Abstract
The c-Myc protein functions as a transcription factor to facilitate oncogenic transformation; however, the biochemical and genetic pathways leading to transformation remain undefined. We demonstrate here that the recently described c-Myc cofactor TRRAP recruits histone acetylase activity, which is catalyzed by the human GCN5 protein. Since c-Myc function is inhibited by recruitment of histone deacetylase activity through Mad family proteins, these opposing biochemical activities are likely to be responsible for the antagonistic biological effects of c-Myc and Mad on target genes and ultimately on cellular transformation.
The c-Myc oncoprotein functions as a transcription factor and exerts a variety of biological effects in both normal and malignant cells (reviewed in reference 12). c-Myc functions include the ability to transform primary rodent fibroblasts in cooperation with an activated allele of p21H-ras, the induction of apoptosis in growth factor-deprived cells, the ability to block the terminal differentiation of cells in several in vitro differentiation models, and the ability to promote S-phase induction when expressed in otherwise quiescent cells (reviewed in reference 4). The most widely accepted model of c-Myc function suggests that c-Myc–Max heterodimers bind to CACGTG consensus sites within regulatory regions of specific cellular genes (11). In support of this model, the C-terminal DNA binding domain (DBD) of c-Myc is essential for all its biological activities (25, 35, 40). Many potential c-Myc target genes have been reported (32), although none of the proposed targets appear to mediate the potent effects of c-Myc on cell cycle progression (reviewed in reference 10).
c-Myc function is antagonized by another heterodimeric transcription factor complex containing one of the Mad family proteins in association with Max (1, 20, 23, 47, 50). Mad-Max dimers block c-Myc–Max function by recruitment of either of two large nuclear proteins, mSin3A or mSin3B (2, 36), which in turn recruit one of the two histone deacetylases, HDAC1 and HDAC2 (18, 22, 51). Since Myc-Max and Mad-Max dimers are reported to have indistinguishable DNA binding specificities (1, 50), current models suggest that repression of c-Myc function by the Mad-Max-mSin3-HDAC complex results from the deacetylation of nucleosomal histones at shared target sites within the genome (12).
Studies from our lab and others suggest that defining the function of the c-Myc N terminus may provide insight into the role of c-Myc in oncogenic transformation and normal cell cycle progression (6, 14, 25, 29). While the precise biochemical mechanism remains unknown, the c-Myc N terminus has been shown to be required for transcriptional activation (21). All biological activities of c-Myc also require an evolutionarily conserved block of approximately 20 amino acids referred to as MbII (6, 35, 40). However, deletion of MbII has no deleterious effect on transactivation by c-Myc as measured in conventional reporter gene assays (6). We have recently found that the MbII domain is required to recruit a novel nuclear cofactor called TRRAP and that TRRAP is essential for the oncogenic activity of c-Myc (29).
Traditionally, the activation domains of transcription factors have been thought to function either by mediating the recruitment of the basal transcriptional machinery or by the recruitment of complexes capable of altering nucleosomal organization (49). We recently found that the Saccharomyces cerevisiae ortholog of TRRAP, TRA1p, is a component of the SAGA complex (34), which regulates transcription through chromatin remodeling. Within the multiprotein SAGA complex the only known catalytic subunit is GCN5p (15), which mediates the acetylation of histone tails, resulting in an open chromatin configuration and increased transcription. Several distinct mammalian complexes with characteristics similar to those of the S. cerevisiae SAGA complex have recently been identified (28, 30, 46). The present study was undertaken to determine whether c-Myc, through its association with TRRAP, recruits a mammalian histone acetylase complex, thereby providing a potential mechanism for the antagonistic functions of c-Myc–Max and Mad-Max dimers on target gene expression and cellular transformation.
MATERIALS AND METHODS
Purification of HeLa cell nuclear proteins interacting with the c-Myc N terminus.
FLAG epitope-tagged GAL4 and GAL4–c-Myc fusion proteins were produced by baculovirus infection of insect cells as previously described (29). Nuclear extracts from HeLa cells were generated and then mixed with the baculovirus-produced proteins as described elsewhere (29).
Immunoprecipitation.
293 cell lysates were prepared as described elsewhere (29) and subjected to immunoprecipitation with rabbit antisera directed against c-Myc (N262; Santa Cruz Biotechnology), Max (C-17; Santa Cruz Biotechnology), or hGCN5 (generous gift from Nickolai Barlev and Shelley Berger) (8) or with a mouse monoclonal antibody against c-Myc (C-33; Santa Cruz Biotechnology). Precipitated proteins were either resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) on an 8% gel and immunoblotted by using standard techniques or subjected to a histone acetyltransferase (HAT) assay as described below.
HAT assay.
c-Myc-associated HeLa cell proteins or 293 immunoprecipitates were assayed for HAT activity essentially as described elsewhere (7). Purified histones (type II-A; Sigma) and radiolabeled acetyl coenzyme A (acetyl-CoA) (TRK688; Amersham) were purchased from the suppliers indicated.
Transformation assay.
Primary rat embryo fibroblasts were transfected as described previously (29). Transfections included an activated allele of p21H-ras (1 μg) in conjunction with the equal amounts of the c-Myc expression vectors indicated. Cells were maintained in Dulbecco modified Eagle medium (Gibco BRL) with 4% fetal calf serum for 2 weeks, at which time transformed foci were counted.
RESULTS
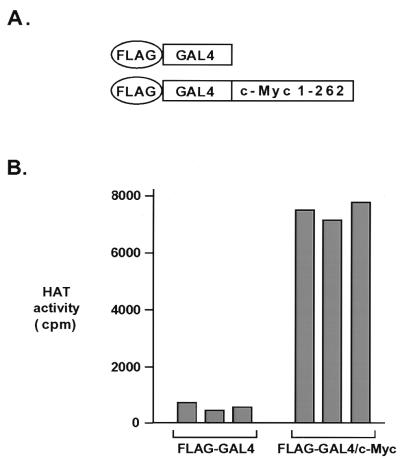
Since c-Myc oncoprotein function can be antagonized by Mad and the recruitment of histone deacetylases, we were interested in determining whether c-Myc itself could recruit HAT activity. In particular, we focused on the N-terminal domain of c-Myc, which is required for cell transformation but not for DNA binding. We have recently described an approach for the affinity purification of nuclear cofactors associated with the c-Myc N terminus (29). Briefly, a FLAG epitope-tagged protein containing murine c-Myc (amino acids 1 to 262) fused to the GAL4 DBD was produced by baculovirus infection of insect cells and then used as an affinity reagent to isolate c-Myc N-terminus-interacting complexes from HeLa cell nuclear extracts. This protocol was previously used for the isolation of the novel c-Myc cofactor TRRAP from prefractionated nuclear extracts. In the present experiments, unfractionated nuclear extracts were mixed with the FLAG-GAL4–c-Myc fusion protein and then immunoprecipitated with antibodies to the FLAG epitope. Precipitates were washed, and the captured proteins were eluted from the antibody by incubation with FLAG peptide. This protocol allows the isolation of GAL4–c-Myc and any associated proteins in their native configuration such that they can be assayed for both protein composition and biochemical activity. An epitope-tagged protein containing only the GAL4 DBD served as a control. Nuclear proteins from HeLa cells which were recruited to the c-Myc N terminus were assayed for HAT activity by assessing their ability to transfer radiolabeled acetyl groups from acetyl-CoA to purified histones (Fig. 1). The results of this assay firmly establish that the c-Myc N terminus recruits HeLa nuclear protein(s) capable of significant histone acetylation (Fig. 1), whereas the control affinity matrix (FLAG-GAL4) recruits negligible activity in comparison.
FIG. 1.
The c-Myc N terminus recruits a HAT. (A) FLAG epitope-tagged GAL4 DBD or a FLAG epitope-tagged GAL4 DBD–c-Myc (amino acids 1 to 262) fusion was used as an affinity matrix to isolate proteins from HeLa cell nuclear extracts. (B) Proteins recruited by each matrix were incubated with purified histones in the presence of radiolabeled acetyl-CoA. Following filter binding and washing, the degree of histone acetylation in each sample was quantitated by scintillation counting. Assays were performed in triplicate. Values for individual samples after subtraction of background are reported.
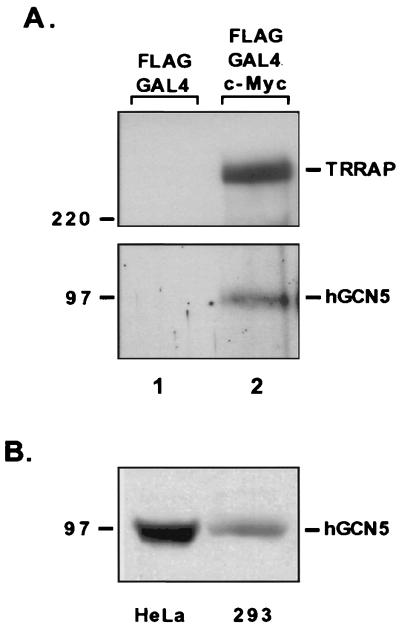
Among the mammalian nuclear cofactors with well-documented histone acetylase activity is the ortholog of the yeast protein GCN5p, termed hGCN5 in humans (45). We have recently shown that in yeast, TRA1p is a component of the GCN5p-containing SAGA complex (34). This observation prompted an examination of whether hGCN5 could account for the HAT activity associated with the c-Myc N terminus. Western blots of affinity-purified proteins were probed with anti-hGCN5 serum, revealing that the c-Myc N terminus formed a specific interaction with hGCN5 from HeLa cell nuclear extracts (Fig. 2A, lower panel). The hGCN5 protein detected by the antiserum migrates with an apparent molecular mass of 100 kDa, consistent with the full-length form of the protein. While some investigators have reported the existence of a 50-kDa form of hGCN5 (termed hGCN5-S) (8, 37), no such species was apparent in our studies. The specificity of the hGCN5 interaction with c-Myc was demonstrated by the absence of detectable hGCN5 in eluates from GAL4 DBD affinity purification. As previously reported, the c-Myc N terminus also specifically recruits the nuclear cofactor TRRAP (Fig. 2A, upper panel). By comparing the signal for hGCN5 in our captured material to the signal obtained from a known quantity of the starting extract (HeLa nuclear extracts [Fig. 2B]), we have determined that less than 0.1% of the total hGCN5 available is recruited in our in vitro binding reactions. However, this estimate may be artificially lower than the actual percentage of hGCN5 recruited into the c-Myc–TRRAP complex, since the in vitro purification scheme requires that the complex remain stable through both a 16-h binding reaction and an extensive series of wash steps.
FIG. 2.
The c-Myc N terminus recruits the HAT hGCN5. (A) Proteins captured by the GAL4 DBD or GAL4 DBD–c-Myc fusion were resolved by SDS-PAGE (8% gel) and immunoblotted for either the c-Myc cofactor TRRAP (A) or the recently described mammalian HAT hGCN5 (B). Antibodies to hGCN5 were the generous gift from Nickolai Barlev and Shelley Berger. Numbers at left (in kilodaltons) indicate positions of size markers. (B) Western blotting for hGCN5 was performed with HeLa cell nuclear extracts and whole-cell lysates from 293 cells.
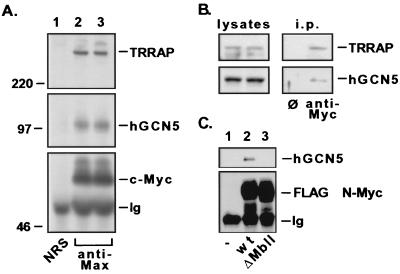
The in vitro binding of hGCN5 to the c-Myc N terminus prompted an examination of whether this interaction could be observed in vivo with endogenous proteins. c-Myc–Max heterodimers were immunoprecipitated from 293 cell lysates by using antibodies specific for Max (Fig. 3A, lower panel). Western blots of the precipitates showed that hGCN5 specifically associates with the c-Myc–Max dimer (Fig. 3A, middle panel). Control antiserum did not precipitate either c-Myc or hGCN5 (Fig. 3A, lane 1). TRRAP was also specifically coprecipitated with the c-Myc–Max heterodimers as previously demonstrated (Fig. 3A, upper panel). Identical results were obtained when anti-c-Myc antibodies were used for immunoprecipitation (Fig. 3B).
FIG. 3.
Myc family oncoproteins recruit hGCN5 in human cells. (A) 293 cells were lysed under nondenaturing conditions, and c-Myc–Max dimers were immunoprecipitated with antisera against Max (lanes 2 and 3). A parallel precipitation was performed with nonimmune rabbit serum (NRS) as a control (lane 1). Precipitated proteins were resolved by SDS-PAGE (8% gel) and subjected to immunoblotting for either c-Myc (lower panel), hGCN5 (middle panel), or TRRAP (upper panel). Numbers at the left (in kilodaltons) indicate positions of size markers. The position of the heavy-chain polypeptide from the precipitating antibody is indicated (Ig). (B) 293 cells were lysed and subjected to immunoprecipitation with either a control monoclonal antibody (ø) or a monoclonal antibody directed against c-Myc. Immunoprecipitates (i.p.) were blotted and probed for TRRAP (upper panel) and hGCN5 (lower panel). (C) 293 cells were transiently transfected with a cytomegalovirus-driven expression vector encoding FLAG epitope-tagged versions of either wild-type (wt) murine N-Myc (lane 2) or a mutant lacking amino acids 100 to 116 of the MbII domain (lane 3). Following transfection, cells were lysed as for panel A, immunoprecipitations were performed with anti-FLAG antibody; precipitates were resolved by SDS-PAGE and Western blotted for either the FLAG epitope (lower panel) or hGCN5 (upper panel). Mock transfected 293 cells served as a control (lane 1).
Recruitment of hGCN5 to c-Myc is presumed to be mediated by TRRAP, since TRRAP and GCN5 exist in the SAGA complex in S. cerevisiae where no Myc protein is present. We have previously demonstrated that recruitment of TRRAP to the transactivation domain of Myc family proteins requires the highly conserved MbII domain (29; S. B. McMahon et al., unpublished data). TRRAP-mediated recruitment of hGCN5 to Myc family proteins should therefore require the presence of MbII. To test this hypothesis, 293 cells were transiently transfected with expression vectors encoding FLAG epitope-tagged versions of either wild-type or MbII-deleted N-Myc. The N-Myc protein appears to have biological activities indistinguishable from those of c-Myc in terms of transformation potential and transactivation (S. B. McMahon and M. D. Cole, unpublished observations). Following lysis, these proteins were immunoprecipitated and the precipitates were analyzed by Western blotting (Fig. 3C). In keeping with a TRRAP-dependent mechanism of hGCN5 recruitment, hGCN5 is recruited only to wild-type N-Myc and not to the mutant of N-Myc lacking MbII (Fig. 3C, upper panel). Probing for the common FLAG epitope revealed that both wild-type and mutant proteins were expressed and precipitated in similar quantities (Fig. 3C, lower panel).
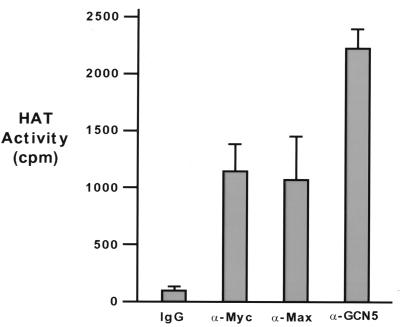
We next addressed whether c-Myc-associated HAT activity could be observed by direct immunoprecipitation of endogenous c-Myc from human cells. For this study, lysates prepared from serum-stimulated 293 cells were subjected to immunoprecipitation by antibodies directed against c-Myc, Max, or hGCN5. Precipitates were assayed for HAT activity, and the results are displayed in Fig. 4. Precipitation of c-Myc, either directly or as a dimer with Max, resulted in coprecipitation of significant HAT activity relative to control precipitates (Fig. 4). As expected, direct precipitation of hGCN5 also resulted in significant HAT activity.
FIG. 4.
The c-Myc oncoprotein associates with HAT activity in vivo. Immunoprecipitation of 293 cell lysates for either c-Myc, Max, or hGCN5 were subjected to an in vitro HAT assay in the presence of purified histones and radiolabeled acetyl-CoA. For each precipitation, lysate was prepared from a single confluent 15-cm-diameter dish of serum-stimulated 293 cells. Precipitations contained approximately 4 mg of total protein and were conducted in triplicate. Bars represent average values (± standard error).
The experiments above demonstrate an in vivo association between the c-Myc N terminus, hGCN5, and TRRAP, but they do not resolve whether hGCN5 is bound to c-Myc directly or through TRRAP. S. cerevisiae TRA1p copurifies with the SAGA complex which includes GCN5p, yet there is no ortholog of c-Myc in yeast. Thus, it seemed likely that human TRRAP would also exist in a complex with hGCN5 independent of c-Myc and that the binding of TRRAP to the c-Myc N terminus would be responsible for recruitment of the hGCN5 protein to the complex. To distinguish between these possibilities, hGCN5 was precipitated from lysates prepared from 293 cells, and the precipitates were probed for hGCN5 (Fig. 5A, lower panel) or TRRAP (Fig. 5A, upper panel). Immunoprecipitation of hGCN5 results in the specific coprecipitation of TRRAP, whereas parallel samples with control nonimmune serum showed no precipitation of either hGCN5 or TRRAP. Probing of these precipitates with antisera specific for c-Myc revealed no detectable c-Myc protein in the hGCN5-TRRAP complex (Fig. 5B), despite the presence of readily detectable c-Myc in 293 lysates when immunoprecipitated directly with anti-Myc antibodies. Coprecipitation of hGCN5 and TRRAP has also been observed in murine fibroblasts that have been rendered genetically deficient for c-Myc (data not shown). The absence of detectable c-Myc protein in the immunoprecipitates suggests that hGCN5 and TRRAP preexist in a complex in mammalian cells, that their interaction does not require c-Myc, and that only a small fraction of the hGCN5-TRRAP complex contains c-Myc. In support of this conclusion is the observation that the HAT activity recruited to the c-Myc N terminus represents less than 0.1% of the total hGCN5-dependent HAT activity in HeLa nuclear extracts in our in vitro binding reactions (Fig. 1 and 2).
FIG. 5.
In vivo association of hGCN5 and TRRAP in the absence of c-Myc. (A) Aliquots of 293 cell lysates were either electrophoresed directly (lane 1) or subjected to immunoprecipitation with nonimmune rabbit serum (NRS; lane 2) or anti-hGCN5 (lane 3). Proteins were resolved by SDS-PAGE (8% gel) and immunoblotted for TRRAP (upper panel) or hGCN5 (lower panel). Numbers at the left (in kilodaltons) indicate positions of molecular weight markers. (B) 293 cell lysates were subjected to immunoprecipitation with antibodies specific for hGCN5 (lane 2), c-Myc (lane 4), or species-matched, nonimmune sera (lanes 1 and 3). Precipitated proteins were resolved by SDS-PAGE (8% gel) and immunoblotted for c-Myc. The position of the heavy-chain polypeptide from the precipitating antibody is indicated (Ig).
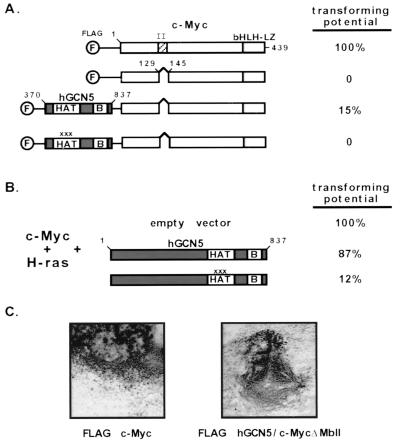
We recently demonstrated that recruitment of TRRAP to c-Myc is essential for transformation of mammalian cells. Coupling this observation with the evidence reported here that TRRAP recruits hGCN5 to c-Myc suggested the possibility that recruitment of hGCN5 is the essential function of TRRAP in c-Myc-mediated transformation. To test this hypothesis, the HAT domain of hGCN5 was directly fused to a mutant of c-Myc which is defective for TRRAP recruitment. This fusion protein was then assayed for its potential to transform primary rat embryo fibroblasts in cooperation with an activated allele of p21H-ras. As evident in Fig. 6A, wild-type c-Myc is a potent transforming agent, while the MbII deletion mutant (which fails to recruit TRRAP) is completely defective for transformation. Fusion of the hGCN5 HAT domain to this transformation-defective c-Myc mutant provides a partial rescue of its transforming potential (Fig. 6A and C). To ensure that this partial rescue was due to the HAT activity of hGCN5, specific point mutations were introduced in three amino acids known to be essential for the enzymatic function of GCN5 proteins (44). Introduction of these mutations blocked transformation by the fusion protein (Fig. 6A), suggesting that the loss of TRRAP recruitment can be rescued only by direct recruitment of an enzymatically active HAT domain. Further support for a role for hGCN5 in c-Myc-mediated transformation comes from studies in which a dominant negative mutant of hGCN5 (which carries the same mutations in the catalytic domain as those described above) blocks transformation by c-Myc (Fig. 6B). Expression of wild-type hGCN5 in this assay had no significant effect.
FIG. 6.
Partial rescue of a nontransforming c-Myc mutant by direct fusion to hGCN5. (A) Primary rat embryo fibroblasts were transfected with expression vectors for an activated allele of p21H-ras and one of the c-Myc constructs schematized at the left. The c-Myc constructs encoded either wild-type mouse c-Myc or a deletion mutant of this protein lacking 17 amino acids from the MbII domain. In addition, two fusion proteins were generated between the c-Myc MbII domain deletion mutant and the catalytic domain of hGCN5 (amino acids 370 to 837). For the first of these fusions, wild-type hGCN5 sequences were fused to c-Myc ΔMbII. The second fusion contained three single amino acid substitutions within the hGCN5 HAT domain (as indicated by x's). These mutations have been shown previously to block both HAT activity and transcriptional activation when introduced into the corresponding residues of S. cerevisiae GCN5. The critical bromodomain of hGCN5 (labeled B) was also included in these fusion proteins. Transforming potential of each of these proteins was determined by examining cells approximately 2 weeks posttransfection. Transforming potential was estimated based on both the total number of transformed foci generated with a given protein and the size of the individual foci obtained. bHLH-LZ, basic helix-loop-helix leucine zipper. (B) A transformation assay was performed as for panel A except that full-length versions of hGCN5 were coexpressed with c-Myc. Both wild-type and catalytically inactive forms of hGCN5 were assayed for their effect on c-Myc mediated transformation. (C) Representative foci obtained by transfection of rat embryo fibroblasts with p21H-ras and either wild-type c-Myc (left) or the MbII mutant-hGCN5 fusion protein (right).
DISCUSSION
All of the biological functions of c-Myc require a unique N-terminal domain whose biochemical function has only recently been determined (29). We show here that one function of this domain is to recruit the HAT hGCN5, which presumably modifies nucleosomal packaging at specific chromosomal targets. The recruitment of hGCN5 by c-Myc is mediated by the recently described nuclear cofactor TRRAP (29). TRRAP and hGCN5 are contained in a complex which associates with the c-Myc N terminus in vivo and after in vitro reconstitution. Furthermore, immunoprecipitation of endogenous c-Myc results in the copurification of a potent HAT activity from human cells. Finally, we show that hGCN5 exists in a complex with TRRAP in vivo that is independent of any association through c-Myc. Since the binding of TRRAP and hGCN5 to Myc is dependent on the essential N-terminal domain, MbII, it appears likely that the recruitment of the TRRAP-hGCN5 complex may also be essential for c-Myc biological activities.
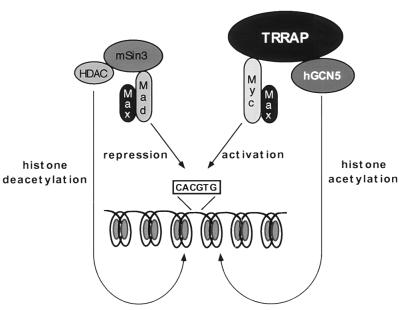
Many recent studies have demonstrated that the acetylation of histones is a critical step in gene activation, and several transcriptional cofactors have been shown to have HAT activity (3, 9, 31, 39, 48). Mad-Max heterodimers inhibit the transforming activity of c-Myc–Max heterodimers, while both heterodimers reportedly possess the same DNA binding specificity (1, 20, 23, 47, 50). In addition, overexpression of Mad leads to a dose-dependent decrease in the ability of c-Myc to transactivate reporter genes (1, 47). Mad-mediated inhibition of c-Myc function requires an interaction between Mad and the mSin3 adapter protein (2, 36), and this complex subsequently recruits the HDAC family of histone deacetylases (18, 22, 51). The current model suggests that Mad-Max dimers bind to c-Myc–Max target sites within the genome, recruiting the mSin3-HDAC complex which in turn deacetylates nearby histones, resulting in a closed chromatin configuration at essential c-Myc target genes (11). The results presented in this study provide a biochemical basis for the antagonistic effects of Myc and Mad by demonstrating that c-Myc recruits the HAT hGCN5, whose activity would be directly opposed by the recruitment of the histone deacetylase HDAC by Mad (Fig. 7).
FIG. 7.
Model of the opposing biochemical functions of c-Myc and Mad. Mad family proteins dimerize with Max and repress the expression of c-myc and max target genes, thereby blocking c-Myc function. Mad-mediated repression requires the recruitment of a multiprotein complex containing histone deacetylases of the HDAC family. The demonstration here that c-Myc, through its essential cofactor TRRAP, recruits the mammalian HAT hGCN5 suggests a potential biochemical basis for antagonistic biological functions of c-Myc and Mad. In this model, c-Myc–Max heterodimers activate transcription of target genes by recruitment of TRRAP and a HAT such as hGCN5. HAT activity results in nucleosomal remodeling at target gene loci, allowing more efficient transcription. Following displacement of Myc-Max dimers from their cognate DNA recognition element, Mad-Max dimers recruit deacetylase activity to these sites, which in turn removes the acetyl groups from histones in nearby nucleosomes. This process facilitates chromatin condensation and consequently transcriptional repression.
In S. cerevisiae, TRA1p is a component of the macromolecular SAGA complex (34). At least four distinct mammalian counterparts of the SAGA complex have now been described (28, 30, 46); two of them, TFTC and STAGA, contain the full-length form of hGCN5. TFTC and STAGA can be distinguished based on the presence and absence, respectively, of the TAFII100 protein. Our preliminary results indicate that the hGCN5-TRRAP complex recruited to the c-Myc N terminus (as defined in Fig. 2) lacks TAFII100 (data not shown). While this result suggests that the hGCN5 complex recruited by c-Myc is STAGA-like, it remains possible that c-Myc recruits an hGCN5 complex which is distinct from those described to date. Moreover, in S. cerevisiae, TRA1 is essential for viability (34) whereas the GCN5 gene is not (13, 27). This observation suggests that the histone acetylase hGCN5 may not be the only critical activity recruited to c-Myc by TRRAP. In support of this suggestion, TRA1p has been reported to be a component of a second multiprotein transcriptional regulatory complex designated NuA4 (16). The NuA4 complex lacks GCN5p but contains another HAT, ESA1p (16); like TRA1, ESA1 is essential for growth of S. cerevisiae (38). The interaction of TRRAP with potential human orthologs of ESA1p is currently under investigation. In addition, it has recently been reported that in human cells, TRRAP is a component of a large multiprotein complex which includes the hGCN5 homolog PCAF (42). Our failure to detect PCAF in our c-Myc complexes (data not shown) presumably results from the low expression level of PCAF in the HeLa and 293 cells used in our study. To date, no functional distinctions have been found between the closely related hGCN5 and PCAF proteins, and current data suggest that TRRAP can enter complexes containing either of these HATs (this study and reference 42). In addition, it remains possible that TRRAP recruits other HAT proteins to c-Myc.
The finding that a majority of the TRRAP-hGCN5 complex lacks c-Myc suggests that this complex also plays a role in the activity of transcription factors other than c-Myc. In support of this, we have recently reported that TRRAP is recruited to the transactivation domain of the E2F1 transcription factor (29). Intriguingly, E2F1 activity, like that of c-Myc, is antagonized by the HDAC family of histone deacetylases (5, 24, 26). Furthermore, another component of the SAGA complex (ADA3) binds to the nuclear retinoic acid receptor (43). Since TRA1p, GCN5p, and ADA3p/NGG1p are all components of SAGA in yeast (15, 19, 33, 34), it seems likely that mammalian TRRAP will associate not only with hGCN5 but also with the other components of the mammalian SAGA complex. This complex is presumably available for recruitment by many mammalian transcription factors in addition to c-Myc.
The partial genetic rescue of transformation by direct fusion of the hGCN5 catalytic domain to the ΔMbII mutant of c-Myc suggests that at least some portion of the essential role of TRRAP as a cofactor for c-Myc relies on its ability to recruit hGCN5 or a similar HAT to c-Myc target genes. The finding that only a partial rescue is achieved by direct recruitment of a HAT may result from the artificial context in which the HAT domain is located in the fusion protein, potentially resulting in suboptimal HAT activity or slightly altered substrate specificity. An alternative explanation for the partial rescue is that the c-Myc N terminus provides essential functions other than recruitment of the TRRAP-hGCN5 complex. In support of this hypothesis, we have recently isolated a second chromatin remodeling complex which is recruited to the c-Myc N terminus and which may function in concert with the HAT activity recruited by TRRAP (46a).
c-Myc is capable of both activation and repression of distinct target genes (32). An implication of the results presented here is that targets of the c-Myc–TRRAP–hGCN5 complex should be restricted to the subset of genes activated by c-Myc. This conclusion is based on current models of chromatin reorganization, which suggest that acetylation of core histone by proteins such as hGCN5 results in an open configuration, thereby allowing more efficient transcription (41). Thus, it is perplexing that MbII deletion mutations in c-Myc that fail to bind TRRAP do not have reduced transactivation activity in most transient reporter assays (6). One explanation is that transiently transfected DNA is not packaged into a chromatin configuration that is responsive to the recruitment of hGCN5-mediated acetylation of histones. As most assays of the role of MbII in transactivation have been performed with episomal plasmids as targets, they may not represent an accurate assessment of the true role of MbII in transactivation by c-Myc.
Current models of HAT protein activity suggest that one hypothesis for the role of hGCN5 in c-Myc's activities might be due to the relaxing of chromatin packaging at target genes following histone acetylation by hGCN5. Alternatively, hGCN5 may be recruited to this complex to acetylate nonhistone proteins such as c-Myc itself. This second scenario has ample precedence, for example, with the acetylation of p53 protein by p300, which results in increased p53 DNA binding activity (17). If hGCN5 acetylates nonhistone proteins when recruited into the c-Myc–TRRAP complex, a simple histone-based model of functional activity may require reevaluation.
ACKNOWLEDGMENTS
We are grateful to Shelley Berger and Nickolai Barlev of the Wistar Institute for generously providing antibodies to human GCN5 and to Elizabeth Moran for advice regarding histone acetylation assays. We also thank Penny Rushton for excellent technical assistance.
S.B.M. is a Special Fellow of the Leukemia Society of America.
REFERENCES
- 1.Ayer D E, Kretzner L, Eisenman R N. Mad: a heterodimeric partner for max that antagonizes myc transcriptional activity. Cell. 1993;72:211–222. doi: 10.1016/0092-8674(93)90661-9. [DOI] [PubMed] [Google Scholar]
- 2.Ayer D E, Lawrence Q A, Eisenman R N. Mad-Max transcriptional repression is mediated by ternary complex formation with mammalian homologs of yeast repressor Sin3. Cell. 1995;80:767–776. doi: 10.1016/0092-8674(95)90355-0. [DOI] [PubMed] [Google Scholar]
- 3.Bannister A J, Kouzarides T. The CBP co-activator is a histone acetyltransferase. Nature. 1996;384:641–643. doi: 10.1038/384641a0. [DOI] [PubMed] [Google Scholar]
- 4.Bouchard C, Staller P, Eilers M. Control of cell proliferation by Myc. Trends Cell Biol. 1998;8:202–206. doi: 10.1016/s0962-8924(98)01251-3. [DOI] [PubMed] [Google Scholar]
- 5.Brehm A, Miska E A, McCance D J, Reid J L, Bannister A J, Kouzarides T. Retinoblastoma protein recruits histone deacetylase to repress transcription. Nature. 1998;391:597–601. doi: 10.1038/35404. [DOI] [PubMed] [Google Scholar]
- 6.Brough D E, Hofmann T J, Ellwood K B, Townley R A, Cole M D. An essential domain of the c-Myc protein interacts with a nuclear factor that is also required for E1A-mediated transformation. Mol Cell Biol. 1995;15:1536–1544. doi: 10.1128/mcb.15.3.1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brownell J E, Allis C D. An activity gel assay detects a single, catalytically active histone acetyltransferase subunit in Tetrahymena macronuclei. Proc Natl Acad Sci USA. 1995;92:6364–6368. doi: 10.1073/pnas.92.14.6364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Candau R, Moore P A, Wang L, Barlev N, Ying C Y, Rosen C A, Berger S L. Identification of human proteins functionally conserved with yeast putative adaptors ADA2 and GCN5. Mol Cell Biol. 1996;16:593–602. doi: 10.1128/mcb.16.2.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen H, Lin R J, Schiltz R L, Chakravarti D, Nash A, Nagy L, Privalsky M L, Nakatani Y, Evans R M. Nuclear receptor coactivator ACTR is a novel histone acetyltransferase and forms a multimeric complex with P/CAF and CBP/p300. Cell. 1997;90:569–580. doi: 10.1016/s0092-8674(00)80516-4. [DOI] [PubMed] [Google Scholar]
- 10.Cole M D, McMahon S B. The Myc oncoprotein: a critical evaluation of transactivation and target gene regulation. Oncogene. 1999;18:2916–2924. doi: 10.1038/sj.onc.1202748. [DOI] [PubMed] [Google Scholar]
- 11.Dang C V. c-Myc target genes involved in cell growth, apoptosis, and metabolism. Mol Cell Biol. 1999;19:1–11. doi: 10.1128/mcb.19.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Facchini L M, Penn L Z. The molecular role of Myc in growth and transformation: recent discoveries lead to new insights. FASEB J. 1998;12:633–651. [PubMed] [Google Scholar]
- 13.Georgakopoulos T, Thireos G. Two distinct yeast transcriptional activators require the function of the GCN5 protein to promote normal levels of transcription. EMBO J. 1992;11:4145–4152. doi: 10.1002/j.1460-2075.1992.tb05507.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goruppi S, Gustincich S, Brancolini C, Lee W M F, Schneider C. Dissection of c-myc domains involved in S phase induction in NIH3T3 fibroblasts. Oncogene. 1994;9:1537–1544. [PubMed] [Google Scholar]
- 15.Grant P A, Duggan L, Cote J, Roberts S M, Brownell J E, Candau R, Ohba R, Owen-Hughes T, Allis C D, Winston F, Berger S L, Workman J L. Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes Dev. 1997;11:1640–1650. doi: 10.1101/gad.11.13.1640. [DOI] [PubMed] [Google Scholar]
- 16.Grant P A, Schieltz D, Pray-Grant M G, Yates III J R, Workman J L. The ATM-related cofactor Tra1 is a component of the purified SAGA complex. Mol Cell. 1998;2:863–867. doi: 10.1016/s1097-2765(00)80300-7. [DOI] [PubMed] [Google Scholar]
- 17.Gu W, Roeder R G. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell. 1997;90:595–605. doi: 10.1016/s0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- 18.Hassig C A, Fleischer T C, Billin A N, Schreiber S L, Ayer D E. Histone deacetylase activity is required for full transcriptional repression by mSin3A. Cell. 1997;89:341–347. doi: 10.1016/s0092-8674(00)80214-7. [DOI] [PubMed] [Google Scholar]
- 19.Horiuchi J, Silverman N, Marcus G A, Guarente L. ADA3, a putative transcriptional adaptor, consists of two separable domains and interacts with ADA2 and GCN5 in a trimeric complex. Mol Cell Biol. 1995;15:1203–1209. doi: 10.1128/mcb.15.3.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hurlin P J, Queva C, Koskinen P J, Steingrimsson E, Ayer D E, Copeland N G, Jenkins N A, Eisenman R N. Mad3 and Mad4: novel Max-interacting transcriptional repressors that suppress c-myc dependent transformation and are expressed during neural and epidermal differentiation. EMBO J. 1995;14:223–232. doi: 10.1002/j.1460-2075.1995.tb00252.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kato G J, Barrett J, Villa-Garcia M, Dang C V. An amino-terminal c-Myc domain required for neoplastic transformation activates transcription. Mol Cell Biol. 1990;10:5914–5920. doi: 10.1128/mcb.10.11.5914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Laherty C D, Yang W M, Sun J M, Davie J R, Seto E, Eisenman R N. Histone deacetylases associated with the mSin3 corepressor mediate Mad transcriptional repression. Cell. 1997;89:349–356. doi: 10.1016/s0092-8674(00)80215-9. [DOI] [PubMed] [Google Scholar]
- 23.Lahoz E G, Xu L, Schreiber-Agus N, DePinho R A. Suppression of Myc, but not E1a, transformation activity by Max-associated proteins, Mad and Mxil. Proc Natl Acad Sci USA. 1994;91:5503–5507. doi: 10.1073/pnas.91.12.5503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Luo R X, Postigo A A, Dean D C. Rb interacts with histone deacetylase to repress transcription. Cell. 1998;92:463–473. doi: 10.1016/s0092-8674(00)80940-x. [DOI] [PubMed] [Google Scholar]
- 25.MacGregor D, Li L H, Ziff E B. Dominant negative mutants of Myc inhibit cooperation of both Myc and adenovirus serotype-5 E1A with Ras. J Cell Physiol. 1996;167:95–105. doi: 10.1002/(SICI)1097-4652(199604)167:1<95::AID-JCP11>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 26.Magnaghi-Jaulin L, Groisman R, Naguibneva I, Robin P, Lorain S, Le Villain J P, Troalen F, Trouche D, Harel-Bellan A. Retinoblastoma protein represses transcription by recruiting a histone deacetylase. Nature. 1998;391:601–605. doi: 10.1038/35410. [DOI] [PubMed] [Google Scholar]
- 27.Marcus G, Silverman N, Berger S, Horiuchi J, Guarente L. Functional similarity and physical association between GCN5 and ADA2: putative transcriptional adaptors. EMBO J. 1994;13:4807–4815. doi: 10.1002/j.1460-2075.1994.tb06806.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martinez E, Kunda T K, Fu J, Roeder R G. A human SPT3-TAFII31-GCN5-L acetylase complex distinct from transcription factor IID. J Biol Chem. 1998;273:23781–23785. doi: 10.1074/jbc.273.37.23781. [DOI] [PubMed] [Google Scholar]
- 29.McMahon S B, VanBuskirk H A, Dugan K A, Copeland T D, Cole M D. The novel ATM-related protein TRRAP is an essential cofactor for the c-Myc and E2F oncoproteins. Cell. 1998;94:363–374. doi: 10.1016/s0092-8674(00)81479-8. [DOI] [PubMed] [Google Scholar]
- 30.Ogryzko V V, Kotani T, Zhang X, Schiltz R L, Howard T, Quin J, Nakatani Y. Histone-like TAFs within the PCAF histone acetylase complex. Cell. 1998;94:35–44. doi: 10.1016/s0092-8674(00)81219-2. [DOI] [PubMed] [Google Scholar]
- 31.Ogryzko V V, Schiltz R L, Russanova V, Howard B H, Nakatani Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 32.Peters M A, Taparowsky E J. Target genes and cellular regulators of the Myc transcription complex. Crit Rev Eukaryot Gene Expr. 1998;8:277–296. doi: 10.1615/critreveukargeneexpr.v8.i3-4.30. [DOI] [PubMed] [Google Scholar]
- 33.Saleh A, Lang V, Cook R, Brandl C J. Identification of native complexes containing the yeast coactivator/repressor proteins NGG1/ADA3 and ADA2. J Biol Chem. 1997;272:5571–5578. doi: 10.1074/jbc.272.9.5571. [DOI] [PubMed] [Google Scholar]
- 34.Saleh A, Schieltz D, Ting N, McMahon S B, Litchfield D W, Yates III J R, Lees-Miller S P, Cole M D, Brandl C J. Tra1p is a component of the yeast ADA/SPT transcriptional regulatory complexes. J Biol Chem. 1998;273:26559–26570. doi: 10.1074/jbc.273.41.26559. [DOI] [PubMed] [Google Scholar]
- 35.Sarid J, Halazonetis T D, Murphy W, Leder P. Evolutionarily conserved regions of the human c-myc protein can be uncoupled from transforming activity. Proc Natl Acad Sci USA. 1987;84:170–173. doi: 10.1073/pnas.84.1.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schreiber-Agus N, Chin L, Chen K, Torres R, Rao G, Guida P, Skoultchi A I, DePinho R A. An amino-terminal domain of Mxi1 mediates anti-Myc oncogenic activity and interacts with a homolog of the yeast transcriptional repressor SIN3. Cell. 1995;80:777–786. doi: 10.1016/0092-8674(95)90356-9. [DOI] [PubMed] [Google Scholar]
- 37.Smith E R, Belote J M, Schiltz R L, Yang X-J, Moore P A, Berger S L, Nakatani Y, Allis C D. Cloning of Drosophila GCN5: conserved features among metazoan GCN5 family members. Nucleic Acids Res. 1998;26:2948–2954. doi: 10.1093/nar/26.12.2948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smith E R, Eisen A, Gu W, Sattah M, Pannuti A, Zhou J, Cook R G, Lucchesi J C, Allis C D. ESA1 is a histone acetyltransferase that is essential for growth in yeast. Proc Natl Acad Sci USA. 1998;95:3561–3565. doi: 10.1073/pnas.95.7.3561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Spencer T E, Jenster G, Burcin M M, Allis C D, Zhou J, Mizzen C A, McKenna N J, Onate S A, Tsai S Y, Tsai M J, O'Malley B W. Steroid receptor coactivator-1 is a histone acetyltransferase. Nature. 1997;389:194–198. doi: 10.1038/38304. [DOI] [PubMed] [Google Scholar]
- 40.Stone J, DeLange T, Ramsay G, Jakobovits E, Bishop J M, Varmus H, Lee W. Definition of regions in human c-myc that are involved in transformation and nuclear localization. Mol Cell Biol. 1987;7:1697–1709. doi: 10.1128/mcb.7.5.1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Struhl K. Histone acetylation and transcriptional regulatory mechanisms. Genes Dev. 1998;12:599–606. doi: 10.1101/gad.12.5.599. [DOI] [PubMed] [Google Scholar]
- 42.Vassilev A, Yamauchi J, Kotani T, Prives C, Avantaggiati M L, Qin J, Nakatani Y. The 400 kDa subunit of the PCAF histone acetylase complex belongs to the ATM superfamily. Mol Cell. 1998;2:869–875. doi: 10.1016/s1097-2765(00)80301-9. [DOI] [PubMed] [Google Scholar]
- 43.vom Baur E, Harbers M, Um S J, Benecke A, Chambon P, Losson R. The yeast Ada complex mediates the ligand-dependent activation function AF-2 of retinoid X and estrogen receptors. Genes Dev. 1998;12:1278–1289. doi: 10.1101/gad.12.9.1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang L, Liu L, Berger S L. Critical residues for histone acetylation by GCN5, functioning in Ada and SAGA complexes, are also required for transcriptional function in vivo. Genes Dev. 1998;12:640–653. doi: 10.1101/gad.12.5.640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang L, Mizzen C, Ying C, Candau R, Barlev N, Brownell J, Allis C D, Berger S L. Histone acetyltransferase activity is conserved between yeast and human GCN5 and is required for complementation of growth and transcriptional activation. Mol Cell Biol. 1997;17:519–527. doi: 10.1128/mcb.17.1.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wieczorek E, Brand M, Jacq X, Tora L. Function of TAF(II)-containing complex without TBP in transcription by RNA polymerase II. Nature. 1998;393:187–191. doi: 10.1038/30283. [DOI] [PubMed] [Google Scholar]
- 46a.Wood, M. A., S. B. McMahon, and M. D. Cole. An ATPase/helicase complex is an essential cofactor for oncogenic transformation by c-Myc. Mol. Cell, in press. [DOI] [PubMed]
- 47.Wu S, Pena A, Korcz A, Soprano D R, Soprano K J. Overexpression of Mxi1 inhibits the induction of the human ornithine decarboxylase gene by the Myc/Max protein complex. Oncogene. 1996;12:621–629. [PubMed] [Google Scholar]
- 48.Yang X-J, Ogryzko V V, Nishikawa J, Howard B H, Nakatani Y. A p300/CBP-associated factor that competes with the adenoviral oncoprotein E1A. Nature. 1996;382:319–324. doi: 10.1038/382319a0. [DOI] [PubMed] [Google Scholar]
- 49.Zawel L, Reinberg D. Common themes in assembly and function of eukaryotic transcription complexes. Annu Rev Biochem. 1995;64:533–561. doi: 10.1146/annurev.bi.64.070195.002533. [DOI] [PubMed] [Google Scholar]
- 50.Zervos A, Gyuris J, Brent R. Mxi1, a protein that specifically interacts with Max to bind Myc-Max recognition sites. Cell. 1993;72:223–232. doi: 10.1016/0092-8674(93)90662-a. [DOI] [PubMed] [Google Scholar]
- 51.Zhang Y, Iratni R, Erdjument-Bromage H, Tempst P, Reinberg D. Histone deacetylases and Sap18, a novel polypeptide, are components of a human Sin3 complex. Cell. 1997;89:357–364. doi: 10.1016/s0092-8674(00)80216-0. [DOI] [PubMed] [Google Scholar]