Abstract
The treatment of coronavirus disease (COVID-19) and COVID-19-associated diarrhea remains challenging. This study aimed to evaluate the efficacy of a multi-strain probiotic in the treatment of COVID-19. This was a randomized, controlled, single-center, open-label trial (NCT04854941). Inpatients with confirmed COVID-19 and pneumonia were randomly assigned to a group that received a multi-strain probiotic (PRO group) or to the control group (CON group). There were 99 and 101 patients in the PRO and CON groups, respectively. No significant differences in mortality, total duration of disease and hospital stay, incidence of intensive care unit admission, need for mechanical ventilation or oxygen support, liver injury development, and changes in inflammatory biomarker levels were observed between the PRO and CON groups among all included patients as well as among subgroups delineated based on age younger or older than 65 years, and subgroups with chronic cardiovascular diseases and diabetes. Diarrhea on admission was observed in 11.5% of patients; it resolved earlier in the PRO group than in the CON group (2 [1–4] vs. 4 [3–6] days; p = 0.049). Hospital-acquired diarrhea developed less frequently in the PRO group than in the CON group among patients who received a single antibiotic (0% vs. 12.5%; p = 0.023) unlike among those who received > 1 antibiotic (10.5% vs. 13.3%; p = 0.696). The studied probiotic had no significant effect on mortality and changes in most biomarkers in COVID-19. However, it was effective in treating diarrhea associated with COVID-19 and in preventing hospital-acquired diarrhea in patients who received a single antibiotic.
Supplementary Information
The online version contains supplementary material available at 10.1007/s12602-021-09858-5.
Keywords: COVID-19, Probiotics, Diarrhea, Mortality, Liver
Introduction
Coronavirus disease 2019 (COVID-19) is an acute respiratory infection with systemic manifestations [1]. Despite intensive research, COVID-19 treatment remains an important challenge [2]. Probiotics are live microorganisms that, when administered in adequate amounts, confer health benefits on the host [3]. They have been reported to show a positive effect on acute respiratory infections by modulating immune responses [4].
Diarrhea is a manifestation of COVID-19, and has been reported in approximately 10% of COVID-19 patients [5]. COVID-19-associated diarrhea has distinct characteristics [6], and may even be the first manifestation of COVID-19 [7]. Although the exact mechanisms of the development of diarrhea in COVID-19 remain unknown, the following factors of its pathogenesis can be suggested. The viral E protein binds to proteins of the tight junctions of enterocytes, which leads to an increase in the permeability of the intestinal barrier, bacterial translocation, and gut inflammation. Moreover, the viral proteins E and Orf3a also disrupt the functioning of ion channels in the enterocyte. In addition, the pathogenesis of diarrhea in COVID-19 patients may also include intestinal damage during a cytokine storm, gut dysbiosis, and Clostridioides difficile superinfection [8]. A number of probiotics have been shown to be effective in the treatment of diarrhea of various etiologies [9–11].
Liver injury develops in an average of 20–25% of COVID-19 patients [12]. Several probiotics have been shown to be effective in the treatment of certain liver diseases [13].
Many experts have suggested the use of probiotics in combination with other drugs for the management of COVID-19 [14–21].
The effect of multi-strain probiotics on COVID-19 has been evaluated in three published retrospective studies [22–24]. Li et al. reported that the administration of a probiotic including Bifidobacterium longum subsp. infantis, Bifidobacterium longum subsp. longum, Lactobacillus acidophilus, Lactobacillus delbrueckii subsp. bulgaricus, Bacillus cereus, Bacillus subtilis, Streptococcus thermophiles, and Enterococcus faecium led to a slight decrease in hospitalization duration and accelerated the clearance of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the causative agent of COVID-19 [22]. Ceccarelli et al. showed that the use of a probiotic including S. thermophilus DSM 32,245, Bifidobacterium lactis DSM 32,246, Bifidobacterium lactis DSM 32,247, L. acidophilus DSM 32,241, L. helveticus DSM 32,242, Lacticaseibacillus paracasei DSM 32,243, L. plantarum DSM 32,244, and L. brevis DSM 27,961 reduced mortality in COVID-19 patients [23]. D’Ettorre et al. described the positive effects of the same probiotic on diarrhea associated with COVID-19 [24].
However, these findings have not been verified in prospective randomized studies. The aim of our study was to evaluate the effect of a multi-strain probiotic on mortality, disease course, respiratory function, diarrhea, and liver injury in COVID-19 patients in a randomized controlled trial.
Materials and Methods
This was a randomized, controlled, single-center, open-label trial. All patients signed an informed consent for study participation and the use of off-label drugs. The study was approved by the local ethics committee (Conclusion №. 34–20 of September 9, 2020) in accordance with the Declaration of Helsinki, and was registered at https://clinicaltrials.gov (NCT04854941). The research protocol can be accessed at this website as well. The study received no funding.
Patients
The study included patients with COVID-19 admitted to the Clinic of Internal Diseases of Sechenov University. COVID-19 was confirmed using polymerase chain reaction on nasopharyngeal and oropharyngeal swabs to detect SARS-CoV-2. The study was conducted from December 2020 to March 2021 and included participants in the age range from 18 to 75 years.
The exclusion criteria were as follows: age over 75 years or under 18 years, consumption of probiotics for 3 months prior to admission, presence or history of intolerance to probiotics or their components, refusal to participate and sign informed consent, pregnant or breastfeeding, cancer or mental illness, and severe renal (glomerular filtration rate less than 50 mL/min) or hepatic (equivalent to cirrhosis class B or C on the Child–Pugh scale) dysfunction at the time of admission.
Patients who prematurely discontinued the consumption of probiotics for reasons not related to the development of side effects were excluded from the study.
Intervention
The patients were randomized to the probiotics group (PRO group) or the control group (CON group). During the hospital stay, patients in the PRO group received the following probiotics three times a day for no more than 14 days: Florasan-D containing ~ 109 colony forming units (CFU) of Lacticaseibacillus rhamnosus PDV 1705, ~ 109 CFU of Bifidobacterium bifidum PDV 0903, ~ 109 CFU of Bifidobacterium longum subsp. infantis PDV 1911, and ~ 109 CFU of Bifidobacterium longum subsp. longum PDV 2301. The end point of the trial was day 14 of hospitalization or the day of the patient’s discharge or death, whichever occurred earlier. The follow-up period lasted from the time of the inclusion until recovery or death.
Controls
The control group consisted of patients who did not receive probiotics.
Patients in both the groups also received dexamethasone and antiviral (favipiravir and/or riamilovir), antibacterial, anticoagulant (enoxaparin in most cases; rivaroxaban and dabigatran were used less frequently), and anticytokine (tocilizumab or/and olokizumab) drugs according to indications and contraindications (Table S1).
Outcomes
Death from any cause, duration of hospitalization, total duration of the disease, incidence of admission to intensive care unit, need for oxygen support or mechanical ventilation, and changes in the values of key biomarkers were considered as the main outcomes. The duration of diarrhea [an increase in the frequency of bowel movements (more than three times per day)], incidence of hospital-acquired diarrhea, progression of pre-existing liver injury, and onset of liver injury were considered as additional outcomes. Liver injury was determined based on the presence of abnormalities in any of the main liver test findings (serum alanine transaminase, aspartate transaminase, alkaline phosphatase, gamma-glutamyl transferase, total bilirubin, and albumin levels).
The volume of the affected lungs was measured using chest computed tomography (CT); it included the sum of ground glass and consolidation volumes.
We performed per-protocol analysis as there was no final point to perform intention-to-treat analysis.
Statistics
Results are presented as median [interquartile range]. The groups were compared using the Mann–Whitney test for continuous data and chi-square test for categorical data. Wilcoxon test was used to assess changes in continuous biomarker values. Mortality was assessed using the Kaplan–Meier estimator and Cox’s test. A p value ≤ 0.05 was considered as the criterion for significance. Statistical calculations were performed using STATISTICA 10 (TIBCO Software, Palo Alto, CA).
Results
Characteristics of the Included Patients by Groups
The study included 99 patients in the PRO group and 101 patients in the CON group (Fig. 1). COVID-19 pneumonia was confirmed in all patients using a chest CT scan. There was no significant difference in age, sex distribution, body mass index, body temperature at admission, symptoms of COVID-19, incidence of co-morbidities, and use of other drugs for the treatment of COVID-19 between the groups (Table 1; Table S1).
Fig. 1.
CONSORT 2010 flow diagram
Table 1.
Main characteristics and outcomes of patients who received probiotics (PRO group) and who did not (CON group)
| Group PRO (n = 99) | Group CON (n = 101) | p | |
|---|---|---|---|
| Age, years | 65 (59–71) | 64 (54–70) | 0.283 |
| Male/female | 44/55 | 48/53 | 0.662 |
| Body temperature at admission, °C | 37.3 (36.9–37.7) | 37.2 (36.8–37.6) | 0.657 |
| Body mass index, kg/m2 | 30.5 (27.4–35.3) | 31.2 (27.1–33.5) | 0.910 |
| Time from symptom onset to inclusion, days | 8 (6–12) | 9 (7–11) | 0.504 |
| Length of hospital stay, days | 11 (10–14) | 11 (9–14) | 0.440 |
| Total duration of disease, days | 20 (18–24) | 21 (18–25) | 0.471 |
| Death, n (%) | 4 (4.0%) | 4 (4.0%) | 0.491 |
| Admission to intensive care unit, n (%) | 5 (5.1%) | 7 (6.9%) | 0.576 |
| The need for mechanical ventilation, n (%) | 4 (4.0%) | 5 (5.0%) | 0.976 |
| Oxygen support, n (%) | 47 (47.5%) | 44 (43.6%) | 0.579 |
| Duration of oxygen support, days | 6 (2–11) | 7 (1–11) | 0.513 |
| Patients with diarrhea on admission, n (%) | 12 (12.1%) | 11 (10.9%) | 0.785 |
| Duration of diarrhea on admission, days | 2 (1–4) | 4 (3–6) | 0.049 |
| Patients with hospital-acquired diarrhea, n (%) | 4 (4.0%) | 10 (9.9%) | 0.109 |
| Duration of hospital-acquired diarrhea, days | 4 (4–6) | 5 (3–6) | 0.777 |
Main Outcomes
No significant differences in the total duration of disease, length of hospital stay, incidence of intensive care unit admission, need for mechanical ventilation or oxygen support (Table 1), and the changes in the volume of the affected lungs and serum levels of biomarkers of systemic inflammation [C-reactive protein (CRP), erythrocyte sedimentation rate, ferritin, fibrinogen, white blood cells, neutrophils, and lymphocytes], renal function (creatinine), and liver function (alanine aminotransferase, aspartate aminotransferase, albumin, and total bilirubin) were observed between the groups (Tables 2 and 3).
Table 2.
Change in the values of the main biomarkers between the beginning (point 1) and end (point 2) of the trial in patients who received probiotics (PRO group) and those who did not (CON group)
| Group | PRO group (n = 99) | CON group (n = 101) | p** | p** | ||||
|---|---|---|---|---|---|---|---|---|
| Biomarker | 1 | 2 | p* | 1 | 2 | p* | 1 | 2 |
| Lung lesion volume, % | 50 [50–75] | 50 [50–75] | 0.453 | 50 [50–75] | 50 [50–75] | 0.547 | 0.393 | 0.570 |
| C-reactive protein, mg/L | 66 [24–116] | 3 [1–5] | < 0.001 | 58 [28–108] | 3 [1–5] | < 0.001 | 0.784 | 0.680 |
| White blood cells, 109/L | 5.5 [4.0–7.7] | 8.2 [6.7–10.0] | < 0.001 | 6.1 [4.6–9.7] | 8.6 [6.3–11.9] | < 0.001 | 0.035 | 0.210 |
| Neutrophils, 109/L | 4.1 [2.6–5.9] | 6.2 [4.2–7.4] | 0.002 | 4.6 [2.9–8.5] | 6.3 [4.5–9.5] | 0.019 | 0.053 | 0.231 |
| Lymphocytes, 109/L | 1.0 [0.7–1.4] | 1.4 [1.0–1.9] | < 0.001 | 1.0 [0.7–1.3] | 1.2 [0.9–1.8] | < 0.001 | 0.740 | 0.656 |
| Platelets, 109/L | 226 [171–272] | 311 [250–392] | < 0.001 | 236 [168–316] | 316 [227–398] | 0.001 | 0.697 | 0.615 |
| ESR, mm/L | 25 [21–28] | 20 [14–25] | 0.001 | 24 [20–27] | 20 [14–24] | 0.001 | 0.446 | 0.903 |
| Creatinine, μmol/L | 96 [80–110] | 83 [76–94] | < 0.001 | 96 [84–109] | 83 [75–100] | < 0.001 | 0.632 | 0.752 |
| ALT, U/L | 30 [20–42] | 40 [32–64] | < 0.001 | 31 [22–46] | 37 [22–82] | < 0.001 | 0.367 | 0.516 |
| AST, U/L | 35 [29–49] | 36 [25–51] | 0.626 | 35 [28–48] | 43 [22–53] | 0.404 | 0.548 | 0.530 |
| Albumin, g/L | 41 [39–44] | 37 [31–43] | 0.450 | 41 [40–44] | 35 [30–37] | 0.221 | 0.230 | 0.270 |
| Total bilirubin, μmol/L | 9 [6–11] | 8 [6–10] | 0.605 | 10 [7–12] | 11 [9–14] | 0.579 | 0.386 | 0.051 |
| LDH, U/L | 541 [453–687] | 453 [392–564] | < 0.001 | 533 [410–698] | 441 [384–568] | < 0.001 | 0.578 | 0.538 |
| Ferritin, μg/L | 442 [224–639] | 469 [347–793] | 0.617 | 436 [208–749] | 501 [211–779] | 0.067 | 0.923 | 0.593 |
| Fibrinogen, g/L | 6.0 [5.1–7.4] | 3.5 [2.8–3.9] | < 0.001 | 5.8 [4.7–7.4] | 3.5 [2.9–4.5] | < 0.001 | 0.283 | 0.376 |
| Potassium, mmol/L | 4.5 [4.1–4.9] | 5.1 [4.5–5.5] | 0.037 | 4.4 [4.1–4.7] | 4.9 [4.6–5.4] | 0.034 | 0.433 | 0.849 |
ESR erythrocyte sedimentation rate, ALT alanine aminotransferase, AST aspartate aminotransferase, LDH lactate dehydrogenase
*Difference between the beginning (point 1) and end (point 2) of the trial within the groups
**Difference between groups at the beginning (point 1) and end (point 2) of the trial
Table 3.
Maximum or minimum values of the main biomarkers during the trial
| PRO group (n = 99) | CON group (n = 101) | p | |
|---|---|---|---|
| The maximum value during the trial | |||
| Lung lesion volume, % | 50 [50–75] | 50 [50–75] | 0.531 |
| C-reactive protein, mg/L | 83 [53–125] | 74 [45–128] | 0.398 |
| White blood cells, 109/L | 8.9 [7.5–12.0] | 10.0 [7.3–14.2] | 0.268 |
| Neutrophils, 109/L | 7.0 [5.3–10.0] | 7.6 [5.0–11.6] | 0.500 |
| ESR, mm/L | 28 [24–32] | 27 [22–30] | 0.132 |
| Creatinine, μmol/L | 96 [83–110] | 99 [84–112] | 0.509 |
| ALT, U/L | 38 [23–85] | 43 [33–74] | 0.964 |
| AST, U/L | 43 [30–59] | 44 [29–58] | 0.396 |
| Total bilirubin, mmol/l | 10 [7–12] | 11 [9–14] | 0.313 |
| LDH, U/L | 652 [542–793] | 633 [464–830] | 0.394 |
| Ferritin, μg/L | 489 [321–762] | 518 [212–988] | 0.753 |
| Fibrinogen, g/L | 6.3 [5.4–7.6] | 6.3 [4.9–7.7] | 0.504 |
| The minimum value during the trial | |||
| Albumin, g/L | 37 [31–42] | 34 [30–37] | 0.057 |
| Lymphocytes, 109/L | 0.7 [0.6–1.1] | 0.9 [0.5–1.2] | 0.189 |
ESR erythrocyte sedimentation rate, ALT alanine aminotransferase, AST aspartate aminotransferase, LDH lactate dehydrogenase
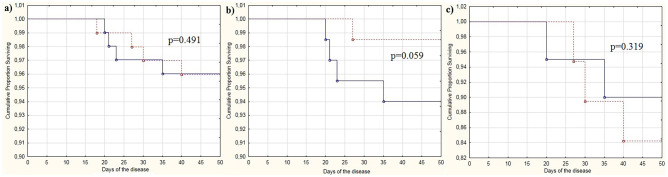
The patients in the PRO and CON groups had similar survival rates (p = 0.491) (Table 1; Fig. 2a). Two patients who stopped taking the probiotic prematurely also survived.
Fig. 2.
Survival curves for patients with coronavirus disease (COVID-19) who received the probiotic (dotted line) and those who did not (control group) (solid line): (a) all patients, (b) patients with cardiovascular diseases, and (c) patients with diabetes mellitus
As the presence of cardiovascular diseases or diabetes is a risk factor for poor prognosis in COVID-19 [25], an analysis was performed for these subgroups of patients. The administration of the probiotics had no significant effects on the course of COVID-19 in the aforementioned patients, except for a tendency towards a decrease in mortality rate in patients with cardiovascular diseases (Table S2; Fig. 2b, c).
Probiotic administration did not exert significant effects on the course of COVID-19 in the subgroups of patients who had < 25%, 25–50%, and > 50% lung involvement (Table S3), except for a decrease in frequency of admission to the intensive care unit in the group of patients with 25–50% lung involvement (0.0% vs. 10.9%; p = 0.024).
The administration of the probiotic did not exert a significant effect on the course of COVID-19 in subgroups of patients delineated based on age younger or older than 65 years (Table S4). In addition, no significant differences were observed between patients with serum CRP levels less or greater than 60 mg/L on admission (Table S5) (this is the cut-off value for severe systemic inflammation according to the local guidelines [26]). However, a more pronounced decrease in lymphocyte count was observed in the PRO group among patients with CRP level less than 60 mg/L.
There were no cases of infections caused by these probiotic strains.
Diarrhea in COVID-19
Diarrhea on admission was observed in 23 (11.5%) patients; no significant difference was observed between the PRO and CON groups in this regard (Table 1). It was observed that diarrhea ceased earlier in patients who received the probiotic than in those who did not (Table 1).
Hospital-acquired diarrhea developed in 14 (7.0%) patients, and only in those who received antibiotics (14/154 (9.1%) vs. 0/46 (0.0%); p = 0.034). Probiotic intake in the general cohort tended to prevent the development of hospital-acquired diarrhea. However, a significant difference was observed in the group that received only one antibiotic [n = 71; hospital-acquired diarrhea incidence: 0 vs. 4 (12.5%); p = 0.023]. The probiotic was not found to be effective in preventing hospital-acquired diarrhea in patients who received more than one antibiotic simultaneously or sequentially (n = 83; hospital-acquired diarrhea incidence: 4 (10.5%) vs. 6 (13.3%); p = 0.696).
Intake of the probiotic did not significantly affect the duration of hospital-acquired diarrhea (Table 1).
Liver Injury in COVID-19
Signs of liver injury were observed in 95 (47.5%) patients on admission, with no significant difference between the PRO and CON groups. Liver injury progressed during hospitalization in 31 (15.5%) of these patients, and developed after admission in 55 (27.5%) other patients; probiotic intake did not prevent liver injury in any of these cases (Table 4).
Table 4.
Patients with abnormal liver biomarker values
| Group PRO (n = 99) | Group CON (n = 101) | p | |
|---|---|---|---|
| Patients with abnormal liver biomarkers on admission | |||
| ALT > ULN (45 U/L) on admission, n (%) | 21 (21.2%) | 26 (25.7%) | 0.450 |
| ALT > 3ULN on admission, n (%) | 2 (2.0%) | 1 (1.0%) | 0.549 |
| AST > ULN (45U/L) on admission, n (%) | 27 (27.3%) | 27 (26.7%) | 0.932 |
| AST > 3ULN on admission, n (%) | 3 (3.0%) | 1 (1.0%) | 0.303 |
| Total bilirubin > ULN (21 μmol/L) on admission, n (%) | 4 (4.0%) | 4 (4.0%) | 0.977 |
| Serum albumin < ULN (35 g/L) on admission, n (%) | 10 (10.1%) | 4 (4.0%) | 0.889 |
| ALP > ULN (360 U/L) on admission, n (%) | 3 (3.0%) | 3 (3.0%) | 0.980 |
| GGT > ULN (60 U/L) on admission, n (%) | 11 (11.1%) | 15 (14.9%) | 0.432 |
| Any abnormal liver biomarker, n (%) | 45 (45.5%) | 50 (49.5%) | 0.566 |
| Patients with progression of pre-admission liver injury | |||
| Increase in the ALT level during the trial in those with high ALT levels at admission, n (%) | 12 (12.1%) | 12 (11.9%) | 0.958 |
| Increase in the AST level during the trial in those with high AST levels at admission, n (%) | 4 (4.0%) | 10 (9.9%) | 0.105 |
| Any progression of pre-admission liver injury, n (%) | 15 (15.2%) | 16 (15.8%) | 0.893 |
| Patients who developed liver injury after admission | |||
| ALT > ULN during the trial, n (%) | 25 (25.3%) | 15 (14.9%) | 0.066 |
| ALT > 3ULN during the trial, n (%) | 5 (5.1%) | 11 (10.9%) | 0.128 |
| AST > ULN during the trial, n (%) | 18 (18.2%) | 9 (8.9%) | 0.055 |
| AST > 3ULN during the trial, n (%) | 1 (1.0%) | 4 (4.0%) | 0.182 |
| Total bilirubin > ULN during the trial, n (%) | 0 (0.0%) | 2 (2.0%) | 0.159 |
| Serum albumin < ULN during the trial, n (%) | 5 (5.1%) | 6 (5.9%) | 0.783 |
| ALP > ULN during the trial, n (%) | 0 (0.0%) | 2 (2.0%) | 0.159 |
| GGT > ULN during the trial, n (%) | 3 (3.0%) | 5 (5.0%) | 0.488 |
| Any liver injury after admission, n (%) | 32 (32.3%) | 23 (22.8%) | 0.130 |
ALT alanine aminotransferase, ULN upper limit of normal, AST aspartate aminotransferase, ALP alkaline phosphatase, GGT gamma-glutamyl transferase
Discussion
The aim of our study was to evaluate the effect of a multi-strain probiotic on mortality, disease course, respiratory function, diarrhea, and liver injury in COVID-19 patients in a randomized controlled trial. We could not confirm the findings of an Italian retrospective study which reported that probiotic supplementation could reduce mortality in a cohort of COVID-19 patients [23]. It should be noted that the probiotic composition we used was different from that used by the Italian research group, though it is unlikely that this difference in composition can explain the difference in study outcomes. Mortality was significantly higher in the above mentioned study than in the present study (22% vs. 4%). This significant difference in mortality between the two studies may be explained by the different strategies used for COVID-19 management. The Italian researchers mainly administered hydroxychloroquine, lopinavir, azithromycin, and tocilizumab in accordance with the clinical recommendations of that time (the study was performed between March and April 2020). In the present study, lopinavir was not used, azithromycin and hydroxychloroquine were used less frequently, and almost all patients were administered with dexamethasone and anticoagulants.
The second factor responsible for the difference in mortality may be the differences in ethnicity, resulting in differences in the interaction between the virus and the host, as well as the differences in the strain composition in different countries. A third factor may be selection bias that is characteristic of retrospective studies. In addition, the severity of disease could have differed among patients in the two studies.
The differences in mortality between the patients who received and did not receive probiotics almost reached the limits of significance in the subgroup of patients with chronic cardiovascular diseases (1.5% vs. 6.0%; p = 0.059). Notably, there were no patients with chronic cardiovascular diseases in the above mentioned study by Ceccarelli et al. [23]. It is interesting that gut microbiota has been shown to play a role in the development of heart failure [27]. A study with a larger sample size should be conducted to examine the effect of probiotics on mortality in this subgroup of patients and to test the hypothesis that probiotics may reduce mortality in patients with chronic cardiovascular diseases.
The probiotic did not have a significant effect on the course of the disease, inflammatory biomarkers, and renal dysfunction in our study. Moreover, this finding was observed not only in the general cohort of patients, but also in the following subgroups: subgroups of patients with chronic cardiovascular diseases, diabetes, and different volumes of lung damage; subgroups delineated based on age younger or older than 65 years; and subgroups defined based on the presence or absence of severe systemic inflammation on admission.
In our study, the administration of probiotics shortened the duration of COVID-19-associated diarrhea. This result is in agreement with the results of a retrospective study by d’Ettorre et al. [24]. It should be noted that the combination of probiotics used in the two studies differed. The causes of diarrhea in COVID-19 patients were heterogeneous [6]; viral diarrhea which develops in the early days of the disease as well as antibiotic-associated diarrhea which develops later and often in the hospital (hospital-acquired diarrhea) were observed. We also studied the effect of the multi-strain probiotic on the incidence and duration of hospital-acquired diarrhea. Hospital-acquired diarrhea developed only in those patients who received antibiotics, which may confirm its antibiotic-associated nature. Although hospital-acquired diarrhea was less common among patients who received the probiotic than among those who did not, this difference was not significant in the general cohort of patients. However, it was significant in the subgroup of patients who received only one antibiotic. Additionally, the administration of the probiotic did not affect the duration of hospital-acquired diarrhea.
Thus, in our study, probiotics for COVID-19 showed small but distinctly positive effects; these effects included a shortening of the duration of viral diarrhea by an average of 2 days, and the prevention of hospital-acquired diarrhea in patients who received a single antibiotic.
The bacteria included in the probiotic therapy used in this study have been reported to be useful in the treatment of antibiotic-associated diarrhea [28] and acute viral diarrhea [29, 30] in previous trials. These effects may be associated with their ability to form biofilms on the surface of the gut epithelium, which prevents colonization by Clostridioides difficile [31]. This in turn inhibits the growth and toxinogenesis of these and other pathogenic bacteria [32]. Intestinal viral infections cause increased permeability of the intestinal barrier [33]; probiotic bacteria have been reported to reduce the permeability of the intestinal barrier, and normalize the functioning of ion transporters in the epithelial cell membrane [34]. Thus, our results in patients with COVID-19 are consistent with the findings from the studies cited above.
The main strength of our study is that this is the first randomized controlled study investigating the effect of probiotics on a wide range of indicators in COVID-19. In addition, we analyzed these effects not only in the general cohort of patients, but also in subgroups within the cohort. We also performed a detailed analysis of the effects of probiotic supplementation on liver injury associated with COVID-19. However, there are several limitations to our study. This was a single-center open-label study, and did not include a placebo group. We used a probiotic that was different from those used in earlier studies, which may also be interpreted as a limitation. In addition, it was not possible to differentiate between the adverse effects of the probiotic and COVID-19-related signs due to the pronounced polymorphism of the manifestations of this disease. However, there were no cases of infections caused by these probiotic strains in our study.
New randomized controlled trials that include populations from different countries are needed to confirm our findings.
Conclusion
In conclusion, the studied probiotic did not have a significant effect on mortality and changes in most biomarkers in COVID-19 patients. However, the probiotic showed potential as treatment for diarrhea associated with COVID-19, and for the prevention of diarrhea in patients who receive a single antibiotic as part of COVID-19 treatment. A larger study on the effect of probiotics in COVID-19 patients with chronic cardiovascular diseases should be conducted to test the hypothesis that probiotics may reduce mortality in this cohort of patients.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
We would like to express our gratitude to the doctors, nurses, and other staff of the Tareev Clinic of Internal Diseases who took part in this study.
Author Contribution
The idea and design of the study were developed by Vladimir Ivashkin and Elena Poluektova. Material preparation, data collection, and analysis were performed by all authors. The first draft of the manuscript was written by Roman Maslennikov, and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Availability of Data and Material
The datasets analyzed during the current study may be available from the corresponding author on reasonable request.
Declarations
Ethics Approval
The study was approved by the local ethics committee (Conclusion №. 34-20 of September 9, 2020) in accordance with the Declaration of Helsinki.
Consent to Participate
Informed consent was obtained from all individual participants included in the study.
Consent for Publication
Patients’ personal data are not published.
Conflicts of Interest
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Mohamadian M, Chiti H, Shoghli A, et al. COVID-19: virology, biology and novel laboratory diagnosis. J Gene Med. 2021;23:e3303. doi: 10.1002/jgm.3303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Umakanthan S, Sahu P, Ranade AV, et al. Origin, transmission, diagnosis and management of coronavirus disease 2019 (COVID-19) Postgrad Med. 2020;96:753–758. doi: 10.1136/postgradmedj-2020-138234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Trush EA, Poluektova EA, Beniashvilli AG, et al. The evolution of human probiotics: challenges and prospects. Probiotics Antimicrob Proteins. 2020;12:1291–1299. doi: 10.1007/s12602-019-09628-4. [DOI] [PubMed] [Google Scholar]
- 4.Zolnikova O, Komkova I, Potskherashvili N, et al. Application of probiotics for acute respiratory tract infections. Ital J Med. 2018;12:32–38. doi: 10.4081/itjm.2018.931. [DOI] [Google Scholar]
- 5.Tariq R, Saha S, Furqan F, et al. Prevalence and mortality of COVID-19 patients with gastrointestinal symptoms: a systematic review and meta-analysis. Mayo Clin Proc. 2020;95:1632–1648. doi: 10.1016/j.mayocp.2020.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maslennikov R, Poluektova E, Ivashkin V, et al. Diarrhoea in adults with coronavirus disease-beyond incidence and mortality: a systematic review and meta-analysis. Infect Dis (Lond) 2021;53:348–360. doi: 10.1080/23744235.2021.1885733. [DOI] [PubMed] [Google Scholar]
- 7.Maslennikov R, Ivashkin V, Ufimtseva A, Poluektova E (2021) A clinical variant of coronavirus disease 2019 with diarrhoea as the initial symptom compared with other variants. Minerva Gastroenterol (Torino). 10.23736/S2724-5985.21.02827-0 [DOI] [PubMed]
- 8.Megyeri K, Dernovics Á, Al-Luhaibi ZII, Rosztóczy A. COVID-19-associated diarrhea. World J Gastroenterol. 2021;27:3208–3222. doi: 10.3748/wjg.v27.i23.3208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Agamennone V, Krul CAM, Rijkers G, et al. A practical guide for probiotics applied to the case of antibiotic-associated diarrhoea in the Netherlands. BMC Gastroenterol. 2018;18:103. doi: 10.1186/s12876-018-0831-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cangemi DJ, Lacy BE. Management of irritable bowel syndrome with diarrhoea: a review of nonpharmacological and pharmacological interventions. Ther Adv Gastroenterol. 2019;12:1-19. doi: 10.1177/1756284819878950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Isolauri E. Probiotics for infectious diarrhoea Gut. 2003;52:436–437. doi: 10.1136/gut.52.3.436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wijarnpreecha K, Ungprasert P, Panjawatanan P, et al. COVID-19 and liver injury: a meta-analysis. Eur J Gastroenterol Hepatol. 2021;33:990–995. doi: 10.1097/MEG.0000000000001817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maslennikov R, Ivashkin V, Efremova I, Poluektova E, Shirokova E (2021) Probiotics in hepatology: an update. World J Hepatol 13:1154–1166 10.4254/wjh.v13.i9.1154 [DOI] [PMC free article] [PubMed]
- 14.Mirzaei R, Attar A, Papizadeh S, et al. The emerging role of probiotics as a mitigation strategy against coronavirus disease 2019 (COVID-19) Arch Virol. 2021;166:1819–1840. doi: 10.1007/s00705-021-05036-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Singh K, Rao A. Probiotics: a potential immunomodulator in COVID-19 infection management. Nutr Res. 2021;87:1–12. doi: 10.1016/j.nutres.2020.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Patra S, Saxena S, Sahu N, et al. Systematic network and meta-analysis on the antiviral mechanisms of probiotics: a preventive and treatment strategy to mitigate SARS-CoV-2 infection. Probiotics Antimicrob Proteins. 2021;13:1138–1156. doi: 10.1007/s12602-021-09748-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mullish BH, Marchesi JR, McDonald JAK, et al. Probiotics reduce self-reported symptoms of upper respiratory tract infection in overweight and obese adults: should we be considering probiotics during viral pandemics? Gut Microbes. 2021;13:1–9. doi: 10.1080/19490976.2021.1900997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Manna S, Chowdhury T, Chakraborty R, Mandal SM. Probiotics-derived peptides and their immunomodulatory molecules can play a preventive role against viral diseases including COVID-19. Probiotics Antimicrob Proteins. 2021;13:611–623. doi: 10.1007/s12602-020-09727-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Olaimat AN, Aolymat I, Al-Holy M, et al. The potential application of probiotics and prebiotics for the prevention and treatment of COVID-19. NPJ Sci Food. 2020;4:17. doi: 10.1038/s41538-020-00078-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mahooti M, Miri SM, Abdolalipour E, Ghaemi A. The immunomodulatory effects of probiotics on respiratory viral infections: a hint for COVID-19 treatment? Microb Pathog. 2020;148:104452. doi: 10.1016/j.micpath.2020.104452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bozkurt HS, Quigley EM. The probiotic Bifidobacterium in the management of Coronavirus: a theoretical basis. Int J Immunopathol Pharmacol. 2020;34:2058738420961304. doi: 10.1177/2058738420961304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li Q, Cheng F, Xu Q, et al. The role of probiotics in coronavirus disease-19 infection in Wuhan: a retrospective study of 311 severe patients. Int Immunopharmacol. 2021;95:107531. doi: 10.1016/j.intimp.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ceccarelli G, Borrazzo C, Pinacchio C, et al. Oral bacteriotherapy in patients with COVID-19: a retrospective cohort study. Front Nutr. 2021;7:613928. doi: 10.3389/fnut.2020.613928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.d’Ettorre G, Ceccarelli G, Marazzato M, et al. Challenges in the management of SARS-CoV2 infection: the role of oral bacteriotherapy as complementary therapeutic strategy to avoid the progression of COVID-19. Front Med (Lausanne) 2020;7:389. doi: 10.3389/fmed.2020.00389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li B, Yang J, Zhao F, et al. Prevalence and impact of cardiovascular metabolic diseases on COVID-19 in China. Clin Res Cardiol. 2020;109:531–538. doi: 10.1007/s00392-020-01626-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kamkin E et al. (2020) Interim guidelines for the prevention, diagnosis and treatment of new coronaviral infection (COVID-19). https://static-0.minzdrav.gov.ru/system/attachments/attaches/000/051/777/original/030902020_COVID-19_v8.pdf. Accessed 03 Sept 2020
- 27.Ivashkin V, Fadeeva M, Skhirtladze M, et al. Intestinal microbiota in the pathogenesis of chronic heart failure. Ital J Med. 2020;14:1–8. doi: 10.4081/itjm.2020.1185. [DOI] [Google Scholar]
- 28.Szajewska H, Kołodziej M. Systematic review with meta-analysis: Lactobacillus rhamnosus GG in the prevention of antibiotic-associated diarrhoea in children and adults. Aliment Pharmacol Ther. 2015;42:1149–1157. doi: 10.1111/apt.13404. [DOI] [PubMed] [Google Scholar]
- 29.Li YT, Xu H, Ye JZ. Efficacy of Lactobacillus rhamnosus GG in treatment of acute pediatric diarrhea: a systematic review with meta-analysis. World J Gastroenterol. 2019;25:4999–5016. doi: 10.3748/wjg.v25.i33.4999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Di JB, Gai ZT (2020) Protective efficacy of probiotics on the treatment of acute rotavirus diarrhea in children: an updated meta-analysis. Eur Rev Med Pharmacol Sci 24:9675–9683. 10.26355/eurrev_202009_23057 [DOI] [PubMed]
- 31.Rasinkangas P, Tytgat HLP, Ritari J, et al. Characterization of highly mucus-adherent non-GMO derivatives of Lacticaseibacillus rhamnosus GG. Front Bioeng Biotechnol. 2020;8:1024. doi: 10.3389/fbioe.2020.01024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Valdés-Varela L, Hernández-Barranco AM, Ruas-Madiedo P, Gueimonde M. Effect of Bifidobacterium upon Clostridium difficile growth and toxicity when co-cultured in different prebiotic substrates. Front Microbiol. 2016;7:738. doi: 10.3389/fmicb.2016.00738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kawahara T, Makizaki Y, Oikawa Y, et al. Oral administration of Bifidobacterium bifidum G9–1 alleviates rotavirus gastroenteritis through regulation of intestinal homeostasis by inducing mucosal protective factors. PLoS ONE. 2017;12:e0173979. doi: 10.1371/journal.pone.0173979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kumar A, Hecht C, Priyamvada S. Probiotic Bifidobacterium species stimulate human SLC26A3 gene function and expression in intestinal epithelial cells. Am J Physiol Cell Physiol. 2014;307:C1084–C1092. doi: 10.1152/ajpcell.00194.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets analyzed during the current study may be available from the corresponding author on reasonable request.