryol Exp Morphol. 1973;30:561–572. [PubMed] [Google Scholar]
- 18.Guillemot F, Nagy A, Auerbach A, Rossant J, Joyner A L. Essential role of Mash-2 in extraembryonic development. Nature. 1994;371:333–336. doi: 10.1038/371333a0. [DOI] [PubMed] [Google Scholar]
- 19.Hollenberg S M, Sternglanz R, Cheng P F, Weintraub H. Identification of a new family of tissue-specific basic helix-loop-helix proteins with a two-hybrid system. Mol Cell Biol. 1995;15:3813–3822. doi: 10.1128/mcb.15.7.3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hunter P J, Swanson B J, Haendel M A, Lyons G E, Cross J C. Mrj encodes a DnaJ-related co-chaperone that is essential for murine placental development. Development. 1999;126:1247–1258. doi: 10.1242/dev.126.6.1247. [DOI] [PubMed] [Google Scholar]
- 21.Jackson L L, Colosi P, Talamantes F, Linzer D I. Molecular cloning of mouse placental lactogen cDNA. Proc Natl Acad Sci USA. 1986;83:8496–8500. doi: 10.1073/pnas.83.22.8496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jen Y, Manova K, Benezra R. Each member of the Id gene family exhibits a unique expression pattern in mouse gastrulation and neurogenesis. Dev Dyn. 1997;208:92–106. doi: 10.1002/(SICI)1097-0177(199701)208:1<92::AID-AJA9>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 23.Johnson J E, Birren S J, Saito T, Anderson D J. DNA binding and transcriptional regulatory activity of mammalian achaete-scute homologous (MASH) proteins revealed by interaction with a muscle-specific enhancer. Proc Natl Acad Sci USA. 1992;89:3596–3600. doi: 10.1073/pnas.89.8.3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johnson M H. Relationship between inner cell mass derivatives and trophoblast proliferation in ectopic pregnancy. J Embryol Exp Morphol. 1972;28:306–312. [Google Scholar]
- 25.Kraut N, Snider L, Amy Chen C-M, Tapscott S J, Groudine M. Requirement of the mouse I-mfa gene for placental development and skeletal patterning. EMBO J. 1998;17:6276–6288. doi: 10.1093/emboj/17.21.6276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lassar A B, Davis R L, Wright W E, Kadesch T, Murre C, Voronova A, Baltimore D, Weintraub H. Functional activity of myogenic HLH proteins requires hetero-oligomerization with E12/47-like proteins in vivo. Cell. 1991;66:305–315. doi: 10.1016/0092-8674(91)90620-e. [DOI] [PubMed] [Google Scholar]
- 27.Lenormand J L, Benayoun B, Guillier M, Vandromme M, Leibovitch M P, Leibovitch S A. Mos activates myogenic differentiation by promoting heterodimerization of MyoD and E12 proteins. Mol Cell Biol. 1997;17:584–593. doi: 10.1128/mcb.17.2.584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lescisin K R, Varmuza S, Rossant J. Isolation and characterization of a novel trophoblast-specific cDNA in the mouse. Genes Dev. 1988;2:1639–1646. doi: 10.1101/gad.2.12a.1639. [DOI] [PubMed] [Google Scholar]
- 29.Markus M, Benezra R. Two isoforms of protein disulfide isomerase alter the dimerization status of E2A proteins by a redox mechanism. J Biol Chem. 1999;274:1040–1049. doi: 10.1074/jbc.274.2.1040. [DOI] [PubMed] [Google Scholar]
- 30.Millen K, Hui C. Radioactive hybridization of tissue sections. In: Krieg P, editor. A laboratory guide to RNA: isolation, analysis and synthesis. New York, N.Y: Wiley-Liss; 1996. pp. 339–355. [Google Scholar]
- 31.Mitsui K, Shirakata M, Paterson B M. Phosphorylation inhibits the DNA-binding activity of MyoD homodimers but not MyoD-E12 heterodimers. J Biol Chem. 1993;268:24415–24420. [PubMed] [Google Scholar]
- 32.Nakayama H, Liu Y, Stifani S, Cross J C. Developmental restriction of Mash-2 expression in trophoblast correlates with potential activation of the NOTCH-2 pathway. Dev Genet. 1997;21:21–30. doi: 10.1002/(SICI)1520-6408(1997)21:1<21::AID-DVG3>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 33.Nakayama H, Scott I C, Cross J C. The transition to endoreduplication in trophoblast giant cells is regulated by the Sna zinc-finger transcription factor. Dev Biol. 1998;199:150–163. doi: 10.1006/dbio.1998.8914. [DOI] [PubMed] [Google Scholar]
- 34.Neuhold L A, Wold B. HLH forced dimers: tethering MyoD to E47 generates a dominant positive myogenic factor insulated from negative regulation by Id. Cell. 1993;74:1033–1042. doi: 10.1016/0092-8674(93)90725-6. [DOI] [PubMed] [Google Scholar]
- 35.Neuman T, Keen A, Knapik E, Shain D, Ross M, Nornes H O, Zuber M X. ME1 and GE1: basic helix-loop-helix transcription factors expressed at high levels in the developing nervous system and in morphogenetically active regions. Eur J Neurosci. 1993;5:311–318. doi: 10.1111/j.1460-9568.1993.tb00498.x. [DOI] [PubMed] [Google Scholar]
- 36.Nielsen A L, Pallisgaard N, Pedersen F S, Jorgensen P. Murine helix-loop-helix transcriptional activator proteins binding to the E-box motif of the Akv murine leukemia virus enhancer identified by cDNA cloning. Mol Cell Biol. 1992;12:3449–3459. doi: 10.1128/mcb.12.8.3449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Quong M W, Massari M E, Zwart R, Murre C. A new transcriptional-activation motif restricted to a class of helix-loop-helix proteins is functionally conserved in both yeast and mammalian cells. Mol Cell Biol. 1993;13:792–800. doi: 10.1128/mcb.13.2.792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Riley P, Anson-Cartwright L, Cross J C. The Hand1 helix-loop-helix transcription factor is essential for placentation and cardiac morphogenesis. Nat Genet. 1998;18:271–275. doi: 10.1038/ng0398-271. [DOI] [PubMed] [Google Scholar]
- 39.Rinkenberger J, Cross J C, Werb Z. Molecular genetics of implantation in the mouse. Dev Genet. 1997;21:6–20. doi: 10.1002/(SICI)1520-6408(1997)21:1<6::AID-DVG2>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 40.Roberts V J, Steenbergen R, Murre C. Localization of E2A mRNA expression in developing and adult rat tissues. Proc Natl Acad Sci USA. 1993;90:7583–7587. doi: 10.1073/pnas.90.16.7583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rossant J, Guillemot F, Tanaka M, Latham K, Gertsenstein M, Nagy A. Mash2 is expressed in oogenesis and preimplantation development but is not required for blastocyst formation. Mech Dev. 1998;73:138–191. doi: 10.1016/s0925-4773(98)00051-3. [DOI] [PubMed] [Google Scholar]
- 42.Rossant J, Ofer L. Properties of extra-embryonic ectoderm isolated from postimplantation mouse embryos. J Embryol Exp Morphol. 1977;39:183–194. [PubMed] [Google Scholar]
- 43.Rossant J, Tamura-Lis W. Effect of culture conditions on diploid to giant-cell transformation in postimplantation mouse trophoblast. J Embryol Exp Morphol. 1981;62:217–227. [PubMed] [Google Scholar]
- 44.Rutherford M N, LeBrun D P. Restricted expression of E2A protein in primary human tissues correlates with proliferation and differentiation. Am J Pathol. 1998;153:165–173. doi: 10.1016/S0002-9440(10)65557-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Press; 1989. [Google Scholar]
- 46.Soares M J, Chapman B M, Kamei T, Yamamoto T. Control of trophoblast cell differentiation: lessons from genetics of early pregnancy loss and trophoblast neoplasia. Dev Growth Differ. 1995;37:355–364. doi: 10.1046/j.1440-169X.1995.t01-3-00001.x. [DOI] [PubMed] [Google Scholar]
- 47.Soosaar A, Chiaramello A, Zuber M X, Neuman T. Expression of basic-helix-loop-helix transcription factor ME2 during brain development and in the regions of neuronal plasticity in the adult brain. Brain Res Mol Brain Res. 1994;25:176–180. doi: 10.1016/0169-328x(94)90297-6. [DOI] [PubMed] [Google Scholar]
- 48.Stewart H J S, Zoidl G, Rossner M, Brennan A, Zoidl C, Nave K-A, Mirsky R, Jessen K R. Helix-loop-helix proteins in Schwann cells: a study of regulation and subcellular localization of Ids, REB, and E12/47 during embryonic and postnatal development. J Neurosci Res. 1997;50:684–701. doi: 10.1002/(SICI)1097-4547(19971201)50:5<684::AID-JNR6>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 49.Swanson H I, Chan W K, Bradfield C A. DNA binding specificities and pairing rules of the Ah receptor, ARNT, and SIM proteins. J Biol Chem. 1995;270:26292–26302. doi: 10.1074/jbc.270.44.26292. [DOI] [PubMed] [Google Scholar]
- 50.Tanaka M, Gertsenstein M, Rossant J, Nagy A. Mash2 acts cell autonomously in mouse spongiotrophoblast development. Dev Biol. 1997;190:55–65. doi: 10.1006/dbio.1997.8685. [DOI] [PubMed] [Google Scholar]
- 51.Tanaka S, Kunath T, Hadjantonakis A-N, Nagy A, Rossant J. Promotion of trophoblast stem cell proliferation by FGF4. Science. 1998;282:2072–2075. doi: 10.1126/science.282.5396.2072. [DOI] [PubMed] [Google Scholar]
- 52.Voronova A, Baltimore D. Mutations that disrupt DNA binding and dimer formation in the E47 helix-loop-helix protein map to distinct domains. Proc Natl Acad Sci USA. 1990;87:4722–4726. doi: 10.1073/pnas.87.12.4722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Watada H, Kajimoto Y, Umayahara Y, Matsuoka T, Morishima T, Yamasaki Y, Kawamori R, Kamada T. Ubiquitous, but variable, expression of two alternatively spliced mRNAs encoding mouse homologues of transcription factors E47 and E12. Gene. 1995;153:255–259. doi: 10.1016/0378-1119(94)00765-k. [DOI] [PubMed] [Google Scholar]
- 54.Zybina E V, Zybina T G. Polyteny and endomitosis in supergiant trophoblast cells of the gray vole Microtus subarvalis. Tsitologiia. 1985;27:402–410. [PubMed] [Google Scholar]