Abstract
miR-155 plays key promoting roles in several cancers and emerges as an important anti-cancer therapeutic target. However, the discovery of small molecules that target RNAs is challenging. Peptidomimetics have been shown to be a rich source for discovering novel ligands to regulate cellular proteins. However, the potential of using peptidomimetics for RNA targeting is relatively unexplored. To this end, we designed and synthesized members of a novel 320,000 compound macrocyclic peptidomimetic library. An affinity-based screening protocol led to the identification of a pre-miR-155 binder that inhibits oncogenic miR-155 maturation in vitro and in cell, and that induces cancer cell apoptosis. The results of this investigation demonstrate that macrocyclic peptidomimetics could serve as a new scaffold for RNA targeting.
Keywords: peptidomimetics, combinatorial chemistry, microRNA, inhibitor, antitumor agents
Graphical Abstract
INTRODUCTION
Peptidomimetics contain unnatural polyamide backbones and can be equipped with diverse side chains. Compared to natural peptides, these molecules possess a high degree of chemical diversity, protease resistance and bioavailability.1 Consequently, peptidomimetics represent a class of compounds that serves as a rich source for discovering novel ligands for biological molecules. Head to tail backbone cyclization of peptidomimetics can be utilized to further improve exopeptidase resistance and enhance structural rigidity. Thus, the development and applications of cyclic peptidomimetics have become emerging areas of interest.2–5 Several reports have shown that cyclic peptidomimetics can provide biologically active ligands for protein binding or blocking protein-protein interactions,2–4 however, using cyclic peptidomimetics for RNA targeting,6 especially microRNA (miRNA), is underexplored.
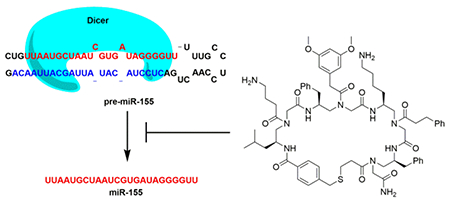
miRNAs are short noncoding RNAs that play key regulatory roles in gene expression and cellular processes.7 They are generated through a series of maturation steps in cells, in which the primary transcript of a miRNA gene (pri-miRNA) is processed by the ribonuclease Drosha to produce the shorter hairpin precursor miRNA, pre-miRNA. A pre-miRNA is further processed by the ribonuclease Dicer to eventually yield the mature miRNA.8 Aberrant expression of miRNAs is linked to various human diseases including cancers.9 For example, oncogenic miR-155 is overexpressed in several cancers, its downregulation has been shown to induce cancer cell apoptosis and to block tumor growth in mice.10 Thus, miR-155 is a promising anti-cancer therapeutic target.
Both small molecules and antisense oligonucleotides have been used to regulate miRNA functions. In general, antisense molecules are limited by the difficulty in cellular delivery, the off-target inhibition, and poor pharmacokinetics and biodistribution.11 Alternatively, small molecules have been discovered or developed through screening or rational designs that achieve the inhibition of the biogenesis of miRNAs.12–25 Many of the currently developed miRNA inhibitors, or RNA ligands in general, are comprised of polycationic scaffolds (e.g. aminoglycosides),14, 16–18 which can suffer from non-specific targeting and poor cell permeability.26–29 Although several alternative small molecule structures have been reported to target RNAs,15, 20, 30 the development of cell permeable small molecules that target miRNA precursors with biological activity, specificity and good cell permeability remains a challenging task.
A few peptides19, 22, 25 and linear peptoids,24 many contain multiple cationic side chains, have recently been found to bind and inhibit processing of pre-miRNAs. Inspired by these findings, we envision that cyclic peptidomimetics may serve as a type of scaffold for targeting pre-miRNA. The structural diversity, enhanced stability and cell permeability of cyclic peptidomimetics can potentially address the limitation faced by current approaches. To explore the potential of using cyclic peptidomimetics to target and inhibit miRNAs, we focused on the identification of biologically active pre-miR-155 ligands from a novel cyclic peptidomimetics library.
EXPERIMENTAL SECTION
In vitro Transcription of pre-miR-155.
The sequence of pre-miR-155 was obtained from miRBase (http://www.mirbase.org/).31 The DNA template used for making pre-miR-155 RNA was generated by PCR primer extension. Briefly, forward primer and reverse primer (Table S1), 0.8 μM of each were subjected to primer extension using Taq polymerase (Ambion) per the manufacture’s protocol. The extended dsDNA was purified with NucleoSpin gel and PCR cleanup kit (Macherey-Nagel). The hybrid DNA template with T7 promoter was used for in vitro transcription using T7 polymerase following manufacturer’s instructions (New England Biolabs). 32 The reaction mixture was then treated with DNase and extracted with phenol/chloroform/isoamyl alcohol (25:24:1) (pH 6.7). The RNA was finally precipitated by ethanol and resuspended in water for storage at −20 °C. Right before use, the RNA was allowed to refold as follows: RNA was heated to 94 °C for 2 min and then cooled to 4 °C at a rate of 1 °C/s.
Preparation of Fluorophore Labeled pre-miR-155.
5’-GMPS primed pre-miR-155 was first prepared by in vitro transcription as described above except that 5’-GMPS (Biolog) :GTP:ATP:CTP:UTP (8:1:1:1:1 mM) was used.33 The RNA was purified by phenol/chloroform extraction and ethanol precipitation. ATTO 488-Iodoacetamide (ATTO-TEC) was then conjugated onto RNA via the 5’ thiol group following a reported method.34
Library Synthesis and Screening.
The library was prepared using split and pool method as reported previously (Figure S1 & S2).3 A detailed protocol was included in Supporting Information. Library beads were incubated with 1% BSA and 1000-fold excess of tRNAs (based on 20 nM of ATTO 488 labeled pre-miR-155) in Tris buffer for 1 h and then washed. ATTO 488 labeled pre-miR-155 (20 nM) was then incubated with the beads in the presence of 1000-fold excess of tRNAs for another 4 h. After thorough wash, beads were observed under the fluorescence microscope and the ones emitting green fluorescence were picked up as the putative positive hits. Three beads with strong green fluorescence were picked out and the structures of two of them were decoded unambiguously by MALDI MS/MS as reported previously.3
Fluorescence Polarization Assay.
The affinity of 1-Fl and 2-Fl (see Supporting Information for synthesis details) to pre-miR-155 was determined as follows. Fluorescent compound (10 nM) was incubated with various concentrations of in vitro transcribed pre-miR-155 in cacodylate buffer (10 mM, pH 7.4, 0.01% Triton X-100) at room temperature for 30 min. The polarization values were obtained with a microplate reader (SpectraMax i3X, Molecular Devices) equipped with a fluorescence polarization detection cartridge. The polarization units (mP) were plotted against pre-miR-155 concentrations and fit in the following equation to determine Kd:
where P0 is the polarization of free fluorescent small molecule, P is the measured polarization at each pre-miR-155 concentration [RNA]total, ΔP is the total change of polarization upon saturation, [F]total is the total concentration of fluorescent compound.35 The experiment was performed in triplicate.
Electrophoretic Mobility Shift Assay.
32P-labeled pre-miR-155 was prepared by in vitro transcription as described above except that [α-P32] Uridine 5’-triphosphate (UTP) (Perkinelmer) was used to take the place of regular UTP. A 10 μL of binding mixture was made by incubating 32P-labeled pre-miR-155 (1 μL, ~20 ng) with various concentrations of tested compound in buffer (HEPES 24 mM, NaCl 200 mM, EDTA 0.04 mM, MgCl2 2.5 mM, pH 7.5) at 37 °C for 20 min. Gel loading buffer (10X, Sucrose 40%, Xylene Cyanol 0.17%, Bromophenol Blue 0.17%) was then added. The bound RNA was resolved from the free RNA using 12% non-denaturing polyacrylamide gel at 4 °C and visualized by phosphor imaging analyzed by Quantity One software (Bio-rad).
Dicer-mediated pre-miR-155 Cleavage Assay.
Dicer enzyme was expressed as reported before.36 A 10 μL of the reaction mixture was made by incubating 32P-labeled pre-miR-155 (1 μL, ~20 ng) with Dicer enzyme (3 μL) and various concentrations of tested compound in buffer (HEPES 24 mM, NaCl 200 mM, EDTA 0.04 mM, MgCl2 2.5 mM, ATP 1 mM, pH 7.5) at 37 °C for 2.5 h. The reaction was stopped by boiling with equal volume of Gel Loading Buffer II (ThermoFisher Scientific) for 5 min. The non-cleaved pre-miR-155 and the processed miR-155 were resolved by 18% denaturing polyacrylamide gel. The gel was analyzed by phosphor imaging.
Cell Culture and Fluorescence Microscope Imaging.
HEK293T and MCF-7 cells were cultured in DMEM medium (Gibco) without antibiotics, supplemented with 10% FBS and 2 mM GlutaMAX (Life Technologies) at 37 °C in a humidified atmosphere containing 5% CO2. To assess the cell permeability of the compounds, HEK293T or MCF-7 cells were plated into 24-well plates and grown overnight to 50% confluency. The cells were then treated with fluorescent compound (5 μM) or DMSO (0.2% v/v) for 7 h. After removing the medium, the cells were washed with PBS and imaged by fluorescence microscope (Axio Observer, Zeiss) under the GFP channel.
miR-155 Inhibition in Cell.
HEK293T cells were plated in 24-well plates and grown overnight to 60% confluency. The cells were transfected with a plasmid DNA (200 ng/well) coding miR-155 (Addgene plasmid # 78126)37 using Lipo3000 transfection reagent (Invitrogen) per the manufacture’s protocol and treated with tested compound. The cells were harvested for RNA extraction after 24 h incubation. MCF-7 cell was used for studying the activity of compound in inhibiting endogenous miR-155 in cell. The cells were plated in 24-well plates and grown overnight to 60% confluency. The cells were then treated with tested compound (30 μM), DMSO (0.2% v/v) or anti-miR-155 (30 nM, Integrated DNA Technologies). The medium was replaced with fresh one containing the corresponding small molecule every 24 h. The cells were harvested after 4-day incubation for the following RT-qPCR, Western blotting, and flow cytometry analysis.
RNA Extraction and RT-qPCR.
Total RNA was extracted using miRNeasy Mini Kit (Qiagen) per the manufacturer’s protocol. miRNA reverse transcription reactions were completed using a Taqman MicroRNA RT Kit (Applied Biosystems). 10 ng of total RNA was used for U6, miR-25, miR-29a, miR-29b and miR-221, 300 ng was used for miR-155, miR-214 and miR-519d. The qPCR was performed on 7900HT Fast Real-Time PCR System (Applied Biosystems) using Taqman Universal PCR Master Mix (Applied Biosystems) and Taqman miRNA assays (Applied Biosystems) per the manufacturer’s protocol. 0.5 μL of the RT product was used for a 10 μL qPCR reaction. The reverse transcription reaction for mRNA (FOXO3A and GAPDH) and miR-155 precursors was completed with 300 ng of total RNA using High Capacity cDNA Reverse Transcription Kit (Applied Biosystems) per the manufacturer’s protocol. 0.5 μL of the reaction mixture was used for qPCR assay (10 μL) with PowerUp SYBR Green Master Mix (Applied Biosystems) per the manufacturer’s protocol (See Table S1 for primer sequences). The triplicate threshold cycles (Ct) obtained for each treatment were used to determine the level of miRNA (normalized to U6 small nuclear RNA) and mRNA (normalized to GAPDH) using the 2−ΔΔCt method.38 The results presented were based on three independent assays.
Western Blotting.
Total protein was extracted using RIPA lysis buffer (EMD Millipore) and quantified using Protein Assay (Bio-Rad). 20 μg of total protein was resolved on a 4-15% SDS-polyacrylamide gel, and then transferred to a PVDF membrane. The membrane was briefly washed with 1× Tris-buffered saline (TBS), and then blocked in 5% milk dissolved in 1× TBST (1× TBS containing 0.1% Tween-20) for 1 h at room temperature. The membrane was then incubated with 1:1000 FOXO3A primary antibody (Cell Signaling Technology, FoxO3a (D19A7) Rabbit mAb) or GAPDH primary antibody (Cell Signaling Technology, GAPDH (D16H11) XP® Rabbit mAb) in 1 × TBST containing 3% (w/v) BSA overnight at 4 °C. The membrane was washed with 1 × TBST and incubated with 1:10000 anti-rabbit IgG horseradish-peroxidase conjugate in 1 × TBST for 1 h at room temperature. After washing with 1× TBST, protein expression was quantified using Clarity Western ECL Substrate (Bio-Rad).
Flow Cytometry Assay.
MCF-7 cells were treated with different compounds as described above for 4 days. After washing with PBS twice, the cells were collected by trypsinizing and centrifugation. The cells were then stained with a mixture of fluorescein-annexin V and propidium iodide (PI) using an apoptosis kit (Invitrogen, eBioscience Annexin V-FITC Apop Kit) per the manufacture’s protocol. Flow cytometry was performed using a BD Accuri C6 Plus flow cytometer (BD Biosciences). The quantification data provided were based on three independent assays.
Cell Viability Assay.
The effect of compound on MCF-7 cell viability were tested by MTT assay using 96-well plates. After 4 day treatment as described above, the medium was removed and 10 μL of MTT stock solution (12 mM) was added to wells along with 100 μL of phenol free DMEM. The cells were incubated at 37 °C for 4 h. The MTT containing medium was replaced with 50 μL of DMSO. The solution was mixed thoroughly by pipetting and incubated at 37 °C for 10 min. The absorbance at 540 nm was recorded with a plate reader (SpectraMax i3X, Molecular Devices). The relative viability of the cells was calculated based on 6 parallel tests by comparing to the DMSO control. The result presented was based on three independent assays.
Statistical Analysis.
Data are presented as the mean ± SEM of three independent experiments. Student’s t-test for significance was carried out using Microsoft Excel 2010 software. *p < 0.05, **p < 0.005 verses control sample were considered significant.
RESULTS AND DISCUSSION
Library Design, Synthesis and Screening.
Oligomers of γ-substituted-N-acylated-N-aminoethylamino acids (γ-AA) have been used earlier to construct one-bead–one-compound (OBOC) combinatorial libraries comprised of linear, γ-AA derived, peptides (γ-AApeptides). Members of these libraries were found to be remarkably stable and to have great functional diversity.39 We recently reported a new strategy for preparing cyclic γ-AApeptide libraries that employs a thioether-bridge approach.3 In the current effort, we utilized this strategy to prepare an unbiased library of novel macrocyclic γ-AApeptides (Figure 1a, S1 & S2), which contains four γ-AA building blocks and bears much enhanced functional diversity. We believed that these macrocyclic γ-AApeptides would provide appropriate small-molecule/pre-miRNA binding interface for recognition and compete with the Dicer/pre-miRNA interaction. Furthermore, a diverse set of hydrophobic, cationic and negatively charged side chains were incorporated into the γ-AApeptides to ensure that the library is unbiased and has a theoretical library diversity of 320,000.
Figure 1.
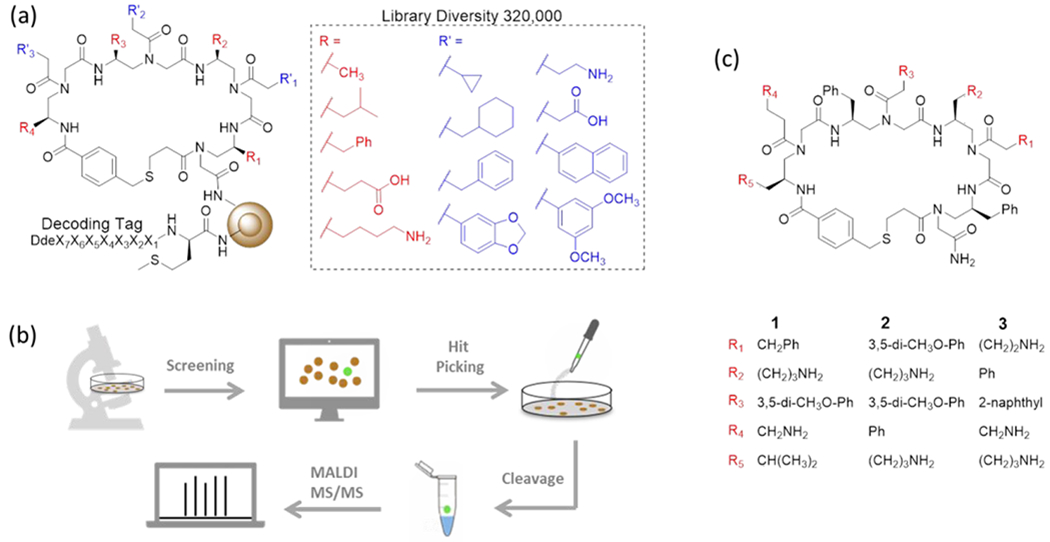
(a) Structure and chemical diversity of the library. (b) A schematic illustration of the screening method. (c) Structures of cyclic peptidomimetics studied in this work.
With the γ-AApeptide library in hand, we conducted affinity based screening against pre-miR-155 (Figure 1b). For this purpose, the on-bead library was incubated with ATTO 488 labeled pre-miR-155 (Figure S3) under highly stringent screening condition (i.e. with 1,000-fold excess of tRNAs), and then subjected to stringent washing conditions. Under fluorescence microscope, the brightest beads, resulted from strong binding to pre-miR-155, were manually picked up (Figure S4). The compounds on the bead were then cleaved off and analyzed by MALDI MS/MS. The structures of two hits (Compound 1 and 2, Figure 1c) were unambiguously decoded. Finally, the compounds were resynthesized in large scale for use in the studies described below.
Binding Affinity of the Hits to pre-miR-155.
Binding of the hits to pre-miR-155 was first validated using the fluorescence polarization (FP) assay, which is commonly used for probing molecular interaction. Typically, when a fluorescent compound forms a complex with a macromolecule, it displays an increased FP value owing to reduction in the rate of fluorophore tumbling. Fluorescein-tagged derivatives of 1 (1-F1) and 2 (2-F1) were prepared for use in the FP assay (Figure S5). The results show that the fluorescence polarization of both 1-F1 and 2-F1 increases with increasing concentrations of pre-miR-155 (Figure S5). The data arising from the saturation binding curves were used to determine that the binding affinity (Kd) of 1-F1 and 2-F1 toward pre-miR-155 are 515 (±186) and 213 (±96) nM respectively. In contrast, fluorescein alone does not have an effect on the FP of pre-miR-155.
miR-155 Inhibition in Cell.
High cell permeability is a prerequisite for the activities of miRNA inhibitors in cells. To evaluate the cellular uptake efficiency of the hits, HEK293T and MCF-7 cells were treated with 1-F1 and 2-F1. The results of fluorescent microscopy showed that after incubation for 7 h very strong fluorescence emanates from both cell lines (Figure S6), indicating that the hits are readily cell permeable. The inhibitory effects on miR-155 maturation in cells were assessed next.
To evaluate the activity of 1 and 2 in inhibiting miR-155 maturation, HEK293T cells were transiently transfected with a plasmid encoding pre-miR-155. Mature miR-155 levels were then determined when cells were treated with different compound (30 μM) by using RT-qPCR. Compound 3 (Figure 1c), a randomly chosen compound from the library that bears more amines (more cationic) than 1 and 2 and did not show binding in the screening, was used as a negative control. As shown in Figure 2a, 3 does not reduce miR-155 level. In contrast, 1 decreases the expression level of miR-155 by 70%, indicating a strong inhibitory effect. Surprisingly, 2 does not cause significant change in miR-155 level, though it shows strong binding to pre-miR-155. It is possible that 2 occupies a site not critical to pre-miR-155 processing, which is not an uncommon phenomenon.14, 40
Figure 2.
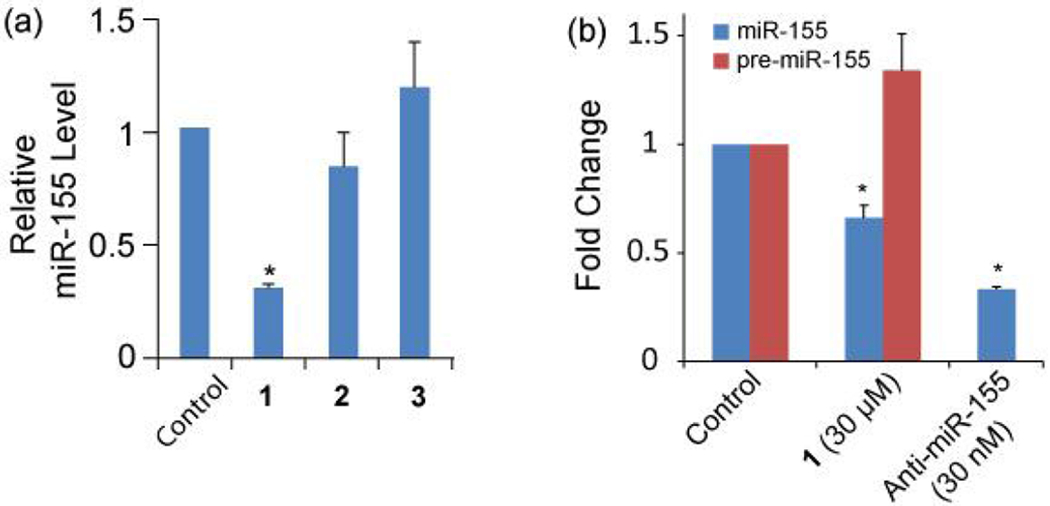
(a) Inhibition of miR-155 production by different compound (30 μM, 24 h treatment) in HEK293T cells overexpressing miR-155. (b) 1 inhibits endogenous miR-155 formation after 96 h treatment of MCF-7 cells. The error bars represent the standard error of mean (N = 3, *p < 0.05).
As 1 shows promising activity in cells, the following studies focus on 1. We examined if 1 inhibits endogenous miR-155 production in cells. Because miR-155 is upregulated in many cancer cells, the breast cancer cell line MCF-7 was used in this study. We found that treating MCF-7 cells with 1 (30 μM) for 96 h lead to a 38% decrease in miR-155 level (Figure 2b), a reduction level comparable to previous reports.14, 17 Meanwhile, miR-155 precursors were found to accumulate in cells (Figure 2b), indicating that the reduction in miR-155 formation is caused by blocking pre-miR-155 cleavage. To further confirm this inhibition mechanism, we performed additional in vitro assays.
pre-miR-155 Interaction in vitro.
Electrophoretic mobility shift assay was carried out as an alternative method to confirm the binding of 1, without any modification, to pre-miR-155. The electrophoretic mobility of 32P-labeled pre-miR-155, prepared by using in vitro transcription, in the presence of various concentrations of 1 was determined by using a non-denaturing polyacrylamide gel and phosphor imaging. The observation of a clear reduction in the mobility rate of pre-miR-155 on the gel with increasing concentrations of 1 (Figure S7) showed that a complex between 1 and pre-miR-155 was formed.
Next we assessed whether binding of 1 to pre-miR-155 disrupts the interaction between pre-miR-155 and Dicer leading to the inhibition of miR-155 maturation. Applying an in vitro pre-miR-155 processing assay, 32P-labeled pre-miR-155 was incubated with recombinant human Dicer in the absence or presence of 1. The Dicer processing reaction was then monitored by using denaturing polyacrylamide gel electrophoresis along with phosphor imaging. In the absence of 1, Dicer efficiently cleaves pre-miR-155 to form mature miR-155 (Figure 3). However, in the presence of increasing concentrations of 1, Dicer-mediated miR-155 processing is disrupted, as reflected in a 63% reduction in the level of mature miR-155 formation caused by 30 μM of 1. An apparent half maximal inhibitory concentration (IC50) of 22 μM was obtained. The combined results demonstrate that 1 inhibits miR-155 maturation by binding to pre-miR-155 and blocking Dicer processing.
Figure 3.
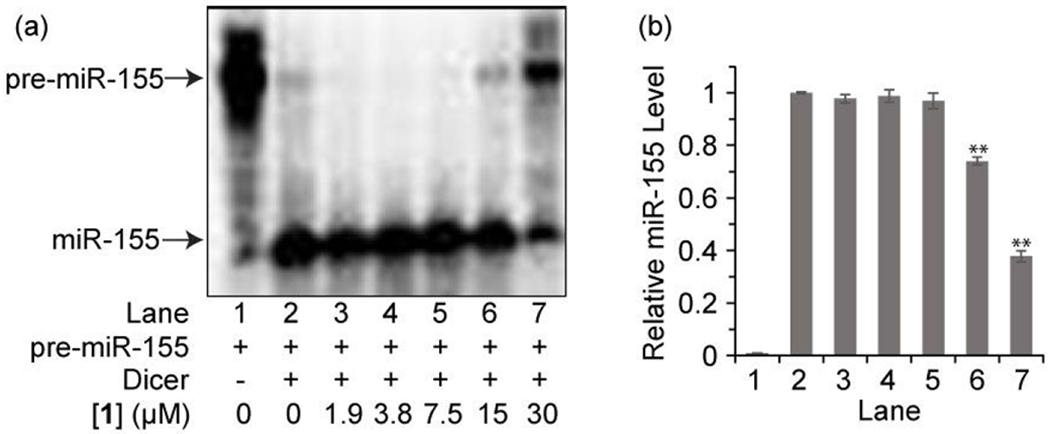
(a) Representative image from electrophoresis analysis of Dicer-mediated pre-miR-155 cleavage in the presence of varying concentrations of 1. (b) Densitometric quantitative analysis of miR-155 for images in panel a arising from three independent assays. Error bars represent the standard error of mean (N = 3).
Selectivity of 1 to miR-155.
To assess the selectivity of 1 toward pre-miR-155, we examined if changes in the expression levels of several other random chosen miRNAs in MCF-7 occur when treating cells with 1 for 96 h. As shown in Figure 4, 1 displays little or no inhibitory effects on other tested miRNAs except miR-155, indicating certain selectivity can be achieved among miRNAs, although further analysis is required to comprehensively investigate the selectivity of 1 against different RNA species in cells and to understand the detailed recognition mechanism contributing to the selectivity.
Figure 4.
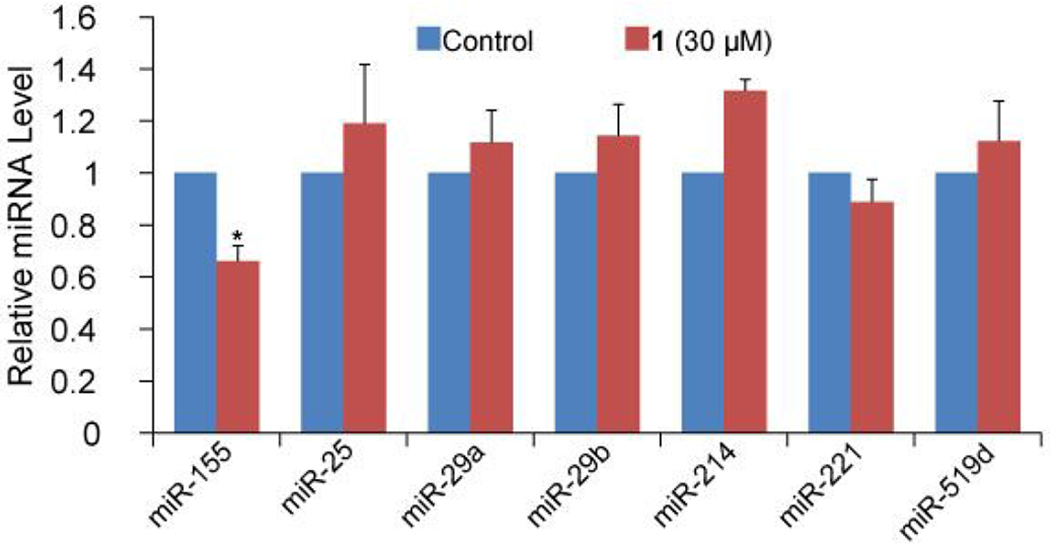
The selectivity of 1 to miR-155 over other miRNAs in MCF-7 cells. The error bars represent the standard error of mean (N = 3).
Downstream Effects of miR-155 Inhibition by 1.
To study the downstream effect of miR-155 inhibition in cells caused by 1, we examined the changes in the expression of FOXO3A, a tumor suppressor gene and a direct target of miR-155 whose loss in activity increases the resistance of cancer cells to apoptosis, which results in cell-cycle progression.41–42 We expect to observe FOXO3A derepression after miR-155 inhibition occurs. In line with expectations, we observed, using RT-qPCR (Figure 5a) and immunoblotting (Figure 5b and c), that the inhibition of miR-155 by 1 in MCF-7 cells leads to increases in the expression of FOXO3A mRNA and protein. To assess whether 1 induces cell apoptosis as a result of miR-155 inhibition, MCF-7 cells were incubated with 1 for 96 h and then stained with propidium iodide (PI) and fluoresceinlabeled annexin V. Flow cytometry analysis showed that 1 promotes an increase in the population of cells undergoing both early apoptosis (Annexin+/PI−) and late apoptosis (Annexin+/PI+) (Figure S8). The cell apoptosis rate is increased by 12%, which is at a similar level as that induced by treatment with an anti-miR-155 oligonucleotide (Figure 5d). Characteristic morphological changes of apoptotic cells including cell shrinkage, rounding and membrane blebbing43 were also observed to take place using microscopy (Figure S9). These results demonstrate that 1 promotes the upregulation of a miR-155 target gene and induces apoptosis.
Figure 5.
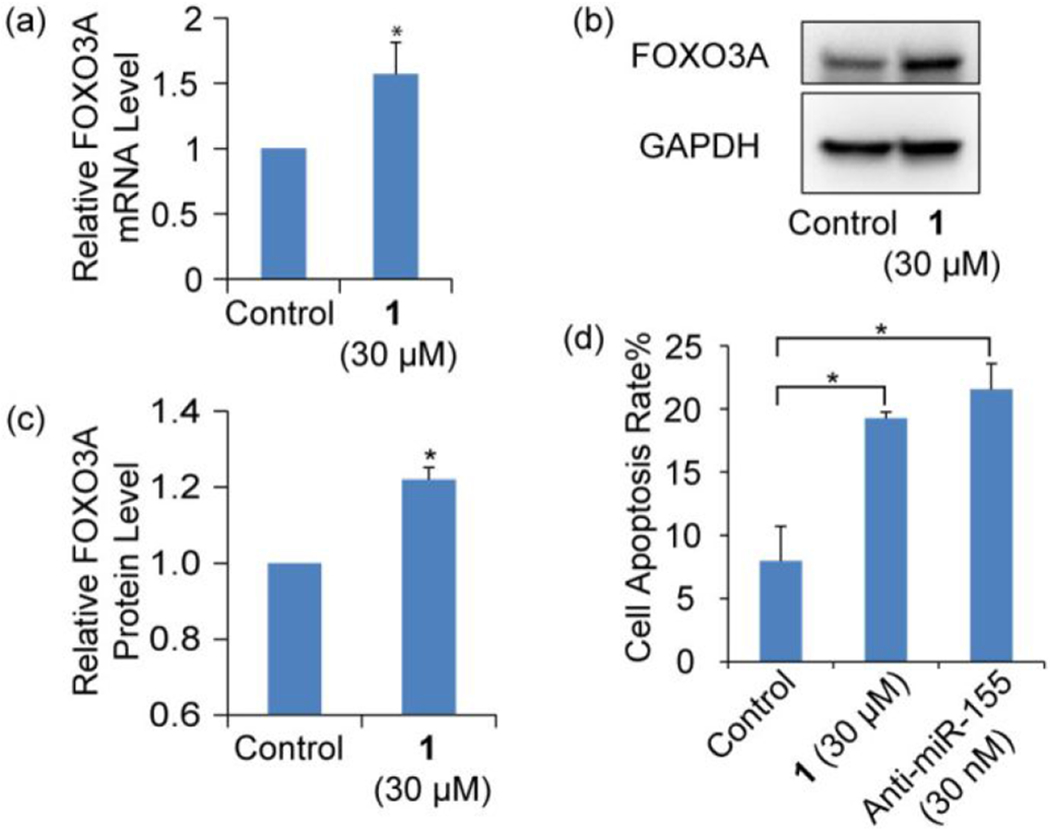
(a) RT-qPCR analysis of mRNA and (b) Western blotting analysis of protein levels of FOXO3A in MCF-7 cells either treated or not treated with 1. (c) Densitometric quantitative analysis of FOXO3A as shown in panel b from three independent assays. (d) Flow cytometry analysis of apoptosis of MCF-7 cell induced by 1 and anti-miR-155. Error bars represent the standard error of mean (N = 3, *p < 0.05).
Finally, because miR-155 inhibition reduces cancer cell proliferation,44 it was anticipated that 1 would cause a decrease in the growth of MCF-7 cells. To test this, cell viability assays were conducted using 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT). The observation of a 70% reduction in the viability of MCF-7 cells after incubation with 1 (30 μM) for 96 h (Figure S10) demonstrates that this macrocyclic peptidomimetic inhibits cancer cell growth as expected.
CONCLUSION
In summary, in the effort described above, we designed and synthesized a novel combinatorial macrocyclic γ-AApeptide library for miRNA targeting. A screen of this library against pre-miR-155 led to the identification of new pre-miR-155 binder 1. We found that 1 inhibits Dicer-mediated pre-miR-155 processing in vitro and in MCF-7 cells, and consequently, induces apoptosis and reduced growth of these cancer cells. Because of the modular nature along with the high diversity that can be introduced through side chain alteration, macrocyclic peptidomimetics should be amenable to systematic structural optimization to obtain more potent and specific binding ligands for pre-miRNAs. This work also demonstrated that macrocyclic peptidomimetics could serve as an alternative RNA targeting scaffold.
Supplementary Material
ACKNOWLEDGMENT
This research was supported by NIH 1R01AG056569 (J.C.), NIH R01GM112652 (J.C), NIH R21CA202831 (F.-S.L.), and an Institutional Development Award (IDeA) from the National Institute of General Medical Sciences of the National Institutes of Health (P20GM103451) (F.-S.L.).
Footnotes
Supporting Information
The supporting information is available free of charge.
Supplementary figures, table and experimental details (PDF).
The authors declare no competing financial interest.
REFERENCES
- (1).Vagner J; Qu H; Hruby VJ, Peptidomimetics, a synthetic tool of drug discovery. Curr. Opin. Chem. Biol. 2008, 12 (3), 292–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Oh M; Lee JH; Moon H; Hyun YJ; Lim HS, A Chemical Inhibitor of the Skp2/p300 Interaction that Promotes p53-Mediated Apoptosis. Angew. Chem. Int. Ed. 2016, 55 (2), 602–606. [DOI] [PubMed] [Google Scholar]
- (3).Shi Y; Challa S; Sang P; She F; Li C; Gray GM; Nimmagadda A; Teng P; Odom T; Wang Y; van der Vaart A; Li Q; Cai J, One-Bead–Two-Compound Thioether Bridged Macrocyclic γ-AApeptide Screening Library against EphA2. J. Med. Chem. 2017, 60 (22), 9290–9298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Simpson LS; Kodadek T, A cleavable scaffold strategy for the synthesis of one-bead one-compound cyclic peptoid libraries that can be sequenced by tandem mass spectrometry. Tetrahedron Lett. 2012, 53 (18), 2341–2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Rhodes CA; Dougherty PG; Cooper JK; Qian Z; Lindert S; Wang Q-E; Pei D, Cell-Permeable Bicyclic Peptidyl Inhibitors against NEMO-IκB Kinase Interaction Directly from a Combinatorial Library. J. Am. Chem. Soc. 2018, 140 (38), 12102–12110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Shi Y; Parag S; Patel R; Lui A; Murr M; Cai J; Patel NA, Stabilization of lncRNA GAS5 by a Small Molecule and Its Implications in Diabetic Adipocytes. Cell Chemical Biology 2018, 10.1016/j.chembiol.2018.11.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Ameres SL; Zamore PD, Diversifying microRNA sequence and function. Nat. Rev. Mol. Cell Biol. 2013, 14 (8), 475–488. [DOI] [PubMed] [Google Scholar]
- (8).Ha M; Kim VN, Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 2014, 75, 509–524. [DOI] [PubMed] [Google Scholar]
- (9).Li ZH; Rana TM, Therapeutic targeting of microRNAs: current status and future challenges. Nat. Rev. Drug Discov. 2014, 13 (8), 622–638. [DOI] [PubMed] [Google Scholar]
- (10).Cheng CJ; Bahal R; Babar IA; Pincus Z; Barrera F; Liu C; Svoronos A; Braddock DT; Glazer PM; Engelman DM; Saltzman WM; Slack FJ, MicroRNA silencing for cancer therapy targeted to the tumor microenvironment. Nature 2015, 518 (7537), 107–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Juliano R; Bauman J; Kang H; Ming X, Biological Barriers to Therapy with Antisense and siRNA Oligonucleotides. Mol. Pharmaceutics 2009, 6 (3), 686–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Gumireddy K; Young DD; Xiong X; Hogenesch JB; Huang QH; Deiters A, Small-molecule inhibitors of microRNA miR-21 function. Angew. Chem. Int. Ed. 2008, 47 (39), 7482–7484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Velagapudi SP; Gallo SM; Disney MD, Sequence-based design of bioactive small molecules that target precursor microRNAs. Nat. Chem. Biol. 2014, 10 (4), 291–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Childs-Disney JL; Disney MD, Small Molecule Targeting of a MicroRNA Associated with Hepatocellular Carcinoma. ACS Chem. Biol. 2016, 11 (2), 375–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Costales MG; Haga CL; Velagapudi SP; Childs-Disney JL; Phinney DG; Disney MD, Small Molecule Inhibition of microRNA-210 Reprograms an Oncogenic Hypoxic Circuit. J. Am. Chem. Soc. 2017, 139 (9), 3446–3455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Bose D; Jayaraj G; Suryawanshi H; Agarwala P; Pore SK; Banerjee R; Maiti S, The Tuberculosis Drug Streptomycin as a Potential Cancer Therapeutic: Inhibition of miR-21 Function by Directly Targeting Its Precursor. Angew. Chem. Int. Ed. 2012, 51 (4), 1019–1023. [DOI] [PubMed] [Google Scholar]
- (17).Vo DD; Staedel C; Zehnacker L; Benhida R; Darfeuille F; Duca M, Targeting the Production of Oncogenic MicroRNAs with Multimodal Synthetic Small Molecules. ACS Chem. Biol. 2014, 9 (3), 711–721. [DOI] [PubMed] [Google Scholar]
- (18).Nahar S; Ranjan N; Ray A; Arya DP; Maiti S, Potent inhibition of miR-27a by neomycin-bisbenzimidazole conjugates. Chem. Sci. 2015, 6 (10), 5837–5846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Pai J; Hyun S; Hyun JY; Park SH; Kim WJ; Bae SH; Kim NK; Yu J; Shin I, Screening of Pre-miRNA-155 Binding Peptides for Apoptosis inducing Activity Using Peptide Microarrays. J. Am. Chem. Soc. 2016, 138 (3), 857–867. [DOI] [PubMed] [Google Scholar]
- (20).Connelly CM; Boer RE; Moon MH; Gareiss P; Schneekloth JS, Discovery of Inhibitors of MicroRNA-21 Processing Using Small Molecule Microarrays. ACS Chem. Biol. 2017, 12 (2), 435–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Shi Z; Zhang J; Qian X; Han L; Zhang K; Chen L; Liu J; Ren Y; Yang M; Zhang A; Pu P; Kang C, AC1MMYR2, an Inhibitor of Dicer-Mediated Biogenesis of Oncomir miR-21, Reverses Epithelial-Mesenchymal Transition and Suppresses Tumor Growth and Progression. Cancer Res. 2013, 73 (17), 5519–5531. [DOI] [PubMed] [Google Scholar]
- (22).Shortridge MD; Walker MJ; Pavelitz T; Chen Y; Yang W; Varani G, A Macrocyclic Peptide Ligand Binds the Oncogenic MicroRNA-21 Precursor and Suppresses Dicer Processing. ACS Chem. Biol. 2017, 12 (6), 1611–1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Yan H; Bhattarai U; Guo Z-F; Liang F-S, Regulating miRNA-21 Biogenesis By Bifunctional Small Molecules. J. Am. Chem. Soc. 2017, 139 (14), 4987–4990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Diaz JP; Chirayil R; Chirayil S; Tom M; Head KJ; Luebke KJ, Association of a peptoid ligand with the apical loop of pri-miR-21 inhibits cleavage by Drosha. RNA 2014, 20 (4), 528–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Bose D; Nahar S; Rai MK; Ray A; Chakraborty K; Maiti S, Selective inhibition of miR-21 by phage display screened peptide. Nucleic Acids Res. 2015, 43 (8), 4342–4352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Tor Y, Targeting RNA with small molecules. Chembiochem 2003, 4 (10), 998–1007. [DOI] [PubMed] [Google Scholar]
- (27).Luedtke NW; Carmichael P; Tor Y, Cellular uptake of aminoglycosides, guanidinoglycosides, and poly-arginine. J. Am. Chem. Soc. 2003, 125 (41), 12374–12375. [DOI] [PubMed] [Google Scholar]
- (28).Thomas JR; Hergenrother PJ, Targeting RNA with small molecules. Chem. Rev. 2008, 108 (4), 1171–1224. [DOI] [PubMed] [Google Scholar]
- (29).Eubanks CS; Forte JE; Kapral GJ; Hargrove AE, Small Molecule-Based Pattern Recognition To Classify RNA Structure. J. Am. Chem. Soc. 2017, 139 (1), 409–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Lorenz DA; Kaur T; Kerk SA; Gallagher EE; Sandoval J; Garner AL, Expansion of cat-ELCCA for the Discovery of Small Molecule Inhibitors of the Pre-let-7-Lin28 RNA–Protein Interaction. ACS Medicinal Chemistry Letters 2018, 9 (6), 517–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Kozomara A; Griffiths-Jones S, miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014, 42 (D1), D68–D73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Huang C; Yu Y-T, Synthesis and Labeling of RNA In Vitro. In Curr. Protoc. Mol. Biol, John Wiley & Sons, Inc.: Hoboken, New Jersey, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Wu C-W; Eder PS; Gopalan V; Behrman EJ, Kinetics of Coupling Reactions That Generate Monothiophosphate Disulfides: Implications for Modification of RNAs. Bioconjugate Chem. 2001, 12 (6), 842–844. [DOI] [PubMed] [Google Scholar]
- (34).Zhang L; Sun L; Cui Z; Gottlieb RL; Zhang B, 5’-Sulfhydryl-Modified RNA: Initiator Synthesis, in Vitro Transcription, and Enzymatic Incorporation. Bioconjugate Chem. 2001, 12 (6), 939–948. [DOI] [PubMed] [Google Scholar]
- (35).Kirk SR; Luedtke NW; Tor Y, Neomycin-acridine conjugate: A potent inhibitor of Rev-RRE binding. J. Am. Chem. Soc. 2000, 122 (5), 980–981. [Google Scholar]
- (36).Yan H; Bhattarai U; Song Y; Liang F-S, Design, synthesis and activity of light deactivatable microRNA inhibitor. Bioorg. Chem. 2018, 80, 492–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Eichelser C; Stückrath I; Müller V; Milde-Langosch K; Wikman H; Pantel K; Schwarzenbach H, Increased serum levels of circulating exosomal microRNA-373 in receptor-negative breast cancer patients. Oncotarget 2014, 5 (20), 9650–9663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Livak KJ; Schmittgen TD, Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCt method. Methods 2001, 25 (4), 402–408. [DOI] [PubMed] [Google Scholar]
- (39).Shi Y; Teng P; Sang P; She F; Wei L; Cai J, γ-AApeptides: Design, Structure, and Applications. Acc. Chem. Res. 2016, 49 (3), 428–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Maiti M; Nauwelaerts K; Herdewijn P, Pre-microRNA binding aminoglycosides and antitumor drugs as inhibitors of Dicer catalyzed microRNA processing. Bioorg. Med. Chem. Lett. 2012, 22 (4), 1709–1711. [DOI] [PubMed] [Google Scholar]
- (41).Yamamoto M; Kondo E; Takeuchi M; Harashima A; Otani T; Tsuji-Takayama K; Yamasaki F; Kumon H; Kibata M; Nakamura S, miR-155, a Modulator of FOXO3a Protein Expression, Is Underexpressed and Cannot Be Upregulated by Stimulation of HOZOT, a Line of Multifunctional Treg. PLOS ONE 2011, 6 (2), e16841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Myatt SS; Lam EWF, The emerging roles of forkhead box (Fox) proteins in cancer. Nat. Rev. Cancer 2007, 7, 847–859. [DOI] [PubMed] [Google Scholar]
- (43).Elmore S, Apoptosis: A Review of Programmed Cell Death. Toxicol. Pathol. 2007, 35 (4), 495–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Chen J; Wang BC; Tang JH, Clinical significance of MicoRNA-155 expression in human breast cancer. J. Surg. Oncol. 2012, 106 (3), 260–266. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


