Abstract
Stool, gastric biopsy, and serum samples were collected from 22 subjects. DNA from stool was extracted, amplified, and hybridized with primers specific for the 16S rRNA gene of Helicobacter pylori. DNA from gastric biopsy specimens was analyzed similarly for comparison. Universal primers were used to confirm successful extraction of DNA from samples. Histologic, serologic, and DNA analyses were scored in a blinded fashion. Universal primer amplification verified successful DNA extraction from all stool and gastric tissue specimens. The gastric tissue DNA assay was positive for H. pylori in 11 of the 22 subjects, correlating completely with histologic and serologic results. Stool DNA was positive for H. pylori by our molecular assay in 8 of these 11 H. pylori-positive subjects. All subjects who were negative by histologic, serologic, and gastric tissue DNA analyses were also negative by stool DNA analysis. Compared to histology, serology, and gastric tissue DNA analyses, the sensitivity of our stool DNA assay was 73%, with a specificity of 100%.
Helicobacter pylori, first isolated by Warren and Marshall in 1983 (12), has been shown to play an important role in gastritis, peptic ulcer disease, and gastric malignancies (14, 25). Recent studies suggest that certain H. pylori genotypes, such as strains possessing the cagA gene, may be more virulent than others (2). Several diagnostic tests are available for determining the presence of H. pylori infection (19). Tests that require endoscopy include the biopsy urease test, histology, culture, PCR-based methods, and phase-contrast microscopy of gastric tissue. Diagnostic tests that do not require endoscopy include [13C] and [14C]urea breath tests, serology, the string test, and stool antigen enzyme immunoassay (EIA).
Molecular methods such as PCR and Southern blot hybridization have the capability to sensitively and accurately determine both the presence of infection and the genotype of bacteria. These techniques have been used successfully to detect H. pylori DNA in gastric tissue by amplifying genes such as the adhesin gene (7), the urease gene (5), and the 16S rRNA gene (8). The 16S rRNA gene of H. pylori is a highly specific target for amplification and has been used previously to help reclassify the organism. Weiss et al. demonstrated the specificity of unique H. pylori 16S rRNA gene primers to identify the organism in paraffin-embedded gastric biopsy specimens (24).
Stool analysis would provide a noninvasive means of detecting H. pylori. PCR analysis of stool has been used successfully to diagnose several infections, including rotovirus (22), microsporidia (6), Vibrio cholerae (1), verotoxin-producing Escherichia coli (16), and Salmonella (4) infections. PCR analysis of stool has even detected mutations of K-ras from tumor cells shed from colonic neoplasms (18).
Previous reports of PCR analysis of stool for H. pylori have shown low sensitivity (23). Culturing stool samples allowed detection of the urease gene by PCR (9), but the sensitivity of this assay was low and the ability to routinely culture stools for this purpose was unproven. The difficulty in direct PCR amplification of DNA from stool samples is generally thought to be related to the presence of enzyme inhibitors. We sought to develop a novel stool DNA extraction process which could consistently generate amplifiable DNA for detection purposes. Our results herein provide evidence for the routinely successful detection of H. pylori DNA in stool samples from the majority of patients infected with this organism.
MATERIALS AND METHODS
Patients undergoing upper endoscopy were recruited consecutively between August 1996 and December 1996 after giving informed consent according to our institution’s internal review board approval. Esophagogastroduodenoscopy was performed on all subjects with endoscopes that had been sterilized by a Steris (Mentor, Ohio) machine. Autoclaved biopsy forceps were used in obtaining gastric biopsy specimens from the antrum for rapid urease testing (CLOtest). Gastric tissue was also obtained from the antrum, incisura, and body of the stomach for histologic examination and for DNA analysis. Stool specimens were collected within 2 weeks of the time of endoscopy in sterile containers and kept at −80°C until analysis. Blood from all patients was collected, and the serum was stored at −20°C until the EIA was performed with a Food and Drug Administration-approved, commercially available kit (HM-CAP EIA kit; Enteric Products, Stonybrook, N.Y.) which detects immunoglobulin G antibody to H. pylori. Zinc formalin-fixed paraffin-embedded biopsy specimens were stained with hematoxylin and eosin and Giemsa. A single pathologist (H.F.F.) scored all gastric biopsy specimens without knowledge of the results of the other tests. The number of H. pylori organisms was semiquantitatively scored as 0 (none), 1 (few; organisms were present but difficult to find and rare in 400× fields), 2 (moderate; organisms were readily identified upon microscopic examination and present in most 400× fields), and 3 (numerous; organisms were present in virtually all 400× fields).
DNA extraction.
One gram of stool from each patient was dissolved in 100% ethanol and chloroform and then centrifuged at 2,135 × g and rinsed with acetone. The sample was then mixed with 8 M urea containing 1% sodium dodecyl sulfate, 20 mM Tris-HCl (pH 8.0), 100 mg of Chelex (Bio-Rad, Hercules, Calif.) and 50 mg for of polyvinylpyrrolidone subsequent incubation at 60°C. The samples were then boiled and centrifuged at 469 × g. The supernatant was organically extracted, precipitated with alcohol, and redissolved with 0.7 M sodium chloride and 1% hexadecyltrimethylammoniumbromide (CTAB) (Sigma) for incubation at 65°C. Organic extraction and alcohol precipitation were performed for subsequent RNase A (1 mg/ml; Sigma) and proteinase K (0.5 mg/ml; Bio-Rad) incubation at 58°C for 2 h. Another round of organic extraction and alcohol precipitation was preformed with reconstitution in a solution of 3 mM Tris-HCl (pH 7.5) and 0.2 mM EDTA.
Gastric tissue DNA extraction from paraffin-embedded specimens was performed using xylene and 0.1% sodium dodecyl sulfate–proteinase K (0.5 mg/ml; Bio-Rad) on two 5-μm thick sections as previously described (13). Cultured H. pylori DNA extraction was conducted with an H. pylori isolate from a human subject who was confirmed to have this infection. H. pylori cultured on horse blood agar plates was scraped into 1 ml of phosphate-buffered saline. An aliquot of this suspension was then incubated overnight with proteinase K (0.5 mg/ml; Bio-Rad) prior to organic extraction and alcohol precipitation. The optical density was measured in the redissolved pellet for quantitation and subsequent serial dilutions of H. pylori DNA. Concentrations as low as 1 fg of DNA per μl were generated. A single bacterial genome was considered equivalent to 1.6 fg of DNA (21).
PCR amplification. (i) Universal primers.
PCR amplification with nonspecific, universal primers was performed in 25-μl reaction volumes containing 10 mM Tris-HCl (pH 8.3), 1.5 mM MgCl2, 50 mM KCl, a 200 μM concentration of each deoxynucleoside triphosphate, 10 pmol of each primer, 2.5 U of Taq polymerase (Perkin-Elmer Cetus, Norwalk, Conn.), and 1.5 μl of DNA template prepared as described above. The universal primers for stool analysis consisted of two nonspecific 16S rRNA oligonucleotides, designated USA-1 (AGAATGCCACGGTGAATACG) and USA-2 (CCTACGGTTACCTTGTTACG). Forty thermocycles were performed, with each cycle consisting of a 30-s denaturation step at 95°C, a 60-s annealing step at 50°C, and a 60-s extension step at 70°C. The universal primers for gastric tissue DNA analysis consisted of two oligonucleotides designed to amplify exon 7 of the Smad4 gene. The forward primer TGAAAGTTTTAGCATTAGACAAC and the reverse primer TGTACTCATCTCAGAAGTGAC were used in a PCR similar to that described above with an annealing temperature of 50°C. These primers have previously been proven to successfully amplify this exon for subsequent sequencing (15).
(ii) H. pylori-specific primers.
PCR amplification with H. pylori-specific primers was performed in a manner similar to the universal primer amplifications in 25-μl reaction volumes, with the same buffer, deoxyribonucleoside triphosphate, and Taq polymerase concentrations. The thermocycle conditions were similar, with the exception of an annealing temperature of 60°C. The primers consisted of two specific 16S rRNA oligonucleotides, designated HPF (5′ GCG ACC TGC TGG AAC ATT AC 3′) and HPR (5′ CGT TAG CTG CAT TAC TGG AGA 3′), which generated a 138-bp product. In all PCR amplifications, the final cycle was a 5-min extension step at 70°C to allow full product extension. Each experiment included a negative control template consisting of water and a positive control consisting of 100 fg of cultured H. pylori DNA.
Southern blot hybridization.
Half of the PCR products were electrophoresed on 2% agarose gels, transferred to nylon membranes (Bio-Rad), and hybridized with an end-labeled (106 cpm/ml) probe in a standard Southern blot fashion. The probe was a 16-bp oligonucleotide (CGCTGATTGCGCGAAA) designed specifically for a region within the 16S rRNA gene of H. pylori as previously described (24) and was labeled in a [32P]dATP T4 kinase (10 U/ml; New England Biolab) reaction according to the manufacturer’s instructions. Autoradiographs generated clear signals after an overnight exposure at room temperature. The autoradiographs were scored by two independent observers, each of whom was blinded to the results of the other tests, with a signal in the expected location recorded as either present or absent.
RESULTS
Detection of Helicobacter pylori DNA.
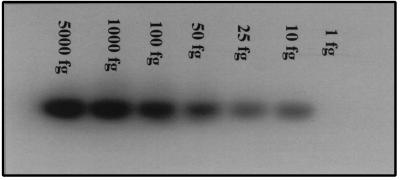
We first assessed the sensitivity of our molecular assay by determining the threshold for H. pylori DNA detection. A clinical isolate of H. pylori was grown in culture, and DNA was extracted and quantitated. Dilutions of this cultured H. pylori DNA were PCR amplified with specific H. pylori 16S rRNA primers prior to subsequent hybridization. Figure 1 shows specific H. pylori signals with DNA amounts as low as 10 fg, which corresponds to less than seven genome equivalents of H. pylori (21).
FIG. 1.
Molecular assay sensitivity. DNA was extracted from cultured H. pylori and then diluted to amounts ranging from 5,000 to 1 fg of bacterial DNA and amplified with primers specific for the 16S rRNA gene of H. pylori. The PCR products were then hybridized with a 32P-labeled H. pylori-specific oligomer. The lowest amount that produced a signal was 10 fg, which corresponds to less than seven bacterial genomes.
H. pylori status in human subjects.
From 22 individuals presenting for upper endoscopy examination, the following specimens were collected: gastric biopsy tissue for CLOtest, histology, and PCR amplification; serum for EIA; and stool for PCR amplification. Endoscopic findings, medication use, and results of conventional tests for H. pylori (i.e., serology, histology, and CLOtest) are summarized in Table 1. Half of the subjects tested positive by serology and histology, whereas the CLOtest failed to give a positive result for one of these individuals (subject 22) who was on 40 mg of omeprazole daily.
TABLE 1.
Clinical features of subjects in this study
| Subject no. | Acid suppression medication (dosage [mg])a | Endoscopic finding(s) | Result of:
|
||
|---|---|---|---|---|---|
| CLOtest | Serology | Histology (grade)b | |||
| 1 | None | Normal | Positive | Positive | Positive (1) |
| 2 | Ranitidine (150 bid) | Gastritis | Negative | Negative | Negative (0) |
| 3 | Omeprazole (20 bid) | Normal | Negative | Negative | Negative (0) |
| 4 | None | Duodenal dilatation | Negative | Negative | Negative (0) |
| 5 | Famotidine (20 qd) | Esophagitis | Negative | Negative | Negative (0) |
| 6 | Nizatidine (150 bid) | Duodenitis | Negative | Negative | Negative (0) |
| 7 | Nizatidine (150 qd) | Antritis | Positive | Positive | Positive (2) |
| 8 | Ranitidine (150 bid) | Duodenitis | Positive | Positive | Positive (1) |
| 9 | None | Duodenitis | Negative | Negative | Negative (0) |
| 10 | Ranitidine (150 bid) | Normal | Positive | Positive | Positive (2) |
| 11 | Omeprazole (20 qd) | Antritis, esophagitis | Negative | Negative | Negative (0) |
| 12 | None | Esophageal nodule | Negative | Negative | Negative (0) |
| 13 | Nizatidine (150 bid) | Gastritis | Negative | Negative | Negative (0) |
| 14 | Nizatidine (150 bid) | Duodenitis | Positive | Positive | Positive (2) |
| 15 | None | Schatzki’s ring, antritis | Positive | Positive | Positive (3) |
| 16 | Nizatidine (150 bid) | Duodenal ulcer | Negative | Negative | Negative (0) |
| 17 | Famotidine (20 qd) | Erosive antritis, duodenitis | Negative | Negative | Negative (0) |
| 18 | None | Erosive esophagitisitis | Positive | Positive | Positive (2) |
| 19 | None | Prepyloric ulcer | Positive | Positive | Positive (2) |
| 20 | None | Gastric ulcer | Positive | Positive | Positive (2) |
| 21 | None | Gastritis, duodenitis | Positive | Positive | Positive (2) |
| 22 | Omeprazole (20 bid) | Esophagitis, antritis | Negative | Positive | Positive (2) |
Medication taken prior to specimen accrual. Abbreviations: qd, once daily; bid, twice daily.
Semiquantitation of the number of H. pylori organisms: O = none; 1 = few; 2 = moderate; 3 = numerous.
Gastric biopsy DNA analysis.
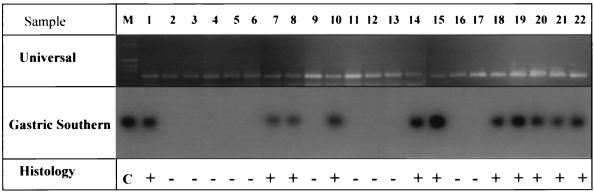
Amplifiable DNA extracted from the gastric biopsy specimens was obtained from each of the 22 study subjects. A 224-bp PCR product from exon 7 of Smad4 was successfully generated and seen on ethidium bromide-stained agarose gels for each subject (Fig. 2). The same DNA templates were amplified and hybridized with oligonucleotides specific for H. pylori’s 16S rRNA gene, generating positive signals in 11 of the 22 patients (Fig. 2). Independent PCR amplification and hybridization experiments confirmed these results. The autoradiographic signals were easily scored with 100% agreement by each of the two observers. These DNA analysis results correlated perfectly with the histologic and serologic findings for these same subjects.
FIG. 2.
Gastric tissue DNA analysis. The ethidium bromide-stained agarose gel in the top panel demonstrates amplification of exon 7 of Smad4 in gastric tissue DNAs of all 22 subjects, confirming the presence of amplifiable DNA. Lane M displays DNA size markers (Bethesda Research Laboratories 1-kb ladder). Specific H. pylori amplification produced an expected 139-bp PCR product with samples from infected subjects, seen as hybridization signals in the middle panel, with a 32P-labeled H. pylori-specific oligomer used as a probe. The results of histologic analysis (+, positive; −, negative) are presented in the bottom panel for comparison. The positive control lane (C) used DNA extracted from cultured H. pylori as a template.
The sensitivity of this molecular assay was further illustrated by gastric tissue testing of 24 additional subjects. For two subjects, the molecular assay detected a clear but weaker signal when chronic gastritis was present, histologically consistent with infection, but no demonstrable organisms were found even on analysis of additional stained sections. Both of these subjects were H. pylori positive by serology testing but had negative CLOtest results. In fact, both of these subjects had been treated with antiacid secretory therapy prior to examination, one with omeprazole (20 mg daily) and the other with ranitidine (300 mg daily). No prior antibiotic use was noted. Complete agreement on histology, serology, and DNA analysis for the other 22 gastric biopsy subjects was found (data not shown).
Stool DNA analysis.
Several methods of stool DNA extraction from subjects with and without H. pylori infection as shown by conventional testing were tested. The amount of DNA recovered varied depending on the protocol used. A novel method of extraction using lipid solubilizers, ionic and nonionic detergents, chelators, and organic solvents, was developed and routinely produced amplifiable DNA (see Materials and Methods). One gram of stool consistently generated approximately 10 μg of DNA. Identifiable signals were repeatedly generated when 200 ng of DNA was used as a PCR template in our assay. At least three independent experiments of stool DNA extraction, PCR amplification, and hybridization for 10 subjects (5 H. pylori positive and 5 H. pylori negative) consistently identified the presence or absence of H. pylori DNA, confirming the reproducibility and accuracy of our molecular assay.
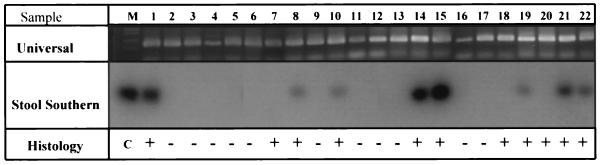
We then sought to determine the sensitivity and specificity of this assay in identifying H. pylori infection. Frozen samples of stool collected from 22 study subjects were subjected to DNA extraction according to our newly developed protocol and tested for H. pylori DNA to compare the results of conventional testing and gastric tissue DNA analysis. Stool sample extracts were first amplified with universal primers for the 16S rRNA gene to demonstrate the presence of amplifiable DNA. A clearly visible PCR product of expected size (148 bp) was generated in all samples, as seen by ethidium bromide staining of agarose gels (Fig. 3). Subsequent amplification and hybridization of the same stool DNA templates with primers specific for the H. pylori 16S rRNA gene resulted in unambiguous positive signals in 8 subjects and negative signals in the other 14 subjects (Fig. 3). Independent PCR amplification and hybridization of all DNA samples confirmed these results. Compared with histology, serology, and gastric DNA analysis, the molecular stool DNA assay had a sensitivity of 73% and a specificity of 100%.
FIG. 3.
Stool DNA analysis. The ethidium bromide-stained agarose gel in the top panel illustrates amplification from the stool DNA of all 22 subjects of an expected 148-bp PCR product with universal 16S rRNA gene primers, confirming the presence of amplifiable DNA. Lane M displays DNA size markers (Bethesda Research Laboratories 1-kb ladder). Specific H. pylori amplification primers generated an expected 139-bp PCR product with samples from infected subjects, seen as hybridization signals in the middle panel, with a 32P-labeled H. pylori-specific oligomer used as a probe. The results of histologic analysis (+, positive; −, negative) are presented in the bottom panel for comparison. The positive control lane (C) used DNA extracted from cultured H. pylori as a template.
DISCUSSION
Currently available tests for the diagnosis of H. pylori infection have relatively high sensitivities and specificities, but each has its limitations in clinical application. Urease-based biopsy tests require endoscopy and are not reliable in patients taking proton pump inhibitors. Histologic examination also follows endoscopy and is subject to sampling error, and its accuracy is dependent on the stain selected and on the pathologists’ skill. Serology is inexpensive but is not reliable in determining the presence of active infection.
Our successful amplification and specific detection of H. pylori DNA directly from stool samples in the majority of infected subjects indicates that this approach is feasible and demonstrates it has true potential in aiding the diagnosis and management of patients with H. pylori infection. The sensitivity of H. pylori DNA detection of 73% from uncultured stool specimens exceeds that in previous attempts (9, 23). This higher sensitivity most likely is the result of our extraction method, which removes from the stool samples impurities that are known to contain PCR inhibitors (18).
The sensitivity of PCR amplification is well known and is exhibited by our ability to easily detect DNA from less than seven H. pylori bacterial organisms. Other investigators have used PCR of gastric tissue specimens to identify H. pylori DNA (17). We demonstrated the extreme sensitivity and specificity of our molecular assay in analyzing gastric tissue biopsy samples in our study subjects as well as in an additional set of subjects. In two of these additional subjects in which infection was felt to be present serologically and histologically by the pattern of gastritis, H. pylori DNA was detectable, but organisms could not be identified microscopically. Notably, these two patients had had prior antiacid secretory therapy, but no prior antibiotic use was noted.
Our findings indicate that this molecular assay is specific for H. pylori DNA in human stool and gastric specimens. The H. pylori 16S rRNA primers used were designed from those tested by Weiss et al. (24), who demonstrated the specificity of their primers in detecting H. pylori DNA in paraffin-embedded gastric tissue. Furthermore, they found that these primers did not cross-react with many other common microorganisms, such as E. coli, various Campylobacter species, and other Helicobacter species, including H. cinnaedi, H. fenelliae, and H. muselae. Recently, Scholte et al. (17) have tested similar primers designed to amplify this unique region of 16S rRNA on 38 different bacteria, including 10 Helicobacter species, and found 100% sensitivity and specificity.
Our DNA analyses of human stool and gastric biopsy samples using these specific oligonucleotides correlated well with conventional tests for H. pylori infection. We chose histology as the “gold standard” in this study due to the expertise of our pathologist (H.F.F.). These results also suggest that sampling error is not a major obstacle in the detection of H. pylori DNA in stool samples. Small portions of spontaneously passed stool yielded consistent results on repeated analyses and identified the majority of infections. The reason for the failure to identify H. pylori DNA in three stool samples from individuals known to have H. pylori organisms and DNA in their gastric tissue is uncertain. Sampling error certainly could have played a role in these three cases, as could have degradation of DNA and organisms during intestinal transit. Additional studies to further analyze these samples are planned.
The molecular detection of H. pylori DNA has the added benefits of being able to genotype the infecting strains and to provide useful information about the presence or absence of potential virulence factors for a particular infection. For instance, once virulent components of H. pylori are characterized, PCR primers can be designed to specifically identify these important variants. Potentially important virulence factors include vacA gene variants and the cagA gene. The presence of certain genotypes of vacA and cagA in infecting strains has been shown to be associated with a more dense inflammatory gastritis and peptic ulcer disease (2).
The finding of detectable H. pylori DNA in the stool of the majority of infected subjects has important implications for transmission of this microorganism. These data support a fecal-oral route of transmission, although viable bacteria were not sought in this study. While it is known that H. pylori can be found in and cultured from the stool of infected individuals (9, 11, 20), the present study provides further supporting evidence that this may be a frequent occurrence. Makristathis et al. identified H. pylori’s species-specific protein antigen in stool samples by a seminested PCR method in 93.7% of patients with duodenal ulcers (10). A study of Bangladeshi children found 60% had H. pylori in their stool upon testing with immunomagnetic separation and ureA gene PCR amplification (3).
Our noninvasive assay to detect H. pylori DNA in spontaneously passed stool is currently labor-intensive, mostly due to the extensive DNA extraction procedure. Further development of the assay will focus on simplifying the DNA extraction process. Significant advances in technology to neutralize or remove impurities from DNA efficiently are eagerly awaited to make this assay more clinically applicable. Due to the cumbersome technical procedures required to handle radioactivity, the incorporation of chemiluminescence detection methods will be explored. Further testing will be required to determine if the method can be modified for stool card tests analogous to guaiac testing. Moreover, additional testing of stools after treatment of H. pylori infection is planned to determine if the assay is helpful in determining eradication of the microorganism.
ACKNOWLEDGMENTS
We are grateful to Richard Zoltec for facilitating the H. pylori serology testing and Mark Clem for technical assistance in processing histologic specimens.
This work was supported in part by NIH grant CA67900-04.
REFERENCES
- 1.Albert M J, Islam D, Nahar S, Qadri F, Falklind S, Wintraub A. Rapid detection of Vibrio cholerae O139 Bengal from stool specimens by PCR. J Clin Microbiol. 1997;35:1633–1635. doi: 10.1128/jcm.35.6.1633-1635.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Atherton J C, Peek R M, Jr, Tham K T, Cover T L, Blaster M J. Clinical and pathological importance of heterogeneity in VacA, the vacuolating cytotoxin gene of helicobacter pylori. Gastroenterology. 1997;112:92–99. doi: 10.1016/s0016-5085(97)70223-3. [DOI] [PubMed] [Google Scholar]
- 3.Casswal T H, Nilsson H, Bergstrom M, Aleljung P, Wadstrom T, Dahlstrom A K, Albert M J, Sarker S A. Evaluation of serology, 13C-urea breath test, and polymerase chain reaction of stool samples to detect Helicobacter pylori in Bangladeshi children. J Pedicatr Gastroenterol Nutr. 1999;28:31–36. doi: 10.1097/00005176-199901000-00009. [DOI] [PubMed] [Google Scholar]
- 4.Chiu C, Ou J T. Rapid identification of Salmonella serovars in feces by specific detection in virulence genes, invA and spvC, by an enrichment broth culture-multiplex PCR combination assay. J Clin Microbiol. 1996;34:2619–2622. doi: 10.1128/jcm.34.10.2619-2622.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clayton C L, Kleanthous H, Coates P J, Morgan D D, Tabaqchali S. Sensitive detection of Helicobacter pylori by using polymerase chain reaction. J Clin Microbiol. 1992;30:192–200. doi: 10.1128/jcm.30.1.192-200.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Da Silva A J, Bornay-Linares F J, De la Puente C A, Moura H, Peralta J M, Sobottka I, Schwartz D A, Visvesvara G S, Slemenda S B, Pieniazek N J. Diagnosis of Enterocytozoon bieneusi (Microsporidia) invections by polymerase chain reaction in stool samples using primers based on the region coding for small-subunit ribosomal RNA. Arch Pathol Lab Med. 1997;121:874–879. [PubMed] [Google Scholar]
- 7.Evans D E, Evans D J, Lampert H C, Graham D Y. Restriction fragment length polymorphism in the adhesin gene hpaA of Helicobacter pylori. Am J Gastroenterol. 1995;90:1282–1288. [PubMed] [Google Scholar]
- 8.Ho S, Hoyle J, Lewis F, Secker A, Cross D, Mapstone N, Dixon M, Wyatt J, Tompkins D, Taylor G, Quirke P. Direct polymerase chain reaction test for detection of Helicobacter pylori in humans and animals. J Clin Microbiol. 1991;29:2543–2549. doi: 10.1128/jcm.29.11.2543-2549.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kelly S, Pitcher M, Farmery S, Gibson G. Isolation of Helicobacter pylori from feces of patients with dyspepsia in the United Kingdom. Gastroenterology. 1994;107:1671–1674. doi: 10.1016/0016-5085(94)90806-0. [DOI] [PubMed] [Google Scholar]
- 10.Makristathis A, Pasching E, Schutze K, Wimmer M, Rotter M L, Hirschl A M. Detection of Helicobacter pylori in stool specimens by PCR and antigen enzyme immunoassay. J Clin Microbiol. 1998;36:2772–2774. doi: 10.1128/jcm.36.9.2772-2774.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mapstone N, Lewis F, Tompkins D, Lynch D, Axon A, Dixon M, Quirke P. PCR identification of Helicobacter pylori in faeces from gastritis patients. Lancet. 1993;341:447. doi: 10.1016/0140-6736(93)93053-4. [DOI] [PubMed] [Google Scholar]
- 12.Marshall B, Warren J. Unidentified curved bacillus on gastric epithelium in active chronic gastritis. Lancet. 1983;i:1233–1235. [PubMed] [Google Scholar]
- 13.Moskaluk C A, Kern S E. Microdissection and polymerase chain reaction amplification of genomic DNA from histological tissue sections. Am J Pathol. 1997;150:1547–1549. [PMC free article] [PubMed] [Google Scholar]
- 14.Parsonnet J, Friedman G, Vandersteen D, Chang Y, Vogelman H, Orientreich N, Sibley R. Helicobacter pylori infection and the risk of gastric carcinoma. N Engl J Med. 1991;325:1127–1131. doi: 10.1056/NEJM199110173251603. [DOI] [PubMed] [Google Scholar]
- 15.Powell S M, Harper J, Hamilton S, Robinson C, Cummings O W. Inactivation of Smad4 in gastric carcinomas. Cancer Res. 1997;57:4221–4224. [PubMed] [Google Scholar]
- 16.Ramotar K, Waldhart B, Church D, Szumski R, Louie T J. Direct detection of verotoxin-producing Escherichia coli in stool samples by PCR. J Clin Microbiol. 1995;33:519–524. doi: 10.1128/jcm.33.3.519-524.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Scholte G H, van Doorn L J, Quint W G, Lindeman J. Polymerase chain reaction for the detection of Helicobacter pylori in formaldehyde-sublimate fixed, paraffin-embedded gastric biopsies. Diagn Mol Pathol. 1997;6:238–243. doi: 10.1097/00019606-199708000-00008. [DOI] [PubMed] [Google Scholar]
- 18.Sidransky D, Tokino T, Hamilton S, Kinzler K, Levin B, Frost P, Vogelstein B. Identification of ras oncogene mutations in the stool of patients with curable colorectal tumors. Science. 1992;256:102–105. doi: 10.1126/science.1566048. [DOI] [PubMed] [Google Scholar]
- 19.Thijs J C, van Zwet A A, Thijs W J, Oey H B, Karrenbeld A, Stellaard F, Luijt D S, Meyer B C, Kleibeuker J H. Diagnostic tests for Helicobacter pylori: a prospective evaluation of their accuracy, without selecting a single test as the gold standard. Am J Gastroenterol. 1996;91:2125–2129. [PubMed] [Google Scholar]
- 20.Thomas J, Gibson G, Darboe M, Dale A, Weaver L. Isolation of Helicobacter pylori from human feces. Lancet. 1992;340:1194–1195. doi: 10.1016/0140-6736(92)92894-l. [DOI] [PubMed] [Google Scholar]
- 21.Tomb J F, White O, Kerlavage A R, Clayton R A, Sutton G G, Fleischmann R D, Ketchum K A, Klenk H P, Gill S, Dougherty B A, Nelson K, Quackenbush J, Zhou L, Kirkness E F, Peterson S, Loftus B, Richardson D, Dodson R, Khalak H G, Glodek A, McKenney K, Fitzgerald L M, Lee N, Adams M D, Venter J C. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature. 1997;388:539–547. doi: 10.1038/41483. [DOI] [PubMed] [Google Scholar]
- 22.Ushijima H, Koike H, Mukoyama A, Hasegawa A, Nishimura S, Gentsch J. Detection and serotyping of totaviruses in stool specimens by using reverse transcription and polymerase chain reaction amplification. J Med Virol. 1992;38:292–297. doi: 10.1002/jmv.1890380412. [DOI] [PubMed] [Google Scholar]
- 23.van Zwet A A, Thijs J C, Kooistra-Smid A M D, Schirm J, Snijder J A M. Use of PCR with feces for detection of Helicobacter pylori infections in patients. J Clin Microbiol. 1994;32:1346–1348. doi: 10.1128/jcm.32.5.1346-1348.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weiss J, Mecca J, Silva E, Gassner D. Comparison of PCR and other diagnostic techniques for detection of Helicobacter pylori infection in dyspeptic patients. J Clin Microbiol. 1994;32:1663–1668. doi: 10.1128/jcm.32.7.1663-1668.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wotherspoon A C, Ortiz-Hidalgo C, Falzon M F, Isaacson P G. Helicobacter pylori-associated gastritis and primary B-cell lymphoma. Lancet. 1993;338:1175–1176. doi: 10.1016/0140-6736(91)92035-z. [DOI] [PubMed] [Google Scholar]