Abstract
SBF (Swi4/Swi6 Binding Factor) complex is a crucial regulator of G1/S transition in Saccharomyces cerevisiae. Here, we show that SBF complex is required for myriocin resistance, an inhibitor of sphingolipid synthesis. This phenotype was not shared with MBF complex mutants nor with deletion of the Swi4p downstream targets, CLN1/CLN2. Based on data mining results, we selected putative Swi4p targets related to sphingolipid metabolism and studied their gene transcription as well as metabolite levels during progression of the cell cycle. Genes which encode key enzymes for the synthesis of long chain bases (LCBs) and ceramides were periodically transcribed during the mitotic cell cycle, having a peak at G1/S, and required SWI4 for full transcription at this stage. In addition, HPLC-MS/MS data indicated that swi4Δ cells have decreased levels of sphingolipids during progression of the cell cycle, particularly, dihydrosphingosine (DHS), C24-phytoceramides and C24-inositolphosphoryl ceramide (IPC) while it had increased levels of mannosylinositol phosphorylceramide (MIPC). Furthermore, we demonstrated that both inhibition of de novo sphingolipid synthesis by myriocin or SWI4 deletion caused partial arrest at the G2/M phase. Importantly, our lipidomic data demonstrated that the sphingolipid profile of WT cells treated with myriocin resembled that of swi4Δ cells, with lower levels of DHS, IPC and higher levels of MIPC. Taken together, these results show that SBF complex plays an essential role in the regulation of sphingolipid homeostasis, which reflects in the correct progression through the G2/M phase of the cell cycle.
Keywords: cell cycle, ceramide, Swi4p transcription factor, sphingolipid, dihydroceramide, mannosylinositol phosphorylceramide
Graphical Abstract
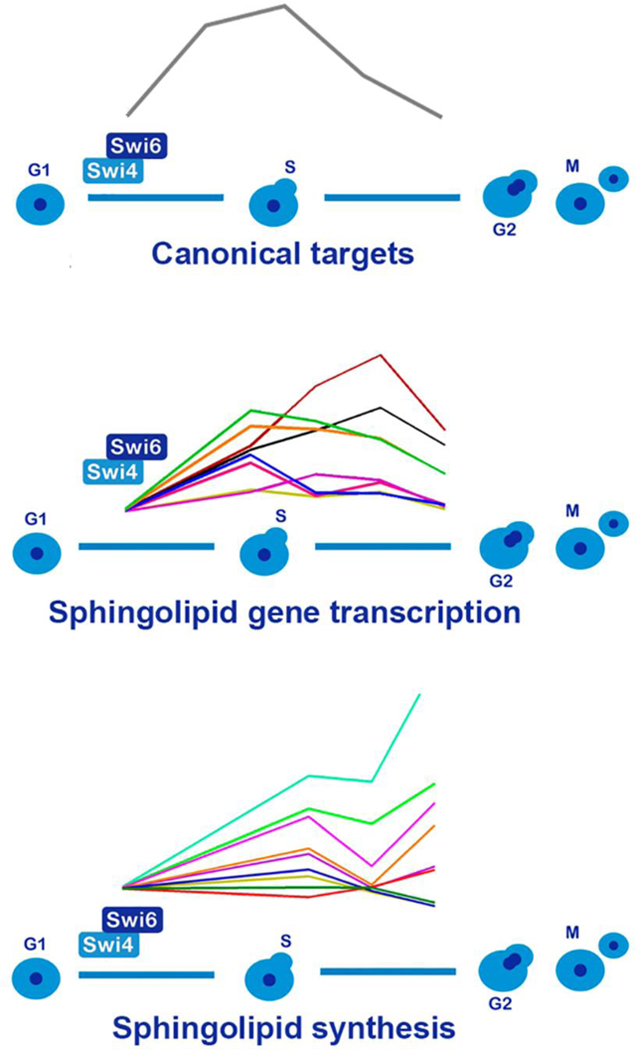
Interaction between cell cycle progression and sphingolipid synthesis.
Transcriptional activation of sphingolipid genes during G1 to S transition, followed by oscillation of sphingolipid metabolites during progression of the cell cycle, in a Swi4p dependent manner.
1. INTRODUCTION
Sphingolipids are versatile molecules that can have a structural and a bioactive function. As bioactive molecules they are considered potential targets for cancer treatment, since changes in sphingolipid homeostasis are particularly observed in cells that do not respond to cell cycle checkpoint signals [1–5]. Sphingolipid signaling is diverse and can have antagonizing effects during cell division. For example, in human cell lines, dihydroceramide accumulation leads to autophagy, endoplasmic reticulum stress and delayed G1/S transition [6]. In contrast, sphingosine-1-phosphate is a lipid second messenger that signals cellular proliferation [7,8] and loss of sphingosine kinase (Sphk1) inhibits chromosome segregation, through regulation of PI3-AKT pathway and thus delays mitotic progression [9,10]. In mammals, ceramide has a potent antiproliferative effect mediated by protein phosphatases that regulate the cell cycle modulators retinoblastoma protein (Rb) and cell cycle dependent kinases [11–13] which can be suppressed by sphingosine-1-phosphate [10]. In yeast, survival during heat stress is mediated by the induction of phosphorylated sphingoid bases, which cause a cell cycle arrest at G1 [14], while synthesis of complex ceramides is crucial for correct cytokinesis [15]. These reports indicate that long chain bases (LCBs) and ceramides are signals that govern the correct progression of the cell cycle. However, knowledge on the regulation of sphingolipid metabolism by the cell cycle remains to be studied. Cerbón and colleagues [16] showed that inhibition of the first step in sphingolipid synthesis, catalyzed by the serine-palmitoyl transferase complex (SPT) (Fig. 1), delays G1/S cell cycle progression. They also demonstrated that entry into G1 phase reduces the levels of dihydrosphingosine (DHS) while the level of phytosphingosine (PHS) and inositol phosphoceramide is increased [16]. A recent work showed that Swe1p kinase, which regulates G2/M transition and is a morphogenesis checkpoint, regulates sphingolipid biosynthesis [17] which might link cell cycle with sphingolipid homeostasis.
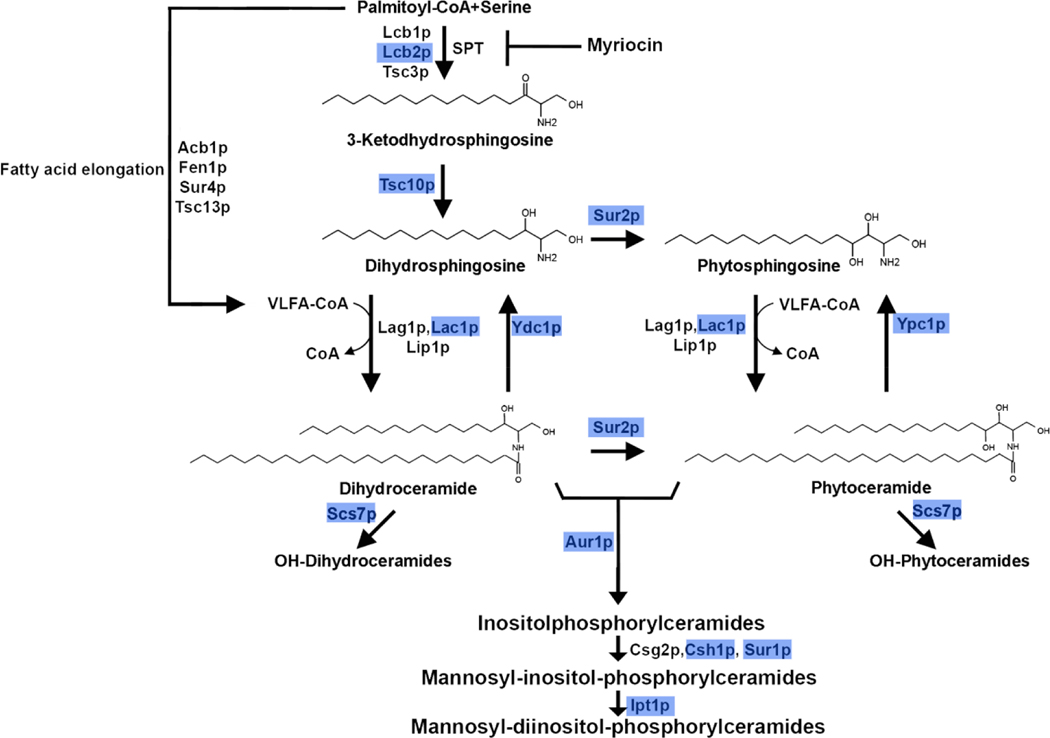
Fig. 1. Major pathway of sphingolipid synthesis in Saccharomyces cerevisiae.
Scheme shows the enzymes of sphingolipid synthesis in S. cerevisiae (Dickson et al, 1998) [63]. The first step is catalyzed by serine-palmitoyl transferase complex (SPT), which catalyzes the condensation of a L-serine and palmitoyl-CoA to 3-ketodihydrosphingosine, CoA and CO2 (Pinto et al, 1992) [64]. This reaction takes place in the endoplasmic reticulum [63] and its activity is inhibited by myriocin [65]. 3-ketodihydrosphingosine is the precursor of the long chain bases (LCBs) dihydrosphingosine (DHS) and phytosphingosine (PHS). These are N-acylated to form either dihydro- or phytoceramide. The novo pathway is catalyzed by the ceramide synthases Lag1p, Lac1p and Lip1p [66]. Sphingoid bases can also be formed by degradation of ceramides by activation of Ydc1p and Ypc1p ceramidases [63]. These first group of reactions are conserved during evolution among the eukaryotic kingdom [67,68] but the formation of dihydroceramide and phytoceramide is different, as hydroxylation of DHS to form PHS and phytoceramide catalyzed by Sur2p is unique to yeast [69]. In S. cerevisiae, unlike mammalian cells, inositol phosphate and mannose are incorporated to form complex ceramides, inositol phosphoceramides (IPC), mannosylinositolphosphoceramide (MIPC) and mannosyldiinositolphosphoceramide (M(IP)2C). We highlighted in blue boxes the enzymes which were found to be transcriptionally regulated in swi4Δ cells (see Fig. 4) which are Lcb2p (subunit of SPT complex), Tsc10p (3-ketosphinganine reductase), Sur2p (sphinganine C4-hydroxylase), Lac1p (ceramide synthase component), , Ydc1p (alkaline dihydroceramidase), Aur1p (phosphatidylinositol: ceramide phosphoinositol transferase), Scs7p (sphingolipid α-hydroxylase and desaturase), Sur1p (mannosylinositol phosphorylceramide synthase subunit), Csh1p (mannosylinositol phosphorylceramide synthase subunit) and Ipt1p (inositol phosphotransferase).
Based on these studies, it was tempting to study the role of the cell cycle G1 checkpoint on the synthesis of sphingolipids. During the mitotic cell cycle in yeast, two major transcription factor complexes coordinate the G1/S checkpoint: the SBF (Swi4p Binding Factor) and MBF (Mbp1p Binding Factor) composed by the transcription factors Swi4p and Mbp1p, respectively. Both transcription factors have a common binding partner, Swi6p [18,19]. Although these complexes regulate an overlapping group of genes, they own specific functions [20,21]. For example, transcription of genes related to morphogenesis such as the septin ring are specifically regulated by the SBF complex (CRCGAAA binding site), while transcription of genes involved in DNA repair and replication are regulated by the MBF complex (ACGCGT binding site) [22]. The activity of yeast and mammalian G1/S complexes is regulated by Whi5p or retinoblastoma protein (Rb), respectively, which bind to SBF or E2F1 complexes, inhibiting their activity [23]. Despite lacking structural similarity, E2F1, the mammalian orthologue of Swi4p, regulates a similar group of genes, indicating a high degree of conservation of the regulatory mechanism of the cell cycle progression [19,24]. As both complexes are functionally conserved, we chose yeast as a model to investigate the molecular mechanisms that link the synthesis of ceramides with the mitotic cell cycle. In this study, we provide evidence that SBF transcription factor regulates genes involved in the sphingolipid synthesis, playing an essential role in the regulation of sphingolipid homeostasis during cell cycle.
2. MATERIALS AND METHODS
2.1. Materials.
The chemicals used were obtained from the following sources: myriocin (Sigma-Aldrich), PHS (Avanti lipids), high-capacity cDNA reverse transcription kit (Applied Biosystems) and Power SYBR™ Green master mix (Thermo Fisher Scientific). Primers were obtained from Exxtend Biotecnologia (Brazil). All other reagents were obtained from Sigma-Aldrich.
2.2. Strains and growth medium.
S. cerevisiae strain BY4741 (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0) and the MATa deletion library were obtained from Open Biosystems. To obtain the double mutant cln1Δ/cln2Δ, the CLN1 gene was deleted in a cln2Δ background, by homologous recombination using the His3MX6 module [25]. The primers used are listed (Supplementary Information, Table S1). Yeast transformation was performed following the protocol described by Gietz and Woods [26]. Yeast cells were grown in YPD liquid medium (1% yeast extract, 2% peptone, 2% dextrose) or synthetic dextrose medium (SD) (2% glucose, 6.7 g/L yeast nitrogen base and the needed supplements according to the strain auxotrophic markers). 2% agarose was added when solid media was prepared.
2.3. Growth assays in the presence of myriocin.
Cells were grown in YPD media to early stationary phase (24 hours), and serially diluted in water (ABS600 nm =0.3, 0.03 and 0.003), spotted with a 48-pin replicator in solid SD medium in the absence or presence of the indicated concentrations of myriocin (dissolved in DMSO) or PHS (dissolved in ethanol). Cells were also treated with serine or palmitate, both at concentration of 0.4% w/v (palmitate was dissolved in 1%Tween 40, Fig. 2D). Plates were incubated at 30 °C for 3 days and then photographed using a Canon 20D camera. Images were processed using Adobe Photoshop software. For lipidomic experiments in the presence of myriocin, cells were grown in YPD to stationary phase (24 hours) and then an aliquot of approximately 108 cells was inoculated in SD medium in the absence or presence of 1 μg. mL−1 of myriocin. Cells were then analyzed both by FACS and HPLC-MS/MS after 5 hours of growth at 30 °C.
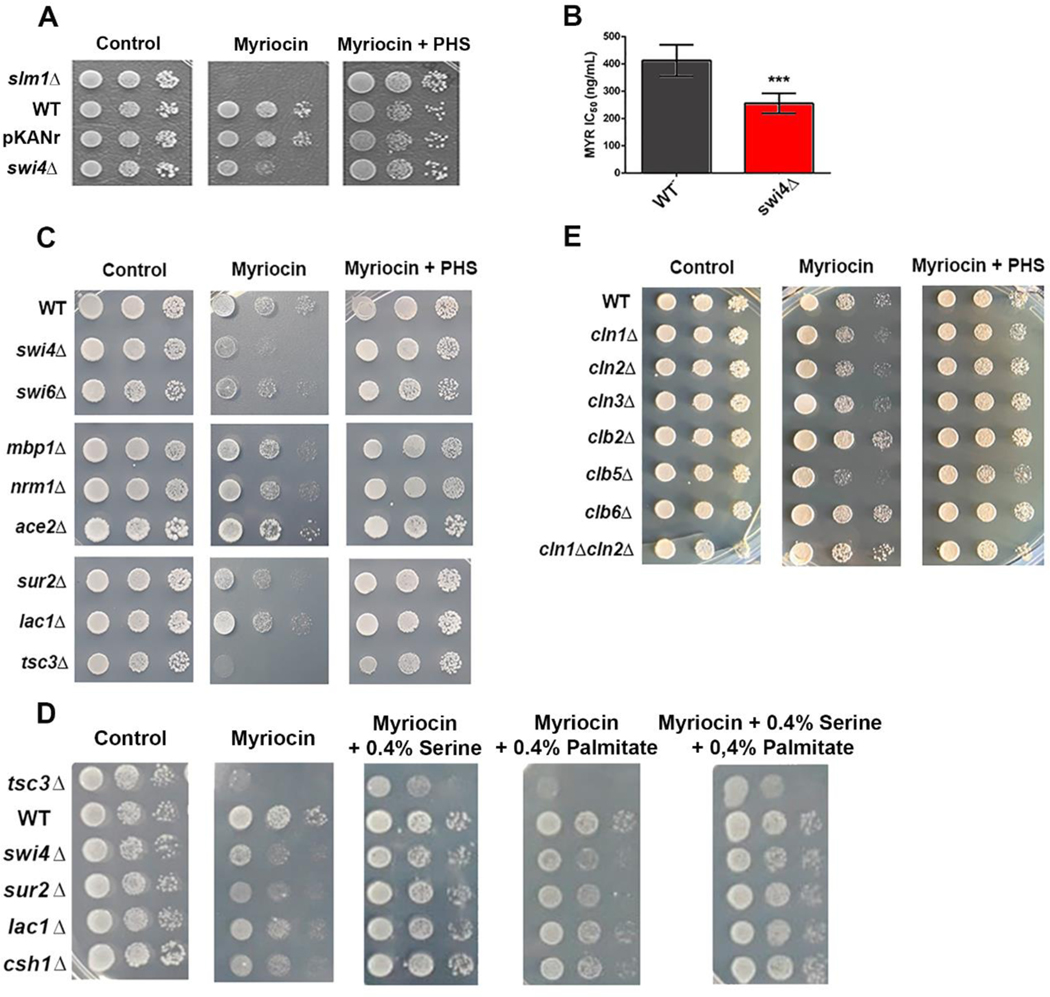
Fig. 2. Growth inhibition of SBF mutants by myriocin.
(A) Strains slm1Δ, WT (BY4741), WTtransformed with plasmid containing geneticin resistance (pKANr), and swi4Δ were spotted in serial dilutions and grown in synthetic minimal medium (SD) at 30ºC for 2 days (control). 15μM PHS and 0. 6μg.ml−1 myriocin were added where indicated. (B) The myriocin IC50 values for WT and swi4Δ strains were determined in SD medium. (C) Strains related to SBF (swi4Δ, swi6Δ,), MBF (mbp1Δ, nrm1Δ) M/G1 regulator (ace2Δ) and ceramide synthesis (sur2Δ, lac1Δ and tsc3Δ) were grown as in A. (D) Reversion of myriocin growth phenotype by the addition of 0.4% serine and 0.4% palmitate. (E) Strains related to G1 (cln1Δ, cln2Δ, cln3Δ), S (clb5Δ, clb6Δ) and G2/M cyclins (clb2Δ) were grown in the presence of myriocin as in A.
2.4. Search for putative Swi4 target genes.
The YEASTRACT database [27] was used to find putative Swi4p target genes using three parameters: 1) genes with the Swi4p binding sequences reported in their promoter regions CACGAAA or CGCGAAA [22]. 2) genes with documented “binding evidence”; or 3) genes with documented “expression regulation evidence” (Last access in November 2019). Genes related to the cellular process “Lipid metabolic processes” were selected using the gene ontology Slim Mapper tool at Saccharomyces Genome Database (https://www.yeastgenome.org/goSlimMapper). The complete list of target genes is found in Supplementary Information, Table S2.
2.5. Cell cycle synchronization and DNA content analysis.
To study gene transcription and lipid metabolite levels during cell cycle progression, cells were grown to stationary phase. After this, cells were seeded in YPD and grown to mid-log phase (5 hours) and then treated for 2 hours with alpha-factor (4 μg. mL−1 for WT cells and 6 μg. mL−1 for swi4Δ cells). To remove alpha-factor, cells were rapidly washed three times with distilled cold water by centrifugation and seeded in fresh medium at a density of approximately 7 × 106 cells/ml at 30º C. Aliquots for DNA content, gene transcription and lipidomic analysis were taken at 0, 45, 60 and 75 minutes from the same experiment and kept at −70˚ C. Analysis of the DNA content was performed as in[28]. Briefly ~7 × 106 cells were fixed overnight with 70% ethanol at −20°C. After fixation, cells pellets were resuspended in 50μl of 50mM sodium citrate buffer, pH 7.0, followed by treatment for 2 h at 40°C with 200μg. mL−1 RNAse diluted in sodium citrate buffer and then treated at 50°C for 2 h with 2mg. mL−1 proteinase K diluted in 50μM Tris-HCl, 1mM CaCl2, pH 8.0. Cells were stained with 5μg. mL−1propidium iodide. DNA content was analyzed with a BD FACS Calibur (BD Biosciences) cytometer and data was analyzed using the Flowing software v.2 (Cell Imaging Core, Turku Centre for Biotechnology).
2.6. Gene transcription analysis.
Total RNA was isolated using the phenol-chloroform extraction method [29]. cDNA was synthesized using the high-capacity cDNA reverse-transcription kit according to the manufactureŕs guidelines (Applied Biosystems). Transcriptional levels were analyzed by quantitative PCR (qPCR) using the Power SYBR™ Green master mix (Thermo Fisher Scientific) and a 7500 real-time PCR system (Applied Biosystems) instrument. The ABI 7500 SDS software was used to analyze the data. Gene transcription was normalized to the transcription of the housekeeping ACT1 gene, and the relative mRNA levels were calculated using the 2-ΔCt method [30]. The primers used for each gene are listed in Supplementary Information, Table S3).
2.7. Sphingolipid analysis by HPLC-MS/MS.
Aliquots of approximately 108 cells were harvested, washed two times with distilled water and then lyophilized before lipid extraction. Prior to cell lysis, C17-sphingolipids standards were added to the lyophilized pellets [31,32], and Mandala extraction was carried out as described previously [33], followed by Bligh and Dyer Extraction [34]. A quarter of each sample obtained from the Bligh and Dyer extraction was reserved for inorganic phosphate (Pi) determination, so the relative sphingolipid signal was normalized by the Pi abundance. The organic phase was transferred to a new tube and submitted for alkaline hydrolysis of phospholipids [35]. Finally, the organic phase was dried and used for mass spectrometry analysis [36].
2.8. Statistics.
Experiments were repeated at least three times. Data are shown as mean ± S.D., and significant differences were analyzed usually using one-way or two-way analysis of variance (ANOVA). Analyses for mRNA levels during cell cycle were performed with two-way ANOVA, using Dunnett’s and Sidak’s post-test. Analyses for sphingolipid content were performed with paired T-test. Cell cycle analyses were performed with Sidak’s post-test. All tests were performed in GraphPrism 6.0 software and significant differences were considered if p < 0.05.
3. Results
3.1. Myriocin induced growth defect in SBF mutants.
To investigate the possible role of the G1/S transcription factor Swi4p in the regulation of sphingolipid metabolism, we compared growth of WT and swi4Δcells in the presence of 0.6 μg. mL−1 myriocin (Fig.2). Growth impairment by myriocin was notably higher in swi4Δcells when compared to WT (Fig. 2A). As a control, we included the myriocin-hypersensitive strain slm1Δ, [37] whose growth was completely inhibited by the drug. We also included the WT strain transformed with the plasmid pKANr, which confers geneticin resistance, to test whether the presence of this marker could lead to pleiotropic drug resistance, but it was unaffected. The growth defect of swi4Δcells was related to sphingolipid synthesis inhibition, since addition of exogenous PHS, an intermediary metabolite of the sphingolipid synthesis pathway (Fig. 1), reverted myriocin-induced growth arrest.Next, we calculated the IC50 values in liquid synthetic minimal liquid and confirmed that swi4Δ was more sensitive to myriocin than WT (0.26 μg. mL−1 and 0.37 μg. mL−1 respectively. Fig.2B).
To study whether other mutants related to G1/S and M/G1 transition were also more sensitive to myriocin, we tested myriocin sensitivity of swi6Δ, mbp1Δ, nrm1Δ (G1/S transition) and ace2Δ (M/G1 transition) strains (Fig. 2C). Results show that only swi4Δ and swi6Δ strains were hypersensitive to myriocin, suggesting that SBF complex is specifically involved with sphingolipid synthesis regulation.
We next compared growth in the presence of myriocin between SBF mutants and the sphingolipid metabolism mutants tsc3Δ, sur2Δ and lac1Δ (Fig. 2C) in which the first reaction of sphingolipid synthesis, C4 hydroxylation of DHS and dihydroceramides and partial ceramide synthesis is inhibited, respectively (Fig. 1). The tsc3Δ and sur2Δ mutants showed a similar growth defect comparable to swi4Δ, while lac1Δ strain was not more sensitive than WT, probably because its homolog, Lag1p, has a similar enzymatic function (Fig. 1). As serine also has a regulatory role in sphingolipid homeostasis [38], we also tested if serine or palmitate could rescue the growth phenotype observed during myriocin treatment. Results show that addition of 0.4% serine rescued growth of WT as well as swi4Δ, lac1Δ, sur2Δ, and to a lower extent that of tsc3Δ strain, while 0.4% palmitate partially rescued growth. However, treatment with both serine and palmitate did not have additional effects on growth rescue. These results show that the effect of myriocin was specific for inhibition of SPT and not to a secondary effect (Fig. 2D).
Next, to evaluate whether swi4Δ growth impairment by myriocin was associated to decreased transcription of cyclins (known SBF targets), we tested the myriocin tolerance of null mutants of G1 cyclins (CLN1, CLN2, CLN3), S cyclins (CLB5, CLB6) and G2/M cyclins (CLB2) (Fig. 2E). Of these strains, only clb5Δ showed increased sensitivity to myriocin compared to the WT strain, while the mutant of CLB2 gene, an inhibitor of SBF at M phase, was more resistant to myriocin. Altogether, these results show that the inhibition of the sphingolipid biosynthetic pathway induces a growth defect of SBF mutants and suggest a direct role for SBF on the regulation of sphingolipid biosynthesis.
3.2. Identification of putative Swi4p target genes involved in sphingolipid biosynthetic pathway.
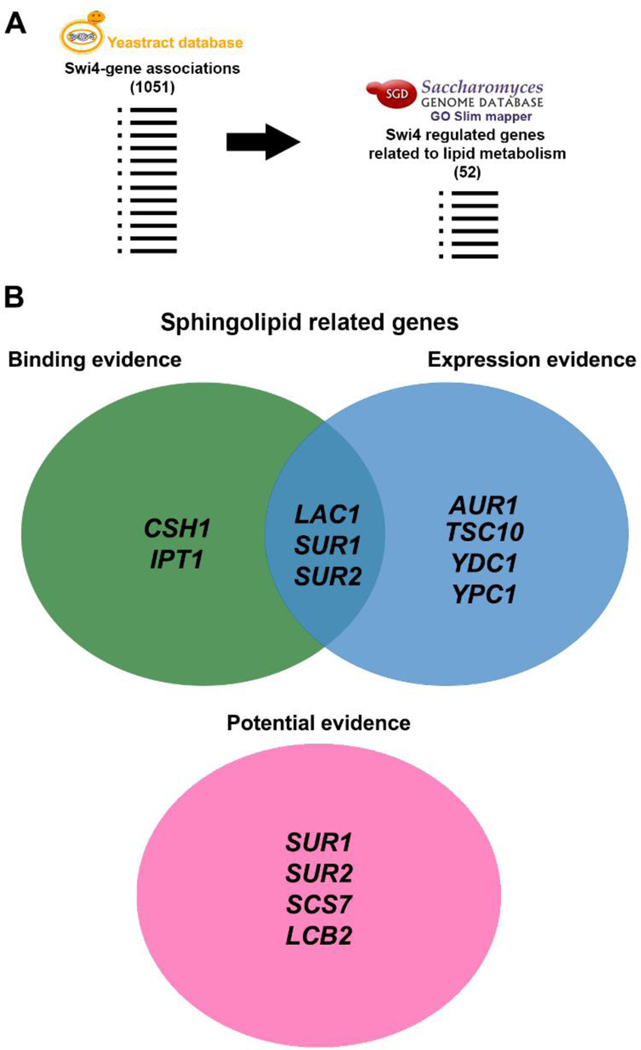
To identify whether Swi4p target genes are related to sphingolipid biosynthetic pathway, we sought for Swi4p regulated genes in YEASTRACT (http://www.yeastract.com/formfindregulated.php), a database that associates transcription factors to target genes in Saccharomyces cerevisiae [27,39]. These associations are based on three categories depending on the experimental evidence: (i) “Binding evidence”, which includes data on binding of transcription factors to promoter regions of their targets, (ii) “Expression evidence”, which includes genes differentially expressed after transcription factordeletion or overexpression, and (iii) “Potential evidence” which includes genes with the transcription factors target sequence on its promoter region, but that have not been proven to be controlled by Swi4p. After gathering the results for Swi4p targets in the three classifications, they were grouped by GO process (https://www.yeastgenome.org/goSlimMapper) (Fig. 3A). We found 52 genes related to lipid metabolism (out of 1051) as putative Swi4p targets, which represent only 5.28% of all genes related to lipid metabolism (Supplementary Information, Table S2). Out of these, 9 genes are directly associated with sphingolipid biosynthesis (LCB2, TSC10, SUR2, LAC1, SCS7, AUR1, CSH1, SUR1 and IPT1) and two with ceramide degradation (YDC1 and YPC1) corresponding to almost 80% of the total sphingolipid synthetic metabolism (Fig. 1 and 3B). CSH1 and IPT1 promoters are reported to bind Swi4p, while AUR1, TSC10, YDC1 and YPC1are differentially expressed when SWI4 transcription is altered. LAC1, SUR1 and SUR2 are reported in both categories (Fig. 3B) [21,40,41]. Additionally, LCB2, SUR2, SCS7 and SUR1 have Swi4 target sequence located in their promoters (Fig 3B). SUR1 and SUR2 are the genes with strongest association with Swi4p, since they are placed in all groups (Fig 3B). Overall, these results support the hypothesis that Swi4p may control transcriptionally the sphingolipid biosynthetic pathway (Fig 1).
Fig. 3. Regulatory associations between Swi4 and sphingolipid synthesis genes.
(A) The potential Swi4p putative targets, derived from YEASTRACT database (http://www.yeastract.com/formfindregulated.php) were grouped according to their function, by using Gene Ontology Slim Mapper tool (http://www.yeastgenome.org/). Among 1051 genes, 52 were associated with lipid metabolism. (B) From those target genes, 11 were directly associated with sphingolipid metabolism, from which YEASTRACT database provided binding, expression and potential evidence, displayed on a Venn diagram.
3.3. Cell cycle-dependent transcriptional regulation of sphingolipid synthesis related genes and metabolite levels.
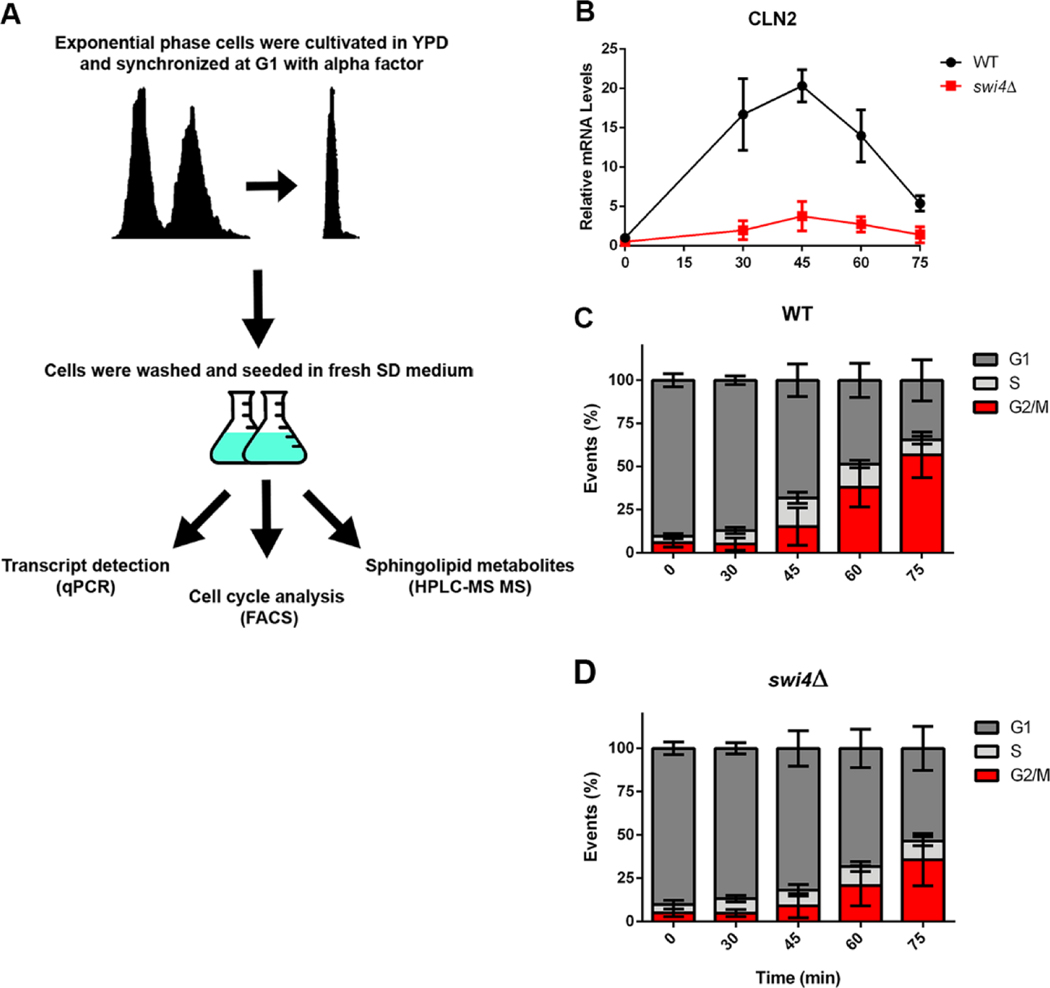
To confirm whether Swi4p is a positive regulator of the sphingolipid biosynthetic pathway, we analyzed cell cycle progression, gene transcription and sphingolipid metabolites in alpha-factor synchronized cells. For this purpose, cells were grown to exponential phase in YPD medium and synchronized at G1 for two hours with alpha-factor. Cells were then released in minimal medium (SD medium), to avoid possible sphingolipid contamination from YPD medium. Three different aliquots were taken at different time points for mRNA transcription, cell cycle and metabolite analysis (Fig 4A). We followed the transcription of the CLN2 gene, a canonical target of Swi4p [24] during cell cycle progression and observed a wave with a peak at 45 minutes, which ended after 75 minutes of growth. Analysis of the DNA content revealed that after pheromone treatment WT and swi4Δ cells were arrested at G1, and that after 75 minutes 50% and 30% of their content was at the G2/M stage respectively (Fig 4C–D). Aliquots of each time point were analyzed for RNA transcription by q-PCR and different classes of sphingolipids identified by HPLC-MS/MS.
Fig. 4. Designed assay for cell synchronization and cell cycle studies.
(A) Cells were cultivated to exponential phase in YPD medium and G1 synchronized with alpha factor. YPD was used for synchronization as swi4Δ cells do not synchronize after two hours in SD medium. Cells then were seeded in fresh minimal medium SD and three different aliquots were taken at the time points indicated to measure cell cycle stage, mRNA transcripts, and sphingolipid metabolites. (B) Transcription of CLN2, canonical target of SBF during cell cycle. WT (●) and swi4Δ cells (■). (C, D) DNA content (FACS), indicating the % of cells at each stage of the cell cycle, relative to total cells.
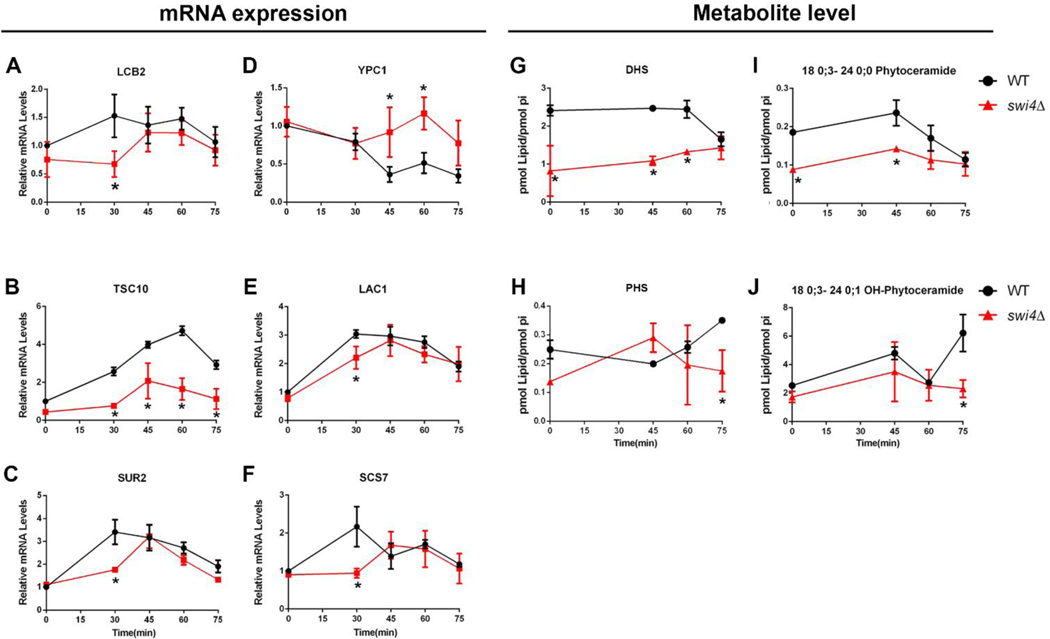
The transcriptional analysis was focused on the genes previously identified in the bioinformatic analysis (Fig. 3). We observed a cell cycle-dependent transcription of the sphingoid base synthesis genes, LCB2, TSC10 and SUR2, with LCB2 and SUR2 being induced in early and late G1 phase and TSC10 at late G1 (Fig. 5). From these, TSC10 transcription (Fig 5B) was markedly reduced in swi4Δ cells, while transcription of LCB2 and SUR2 was delayed in swi4Δ cells (Fig 5A–C). However, in swi4Δ cells, DHS levels were significantly lower throughout the cell cycle and PHS content was lower only after 75 minutes (Fig. 5 G and H). We also measured transcription of phytoceramidase gene YPC1 and results showed that it was repressed in WT cells while this repression failed to occur in the swi4Δ strain (Fig 5D).
Fig. 5. SWI4 deletion reduces transcriptional activation and metabolite levels of LCBs and ceramides.
Aliquots of WT (●) and swi4Δ cells (■) were harvested at indicated time points after alpha factor release, as shown in Fig. 4A. (A-F) Relative mRNA changes during cell cycle. Gene transcription was normalized to actin. *p≤0.05; n=2. (G-J) Metabolite changes during cell cycle, * p≤0.05. n=2. Data were normalized by phosphate levels (pi).
Next, we analyzed the transcription of LAC1 and SCS7 genes which encode enzymes responsible for ceramide synthesis and ceramide hydroxylation, respectively. Like SUR2 and LCB2, both were induced at early G1 phase. LAC1 transcription was only modestly repressed in swi4Δ cells unlike SCS7 which was not induced after 30 minutes (Fig 5 E, F). At the metabolite level, we could not find differences in the total levels of ceramides, but the lack of difference was driven by C26 OH-phytoceramide, the most abundant ceramide in our analysis (Supplemental figure 2B). However, most of the minor ceramides and OH-ceramides species were regulated during the progression of the cell cycle. Particularly, we found that swi4Δ strain has reduced levels of C24 phytoceramide during G1 and S stages, while the level of its hydroxylated derivate was lower at G2/M, when compared to the WT strain (Fig 5I, J).
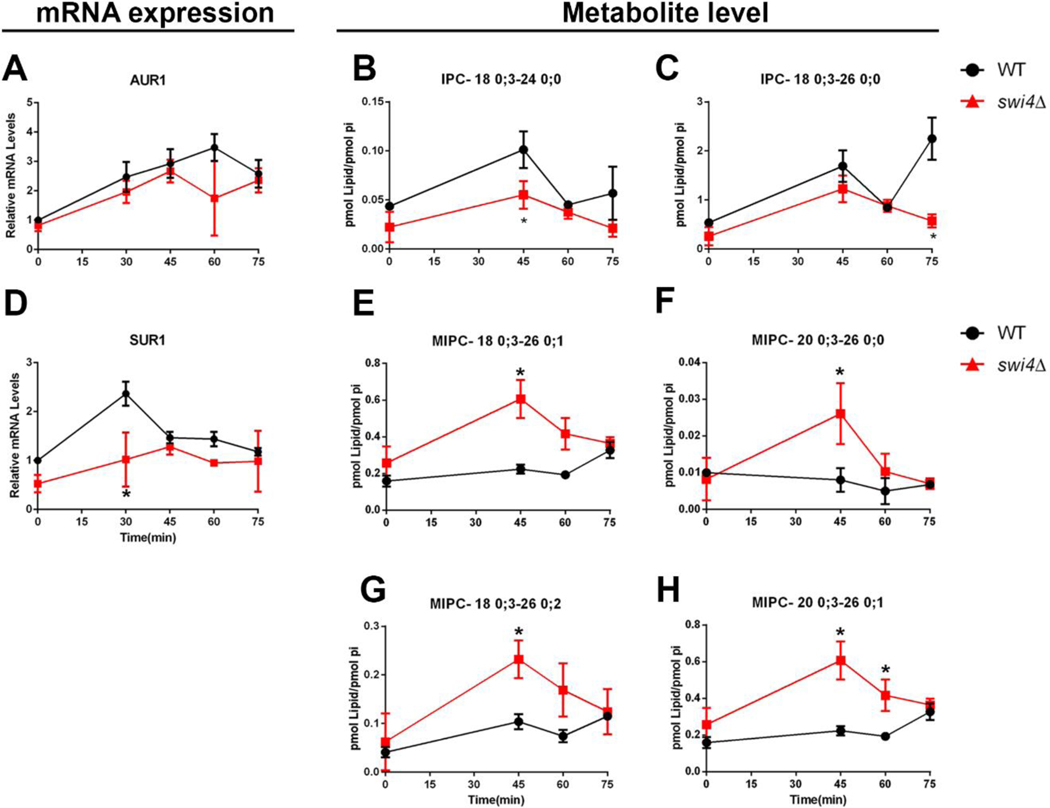
Next, we analyzed the transcription of genes involved in complex ceramide synthesis (AUR1, CHS1, SUR1 and IPT1). Interestingly, we observed induction of their transcription at G1, however only SUR1 transcription was repressed in swi4Δ strain (Fig 6D). The only gene involved in the synthesis of complex ceramides that was repressed during cell cycle progression in the WT strain was IPT1 gene, and it showed a significative lower transcription in the first time point on the swi4Δ strain (Supplemental figure 1). The levels of IPCs were upregulated by the cell cycle, especially the ones derived from very long chain phytoceramides which were less abundant in the swi4Δ mutant, despite the lack of difference in AUR1 transcripts (Fig 6A–C and supplemental figure 3). Surprisingly, the metabolite levels of MIPCs were higher in swi4Δcells, irrespective of the lower transcription of SUR1 gene (Fig 6D–H). Altogether these results are consistent with the notion that there might be a coordinate, G1/S up-regulation of sphingolipid biosynthesis genes by Swi4p.
Fig. 6. SWI4 deletion reduces transcriptional activation and metabolite levels of complex ceramides.
Aliquots of WT (●) and swi4Δ cells (■) were harvested at indicated time points after alpha factor release, as shown in Fig. 4A. (A-E) Relative mRNA changes during cell cycle. Gene transcription was normalized to actin. *p≤0.05. N=3. (F-I) Metabolite changes during cell cycle. Data were normalized by phosphate levels (pi). * p≤0.05. N=2.
3.4. Myriocin induced arrest at G2/M transition.
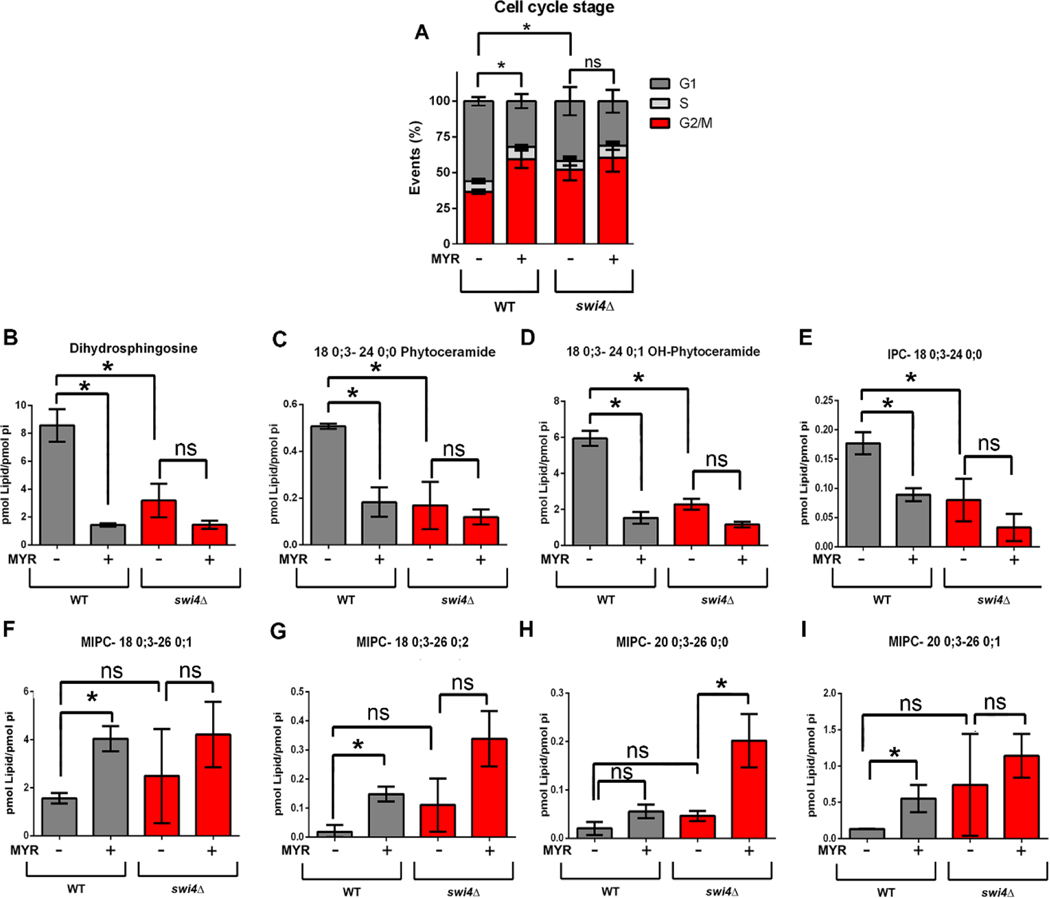
Our results so far indicate that transcription of sphingolipid-related genes are regulated during cell cycle progression, some in a Swi4p dependent manner. These data are supported by our sphingolipid measurements, which demonstrate that swi4Δ cells are also unable to properly control sphingolipid levels during cell cycle. Based on the fact that PHS reverses the myriocin induced growth defect of swi4Δ cells (Fig. 2), we tested whether reversion of low sphingolipid levels by PHS or C12-phytoceramide could result in reversion of swi4Δ induced G1/S arrest or/and of its partial G2/M arrest [42]. Results showed that addition of PHS or C12-phytoceramide did not reverted the defect in G1/S progression of the swi4Δ strain (Supplemental figure 4). We also assayed mitotic cell cycle progression in sur2Δ and lac1Δ cells and confirmed they do not have a significative block at G1/S (Supplemental figure 4). We also found that myriocin did not affect progression of the cell cycle in G1 synchronized cells (Supplemental figure 5 A, B). In an effort to find other viable mutants of sphingolipid metabolism that displayed altered cell cycle progression, we cultivated cells to mid-logarithmic phase and analyzed their DNA content using an approach similar to the one used by Polymenis and colleagues [43]. However, none of these strains presented significative differences (Supplemental figure 5C). These results led us to study whether sphingolipid synthesis impacts progression at a later stage of the cell cycle. To investigate the contribution of sphingolipids in G2/M progression, exponential growing cells were grown in the presence of myriocin (1μg.mL−1) during 5 hours (Fig. 7). Results showed that after 5 hours, myriocin treatment increased 2-fold the G2/M population in WT cells, different from what was observed on swi4Δ strain, (Fig 7A). It must be noted that swi4Δ strain presented 40% more cells arrested at G2/M without myriocin treatment at exponential phase (Fig 7A). This effect was only possible to observe after at least more than one round of cell division where G2/M to G1 transition occurred, differently from results in Figs. 4C and D where only one cell cycle was completed and the transition G2/M to G1 was not observed. These results suggest that both SWI4 deletion and myriocin treatment arrest cells at G2/M.
Fig. 7. Effects of myriocin treatment on cell cycle progression and metabolites related to sphingolipids.
WT and swi4Δ cells were pre-grown in YPD to stationary phase (24 hours) and seeded in SD medium in the absence or presence of 1 μg. mL−1 of myriocin for five hours at 30 °C. (A) G2/M arrest caused by myriocin treatment in both WT and swi4Δ. Cells were then analyzed by FACS * p<0. 05. N=3. (B-I) Changes in the sphingolipid levels after myriocin treatment. HPLC-MS/MS lipidomic analysis. Data were normalized by phosphate levels (pi). Gray bars indicate WT cells and red bars swi4Δ strain. * p<0. 05. N=2.
To identify which class of sphingolipids are modified by myriocin treatment or SWI4 deletion, we performed sphingolipid analysis in these same growth conditions (Fig 7B–I). We found that either myriocin treatment or SWI4 deletion reduce DHS levels (Fig 7B) and that myriocin treatment does not further reduce DHS levels in swi4Δ strain (Fig 7B). Similar results were found for C24 species of phytoceramide and inositol phosphorylceramide (Fig 7C–E). Some MIPC levels increased after myriocin treatment and were particularly higher in swi4Δ cells (Fig 7F–I), in a similar pattern found in G1/S lipidomic analysis (Fig 6F–H). These results show that swi4Δ strain behaves similarly to myriocin treated WT cells, strongly suggest that the sphingolipid biosynthetic pathway is regulated during cell cycle progression at G1 phase in a Swi4p-dependent fashion, and that reduction of sphingolipid levels are associated to a block in G2/M stage.
4. Discussion.
Sphingolipids are important components of the plasma membrane, affecting its biophysical properties and forming enriched microdomains, which are important for vesicle transport [44]. Besides its structural role their involvement in cell proliferation has been well established in mammalian cells [9], while in yeast several studies have shown that sphingolipids play an essential role in endocytosis [45] and cell cycle arrest induced by the heat stress response [46]. However, the regulation of sphingolipid synthesis during cell cycle progression has not been well characterized in yeast. For this purpose, we have studied the involvement of the master regulator of G1/S transition, Swi4p, on the regulation of transcription of sphingolipid biosynthesis related genes and on the dynamics of sphingolipids.
We found that deletion of SBF components caused a growth defect in the presence of myriocin (Fig. 2). This effect could be directly related to inhibition of SPT or to a pleiotropic effect, but results suggest that growth defect is related to SPT inhibition as addition of PHS, an intermediate of sphingolipid metabolism, could suppress myriocin effects. Furthermore, addition of serine and palmitate, substrates of SPT, could also suppress myriocin effects. The suppressive effect of SPT substrates may be counterintuitive since myriocin should be inhibiting SPT. However, we believe that at the concentration used, myriocin is not completely inhibiting SPT activity, because it is not lethal, as deletion of SPT. So, partial inhibition of SPT would allow the suppressive effect of the substrates on myriocin effect (Fig 2D).
Cell cycle studies use different strategies for cell synchronization at G1 (e.g., alpha-factor treatment or elutriation). The first and more conventional is alpha-factor treatment using bar1Δ background strains. This strategy is advantageous because it reduces the amount of pheromone needed for synchronization and could potentially avoid the problem of changes in sphingolipid levels duo to the pheromone treatment [47]. However, the generation time of the double deleted strain bar1Δ/swi4Δ was 3-fold higher than the WT strain, which impaired the use of bar1Δ background (data not shown). It is important to mention that to attain G1 synchronization, we used 1.5-fold more pheromone in swi4Δ than in WT, probably because ceramides, which levels are lower in swi4Δcells (Fig 5 and Supplemental figure 2), are required for alpha-factor signaling [47,48]. The second strategy is to elutriate cells, sorting them according to their size, in a way that newborn cells are smaller than late G2/M cells [49]. However, since swi4Δ strain is 2-fold larger that WT (data not shown), it turns the comparison of these two cells difficult during elutriation experiments. For these reasons, we decided to use alpha-factor treatment in BY4741 background, except for experiments shown in supplementary figure 5.
After setting the cell cycle synchronization methodology, we reported that transcription of the sphingoid base related genes LCB2,TSC10 and SUR2 (related also to phytoceramide synthesis) are differentially expressed in swi4Δ cells. These results correlated to the lower levels of DHS and C24 and C26 phytoceramides (Fig. 5 and Supplemental figure 2) observed in swi4Δ strain. However, although we observed an oscillatory transcription of these genes in WT, the levels of sphingoid bases were not oscillatory. These results are in contrast to a recent report where both mRNA transcription and the metabolites 3-ketodihydrosphingosine, DHS and PHS bases peaked at S phase [50]. This discrepancy might be due to a slight difference in cell cycle synchronization as they used a bar1Δ background [50]. Although we did not observe a cyclic oscillation of LCB metabolites, we provided evidence that Swi4p transcription factor plays a key role in DHS homeostasis.
Different from DHS and PHS, we observed oscillation of very long chain phytoceramides along the cell cycle that matches the transcription pattern of LAC1 gene (Fig 5E, I and Supplemental figure 2). Our results also show that in WT cells, the C24 phytoceramide decreases after G1 (45 min), while the levels of PHS increase. This balance could act as a switch for G1/S progression of the cell cycle as it has been shown that ceramide induces G1 phase arrest of cells via a Sit4p protein phosphatase-dependent pathway [51]. Thus, lowering the ceramide levels could promote G1/S cell cycle progression. Together with these findings, we showed that swi4Δcells have a delayed transcriptional inhibition of YPC1 ceramidase and a delay in activation of LAC1 transcription, which combined may reflect in the metabolite levels of PHS as well as phytoceramides (Fig 5), which could explain, at least in part G1/S arrest in swi4Δ strain.
We also reported that Swi4p regulates the levels of IPCs, independently of a transcriptional control of AUR1 gene. The influence of Swi4p regulation was particularly noticed in IPCs derived from very-long chain phytoceramides, whose levels also oscillate following an AUR1 transcription pattern (Figure 6 A–C, Supplemental figure 3). Supporting our findings, it was already reported that IPCs are synthetized at the G1/S phase of the cell cycle, which also increases the levels of diacylglycerol, a product of their synthesis [16].
A miscorrelation between transcripts and the corresponding metabolite levels was especially found between SUR1 gene and MIPĆs levels as SUR1 transcription is activated at early G1 in WT and delayed in swi4Δ cells but the level of MIPĆs in swi4Δ cells is significantly higher than WT (Fig 6D–H). An increase in MIPĆs was also found when WT cells were treated with myriocin (Fig. 7). These results suggest that inhibition of SPT contributes to altered levels of MIPCs and could be linked to a stress response induced by the drug. Interestingly, MIPCs are involved in the maintenance of cell wall integrity pathway [52], and upon cell wall stress, the SBF complex is activated through MAP kinase signaling [53]. As swi4Δ cells have a defective cell wall, the increased levels of MIPC might partially be explained as response to this phenotype [53].
Furthermore, it was investigated whether sphingolipids influence cell cycle progression. We were unable to show that addition of PHS or phytoceramide to swi4Δ cells could revert G1/S blockage. These results might be explained by the fact that swi4Δ cells are able to synthetize PHS and also that deletion of SWI4 impacts the levels of very long chain ceramides, not C12 phytoceramide, that was used in our experiments. We also observed that myriocin treatment did not affect the first round of cell division (Supplemental figure 5), however after five hours of drug treatment, it caused a G2/M arrest in WT cells (Fig.7A). These results suggest that myriocin only affects sphingolipid levels significantly after two or more rounds of cell division. One explanation is that cells can maintain sphingolipid homeostasis through degradation of complex ceramides [54]. In fact, after 5 hours of growth several changes were observed, mainly at DHS, C24 phytoceramide, C24 OH-phytoceramide and C24 inositol phosphorylceramide levels, that were reduced after myriocin treatment, while different classes of MIPC were accumulated (Fig.7 B–I). These results indicate that cell cycle progression requires more than just one sphingolipid species and therefore, a single mutation is not sufficient to disrupt cell division (Fig. 7 and Supplemental figure 5C). The fact that DHS, C24 phytoceramide, C24 OH-phytoceramide and C24 inositol phosphorylceramide are both regulated by Swi4p during cell cycle and myriocin treatment, sheds light upon their specific role in cell division. Phytoceramides are important for proper formation of nuclear envelope diffusion barrier, which controls daughter cell heritage and allows to receive only healthy or new synthesized components, with direct implications on cell longevity [55,56]. C24 and C26 ceramides also constitute membrane microdomains that are important for protein sorting and improves membrane bending by interdigitation [57–59]. Lysosphingolipids as well as diacylglycerol, product of IPC synthesis, modulate the activity of Protein Kinase C (Pkc1) in mammals and some fungi species, such as C. neoformans [60,61]. Pkc1 targets are involved in process such as cell division and response to stress [62], giving a perspective of future research to establish the targets of sphingolipid signaling during cell cycle.
Overall, Swi4p transcription factor is an important regulator of sphingolipid homeostasis and future research will help to discern whether changes in sphingolipid levels during G1/S stage act as signaling events and/or structural components of the plasma membrane.
Supplementary Material
Highlights.
G1/S cell cycle checkpoint mutants are sensitive to myriocin
Swi4p transcription factor regulates synthesis of sphingoid bases and ceramides
Reduction of sphingolipid levels are associated to a block in G2/M stage
Acknowledgements.
This work was supported by grants from Conselho Nacional de Ciência e Tecnologia (CNPq – Universal to CAM), Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES) and Fundação de Amparo à Pesquisa do Rio de Janeiro (FAPERJ-Cientistas do Nosso Estado to MML, APQ1 to CAM and Auxílio Instalação to BBM). This work was also supported by NIH grants AI136934, AI116420, and AI125770, and by the Merit Review Grant I01BX002924 from the Veterans Affairs Program to MDP. BBM was recipient of a PNPD-CAPES- fellowship (Programa Nacional de Pós Doutorado da CAPES). GSM, was recipient of a CNPq fellowship. We gratefully acknowledge Bruno Bozaquel-Morais, Analice M Teixeira da Silva, Suyane Marculino de Souza and Barbara Teixeira for their assistance in drug sensibility tests and cell cycle experiments and Platoforma de Imunoanálise (PIA) from Instituto de Biofísica Carlos Chagas Filho for the training in flow cytometry analysis.
Footnotes
Declaration of competing interest
Maurizio Del Poeta is a Co-Founder and Chief Scientific Officer (CSO) of MicroRid Technologies Inc. All other authors declare no conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Bibliography
- [1].Ogretmen B, Hannun YA, Biologically active sphingolipids in cancer pathogenesis and treatment., Nat. Rev. Cancer. 4 (2004) 604–616. 10.1038/nrc1411. [DOI] [PubMed] [Google Scholar]
- [2].Pyne NJ, Pyne S, Sphingosine 1-phosphate and cancer, Nat. Rev. Cancer. 10 (2010) 489–503. 10.1038/nrc2875. [DOI] [PubMed] [Google Scholar]
- [3].Kraveka JM, Li L, Szulc ZM, Bielawski J, Ogretmen B, Hannun YA, Obeid LM, Bielawska A, Involvement of dihydroceramide desaturase in cell cycle progression in human neuroblastoma cells, J. Biol. Chem. 282 (2007) 16718–16728. 10.1074/jbc.M700647200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Lin CL, Mao C, Role of sphingolipids in non-melanoma skin cancer, Bioactive Sphingolipids in Cancer Biology and Therapy 2015, 107–122. 10.1007/978-3-319-20750-6_5. [DOI] [Google Scholar]
- [5].Molino S, Tate E, McKillop W, Medin JA, Sphingolipid pathway enzymes modulate cell fate and immune responses, Immunotherapy. 9 (2017) 1185–1198. 10.2217/imt-2017-0089. [DOI] [PubMed] [Google Scholar]
- [6].Gagliostro V, Casas J, Caretti A, Abad JL, Tagliavacca L, Ghidoni R, Fabrias G, Signorelli P, Dihydroceramide delays cell cycle G1/S transition via activation of ER stress and induction of autophagy, Int. J. Biochem. Cell Biol. 44 (2012) 2135–2143. 10.1016/j.biocel.2012.08.025. [DOI] [PubMed] [Google Scholar]
- [7].Olivera A, Spiegel S, Sphingosine-1-phosphate as second messenger in cell proliferation induced by PDGF and FCS mitogens, Nature. 365 (1993) 557–560. 10.1038/365557a0. [DOI] [PubMed] [Google Scholar]
- [8].Van Brocklyn JR, Lee MJ, Menzeleev R, Olivera A, Edsall L, Cuvillier O, Thomas DM, Coopman PJP, Thangada S, Liu CH, Hla T, Spiegel S, Dual actions of sphingosine-1-phosphate: Extracellular through the G(i)- coupled receptor Edg-1 and intracellular to regulate proliferation and survival, J. Cell Biol. 142 (1998) 229–240. 10.1083/jcb.142.1.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Andrieu G, Ledoux A, Branka S, Bocquet M, Gilhodes J, Walzer T, Kasahara K, Inagaki M, Sabbadini RA, Cuvillier O, Hatzoglou A, Sphingosine 1-phosphate signaling through its receptor S1P 5 promotes chromosome segregation and mitotic progression, Science (80-. ). 1 (2017) 1–12. 10.1126/scisignal.aah4007. [DOI] [PubMed] [Google Scholar]
- [10].Cuvillier O, Pirianov G, Kleuser B, Vanek PG, Coso OA, Gutkind JS, Spiegel S, Suppression of ceramide-mediated programmed cell death by sphingosine1-phosphate, Nature. 381 (1996) 800–803. 10.1038/381800a0. [DOI] [PubMed] [Google Scholar]
- [11].Lee JY, Bielawska AE, Obeid LM, Regulation of cyclin-dependent kinase 2 activity by ceramide, Exp. Cell Res. 261 (2000) 303–311. 10.1006/excr.2000.5028. [DOI] [PubMed] [Google Scholar]
- [12].Lee JY, Leonhardt LG, Obeid LM, Cell-cycle-dependent changes in ceramide levels preceding retinoblastoma protein dephosphorylation in G2/M, Biochem. J, (334) 1998, 457–461. 10.1042/bj3340457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Rani CS, Abe A, Chang Y, et al. Cell cycle arrest induced by an inhibitor of glucosylceramide synthase. Correlation with cyclin-dependent kinases. J Biol Chem. 1995;270(6):2859–2867. doi: 10.1074/jbc.270.6.2859. [DOI] [PubMed] [Google Scholar]
- [14].Jenkins GM, Hannun YA, Role for de Novo Sphingoid Base Biosynthesis in the Heat-induced Transient Cell Cycle Arrest of Saccharomyces cerevisiae, J. Biol. Chem. 276 (2001) 8574–8581. 10.1074/jbc.M007425200. [DOI] [PubMed] [Google Scholar]
- [15].Epstein S, Castillon GA, Qin Y, Riezman H, An essential function of sphingolipids in yeast cell division, 84 (2012) 1018–1032. 10.1111/j.1365-2958.2012.08087.x. [DOI] [PubMed] [Google Scholar]
- [16].Cerbón J, Falcon A, Hernández-Luna C, Segura-Cobos D, Inositol phosphoceramide synthase is a regulator of intracellular levels of diacylglycerol and ceramide during the G1 to S transition in Saccharomyces cerevisiae, Biochem. J. 388 (2005) 169–176. 10.1042/BJ20040475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Chauhan N, Han G, Somashekarappa N, Gable K, Dunn T, Kohlwein SD, Regulation of sphingolipid biosynthesis by the morphogenesis checkpoint kinase Swe1, J. Biol. Chem. 291 (2016) 2524–2534. 10.1074/jbc.M115.693200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Nair M, McIntosh PB, Frenkiel T. a., Kelly G, Taylor I. a., Smerdon SJ, Lane AN, NMR structure of the DNA-binding domain of the cell cycle protein Mbp1 from Saccharomyces cerevisiae, Biochemistry. 42 (2003) 1266–1273. 10.1021/bi0205247. [DOI] [PubMed] [Google Scholar]
- [19].Breeden LL, Periodic transcription: A cycle within a cycle, Curr. Biol. 13 (2003) 31–38. 10.1016/S0960-9822(02)01386-6. [DOI] [PubMed] [Google Scholar]
- [20].Hendler A, Medina EM, Buchler NE, de Bruin RAM, Aharoni A, The evolution of a G1/S transcriptional network in yeasts, Curr. Genet. (2017) 1–6. 10.1007/s00294-017-0726-3. [DOI] [PubMed] [Google Scholar]
- [21].Iyer VR, Horak CE, Scafe CS, Botstein D, Snyder M, Brown PO, Genomic binding sites of the yeast cell-cycle transcription factors SBF and MBF., Nature. 409 (2001) 533–538. 10.1038/35054095. [DOI] [PubMed] [Google Scholar]
- [22].Doncic A, Falleur-Fettig M, Skotheim JM, Distinct Interactions Select and Maintain a Specific Cell Fate, Mol. Cell. 43 (2011) 528–539. 10.1016/j.molcel.2011.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Bertoli C, Skotheim JM, de Bruin RAM, Control of cell cycle transcription during G1 and S phases, Nat. Rev. Mol. Cell Biol. 14 (2013) 518–528. 10.1038/nrm3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Wittenberg C, Reed SI, Cell cycle-dependent transcription in yeast: promoters, transcription factors, and transcriptomes., Oncogene. 24 (2005) 2746–2755. 10.1038/sj.onc.1208606. [DOI] [PubMed] [Google Scholar]
- [25].Longtine MS, McKenzie A, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR, Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae, Yeast. 14 (1998) 953–961. . [DOI] [PubMed] [Google Scholar]
- [26].Gietz D, St Jean A, Woods RA, Schiestl RH, Improved method for high efficiency transformation of intact yeast cells., Nucleic Acids Res. 20 (1992) 1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Teixeira MC, Monteiro PT, Guerreiro JF, Gonçalves JP, Mira NP, Dos Santos SC, Cabrito TR, Palma M, Costa C, Francisco AP, Madeira SC, Oliveira AL, Freitas AT, Sá-Correia I, The YEASTRACT database: An upgraded information system for the analysis of gene and genomic transcription regulation in Saccharomyces cerevisiae, Nucleic Acids Res. 42 (2014) 161–166. 10.1093/nar/gkt1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Whitby M, Osman F, Monitoring HR following rep. fork pertubation in S.pombe (MCW methods), Methods. 521 (2009) 437–448. 10.1007/978-1-60327-817-5. [DOI] [PubMed] [Google Scholar]
- [29].Schmitt ME, Brown TA, Trumpower BL, A rapid and simple method for preparation of RNA from Saccharomyces cerevisiae, Nucleic Acids Res 18 (1990) 3091–3092. 10.1093/nar/18.10.3091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Livak KJ, Schmittgen TD, Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method, Methods. 25 (2001) 402–408. 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- [31].Singh A, Del Poeta M, Sphingolipidomics: An important mechanistic tool for studying fungal pathogens, Front. Microbiol. 7 (2016) 1–14. 10.3389/fmicb.2016.00501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Raj S, Nazemidashtarjandi S, Kim J, Joffe L, Zhang X, Singh A, Mor V, Desmarini D, Djordjevic J, Raleigh DP, Rodrigues ML, London E, Del Poeta M, Farnoud AM, Changes in glucosylceramide structure affect virulence and membrane biophysical properties of Cryptococcus neoformans, Biochim. Biophys. Acta - Biomembr. 1859 (2017) 2224–2233. 10.1016/j.bbamem.2017.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Martín I, Peláez F, Harris GH, Curotto JE, Rozdilsky W, Kurtz MB, Giacobbe RA, Bills GF, Cabello MA, The Discovery of Australifungin, a Novel Inhibitor of Sphinganine N-Acyltransferase from Sporormiella australis Producing Organism, Fermentation, Isolation, and Biological Activity, J. Antibiot. (Tokyo). 48 (1995) 349–356. 10.7164/antibiotics.48.349. [DOI] [PubMed] [Google Scholar]
- [34].Bligh WJ, and Dyer EG, A rapid method of total lipid extraction and purification, Canadian Journal of Biochemistry and Physiology, Can. J. Biochem. Physiol. 37 (1959). 10.1139/o59-099 [DOI] [PubMed] [Google Scholar]
- [35].Clarke NG, Dawson RMC, Alkaline O→N-transacylation. A new method for the quantitative deacylation of phospholipids, Biochem. J. 195 (1981) 301–306. 10.1042/bj1950301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Singh A, MacKenzie A, Girnun G, Del Poeta M, Analysis of sphingolipids, sterols, and phospholipids in human pathogenic Cryptococcus strains, J. Lipid Res. 58 (2017) 2017–2036. 10.1194/jlr.M078600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Daquinag A, Fadri M, Jung SY, Qin J, Kunz J, The Yeast PH Domain Proteins Slm1 and Slm2 Are Targets of Sphingolipid Signaling during the Response to Heat Stress, Mol. Cell. Biol. 27 (2007) 633–650. 10.1128/mcb.00461-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Esch BM, Limar S, Bogdanowski A, Gournas C, More T, Sundag C, Walter S, Heinisch JJ, Ejsing CS, André B, Fröhlich F, Uptake of exogenous serine is important to maintain sphingolipid homeostasis in Saccharomyces cerevisiae, PLoS Genet. 16 (2020) 1–29. 10.1371/JOURNAL.PGEN.1008745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Monteiro PT, Oliveira J, Pais P, Antunes M, Palma M, Cavalheiro M, Galocha M, Godinho CP, Martins LC, Bourbon N, Mota MN, Ribeiro RA, Viana R, Sá-Correia I, Teixeira MC, YEASTRACT+: A portal for cross-species comparative genomics of transcription regulation in yeasts, Nucleic Acids Res. 48 (2020) D642–D649. 10.1093/nar/gkz859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Chua G, Morris QD, Sopko R, Robinson MD, Ryan O, Chan ET, Frey BJ, Andrews BJ, Boone C, Hughes TR, Identifying transcription factor functions and targets by phenotypic activation., Proc. Natl. Acad. Sci. U. S. A. 103 (2006) 12045–50. 10.1073/pnas.0605140103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Hendler A, Medina EM, Kishkevich A, Abu-Qarn M, Klier S, Buchler NE, de Bruin RAM, Aharoni A, Gene duplication and co-evolution of G1/S transcription factor specificity in fungi are essential for optimizing cell fitness, PLoS Genet. 13 (2017) 1–24. 10.1371/journal.pgen.1006778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].White MA, Riles L, Cohen BA, A systematic screen for transcriptional regulators of the yeast cell cycle, Genetics. 181 (2009) 435–446. 10.1534/genetics.108.098145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Blank HM, Callahan M, Pistikopoulos IPE, Polymenis AO, Polymenis M, Scaling of G1 duration with population doubling time by a cyclin in Saccharomyces cerevisiae, BioRxiv. 210 (2017) 895–906. 10.1101/240713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Grassmé H, Riethmüller J, Gulbins E, Biological aspects of ceramide-enriched membrane domains, Prog. Lipid Res. 46 (2007) 161–170. 10.1016/j.plipres.2007.03.002. [DOI] [PubMed] [Google Scholar]
- [45].Friant S, Lombardi R, Schmelzle T, Hall MN, Riezman H, Sphingoid base signaling via Pkh kinases is required for endocytosis in yeast, EMBO J. 20 (2001) 6783–6792. 10.1093/emboj/20.23.6783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Jenkins GM, Richards A, Wahl T, Mao C, Obeid L, Hannun Y, Involvement of yeast sphingolipids in the heat stress response of Saccharomyces cerevisiae, J. Biol. Chem. 272 (1997) 32566–32572. 10.1074/jbc.272.51.32566. [DOI] [PubMed] [Google Scholar]
- [47].Villasmil ML, Francisco J, Gallo-Ebert C, Donigan M, Liu HY, Brower M, Nickels JT, Ceramide signals for initiation of yeast mating-specific cell cycle arrest, Cell Cycle. 15 (2016) 441–454. 10.1080/15384101.2015.1127475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Villasmil ML, Gallo-Ebert C, Liu HY, Francisco J, Nickels JT, A link between very long chain fatty acid elongation and mating-specific yeast cell cycle arrest, Cell Cycle. 16 (2017) 2192–2203. 10.1080/15384101.2017.1329065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Spellman PT, Sherlock G, Zhang MQ, Iyer VR, Anders K, Eisen MB, Brown PO, Botstein D, Futcher B, Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization, Mol. Biol. Cell. 9 (1998) 3273–3297. 10.1091/mbc.9.12.3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Campbell Kate, Westholm Jakub, Kasvandik Sergo, Francesca Di Bartolomeo Maurizio Mormino, Nielsen Jens. Proceedings of the National Academy of Sciences Apr 2020, 117 (14) 7575–7583; DOI: 10.1073/pnas.1919535117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Nickels JT, Broach JR, A ceramide-activated protein phosphatase mediates ceramide-induced G1 arrest of Saccharomyces cerevisiae, Genes Dev. 10 (1996) 382–394. 10.1101/gad.10.4.382. [DOI] [PubMed] [Google Scholar]
- [52].Tanaka S, Tani M, Mannosylinositol phosphorylceramides and ergosterol coodinately maintain cell wall integrity in the yeast Saccharomyces cerevisiae, FEBS J. 285 (2018) 2405–2427. 10.1111/febs.14509. [DOI] [PubMed] [Google Scholar]
- [53].Levin DE, Cell Wall Integrity Signaling in Saccharomyces cerevisiae, Microbiol. Mol. Biol. Rev. 69 (2005) 262–291. 10.1128/mmbr.69.2.262-291.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Hannun YA, Obeid LM, Principles of bioactive lipid signalling : lessons from sphingolipids, Nat Rev Mol Cell Biol (9) 2008, 139–150. 10.1038/nrm2329. [DOI] [PubMed] [Google Scholar]
- [55].Clay L, Caudron F, Denoth-Lippuner A, Boettcher B, Frei SB, Snapp EL, Barral Y, A sphingolipid-dependent diffusion barrier confines ER stress to the yeast mother cell, Elife. 2014, 1–23. 10.7554/eLife.01883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Higuchi-Sanabria R, Pernice WMA, Vevea JD, Alessi Wolken DM, Boldogh IR, Pon LA, Role of asymmetric cell division in lifespan control in Saccharomyces cerevisiae, FEMS Yeast Res. 14 (2014) 1133–1146. 10.1111/1567-1364.12216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Rodriguez-Gallardo S, Kurokawa K, Sabido-Bozo S, Cortes-Gomez A, Ikeda A, Zoni V, Aguilera-Romero A, Perez-Linero AM, Lopez S, Waga M, Araki M, Nakano M, Riezman H, Funato K, Vanni S, Nakano A, Muñiz M, Ceramide chain length-dependent protein sorting into selective endoplasmic reticulum exit sites, Sci. Adv. 6 (2020) 1–12. 10.1126/sciadv.aba8237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Pinto SN, Silva LC, Futerman AH, Prieto M, Effect of ceramide structure on membrane biophysical properties: The role of acyl chain length and unsaturation, Biochim. Biophys. Acta - Biomembr. 1808 (2011) 2753–2760. 10.1016/j.bbamem.2011.07.023. [DOI] [PubMed] [Google Scholar]
- [59].Ventura AE, Varela ARP, Dingjan T, Santos TCB, Fedorov A, Futerman AH, Prieto M, Silva LC, Lipid domain formation and membrane shaping by C24-ceramide, Biochim. Biophys. Acta - Biomembr. 1862 (2020) 183400. 10.1016/j.bbamem.2020.183400. [DOI] [PubMed] [Google Scholar]
- [60].Hannun YA, Bell RM, Regulation of protein kinase C by sphingosine and lysosphingolipids, Clin. Chim. Acta. 185 (1989) 333–345. 10.1016/0009-8981(89)90224-6. [DOI] [PubMed] [Google Scholar]
- [61].Heung LJ, Luberto C, Plowden A, Hannun YA, Del Poeta M, The sphingolipid pathway regulates Pkc1 through the formation of diacylglycerol in cryptococcus neoformans, J. Biol. Chem. 279 (2004) 21144–21153. 10.1074/jbc.M312995200. [DOI] [PubMed] [Google Scholar]
- [62].Heinisch JJ, Rodicio R, Protein kinase C in fungi-more than just cell wall integrity, FEMS Microbiol. Rev. 42 (2018) 22–39. 10.1093/femsre/fux051. [DOI] [PubMed] [Google Scholar]
- [63].Dickson RC, Sphingolipid functions in Saccharomyces cerevisiae : Comparison to Mammals, Annu. Rev. Biochem. 67 (1998) 27–48. 10.1146/annurev.biochem.67.1.27. [DOI] [PubMed] [Google Scholar]
- [64].Pinto WJ, Wells GW, Lester RL, Characterization of enzymatic synthesis of sphingolipid long-chain bases in Saccharomyces cerevisiae: mutant strains exhibiting long-chain-base auxotrophy are deficient in serine palmitoyltransferase activity., J. Bacteriol. 174 (1992) 2575 LP – 2581. 10.1128/jb.174.8.25752581.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Fujita T, Inoue K, Yamamoto S, Ikumoto T, Sasaki S, Toyama R, Chiba K, Hoshino Y, Okumoto T, Fungal metabolites. Part 12. Potent immunosuppressant, 14-deoxomyriocin, (2S, 3R, 4R)-(e)-2-amino-3,4-dihydroxy-2-hydroxymethyleicos-6-enoic acid and structure-activity relationships of myriocin derivatives, J. Antibiot. (Tokyo). 47 (1994) 216–224. 10.7164/antibiotics.47.216. [DOI] [PubMed] [Google Scholar]
- [66].Vallée B, Riezman H, Lip1p: A novel subunit of acyl-CoA ceramide synthase, EMBO J. 24 (2005) 730–741. 10.1038/sj.emboj.7600562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Marquês JT, Marinho HS, de Almeida RFM, Sphingolipid hydroxylation in mammals, yeast and plants – An integrated view, Prog. Lipid Res. 71 (2018) 18–42. 10.1016/j.plipres.2018.05.001. [DOI] [PubMed] [Google Scholar]
- [68].Rego A, Trindade D, Chaves SR, Manon S, Costa V, Sousa MJ, Côrte-Real M, The yeast model system as a tool towards the understanding of apoptosis regulation by sphingolipids, FEMS Yeast Res. 14 (2014) 160–178. 10.1111/1567-1364.12096. [DOI] [PubMed] [Google Scholar]
- [69].Grilley MM, Stock SD, Dickson RC, Lester RL, Takemoto JY, Syringomycin action gene SYR2 is essential for sphingolipid 4-hydroxylation in Saccharomyces cerevisiae., J. Biol. Chem. 273 (1998) 11062–11068. 10.1074/jbc.273.18.11062. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.