Abstract
The G1 cyclins of budding yeast drive cell cycle initiation by different mechanisms, but the molecular basis of their specificity is unknown. Here we test the hypothesis that the functional specificity of G1 cyclins is due to differential subcellular localization. As shown by indirect immunofluorescence and biochemical fractionation, Cln3p localization appears to be primarily nuclear, with the most obvious accumulation of Cln3p to the nuclei of large budded cells. In contrast, Cln2p localizes to the cytoplasm. We were able to shift localization patterns of truncated Cln3p by the addition of nuclear localization and nuclear export signals, and we found that nuclear localization drives a Cln3p-like functional profile, while cytoplasmic localization leads to a partial shift to a Cln2p-like functional profile. Therefore, forcing Cln3p into a Cln2p-like cytoplasmic localization pattern partially alters the functional specificity of Cln3p toward that of Cln2p. These results suggest that there are CLN-dependent cytoplasmic and nuclear events important for cell cycle initiation. This is the first indication of a cytoplasmic function for a cyclin-dependent kinase. The data presented here support the idea that cyclin function is regulated at the level of subcellular localization and that subcellular localization contributes to the functional specificity of Cln2p and Cln3p.
Key events in the eukaryotic cell cycle are regulated by the serine-threonine cyclin-dependent kinase (cdk). The binding of regulatory cyclin subunits to the cdk enzyme induces structural changes that are required for kinase activity. In addition to the activation of the cdk, cyclin molecules are thought to serve targeting functions, conferring functional specificity to different cyclin-cdk complexes (37). In Saccharomyces cerevisiae the cdk encoded by the CDC28 gene, Cdc28p, regulates the cell division cycle when bound to one of nine different cyclins: CLN1 to CLN3, and CLB1 to CLB6.
During the postmitotic growth period prior to DNA replication, haploid cells are able to respond to mating pheromone or initiate a round of cell division. Initiation of the cell division cycle follows a critical cell size threshold and requires the accumulation of Clbp-Cdc28p kinase activity, which is required for DNA replication and subsequent mitosis. Each of these events (cell size, mating pathway activation, and Clbp activation) are regulated by the Cln proteins.
Deletion of all three CLN genes results in a cell cycle arrest as unbudded cells with 1C DNA content. Genetic characterization of the CLN genes shows that CLN1, CLN2, and CLN3 are functionally redundant, since any one CLN gene is able to complement the arrest caused by the triple cln1 cln2 cln3 deletion. However, there are significant differences in expression, primary sequence, and functional specificity of CLN2 (representative of the highly homologous CLN1-CLN2 gene pair) and CLN3. CLN2 transcription is cell cycle regulated, peaking at late G1 (59, 64). CLN3 transcription is relatively constitutive, with a two- to threefold peak during the time in the cell cycle where wild-type cells finish mitosis and begin G1, prior to CLN2 expression (16, 33, 41, 59, 64). The sequence similarity between these two classes of CLN genes is low and confined to the cyclin box, a region of the cyclin involved in physical interactions with Cdc28p (19).
Functional analysis of CLN2 and CLN3 indicate that CLN3 is primarily required for the expression of genes during late G1. The Swi4p-Swi6p complex, called SBF, is required for the CLN3-dependent expression of CLN1-CLN2 (26). The BCK2 gene product also triggers transcription of CLN1-CLN2. The BCK2-dependent activation of CLN1-CLN2 most likely occurs through the SBF complex, working in parallel with Cln3p to activate CLN1-CLN2 (9, 13). CLN3 is unable to complement the triple cln1 cln2 cln3 deletion in the absence of SWI4 or SWI6, indicating that this transcription complex is crucial for Cln3p-dependent viability. Cln2p-dependent viability in the triple cln1 cln2 cln3 deletion strain does not require Swi4p or Swi6p (27). The Mbp1p-Swi6p complex, called MBF, may also be responsive to Cln3p. Transcripts potentially regulated by Cln3p through the MBF include the CLB5 and CLB6 cyclins (50), as well as other genes involved in DNA replication. It is important to note that the Cln2p-Cdc28p complex is able to activate SBF- and MBF-dependent transcription, although not as efficiently as the Cln3p-Cdc28p complex (12, 56). For this reason, the Cln3p-Cdc28p complex is thought to be the physiologically relevant cdk complex regulating transcription in G1.
Since Cln2p supports viability in the cln− swi4− and cln− swi6− strains, while Cln3p does not, it is possible that Cln3p induces expression of proteins that carry out one of several events normally triggered more directly by Cln2p. The gene pairs CLB5-CLB6 (50) and PCL1-PCL2 (34, 42) are essential for Cln3p-dependent viability in the triple cln1 cln2 cln3 deletion strain (27). Each of these genes encodes a cyclin homologue that can bind to and activate the cdk Cdc28p or Pho85p, respectively. PHO85 is also essential for viability in cln1 cln2 cells (14), and Pho85p contributes to phosphorylation of the Sic1p inhibitor of Clbp-Cdc28p complexes (40). The molecular basis for lethality of cln1 cln2 pcl1 pcl2 or cln1 cln2 pho85 strains is not well established.
The activation of Clb cyclins results from a combination of cellular events that are regulated by Clnp-Cdc28p protein kinase activity. As described above, Cln3p may regulate the MBF-dependent transcription of CLB5-CLB6. In addition to this, Clnp-Cdc28p-dependent phosphorylation leads to the SCF-dependent ubiquitination and degradation of Sic1p (15, 61). Sic1p is a stoichiometric inhibitor of Clbp-Cdc28p protein kinase activity (35, 49). Overexpression of Sic1p results in growth arrest, due to inactivation of Clbp-Cdc28p activity, and Cln2p is able to suppress this growth arrest (27, 58). Finally, the Clnp-Cdc28p activity may be involved in regulating the phosphorylation state of Hct1p. Unphosphorylated Hct1p promotes the activation of the anaphase-promoting complex toward Clb2p (22, 48, 62, 68). Anaphase-promoting complex activity leads to ubiquitination and subsequent degradation of Clb2p by the proteosome (67).
Events required for proper cell cycle progression that may be regulated by Cln2p, rather than Cln3p, include cell polarization (29) and bud emergence (3, 8). The ability of CLN3 to complement the triple cln1 cln2 cln3 deletion is strongly reduced in the absence of the BUD2 gene due to failure of bud emergence, indicating that this gene product is important for Cln3p-dependent morphogenesis. Cln2p-dependent viability in the triple cln1 cln2 cln3 deletion strain is not influenced by deletion of BUD2 (27). In wild-type cells, Bud2p is not essential and is involved in bud site selection (5).
Intrinsic qualitative differences between Cln2p and Cln3p (as opposed to simple quantitative or timing differences in expression or associated kinase activity) have been demonstrated through a comparison of CLN3::CLN2 (CLN2 under the control of the CLN3 promoter), CLN3, and cln2 mutants by using strains that distinguish genetically between Cln2p and Cln3p activity (27). However, this study did not establish a molecular basis for the functional specificity. We address here two possible mechanisms. First, the cyclin molecule might confer structural differences to the cdk active site, changing the substrate specificity of the kinase. Second, differences in subcellular localization might bring Clnp-Cdc28p complexes into contact with different subsets of Clnp-Cdc28p protein kinase substrates.
Studies of higher eukaryotic cyclin molecules suggest that localization is important for cyclin function. Cyclins E, A, D1, and B1 have been implicated in nuclear events and show nuclear localization when the cyclin-cdk activity is required for function (2, 4, 11, 20, 25, 31, 32, 43, 45). The correlation is extended by the fact that a cyclin D1 mutant that is unable to enter the nucleus is unable to activate DNA replication in fibroblasts (11). The ability of cyclin B1 to promote mitosis in frog eggs is abolished in mutant cyclin B1 that does not localize to the nucleus (30). Also, cyclin B1 that is targeted to the nucleus through addition of a nuclear localization signal (NLS) or through disruption of the cyclin B1 nuclear export signal (NES) shows a defect in DNA damage-induced G2 arrest (23, 57). Different cyclins may be localized by distinct mechanisms. The redistribution of cyclin D1 to the cytoplasm is triggered by a glycogen synthase kinase 3β-dependent phosphorylation event (10). The distinct localization pattern of cyclin B1, which accumulates in the nucleus during mitosis, results from a balance between CRM1-mediated nuclear export and nuclear import most likely mediated by the importin α/β complex (36, 44, 66).
In the study presented here, we analyze differential localization of Cln2p and Cln3p in Saccharomyces cerevisiae. The localization patterns are distinct, and our data support the idea that these distinct patterns contribute to cyclin functional specificity.
MATERIALS AND METHODS
The relevant genotypes of the yeast strains used are shown in Table 1. All strains are congenic with BF264-15D (46). YPDex, YPGal, and synthetic complete (SC) media for yeast growth were prepared as described earlier (51). Yeast cells were transformed with plasmid DNA as described previously (18).
TABLE 1.
Strains used in this study
| Strain | Relevant genotypea |
|---|---|
| 1255-5C | MATa CLN1 CLN2 CLN3 |
| 1607-2D | MATa cln1-del cln2-del cln3-del leu2::LEU2::GAL1::CLN3 |
| 1446-10B | cln1-del cln2-del cln3-del bck2::ARG4 leu2::LEU2::GAL1::CLN3 arg4 |
| 1456-10 | MATa cln1-del cln2-del cln3-del swi4::LEU2 pURA3/GAL1::CLN1 |
| 2866-9D | MATa cln1-del cln2-del cln3-del swi6::LEU2 pURA3/GAL1::CLN1 |
| 1587-7 | MATα cln1-del cln2-del CLN3+ clb5::ARG4 clb6::ADE1 leu2::LEU2::GAL1::CLN1 arg |
| 1238-11A-7 | MATa cln1-del cln2-del CLN3+ URA3::GAL1::SIC1 |
| 1591-10D | MATa cln1-del cln2-del cln3-del pcl1::HIS3 pcl2::URA3 leu2::LEU2::GAL1::CLN1 |
| 1472-24D | MATa cln1-del cln2-del cln3-del bud2::URA3 leu2::LEU2::GAL1::CLN1 |
| 1492-21D | CLN1 cln2 cln3 LEU2::GAL1::CLN3 |
| 2195-5c | MATa CLN1 CLN2 CLN3 LEU2::GAL1::CLN2ha |
| 1022 | MATa CLN1 CLN2 CLN3 LEU2::GAL1::CLN3ha |
| MMY1 | MATa CLN1 CLN2mycCLN3 LEU2 |
| MMY2 | MATa CLN1 CLN2 CLN3 leu2::LEU2::GAL1::CLN3hamyc |
| MMY6-7D | MATa cln1 cln2 CLN3 swi4::LEU2 |
All strains are congenic with BF264-15D (MATa trp1 leu2 ura3 ade1 his2).
Plasmids.
The 9X myc epitope-tagged GAL1::CLN2myc (AG4) and GAL1::CLN3hamyc (AG2) were derived from the plasmids KH100 and KL002-4 (27) by A. Gartner. The 9X myc tag is engineered at the C terminus of the cyclin proteins in these constructs. The Cln2myc integration plasmid (pMM56) was constructed by replacing the NotI cassette containing the 3× hemagglutinin (HA) epitope tag of MT104 with the NotI cassette containing the 9X myc epitope tag of AG2. This construct contains a CLN2 promoter driven CLN2myc and integrates at the endogenous CLN2 locus. The GAL1::CLN3hamyc integration plasmid (pMM64) was constructed by inserting the NotI cassette containing the 9X myc epitope tag of AG2 into the NotI site of pKL036. pKL036 results from the insertion of the SalI-SacII fragment of pKL002-4 (27) into the polylinker of pRS404 (52). The pMM64 construct was used for ectopic integration of the GAL1::CLN3hamyc cassette at the URA3 locus.
The CLN2myc and CLN3hamyc plasmids (pMM82 and pMM45, respectively) each contain a 9X myc epitope-tagged CLN gene under the control of the CLN3 promoter in a low-copy-number TRP1 vector and were constructed as follows. The SalI-ClaI GAL1 promoter cassette of AG2 and AG4 were replaced with the SalI-ClaI CLN3 promoter cassette of pKL001 (27). Sequencing shows a base change within the carboxy-terminal tail of CLN3 used in these studies. This base change results in an amino acid change of proline 462 to glutamic acid (P462Q). Cln3 constructs that contain a proline at position 462 were constructed and tested for function to ensure that this base change did not alter Cln3p function. No differences in protein levels, the presence of the 35-kDa Cln3p species (see below), localization patterns, or CLN function (as defined by cell volume assay, cln-complementation assay, or mutant strain rescue assay as described here) were observed between strains containing pMM45 (CLN3 with P462Q) and pMM99 (CLN3 with P462) (data not shown). The pMM99 construct contains a 9X myc epitope-tagged CLN3 gene (with no HA epitope tag) under the control of the CLN3 promoter in a low-copy-number TRP1 vector. The pMM99 was constructed by PCR amplification of the CLN3 coding sequence by using the plasmid pBSI6 as a template. The 3′ primer for the CLN3 amplifications was 5′-CTAGCGGCCGCTGCGAGTTTTCTTGAGGTTGCTACTATC-3′ and the 5′ primer was 5′-CGAATTGCCCGAGTAGTCTCC-3′. PCR products were subsequently cloned into TOPOII vector with topoisomerase-based ligations (Invitrogen). EcoRI-NotI cassettes from the TOPOII clones were used to replace the EcoRI-NotI CLN3 sequences of pMM45 to give rise to pMM97. The 9X myc epitope tag NotI cassette from pAG4 was cloned into the NotI site following the CLN3 sequences in pMM97 to give rise to pMM99. The pMM99 plasmid was sequenced to confirm that CLN3 coding sequences were followed in frame by the 9X myc epitope tag, and no errors were introduced during the PCR amplification.
The 9X myc epitope-tagged CLN3-1hamyc construct (pMM55) was made by inserting the 9X myc epitope NotI fragment of AG2 at the NotI site immediately following the 3X HA tag of pKL037. This results in CLN3-1hamyc driven from the CLN3 promoter on a low-copy TRP1 vector. The pKL037 plasmid was made by replacing the BamHI-EcoRI fragment of pKL001 with the BamHI-EcoRI fragment of MT9. Constructs were sequenced to confirm that the NotI fragment insertion gave rise to the 9X myc tag expressed in frame with the CLN3-1ha coding sequence.
NLS-CLN2myc (pMM60), mnls-CLN2myc (pMM61), NES-CLN2myc (pMM92), and mnes-CLN2myc (pMM93) were constructed by PCR amplification of AG4 template sequences with a 5′ primer that encodes a ClaI restriction endonuclease site followed by the NLS, nls (39), NES, or nes (17, 54, 63) localization signals in frame with CLN2. PCR products were subsequently cloned into TOPOII vector with topoisomerase-based ligations (Invitrogen). ClaI-SpeI cassettes from the TOPOII clones were used to replace the ClaI-SpeI cassette of pMM82. The 5′ primers for the CLN2 PCR amplifications are as follows (with the ClaI restriction site indicated in brackets, the localization signal underlined, and the bases changed in mutant nls or nes sequences in lowercase letters): NLSCLN2, 5′- GG[ATCGAT]CCGACAGACAATGCCCAAGAAGAAGCGGAAGGTCGC TAGTGCTGAACCAAGACCCCG-3′; nlsCLN2, 5′-GG[ATCGAT]CCGACA GACAATGCCCAAGAcGAAGCGGAAGGTCGCTAGTGCTGAACCAAG ACCCCG; NESCLN2, 5′-CC[ATCGAT]CGACAGACAATGGAATTAGCCT TGAAATTAGCAGGTCTTGATATCAACAAGACAGCTAGTGCTGAACC AAGACCCCG-3′; and nesCLN2, 5′-CC[ATCGAT]CGACAGACAATGGAATTAGCCTTGAAATTAGCAGGTgcTGATATCAACAAGACAGCTAGTGCTGAACCAAGACCCCG-3′. The 3′ primer for the CLN2 PCR reactions is 5′-CTGAGCAGCGTAATCT-3′. The NLS-CLN3hamyc (pMM74), nes-CLN3hamyc (pMM75), NES-CLN3hamyc (pMM76), and nes-CLN3hamyc (pMM77) plasmids were constructed by the same method as that used for the CLN2 plasmids. In this case, the template for PCR reactions was the pKL001 plasmid and the ClaI-EcoRI cassettes from the TOPOII clones were used to replace the ClaI-EcoRI cassette of pMM45. The 5′ primers for these PCR amplifications are as follows: NLSCLN3, 5′-GG[ATCGAT]CCGACAGACAATGCCCAAGAAGAAGCGGAAGGTCGCCATATTGAAGGATACCATAATT-3′; nlsCLN3, 5′-GG[ATCG AT]CCGACAGACAATGCCCAAGAcGAAGCGGAAGGTCGCCATATTG AAGGATACCATAATT-3′; NESCLN3, 5′-CC[ATCGAT]TTTCTGTACGAT GGAATTAGCCTTGAAATTAGCAGGTCTTGATATCAACAAGACAGCC ATATTGAAGGATACCATAATT-3′; and nesCLN3, 5′-CC[ATCGAT]TTTC TGTACGATGGAATTAGCCTTGAAATTAGCAGGTgcTGATATCAACAA GACAGCCATATTGAAGGATACCATAATT-3′. The 3′ primer for the CLN3 PCR amplifications was 5′-GATGGTTTCCAATGCTTGTGACGCGTAGAATTCTTC-3′. All constructs were sequenced to confirm that no errors were introduced into coding sequences during the PCR amplifications.
The NES-CLN3-1hamyc (pMM83), nes-CLN3-1hamyc (pMM84), NLS-CLN3- 1hamyc (pMM85), and nls-CLN3-1hamyc (pMM86) were constructed by replacing the ClaI-EcoRI fragment of pMM55 (CLN3-1hamyc) with the EcoRI-NotI fragment of pMM74 to pMM77 for pMM83 to pMM86, respectively.
Indirect immunolocalization.
Exponentially growing cultures expressing the myc-tagged Cln proteins were fixed in growth medium with formaldehyde (1:10 dilution) for 90 min at 30°C with rotation. Cells were washed with phosphate-buffered saline (PBS), briefly sonicated, washed again in PBS, and then washed in sorbitol citrate buffer (0.1 M K2HPO4, 33 mM citric acid, 1.2 M sorbitol, 2 mM dithiothreitol [DTT]). Cells walls were removed by digestion with a 1:10 volume of glusalase, a 1:100 volume of a 20-mg/ml concentration of zymolyase, and 1 mM DTT for 2 h at 30°C with rotation. Cells were washed four times with sorbitol citrate buffer. Cells were fixed to polylysine-coated slides by placing the cell suspension in wells for 3 min and aspirating off the suspension until they were dry. The slides were placed in 100% Methonal for 5 min, 100% acetone for 5 min, and then air dried. Fixed cells were then rehydrated and blocked by incubation with 2% nonfat dry milk in PBS-Tween solution (1× PBS, 0.2% Tween 20) overnight at 4°C. Primary monoclonal antibody 9E10 (Santa Cruz Biotechnology) was diluted 1:200 in blocking solution, cleared for 2 min by centrifugation in a microfuge, and incubated on the cells for 2 h. The wells were washed quickly four times, followed by three 5-min washes. Secondary antibody (Cy3-conjugated anti-mouse immunoglobulin G; Jackson Immunochemicals) was diluted 1:200 in blocking solution, cleared, and incubated on the wells for 2 h. Cells were again washed, and mounting solution (10% 1× PBS in glycerol with 0.02 μg of DAPI [4′,6′-diamidino-2-phenylindole] and 1 mg of phenylenediamine per ml) was added to the wells. The negative control for integrated myc-tagged Cln proteins was the wild-type strain 1255-5C. The negative control for plasmid-based expression of myc-tagged Cln proteins was the vector control, pRS414 (52).
Immunofluorescence was done on an Axioplan Universal Microscope (Carl Zeiss, Inc.) by using a Hamamatsu digital camera. Images were collected by using Openlab software version 1.2 (Improvision) and processed with Photoshop version 4.0 (Adobe Systems). All images presented in a single figure were captured and processed in parallel by identical means.
Biochemical fractionation.
Biochemical fractionation was carried out as described elsewhere (24, 47), with the following modifications. Culture volumes were scaled down to 1 liter, and cells were allowed to recover from spheroplasting for 1 h in YPDex (for CLN2myc) or YPGal (for GAL1::CLN3hamyc) before the cells were harvested and lysed by Polytron shearing. Western blots of different fractions from these preparations were probed with the anti-myc polyclonal antibody A-14 (Santa Cruz Biotechnologies) and anti-Pgk1p polyclonal antibody (Molecular Probes) to visualize the cytoplasmic fractions and with anti-Nop1p antibody (M. Rout) to visualize the nuclear fractions.
α-factor synchronization.
α-Factor was added to 500 ml of exponentially growing cells to a final concentration of 0.6 mM until they were synchronized, as determined by morphology (2 h for CLN2myc strain grown in YPDex). α-Factor was washed out with 500 ml of medium, then briefly sonicated, and finally washed again. Washing consisted of collecting cells by filtration and resuspending cells in a 500 ml of medium. After the second wash, cells were resuspended in 30°C YPDex medium. Samples were collected every 15 min and assayed by indirect immunofluorescence and Western blot analysis. Synchrony was confirmed by determining the budding index.
Cell volume assay.
Cell volume assays were carried out on a cln2 cln3 GAL1::CLN3 strain grown on glucose-containing medium that had been transformed with the following plasmids: pMM45, pMM55, pMM82, pMM60, pMM61, pMM92, pMM93, pMM83, pMM84, pMM85, pMM86, and pRS416. Subsequent experiments were done with this strain transformed with pMM99 and pRS416. All plasmids are episomal centromeric low-copy plasmids and carry the TRP1 gene. For each strain, three individual transformants were assayed as described earlier (27), with the following modifications. Overnight cultures grown in defined (SC) medium containing 3% galactose and lacking tryptophan (SCGal-trp) were used to inoculate SC medium containing 2% glucose lacking tryptophan (SCDex-trp) and grown overnight at 30°C to an optical density at 660 nm (OD660) of approximately 1.0. These cultures were then diluted to an OD660 of approximately 0.2 in SCDex-trp and allowed to grow at 30°C to an OD660 of 0.8. Cells were then fixed in 1% formaldehyde and sonicated to disperse the clumps. Cell volume analysis was done with a Coulter Channelyzer 256. The data shown are representative of at least two independent experiments for each strain.
Cell viability assays.
Mutant strains (see Table 1) were transformed with the following plasmids: pMM45, pMM55, pMM82, pMM60, pMM61, pMM92, pMM93, pMM83, pMM84, pMM85, pMM86, pMM99, and pRS416. For each transformant strain, 10-fold serial dilutions were prepared for two pools of transformants (5 to 10 colonies), and 5 μl of each dilution were plated onto a YPDex (dextrose) or YPGal (galactose) plate and assayed for growth at 30 and 38°C after 2 to 3 days. The maximum decrease in viability that can be determined from this assay is 1,000-fold. Plates were replica plated to SCGal-trp and SCDex-trp to confirm that colonies forming on YPDex and YPGal plates maintained the test plasmids. The data presented are representative of data from at least two independent experiments.
Protein extraction and immunoblotting.
Total cellular protein lysates were obtained as follows: 10 ml of exponentially growing yeast cultures (OD660 between 0.5 and 0.9) were quick chilled by pouring over ice. The remaining steps were done at 4°C. The cells were spun, and the pellet was resuspended in 1 ml of 1× TE (pH 7.4; 10 mM Tris, 1 mM EDTA) and transferred to a 1.5-ml microfuge tube. Cells were spun for 5 s in a microcentrifuge, and the pellets were resuspended in 100 μl of extraction buffer with inhibitors (0.6% sodium dodecyl sulfate [SDS]; 10 mM Tris, pH 7.4; 10% aprotinin; 1:1,000 [vol/vol] 0.5 M phenylmethylsulfonyl fluoride; 1:100 [vol/vol] 1 mg of leupeptin-pepstatin per ml; 1:10 [vol/vol] 100 mM NaPPi, pH 7.3). An equivalent of 150 μl of glass beads was added, and the bead slurry was vortexed for 3 min. Then, 100 μl of 2× SDS sample buffer was added, and the lysates were analyzed by immunoblotting. Cellular lysates containing the 35-kDa Cln3hamycp were also prepared by lysis in 1.85 N NaOH and 7.4% β-mercaptoethanol, followed by trichloroacetic acid precipitation. Identical results were obtained by using both lysis protocols. Lysates were run on SDS-polyacrylamide gels and transferred to immunoblots as described previously (7). Immunoblotting was performed as described previously (27). The antibody used to detect the myc epitope was polyclonal α-myc A-14 (Santa Cruz Biotechnology), and the antibody used to detect the HA epitope in the peptide kinase assays was polyclonal antibody 12CA5 (Babco). Detection was done by enhanced chemiluminescence (ECL) with the super signal ECL Kit (Pierce).
Immunoprecipitation and peptide kinase assays.
Immunoprecipitations were performed as described earlier (27), with the following modifications. The HA epitope-tagged Cln strains (2195-5c for Cln2hap and 1022 for Cln3hap expressed from the GAL1 promoter) were scaled up to 500 ml and resuspended in a final volume of 300 μl of kinase buffer. Extraction buffer N (50 mM Tris-HCl, pH 7.5; 100 mM NaCl; 0.1 mM EDTA, pH 8.0; 10 mM NaF; 60 mM β-glycerophosphate; 0.1% NP-40) was used instead of TNN buffer. Immunoprecipitates were washed with extraction buffer five times instead of three times. Then, 15 μl of the final suspension was used for each peptide kinase assay. The cell suspension was incubated with various concentrations of peptide substrate in a volume of 2 μl, with 2 μl of 50 mM ATP and 1 μl of [γ-32P]ATP (NEN). The kinase reaction mixtures were incubated at 30°C for 5 min. The reactions were stopped by adding 18 μl of the reaction mixture to 2 μl of 0.5 M EDTA, followed by a 10-min incubation at 65°C. The reaction was then spotted onto phosphocellulose paper disks and processed as described elsewhere (53). D. Lawrence kindly provided the peptides. Each assay was performed in duplicate, and assays were done from at least three separate immunoprecipitations. The apparent Km (± the standard error of the mean [SEM]) and Vmax (± SEM) values were determined from initial rate experiments that included five different peptide concentrations over a 10-fold range encompassing the Km. These data were plotted by using the Lineweaver-Burke procedure; the corresponding plots proved to be linear.
RESULTS
Enzyme kinetics of Cln2p-Cdc28p and Cln3p-Cdc28p kinase activities.
To investigate the possibility that the functional specificity observed between Cln2p and Cln3p reflects differences in substrate specificity, we measured the phosphorylation kinetics of Cln2p-Cdc28p and Cln3p-Cdc28p with previously characterized peptide substrates (53). The amino acid sequence of these peptides is based on the minimal necessary sequence for substrate recognition by cyclin-dependent kinase, S/T-P-X-Z, where Z is typically basic (see Table 2 for peptide sequences). To allow analysis of Cln2p- and Cln3p-associated Cdc28p activity, immunoprecipitated complexes of HA epitope-tagged cyclin were used as the source of cyclin-associated kinase activity. The Vmax is expressed relative to the amount of Cdc28p present in coimmunoprecipitation, as determined by Western blot analysis (see Materials and Methods).
TABLE 2.
Km, Vmax, and Kcat/Km valuesa
| Peptide (sequence) | CLN type | Km (μM) | Vmax (μmol min−1 mg−1) | Kcat/Km (min−1 μM−1) |
|---|---|---|---|---|
| P (Ac-Ser-Pro-Gly-Arg-Arg-Arg-Arg-Lys-amide) | CLN2 | 1.63 ± 0.22 | 646 ± 123 | 13,839 ± 3,583 |
| CLN3 | 2.32 ± 0.37 | 777 ± 62 | 13,190 ± 3,675 | |
| P-1 (Ac-Asp-Ser-Pro-Gly-Arg-Arg-Arg-Arg-Lys-amide) | CLN2 | 51.35 ± 7.8 | 735 ± 105 | 511 ± 71 |
| CLN3 | 55.57 ± 7.5 | 1,189 ± 312 | 661 ± 250 | |
| P+2 (Ac-Ser-Pro-Arg-Arg-Arg-Arg-Arg-Lys-amide) | CLN2 | 2.40 ± 0.39 | 793 ± 137 | 11,550 ± 3,383 |
| CLN3 | 3.33 ± 0.60 | 648 ± 108 | 6,725 ± 1,691 |
Kinetic constants were determined as described in Materials and Methods section. Values are given as the mean ± the SEM.
To determine if the Cln2p- and Cln3p-associated kinase activity measured in these assays are due to associated Cdc28p, we measured the Cln2p- and Cln3p-dependent phosphorylation of peptide substrate P from a strain containing a mutant Cdc28p. This mutant Cdc28csr1-1p (28) is defective for binding to Cln2p and Cln3p. Cln2p and Cln3p immunoprecipitates from cdc28csr1-1 cells show reduction in peptide phosphorylation that is consistent with the reduction in Clnp-Cdc28p complex formation observed by Western blot analysis (data not shown).
Peptide P is an efficient substrate of cyclin B-cdc2 from Pisaster ochraceus, with a Km of 1.5 ± 0.04 μM and a Vmax of 2 ± 0.18 μmol/min/mg (53). We find that this peptide also serves as an efficient substrate for the yeast Cln2p and Cln3p-Cdc28p (Table 2). We find no significant difference between Cln2p and Cln3p complexes in binding affinity (Km), rate of catalysis (Vmax), or overall efficiency (Kcat/Km) for this peptide (Table 2). The presence of an aspartic acid residue at the −1 position relative to the phosphorylated serine (P-1) reduced binding affinity approximately 15-fold, with no changes in the catalysis rate (Table 2, P-1). These results are similar to those found by using cyclin B-cdc2, which showed an approximate 10-fold reduction in Km (53). When the P+2 glycine was substituted with arginine, no significant differences were observed when compared to peptide P (Table 2).
Although limited, these data did not suggest that the catalytic core of Cdc28p was folded very differently when in complex with Cln2p compared to Cln3p and prompted us to look elsewhere for the source of functional specificity. Data on mammalian cyclins have similarly shown little specific effect on peptide substrate specificity due to the cyclin subunit associated with a given Cdk (21).
Cellular localization of Cln2p.
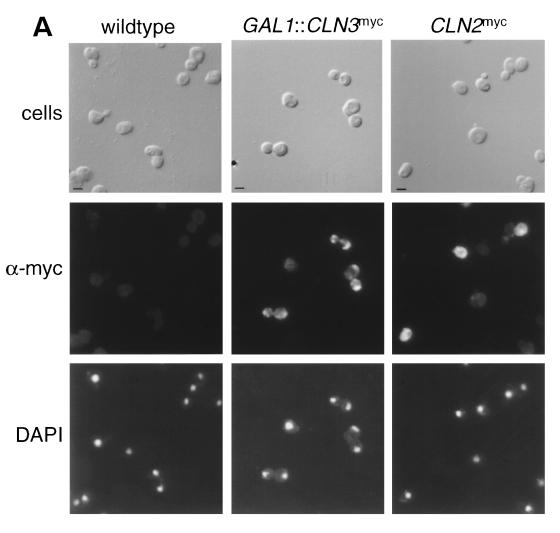
The functional specificity of Cln2p-Cdc28p and Cln3p-Cdc28p complexes may result from differential localization of cyclins, making distinct subsets of substrates or cyclin-cdk regulators accessible to different cyclin-Cdc28p complexes. Cellular localization of Cln2p and Cln3p were determined by using indirect immunofluorescence of 9X myc epitope-tagged proteins. We myc epitope tagged the carboxy terminus of CLN2 at the endogenous CLN2 locus (see Materials and Methods). The integration results in the expression of myc epitope-tagged Cln2p under the control of the endogenous CLN2 promoter (Cln2mycp). The staining pattern for Cln2mycp appears to be primarily cytoplasmic and somewhat punctate, with the nucleus appearing understained relative to the cytoplasm (Fig. 1A). This signal was specific to Cln2mycp and occurred in unbudded and small budded cells, a finding consistent with the cell cycle-regulated expression pattern of CLN2. To confirm that the signal observed by indirect immunofluorescence of asynchronous cultures was due to Cln2mycp, we assayed for localization by using a synchronized culture. In a culture synchronized by α-factor block and release, cytoplasmic Cln2mycp signal was detected by indirect immunofluorescence almost uniformly in the population at the same time (at or just prior to bud emergence) that Cln2myc protein peaked, as determined by Western blot analysis (data not shown).
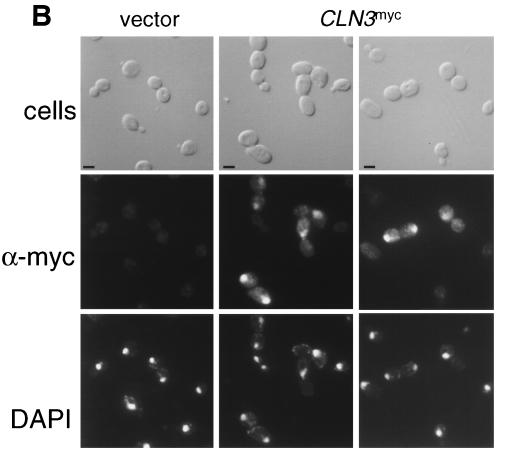
FIG. 1.
Indirect immunolocalization of Cln2p and Cln3p. (A) Strains expressing Cln2mycp from the CLN2 promoter (strain MMY1) and Cln3hamycp from the GAL1 promoter (strain MMY2). (B) Cln3hamycp from the CLN3 promoter (1255-5C with pMM45). Strains were assayed by indirect immunofluorescence as described in Materials and Methods. The first row shows DIC images (cells); the second row shows indirect immunofluorescence where monoclonal anti-myc 9E10 (Santa Cruz Biotechnology) was used as the primary antibody (α-myc), and the third row shows the DAPI staining of DNA. The myc-tagged cyclins expressed are indicated at the top of each set. The Cln2mycp signal appeared to be punctate and cytoplasmic. At a low frequency, spots of Cln2mycp signal are visible but do not lie within the area of the nucleus as defined by the DAPI stain. Nuclear Cln3-1hamycp staining was usually throughout the area of the nucleus, but on rare occasions (<0.5% of the cells with overexpressed Cln3-1hamycp) the staining appeared along the edge of the nucleus. Bar, 5 μm.
Cellular localization of Cln3p.
The localization pattern of Cln3p was visualized by using the 9X myc epitope placed in frame at the carboxy terminus of the CLN3hap. Since the expression level of Cln3p is significantly lower than the peak expression of cell cycle-regulated Cln2p, we first characterized the localization of overexpressed myc epitope-tagged Cln3p by using the galactose-inducible GAL1 promoter (GAL1::CLN3hamyc). The GAL1::CLN3hamyc construct was integrated at the URA3 locus in strain 1255-5c, and cells grown in the presence of galactose were assayed by indirect immunofluorescence. Overexpressed Cln3hamycp is present throughout the cell, with a concentration of Cln3hamycp in the nucleus of some cells (Fig. 1A). The cells that accumulate Cln3hamycp in the nucleus consist primarily of budded cells.
To determine if the galactose-induced overexpression influenced the localization pattern of Cln3hamycp, indirect immunofluorescence was done on a wild-type strain containing a low-copy-number plasmid that encodes CLN3hamyc under the control of the CLN3 promoter. Cln3hamycp expressed at endogenous levels localizes to the nuclei of large budded cells. On occasion (<10%), large unbudded cells show nuclear accumulation of Cln3hamycp, but cells with small buds or small unbudded cells do not show Cln3hamycp signal (Fig. 1B and Fig. 4). It is unclear if Cln3hamycp is below the level of detection or absent from these populations of cells. It is interesting to note that the time of nuclear accumulation of Cln3p may roughly coincide with the early cell cycle box-, or ECB-, dependent peak in CLN3 transcription (16, 33, 41, 59, 64), although a similar pattern was observed with the cell-cycle-constitutive GAL1 promoter.
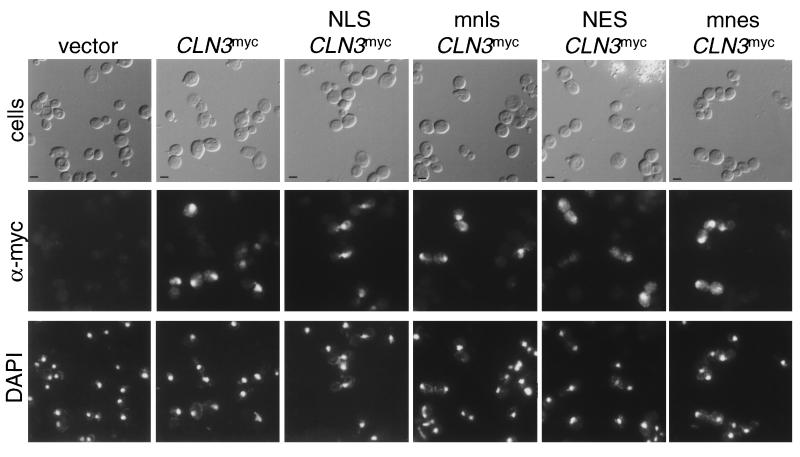
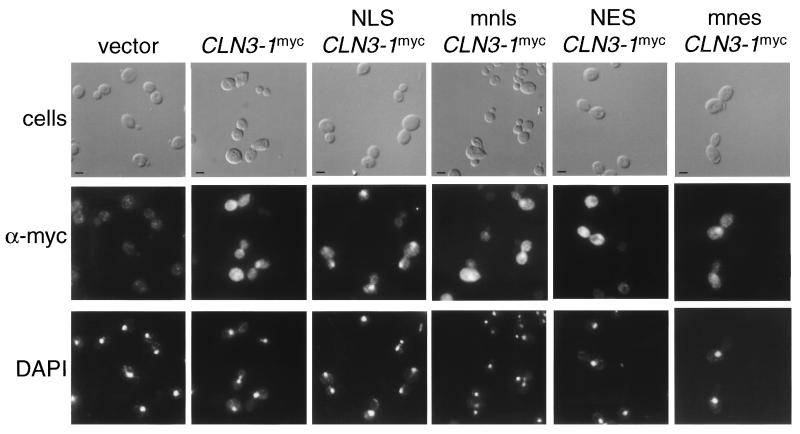
FIG. 4.
Indirect immunolocalization of NLS- and NES-Cln3mycp. Wild-type cells (1255-5c) were transformed with plasmids pRS414 (vector), pMM99 (Cln3mycp), pMM100 (NLS-Cln3mycp), pMM101 (mnls-Cln3mycp), pMM102 (NES-Cln3mycp), and pMM103 (mnes-Cln3mycp). All plasmids are episomal CEN (low copy number) and express the myc-tagged Cln3 protein from the CLN3 promoter. Transformants were assayed by indirect immunofluorescence as described in Materials and Methods. The first row shows DIC images (cells), the second row shows the indirect immunofluorescence with monoclonal anti-myc antibody 9E10 (α-myc), and the third row shows DAPI staining of DNA. The myc-tagged cyclins expressed are indicated at the top of each set. Bar, 5 μm.
Cellular fractionation of Cln2p and Cln3p.
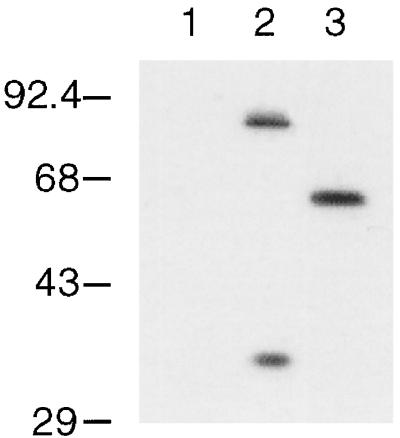
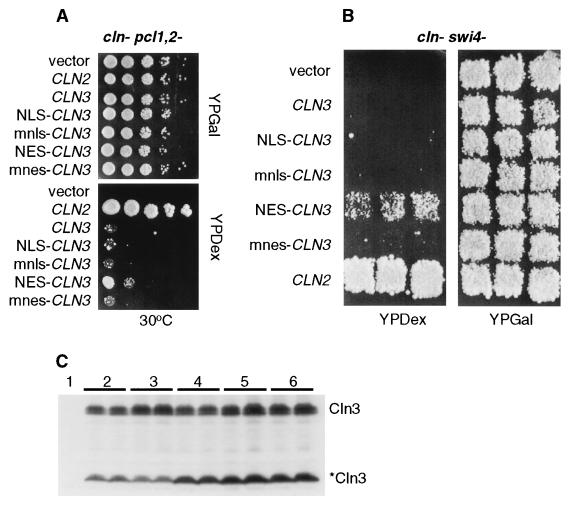
Western blot analysis with antibodies against the myc epitope indicate that a significant amount of the immunoreactive myc in lysates from cells expressing Cln3hamycp comes from a species of approximately 35 kDa (Fig. 2). An estimated 18 kDa of this protein is due to the 3X HA and 9X myc epitopes, leaving 17 kDa of Cln3p. This product is consistent with a cleavage event internal to Cln3p, one approximately 150 amino acids from the carboxy-terminal end of the protein. The 35-kDa species is observed in both anti-myc immunoprecipitations and crude cellular lysates. It is present when Cln3p is expressed from the GAL1 and CLN3 promoters. A similar species is observed when the protein A epitope is engineered at the carboxy terminus of CLN3 (data not shown). We have been unable to detect a faster-migrating species with the 3X HA epitope-tagged Cln3p, possibly because of the smaller size of this epitope. Experiments that use anti-Cln3p serum to detect untagged Cln3p on Western blots show a nonfunctional species shorter than full-length Cln3p (7), the size of which might result from the loss of C-terminal sequences. It remains unclear if the cleavage of Cln3hamycp resulting in the approximately 35-kDa species occurs in vivo or in vitro.
FIG. 2.
Immunoblot analysis of Cln3hamycp and Cln3-1hamycp. A wild-type strain (1255-5c) was transformed with plasmids pRS414 (vector, lane 1), pAG2 (Cln3hamycp expressed from the GAL1 promoter, lane 2), and pMM55 (Cln3-1hamycp, lane 3). All plasmids are episomal CEN (low copy number). Cellular lysates were separated by SDS–12% polyacrylamide gel electrophoresis (PAGE) and analyzed by Western blotting as described in Materials and Methods. Proteins were visualized by using the polyclonal anti-myc A-14 antibody (Santa Cruz Biotechnology). Molecular size markers are listed to the left of the blot in kilodaltons.
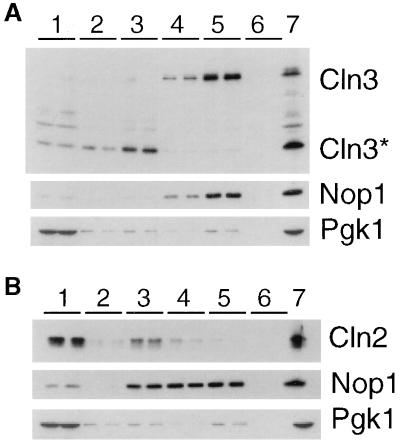
The immunofluorescence data described above may reflect the localization of both full-length and 35-kDa Cln3hamycp. To confirm our indirect immunofluorescence results and to determine where full-length Cln3hamycp localizes, we did cellular fractionation experiments to separate the cytoplasmic and nuclear components of the cell. Consistent with the indirect immunofluorescence studies, the Cln2myc protein cofractionates with cytoplasmic marker Pgk1p (Fig. 3B), while full-length Cln3hamycp cofractionates with the nuclear marker, Nop1p (Fig. 3A). The 35-kDa Cln3hamycp does not cofractionate with nuclear markers. The 35-kDa species is found primarily at the interface between the nuclear and cytoplasmic fractions, with low levels visible in the cytoplasmic fractions (see fraction 3, Fig. 3A). The interface between nuclear and cytoplasmic fractions usually contains the larger organelles of the cell, including the endoplasmic reticulum and Golgi (55). It is clear from these results that full-length Cln3hamycp is localized to the nucleus, while Cln2mycp is localized primarily to the cytoplasm. These results do not rule out the possibility that a fraction of Cln3hamycp is located in the cytoplasm, since it is possible that full-length Cln3p is degraded to the 35-kDa form after cell breakage.
FIG. 3.
Localization of Cln3p and Cln2p by subcellular fractionation. Fractions were prepared as described in Materials and Methods. Duplicate samples of each fraction were separated by SDS–12% PAGE and analyzed by Western blotting. Fractions are indicated at the top of each blot. Fraction 1 corresponds to the crude cytoplasmic fraction, fraction 2 corresponds to the surface of the sucrose gradient, fraction 3 corresponds to the surface-2.01 M sucrose interface, fraction 4 corresponds to the 2.01 to 2.1 M sucrose interface, fraction 5 corresponds to the 2.1 to 2.3 M sucrose interface, and fraction 6 corresponds to the remaining pellet of the sucrose gradient. The total protein isolated from cells prior to polytron shearing is indicated in the lane marked 7. (A) Immunoblot analysis of fractions from a strain expressing Cln3hamycp from the GAL1 promoter (MMY2). The position of full-length Cln3hamycp is indicated by Cln3, and the 35-kDa species is indicated by Cln3*. Corresponding immunoblot analysis of the Nop1p and Pgk1p fractionation is shown below the Cln3p blot. The nucleolar protein Nop1p detected with the monoclonal antibody D77 (1) indicates which fractions contain nuclear proteins. The cytoplasmic protein Pgk1p detected with the anti-Pgk1p polyclonal antibody (Molecular Probes) indicates which fractions contain cytoplasmic proteins. (B) Immunoblot analysis of fractions from a strain expressing Cln2mycp from the CLN2 promoter (MMY1). The Cln2p, Pgk1p, and Nop1p immunoblots are shown. A moderate level of nuclear breakage apparently occurred in this fractionation, as indicated by leakage of Nop1p into the intermediate and cytosolic fractions.
Cellular localization of Cln3-1p.
If the 35-kDa Cln3hamycp results from a cleavage within the C-terminal tail of Cln3p, then a mutant Cln3p lacking the C-terminal tail would be missing the presumed site of cleavage and would not produce the 35-kDa Cln3hamycp. To test this idea, we 9X myc epitope tagged the truncated allele of CLN3ha, CLN3-1ha. CLN3-1 encodes a stable Cln3-1p that is more active than full-length Cln3p, as measured by cell size experiments, presumably because of the increase in steady-state levels of Cln3-1p (7, 60, 65). Protein lysates from a strain expressing Cln3-1hamycp do not show faster-migrating species (Fig. 2, lane 3), a result consistent with the idea that a cleavage site falls within the C-terminal tail of Cln3. Indirect immunofluorescence of Cln3-1hamycp shows a staining pattern similar to that of overexpressed Cln3hamycp, with localization to the nucleus in budded cells and localization throughout the cell in unbudded cells (see Fig. 5). Comparison of Cln3-1p localization and Cln3p localization shows an increase in cytoplasmic staining with Cln3-1hamycp (see Fig. 1 and 5).
FIG. 5.
Indirect immunolocalization of NLS– and NES–Cln3-1hamycp. Wild-type cells (1255-5c) were transformed with plasmids pRS414 (vector), pMM55 (Cln3-1hamycp), pMM83 (NLS–Cln3-1hamycp), pMM84 (nmls–Cln3-1hamycp), pMM85 (NES–Cln3-1hamycp), and pMM86 (mnes–Cln3-1hamycp). All plasmids are episomal CEN (low copy number) and express the myc-tagged Cln protein from the CLN3 promoter. Transformants were assayed by indirect immunofluorescence as described in Materials and Methods. The first row shows DIC images (cells), the second row shows the indirect immunofluorescence with monoclonal anti-myc antibody 9E10 (α-myc), and the third row shows DAPI staining of DNA. The myc-tagged cyclins expressed are indicated at the top of each set. Bar, 5 μm.
Altering cellular localization of Cln3p.
To determine the functional relevance of Clnp localization patterns, we attempted to redirect localization by engineering localization signals on the N terminus of Cln3p. To facilitate exit from the nucleus, the PKI NES was used. The NES consists of a leucine-rich sequence that is necessary and sufficient to mediate nuclear export (63). This export signal is functional in S. cerevisiae (17, 54). We also engineered the simian virus 40 large T NLS onto Cln3p. The NLS consists of a short lysine-rich sequence that is sufficient to mediate nuclear import of a heterologous protein in yeast cells (39). Point mutants of the NLS (mnls) and NES (mnes) that disrupt localization activity of these sequences were used to control for nonspecific effects of the additional sequences at the N terminus of the proteins. Indirect immunofluorescence shows no changes in the cellular localization of the proteins. Indirect immunofluorescence shows no changes in the cellular localization of the NLS-Cln3mycp expressed from the CLN3 promoter compared to the Cln3mycp or mnls-Cln3mycp patterns. A slight increase in cytoplasmic staining was observed with the NES-Cln3mycp compared to Cln3mycp or mnes-Cln3mycp (Fig. 4 and data not shown).
In contrast, when we placed the NLS, mnls, NES, and mnes at the N terminus of Cln3-1hamycp, we detected clear NLS- and NES-dependent changes in the cellular localization of Cln3-1hamycp. NES-Cln3-1hamycp is localized primarily to the cytoplasm, with lighter staining in the region of the nucleus compared to Cln3-1hamycp and mnes-Cln3-1hamycp. NLS-Cln3-1hamycp localizes almost entirely to the nucleus in both budded and unbudded cells compared to Cln3-1hamycp or mnls-Cln3-1hamycp (Fig. 5). The mnls- and mnes-Cln3-1hamycp localization patterns most resemble those of wild-type Cln3-1hamycp, although a slight shift of signal into the nucleus with mnls and into the cytoplasm with mnes is detectable. In all cases, the immunofluorescence data shown is representative of the population of cells and is reproducible in different experiments. These data show that addition of the NLS shifts the Cln3-1p localization to a pattern that more resembles that of Cln3p and that addition of the NES shifts Cln3-1p to a localization pattern that more resembles that of Cln2p.
The increased ability to mislocalize Cln3p when C-terminal sequences are deleted suggests that the Cln3p C-terminal third may include endogenous localization signals, a possibility that we are exploring. This idea is consistent with the significant loss of tight nuclear localization when Cln3-1p is compared with Cln3p (Fig. 5). Alternatively, the differences in the localization patterns of Cln3p and Cln3-1p may be due to differences in the turnover rates of these two proteins.
Altering cellular localization of Cln2p.
Attempts to change the localization pattern of Cln2mycp were carried out as described for Cln3hamycp. Sequences comprising the NLS, mnls, NES, and mnes sequences were engineered at the N terminus of Cln2mycp and assayed for localization by indirect immunofluorescence. Expression of the NLS-Cln2mycp does not significantly change the localization pattern compared to mnls-Cln2mycp or Cln2mycp. Some nuclear accumulation of NLS-Cln2mycp (but not of Cln2mycp and mnls-Cln2mycp) is observed when overexpressed from the GAL1 promoter, but cytoplasmic staining is still obvious (data not shown). Thus, while the NLS is functional, it is unable to confer efficient nuclear localization to Cln2mycp. As for Cln3p, this may be due to efficient endogenous regulation of Cln2p localization. NES-Cln2mycp shows no evident difference in localization patterns from wild-type Cln2mycp and mnes-Cln2mycp, although the cytoplasmic localization of Cln2p already observed would make any increased cytoplasmic accumulation difficult to detect (data not shown).
Localization-mediated functional specificity of Cln2p, Cln3-1p, and Cln3p.
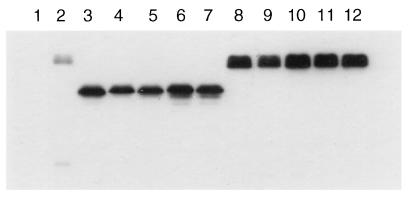
Strains containing the NLS- or NES-CLN2myc and the NLS- or NES-CLN3-1hamyc constructs were assayed for CLN function. Both CLN2 and CLN3-1 are expressed from the constitutive CLN3 promoter to control for differences in the expression of CLN2 and CLN3. Western blot analysis of the NES/mnes/NLS/mnls–Cln3-1hamycp and NES/mnes/NLS/mnls-Cln2mycp show similar steady-state protein levels (Fig. 6). In all of the assays described here, we assigned functional defects to changes in localization exclusively based on differences between Cln proteins that contain the wild-type (NLS or NES) and mutant (nls or nes) localization signals. The mnls and mnes control for nonspecific alterations in Clnp function that may result from the addition of sequences to the N terminus of the protein. In cases where indirect immunofluorescence shows little to no detectable alterations in localization patterns, the mnls and mnes will allow us to attribute any detectable functional defects to the localization activity of the NLS or NES.
FIG. 6.
Comparison of myc-tagged Cln proteins. Wild-type strain (1255-5c) was transformed with plasmids pRS414 (vector, lane 1), pMM45 (Cln3, lane 2), pMM55 (Cln3-1, lane 3), pMM83 (NLS–Cln3-1, lane 4), pMM84 (mnls–Cln3-1, lane 5), pMM85 (NES–Cln3-1, lane 6), pMM86 (mnes–Cln3-1, lane 7), pMM82 (Cln2, lane 8), pMM60 (NLS-Cln2, lane 9), pMM61 (mnls-Cln2, lane 10), pMM92 (NES-Cln2, lane 11), and pMM93 (mnes-Cln2, lane 12). All plasmids are episomal CEN (low copy number) and express the myc-tagged Cln protein from the CLN3 promoter. Cellular lysates were separated by SDS–12% PAGE and analyzed by Western blotting as described in Materials and Methods.
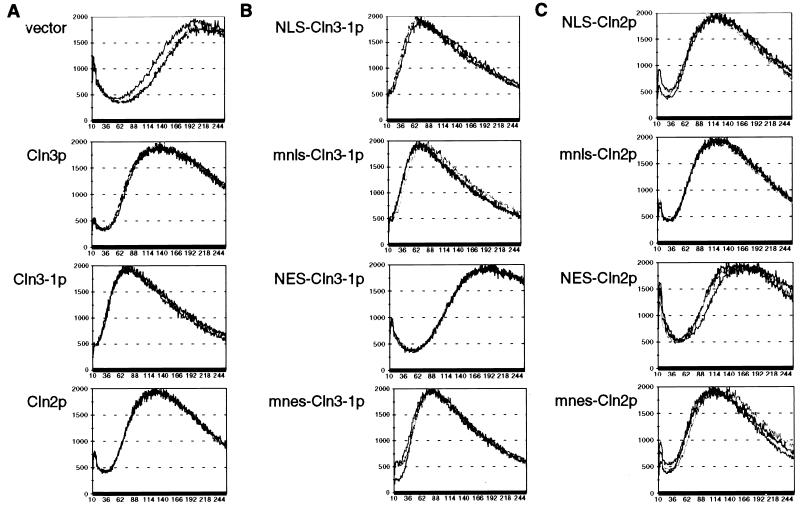
Initially, we assayed for CLN activity by monitoring cell volume. Initiation of the cell cycle requires CLN activity for bud emergence and activation of Clbp-Cdc28p complexes, as described above. Cells with reduced CLN activity will have a delay in these events, resulting in a population of large cells due to a longer period of cell growth after cell division and before cell cycle initiation. Conversely, high CLN activity results in a population of small cells (6, 38). Plasmids encoding the various mutant Cln proteins were introduced into a cln2 cln3 strain, and transformants were assayed for cell volume as described in Materials and Methods. As expected, expression of Cln2mycp and Cln3hamycp confer smaller cell volumes than the vector control. Cln3-1hamycp confers a smaller cell volume than do Cln2p and Cln3p, most likely because of the increased steady-state levels of Cln3-1p (7, 60, 65) (Fig. 7A). In contrast, we find that the expression of NES-Cln3-1hamycp and NES-Cln2mycp (but not the mnes controls) produces populations of cells with large cell volumes, as with the vector transformants (Fig. 7B and C). These data show a reduction of CLN activity when Cln3-1p (and perhaps Cln2mycp) is exported out of the nucleus and into the cytoplasm via the NES.
FIG. 7.
Cell size assay for CLN gene function. CLN1 cln2 cln3 cells (1421-21D) were transformed with plasmids containing various CLN2 and CLN3 constructs (see Fig. 5 legend). Cell size analysis was carried out as described in Materials and Methods. The data for three transformants are shown on each graph. The x and y axis scales for all of the graphs in this figure are identical.
Thus, in the cell volume assay for CLN activity, a functional NES appears to significantly reduce the biological activity of both Cln3-1p and Cln2p. Despite the fact that the NES did little to the observed distribution of Cln2p, the result appears to be reliable since the effect was eliminated by a point mutation of the NES. This result suggests that nuclear Cln protein may be required at least transiently to efficiently drive cell cycle initiation. The NLS had no effect on either Cln2p or Cln3-1p function. In the case of Cln3-1p, which was efficiently localized to the nucleus by this sequence, this result suggests that sufficient nuclear localization of Cln3-1p to drive initiation of the cell cycle may occur even without the NLS (Fig. 7). NES–Cln3-1-expressing cells show a wider distribution of slightly larger cells than the NES-Cln2p-expressing cells, for unknown reasons.
The second assay used to address CLN function is the ability of the various Clnp mutants to complement a triple cln deletion mutant (cln−). This experiment was done by using a cln1 cln2 cln3 GAL1::CLN3 strain. The GAL1 promoter is induced by growth on medium containing galactose, resulting in the expression of Cln3p. Growth on glucose shuts off the GAL1 promoter, resulting in growth arrest due to the lack of CLN function. We are able to test our CLN constructs by introducing into this strain plasmids encoding the various mutants, all expressed from the CLN3 promoter, and then comparing the ability of these plasmid-bearing strains to form colonies on galactose- and glucose-containing media. All CLN2 and CLN3 plasmids tested were able to support viability in the cln− strain at 30°C. However, at 38°C the plasmids expressing NES–Cln3-1hamycp and NES-Cln2mycp were unable to rescue viability as well as the plasmids expressing mnes–Cln3-1hamycp and mnes-Cln2mycp (Fig. 8A). The NLS and mnls had no effect in this assay. These data support the results from the cell volume assay.
FIG. 8.
Viability assay for NLS- and NES–CLN2 and CLN3-1 strains. Mutant strains, each containing a GAL1::CLN for viability, were transformed with CLN plasmids. For each transformant strain, 10-fold serial dilutions were prepared from independent pools of transformants (5 to 10 colonies), and 3 μl of each dilution was plated onto both YPDex and YPGal plates. Plates were incubated at 30 or 38°C as indicated. CLN plasmids used are those listed in the legend to Fig. 5. (A) cln− and cln− bck2− strains. (B) cln− swi4− and CLN3 cln1,2− GAL::SIC1 strains. (C) CLN3 cln1,2− clb5,6− strain. (D) cln− pcl1,2− and cln− bud2− strains. (E) CLN3 cln1,2− swi4− strain.
The third assay used to test for CLN activity utilizes a series of mutant strains that allow us to differentiate between CLN2 and CLN3 function. In the strains assayed here, except for the cln− bck2− strain, wild-type CLN3 is unable to rescue in the absence of additional CLN genes, but wild-type CLN2 does rescue (see the introduction). These assays were performed as described for the cln− strain, except in the case of the GAL1::SIC1 strain. In this case, growth on galactose results in overexpression of Sic1p and growth arrest, so this strain is not viable on galactose-containing medium and viable on glucose-containing medium. Expression of CLN2 from the CLN3 promoter suppresses GAL1::SIC1 lethality (27, 58).
We found that in strains where Cln3hamycp is unable to rescue, Cln3-1hamycp is able to rescue to various degrees, although not to the level of Cln2mycp (Fig. 8). In the case of the cln− clb5,6− strain and the cln− swi4− strain, this may in part reflect higher steady-state levels of Cln3-1p versus Cln3p in the cell (27). Additionally, it may be due to the increase in cytoplasmic localization observed with CLN3-1hamycp compared to Cln3hamycp (compare Fig. 1 and Fig. 5). The latter idea is supported by the observation that Cln3-1p function is altered by mislocalization with the NLS or NES but not with the control mnls or mnes (see below).
Analysis of rescue by the NLS- and NES-mislocalized cyclins and the mnls and mnes controls strongly suggested that subcellular localization is a major determinant of cyclin specificity. In the cln− bck2− strain, the NES addition (but not the mnes, NLS, or mnls addition) to Cln2p or Cln3-1p yielded a partial defect in rescue at 30°C (Fig. 8A). Since BCK2 may activate a set of transcripts similar to those that are regulated by CLN3 activity (9, 13), this phenotype may reflect a nuclear requirement for Clnp that involves activation of transcription. These data are consistent with the findings in the cell volume and cln− complementation assays, suggesting an important functional role for nuclear localization of Cln proteins.
A different pattern was observed with the GAL1::SIC1 and cln− swi4− strains: rescue by Cln3-1p was hampered by either the NLS or the NES (but not by the mnls or the mnes) (Fig. 8B). These data suggest that there are CLN requirements in both the nucleus and cytoplasm for rescue in this context. If so, the NES-Cln3-1p should rescue in the presence of wild-type Cln3p. The cytoplasmic Cln3p activity would be provided by NES–Cln3-1p, and the nuclear Cln3p activity would be provided by wild-type Cln3p. To test this idea, we introduced the NES–Cln3-1p into a CLN3 cln1,2− swi4− strain and assayed for rescue as described for the cln− swi4− strain. Consistent with a requirement of Cln3p activity in both the nucleus and cytoplasm in this strain background, NES–Cln3-1p is able to rescue as well as mnes–Cln3-1p and Cln3-1p in the presence of wild-type Cln3p (Fig. 8E). Thus, wild-type (nuclear) Cln3p and cytoplasmic NES–Cln3-1p complement each other for the rescue of the cln1,2 swi4 mutant strain, a result consistent with CLN requirements in both compartments for rescue.
In the cln− clb5,6− strain, NES (but not mnes) addition to Cln3-1p or Cln2p strongly reduced rescue, but NLS (and not mnls) addition yielded a reproducible 5- to 10-fold increase in viability (Fig. 8C). These data suggest an important role for the efficient nuclear localization of Cln protein.
In the cln− bud2− and cln− pcl1,2− strains, the NLS (but not the mnls) strongly reduced rescue by Cln3-1p, while the NES (but not the mnes) strongly increased rescue (Fig. 8D). These data suggest an important cytoplasmic CLN function.
Despite the low impact on full-length Cln3p localization with the addition of NES or NLS sequences, we found that NES addition (but not mnes, NLS, or mnls addition) resulted in partial Cln3p rescue of the cln− pcl1,2− and cln− swi4− strains (Fig. 9A and B). These data are consistent with the interpretation above that cytoplasmic Cln3p may gain some ability to function in assays normally restricted to Cln2p.
FIG. 9.
Viability assays and Western blot for NLS- and NES-CLN3 strains. (A and B) Mutant strains, each containing a GAL1::CLN for viability, were transformed with plasmids pRS414 (vector), pMM45 (CLN3), pMM83 (NLS-CLN3), pMM84 (mnls-CLN3), pMM85 (NES-CLN3), and pMM86 (mnes-CLN3). For each transformant strain, 10-fold serial dilutions were prepared from independent pools of transformants (5 to 10 colonies), and 3 μl of each dilution was plated onto both YPDex and YPGal plates. Plates were incubated at 30 or 38°C as indicated. (A) cln− pcl1,2− strain. (B) cln− swi4− strain. (C) Wild-type strain 1255-5C was transformed with the plasmids listed above. Cellular lysates from two independent transformants were prepared in parallel for each plasmid-bearing strain (except pRS414). All plasmids are episomal CEN (low copy number) and express the myc-tagged Cln protein from the CLN3 promoter. Cellular lysates were separated by SDS–10% PAGE and analyzed by Western blotting as described in Materials and Methods. Proteins were visualized by using the polyclonal anti-myc A-14 antibody (Santa Cruz Biotechnology). Lanes: 1, vector; 2, CLN3; 3, NLS-CLN3; 4, mnls-CLN3; 5, NES-CLN3; 6, mnes-CLN3. The position of full-length Cln3hamycp is indicated by Cln3, and the 35-kDa species is indicated by *Cln3.
Previous studies have established that an increase in steady-state protein levels of Cln3p is sufficient for partial rescue of the cln− swi4− strain (27). Western blot analysis was done to ensure that the ability of NES-Cln3p to rescue the cln− swi4− strain is not due to an NES-dependent increase in Cln3p protein levels. A comparison of NES-Cln3p and mnes-Cln3p showed no significant difference in steady-state protein levels (Fig. 9).
DISCUSSION
Enzyme kinetics of Cln2p-Cdc28p and Cln3p-Cdc28p.
No significant differences in the binding affinity, catalysis rate, or overall efficiency of the Cln2p-Cdc28p and Cln3p-Cdc28p complexes were observed in our analysis. These data are consistent with an overall similarity of the active site of Cdc28p when activated by the two different cyclins. We note that these studies utilize a limited set of three short peptides. For this reason, these data are not definitive, and kinetic differences may exist with additional peptide substrates. However, the fact that the enzyme kinetics in this limited study showed no differences between Cln2p- and Cln3p-associated Cdc28 kinase activity prompted us to look for other mechanisms to explain the cyclin functional specificity.
Subcellular localization of Cln2p and Cln3p.
Cln2p and Cln3p have distinct localization patterns. Cln2mycp is localized primarily to the cytoplasm, as demonstrated by indirect immunofluorescence and biochemical fractionation of cells. While the majority of Cln2mycp is clearly cytoplasmic, it is likely that some proportion of Cln2p also localizes to the nucleus, since limiting the ability of Cln2p to reside in the nucleus with an NES inhibits some Cln2p functions (Fig. 7 and 8). The point mutant mnes has no effect, indicating that this is probably specific to nuclear export, not to addition of a nonspecific sequence to Cln2p. In contrast to the primarily cytoplasmic localization of Cln2mycp, Cln3mycp accumulates in the nuclei of large budded cells. Nuclear accumulation of Cln3p appears gradual, in that we cannot correlate a morphological event (such as bud emergence or nuclear division) with efficient Cln3p nuclear localization. Constitutively expressed Cln2p remains cytoplasmic in large budded cells (data not shown), while Cln3p localizes to the nuclei. These data indicate distinctly regulated cellular localization of Cln2p and Cln3p, providing a possible mechanism to confer substrate specificity to Clnp-Cdc28p complexes.
Cln3p function and cellular localization.
The mechanism by which Cln3p activity results in the activation of late G1 transcripts, such as those regulated by the SBF or MBF transcription factors, is not understood. However, one might speculate that Cln3p acts directly on the components of the transcriptional machinery, possibly within the nucleus of the cell. We demonstrate that the efficient nuclear localization of Cln3-1p is required for proper CLN function in the cln− bck2− strain, which is specifically defective in SBF-dependent transcription (27). These data suggest the likelihood that Cln3p acts directly within the nucleus to trigger transcription of late G1 transcripts. This raises the speculation that direct interactions occur between Cln3p and the transcriptional machinery; testing this idea further is beyond the scope of this study.
The localization of Cln3p in the nuclei of large budded cells introduces interesting possibilities for the regulation of Cln3p function. In a wild-type cell, Cln3p-responsive transcription is triggered just prior to DNA replication, in late G1. Cln3p does not appear to be tightly localized to the nucleus during this time. The maximal accumulation of nuclear Cln3p occurs when Cln3p-dependent transcription is off (large budded cells with one or two nuclei). Assuming that Cln3p must localize to the nucleus to trigger transcription of late-G1 transcripts (see paragraph above), one might expect for the peak nuclear accumulation of Cln3p to coincide with the Cln3p activity. In this case, Cln3p may function during the preceding cell cycle, during late mitosis or early G1, when Cln3p localizes most clearly to the nucleus, to create an environment in the cell which is responsive to SCB- and MCB-mediated transcription in late G1. Alternatively, the relatively low amount of Cln3p that localizes to the nucleus of cells in late G1 may be sufficient to trigger the transcription of genes regulated by Cln3p.
Functional significance of Cln2p and Cln3p localization.
If the localization patterns of Cln2p and Cln3p reflect significant regulatory mechanisms, then mislocalization of the cyclins should alter CLN function. We found NES-dependent decreases in Cln2p and Cln3-1p activity in both cln complementation and cell size assays, indicating that the presence of Cln proteins in the nucleus is critical for some CLN functions (see Fig. 7 and 8). We also saw consequences of the addition of NES to Cln2p in most of the mutant strains tested (Fig. 8). These data suggest that the localization of Cln protein to the nucleus of the cell is important for the efficient function of both Cln2p and Cln3p. The lack of effect of the NLS on Cln2p function in most assays is difficult to evaluate given that the NLS has very little cytologically detectable effect on Cln2p localization. Additionally, as described below, specific NES– and NLS–Cln3-1p-dependent changes in Cln protein activity were observed in the series of mutant strains tested. These results suggest that there are CLN-dependent cytoplasmic and CLN-dependent nuclear events that are important for cell cycle initiation.
Cln3-1p shows an intermediate localization pattern between Cln2p and Cln3p, with an increase in cytoplasmic localization and accumulation in the nuclei of large budded cells (Fig. 5), suggesting that some signals for correct Cln3p nuclear localization are present in the C-terminal third of the protein. Correlating with this loss of localization specificity, Cln3-1p shows some loss of biological specificity: in the mutant strains where Cln2p is able to rescue viability and Cln3p is not able to rescue, Cln3-1p shows intermediate degrees of rescue (Fig. 8). Addition of the NLS (but not mnls) to Cln3-1p moves the Cln3-1p into the nucleus and results in the loss of Cln3-1p-dependent rescue in four of the five mutant strains tested. The exception to this is the cln− clb5,6− strain, which is rescued by Cln3p when overexpressed (27), and it is known that Cln3-1p accumulates to higher levels than Cln3p due to the loss of C-terminal PEST sequences (60). These data suggest that once the partial defect in nuclear localization is corrected, Cln3-1p functions similarly to overexpressed Cln3p.
In contrast, addition of the NES (but not the mnes) to Cln3-1p results in an increase in rescue in the cln− pcl1,2− and cln− bud2− strains normally rescued by Cln2p but not by Cln3p. Therefore, shifting Cln3-1p into a more Cln2p-like localization pattern allows a more Cln2p-like activity. This finding may not be restricted to the Cln3-1p truncation since, consistent with the Cln3-1p results, we found that the NES (but not mnes)-Cln3p provides weak rescue of the cln pcl1,2− and cln− swi4− strains. Therefore, the ectopic presence of Cln3p in the cytoplasm allows Cln3p or Cln3-1p some ability to perform functions normally restricted to Cln2p. Overall, it appears that Cln2p is required in both the cytoplasm and the nucleus of the cell for maximal function. Cln3p, on the other hand, appears to function primarily in the nucleus of the cell, although Cln3p is capable of Cln2p-like activity when forced into the cytoplasm (Fig. 8D and 9). These data are the first to suggest a cytoplasmic function for a cyclin-Cdk complex. Although the putative cytoplasmic function remains poorly defined, it is likely to involve some aspect of cell polarity determination or bud emergence (29).
We conclude that CLN activity is influenced by Clnp localization and that the functional specificity of Cln proteins is determined, at least in part, by subcellular localization. These data suggest that investigation of mechanisms that regulate cyclin localization will provide information about the regulation of cyclin specificity.
ACKNOWLEDGMENTS
We thank Mike Rout and David Lawrence for advice and helpful discussions in these studies. We thank Ray Deshaies for providing the anti-Cdc28p antibody, Mike Rout for providing the anti-Nop1p antibody, David Lawrence for providing peptide substrates, and Tony Gartner and Mike Tyers for providing plasmids. We also thank Mary Koszelak, Kristi Levine, Kimberly Huang, Maria Yuste Rojas, and David W. Miller for helpful discussions.
This work was supported by NIH grant GM47238 to F.R.C. M.E.M. was supported by an NRSA GM18782.
REFERENCES
- 1.Aris J P, Blobel G. Identification and characterization of a yeast nucleolar protein that is similar to a rat liver nucleolar protein. J Cell Biol. 1988;107:17–31. doi: 10.1083/jcb.107.1.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baldin V, Lukas J, Marcote M J, Pagano M, Draetta G. Cyclin D1 is a nuclear protein required for cell cycle progression in G1. Genes Dev. 1993;7:812–821. doi: 10.1101/gad.7.5.812. [DOI] [PubMed] [Google Scholar]
- 3.Benton B K, Tinkelenberg A, Gonzalez I, Cross F R. Cla4p, a Saccharomyces cerevisiae Cdc42p-activated kinase involved in cytokinesis, is activated at mitosis. Mol Cell Biol. 1997;17:5067–5076. doi: 10.1128/mcb.17.9.5067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cardosa M C, Leonhardt H, Nada-Ginard B. Reversal of terminal differentiation and control of DNA replication cyclin A and Cdk2 specifically localize at subnuclear sites of DNA replication. Cell. 1993;74:979–992. doi: 10.1016/0092-8674(93)90721-2. [DOI] [PubMed] [Google Scholar]
- 5.Chant J, Corrado K, Pringle J R, Herskowitz I. Yeast BUD5, encoding a putative GDP-GTP exchange factor, is necessary for bud site selection and interacts with bud formation gene BEM1. Cell. 1991;65:1213–1224. doi: 10.1016/0092-8674(91)90016-r. [DOI] [PubMed] [Google Scholar]
- 6.Cross F R. DAF1, a mutant gene affecting size control, pheromone arrest, and cell cycle kinetics of Saccharomyces cerevisiae. Mol Cell Biol. 1988;8:4675–4684. doi: 10.1128/mcb.8.11.4675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cross F R, Blake C M. The yeast Cln3 protein is an unstable activator of Cdc28. Mol Cell Biol. 1993;13:3266–3271. doi: 10.1128/mcb.13.6.3266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cvrcková F, Nasmyth K. Yeast G1 cyclins CLN1 and CLN2 and a GAP-like protein have a role in bud formation. EMBO J. 1993;12:5277–5286. doi: 10.1002/j.1460-2075.1993.tb06223.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Di Como C J, Chang H, Arndt K T. Activation of CLN1 and CLN2 G1 cyclin gene expression by BCK2. Mol Cell Biol. 1995;15:1835–1846. doi: 10.1128/mcb.15.4.1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Diehl J A, Cheng M, Roussel M F, Sherr C J. Glycogen synthase kinase 3beta regulates cyclin D1 proteolysis and subcellular localization. Genes Dev. 1998;15:3499–3511. doi: 10.1101/gad.12.22.3499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Diehl J A, Sherr C J. A dominant-negative cyclin D1 mutant prevents nuclear import of cyclin-dependent kinase 4 (CDK4) and its phosphorylation by CDK-activating kinase. Mol Cell Biol. 1997;17:7362–7374. doi: 10.1128/mcb.17.12.7362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dirick L, Böhm T, Nasmyth K. Roles and regulation of Cln-Cdc28 kinases at the start of the cell cycle of Saccharomyces cerevisiae. EMBO J. 1995;14:4803–4813. doi: 10.1002/j.1460-2075.1995.tb00162.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Epstein C B, Cross F R. Genes that can bypass the CLN requirement for Saccharomyces cerevisiae cell cycle START. Mol Cell Biol. 1994;14:2041–2047. doi: 10.1128/mcb.14.3.2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Espinoza F H, Ogas J, Herskowitz I, Morgan D O. Cell cycle control by a complex of the cyclin HCS26 (PCL1) and the kinase PHO85. Science. 1994;266:1388–1391. doi: 10.1126/science.7973730. [DOI] [PubMed] [Google Scholar]
- 15.Feldman R M R, Correll C C, Kaplan K B, Deshaies R J. A complex of Cdc4p, Skp1p, and Cdc53p/Cullin catalyzes ubiquitination of the phosphorylated CDK inhibitor Sic1p. Cell. 1997;91:221–230. doi: 10.1016/s0092-8674(00)80404-3. [DOI] [PubMed] [Google Scholar]
- 16.Fernandez-Sarabia M J, Sutton A, Zhong T, Arndt K T. SIT4 protein phosphatase is required for the normal accumulation of SWI4, CLN1, CLN2, and HCS26 RNAs during late G1. Genes Dev. 1992;6:2417–2428. doi: 10.1101/gad.6.12a.2417. [DOI] [PubMed] [Google Scholar]
- 17.Fritz C C, Green M R. HIV Rev uses a conserved cellular protein export pathway for the nucleocytoplasmic transport of viral RNAs. Curr Biol. 1996;6:848–854. doi: 10.1016/s0960-9822(02)00608-5. [DOI] [PubMed] [Google Scholar]
- 18.Gietz R D, Schiestl R H. Applications of high efficiency lithium acetate transformation of intact yeast cells using single-stranded nucleic acids as carrier. Yeast. 1991;7:253–263. doi: 10.1002/yea.320070307. [DOI] [PubMed] [Google Scholar]
- 19.Hadwiger J A, Wittenberg C, Mendenhall M D, Reed S I. The Saccharomyces cerevisiae CKS1 gene, a homolog of the Schizosaccharomyces pombe suc1+ gene, encodes a subunit of the Cdc28 protein kinase complex. Mol Cell Biol. 1989;9:2034–2041. doi: 10.1128/mcb.9.5.2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hagting A, Karlsson C, Clute P, Jackman M, Pines J. MPF localization is controlled by nuclear export. EMBO J. 1998;17:4127–4138. doi: 10.1093/emboj/17.14.4127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Holmes J K, Solomon M J. A predictive scale for evaluating cyclin-dependent kinase substrates. A comparison of p34cdc2 and p33cdk2. J Biol Chem. 1996;11:25240–25246. doi: 10.1074/jbc.271.41.25240. [DOI] [PubMed] [Google Scholar]
- 22.Jaspersen S L, Charles J F, Morgan D O. Inhibitory phosphorylation of the APC regulatory hct1 is controlled by the kinase cdc28 and the phosphatase cdc14. Curr Biol. 1999;9:227–236. doi: 10.1016/s0960-9822(99)80111-0. [DOI] [PubMed] [Google Scholar]
- 23.Jin P, Hardy S, Morgan D O. Nuclear localization of cyclin B1 controls mitotic entry after DNA damage. J Cell Biol. 1998;18:875–885. doi: 10.1083/jcb.141.4.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kilmartin J V, Fogg J. Partial purification of yeast spindle pole bodies. In: Cappucinelli P, Morris N R, editors. Microtubules and microorganisms. New York, N.Y: Marcel Dekker, Inc.; 1982. pp. 157–170. [Google Scholar]
- 25.Knoblich J A, Sauer K, Jones L, Richardson H, Saint R, Lehner C F. Cyclin E controls S phase progression and its down regulation during Drosophila embryogenesis is required for the arrest of cell proliferation. Cell. 1994;77:107–120. doi: 10.1016/0092-8674(94)90239-9. [DOI] [PubMed] [Google Scholar]
- 26.Koch C, Nasmyth K. Cell cycle regulated transcription in yeast. Curr Opin Cell Biol. 1994;6:451–459. doi: 10.1016/0955-0674(94)90039-6. [DOI] [PubMed] [Google Scholar]
- 27.Levine K, Huang K, Cross F R. Saccharomyces cerevisiae G1 cyclins differ in their intrinsic functional specificities. Mol Cell Biol. 1996;16:6794–6803. doi: 10.1128/mcb.16.12.6794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Levine K, Oehlen L J W M, Cross F R. Isolation and characterization of new alleles of the cyclin-dependent kinase gene CDC28 with cyclin-specific functional and biochemical defects. Mol Cell Biol. 1998;18:290–302. doi: 10.1128/mcb.18.1.290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lew D J, Reed S I. Morphogenesis in the yeast cell cycle: regulation by Cdc28 and cyclins. J Cell Biol. 1993;120:1305–1320. doi: 10.1083/jcb.120.6.1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li J, Meyer A N, Donoghue D. Nuclear localization of cyclin B1 mediates ints biological activity and is regulated by phosphorylation. Proc Natl Acad Sci USA. 1997;94:502–507. doi: 10.1073/pnas.94.2.502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lukas J, Pagano M, Staskova Z, Draetta G, Bartek J. Cyclin D1 protein oscillates and is essential for cell cycle progression in human tumour cell lines. Oncogene. 1994;9:707–718. [PubMed] [Google Scholar]
- 32.Maridor G, Gallant P, Golsteyn R, Nigg E A. Nuclear localization of vertebrate cyclin A correlates with its ability to form complexes with cdk catalytic subunits. J Cell Sci. 1993;106:535–544. doi: 10.1242/jcs.106.2.535. [DOI] [PubMed] [Google Scholar]
- 33.McInerny C J, Partridge J F, Mikesell G E, Creemer D P, Breeden L L. A novel Mcm1-dependent element in the SWI4, CLN3, CDC6, and CDC47 promoters activates M/G1-specific transcription. Genes Dev. 1997;11:1277–1288. doi: 10.1101/gad.11.10.1277. [DOI] [PubMed] [Google Scholar]
- 34.Measday V, Moore L, Ogas J, Tyers M, Andrews B. The PCL2 (ORFD)-PHO85 cyclin-dependent kinase complex: a cell cycle regulator in yeast. Science. 1994;266:1391–1395. doi: 10.1126/science.7973731. [DOI] [PubMed] [Google Scholar]
- 35.Mendenhall M D. An inhibitor of p34CDC28 protein kinase activity from Saccharomyces cerevisiae. Science. 1993;259:216–219. doi: 10.1126/science.8421781. [DOI] [PubMed] [Google Scholar]
- 36.Moore J D, Yang J, Truant R, Kornbluth S. Nuclear import of Cdk/cyclin complexes: identification of distinct mechanisms for import of Cdk2/cyclin E and Cdc2/cyclin B1. J Cell Biol. 1999;144:213–224. doi: 10.1083/jcb.144.2.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Morgan D O. Principles of CDK regulation. Nature. 1995;374:131–134. doi: 10.1038/374131a0. [DOI] [PubMed] [Google Scholar]
- 38.Nash R, Tokiwa G, Anand S, Erickson K, Futcher A B. The WHI1+ gene of Saccharomyces cerevisiae tethers cell division to cell size and is a cyclin homolog. EMBO J. 1988;7:4335–4346. doi: 10.1002/j.1460-2075.1988.tb03332.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nelson M, Silver P. Context affects nuclear protein localization in Saccharomyces cerevisiae. Mol Cell Biol. 1989;9:384–389. doi: 10.1128/mcb.9.2.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nishizawa M, Kawasumi M, Fujino M, Toh-e A. Phosphorylation of sic1, a cyclin-dependent kinase (Cdk) inhibitor, by Cdk including Pho85 kinase is required for its promp degradation. Mol Biol Cell. 1998;9:2395–2405. doi: 10.1091/mbc.9.9.2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Oehlen L J W M, Cross F R. G1 cyclins CLN1 and CLN2 repress the mating factor response pathway at Start in the yeast cell cycle. Genes Dev. 1994;8:1058–1070. doi: 10.1101/gad.8.9.1058. [DOI] [PubMed] [Google Scholar]
- 42.Ogas J, Andrews B J, Herskowitz I. Transcriptional activation of CLN1, CLN2, and a putative new G1 cyclin (HCS26) by SWI4, a positive regulator of G1-specific transcription. Cell. 1991;66:1015–1026. doi: 10.1016/0092-8674(91)90445-5. [DOI] [PubMed] [Google Scholar]
- 43.Ohtsubo M, Theodoras A M, Schumacher J, Roberts J M, Pagano M. Human cyclin E, a nuclear protein essential for the G1-to-S phase transition. Mol Cell Biol. 1995;15:2612–2524. doi: 10.1128/mcb.15.5.2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pines J, Hunter T. The differential localization of human cyclins A and B is due to a cytoplasmic retention signal in cyclin B. EMBO J. 1994;13:3772–3781. doi: 10.1002/j.1460-2075.1994.tb06688.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pines J, Hunter T. Human cyclins A and B1 are differentially located in the cell and undergo cell cycle-dependent nuclear transport. J Cell Biol. 1991;115:1–17. doi: 10.1083/jcb.115.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Richardson H E, Wittenberg C, Cross F, Reed S I. An essential G1 function for cyclin-like proteins in yeast. Cell. 1989;59:1127–1133. doi: 10.1016/0092-8674(89)90768-x. [DOI] [PubMed] [Google Scholar]
- 47.Rout M P, Kilmartin J V. Components of the yeast spindle and spindle pole body. J Cell Biol. 1990;111:1913–1927. doi: 10.1083/jcb.111.5.1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schwab M, Lutum A S, Seufert W. Yeast Hct1 is a regulator of Clb2 cyclin proteolysis. Cell. 1997;90:683–693. doi: 10.1016/s0092-8674(00)80529-2. [DOI] [PubMed] [Google Scholar]
- 49.Schwob E, Böhm T, Mendenhall M D, Nasmyth K. The B-type cyclin kinase inhibitor p40SIC1 controls the G1 to S transition in S. cerevisiae. Cell. 1994;79:233–244. doi: 10.1016/0092-8674(94)90193-7. [DOI] [PubMed] [Google Scholar]
- 50.Schwob E, Nasmyth K. CLB5 and CLB6, a new pair of B cyclins involved in DNA replication in Saccharomyces cerevisiae. Genes Dev. 1993;7:1160–1175. doi: 10.1101/gad.7.7a.1160. [DOI] [PubMed] [Google Scholar]
- 51.Sherman F, Fink G R, Hicks J B. Laboratory course manual for methods in yeast genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 52.Sikorski R S, Hieter P. A system for shuttle vectors and yeast strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Srinivasan J, Koszelak M, Mendelow M, Kwon Y G, Lawrence D S. The design of peptide based substrates for the cdc2 protein kinase. Biochem J. 1995;309:927–931. doi: 10.1042/bj3090927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stade K, Ford C S, Guthrie C, Weis K. Exportin 1 (Crm1p) is an essential nuclear export factor. Cell. 1997;90:1041–1050. doi: 10.1016/s0092-8674(00)80370-0. [DOI] [PubMed] [Google Scholar]
- 55.Strambio-de-Castillia C, Blobel G, Rout M P. Isolation and characterization of nuclear envelopes from the yeast Saccharomyces. J Cell Biol. 1995;131:19–31. doi: 10.1083/jcb.131.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stuart D, Wittenberg C. CLN3, not positive feedback, determines the timing of CLN2 transcription in cycling cells. Genes Dev. 1995;9:2780–2794. doi: 10.1101/gad.9.22.2780. [DOI] [PubMed] [Google Scholar]
- 57.Toyoshima F, Moriguchi T, Wada A, Fukuda M, Nishida E. Nuclear export of cyclin B1 and its possible role in the DNA damage-induced G2 checkpoint. EMBO J. 1998;17:2728–2735. doi: 10.1093/emboj/17.10.2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tyers M. The cyclin-dependent kinase inhibitor p40SIC1 imposes the requirement for CLN G1 cyclin function at Start. Proc Natl Acad Sci USA. 1996;93:7772–7776. doi: 10.1073/pnas.93.15.7772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tyers M, Tokiwa G, Futcher B. Comparison of the Saccharomyces cerevisiae G1 cyclins: Cln3 may be an upstream activator of Cln1, Cln2 and other cyclins. EMBO J. 1993;12:1955–1968. doi: 10.1002/j.1460-2075.1993.tb05845.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tyers M, Tokiwa G, Nash R, Futcher B. The Cln3-Cdc28 kinase complex of S. cerevisiae is regulated by proteolysis and phosphorylation. EMBO J. 1992;11:1773–1784. doi: 10.1002/j.1460-2075.1992.tb05229.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Verma R, Annan R S, Huddleston M J, Carr S A, Reynard G, Deshaies R J. Phosphorylation of Sic1p by G1 Cdk required for its degradation and entry into S phase. Science. 1997;278:455–460. doi: 10.1126/science.278.5337.455. [DOI] [PubMed] [Google Scholar]
- 62.Visintin R, Prinz S, Amon A. CDC20 and CDH1: a family of substrate-specific activators of APC-dependent proteolysis. Science. 1998;278:460–463. doi: 10.1126/science.278.5337.460. [DOI] [PubMed] [Google Scholar]
- 63.Wen W, Meinkoth J L, Tsein R Y, Taylor S S. Identification of a signal for rapid export of proteins from the nucleus. Cell. 1995;82:463–473. doi: 10.1016/0092-8674(95)90435-2. [DOI] [PubMed] [Google Scholar]
- 64.Wittenberg C, Sugimoto K, Reed S I. G1-specific cyclins of S. cerevisiae: cell cycle periodicity, regulation by mating pheromone, and association with the p34CDC28 protein kinase. Cell. 1990;62:225–237. doi: 10.1016/0092-8674(90)90361-h. [DOI] [PubMed] [Google Scholar]
- 65.Yaglom J, Linskens M H K, Sadis S, Rubin D M, Futcher B, Finley D. p34Cdc28-mediated control of Cln3 cyclin degradation. Mol Cell Biol. 1995;15:731–740. doi: 10.1128/mcb.15.2.731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yang J, Bardes E S, Moore J D, Brennan J, Powers M A, Kornbluth S. Control of cyclin B1 localization through regulated binding of the nuclear export factor CRM1. Genes Dev. 1998;12:2131–2143. doi: 10.1101/gad.12.14.2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zachariae W, Nasmyth K. TPR proteins required for anaphase progression mediate ubiquitination of mitotic B-type cyclins in yeast. Mol Biol Cell. 1996;7:791–801. doi: 10.1091/mbc.7.5.791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zachariae W, Shevchenko A, Andrews P D, Ciosk R, Galova M, Stark M J R, Mann M, Nasmyth K. Mass spectrometric analysis of the anaphase-promoting complex of yeast: identification of a subunit related to cullins. Science. 1998;279:1216–1219. doi: 10.1126/science.279.5354.1216. [DOI] [PubMed] [Google Scholar]