Highlights
-
•
The expression of PLA1A, DSC3, and DMKN genes function as oncogenes that trigger. melanomagenesis on interaction with miR-563, miR-363, miR-770–5p, and miR-370.
-
•
PLA1A and DMKN genes can be used as a promising diagnosis biomarkers to distinguish melanoma patients.
-
•
The expression of PLA1A and DMNK are regulated by EMT and formation of VE.
-
•
PLA1A/miR-563 and DMNK/miR-770–5p/miR-370 may contribute to melanomagenesis by triggering the EMT signaling pathway and VM formation.
Keywords: mRNA/micro-RNA co-expression, Melanomagenesis, Epithelial–mesenchymal transition, Vasculogenic mimicry, Biomarker
Abbreviations: MM, metastatic melanoma; EMT, epithelial-to-mesenchymal transition; VM, vasculogenic mimicry; WGCNA, weighted gene co-expression network analysis; miRNAs, microRNAs; PLA1A, phosphatidylserine-specific phospholipase A1-alpha; DMKN, dermokine; NCBI, National Center of Biotechnology Information; GEO, gene expression omnibus; PICO, population, intervention, control, and outcomes; TOP, topological overlap matrix; ROC, receiver operating characteristic; GO, gene ontology; FC, fold change; ECM, extracellular matrix; KEGG, Kyoto Encyclopedia of Genes and Genomes; PPI, protein–protein interactive; WHO, world health organization; UICC, union for international cancer control; qRT-PCR, quantitative real-time PCR; ZEB1, zinc-finger E-box-binding-1; SLUG, zinc finger protein SNAI2; NOTCH1, notch homolog 1 translocation-associated; EMT-TFs, epithelial-to-mesenchymal transition-inducing transcription factors; PAS, periodic acid solution; MVD, microvessel density; VMD, vasculogenic mimicry density, DSC3, desmocollin 3; DEMs, differentially expressed mRNAs
Abstract
This study aimed to identify a novel disease-associated differentially co-expressed mRNA-microRNA (miRNA) that is associated with vasculogenic mimicry (VM) and epithelial-to-mesenchymal transition (EMT) network at different stages of melanoma. By applying weighted gene co-expression network analysis, we constructed a VM+EMT biological network with the available microarray dataset downloaded from a public database. Quantitative real-time PCR, immunohistochemical staining, and CD31-periodic acid solution dual staining were performed to confirm the expression of genes associated with EMT and VM formation in subjects with malignant melanoma (n = 18) and primary melanoma (n = 13) and in healthy subjects (n = 10). Our findings suggested that phosphatidylserine-specific phospholipase A1-alpha (PLA1A) and dermokine (DMKN) genes function as oncogenes that trigger VM and EMT processes during melanomagenesis on interaction with miR-370, miR-563, and miR-770–5p. PLA1A and DMKN genes can be considered potential VM+EMT network-based diagnostic biomarkers for distinguishing between melanoma patients. We postulate that a network with altered PLA1A/miR-563 and DMNK/miR-770–5p/miR-370 may contribute to melanomagenesis by triggering the EMT signaling pathway and VM formation. This study provides a potentially valuable approach for the early diagnosis and prognosis of melanoma progression.
Introduction
Metastatic melanoma (MM) is the most aggressive type of skin cancer, with an estimated global incidence of approximately 200,000 new cases per year and 50,000 cancer-related deaths reported in 2018 [1,2]; there is no curative therapy for advanced stages of melanoma. Thus, new prognostic and therapeutic biomarkers are urgently needed for MM. More than 50% of MM patients have been estimated to display cytogenetic abnormalities, including chromosomal translocation, inversion, and deletion, and have significantly heterogeneous outcomes. Some chromosomal abnormalities have a better prognosis than others in MM.
Thus, various research groups have investigated the genetic risk factors of MM with the aim to identify genes associated with the epithelial-to-mesenchymal transition (EMT) network and, tumor cell vasculogenic mimicry (VM) [3]. EMT is defined as the process wherein epithelial cells lose the apical-basal polarity and cell-cell adhesion and transition to invasive mesenchymal cells, which can promote melanoma cell progression and metastasis [4]. EMT and its reverse mesenchymal‐to‐epithelial transition (MET) play critical roles during number of pathological disorders, including fibrosis, inflammation, wound repair, and metastatic dissemination of carcinomas. In carcinoma cells, the EMT/MET plasticity triggers the metastasis and consequently proposes alternative mechanisms for successful dissemination and metastases [5,6]. During various stages of the metastatic cascade, the EMT/MET hybrid and its molecular underpinning phenotype in each tumor are slightly different and unique [5,7,8]. Further, VM is characterized by vascular‐like structures that can mimic the embryonic vascular network pattern to nourish the MM tissue [8]. VM is associated with tumor perfusion and formation of de novo vascular networks in aggressive cancer cells [9]. VM-associated tumor cell plasticity have been introduced as a novel paradigm and potential therapeutic targets in antitumor therapy in association with EMT/MET vascular and hypoxia-related signaling pathways [10,11]. Many studies show that high expression of VM and underlying molecular pathways supporting VM is associated with a high tumor grade, short survival duration, invasion, and MM [8,10,12]. Melanoma cells can form networks of vessel-like fluid conducting channels through EMT-accelerated VM and subsequently remodel the extracellular matrix (ECM) and EMT/MET plasticity, connecting VM channels with host blood vessels and triggering the melanoma tumor perfusion [3,8,13]. These pathways are the novel target approaches for the new treatment of melanoma, and play a crucial role in melanomagenesis by increasing the functional metastatic burden [3,8,11,[13], [14], [15]].
Although previously there was little agreement between transcriptomic studies on melanoma, many currently available EMT and VM biomarkers depend on the differential expression of these biomarkers in different melanoma states [16]. The most common analytical tools used for biomarker identification are clustering methods, artificial neural networks, and differential expression-based analysis. These methods are typically data-driven and do not consider any biological information of the component genes as input; nonetheless, they have the advantage of identifying the distinguishing features without having any biological information about those features. In this regard, weighted gene co-expression network analysis (WGCNA) is a new biological construction method offering an alternative approach to discover new biomarkers for melanoma based on the known functions and interactions of individual molecules [17,18]. Furthermore, WGCNA has the added advantage of combining condition-specific transcriptome data and allowing the understanding of the functional role of individual genes and other biological molecules such as microRNAs (miRNAs), which have the capability of discriminating between diseased and healthy cells or between different melanoma stages [18]. Bioinformatics methods such as differentiation and enrichment analysis, clustering class discovery, mechanistic analysis, and class prediction of targeted mRNAs-miRNAs in VM+EMT network can help us in the discovery of the complexity of the melanomagenesis.
Here, we aimed to re-analyze the available transcriptomic microarray dataset of patients with melanoma to identify comparable biomarkers based on disrupted mRNA-miRNA regulation with the help of VM and EMT network. Using the cooperative mRNA-miRNA network, we aimed to determine whether the network-based exploration of melanoma intervention opportunities can be a rich source for target discovery given the sufficient resolution of EMT and VM correlates, while conducting a systematic search and network meta-analysis. Subsequently, the diagnostic value of the targeted mRNAs-miRNAs was confirmed at different stages in subjects with melanoma. After multiple integrative and large-scale WGCNA, we identified that phosphatidylserine-specific phospholipase A1-alpha (PLA1A) and dermokine (DMKN) oncogenes trigger VM and EMT processes on interaction with miR-370, miR-563, and miR-770–5p during melanomagenesis. We then confirmed the biological function of PLA1A and DMNK in VM+EMT network in subjects with MM and primary melanoma and healthy subjects. This may provide a potentially valuable approach for the genetic characterization of patients with melanoma.
Materials and methods
Ethics statement
This study was granted approval by the “Ethics Review Board” at the Affiliated Hospital of Southwest Medical University (No. KY2019041). Written informed consent that conformed to the tenets of the Declaration of Helsinki (1983 Revision) for use of their clinical and pathology information, as well as for mutation analysis, was obtained from all participants or their guardians before study initiation. Prospective volunteers were informed about study goals and protocols. Additionally, all clinical assessments were performed according to the local Ethics Committee guidelines of Pathology Department and Oncology Department at the Affiliated Hospital of Southwest Medical University in Luzhou, Sichuan, China.
Search strategy and dataset selection
Transcriptomic datasets were retrieved from the National Center of Biotechnology Information (NCBI) Gene Expression Omnibus (GEO) database (NCBI; http://www.ncbi.nlm.nih.gov/geo/), an international public functional data repository including data from two platforms, Affymetrix and Agilent Whole Human Genome microarray platforms, following Preferred Reporting Items for Systematic Reviews and Meta-analysis (PRISMA) statement guideline recommendations. All articles published until May 15, 2020 were included in the analysis [19,20]. Data collection was reviewed according to the population, intervention, control, and outcomes (PICO) framework [21]. The search was performed using a combination of the following keywords: melanoma, basal cell carcinoma, squamous cell carcinoma, cancer, neoplasms, MM, neoplasm, basal-cell skin cancer, squamous-cell skin cancer, skin neoplasms, and skin cancer. Different spellings and synonyms were combined using the Boolean “OR” and main terms were linked using the Boolean “AND” to identify all relevant studies. The search strategies were separately retrieved and screened by two investigators (SI and MM).
Pre-processing data
Raw data were pre-processed using the Limma package in R/Bioconductor (version R 2.14.0). The expression data were normalized using the quantile normalization method, as implemented in the Limma R package. We used the microarray probe sets to match with the corresponding genes using an annotation file released by Affymetrix and Agilent for each platform. Probes mapped to more than one gene were removed, and the average expression level of multiple probes mapped to the same gene was considered. We subsequently performed hierarchical clustering to remove the noisy data.
Gene co-expression network reconstruction
The co-expressed gene modules were identified using unsupervised hierarchical clustering by dynamic branch cut methods (WGCNA: an R package for weighted correlation network analysis) [22,23]. We reconstructed three biological co-expression networks of EMT and VM pathways at the system level based on the expression levels of DEGs among all three group matrices (matrix NP: between histologically non-neoplastic nevus (Nevi) patients and primary melanoma patients, matrix NM: between Nevi patients and MM patients, and matrix PM: between primary melanoma and MM patients). We used the web-based EMTome portal (EMTome: www.emtome.org) for identifying metastasis-related features, exploring EMT-related markers, and analyzing the relevance of EMT and VM signatures in the diagnosis or prevention of cancer metastasis [24,25]. Furthermore, the soft-thresholding power beta parameter was used to adjust the scale-free property of the networks to remove the weakly correlated genes and retain the strongly correlated ones. Following this, the biweight midcorrelation (bicor) measure was calculated for all VM+ and EMT+ genes in the dataset, and the correlation matrices for all pairs of genes were evaluated across all samples. We also considered network type as a parameter. The process produced a weighted network called “adjacent matrix” that was transformed into a topological overlap matrix (TOM) network, a biologically meaningful measure that could evaluate the network connectivity of a gene [23,26]. Other parameters in WGCNA were considered based on their default values. The VM+ and EMT+ module was defined as a cluster of closely interconnected mRNAs based on the cluster dendrogram of expression data. Therefore, in this study, each co-expression network was clustered via average linkage hierarchical clustering in WGCNA to classify genes with similar expression profiles into different modules. In addition, the cutreeDynamic (minModuleSize = 30) function was used to identify the functional modules in each group. Each module was summarized using the module eigengenes after the detection of modules, and similar modules were combined using the eigengene. Subsequently, the module–trait association in each group was analyzed based on the disease state and modules with a correlation of ≥|0.6| were selected. Furthermore, we used the DisGeNET (version 7.0) online database to determine the number of EMT+ and VM+ genes in each group within the modules. Finally, Chi-square test was performed to assess the significance of EMT+ and VM+ module genes in each group (P < 0.05).
Enrichment analysis
The Gene Ontology (GO) function and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses were performed to determine the cellular components, molecular functions, and biological processes among others using ToppGene online database and package clusterProfiler (version 3.2.14) of R (version 3.3.3), as described previously [27,28].
Protein–protein interactive (PPI) network construction
The Search Tool for the Retrieval of Interacting Genes (STRING; https://string-db.org/) was used to construct a PPI network from genes in the enriched modules [29,30]. PPI networks were visualized using Cytoscape (version 3.8.2), as described previously. In the PPI network, a gene with a node connectivity of ≥ 5 was defined as a hub gene.
mRNA–miRNA bipartite network reconstruction
The miRWalk v3.0 tool (http://mirwalk.umm.uni-heidelberg.de/) was used to determine the mRNA-miRNA interaction. We used this tool to obtain the miRNAs of target genes. During this step, the miRNAs, the targets of which were genes of candidate modules, were identified using target prediction tools TargetScan (http://www.targetscan.org; release 5.1), PicTar (http://pictar.mdc-berlin.de), and miRanda (http://www.microrna.org). Only experimentally validated mRNA–miRNA interactions were considered. Consequently, three mRNA–miRNA bipartite networks were reconstructed from the miRNAs and target genes in each group. In the miRNA–miRNA synergistic network, two miRNAs were connected if they significantly coregulated common mRNAs, which were significantly enriched in at least one biological process GO term. We used Cytoscape (version 3.8.2) for visualizing the bipartite network of candidate modules in each group.
Melanoma patient selection
This analytical study was conducted during an 6-month period from April 2020 to September 2020. The participants were adult patients who were referred to the Dermatology and Oncology department of Affiliated Hospital of Southwest Medical University, Luzhou, Sichuan, China. Patients with melanoma who were preliminarily selected for this prospective study were confirmed as having the disease by two expert oncologists (SI and QW), as well as a pathologist (CZ). The diagnosis was made according to the world health organization (WHO) guidelines and the Tumor-Node-Metastasis (TNM)-based staging criteria of the Union for International Cancer Control (UICC) pathological staging criteria in the Department of Dermatology at the Affiliated Hospital of Southwest Medical University, Luzhou, China. Disease stages were classified according to the AJCC 8th edition [31]. The participants were excluded if clinically diagnosed with any significant medical condition, preexisting cardiac conditions, and/or acute gastrointestinal bleeding. Furthermore, we excluded any patient who had undergone a major surgery within 14 days of study enrollment and/or was diagnosed with or treated for another malignancy within 3 years of enrollment. In addition, patients with any evidence of severe or uncontrolled systemic diseases, such as peripheral neuropathy ≥grade 2 or an active infection, requiring chronic maintenance of red blood cell, white blood cell, or granulocyte counts and those with a previous history of seizures were excluded. In our study, all patients were required to have tumors accessible for core biopsy with 5–6 passes of a 16 or 18 gauge needle (defined as at least 1 cm3 tumor/50 mg accessible for biopsy. Furthermore, we considered melanoma, metastatic or locally advanced and unresectable BRAF, and NRAS wild-type melanoma as a minimum inclusion criteria. All included patients were able to comply with the protocol for the duration of the study, including attending scheduled visits, undergoing examinations, undergoing the biopsy procedure, and having their tumor and blood molecularly characterized. A histological section was made for each sample, and diagnosis was confirmed via a different pathologist than the one who made the initial diagnosis. They were suspected to have a primary tumor and the potential local, regional, and distant extension diagnosis of MM was assessed by Fluorine-18-fluorodeoxyglucose positron emission tomography/computed tomography (18F-FDG PET/CT) imaging [28,32]. In all, 41 patients were selected for the study based on the inclusion/exclusion criteria. The participants were divided into three groups; 10 patients with histologically non-neoplastic nevus (melanocytic Nevi, stage 0-I melanoma), 13 with primary melanoma (stage II-III melanoma), and 18 with MM (stage III-IV melanoma). We categorized patients in the non-neoplastic nevus group if they had any abnormal, congenital formation or mark on the skin or neighboring mucosa that did not show neoplastic growth. A total of 41 cases were sequenced from 22 patients (53% female) with a median age at diagnosis of 55 years. Moreover, melanomas were mostly isolated from the lower limb and hip (52%), as well as the trunk and neck (22%). Histologically, the median thickness of melanomas was 3.8 mm in the primary and 5.4 mm in the metastatic group. As expected, the most common clinically apparent sites of distant metastases were lymph nodes (67.8%) and lungs (19.4%) (Supplementary Table S1). Paraffin-embedded or formalin-fixed tissue samples from the tumor biopsies in all three groups were obtained from the pathology sample bank at our hospital. The serum samples used in this study were residual samples that were obtained from all subjects when they did not undergo any treatment, with the exception of palliative care. To obtain serum samples, 2 mL blood specimens were allowed to clot at room temperature for at least 30 min, and then centrifuged at 1200 × g for 8 min. The serum was collected and subsequently divided into 500 µL aliquots, which were then stored at −80 °C until future DNA and RNA extraction. All patients underwent standard treatments according to each treating physician's practice. Serum and melanoma tissue samples were obtained from adult melanoma patients who were referred to the Department of Dermatology and Oncology at the Affiliated Hospital of Southwest Medical University, Luzhou, China. The demographic and histopathological variables of all subjects, including medical, reproductive, family history, tumor site, histological type, treatment, and survival, are described in Supplementary Table S1. Written informed consent to use the clinical and pathology information of patients was obtained from all participants or their guardians before inclusion in the study. This study is in agreement with the requirements specified in the Declaration of Helsinki (1983 Revision). A histological section was made for each sample, and diagnosis was confirmed by a different pathologist than the one who made the initial diagnosis. Detailed information on patient population, clinical assessment, and sample preparation has been reported previously [33].
Quantitative real-time PCR (qRT-PCR)
To quantify and compare the expression of targeted mRNAs-miRNAs, qRT-PCR was performed based on the standard protocols as described previously [33], [34], [35]. The expressions of target genes and mRNA in MM tissues were quantified using predeveloped TaqMan assay kits from Applied Biosystems (Life Technologies, Foster City, CA) [27, [34], [35], [36]]. In a 10-μl RT reaction system, 2 μl of 5 × RT buffer, 1 μl of dNTPs, 0.5 μl of random primer, 0.5 μl of revers. Ace, 0.25 μl of super RI, 0.25 μl of RT-enhancer, 2.25 μl of RNase-free water, and 3.25 μl of RNA (150 ng/μl) were added. The reaction was completed in a thermocycler (Mastercycler gradient, Eppendorf, Germany) with the following steps: 10 min at 30 °C, 30 min at 42 °C, 5 min at 99 °C, and 5 min at 4 °C, followed by final holding at 16 °C. The RT reaction was performed using the TaqMan® MicroRNA Reverse Transcription Kit (PN 4,366,597, 1500 reactions). Following the RT step, 0.8 μl of the RT reaction mixture was combined with 0.5 μl of a TaqMan MicroRNA Assay mix (20 ×; forward primer, reverse primer, and probe) and 5 μl of TaqMan® Universal PCR Master Mix, No AmpErase® UNG (PN 4,324,018) to make the final volume 10 μl. The synthesized cDNAs were then diluted by adding 40 μl ddH2O, and used as templates for quantitative PCR (qPCR). In a 10 μl reaction system, 5 μl of 2 × PCR-probe mix, 0.02 μl of probe, 1 μl of primers, 2 μl of H2O, and 2 μl of cDNA were mixed, and the reaction was completed in StepOne plus Thermocycler (Applied Biosystem) with 40 amplification cycles, according to the manufacturer's protocol. According our previous setting, we used the most reliable reference genes, 18S RNA and RNU6B snRNA from ABI (Cat# 4,427,975), to normalize targeted mRNA and miRNA expression data, respectively [27,34,35,37,38]. After normalization, relative contents of targeted mRNA and miRNA were calculated and expressed according the 2−ΔΔCT method. For the confirmation of EMT+ samples, we examined the expressions for zinc-finger E-box-binding-1 (ZEB1), zinc finger protein SNAI2 (SLUG), and Notch homolog 1, translocation-associated (NOTCH1), as selected epithelial-to-mesenchymal transition-inducing transcription factors (EMT-TFs) responsible for promoting tumor metastasis, by qRT-PCR in MM tissues and their adjacent normal tissues. Correspondingly, the EMT-TF expression levels were compared between group tissues after normalization to β-actin housekeeping gene. Specific forward and reverse primer sequences are listed in Supplementary Table S2.
CD31-periodic acid solution (PAS) dual staining and VM identification
The assays performed to test VM positivity according to standard CD31-PAS dual staining protocols as described previously [3,39]. Endogenous peroxidase activity was blocked with 3% hydrogen peroxide in 50% methanol for 10 min at room temperature. Sections were rehydrated and washed with PBS and then pretreated with citrate buffer (0.01 M citric acid, pH 6.0) for 20 min at 100 °C in a microwave oven. Non-specific binding sites were blocked with 2% normal goat serum in PBS for 20 min at 37 °C. Sections were then incubated overnight at 4 °C with anti-CD34 at a dilution of 1:200 (Dako, USA). Subsequently, sections were rinsed with PBS and incubated with biotinylated goat anti-mouse IgG for 20 min at 37 °C, followed by incubation with 3,3′-diaminobenzidine (DAB) chromogen for 10 min at room temperature. Sections were then rinsed with water for 1 min to stop the DAB-staining reaction. Formalin and melanin granules were then removed following the abovementioned methods. Finally, the sections were treated with 0.5% PAS for 8–10 min and rinsed with distilled water for 3 min. All of the mucous sites were stained cherry red, which proved the reliability of the quality of the PAS reagents. After rinsing with distilled water, the sections were counterstained with hematoxylin, and five fields at a magnification of × 200 were selected for counting of the number of endothelial-dependent vessels and VM. Microvessels were defined as any CD31+ endothelial cell cluster with or without a viable lumen. Also, large anastomosing sinusoidal vessels were considered as single vessels. According the standard method, VM is indicated by CD31 and PAS staining, with CD31-negative and PAS-positive vascular-like patterns [40]. The average number of endothelial-dependent vessels and VM, which quantify the microvessel density (MVD) and the vasculogenic mimicry density (VMD) in tumors, were calculated blindly, according to consensus guidelines [41,42]. The average of the 10 counts was considered as the final expression of MVD and VMD.
Immunohistochemistry
Melanoma tissues were sampled from patients at the Pathology Department and Oncology Department, The Affiliated Hospital of Southwest Medical University in Luzhou, Sichuan, China. Expert pathologist (WQ) and dermatologist (CW) resected the primary tumors and the metastases and confirmed clear margins on the samples. For the analysis of PLA1A, desmocollin 3 (DSC3), and DMKN, the representative tissue sections from all three groups were transferred into 10% neutral buffered formalin; fixed and embedded in Tissue Tek II OCT (Miles Scientific, Naperville, IL, USA); frozen for 15 min in isopentane; pre-cooled in liquid nitrogen; and finally stored at −80 °C for future hematoxylin and eosin (H&E) staining and immunostaining assays. Hematoxylin (HHS16, Sigma-Aldrich) and eosin (HT110232, Sigma-Aldrich) staining of samples was performed according to the manufacturer's instructions. The best frozen sample was used for immunostaining of PLA1A, DSC3, and DMKN, which was performed following the streptavidin-biotin alkaline phosphatase complex method using the Vectastain ABC-AP standard kit (Vector Laboratories; Burlingame, CA) as previously described (59,60). Complete information about the primary monoclonal antibodies and secondary monoclonal antibodies has been provided in Supplementary Table S3.
Quality assessment
Pearson's correlation coefficients were calculated for all genes in the dataset and the correlation matrices for all pair-wise comparisons of the known VM+ and EMT+ genes were evaluated across samples. Power β was used to remove the weakly correlated genes and retain the strongly correlated ones. The gene modules were represented by different colors, and the gray module indicated genes that could not be merged into any other module. Median powers of β = 2.5, 20, and 20 (scale-free R2 = 0.9) were used to construct the scale-free network modules between Nevi and MM patients, Nevi and primary melanoma patients, and primary melanoma and MM patients, respectively. Furthermore, a threshold of ≥ 0.7 was applied to export the network to Cytoscape. The process produced a weighted network called "adjacent matrix" that was transformed into a TOM network, a biologically meaningful VM and EMT measure that can determine the network connectivity of a gene [26]. Subsequently, average linkage hierarchical clustering was performed to classify genes with similar expression profiles into different modules. Using quantile normalization, pre-processor, and SVA packages in "R" statistical software (version 3.3.3), we removed the batch effects [43,44]. If multiple probes corresponded to one gene, the average expression value was considered as the gene expression value. Subsequently, DEGs were extracted following the Linear Models for Microarray Data (LIMMA) method, and the gene with |log2 fold change (FC)|>1 and adjusted P value of < 0.05 was considered to be statistically significant [44]. A heatmap was visualized using the heatmap package of R, based on standard methods [18,44].
Data analysis
All statistical analyses were performed using "R" software (version 3.3.3). For the variables following normal distribution, data are presented as mean ± standard deviation (SD). For non-normally distributed variables, data are reported as medians (ranges). Qualitative variables are presented as numbers and percentages. Data were analyzed via t-test (two-tailed) or one-way analysis of variance (ANOVA). The qRT-PCR data were analyzed using the Applied Biosystem 7500 Software 2.0.5. The expression levels of the genes of interest were calculated following the ΔCt method and normalized to 18SRNA and β-actin housekeeping gene or RNU6B snRNA. For identifying the effect of the cut-off threshold and constructing the receiver operating characteristic (ROC) curve, Spearman's rank correlation coefficient test was performed and the relationship between sensitivities (ordinate) and specificity (abscissa) [45] in all three groups was determined. An adjusted P of <0.05 was considered statistically significant.
Results
Search results and pre-processing data
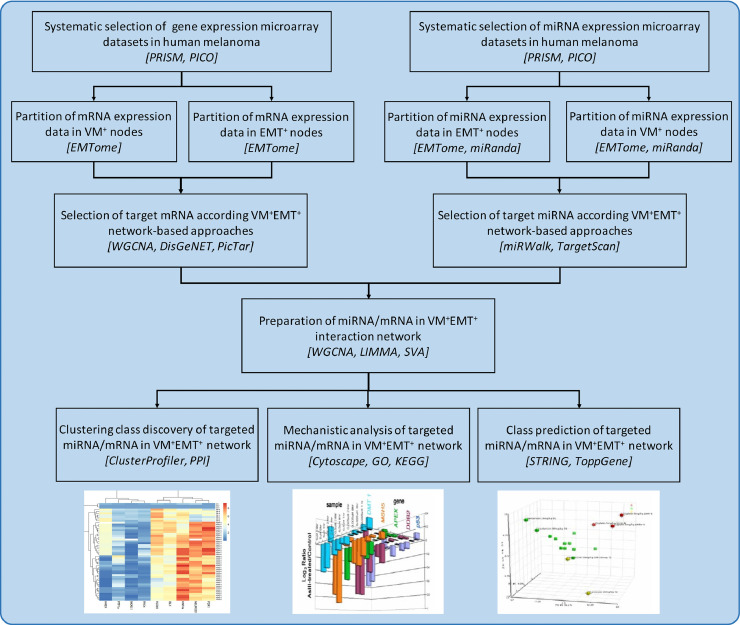
Fig. 1 illustrates the schematic representation of the workflow of the mRNA-miRNA co-expression network analysis in detail. We extracted and analyzed six microarray datasets: GSE34460, GSE18509, GSE24996, GSE7553, GSE15605, and GSE19234. The detailed information of microarray datasets extracted according to PRISMA guidelines with regard to study identification, screening, eligibility, inclusion process, and exclusion process are sorted in Supplementary Table S4. The data analyzed in this study were obtained from the supplementary material of the original article [46], [47], [48], [49], [50], [51]. In all, 244 samples were analyzed in this study, including 44 Nevi melanocyte samples (stage 0-I melanoma), 84 primary melanoma samples (stage II/III), and 116 MM samples (stage IV). All samples were diagnosed more than 2 years ago. The studies were conducted mostly in USA (3 studies, 50%) and Canada (2 studies, 40%), and one study was conducted in Germany (10%). All datasets were publicly available between 2008 and 2013. The Affymetrix and Agilent platforms were the major platforms of included datasets. Furthermore, during the combination of both platforms, no significant bias and unconvertable data were found in any of the selected datasets. The basic information of all included datasets is presented in Supplementary Table S4 in the order of the year of publication.
Fig. 1.
Schematic representation and experimental design of the study. The workflow protocol of the mRNA-miRNA co-expression network analysis among Nevi, primary melanoma, and metastatic melanoma samples. Nevi refers to patients diagnosed with any abnormal, congenital formation or mark on the skin or neighboring mucosa that does not show neoplastic growth. The bioinformatic tools used along the analysis pipeline were indicated by italic square brackets []. miRNAs, microRNAs; MM, malignant melanoma; VM, vascular mimicry; EMT, epithelial-to-mesenchymal transition, WGCNA, weighted gene co-expression network analysis; PRISMA, preferred reporting items for systematic reviews and meta-analysis; PICO, population, intervention, control, and outcomes framework; GO, gene ontology; KEGG, function and kyoto encyclopedia of genes and genomes; STRING, the search tool for the retrieval of interacting genes; PPI, protein–protein interactive.
Differential co-expressions of mRNA during melanomagenesis
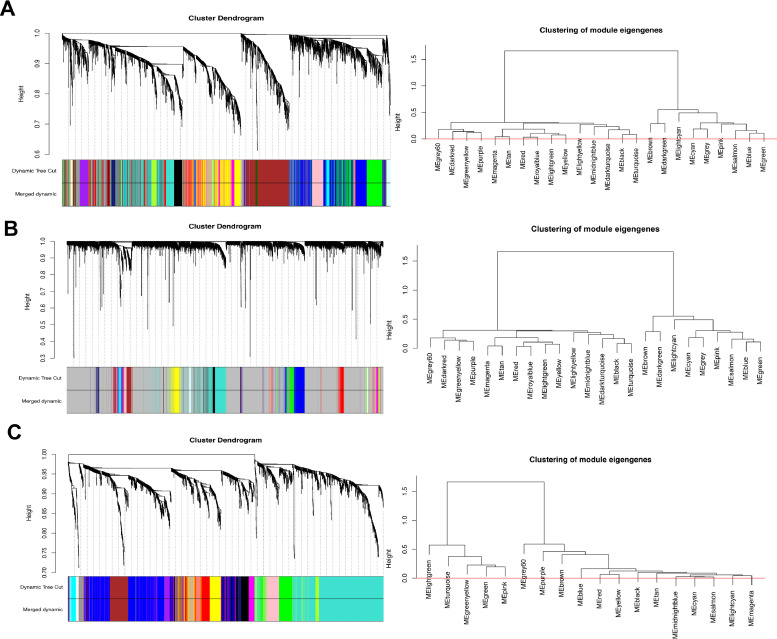
We constructed a VM+, EMT+ biological network at the systems' level of the available microarray data downloaded from the public database. We performed WGCNA with 862 genes, which were the most significant genes from 6543 genes, as key modules of highly positively correlated genes with melanoma (Fig. 2). We reconstructed three networks using group matrices in WGCNA. Optimal β parameters for obtaining a scale-free network were computed as 20, 25, and 10 for Nevi-primary melanoma (NP), Nevi-metastatic melanoma (NM), and primary melanoma-metastatic melanoma (PM), respectively. Modules were extracted in each network, and each module was marked with a unique color. Thus, a total of 24, 22, and 18 modules were obtained for NP, NM, and PM, respectively. Moreover, we extracted a list of melanoma genes from DisGeNET, and performed chi-square test to exclude those modules in three groups (Table 1).
Fig. 2.
Weighted gene correlation network analysis (WGCNA). The average linkage hierarchical clustering and module-trait analysis between Nevi and metastatic melanoma groups (A); Nevi and primary melanoma groups (B), and primary melanoma and metastatic melanoma groups (C). Different colors of the column indicate different hub modules. Nevi refers to patients diagnosed with any abnormal, congenital formation or mark on the skin or neighboring mucosa that does not show neoplastic growth.
Table 1.
The results of DEGs and significant gene-modules among groups.
| Groups (matrices) | Total VM+,EMT+ DEGs No. | Up-regulate No. | Down-regulate No. | Total module | Top module-trait⁎⁎ | Total genes of module | Associated gene-disease⁎⁎⁎ | P-value |
|---|---|---|---|---|---|---|---|---|
| Nevi*-Primary (NP) | 3965 | 1805 | 2160 | 22 | Brown | 533 | 113 | 0.0003 |
| Green | 348 | 75 | 0.0015 | |||||
| Lightcyan | 48 | 13 | 0.0351 | |||||
| Nevi-Metastasis (NM) | 6543 | 3237 | 3306 | 24 | Brown | 199 | 40 | 0.0162 |
| Magenta | 75 | 20 | 0.0047 | |||||
| Primary-Metastasis (PM) | 2380 | 1406 | 974 | 18 | Brown | 139 | 33 | 0.0226 |
Abbreviations: VM, vascular mimicry; EMT, epithelial to mesenchymal transition; DEGs, differentially expressed genes.
Nevi refers to patients diagnosed with any abnormal, congenital formation or mark on the skin or neighbouring mucosa that does not show neoplastic growth.
Top- module-trait suggested the highest correlation in adjacency matrices according to the dimensionless topological network analysis.
Associated gene-disease of EMT+ and VM+ genes in each group were determined by applying on DisGeNET online database.
Key module identification between different stages of melanoma
To identify the potential candidate VM+ and/or EMT+ key genes associated with melanoma tumorigenesis, we performed the dimensionless topological network analysis between all three groups, which was used to scale the adjacency matrix. As indicated in Fig. 2A, the most significant module associated with NM was the magenta module, which comprised 862 mRNAs identified as candidates for further analysis (P = 0.004). Also, the gene co-expression modules between NM groups determined by average linkage hierarchical clustering are illustrated in Fig. 2A. Furthermore, Supplementary Fig. S1 ranks the correlation analysis between modules and clinical characteristics, which were significantly correlated with melanoma tumorigenesis. The heatmap of module-trait analysis in the magenta module suggested the highest correlation between NM groups. Fig. 2B presents the data of WGCNA between NP groups within the melanoma groups. Following WGCNA, we found that 558 of 3965 genes were significantly correlated with melanoma in the light cyan module, which was the key module between NP groups (P = 0.035, Supplementary Fig. S1B). As shown in Fig. 2C, four gene modules were divided by dynamic tree cutting: brown, blue, midnight blue, and light cyan. Each module was independently conformed to the other modules based on the correlation and significance between two modules. After investigating their eigengene co-expression, we merged these dynamic modules into three with a threshold of 0.5, confirming the reliability of the brown module (Supplementary Fig. S1C). The correlation of the brown module with other modules was significant (P = 0.002). This module had 33 most significant genes of the 139 genes that were highly correlated to melanoma.
Differential co-expressions of miRNA during melanomagenesis
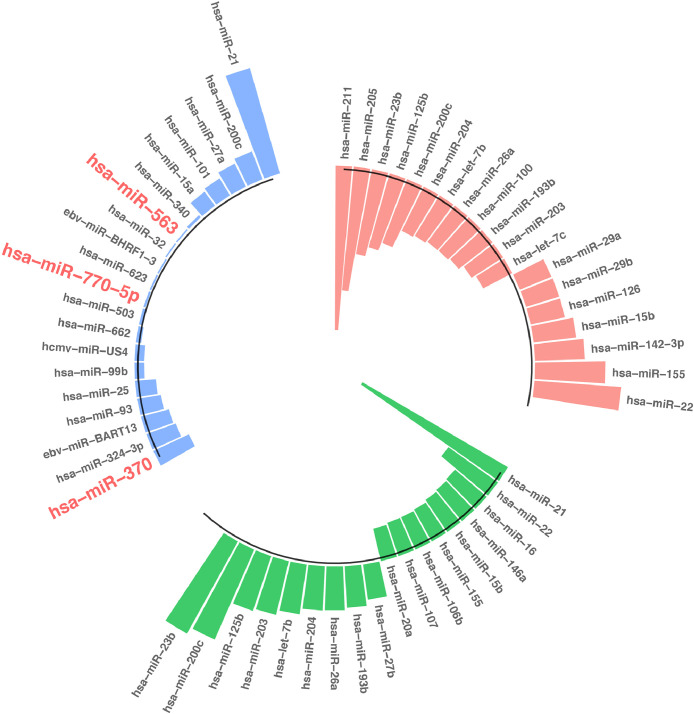
We identified miRNAs of target genes in each group using miRWalk v3.0 tool based on the genes of candidate modules. To reduce analysis complexity and select the most effective miRNAs, the top 20 highest degree (hub) miRNAs between different groups were shortlisted and are listed in Fig. 3. A total of 78 differentially expressed miRNAs (DEmiRNAs), 46 upregulated miRNAs, and 32 downregulated ones, were identified between three groups (Adj-p value < 0.05 and log2FC ≥|0.7|). As presented in Fig. 3, the top significantly regulated miRNAs between NM groups were miR-21 and miR-370 (blue chart). Further, miR-21 was upregulated between PM groups with log2 FC ≥ 0.7 (green chart). Fig. 3 shows that miR-211 and miR-22 were top significantly regulated miRNAs between NP groups with log2 FC ≥ 0.7 (orange chart). However, miR-200c was upregulated between NM and also between PM groups (Fig. 3; Adj-p-value < 0.05 and log2FC ≥ 0.7).
Fig. 3.
The top 20 differentially expressed miRNAs between Nevi and metastatic melanoma groups (blue chart), Nevi and primary melanoma groups (orange chart), and primary melanoma and metastatic melanoma groups (green chart). Nevi refers to patients diagnosed with any abnormal, congenital formation or mark on the skin or neighboring mucosa that does not show neoplastic growth (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.).
The mRNA-miRNA bipartite network during melanomagenesis
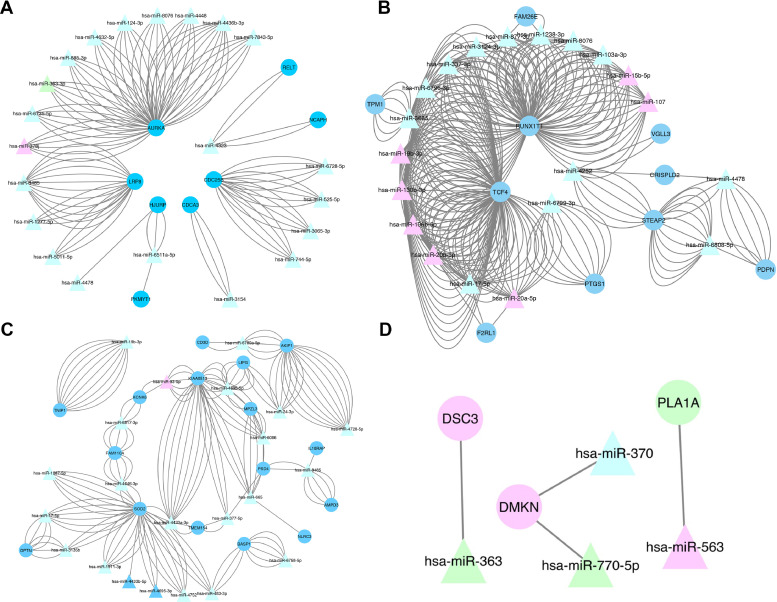
To better understand the role of mRNAs and miRNAs during melanoma tumorigenesis, we constructed an mRNA-miRNA bipartite network between the three groups. This bipartite network can represent the progression of melanogenesis, when naïve pigment melanin is produced in melanosomes by melanocytes and then melanocytes initiate VM and transition from epithelial to mesenchymal forms, finally shifting to aggregative and metastatic stages [52]. We predicted the overlaps (intersection) between mRNA/miRNAs using the miRWalk 3.0 database. These targeted mRNAs were cross-checked against the DEmiRNAs retrieved from the TCGA database. These data indicated that 36 DEMs were involved in the miRNA network. Finally, we constructed a bipartite network based on 19 miRNA nodes and 36 mRNA nodes between NM groups (Fig. 4A), NP groups (Fig. 4B), and PM groups (Fig. 4C). Fig. 4D shows the common mRNAs-miRNAs between the three groups. Notably, PLA1A, DSC3, and DMKN were the novel cancer driver genes in melanoma, exhibiting regulatory functions on interaction with miR-563, miR-363, miR-770–5p, and miR-370. The enriched pathways were reported to exhibit critical roles in the development of other cancers. In contrast, ECM-receptor interaction, cell cycle, cytokine-cytokine receptor interaction, and T cell receptor signaling pathways were found to be enriched in patients with high expression levels of PLA1A, DSC3, and DMKN. Collectively, these data indicated that these three hub genes may play comparable and critical roles in melanoma development and progression. After considering the common mRNAs-miRNAs between all three groups, we presented the relative expression levels of the targeted novel cancer driver mRNAs (Fig. 4D). Together, these data suggested that PLA1A, DSC3, and DMKN genes function as oncogenes that trigger melanomagenesis on interaction with miR-563, miR-363, miR-770–5p, and miR-370.
Fig. 4.
The mRNA-miRNA bipartite network analysis between Nevi and metastatic melanoma groups (A), Nevi and primary melanoma groups (B), and primary melanoma and metastatic melanoma groups (C). (D) Common mRNA-miRNA networks between all three groups. The circle indicates mRNA, and the triangle indicates miRNA. The nodes highlighted in blue and green represent up-regulation and those highlighted in pink represent down-regulation. The nodes highlighted in green and pink represent up-regulation and down-regulation, respectively. The node highlighted in light blue shows no change (logFC=0) (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.).
The pathway enrichment analysis
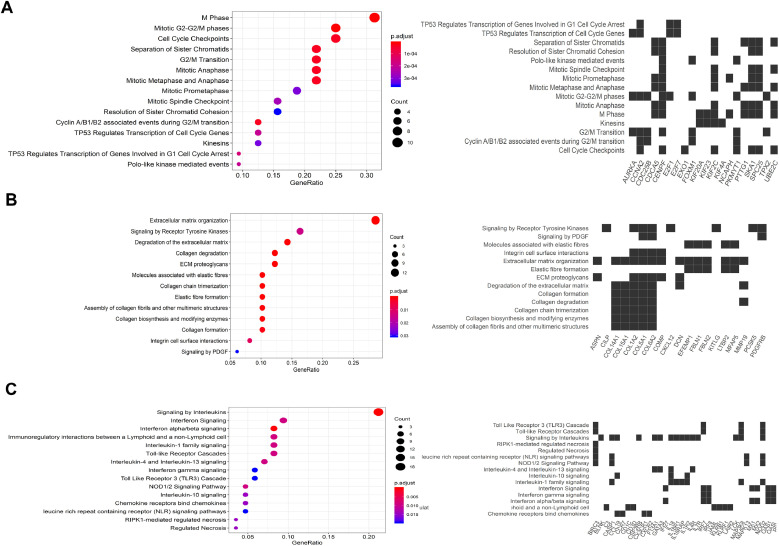
We enriched the EMT+ and VM+ modules obtained in the previous steps to identify the most significant module in each group. The pathway enrichment analysis indicated that the differentially expressed mRNAs (DEMs) predominantly targeted 36 pathways, mainly associated with protein digestion and absorption and ECM-receptor interaction (Fig. 5). Furthermore, top-ranked pathway terms enriched by genes in each module for all three groups are sorted in Table 2. As clearly shown in Fig. 5A, enrichment analysis of the core genes between NM groups revealed that the magenta module was significantly enriched in genes involved in the regulation of cytokinesis (GO:0032465), cell cycle G2/M phase transition (GO:0044839), regulation of mitotic cell cycle phase transition (GO:1901990), mitotic sister chromatid segregation (GO:0000070), and positive regulation of mitotic cell cycle phase transition (GO:1901992). Fig. 5B represents the functional analysis of the light cyan module genes between NP groups. In all, 558 DEMs were significantly enriched during the development of ECM organization (GO:0030198), collagen fibril organization (GO:0030199), regulation of intrinsic apoptotic signaling pathways in response to DNA damage (GO:1902229), negative regulation of calcium ion transmembrane transport (GO:1903170), and regulation of cell migration (GO:0030334). Notably, cytokine-mediated signaling pathway (GO:0019221), regulation of inflammatory response (GO:0050727), inflammatory response (GO:0006954), cellular response to interferon-gamma (GO:0071346), and cellular response to cytokine stimulus (GO:0071345) were the most significant biological functions that exhibited brown module genes in NM groups (Fig. 5C). These three crucial modules, i.e., magenta, light cyan, and brown modules, contained 862, 558, and 139 cancer-driver genes, respectively, that were significantly associated with VM and EMT processes during melanoma progression. Furthermore, our data shows the main pathway networks between all three groups.
Fig. 5.
Pathway enrichment analysis. (A) Enrichment analysis and main pathway networks between Nevi and metastatic melanoma groups in the magenta module. (B) Pathway analysis of the light cyan module genes between Nevi and primary melanoma groups. (C) The significant pathway enrichment analysis of brown module genes between primary melanoma and metastatic melanoma groups.
Table 2.
Top-ranked pathway terms enriched by genes in each module and group.
| Groups | Module | EMT/VM+ cancer driver gene/whole gene (%) | Biological term | P-value | Genes |
|---|---|---|---|---|---|
| Nevi*-Primary (NP) | Lightcyan | 558/3965 (14.1) | Cell cycle | 0.00047 | CCNA2;PTTG1;E2F1;PKMYT1;CDC25B |
| Progesterone-mediated oocyte maturation | 0.03363 | CCNA2;PKMYT1;CDC25B | |||
| Oocyte meiosis | 0.04279 | PTTG1;PKMYT1;AURKA | |||
| Nevi-Metastasis (NM) | Magenta | 862/6543 (13.2) | Protein digestion and absorption | 0.00022 | COL15A1;COL1A2;COL5A1;COL14A1;COL6A2 |
| ECM-receptor interaction | 0.01304 | COMP;COL1A2;COL6A2;CD36 | |||
| Primary-Metastasis (PM) | Brown | 33/139 (23.7) | Cytokine-cytokine receptor interaction | 0.00006 | CX3CR1;IL4R;IL1R2;IL18;CSF2RB;TNF;IL18RAP;CXCR2;CCL19;CCL18;IL7R;IL18R1 |
| NF-kappa B signaling pathway | 0.00546 | LCK;BLNK;CCL19;TNF;BIRC3 | |||
| T cell receptor signaling pathway | 0.00021 | ITK;LCK;MAP3K8;ICOS;TNF;RASGRP1;CD3D;MAPK13 | |||
| Hematopoietic cell lineage | 0.00007 | CD2;IL4R;IL1R2;CD24;IL7R;CD1C;TNF;CD3D |
Nevi refers to patients if they diagnosed with any abnormal, congenital formation or mark on the skin or neighbouring mucosa that does not show neoplastic growth.
Abbreviations: VM, vascular mimicry; EMT, epithelial to mesenchymal transition.
Diagnostic accuracy analysis
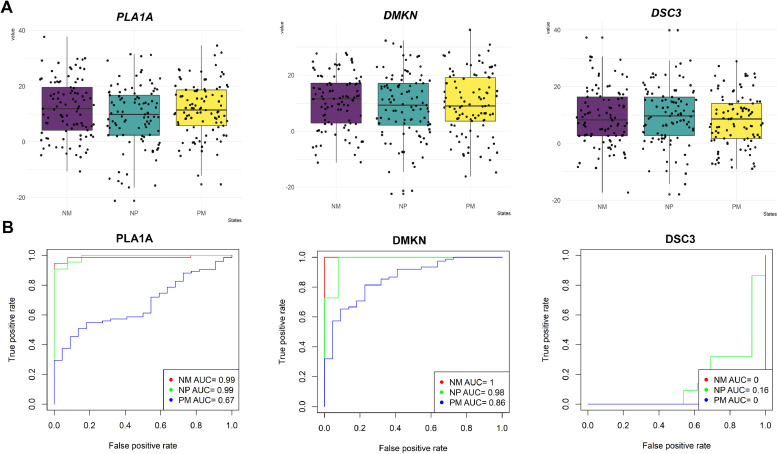
We analyzed the relative expression levels of targeted novel cancer driver mRNAs and miRNAs that have interacted with EMT and VM pathways in Nevi, primary, and MM groups to determine any possible relationship between the expressions of these genes (Fig. 6A). Furthermore, receiver operating characteristic (ROC) curves for PLA1A, DSC3, and DMKN expressions among the NM group (red line), NP group (green line), and PM group (blue line) are presented in the Fig. 6B. The area under the ROC curve (AUC) for PLA1A in between the three groups was 0.99, 0.99, and 0.67, respectively. We also found DMNK to be a significant distinctive biomarker among NM, NP, and PM groups with AUCs of 1, 0.98, and 0.86, respectively. Taken together, these data suggest that PLA1A and DMKN genes can be used as strong diagnosis biomarkers to distinguish melanoma patients, enabling the identification of new, network-based candidate diagnostic biomarkers.
Fig. 6.
Diagnostic accuracy analysis of targeted mRNAs-miRNAs. (A) The boxplot of targeted novel cancer drivers PLA1A, DMKN, and DSC3C between three groups. Collectively, these three genes were overexpressed in the metastatic groups. Similar results were obtained in all three independent experiments. Horizontal bars represent mean ± SD. (B) Receiver operating characteristic (ROC) curve for PLA1A, DMKN, and DSC3C expression between Nevi and metastatic melanoma groups (red line), Nevi and primary melanoma groups (green line), and primary and metastatic melanoma groups (blue line). The diagnostic values were calculated by considering both PLA1A mRNA expression and serum levels. PLA1A, phosphatidylserine-specific phospholipase A1-alpha; DSC3, desmocollin 3; DMKN, dermokine; NM, Nevi and metastatic melanoma; NP, Nevi and primary melanoma; PM, primary melanoma and metastatic melanoma; AUC, area under the curve. Nevi refers to patients diagnosed with any abnormal, congenital formation or mark on the skin or neighboring mucosa that does not show neoplastic growth (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.).
Epithelial-to-mesenchymal transition and formation of vasculogenic mimicry variables in patients with melanoma
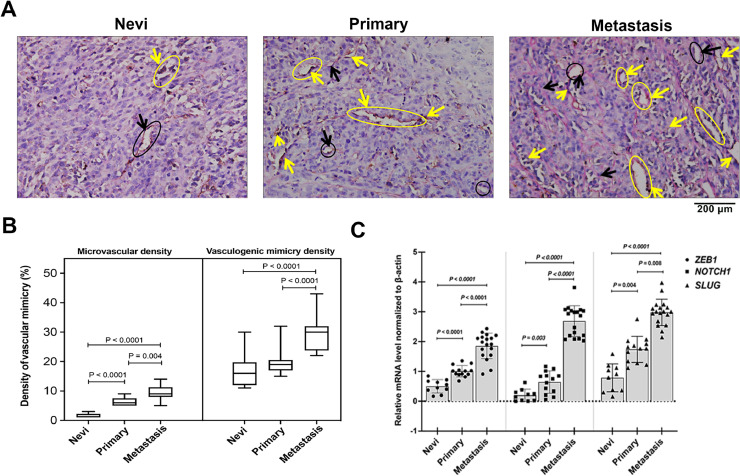
In this study, assays for counting the number of endothelial-dependent vessels were performed following CD31-PAS dual staining methods and quantification of MVD and VMD in tissue samples of 41 melanoma patients. These patients were subdivided into three groups: 10 patients with non-neoplastic nevus (stage 0-I) diagnosed more than 2 years ago, 13 patients with primary melanoma (stage II-III), and 18 patients with MM (stage III-IV). The CD31/PAS and VM expression levels in each case are shown in Supplementary Table S5. VM was observed in 27 of 41 melanoma samples (65.9%). In the Nevi group, the presence of VM was observed in 7 of 10 (70%) patients, whereas in the primary group, VM was detected in 11 of 13 (84.6%) patients (Table 3). Further, among the 18 MM patients, 16 (88.9%) were VM+. As expected, the difference in the presence of VM between NM and NP groups was significant (P < 0.0001). Fig. 7A shows the traditional endothelial-dependent vessels and VM with CD31-PAS double staining among Nevi, primary melanoma, and MM groups, which clearly shows that the VM status significantly increased during melanomagenesis. As is clear in Fig. 7A, the presence of PAS+/CD31+ endothelial-dependent vessels (yellow arrow) was surrounded by PAS+ “patterned structures” that have been speculated to represent VM (black arrow). This in vivo experiment results show that VM tends to increase in patients with a high stage or grade of melanoma. The MVD and VMD levels were significantly higher in patients with late-stage melanoma than in those with Nevi (Table 3). As a shown in Fig. 7B, compared with the primary melanoma group (19.62 ± 4.23, p < 0.05) and the MM group (29.67 ± 5.48, P < 0.001), the Nevi group (16.90 ± 5.74) formed fewer VM channels. To confirm the EMT expression, we determined the relative levels of EMT-TF NOTCH1, SLUG, and ZEB1 by conducting of qRT-PCR (Fig. 7C). Interestingly, the average EMT-TF NOTCH1, SLUG, and ZEB1 expression levels were consistently and significantly higher in MM tissues (n = 18) than in Nevi tissues (n = 10). In addition, EMT-TF levels were significantly higher in primary melanoma tissues (n = 13) than in Nevi tissues. ECM deformation and angiogenesis are the primary pathological pathways when naïve melanocytes initiate VM and transition from epithelial to mesenchymal forms. As excepted, cell cycle and meiosis re-programming are the main phytological pathways that are implicated when naïve cells incline to shift to the primary stage. The tumor microenvironment and misbalancing of immune mediators are the main biological reason leading to mesenchymal cell metastases and preparation of a physical scaffold for extending VM. Thus, our study suggests that both VM and EMT pathways trigger angiogenesis and the formation of VM in during melanomagenesis.
Table 3.
VM and EMT variables in study patients.
| Pathways | Variable | Control group | Melanoma patients | |
|---|---|---|---|---|
| Nevi (n = 10)&& | Primary (n = 13) | Metastasis (n = 18) | ||
| VM | VM n (%)& | |||
| Positive | 7 (60) | 11 (53.8)⁎⁎ | 16 (33.4)⁎⁎,## | |
| Negative | 3 (40) | 2 (46.2) | 2 (66.6) | |
| MVD | 1.80 ± 0.79 | 6.38 ± 1.14⁎⁎ | 9.5 ± 2.3⁎⁎,# | |
| VMD | 16.90 ± 5.74 | 19.62 ± 4.23 | 29.67 ± 5.48⁎⁎,## | |
| EMT | ZEB1 | 5.10 ± 0.23 | 1.00 ± 0.42⁎⁎ | 1.85 ± 0.43⁎⁎,## |
| NOTCH1 | 0.21 ± 0.06 | 0.65 ± 0.37* | 2.69 ± 0.52⁎⁎,## | |
| SLUG | 0.78 ± 0.47 | 1.73 ± 0.44* | 2.98 ± 0.45⁎⁎,# | |
Data are presented as mean ± SD for all others.
& The VM positive assays were performed according to standard CD31-PAS dual staining protocols.
&& Nevi refers to patients if they diagnosed with any abnormal, congenital formation or mark on the skin or neighboring mucosa that does not show neoplastic growth.
Abbreviation: VM, vasculogenic mimicry; EMT, epithelial-to-mesenchymal transition; MVD, microvessel density; VMD, vasculogenic mimicry density; ZEB1, zinc-finger E-box-binding-1; NOTCH1, Notch homolog 1, translocation-associated (Drosophila); SLUG, zinc finger protein SNAI2.
: p < 0.05 and.
: p < 0.001 vs the Nevi group.
: p < 0.05 and.
: p < 0.001 vs the primary group.
Fig. 7.
Characterization of VM formation and EMT pathways in study groups. (A) Representative micrographs of CD31/PAS dual staining of VM during melanomagenesis in non-neoplastic nevus patient No. NN06, primary melanoma patient No. PM08, and metastatic melanoma patient No. MM05 tissues (Supplemental Table S5). The CD31 staining is shown in brown color (yellow arrow) and PAS staining is shown deep blue color (black arrow). Melanoma cross-section shows the presence of a PAS+/CD31+ endothelial-dependent vessels (black cycle) and the presence of PAS+ “patterned structures” that have been speculated to represent VM (yellow cycle). This in vivo experiment results show that VM tends to increase in patients with a high stage or grade of melanoma. Original magnification, × 200. Black arrow: structure of VM; Yellow arrow: structure of endothelial-dependent vessels. (B) The quantification of vasculogenic mimicry density (VMD) in different groups by comparing the microvessel density (MVD) and VMD in tumors. The MVD and VMD levels were significantly higher in patients with late-stage melanoma than in those with non-neoplastic nevus. (C) EMT-TFs, NOTCH1, SLUG, and ZEB1 expression levels were detected in melanoma tissues and matched with the corresponding paired adjacent normal tissues through qRT-PCR. The average EMT-TFs NOTCH1, SLUG, and ZEB1 expression levels were significantly higher in tissues of metastatic melanoma than in tissues of Nevi and primary melanoma. EMT-TF expression levels were compared between primary cancer and paired adjacent tissues in each case after normalization to β-actin housekeeping gene via the 2−ΔΔCT method. Nevi refers to patients diagnosed with any abnormal, congenital formation or mark on the skin or neighboring mucosa that does not show neoplastic growth. ZEB1, zinc-finger E-box-binding-1; SLUG, zinc finger protein SNAI2; NOTCH1, Notch homolog 1, translocation-associated (Drosophila). *p < 0.05 and **p < 0.001 vs the Nevi group; #p < 0.05 and ##p < 0.001 vs the primary group (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.).
In vivo confirmation of targeted mRNAs-miRNAs during melanomagenesis
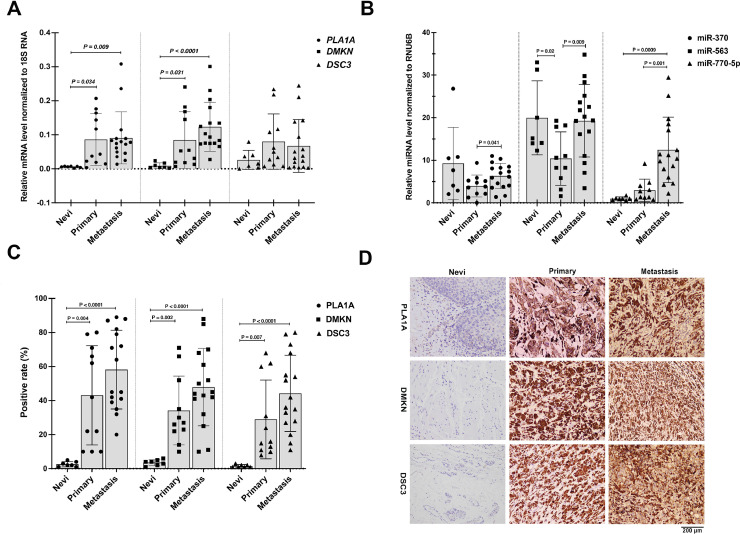
To confirm targeted mRNA-miRNA expression, we determined the relative levels of PLA1A and DMKN genes on interaction with miR-563, miR-770–5p, and miR-370 in 41 melanoma patients (Fig. 8). Also, the details of the expression levels of targeted mRNAs-miRNAs from each patient are shown in Supplementary Table S5. Our previous bioinformatics screenings for PLA1A, DSC3, and DMKN expressions among patients of different sexes and at stages of melanoma revealed that high gene expression levels of PLA1A and DMKN were associated with different clinical stages of melanoma. Therefore, both gene and protein expression levels of targeted PLA1A, DSC3, and DMKN were measured in our collected melanoma tissue and serum samples via qRT-PCR and immunohistochemistry, respectively. In line with ROC findings, PLA1A and DMKN mRNA expression significantly increased during melanomagenesis (Fig. 8A). The average PLA1A and DMNK expression level was consistently and significantly higher in EMT+/VM+ tissues of MM (n = 15) than in tissues of patients with non-neoplastic nevus (n = 10). On conducting immunohistochemical analysis, we found that PLA1A and DMNK levels were significantly higher in EMT+/VM+ melanoma tissues (n = 27) than in Nevi tissues (n = 7) (Fig. 8C and D). Therefore, we conclude that a significant relationship exists between PLA1A and DMNK levels and melanoma stage. As shown in Fig. 8B, the miR-770–5p levels, as a typical onco-miRNA, were significantly increased during melanomagenesis. In contrast, the miR-563 and miR-370 expression levels were significantly lower in patients with late-stage melanoma than in those with Nevi . With regard to the ratio of DMNK to miR-370 and miR-770–5p, the expression of DMNK/miR-770–5p/miR-370 significantly increased during melanomagenesis.
Fig. 8.
Targeted mRNA-miRNA expression in subjects with melanoma. (A) The expression levels of PLA1A, DSC3, and DMKN in EMT+/VM+ melanoma tissues in different groups, Nevi (n = 7), primary melanoma (n = 11), and metastatic melanoma (n = 16). PLA1A and DMKN expression levels were associated with different clinical stages of melanoma. (B) Comparison of miR-563, miR-770–5p, and miR-370 levels among Nevi (n = 7), primary melanoma (n = 11), and metastatic melanoma (n = 16) tissues. The miR-770–5p levels were significantly increased during melanomagenesis. In contrast, the miR-563 and miR-370 expression levels were significantly lower in patients with late-stage melanoma than in those with Nevi. (C) Quantification and (D) representative microscopic images showing the relative expression levels of PLA1A, DSC3, and DMKN in EMT+/VM+ tumor sections in various groups following immunohistochemical staining. Targeted PLA1A, DSC3, and DMKN expressions were increased in EMT+/VM+ melanoma tissues and serum samples. The immunohistochemical staining of PLA1A, DSC3, and DMKN were represented in one specific case; non-neoplastic nevus patient No. NN06, primary melanoma patient No. PM08, and metastatic melanoma patient No. MM05 tissues (Supplemental Table S5). The expressions of target mRNA in all three groups were quantified using predeveloped TaqMan assay kits from Applied Biosystems (Life Technologies, Foster City, CA) and normalized by the 18S RNA housekeeping gene. Also, the expressions of target miRNA were quantified using predeveloped TaqMan® MicroRNA Reverse Transcription Kit (PN 4366597, 1500 reactions) and normalized using the RNU6B. The elative contents of target mRNAs-miRNAs were calculated and expressed according the 2−ΔΔCT method. Nevi refers to patients diagnosed with any abnormal, congenital formation or mark on the skin or neighboring mucosa that does not show neoplastic growth.*p < 0.05 and **p < 0.001 vs the Nevi group; #p < 0.05 and ##p < 0.001 vs the primary group. PLA1A, phosphatidylserine-specific phospholipase A1-alpha; DSC3, desmocollin 3; DMKN, dermokine.
Discussion
In this study, we first performed a multidimensional pipeline of mRNA/miRNA analysis to identify targeted mRNAs-miRNAs that were associated with EMT and formation of VM in patients with MM. In the second approach, we tried to identify new potential candidate biomarkers distinguishing MM from primary melanoma in an unbiased fashion. In this regard, we applied WGCNA to identify the DEGs in the EMT+/VM+ key modules in melanoma that were associated with disease-related variables and melanomagenesis. Additionally, with regard to overall survival validation and mRNA/miRNA expression level, we expanded the scope of comparison range in tumor groups, with primary and metastasis groups against the Nevi group. Following this, we chose overlapping genes between EMT+/VM+ modules and DEGs for identifying biologically relevant modules that may serve as targets for diagnosis and therapeutic modulation in an unbiased fashion. We then confirmed the biological function of targeted mRNAs-miRNAs in VM+EMT network in subjects with MM and primary melanoma and healthy subjects in vivo. We identified a total of 64 co-expression modules constructed by 6543 genes from 75 human melanoma samples, which were used to detect the relationship between EMT and VM formation in melanoma transcriptome and clinical traits. Among these hub genes, we found that PLA1A and DMKN genes function as novel oncogenes that trigger VM and EMT processes during melanomagenesis on interaction with miR-370, miR-563, and miR-770–5p. We showed that PLA1A and DMKN genes can be considered as potential VM+EMT network-based diagnostic biomarkers to distinguish melanoma patients, with high sensitivity and specificity. With regard to personalized melanoma, this study provides a potentially valuable approach for the early diagnosis and prognosis of melanoma progression; besides, the study also unravels the crucial molecular mechanisms underlying MM progression via EMT and VM pathways.
It is generally accepted that WGCNA has many advantages over traditional methods for differential expression analysis and discovery of biologically relevant biomarkers [18,[53], [54], [55]]. WGCNA identifies powerful co-expression patterns that help detect new hub genes that may eventually serve as targets for diagnosis and therapeutic modulation of complex diseases such as MM [54,55]. With this approach, we applied WGCNA to detect novel VM+EMT network-based diagnostic biomarkers for the diagnosis and prognosis of melanoma. In particular, we found that PLA1A can be considered as a potential diagnostic marker for advanced and BRAF-mutated melanomas. This finding may provide a novel insight into the physiological characteristics of melanomagenesis and a potentially valuable approach for the early diagnosis and prognosis of melanoma by unravelling the molecular mechanisms underlying melanomagenesis.
Previous reports have indicated that DMKN was deregulated in metastatic cancer cell lines and correlated with tumor grade in many cancer patients [56, 57]. Huang et al. show that DMKN silencing slightly inhibited proliferation and induced cell cycle arrest in pancreatic cancer cell lines by altering cell cycle-related genes [57]. Their findings suggested that the knockdown of DMKN and PLA1A suppressed human tumor growth and metastasis in vivo [57,58]. In line with our finding, recent analysis of the oncogenic pathway regulated by DMKN shows that DMKN can modulate the ERK1/2/STAT3/EMT and AKT/STAT3/EMT signaling pathways [57]. Importantly, high expression of these genes may enhance the migration and invasion potential in pancreatic cancer cells by regulating partial EMT-associated proteins [57]. In addition, it plays an important role in promoting or inhibiting cancer cell proliferation/migration depending on the origin and type of cancer cell, and the outcome may be associated with expression levels of PLA1A and DMKN in the tumor [59,60]. A literature review revealed that the field is filled with inconsistent findings that make it difficult to ascertain the roles of PLA1A and DMKN in neoplasia and immunity associated with melanoma [37,57,59,61]. Nonetheless, this pilot study warrants a larger analytical study in transcriptomic and post-transcriptomic levels to confirm the findings. To fully determine whether PLA1A and DMKN are involved in melanoma progression, whole sequencing and scanning are needed for assessing expression patterns of circRNA–miRNA–mRNA regulatory networks [62,63]. However, this warrants another comprehensive study in the future with comprehensive in vitro and in vivo studies on the mechanism of PLA1A and DMKN in cancer cell growth suppression.
For the further confirmation of these results, in our recently published study, we detailed the use of PLA1A as a marker for thre effective prediction of advanced melanoma with subcutaneous metastases following BRAF or NRAS mutation [37]. We confirmed that the upregulation of PLA1A in the serum of patients with advanced MM could represent an excellent distinguishing and diagnostic marker in BRAF/NRAS-driven melanomagenesis. Furthermore, PLA1A mRNA, which is upregulated in tissue samples of advanced MM patients, represents a more accurate diagnostic marker [59, 64]. The PLA1 family is not just associated with tumorigenesis but also detectable across most inflammatory diseases, including systemic lupus erythematosus, hepatitis, and hyperthyroidism [65], [66], [67]. Recently many studies have shown that PLA1A is significantly involved in regulating different stages of carcinogenesis, which are associated with angiogenesis, differentiation, proliferation, invasion, apoptosis, and metastasis [68]. PS-PLA1, the sole substrate of LysoPS, is detectable across several physiological states and is restricted to the inner surface of cellular membranes, apoptotic cells, antigen-activated lymphocytes, and immunological escape of melanoma cells [69]. It is clear that the cause of MM is mostly associated with mutagenesis, with the involvement of several oncogenes, including BRAF and NRAS [70]. It is possible that PLA1A, with the highest sensitivity, specificity, and AUC, could be used as a single indispensable biomarker for the diagnosis, screening, and prognosis of BRAF mutation melanoma.
Evidence implies that miR-564 suppresses migration, invasion, and EMT in melanoma cells [71], [72], [73]. Our results hypothesized that PLA1A/miR-563 regulatory pathways may stabilize epithelial and mesenchymal states and suppress the formation of VM. We have consequently revealed that miR-563 is downregulated in MM tissues, and it suppresses cancer metastasis by affecting several malignancy endpoints via the downregulation of multiple EMT-TFs. The epigenetic inactivation of the PLA1A/miR-563 pathway can potentially push the equilibrium of these regulations toward EMT, thereby contributing to metastasis [74], [75], [76]. Thus, in future research investigations of our laboratory, we will conduct luciferase reporter assays by co-transfection of different amount of pre-miR-563 (along with PLA1A wild-type 3′-UTR reporter genes) in advanced melanoma cells to confirm the contribution of PLA1A/miR-563 interaction in the regulation of EMT and VM pathways.
DMNK is a glycoprotein expressed in epithelial cells, as one of spinous layer-specific genes that is abundant in stratified epithelia and in differentiating primary human keratinocytes [77,78]. DMNK encodes three different classes of isoforms, namely α, β, and γ. The pathology of DMNK in tumorigenesis has not yet been revealed fully. However, many studies have demonstrated that DMNK-β activates Rab5 and is involved in early endosomal trafficking, EMT, and tumor invasion [57,79]. Although, DMNK-α and -γ expressions were not observed in keratinocyte-derived tumor cells such as malignant melanoma, keratosis, basal cell carcinoma, and cutaneous T cell lymphoma cells, but researchers found that DMNK-β expressed in cultured human keratinocytes was associated with cell adhesion and inhibited cancer metastasis. A recent finding has suggested that DMNK-β is involved in keratinocyte differentiation through the regulation of the extracellular-signal-regulated signaling pathway [80]. Using cultured normal human epidermal keratinocytes, Higashi et al., revealed that exogenous DMNK-β triggers epidermal terminal differentiation and EMT. The DMNK-β, as a pro-inflammatory marker during melanomagenesis, is regulated by many cytokines and growth factors [78, 80]. For the first time, we demonstrated that DMNK disruption in advanced melanoma increased invasive ductal carcinoma and the proliferation ability of melanoma. Defiantly, DMKN knockdown could reduce the invasion and migration of keratinocyte-derived tumor cells, partially reversing EMT. Although the roles of DMNK as a regulator in cutaneous inflammation or skin tumors have not been fully elucidated, future comprehensive molecular and cellular investigations in a xenograft animal model need to focus on the roles of the DMKN as a malignancy‐associated gene in skin carcinoma [57]. To further explore the potential mechanism, more investigations with regard to the interplay between the overexpression of DMNK and VM and EMT-dependent genes at different stages of melanoma are needed. Meanwhile, in vitro laboratory investigations need to continue to elucidate the inhibitory effects of the EMT pathway in response to the decrease of DMNK. In a single-gene panel model, current research data shows that serum DMNK is a potential diagnosis biomarker for early-stage detection of advanced malignant cancers, such as colorectal cancer and pancreatic carcinoma [81,82]. Our miRNA/mRNA biological network speculates the serological and immunohistochemical markers of DMNK with the highest sensitivity, specificity, and AUC, and can be used as a single indispensable marker for diagnosis, screening and prognosis in advanced melanoma. Having identified novel miRNAs/mRNA network that could be of value for the understanding of melanoma development, we found out that DMNK/miR-770–5p/miR-563 significantly increased during melanomagenesis and this mRNA/miRNA network could be used as a novel network-based candidate marker in the diagnosis and prognosis of melanoma. Of the 46 upregulated and 32 downregulated miRNAs associated with melanoma development and progression, we only identified miR-770–5p and miR-370 to be correspondingly regulated in our data set, with the strongly direct interaction site on DMNK and PLA1A. In contrast, our miRNA expression profiles in human melanoma samples at different stages were in good agreement with the results reported by Wei et al., who used Invitrogen arrays to profile miRNA expression at different stage of melanoma [83]. Based on these findings and our results, miR-770–5p and miR-563 are the novel well-evaluated tumor suppressor-miRNAs that could effectively inhibit the invasion potential and migration of human cancer cells by directly targeting EMT pathways [84], [85], [86], [87]. Many studies have indicated that miR-770–5p and miR-563 could downregulate EMT-TFs, such as Vimentin, ZEB1, and Snail expression levels, while increasing or restoring the expression levels of E-Cadherin, resulting in the inhibition of EMT phenotypes [71,88,89]. Here, we have observed that the expression levels of miR-770–5p were increased in advanced melanoma and negatively correlated with the mRNA expression of DMNK in MM tumor. Moreover, a higher miR-770–5p expression was associated with poorer melanoma patient survival. Collectively, our results revealed a new role of EMT downregulation in MM and suggested that miR-770–5p is a potential target in counteracting MM. Taken together, our findings support the hypothesis that two tumor suppressor miRNAs, miR-770–5p and miR-563, could reduce the tumorigeneses effect of DMNK. Undoubtably, future relative luciferase assays are needed to measure and normalize the downregulation of DMNK by miR-770–5p and miR-563 in clinical practice. Similarly, in vitro experiments using human melanoma cell lines wherein the miRNAs are altered to confirm changes in the targeted genes would significantly benefit the study and support their hypothesis. Considering the advantages and disadvantages of all existing methods for biological network, future analytical study is required to find promising techniques and methods that are all validated the EMT and VM pathways with more specifically genes, availability, and efficiency. Taking into account the different array platforms and confirmation miRBase versions are having different output.
In precision oncology, our results proposed that PLA1A and DMNK, added to the routine diagnostic panel for melanoma, may serve as potent indicators in the prognosis and determination of the metastatic stage of melanoma. Correspondingly, it is important to keep in mind that variable handling conditions of the sample, such as size, homogeneous distribution, and mutation parameters may directly influence the degree of RNA degradation, which in turn can affect expression levels of some miRNAs. Surely, this experimental research should be followed by further evaluation to standardize and characterize the clinical applicability of candidate biomarkers identified in this study.
In conclusion, we here for the first time showed alterations in PLA1A and DMNK expression patterns at different stages of melanoma. In addition, we obtained evidence that their expressions are regulated by EMT signaling pathway and formation of VE. Furthermore, we conclude that altered expression levels of PLA1A/miR-563 and DMNK/miR-770–5p/miR-370 may contribute to metastatic transformation, and may represent an optimal candidate biomarker, which in future, could be used as a non-invasive marker for the diagnosis and therapy of MM. Our findings suggest that PLA1A and DMKN genes function as oncogenes that trigger VM and EMT processes on interaction with miR-370, miR-563, and miR-770–5p during melanomagenesis. This study mining helped us identify potential novel biomarkers for the early diagnosis and better prognosis of MM.
Supplementary materials: Supplementary material associated with this article can be found, in the online version.
CRediT authorship contribution statement
WenFeng He: Investigation, Writing – review & editing. Gang Yang: Investigation, Data curation, Writing – review & editing. Shuya Liu: Investigation, Resources, Funding acquisition, Writing – review & editing. Mazaher Maghsoudloo: Investigation, Resources, Data curation, Writing – review & editing. Marzieh Dehghan Shasaltaneh: Resources, Visualization, Writing – review & editing. Parham Jabbarzadeh Kaboli: Writing – original draft. Cuiwei Zhang: Data curation, Writing – review & editing. JingHeng Zhang: Data curation, Writing – review & editing. Maliheh Entezari: Visualization, Writing – review & editing. Saber Imani: Writing – original draft, Writing – review & editing, Supervision, Project administration, Funding acquisition. QingLian Wen: Visualization, Project administration, Writing – review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgments
The authors would like to express their appreciation to appreciate Department of Oncology and Department of Pathology, Affiliated Hospital of Southwest Medical University, Luzhou, China. We appreciate all coresearchers for their continuous collaborations with us in sharing information.
Funding
This work was supported by the Southwest Medical University (SWMU), Sichuan Province, China grants [No, 18080] that were awarded to Saber Imani.
Data availability statement
The R codes used to pre-process, merge, and correct for batch effects for the generation of cancer type-specific melanomas in all three group can be found on Figshare (https://figshare.com/s/809108f0af88fefcee58). Also, the normalized and combined datasets are available on Figshare (https://figshare.com/s/818b4ee502a888db4cf6). Detailed information on the clinical predictive value of targeted genes and assessment are described in our previous works.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.tranon.2021.101237.
Contributor Information
Saber Imani, Email: saberimani@swmu.edu.cn.
QingLian Wen, Email: wql73115@hotmail.com.
Appendix. Supplementary materials
References
- 1.Tas F. Metastatic behavior in melanoma: timing, pattern, survival, and influencing factors. J. Oncol. 2012;2012 doi: 10.1155/2012/647684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jemal A., Saraiya M., Patel P., Cherala S.S., Barnholtz-Sloan J., Kim J., Wiggins C.L., Wingo P.A. Recent trends in cutaneous melanoma incidence and death rates in the United States, 1992–2006. J. Am. Acad. Dermatol. 2011;65:S17–S25. doi: 10.1016/j.jaad.2011.04.032. e1-3. [DOI] [PubMed] [Google Scholar]
- 3.Zhang Z., Imani S., Shasaltaneh M.D., Hosseinifard H., Zou L., Fan Y., Wen Q. The role of vascular mimicry as a biomarker in malignant melanoma: a systematic review and meta-analysis. BMC Cancer. 2019;19:1134. doi: 10.1186/s12885-019-6350-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lamouille S., Xu J., Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nat. Rev. Mol. Cell. Biol. 2014;15:178–196. doi: 10.1038/nrm3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jolly M.K., Ware K.E., Gilja S., Somarelli J.A., Levine H. EMT and MET: necessary or permissive for metastasis? Mol. Oncol. 2017;11:755–769. doi: 10.1002/1878-0261.12083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee H.M., Hwang K.A., Choi K.C. Diverse pathways of epithelial mesenchymal transition related with cancer progression and metastasis and potential effects of endocrine disrupting chemicals on epithelial mesenchymal transition process. Mol. Cell Endocrinol. 2017;457:103–113. doi: 10.1016/j.mce.2016.12.026. [DOI] [PubMed] [Google Scholar]
- 7.Son H., Moon A. Epithelial-mesenchymal transition and cell invasion. Toxicol. Res. 2010;26:245–252. doi: 10.5487/TR.2010.26.4.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sun B., Zhang D., Zhao N., Zhao X. Epithelial-to-endothelial transition and cancer stem cells: two cornerstones of vasculogenic mimicry in malignant tumors. Oncotarget. 2017;8:30502–30510. doi: 10.18632/oncotarget.8461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kirschmann D.A., Seftor E.A., Hardy K.M., Seftor R.E., Hendrix M.J. Molecular pathways: vasculogenic mimicry in tumor cells: diagnostic and therapeutic implications. Clin. Cancer Res. 2012;18:2726–2732. doi: 10.1158/1078-0432.CCR-11-3237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Seftor R.E., Hess A.R., Seftor E.A., Kirschmann D.A., Hardy K.M., Margaryan N.V., Hendrix M.J. Tumor cell vasculogenic mimicry: from controversy to therapeutic promise. Am. J. Pathol. 2012;181:1115–1125. doi: 10.1016/j.ajpath.2012.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hendrix M.J., Seftor E.A., Seftor R.E., Chao J.T., Chien D.S., Chu Y.W. Tumor cell vascular mimicry: novel targeting opportunity in melanoma. Pharmacol. Ther. 2016;159:83–92. doi: 10.1016/j.pharmthera.2016.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wei X., Chen Y., Jiang X., Peng M., Liu Y., Mo Y., Ren D., Hua Y., Yu B., Zhou Y., Liao Q., Wang H., Xiang B., Zhou M., Li X., Li G., Li Y., Xiong W., Zeng Z. Mechanisms of vasculogenic mimicry in hypoxic tumor microenvironments. Mol. Cancer. 2021;20:7. doi: 10.1186/s12943-020-01288-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lissitzky J.C., Parriaux D., Ristorcelli E., Verine A., Lombardo D., Verrando P. Cyclic AMP signaling as a mediator of vasculogenic mimicry in aggressive human melanoma cells in vitro. Cancer Res. 2009;69:802–809. doi: 10.1158/0008-5472.CAN-08-2391. [DOI] [PubMed] [Google Scholar]
- 14.Liu Q., Qiao L., Liang N., Xie J., Zhang J., Deng G., Luo H., Zhang J. The relationship between vasculogenic mimicry and epithelial-mesenchymal transitions. J. Cell Mol. Med. 2016;20:1761–1769. doi: 10.1111/jcmm.12851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hendrix M.J., Seftor E.A., Hess A.R., Seftor R.E. Molecular plasticity of human melanoma cells. Oncogene. 2003;22:3070–3075. doi: 10.1038/sj.onc.1206447. [DOI] [PubMed] [Google Scholar]
- 16.Wu Q., Wang J., Liu Y., Gong X. Epithelial cell adhesion molecule and epithelial-mesenchymal transition are associated with vasculogenic mimicry, poor prognosis, and metastasis of triple negative breast cancer. Int. J. Clin. Exp. Pathol. 2019;12:1678–1689. [PMC free article] [PubMed] [Google Scholar]
- 17.Nam Y., Jhee J.H., Cho J., Lee J.H., Shin H. Disease gene identification based on generic and disease-specific genome networks. Bioinformatics. 2019;35:1923–1930. doi: 10.1093/bioinformatics/bty882. [DOI] [PubMed] [Google Scholar]
- 18.Yu H., Cheng Y., Li W., Li Z., Wu P., Qiu S., Zeng B., Huang B. A novel lncRNA-miRNA-mRNA competitive endogenous RNA network for uveal melanoma prognosis constructed by weighted gene co-expression network analysis. Life Sci. 2020;260 doi: 10.1016/j.lfs.2020.118409. [DOI] [PubMed] [Google Scholar]
- 19.Knobloch K., Yoon U., Vogt P.M. Preferred reporting items for systematic reviews and meta-analyses (PRISMA) statement and publication bias. J. Craniomaxillofac Surg. 2011;39:91–92. doi: 10.1016/j.jcms.2010.11.001. [DOI] [PubMed] [Google Scholar]
- 20.Panic N., Leoncini E., de Belvis G., Ricciardi W., Boccia S. Evaluation of the endorsement of the preferred reporting items for systematic reviews and meta-analysis (PRISMA) statement on the quality of published systematic review and meta-analyses. PLoS ONE. 2013;8:e83138. doi: 10.1371/journal.pone.0083138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Richardson W.S., Wilson M.C., Nishikawa J., Hayward R.S. The well-built clinical question: a key to evidence-based decisions. ACP J. Club. 1995;123:A12–A13. [PubMed] [Google Scholar]
- 22.Sundararajan Z., Knoll R., Hombach P., Becker M., Schultze J.L., Ulas T. Shiny-Seq: advanced guided transcriptome analysis. BMC Res. Notes. 2019;12:432. doi: 10.1186/s13104-019-4471-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dong J., Horvath S. Understanding network concepts in modules. BMC Syst. Biol. 2007;1:24. doi: 10.1186/1752-0509-1-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ab Razak N.S., Mutalib N.S.Ab, Mohtar M.A., Abu N. Impact of chemotherapy on extracellular vesicles: understanding the chemo-EVs. Front Oncol. 2019;9:1113. doi: 10.3389/fonc.2019.01113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gibbons D.L., Creighton C.J. Pan-cancer survey of epithelial-mesenchymal transition markers across the cancer genome atlas. Dev. Dyn. 2018;247:555–564. doi: 10.1002/dvdy.24485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yip A.M., Horvath S. Gene network interconnectedness and the generalized topological overlap measure. BMC Bioinformatics. 2007;8:22. doi: 10.1186/1471-2105-8-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fan Y., Mu J., Huang M., Imani S., Wang Y., Lin S., Fan J., Wen Q. Epigenetic identification of ADCY4 as a biomarker for breast cancer: an integrated analysis of adenylate cyclases. Epigenomics. 2019;11:1561–1579. doi: 10.2217/epi-2019-0207. [DOI] [PubMed] [Google Scholar]
- 28.Zou L., Imani S., Maghsoudloo M., Shasaltaneh M.D., Gao L., Zhou J., Wen Q., Liu S., Zhang L., Chen G. Genomewide copy number analysis of circulating tumor cells in breast cancer patients with liver metastasis. Oncol. Rep. 2020;44:1075–1093. doi: 10.3892/or.2020.7650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hill J., Hambley M., Forster T., Mewissen M., Sloan T.M., Scharinger F., Trew A., Ghazal P. SPRINT: a new parallel framework for R. BMC Bioinformatics. 2008;9:558. doi: 10.1186/1471-2105-9-558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang Y., Lu Q., Shao X., Mo B., Nie X., Liu W., Chen X., Tang Y., Deng Y., Yan J. Development of a three-gene prognostic signature for hepatitis b virus associated hepatocellular carcinoma based on integrated transcriptomic analysis. J. Cancer. 2018;9:1989–2002. doi: 10.7150/jca.23762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grob J.J., Schadendorf D., Lorigan P., Ascierto P., Larkin J., Nathan P., Robert C., Hauschild A., Weber J., Daud A., Hamid O., Dummer R., Hansson J., Hoeller C., Schachter J., Van Akkooi A.C.J., Garbe C. Eighth American joint committee on cancer (AJCC) melanoma classification: let us reconsider stage III. Eur. J. Cancer. 2018;91:168–170. doi: 10.1016/j.ejca.2017.11.023. [DOI] [PubMed] [Google Scholar]
- 32.Wright C.L., Miller E.D., Contreras C., Knopp M.V. Precision nuclear medicine: the evolving role of PET in melanoma. Radiol. Clin. North Am. 2021;59:755–772. doi: 10.1016/j.rcl.2021.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.G. Yang, S. Liu, M. Maghsoudloo, M. Dehghan Shasaltaneh, P. Jabbarzadeh Kaboli, C. Zhang, Y. Deng, H. Heidari, M. Entezari, S. Fu, Q. Wen, and S. I., PLA1A expression as a diagnostic marker of BRAF-mutant metastasis in melanoma cancer. 2021: Scientific Reports Journal (in press). 2021. [DOI] [PMC free article] [PubMed]
- 34.Khan M.A., Tania M., Wei C., Mei Z., Fu S., Cheng J., Xu J., Fu J. Thymoquinone inhibits cancer metastasis by downregulating TWIST1 expression to reduce epithelial to mesenchymal transition. Oncotarget. 2015;6:19580–19591. doi: 10.18632/oncotarget.3973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Imani S., Wei C., Cheng J., Khan M.A., Fu S., Yang L., Tania M., Zhang X., Xiao X., Zhang X., Fu J. MicroRNA-34a targets epithelial to mesenchymal transition-inducing transcription factors (EMT-TFs) and inhibits breast cancer cell migration and invasion. Oncotarget. 2017;8:21362–21379. doi: 10.18632/oncotarget.15214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C (T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 37.Yang G., Liu S., Maghsoudloo M., Shasaltaneh M.D., Kaboli P.J., Zhang C., Deng Y., Heidari H., Entezari M., Fu S., Wen Q., Imani S. PLA1A expression as a diagnostic marker of BRAF-mutant metastasis in melanoma cancer. Sci. Rep. 2021;11:6056. doi: 10.1038/s41598-021-85595-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jacob F., Guertler R., Naim S., Nixdorf S., Fedier A., Hacker N.F., Heinzelmann-Schwarz V. Careful selection of reference genes is required for reliable performance of RT-qPCR in human normal and cancer cell lines. PLoS ONE. 2013;8:e59180. doi: 10.1371/journal.pone.0059180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu Z.J., Zhou Y.J., Ding R.L., Xie F., Fu S.Z., Wu J.B., Yang L.L., Wen Q.L. In vitro and in vivo apatinib inhibits vasculogenic mimicry in melanoma MUM-2B cells. PLoS ONE. 2018;13 doi: 10.1371/journal.pone.0200845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Imani S., Liu S., Maghsoudloo M., Wen Q., E.M.D. Reis, F. Berti . Springer Scienes; 2021. Histochemical Staining of Vasculogenic Mimicry: Methods and Protocols, in Methods in Molecular Biology; pp. 1–13. Editors. [DOI] [PubMed] [Google Scholar]
- 41.Nowak-Sliwinska P., Alitalo K., Allen E., Anisimov A., Aplin A.C., Auerbach R., Augustin H.G., Bates D.O., van Beijnum J.R., Bender R.H.F., Bergers G., Bikfalvi A., Bischoff J., Bock B.C., Brooks P.C., Bussolino F., Cakir B., Carmeliet P., Castranova D., Cimpean A.M., Cleaver O., Coukos G., Davis G.E., De Palma M., Dimberg A., Dings R.P.M., Djonov V., Dudley A.C., Dufton N.P., Fendt S.M., Ferrara N., Fruttiger M., Fukumura D., Ghesquiere B., Gong Y., Griffin R.J., Harris A.L., Hughes C.C.W., Hultgren N.W., Iruela-Arispe M.L., Irving M., Jain R.K., Kalluri R., Kalucka J., Kerbel R.S., Kitajewski J., Klaassen I., Kleinmann H.K., Koolwijk P., Kuczynski E., Kwak B.R., Marien K., Melero-Martin J.M., Munn L.L., Nicosia R.F., Noel A., Nurro J., Olsson A.K., Petrova T.V., Pietras K., Pili R., Pollard J.W., Post M.J., Quax P.H.A., Rabinovich G.A., Raica M., Randi A.M., Ribatti D., Ruegg C., Schlingemann R.O., Schulte-Merker S., Smith L.E.H., Song J.W., Stacker S.A., Stalin J., Stratman A.N., Van de Velde M., van Hinsbergh V.W.M., Vermeulen P.B., Waltenberger J., Weinstein B.M., Xin H., Yetkin-Arik B., Yla-Herttuala S., Yoder M.C., Griffioen A.W. Consensus guidelines for the use and interpretation of angiogenesis assays. Angiogenesis. 2018;21:425–532. doi: 10.1007/s10456-018-9613-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Feng Y., Song K., Shang W., Chen L., Wang C., Pang B., Wang N. REDD1 overexpression in oral squamous cell carcinoma may predict poor prognosis and correlates with high microvessel density. Oncol. Lett. 2020;19:431–441. doi: 10.3892/ol.2019.11070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen W., Gao C., Liu Y., Wen Y., Hong X., Huang Z. Bioinformatics analysis of prognostic miRNA signature and potential critical genes in colon cancer. Front Genet. 2020;11:478. doi: 10.3389/fgene.2020.00478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Langfelder P., Horvath S. Eigengene networks for studying the relationships between co-expression modules. BMC Syst. Biol. 2007;1:54. doi: 10.1186/1752-0509-1-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zamora J., Abraira V., Muriel A., Khan K., Coomarasamy A. Meta-DiSc: a software for meta-analysis of test accuracy data. BMC Med. Res. Methodol. 2006;6:31. doi: 10.1186/1471-2288-6-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Raskin L., Fullen D.R., Giordano T.J., Thomas D.G., Frohm M.L., Cha K.B., Ahn J., Mukherjee B., Johnson T.M., Gruber S.B. Transcriptome profiling identifies HMGA2 as a biomarker of melanoma progression and prognosis. J. Invest Dermatol. 2013;133:2585–2592. doi: 10.1038/jid.2013.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Riker A.I., Enkemann S.A., Fodstad O., Liu S., Ren S., Morris C., Xi Y., Howell P., Metge B., Samant R.S., Shevde L.A., Li W., Eschrich S., Daud A., Ju J., Matta J. The gene expression profiles of primary and metastatic melanoma yields a transition point of tumor progression and metastasis. BMC Med. Genom. 2008;1:13. doi: 10.1186/1755-8794-1-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bogunovic D., O'Neill D.W., Belitskaya-Levy I., Vacic V., Yu Y.L., Adams S., Darvishian F., Berman R., Shapiro R., Pavlick A.C., Lonardi S., Zavadil J., Osman I., Bhardwaj N. Immune profile and mitotic index of metastatic melanoma lesions enhance clinical staging in predicting patient survival. Proc. Natl. Acad. Sci. U. S. A. 2009;106:20429–20434. doi: 10.1073/pnas.0905139106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sand M., Skrygan M., Sand D., Georgas D., Gambichler T., Hahn S.A., Altmeyer P., Bechara F.G. Comparative microarray analysis of microRNA expression profiles in primary cutaneous malignant melanoma, cutaneous malignant melanoma metastases, and benign melanocytic nevi. Cell Tissue Res. 2013;351:85–98. doi: 10.1007/s00441-012-1514-5. [DOI] [PubMed] [Google Scholar]
- 50.Chen J., Feilotter H.E., Pare G.C., Zhang X., Pemberton J.G., Garady C., Lai D., Yang X., Tron V.A. MicroRNA-193b represses cell proliferation and regulates cyclin D1 in melanoma. Am. J. Pathol. 2010;176:2520–2529. doi: 10.2353/ajpath.2010.091061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sokilde R., Vincent M., Moller A.K., Hansen A., Hoiby P.E., Blondal T., Nielsen B.S., Daugaard G., Moller S., Litman T. Efficient identification of miRNAs for classification of tumor origin. J. Mol. Diagn. 2014;16:106–115. doi: 10.1016/j.jmoldx.2013.10.001. [DOI] [PubMed] [Google Scholar]
- 52.Dragomir M., Mafra A.C.P., Dias S.M.G., Vasilescu C., Calin G.A. Using microRNA networks to understand cancer. Int. J. Mol. Sci. 2018;19 doi: 10.3390/ijms19071871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen J., Wu F., Shi Y., Yang D., Xu M., Lai Y., Liu Y. Identification of key candidate genes involved in melanoma metastasis. Mol. Med. Rep. 2019;20:903–914. doi: 10.3892/mmr.2019.10314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang L.X., Li Y., Chen G.Z. Network-based co-expression analysis for exploring the potential diagnostic biomarkers of metastatic melanoma. PLoS ONE. 2018;13 doi: 10.1371/journal.pone.0190447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhao B., You Y., Wan Z., Ma Y., Huo Y., Liu H., Zhou Y., Quan W., Chen W., Zhang X., Li F., Zhao Y. Weighted correlation network and differential expression analyses identify candidate genes associated with BRAF gene in melanoma. BMC Med. Genet. 2019;20:54. doi: 10.1186/s12881-019-0791-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Naso M.F., Liang B., Huang C.C., Song X.Y., Shahied-Arruda L., Belkowski S.M., D'Andrea M.R., Polkovitch D.A., Lawrence D.R., Griswold D.E., Sweet R.W., Amegadzie B.Y. Dermokine: an extensively differentially spliced gene expressed in epithelial cells. J. Invest Dermatol. 2007;127:1622–1631. doi: 10.1038/sj.jid.5700779. [DOI] [PubMed] [Google Scholar]
- 57.Huang C., Xiang Y., Chen S., Yu H., Wen Z., Ye T., Sun H., Kong H., Li D., Yu D., Chen B., Zhou M. Dermokine contributes to epithelial-mesenchymal transition through increased activation of signal transducer and activator of transcription 3 in pancreatic cancer. Cancer Sci. 2017;108:2130–2141. doi: 10.1111/cas.13347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Uranbileg B., Kurano M., Sato M., Ikeda H., Ishizawa T., Hasegawa K., Kokudo N., Yatomi Y. Possible involvement of PS-PLA1 and lysophosphatidylserine receptor (LPS1) in hepatocellular carcinoma. Sci. Rep. 2020;10:2659. doi: 10.1038/s41598-020-59590-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kurano M., Miyagaki T., Miyagawa T., Igarashi K., Shimamoto S., Ikeda H., Aoki J., Sato S., Yatomi Y. Association between serum autotaxin or phosphatidylserine-specific phospholipase A1 levels and melanoma. J. Dermatol. 2018;45:571–579. doi: 10.1111/1346-8138.14278. [DOI] [PubMed] [Google Scholar]
- 60.Zhao Y., Hasse S., Bourgoin S.G. Phosphatidylserine-specific phospholipase A1: a friend or the devil in disguise. Prog. Lipid. Res. 2021;83 doi: 10.1016/j.plipres.2021.101112. [DOI] [PubMed] [Google Scholar]
- 61.Schlage P., Kockmann T., Sabino F., Kizhakkedathu J.N., Auf dem Keller U. Matrix metalloproteinase 10 degradomics in keratinocytes and epidermal tissue identifies bioactive substrates with pleiotropic functions. Mol. Cell Proteom. 2015;14:3234–3246. doi: 10.1074/mcp.M115.053520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Khan S., Jha A., Panda A.C., Dixit A. Cancer-associated circRNA-miRNA-mRNA regulatory networks: a meta-analysis. Front. Mol. Biosci. 2021;8 doi: 10.3389/fmolb.2021.671309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tang K., Zhang H., Li Y., Sun Q., Jin H. Circular RNA as a potential biomarker for melanoma: a systematic review. Front. Cell Dev. Biol. 2021;9 doi: 10.3389/fcell.2021.638548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nagai Y., Aoki J., Sato T., Amano K., Matsuda Y., Arai H., Inoue K. An alternative splicing form of phosphatidylserine-specific phospholipase A1 that exhibits lysophosphatidylserine-specific lysophospholipase activity in humans. J. Biol. Chem. 1999;274:11053–11059. doi: 10.1074/jbc.274.16.11053. [DOI] [PubMed] [Google Scholar]
- 65.Nakawatari K., Kurano M., Araki O., Nishikawa M., Shimamoto S., Igarashi K., Aoki J., Murakami M., Yatomi Y. Elevated phosphatidylserine-specific phospholipase A1 level in hyperthyroidism. Clin. Chim. Acta. 2020;503:99–106. doi: 10.1016/j.cca.2020.01.011. [DOI] [PubMed] [Google Scholar]
- 66.Sawada T., Kurano M., Shirai H., Iwasaki Y., Tahara K., Hayashi H., Igarashi K., Fujio K., Aoki J., Yatomi Y. Serum phosphatidylserine-specific phospholipase A1 as a novel biomarker for monitoring systemic lupus erythematosus disease activity. Int. J. Rheum Dis. 2019;22:2059–2066. doi: 10.1111/1756-185X.13689. [DOI] [PubMed] [Google Scholar]
- 67.Yang Q., Guo M., Zhou Y., Hu X., Wang Y., Wu C., Yang M., Pei R., Chen X., Chen J. Phosphatidylserine-specific phospholipase A1 is the critical bridge for hepatitis C virus assembly. Virol. Sin. 2019;34:521–537. doi: 10.1007/s12250-019-00123-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Guo M., Pei R., Yang Q., Cao H., Wang Y., Wu C., Chen J., Zhou Y., Hu X., Lu M., Chen X. Phosphatidylserine-specific phospholipase A1 involved in hepatitis C virus assembly through NS2 complex formation. J. Virol. 2015;89:2367–2377. doi: 10.1128/JVI.02982-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yatomi Y., Kurano M., Ikeda H., Igarashi K., Kano K., Aoki J. Lysophospholipids in laboratory medicine. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2018;94:373–389. doi: 10.2183/pjab.94.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zarei S., Voss J.S., Jin L., Jenkins S.M., Bryce A.H., Erickson L.A., Bell D.A., Kipp B.R., Flotte T.J. Mutational profile in vulvar, vaginal, and urethral melanomas: review of 37 cases with focus on primary tumor site. Int. J. Gynecol. Pathol. 2020;39:587–594. doi: 10.1097/PGP.0000000000000636. [DOI] [PubMed] [Google Scholar]
- 71.Zhang X., Li M., Sun G., Bai Y., Lv D., Liu C. MiR-563 restrains cell proliferation via targeting LIN28B in human lung cancer. Thorac. Cancer. 2020;11:55–61. doi: 10.1111/1759-7714.13257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Xu C., Zhang H., Zhou W., Wu H., Shen X., Chen Y., Liao M., Liu Y., Yuan W. MicroRNA-10a, -210, and -563 as circulating biomarkers for ossification of the posterior longitudinal ligament. Spine J. 2019;19:735–743. doi: 10.1016/j.spinee.2018.10.008. [DOI] [PubMed] [Google Scholar]
- 73.Zhang H., Xu C., Liu Y., Yuan W. [MicroRNA-563 promotes the osteogenic differentiation of posterior longitudinal ligament cells by inhibiting SMURF1] Zhonghua Wai Ke Za Zhi. 2017;55:203–207. doi: 10.3760/cma.j.issn.0529-5815.2017.03.008. [DOI] [PubMed] [Google Scholar]
- 74.Ho J.J.D., Man J.H.S., Schatz J.H., Marsden P.A. Translational remodeling by RNA-binding proteins and noncoding RNAs. Wiley Interdiscip Rev. RNA. 2021:e1647. doi: 10.1002/wrna.1647. [DOI] [PubMed] [Google Scholar]
- 75.Fattore L., Costantini S., Malpicci D., Ruggiero C.F., Ascierto P.A., Croce C.M., Mancini R., Ciliberto G. MicroRNAs in melanoma development and resistance to target therapy. Oncotarget. 2017;8:22262–22278. doi: 10.18632/oncotarget.14763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lee J.J., Murphy G.F., Lian C.G. Melanoma epigenetics: novel mechanisms, markers, and medicines. Lab. Invest. 2014;94:822–838. doi: 10.1038/labinvest.2014.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hasegawa M., Higashi K., Yokoyama C., Yamamoto F., Tachibana T., Matsushita T., Hamaguchi Y., Saito K., Fujimoto M., Takehara K. Altered expression of dermokine in skin disorders. J. Eur. Acad. Dermatol. Venereol. 2013;27:867–875. doi: 10.1111/j.1468-3083.2012.04598.x. [DOI] [PubMed] [Google Scholar]
- 78.Hasegawa M., Higashi K., Matsushita T., Hamaguchi Y., Saito K., Fujimoto M., Takehara K. Dermokine inhibits ELR(+)CXC chemokine expression and delays early skin wound healing. J. Dermatol. Sci. 2013;70:34–41. doi: 10.1016/j.jdermsci.2013.01.007. [DOI] [PubMed] [Google Scholar]
- 79.Monkman J.H., Thompson E.W., Nagaraj S.H. Targeting epithelial mesenchymal plasticity in pancreatic cancer: a compendium of preclinical discovery in a heterogeneous disease. Cancers. 2019;11 doi: 10.3390/cancers11111745. (Basel) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Higashi K., Hasegawa M., Yokoyama C., Tachibana T., Mitsui S., Saito K. Dermokine-beta impairs ERK signaling through direct binding to GRP78. FEBS Lett. 2012;586:2300–2305. doi: 10.1016/j.febslet.2012.06.022. [DOI] [PubMed] [Google Scholar]
- 81.Tagi T., Matsui T., Kikuchi S., Hoshi S., Ochiai T., Kokuba Y., Kinoshita-Ida Y., Kisumi-Hayashi F., Morimoto K., Imai T., Imoto I., Inazawa J., Otsuji E. Dermokine as a novel biomarker for early-stage colorectal cancer. J. Gastroenterol. 2010;45:1201–1211. doi: 10.1007/s00535-010-0279-4. [DOI] [PubMed] [Google Scholar]
- 82.Watanabe K., Oochiai T., Kikuchi S., Kumano T., Matsui T., Morimoto K., Yasukawa S., Nakamori S., Sasako M., Yanagisawa A., Otsuji E. Dermokine expression in intraductal papillary-mucinous neoplasm and invasive pancreatic carcinoma. Anticancer Res. 2012;32:4405–4412. [PubMed] [Google Scholar]
- 83.Wei S., Ma W. MiR-370 functions as oncogene in melanoma by direct targeting pyruvate dehydrogenase B. Biomed. Pharmacother. 2017;90:278–286. doi: 10.1016/j.biopha.2017.03.068. [DOI] [PubMed] [Google Scholar]
- 84.Noyan S., Andac Ozketen A., Gurdal H., Gur Dedeoglu B. miR-770-5p regulates EMT and invasion in TNBC cells by targeting DNMT3A. Cell Signal. 2021;83 doi: 10.1016/j.cellsig.2021.109996. [DOI] [PubMed] [Google Scholar]
- 85.Zhang D., Li Y., Sun P. miR-770-5p modulates resistance to methotrexate in human colorectal adenocarcinoma cells by downregulating HIPK1. Exp. Ther. Med. 2020;19:339–346. doi: 10.3892/etm.2019.8221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Noyan S., Gurdal H., Gur Dedeoglu B. Involvement of miR-770-5p in trastuzumab response in HER2 positive breast cancer cells. PLoS ONE. 2019;14 doi: 10.1371/journal.pone.0215894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Jia L., Song Y., Mu L., Li Q., Tang J., Yang Z., Meng W. Long noncoding RNA TPT1-AS1 downregulates the microRNA-770-5p expression to inhibit glioma cell autophagy and promote proliferation through STMN1 upregulation. J. Cell Physiol. 2020;235:3679–3689. doi: 10.1002/jcp.29262. [DOI] [PubMed] [Google Scholar]
- 88.Shukla G.C., Singh J., Barik S. MicroRNAs: processing, maturation, target recognition and regulatory functions. Mol. Cell Pharmacol. 2011;3:83–92. [PMC free article] [PubMed] [Google Scholar]
- 89.Bartel D.P. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The R codes used to pre-process, merge, and correct for batch effects for the generation of cancer type-specific melanomas in all three group can be found on Figshare (https://figshare.com/s/809108f0af88fefcee58). Also, the normalized and combined datasets are available on Figshare (https://figshare.com/s/818b4ee502a888db4cf6). Detailed information on the clinical predictive value of targeted genes and assessment are described in our previous works.