Abstract
Rationale:
During neonatal heart regeneration, the fibrotic response, which is required to prevent cardiac rupture, resolves via poorly understood mechanisms. Deletion of the Hippo pathway gene Sav in adult CMs increases Yap activity and promotes cardiac regeneration, partly by inducing fibrosis resolution. Deletion of Yap in neonatal cardiomyocytes (CMs) leads to increased fibrosis and loss of neonatal heart regeneration, suggesting that Yap inhibits fibrosis by regulating intercellular signaling from CMs to cardiac fibroblasts (CFs).
Objective:
We investigated the role of Wntless (Wls), which is a direct target gene of Yap, in communication between CMs and CFs during neonatal heart regeneration.
Methods and Results:
We generated two mouse models to delete Wls specifically in CMs (Myh6-Cas9 combined with AAV9-Wls-gRNAs, and Myh6cre-ERT2/+; Wlsflox/flox mouse). Reanalysis of single-cell RNA-sequencing data revealed that Wnt ligands are expressed in CMs, whereas Wnt receptors are expressed in CFs, suggesting that Wnt signaling is directional from CMs to CFs during neonatal heart regeneration. Wls deletion in neonatal hearts disrupted Wnt signaling, showing as reduced noncanonical Wnt signaling in non-CMs. Four weeks after neonatal heart infarction, heart function was measured by echocardiography. Wls deletion in neonatal hearts after myocardial infarction impairs neonatal heart regeneration, marked by decreased contractile function and increased fibrosis. Wls mutant hearts display CF activation, characterized by increased extracellular matrix secretion, inflammation, and CF proliferation.
Conclusions:
These data indicate that during neonatal heart regeneration, intercellular signaling from CMs to CFs occurs via noncanonical Wnt signaling to rebuild cardiac architecture after myocardial infarction.
Subject Terms: Animal Models of Human Disease, Basic Science Research, Cell Signaling/ Signal Transduction, Fibrosis, Myocardial Infarction
Graphical Abstract
INTRODUCTION
Signaling interactions between cells are fundamental to tissue development, homeostasis, and injury responses in multicellular organisms.1 Some types of intercellular signaling require direct cell-cell contact that is mediated in large part through adhesion molecules, such as integrins, but also specialized membrane-bound receptor-ligand interactions such as Notch signaling. Other types of intercellular signaling occur distally, such as Wnt and Bmp-signaling that are fundamental to many organ systems including the heart.2, 3 Introduction of tissue injury adds tremendous complexity to intercellular signaling via the recruitment of inflammatory cells that import multiple new signals to the injured organ to maintain both short- and long-term organ function. Although organ homeostasis and injury responses are highly complex, recent single-cell omics experiments have begun to uncover the molecular events of intercellular signaling.4
After myocardial infarction (MI), millions of cardiomyocytes (CMs) die in the adult mammalian heart and fail to regenerate, disrupting tissue architecture. To compensate for CM loss and prevent ventricular wall rupture, cardiac fibroblasts (CFs) undergo a cell state transition, proliferate, and differentiate into myofibroblasts, which deposit an extracellular matrix (ECM) and secrete factors that recruit inflammatory cells into the damaged heart.5 In contrast to the adult heart, the neonatal mouse heart has transient regenerative ability shortly after birth6–8 and total loss of regenerative ability 7 days after birth.8,9 In addition to productive CM proliferation during neonatal heart regeneration, scar resolution allows for the recovery of pump function. However, mechanisms that regulate fibrosis resolution are poorly understood. Further insight into these mechanisms would have great medical value. In this study, we address whether Wnt-mediated intercellular signaling between CMs and CFs has a role in fibrosis resolution.
The Hippo pathway inhibits CM renewal and heart regeneration in the adult heart after MI.10–12 In response to physiologic cues such as increased cell density, the conserved Hippo pathway kinase cascade phosphorylates the transcriptional cofactor Yap to inhibit Yap transcriptional activity.13 When Hippo pathway activity is low, Yap enters the nucleus and forms a complex with TEA domain (Tead) transcription factors to induce the expression of genes that promote CM proliferation and heart regeneration.11, 12, 14 In addition to activating CM proliferation, Hippo pathway inhibition enhances resolution of cardiac fibrosis in both the adult mouse heart after MI via poorly understood mechanisms. Hippo pathway mutants display increased CM proliferation, reduced fibrosis, and increased vascularity.10, 11 In our previous studies, we specifically inhibited the Hippo pathway in CMs and observed the modulation of CFs. Thus, we hypothesized that the Hippo pathway regulates fibrosis through uncharacterized intercellular signaling between CMs and CFs.
Deletion of Yap in neonatal CMs after MI results in extensive fibrosis rather than neonatal heart regeneration, suggesting that uncharacterized Yap target genes, which likely encode signaling factors, promote the inhibition of fibrosis in neonatal heart regeneration.15 The Wnt signaling pathway interacts closely with the Hippo pathway,16and multiple mechanisms connect the Hippo pathway to Wnt. For example, Yap has been shown to directly interact with the canonical Wnt effector β-catenin.14 In addition, Yap interacts with the β-catenin destruction complex to modulate canonical Wnt signaling directly.17 Moreover, other evidence potentially implicating Wnt-signaling in CM-CF signaling interactions include recent single-cell RNA sequencing (scRNA-seq) profiling which revealed the existence of CF subpopulations that express Wnt pathway genes, suggesting a role for Wnt signaling in the CF injury response.18, 19
The Wnt pathway, which includes nineteen Wnt ligands, is categorized into two subclasses: canonical Wnt/β-catenin signaling and β-catenin–independent, noncanonical signaling. Whereas canonical Wnt signaling relies on the transcriptional activity of β-catenin, noncanonical signaling, which encompasses the Wnt/planar cell polarity (PCP) and the Wnt/Ca2+ pathways, regulates gene expression via the transcription factors activator protein 1 (AP-1) and nuclear factor of activated T-cells (NFAT), respectively.20–23 In the canonical pathway, Wnt ligands bind to the extracellular cysteine-rich domain of frizzled (FZD) receptors and LRP5/6 co-receptors to induce intracellular signal transduction. In the noncanonical pathway, Wnt ligands bind to FZD receptors and ROR1/2 co-receptors.22 Here, we found that the Wnt trafficking gene Wls is a direct target gene of Yap in CMs. Furthermore, in the neonatal heart, noncanonical Wnt signaling that originates in CMs suppresses CF activation and promotes neonatal heart regeneration.
METHODS
Data Availability.
All data and supporting materials have been provided with the published article, and all essential research materials are listed in the Major Resources Table in the Data Supplement. The sequencing data in this manuscript have been deposited in the Gene Expression Omnibus under accession number GSE179355. All other data are available from the corresponding author upon reasonable request.
RESULTS
Wls is a Direct Target Gene of Yap in CMs.
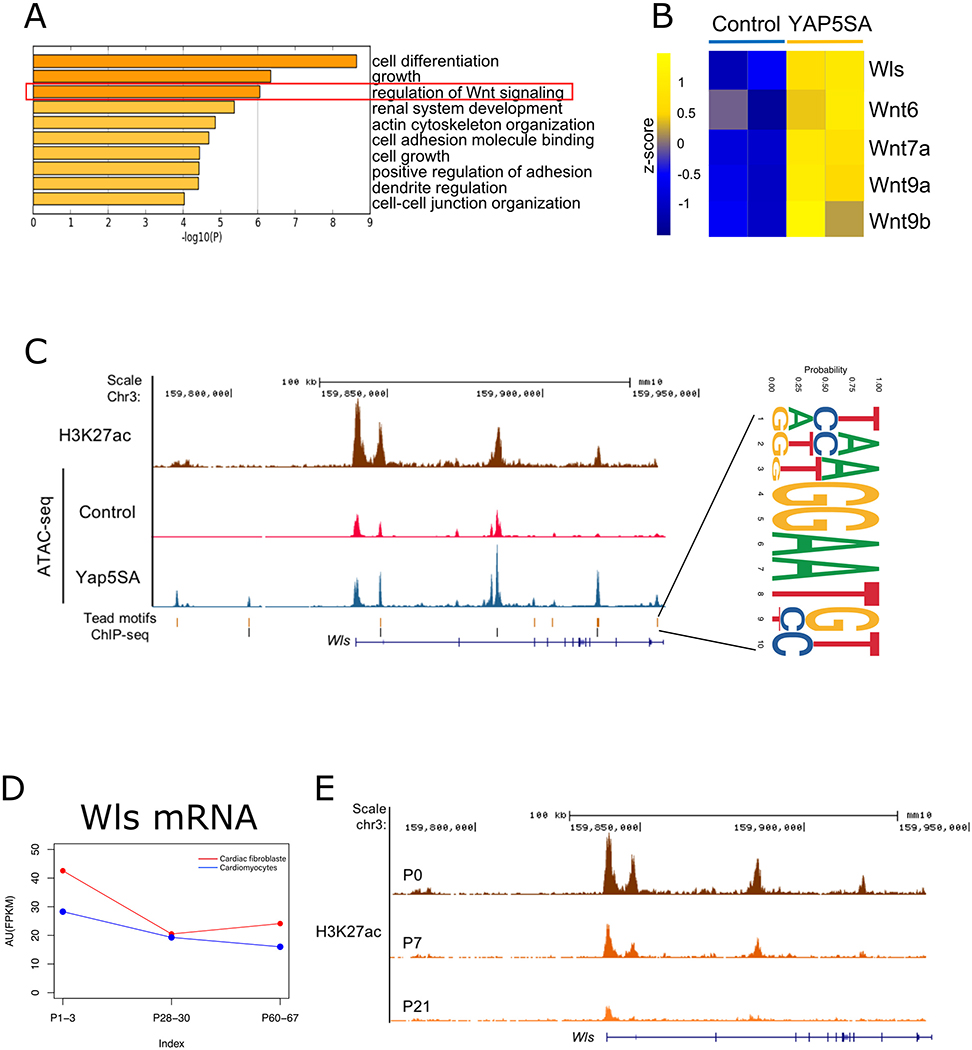
To identify the Yap-regulated factors that in turn regulate signaling from CMs to CFs, we screened for downstream Yap targets in CMs expressing Yap5SA, a version of Yap that contains serine-to-alanine mutations at Lats phosphorylation sites that confer enhanced transcriptional activity.24 We reanalyzed chromatin immunoprecipitation sequencing (ChIP-seq) data, transpose-accessible chromatin with high-throughput sequencing (ATAC-seq) data, and RNA sequencing (RNA-seq) data from Yap5SA adult CMs and identified a Yap-regulated gene program that included the Wnt pathway (Figure 1A).24 Expression levels of Wls and several Wnt ligand genes that are direct targets of Yap were upregulated in Yap5SA CMs (Figure 1B, C). Several Tead motifs residing in accessible chromatin regions and enriched for the active chromatin marker H3K27Ac were found in the Wls locus (Figure 1C). We uncovered Tead motifs both upstream of the transcriptional start site of Wls and in the Wls gene body, supporting the notion that Yap uses Tead binding sites to direct Wls transcription. Furthermore, available Yap ChIP-seq data25 revealed that Yap binds Tead elements in the Wls locus (Figure 1C). YAP activity is increased in CMs after MI,26, 27 which may induce the expression of downstream targets of Yap to promote cardiac regeneration. Given that Wls is essential for outbound Wnt signaling by regulating the sorting and secretion of all Wnt ligands, we focused exclusively on Wls for further study.28, 29
Figure 1. Wls is a direct target of Yap.
A, Gene ontology (GO) analysis of Yap5SA target genes (76 genes that overlapped in ChIP-seq, RNA-seq, and ATAC-seq datasets). The red square highlights genes involved in Wnt signaling regulation. B, Heatmap showing the upregulated expression of Wls and Wnt genes in Yap5SA CMs. C, H3K27ac ChIP-seq and chromatin accessibility mapping of the Wls locus by ATAC-seq. An increase in peaks (open chromatin) is seen in Yap5SA hearts. Tead motifs and enriched Yap ChIP-seq sites are also aligned. The consensus of 13 Tead motifs is shown on the right. D, Wls expression, as shown by RNA-seq. AU, arbitrary units; RPKM, reads per kilobase of transcript per million mapped reads. E, H3K27ac ChIP-seq peaks mark heart-specific enhancers on the Wls locus at different developmental stages.
We investigated the cardiac expression of Wls by using available RNA-seq data30 and observed that Wls expression is expressed in regenerative-stage hearts in both CMs and CFs and that Wls expression is slightly reduced in nonregenerative-stage hearts (Figure 1D). Furthermore, available ChIP-seq data31 revealed that levels of the active histone marker H3K27ac are reduced at the Wls locus in the P21-stage heart (Figure 1E). Collectively, these data suggest that Wls is a direct target of Yap and may be essential for neonatal heart regeneration.
Expression of Wnt Pathway Genes in the Heart.
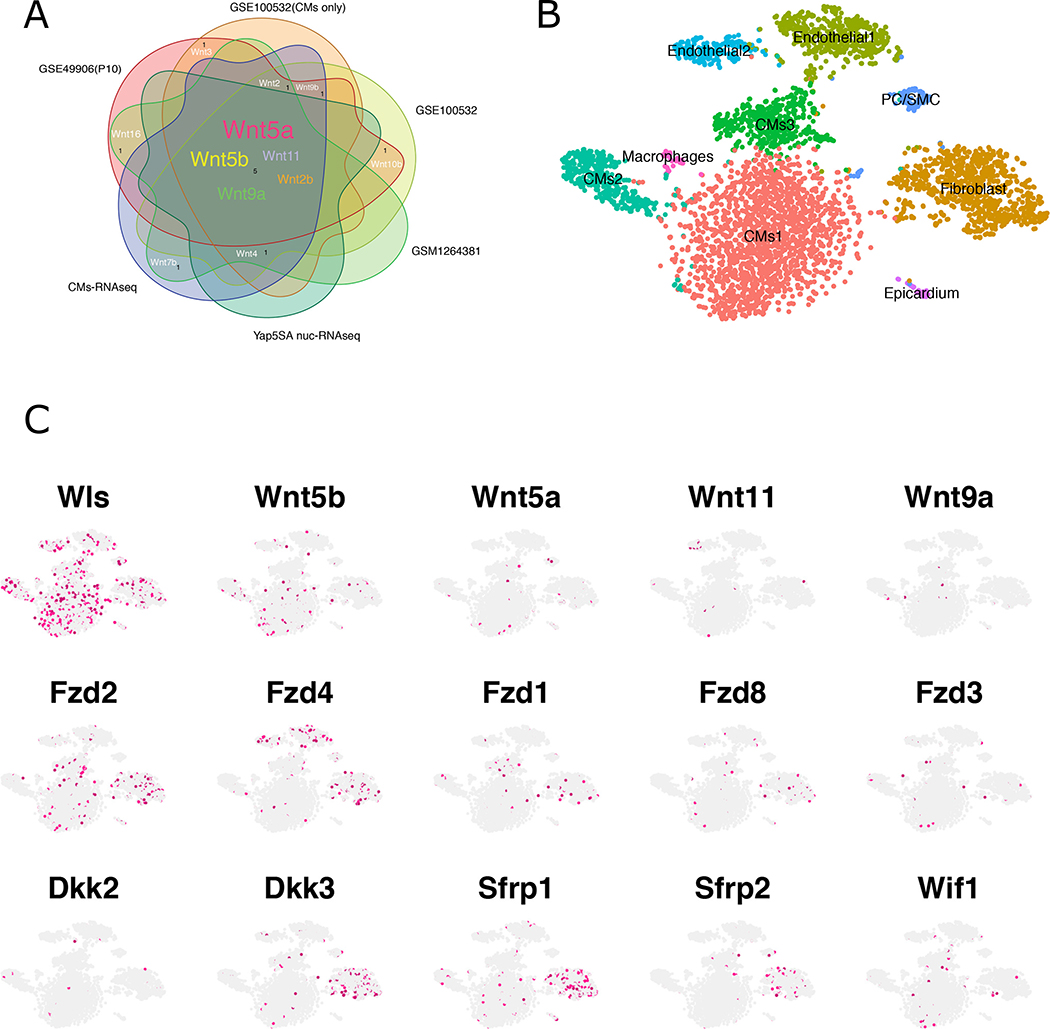
To identify the Wnt ligand gene(s) that are expressed in CMs, we reanalyzed cardiac expression data from previously published RNA-seq datasets.11, 24, 30, 31 Bulk RNA-seq data collected from mouse CMs at different timepoints revealed that a subset of Wnt ligand genes including Wnt5a, Wnt5b, Wnt9a, Wnt11, and Wnt2b was consistently expressed in CMs at higher levels than other Wnt ligands (Figure 2A). To further explore cardiac Wnt ligand gene expression, we reanalyzed previously published scRNA-seq data from embryonic (E) 14.5 hearts32 and single-nuclear RNA-seq (snRNA-seq) data from P6 hearts (Figure 2B,C and Figure IA–D in the Data Supplement).33 Notably, at both stages, Wls was expressed in numerous heart cell types, including CMs, CFs, endothelial cells, and macrophages (Figure 2B, C; Figure IA, B in the Data Supplement). At E14.5, CMs expressed Wnt5a, Wnt11, Wnt16, and Wnt4 (Figure IA, B in the Data Supplement). Several Wnt ligand genes including Wnt5a, Wnt5b, Wnt11, and Wnt9a were expressed in P6 CMs, consistent with the bulk RNA-seq data (Figure 2A–C). These findings indicate that postnatal regenerative-stage CMs express both Wls and Wnt ligand genes. In addition, we observed that many Wnt ligand genes, such as Wnt5a, that were expressed in CMs are Wnts that function in the noncanonical pathway,34–36 suggesting that noncanonical Wnt signaling is important for neonatal heart regeneration (Figure 2B, C; Figure IA, B in the Data Supplement).
Figure 2. Wnt gene expression in the heart.
A, RNA-seq data from multiple sources reveal Wnt ligand expression in the heart. GSE100532, GSE49906, and GSE1264381 from available datasets. CM-RNAseq was performed on isolated WT CMs, and Yap5SA nucRNA-seq was performed on PCM1-enriched nuclei from Yap5SA overexpression heart. B, tSNE visualization of graph-based clustering and classification for P6 neonatal hearts. C, The expression pattern of Wnt components in P6 hearts.
To identify Wnt-responsive cells, we examined the expression of Wnt receptor genes Fzd1-Fzd10 in both E14.5 and P6 datasets. At E14.5 Fzd1, Fzd2, and Fzd3 were expressed in most cell types, whereas Fzd1 and Fzd2 were expressed in CMs and CFs and Fzd3 was more restricted to CFs (Figure IC in the Data Supplement). In the P6 heart, Fzd1, Fzd2, Fzd4, and Fzd8 were expressed in CFs. Additionally, at the P6 stage, Fzd receptor genes were also expressed in other cell types (e.g. Fzd4 was also expressed in endothelial cells, and Fzd2 in CMs) (Figure 2B, C). Approximately 42% of Fzd2-positive cells were CFs, and Fzd2 expression levels were highest in CFs compared with other cell populations (Figure. 2B, C; Figure IIA in the Data Supplement). Interestingly, expression of several Wnt inhibitor genes including Dkk3, Sfrp1, and Sfrp2 was also enriched in CFs at E14.5 and P6 (Figure 2B, C; Figures ID and IIB in the Data Supplement). Wnt inhibitors are secreted matricellular proteins that antagonize Wnt signaling via distinct mechanisms. Although sFRPs interact with all Wnt ligands to block signal transduction, DKK2 and DKK3 inhibit canonical Wnt signaling by specifically interacting with and preventing LRP5/6 activation. Together, these results suggest that Wnt signaling to CFs is tightly regulated. Experiments in mice in which Wnt inhibitor function was modulated support the notion that Wnt signaling must be tightly regulated for normal cardiac homeostasis and in response to injury.37–42
Wls Deletion in CMs Using CRISPR/Cas9 Gene Editing.
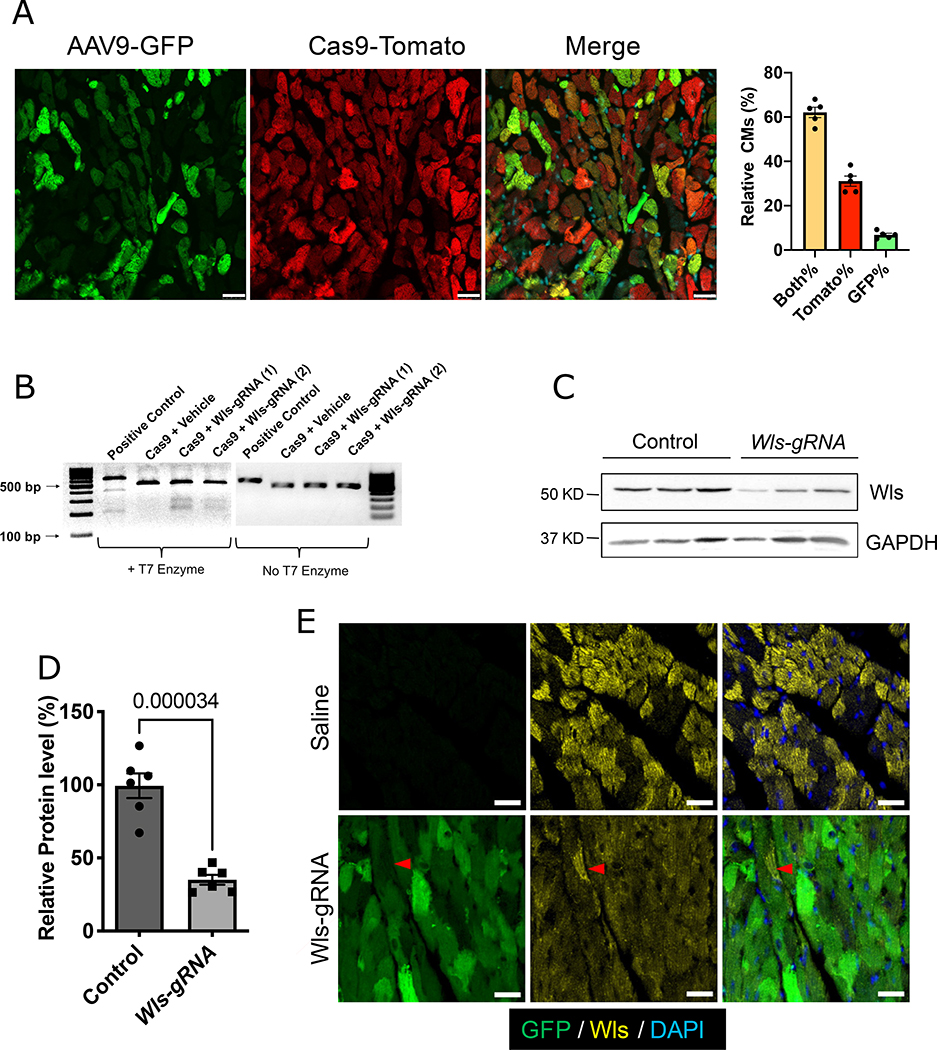
To functionally test the role of directional Wnt signaling from CMs to CFs in vivo, we inactivated Wls in CMs using CRISPR/Cas9 gene editing in a mouse line that expresses Cas9 in CMs (Myh6-Cas9, referred to as CM-Cas9) (Figure IIIA in the Data Supplement).43 Two guide RNAs (gRNAs) with specificity for exon 2 of Wls were delivered via adeno-associated virus 9 (AAV9) infection to regenerative-stage CMs (Figure IIIA in the Data Supplement). After a single injection of AAV9-Wls-gRNA into P3 CM-Cas9 mice, approximately 70% of CMs were infected by AAV9, whereas more than 90% of CMs expressed Cas9 (Figure 3A). CMs isolated from CM-Cas9 mice injected with AAV9-Wls-gRNA exhibited gene editing at the Wls locus (Figure 3B). Consistently, we detected a 60% reduction of Wls expression in CMs from CM-Cas9 mice injected with AAV9-Wls-gRNA compared with nontransgenic controls (Figure 3C, D). Immunofluorescence staining revealed a mosaic pattern of Wls expression in control hearts, with some CMs expressing higher levels of Wls (Figure 3E, and Figure IIIB in the Data Supplement). In hearts of CM-Cas9 mice injected with AAV9-Wls-gRNA, Wls expression was globally decreased; hereafter, we refer to these mice as Wls-knockdown (KD) mice. Together, these results indicate that delivery of AAV9-Wls-gRNA into CM-Cas9 mice is a rapid and efficient way to delete Wls specifically in CMs.
Figure 3. Deletion of Wls in CMs by CRISPR/Cas9.
A, Endogenous GFP (green, reporter of AAV9 infection) and tdTomato (red, reporter of Cas9) expression in CM-Cas9 mice 10 weeks after AAV9-Wls-gRNA injection. Scale bars=25 μm. Bar graph shows the percentages of CMs with both tdTomato and GFP (Both%), tdTomato only (Tomato%), and GFP only expression (GFP%). B, T7 surveyor assay of isolated CMs. Cleaved bands indicate genome editing in exon 2 of Wls. (1) and (2) indicate samples from 2 different mice. C, Western blotting to detect Wls protein in isolated CMs. Quantification of Western blotting is shown in (D) (n=6). Protein expression levels are relative to the GAPDH; the average value of the control group was set to 1. Error bars represent ± standard error of the mean. Mann-Whitney test was used for the comparison. Saline-injected wild-type littermates were used as controls. E, Immunofluorescence staining for Wls (yellow) in Wls-KD mice or saline-injected wild-type mice. Green (GFP), Wls-gRNA. White dashed line indicates uninfected cells that retain Wls expression. Saline indicates saline-injected wild-type littermates. Scale bars=25 μm.
CM-specific Wls Deletion Disrupts Outbound Noncanonical Wnt Signaling.
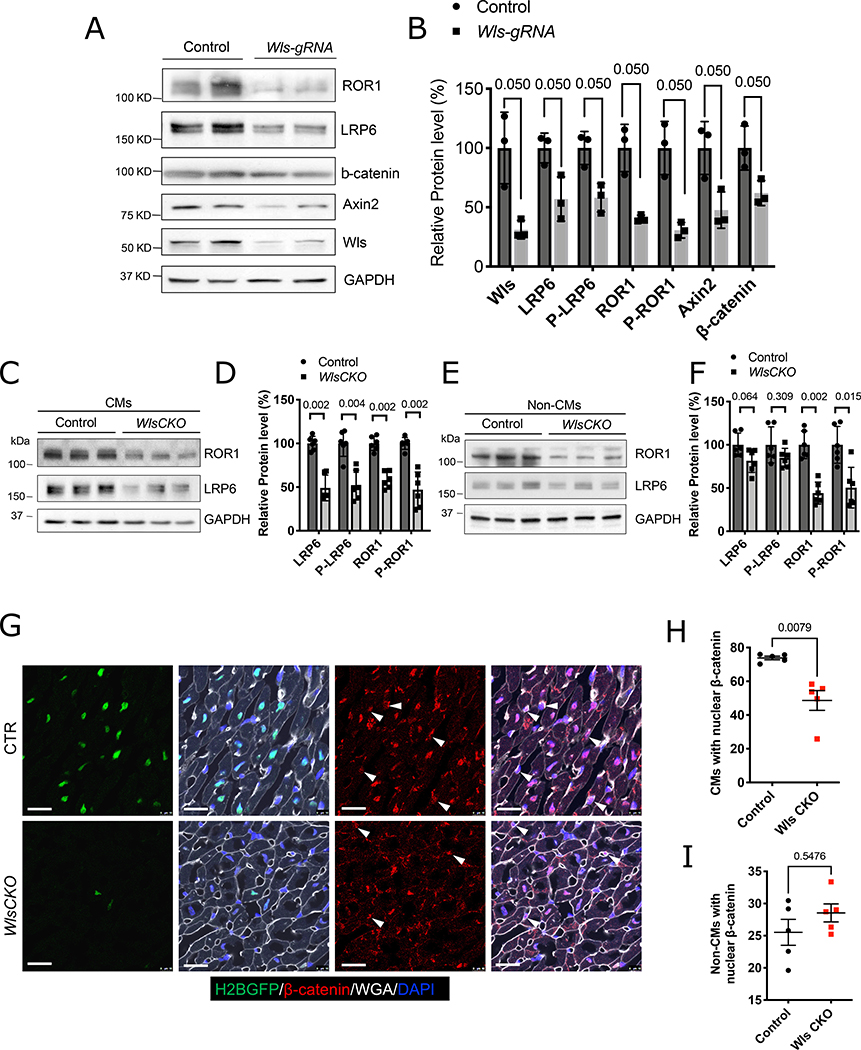
We next sought to determine whether CMs and non-CMs respond to reduced Wnt signaling from CMs via an autocrine or paracrine mechanism(s), respectively. During Wnt signal transduction, canonical Wnt ligands directly induce the phosphorylation of the LRP5/6 co-receptor, whereas noncanonical Wnt ligand/receptor interactions induce phosphorylation of ROR1/2 and RYK.20, 44, 45 We examined the phosphorylation of LRP6 (P-LRP6) and ROR1 (P-ROR1) in isolated CMs of Wls-KD mice. Total protein levels of both LRP6 and ROR1 were decreased in CMs after Wls deletion, consistent with previous reports that Wnt signaling stabilizes LRP6 and ROR146, 47 (Figure 4A, B). In addition, levels of both P-LRP6 and P-ROR1 (super-shifted bands) were decreased in CMs after Wls deletion. This indicated that both canonical and noncanonical signaling was reduced in CMs after Wls deletion in CMs and suggested an autocrine Wnt signaling loop in CMs (Figure 4A, B). Moreover, we observed decreased expression of β-catenin and its target Axin2 in Wls-deleted CMs, providing further evidence that canonical Wnt signaling is reduced in Wls-KD CMs (Figure 4A, B).
Figure 4. CM-specific Wls deletion disrupts Wnt signaling.
A, B, Immunoblotting of the indicated proteins in isolated CM lysates from CM-cas9 mice (A) with quantification (B). C, D, Western blotting of ROR1 and LRP6 proteins in isolated CMs (C) with quantification (D). l.e., long-exposure blot. E, F, Western blotting of ROR1 and LRP6 proteins in isolated non-CMs (E) with quantification (F). Littermate Wlsflox/flox mice were used as controls (CTR). Western blotting of ROR1 and LRP6 consistently detected two bands; the higher molecular weight bands are the phosphorylated proteins. Protein expression levels are relative to GAPDH; the average value of the control group was set to 1. G, Tcf/Lef:H2B-GFP reporter mice exhibited reduced activity of canonical Wnt/β-catenin signaling in CMs of Wls CKO mice. IF staining reveals decreased nuclear intensity of β-catenin in CMs of Wls CKO mice. Arrowhead indicates non-CMs with nuclear β-catenin. Scale bars=20 μm. H, I, Quantification CMs or non-CMs with nuclear β-catenin (n=5). Error bars represent standard error of the mean; B, D, F, H, and I were determined by using the Mann–Whitney test. Each dot represents a single animal.
To validate our CRISPR data, we generated Myh6cre-ERT2/+; Wlsflox/flox conditional knockout (Wls CKO) mice, in which a tamoxifen (TAM)-inducible Cre recombinase is driven by the CM-specific promoter Myh6. Consistent with our Wls-KD data, we observed that LRP6 and ROR1 expression was decreased in CMs after TAM injection, indicating that autocrine canonical and noncanonical Wnt signaling in CMs is disrupted in Wls CKO hearts (Figure 4C, D). We then performed Western blot analysis of non-CMs, which included CFs, endothelial cells, and smooth muscle cells. Interestingly, we observed decreased levels of ROR1 and p-ROR1 but not of LRP6 or p-LRP6 in non-CMs (Figure 4E, F). To validate these data, we used a reporter mouse that detects canonical Wnt signaling (Tcf/Lef:H2B-GFP). Expression of the H2B-GFP reporter and nuclear β-catenin were both reduced in CMs but not in non-CMs of Wls CKO mice (Figure 4G–I). Together, these results suggest that whereas CMs respond to both canonical and noncanonical Wnt signaling, non-CMs respond only to noncanonical Wnt signaling originating from CMs.
Both Wls-KD and Wls CKO mice are healthy and phenotypically indistinguishable from wild-type controls up to four months of age. Cardiac contractile function in 3-month-old mice measured by using echocardiography revealed that Wls-KD mice exhibit cardiac contractility similar to that of controls (Figure IIIC, D in the Data Supplement). In addition, Masson’s trichrome staining revealed that cardiac structure in Wls-KD mice is indistinguishable from that of controls (Figure IIIE in the Data Supplement). Thus, our findings indicate that reducing Wls activity in CMs does not affect postnatal cardiac homeostasis.
Wls is Required for Neonatal Heart Regeneration.
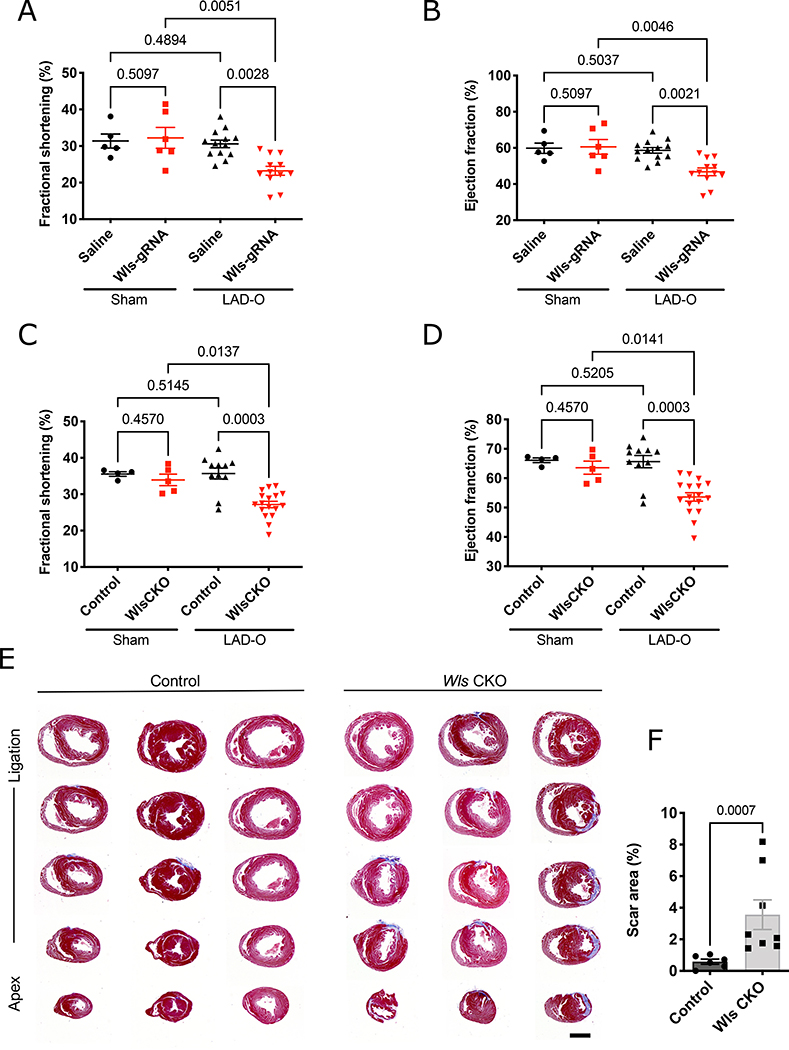
To determine whether Wls is required for neonatal heart regeneration after MI, we performed left anterior descending artery occlusion (LAD-O) in Wls-KD mice at P4, one day after the injection of AAV9-Wls-gRNA. No cardiac contractile function deficiencies were detected in control or Wls-KD sham-treated mice four weeks after treatment (Figure 5A, B and Figure IVA, B in the Data Supplement). Four weeks after LAD-O, cardiac contractile function in Wls-KD mice was significantly reduced compared with that in saline-injected controls (Figure 5A, B). Consistently, Wls-KD mice continued to exhibit decreased contractile function compared with saline-injected controls at six and eight weeks after LAD-O (Figure IVC, D in the Data Supplement). Moreover, we observed increased scarring and fibrosis in Wls-KD hearts (Figure IVE, F in the Data Supplement). Similar to Wls-KD mice, Wls CKO mice exhibited reduced contractile function four weeks after P2 LAD-O (Figure 5C, D), as well as increased scar size and fibrosis eight weeks after MI (Figure 5E, F). These findings indicate that Wls in CMs is essential for neonatal heart regeneration.
Figure 5. Requirement of Wls for neonatal heart regeneration.
A-D, Echocardiography revealed reduced fractional shortening (A, C) and ejection fraction (B, D) 4 weeks after P4 LAD-O in mice with the CRISPR/Cas-mediated deletion of Wls (A, B) or via the Cre/loxP-mediated conditional knockout of Wls (Wls CKO) (C, D) (each dot represents a single animal; n=5 for Saline-Sham; n=6 for Wls-gRNA-Sham; n=13 for Saline-LAD-O; n=12 for Wls-gRNA-LAD-O; n=4 for Control-Sham; n=5 for WlsCKO-Sham; n=11 for Control-LAD-O, n=17 for WlsCKO-LAD-O). Saline-injected wild-type littermates were used as controls for mice with CRISPR/Cas-mediated Wls deletion; Myh6cre-ERT2/+ mice were used as controls for Wls CKO mice. E, Trichrome-stained serial transverse sections of mouse hearts 8 weeks after LAD-O. F, Scar size quantification is shown. Error bars represent standard error of the mean; In A-D, the comparison were determined by using the Kruskal Wallis with two-stage Benjamini, Krieger, & Yekutieli FDR-based multiple comparison, F was determined by using the Mann–Whitney test. Scale bars=1 mm.
Reduced Wls is Tolerated by Neonatal Cardiomyocytes.
To determine the gene expression changes that arise in CMs depleted of Wls, we performed RNA-seq analysis on CMs isolated from control and Wls CKO hearts two weeks after P2-stage MI. Wls expression levels in Wls CKO CMs were reduced by more than 60% compared with control CMs, indicating the efficient deletion of Wls in these CMs (Figure VA in the Data Supplement). Interestingly, only a small number of differentially expressed genes was identified in CMs isolated from Wls CKO and control mice, suggesting that the primary defect after Wls deletion in CMs originates from non-CMs. The genes with the most significantly altered expression levels in Wls CKO CMs, in addition to Wls itself, included Thbs4 and Ces1d, which both had decreased expression levels in Wls CKO CMs (Figure VB in the Data Supplement). Motif analysis of the Thbs4 and Ces1d loci revealed several binding sites for TCF, AP1,23 and NFAT48 (Figure VC, D in the Data Supplement, and see Methods), suggesting that Thbs4 and Ces1d are direct targets of both canonical and noncanonical Wnt signaling in CMs. Notably, however, we detected no significant differences in proliferation and apoptosis rates between control and Wls CKO CMs after MI, suggesting that during neonatal heart regeneration, a 60% reduction in Wls is tolerated by neonatal CMs (Figure VIA–G in the Data Supplement).
Expression of Wnt Inhibitors is Upregulated in Cardiac Fibroblasts after Myocardial Infarction.
To gain insight into Wnt signaling in CFs after injury, we reanalyzed available scRNA-seq data from injured hearts at 3 and 7 days after MI.18 We observed that two main fibroblast populations, F-SL (cells with low expression of Sca represent secretory fibroblasts) and F-SH (cells with high expression of Sca represent cardiac colony-forming mesenchymal stromal cell-like fibroblasts), maintain homeostasis in the heart. These fibroblasts transition to an activated-fibroblast phenotype (F-Act) 3 days after MI and further differentiate into myofibroblasts (MYO) 7 days after MI (Figure VIIA in the Data Supplement). Our analysis revealed that expression of Wnt inhibitors such as Dkk3, Sfrp1, and Sfrp2 are elevated in differentiated myofibroblasts (Figure VIIB in the Data Supplement). We plotted the pseudotime differentiation trajectory (Figure VIIC–E in the Data Supplement) and observed that the expression of Dkk3, Sfrp1, and Sfrp2 increase during the F-Act to MYO transition (Figure VIIE in the Data Supplement). These findings suggest that, albeit transiently, CFs receive less Wnt signal during cell state transitions from F-Act to MYO after MI, suggesting that low Wnt expression is a hallmark of the MYO cell state. Together, these results suggest that the activation of CFs is accompanied by a decrease in the Wnt signaling received. Thus, we hypothesize that Wls deletion in CMs further decreased non canonical Wnt signaling to CFs and leads to CF activation.
Wls Deletion in Cardiomyocytes Promotes Cardiac Fibroblast Activation after Myocardial Infarction.
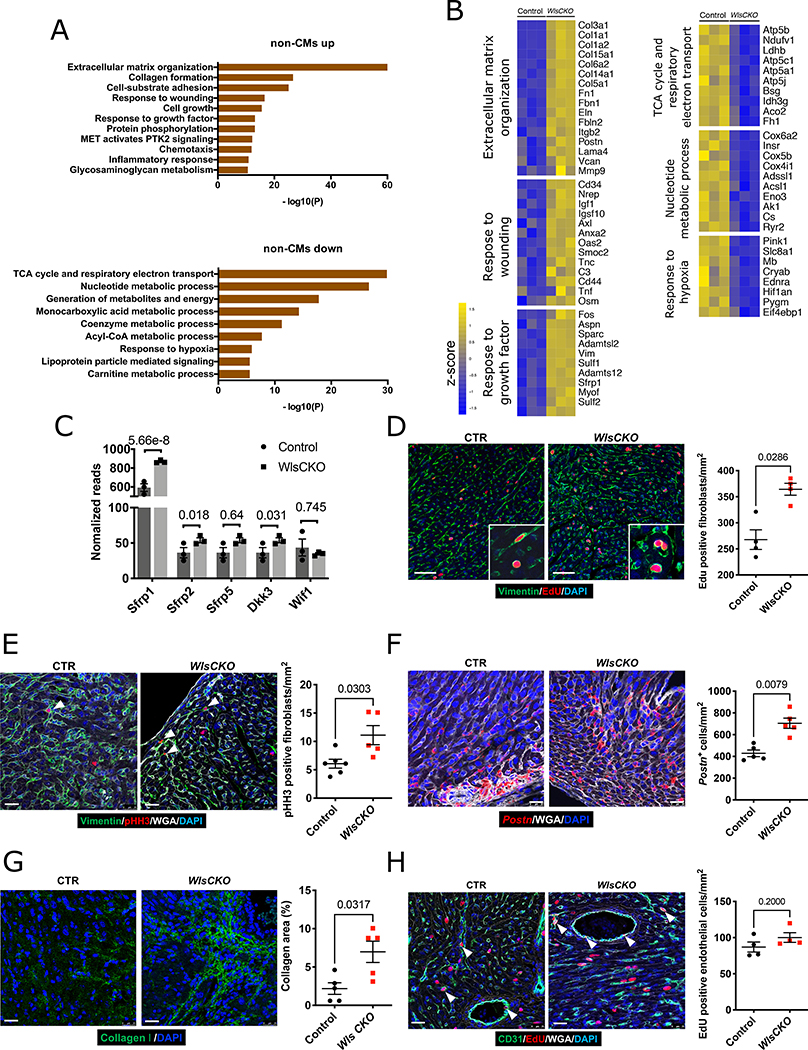
To evaluate the CF response to Wls deletion, we performed RNA-seq analysis on non-CMs isolated from control and Wls CKO hearts 2 weeks after P2-stage MI. In contrast to CMs, non-CMs from Wls CKO hearts showed significant changes in gene expression after MI compared with control hearts (633 genes significantly changed; FDR<0.05). Gene ontology analysis of non-CMs in Wls CKO hearts after MI revealed the downregulated expression of genes associated with metabolism, cytoskeleton organization, cytokine production, and calcium-mediated signaling (Figure 6A) and the significant upregulation of genes associated with extracellular matrix organization, response to wounding, and cell growth (Figure 6A). Genes associated with CF activation such as Postn, Fn1, Mmp9, Tnf, and Osm, as well as genes encoding collagen proteins such as Col1a1, Col1a2, Col3a149–52 were increased in non-CMs from Wls CKO hearts. These data indicate that CF activation is upregulated in Wls CKO hearts (Figure 6B). Motif analysis uncovered several AP1 and NFAT binding motifs in the Col1a2 locus and an AP1 binding motif in the Postn locus, suggesting that these genes are directly regulated by noncanonical Wnt signaling (Figure VIIIA, B in the Data Supplement). Consistent with data showing that the expression of Wnt inhibitor genes increases during CF activation (Figure VIIA–E in the Data Supplement), our RNA-seq results reveal that expression of Wnt inhibitor genes is increased in non-CMs of Wls CKO mice (Figure 6C). Together, these results suggest that noncanonical Wnt signaling from CMs to CFs inhibits fibrosis after MI.
Figure 6. CM-specific deletion of Wls promotes CF activation after injury.
A, GO analysis of 298 downregulated and 335 upregulated genes in non-CMs of Wls CKO mice (n=3). B, Heatmap shows the expression of associated genes in non-CMs of Wls CKO mice. C, RNA-seq shows the expression of Wnt inhibitors in non-CMs of control and Wls CKO mice, statistics were showed as false discovery rate-adjusted p-value. D, EdU incorporation (red) reveals the increased proliferation of fibroblasts with vimentin as a marker (green) in Wls CKO mice. Littermate Wlsflox/flox mice were used as the control (control, n=4; Wls CKO, n=4). Scale bars=50 μm. E, pHH3 immunofluorescence staining (red) of G2/M fibroblasts (green) in Wls CKO mice (control, n=6; Wls CKO, n=5). Scale bars=25 μm. Mice were challenged with LAD-O at P2, and hearts were harvested one week later at P9 for (D) and (E). F, RNA-scope reveals increased Postn-positive cells in Wls CKO mice (n=5 for each genotype). Scale bars=25 μm. G, Increased collagen formation in Wls CKO mice (n=5 for each genotype). Scale bars=25 μm. Mice were challenged with LAD-O at P2, and hearts were harvested two weeks later at P16 for (F) and (G). H, CD31 and EdU staining of endothelial cell proliferation. Mice were challenged with LAD-O at P2, and hearts were harvested one week later at P9. Error bars represent the standard error of the mean; comparisons were determined by using the Mann–Whitney test.
To investigate CF activation in Wls CKO hearts, we examined proliferation and differentiation in CFs.53 Immunostaining for CFs (vimentin) and cells in S-phase (EdU) revealed that the number of EdU-positive fibroblasts is significantly increased in Wls CKO hearts compared with control hearts (Figure 6D). Consistent with this finding, we detected more pHH3-positive fibroblasts in Wls CKO hearts than in control hearts (Figure 6E).
Postn is a marker of activated fibroblasts and has reported roles in tissue regeneration and wound healing.54 RNA-scope staining in the border zone revealed a significant increase in the number of Postn-positive cells in Wls CKO hearts compared with control hearts (Figure 6F). Collagen production in the infarcted zone was also increased in Wls CKO hearts compared with control hearts, indicating an increase in fibrosis (Figure 6G). We also examined the proliferation of endothelial cells in both control and Wls CKO hearts; however, no difference was detected (Figure 6H). Together, these results reveal that Wls deletion in CMs promotes CF proliferation, CF differentiation, and collagen production.
Noncanonical Wnt5a and Wnt9a Suppress Genes Associated with CF Activation.
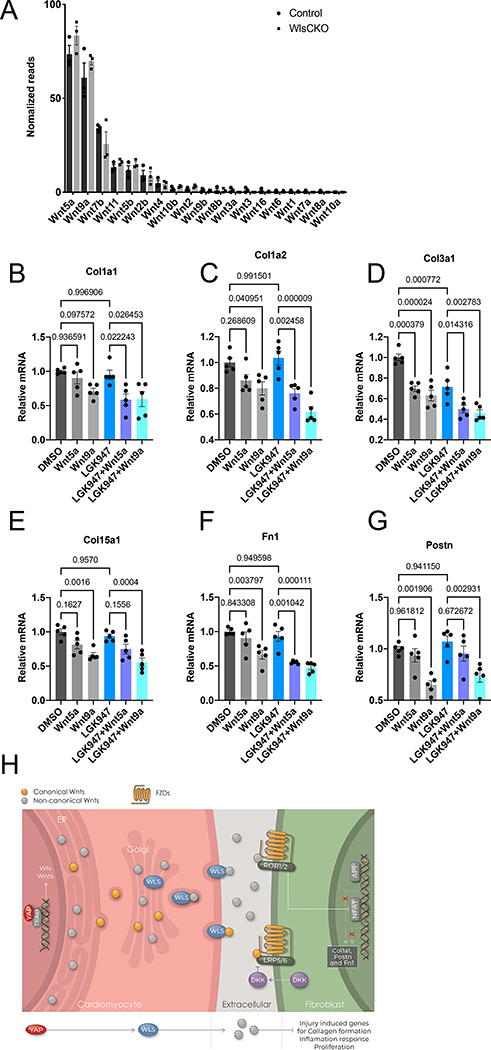
Consistent with the sc-RNA-seq data from uninjured mouse hearts, our RNA-seq analysis showed that Wnt5a, Wnt9a, and Wnt7b were the most highly expressed genes in CMs (Figure 7A). No differences in the expression of Wnt genes in CMs were detected between control hearts and Wls CKO hearts (Figure 7A). To examine how noncanonical Wnt ligands regulate CF activation, we treated human CFs with Wnt5a and Wnt9a, two of the most highly expressed Wnt ligands in CMs. Before Wnt5a or Wnt9a treatment, we pretreated human CFs with LGK-974,55 a porcupine inhibitor, to block autocrine Wnt signaling from human CFs. Interestingly, our results showed that treatment with Wnt5a or Wnt9a suppressed the expression of genes including Col1a1, Col1a2, Col3a1, Col15a1, Fn1, and Postn (Figure 7B–G). Thus, these results indicate that noncanonical Wnt ligands Wnt5a and Wnt9a may directly inhibit CF activation.
Figure 7. Noncanonical Wnt5a and Wnt9a suppress CF activation.
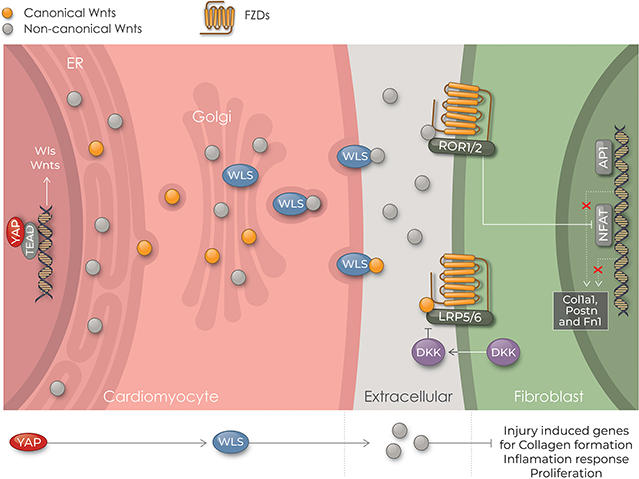
A, RNA-seq analysis of Wnt ligand gene expression in CMs from control and Wls CKO mice (n=3). Littermate Wlsflox/flox mice were used as the control. B-G, qRT-PCR analysis of gene expression in human CFs treated with Wnt5a or Wnt9a (both 100 ng/ml) for 24 hours with or without pretreatment with LGK-974 (1 μM) for 48 hours before harvesting as indicated. One-way ANOVA with Dunnett’s post hoc test was used for the comparison. Data from n=5 independent batches of cells run in triplicate. mRNA expression levels are relative to beta-actin; the average value of samples treated with DMSO was setting to 1. H, Working model in which cardiomyocytes inhibit fibroblast activation through Wls-mediated noncanonical Wnt signaling during neonatal heart regeneration.
DISCUSSION
The Hippo pathway genetically and functionally interacts with Wnt-signaling in multiple contexts. Yap and Taz have been reported to suppress the Wnt/β-catenin pathway by directly binding to either Dishevelled17, 56, 57 or β-catenin.58 Moreover, Wnt ligands such as Wnt5a/b and Wnt3a activate Yap/Taz, which in turn suppress canonical Wnt/β-catenin signaling by inducing the expression of Wnt repressors, such as DKK1, BMP4, and IGFBP4.59 Moreover, our published results reveal that nuclear Yap cooperates with β-catenin to promote the expression of numerous genes involved in heart development.14 This study provides a new mechanism for the interaction between Hippo and Wnt signaling in the heart. We show for the first time that Wls, which encodes a conserved multipass transmembrane protein required for transporting Wnt proteins to the plasma membrane for secretion,28, 29, 60–64 is a direct target of Yap in CMs and regulates noncanonical Wnt signaling between CMs and CFs. Furthermore, we show that the CM-specific deletion of Wls in the neonatal heart results in the loss of regenerative capacity concomitant with increased fibrosis after MI. These findings suggest a model in which Yap in neonatal heart regeneration positively regulates non canonical Wnt signaling to inhibit cardiac fibrosis (Figure 7H).
In the Wls CKO heart, which loses its regenerative capacity and acquires characteristics similar to those of the profibrotic adult heart, we observed the upregulation of Wnt inhibitor genes including Sfrp1 in non-CMs. These findings suggest that the expression of Wnt inhibitors is a marker for the activated profibrotic myofibroblast. Our findings further indicate that noncanonical Wnt ligands from CMs suppress CF activation after injury. This is consistent with another study showing that AAV9-mediated Wnt11 expression in CMs leads to reduced fibrosis and inflammation, although the cell types influenced were not determined.65 A previous study showed that deletion of Sfrp2, which results in increased Wnt signaling in CFs, inhibits collagen deposition after injury.42 Our findings are also in line with those of another study in which the CM-specific overexpression of Sfrp1 in mice, similar to the CM-specific deletion of Wls, led to increased scar size and worsened heart function after injury.41
While our data support a model for outbound directional signaling from CMs to non-CMs, Wnt signaling in the heart is highly complex and is known to also involve autocrine signaling in CFs 66. In cultured CFs, Sfrp2 treatment stimulated cardiac CF proliferation, adding another line of evidence that noncanonical Wnt ligands suppress CF activation.67 We observed highly expressed Wnt inhibitors, which indicates that Wnt signaling is tightly regulated in these cells. CMs are much larger in size than CFs. Considering the distribution of cells in the heart, CFs are almost completely encompassed by CMs. Thus, the effect of paracrine signals from CMs on CFs could be substantial.
In contrast to our data, several studies have revealed the beneficial effects of antagonizing Wnt signaling after injury.68–72 In these studies, inhibitors of Wnt signaling were administered systemically raising the possibility that non-cardiac effects were involved in those experiments. Ruling out non-cardiac effects of systemically administered Wnt inhibitors, such as the response of immune cells, will require more experiments. Moreover, given that Wnt signals can emanate from many additional sources such as inflammatory cells, endothelial cells, CFs and CMs, further study is needed to determine the roles of CM-secreted Wnt ligands during heart regeneration.
By using adult post-MI and sham Axin2 reporter mice, a canonical Wnt reporter mouse, Mizutani et al.73 showed canonical Wnt-responsive cells were localized to epicardium and subepicardial space. In addition, they observed an increased number of Axin2-positive cells, of which only ~8% expressed α-SMA, suggesting that canonical Wnt signaling plays a role in differentiation of a small subset of epicardial cells into myofibroblasts. Consistent with these findings, Xiang et al.74 showed that β-catenin activity in CFs is increased after trans-aortic constriction (TAC), a model of increased afterload, and functions to facilitate cardiac fibrosis in the adult TAC model. Notably, our model holds that in neonatal heart regeneration, noncanonical Wnt signaling inhibits fibrosis. Since it is known that noncanonical Wnt suppresses canonical Wnt signaling,75 we hypothesize that after MI in the context of neonatal heart regeneration, noncanonical Wnt signaling from CMs functions to inhibit canonical Wnt signals emanating from other cell sources, including autocrine Wnts, to suppress fibrosis. Since Wnt inhibitors are targets of canonical Wnt signaling, the reduction of noncanonical Wnt signaling may also explain the increased Wnt inhibitor expression in non-CMs in Wls CKO heart. 76, 77 Further experiments are required to address this hypothesis.
Interestingly, Wang et al.78 found, using single-cell RNA sequence profiling in the neonatal heart and in vitro studies, that Wnt signaling is upregulated in epicardial cells after MI. However, MI-induced Wnt signaling was stage specific. They found that R-Spondin 1, an activator of canonical Wnt signaling, was specifically upregulated in P1 epicardial cells after MI, whereas the noncanonical Wnt5a was only upregulated in P8 epicardial cells after MI. Combined with our findings, the data from Wang et al. raise the possibility that epicardial derived Wnt signaling works together with CM derived noncanonical Wnt signals to finely modulate CF activation after MI.
Regarding the limitations, our study demonstrated there was no significant difference of proliferation and apoptosis between hearts from control and WlsCKO mice. Whether Wls regulates the contractile and conductive abilities of CMs is unclear. In particular, it was reported that noncanonical Wnt signaling regulate Ca2+ transients 22, 79, 80, and canonical Wnt signaling regulates genes associate cardiac electrophysiology such as Gja1 and Scn5 81. Also, Further studies are required to determine how the noncanonical Wnt regulates fibroblast activation through ROR1/2 receptors.
In conclusion, our study provides important evidence that noncanonical Wnt signaling inhibits CF proliferation, collagen formation, and inflammation in neonatal heart regeneration (Figure 7H). Hence, studying the role of Wls and Wnt ligands in an established animal model of heart failure should prove to be valuable. In a previous study, we showed that Hippo deficiency reversed systolic heart failure, promoted CM proliferation, enhanced vascularity, and reduced fibrosis.11 Whether Wls plays a similar role in the adult heart warrants further study. Our firm evidence that outbound noncanonical Wnt ligands from CMs inhibit CF activation after injury has potential clinical implications and places us closer to developing a therapeutic strategy for treating fibrosis in heart failure patients.
Supplementary Material
NOVELTY AND SIGNIFICANCE.
What Is Known?
Neonatal mouse heart retains regenerative ability, and Yap is required for cardiac regeneration in mice
Hippo pathway inhibition promotes cardiac regeneration through stimulating cell autonomous cardiomyocyte proliferation in mice
Cardiac-specific Hippo deletion reverses fibrosis via a non-cell autonomous mechanism in murine cardiac regeneration
What New Information Does This Article Contribute?
Wls is a direct target of Yap in cardiomyocyte, and required for neonatal murine heart regeneration
Cardiac-specific Wls deletion in mice results in cardiac fibroblast activation
Murine cardiomyocytes communicate with cardiac fibroblasts through noncanonical Wnt signaling
The Hippo signal transduction pathway is a kinase cascade that restrains the activity of YAP, a transcriptional cofactor that promotes cell proliferation. Blocking the Hippo pathway specifically in murine cardiomyocytes promotes heart functional recovery after injury, not only by stimulating cardiomyocyte proliferation, but also by preventing fibrosis but the mechanism through which it inhibits fibrosis is unknown. Here we discovered that Wntless (Wls), which is essential for Wnt ligand secretion, is a direct Yap target gene in the murine cardiomyocyte. In neonatal mouse heart regeneration, when Yap activity is high, Wls regulates noncanonical Wnt ligands secretion in the cardiomyocyte, which signals to cardiac fibroblasts and suppresses its activation and transdifferentiation. This work uncovers a previously unknown connection between Hippo and Wnt pathways in cardiac regulation. Importantly, this study reveals how Hippo and Wnt pathways coordinate the crosstalk between cardiomyocytes and fibroblasts during neonatal murine heart regeneration. We also provide new insights into basic cardiac developmental biology, which have important implications in developing novel therapeutic approaches for regenerative medicine.
ACKNOWLEDGMENTS
We thank Tanner Monroe for help with Figure 1A–C; Thuy Tien Tran, Paul Swinton, Ann Bromley and Jong Hwan Kim for the technical supports in Martin Lab.
SOURCES OF FUNDING
The National Institutes of Health (HL 127717, HL 130804, and HL 118761 to J.F.M.) and the Vivian L. Smith Foundation (to J.F.M.). J.F.M. was supported by the LeDucq Foundation’s Transatlantic Networks of Excellence in Cardiovascular Research (14CVD01: “ Defining the Genomic Topology of Atrial Fibrillation” ) and the MacDonald Research Fund Award (16RDM001) and a grant from the Saving Tiny Hearts Society to J.F.M; K01DE026561, R03DE025873, R01DE029014, R56HL142704 and R01HL142704 to J.W.; American Heart Association Postdoctoral Fellowship (18POST34060186) and Career Development Award (849706) to S.J.L.. This work also supported by the Mouse Phenotyping Core, Neuroconnectivity core and Optical Imaging and Vital Microscopy core at Baylor College of Medicine. Nicole Stancel, PhD, ELS(D), of the Department of Scientific Publications at the Texas Heart Institute in Houston, TX, provided editorial support.
Nonstandard Abbreviations and Acronyms:
- ATAC-seq
transpose-accessible chromatin with high-throughput sequencing
- CF
cardiac fibroblasts
- CHIP-seq
chromatin immunoprecipitation sequencing
- CKO
conditional knockout
- ECM
extracellular matrix
- GO
gene ontology
- LAD-O
left anterior descending artery occlusion
- MI
myocardial infarction
- MYO
myofibroblasts
- RNA-seq
RNA sequencing
- scRNA-seq
single-cell RNA sequencing
- snRNA-seq
single-nuclear RNA-seq
- TAM
tamoxifen
Footnotes
DISCLOSURES
JFM is a cofounder of and owns shares in Yap Therapeutics.
SUPPLEMENTAL MATERIALS
Publisher's Disclaimer: This article is published in its accepted form. It has not been copyedited and has not appeared in an issue of the journal. Preparation for inclusion in an issue of Circulation Research involves copyediting, typesetting, proofreading, and author review, which may lead to differences between this accepted version of the manuscript and the final, published version.
Contributor Information
Shijie Liu, Cardiomyocyte Renewal Laboratory, Texas Heart Institute, Houston, TX.
Li Tang, Department of Molecular Physiology and Biophysics, Baylor College of Medicine, One Baylor Plaza, Houston, TX; School of Computer Science and Engineering, Central South University, Changsha, Hunan, China.
Xiaolei Zhao, Department of Pediatrics, McGovern Medical School, The University of Texas Health Science Center at Houston, Houston, TX.
Bao Nguyen, Department of Molecular Physiology and Biophysics, Baylor College of Medicine, One Baylor Plaza, Houston, TX.
Todd R. Heallen, Cardiomyocyte Renewal Laboratory, Texas Heart Institute, Houston, TX
Jianxin Wang, Central South University, Changsha, Hunan, China.
Jun Wang, Department of Pediatrics, McGovern Medical School, The University of Texas Health Science Center at Houston, Houston, TX..
James F. Martin, Cardiomyocyte Renewal Laboratory, Texas Heart Institute, Houston, TX Department of Molecular Physiology and Biophysics, Baylor College of Medicine, One Baylor Plaza, Houston, TX; Cardiovascular Research Institute, Baylor College of Medicine, One Baylor Plaza, Houston, TX.
REFERENCES
- 1.Armingol E, Officer A, Harismendy O and Lewis NE. Deciphering cell–cell interactions and communication from gene expression. Nat Rev Genet. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nusse R and Clevers H. Wnt/β-Catenin Signaling, Disease, and Emerging Therapeutic Modalities. Cell. 2017;169:985–999. [DOI] [PubMed] [Google Scholar]
- 3.Wang J, Greene SB and Martin JF. BMP signaling in congenital heart disease: new developments and future directions. Birth Defects Res A Clin Mol Teratol. 2011;91:441–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tanay A and Regev A. Scaling single-cell genomics from phenomenology to mechanism. Nature. 2017;541:331–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tallquist MD. Cardiac Fibroblast Diversity. Annu Rev Physiol. 2020;82:63–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Notari M, Ventura-Rubio A, Bedford-Guaus SJ, Jorba I, Mulero L, Navajas D, Marti M and Raya A. The local microenvironment limits the regenerative potential of the mouse neonatal heart. Sci Adv. 2018;4:eaao5553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Konfino T, Landa N, Ben-Mordechai T and Leor J. The type of injury dictates the mode of repair in neonatal and adult heart. J Am Heart Assoc. 2015;4:e001320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haubner BJ, Adamowicz-Brice M, Khadayate S, Tiefenthaler V, Metzler B, Aitman T and Penninger JM. Complete cardiac regeneration in a mouse model of myocardial infarction. Aging (Albany NY). 2012;4:966–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Porrello ER, Mahmoud AI, Simpson E, Hill JA, Richardson JA, Olson EN and Sadek HA. Transient regenerative potential of the neonatal mouse heart. Science. 2011;331:1078–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heallen T, Morikawa Y, Leach J, Tao G, Willerson JT, Johnson RL and Martin JF. Hippo signaling impedes adult heart regeneration. Development. 2013;140:4683–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leach JP, Heallen T, Zhang M, Rahmani M, Morikawa Y, Hill MC, Segura A, Willerson JT and Martin JF. Hippo pathway deficiency reverses systolic heart failure after infarction. Nature. 2017;550:260–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang J, Liu S, Heallen T and Martin JF. The Hippo pathway in the heart: pivotal roles in development, disease, and regeneration. Nat Rev Cardiol. 2018;15:672–684. [DOI] [PubMed] [Google Scholar]
- 13.Zheng Y and Pan D. The Hippo Signaling Pathway in Development and Disease. Dev Cell. 2019;50:264–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heallen T, Zhang M, Wang J, Bonilla-Claudio M, Klysik E, Johnson RL and Martin JF. Hippo pathway inhibits Wnt signaling to restrain cardiomyocyte proliferation and heart size. Science. 2011;332:458–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xin M, Kim Y, Sutherland LB, Murakami M, Qi X, McAnally J, Porrello ER, Mahmoud AI, Tan W, Shelton JM, Richardson JA, Sadek HA, Bassel-Duby R and Olson EN. Hippo pathway effector Yap promotes cardiac regeneration. Proc Natl Acad Sci U S A. 2013;110:13839–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Varelas X and Wrana JL. Coordinating developmental signaling: novel roles for the Hippo pathway. Trends Cell Biol. 2012;22:88–96. [DOI] [PubMed] [Google Scholar]
- 17.Azzolin L, Panciera T, Soligo S, Enzo E, Bicciato S, Dupont S, Bresolin S, Frasson C, Basso G, Guzzardo V, Fassina A, Cordenonsi M and Piccolo S. YAP/TAZ incorporation in the beta-catenin destruction complex orchestrates the Wnt response. Cell. 2014;158:157–70. [DOI] [PubMed] [Google Scholar]
- 18.Farbehi N, Patrick R, Dorison A, Xaymardan M, Janbandhu V, Wystub-Lis K, Ho JW, Nordon RE and Harvey RP. Single-cell expression profiling reveals dynamic flux of cardiac stromal, vascular and immune cells in health and injury. Elife. 2019;8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Forte E, Skelly DA, Chen M, Daigle S, Morelli KA, Hon O, Philip VM, Costa MW, Rosenthal NA and Furtado MB. Dynamic Interstitial Cell Response during Myocardial Infarction Predicts Resilience to Rupture in Genetically Diverse Mice. Cell Rep. 2020;30:3149–3163 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Komiya Y and Habas R. Wnt signal transduction pathways. Organogenesis. 2008;4:68–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cadigan KM and Peifer M. Wnt signaling from development to disease: insights from model systems. Cold Spring Harb Perspect Biol. 2009;1:a002881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Niehrs C The complex world of WNT receptor signalling. Nat Rev Mol Cell Biol. 2012;13:767–79. [DOI] [PubMed] [Google Scholar]
- 23.Nishita M, Itsukushima S, Nomachi A, Endo M, Wang Z, Inaba D, Qiao S, Takada S, Kikuchi A and Minami Y. Ror2/Frizzled complex mediates Wnt5a-induced AP-1 activation by regulating Dishevelled polymerization. Mol Cell Biol. 2010;30:3610–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Monroe TO, Hill MC, Morikawa Y, Leach JP, Heallen T, Cao S, Krijger PHL, de Laat W, Wehrens XHT, Rodney GG and Martin JF. YAP Partially Reprograms Chromatin Accessibility to Directly Induce Adult Cardiogenesis In Vivo. Dev Cell. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Croci O, De Fazio S, Biagioni F, Donato E, Caganova M, Curti L, Doni M, Sberna S, Aldeghi D, Biancotto C, Verrecchia A, Olivero D, Amati B and Campaner S. Transcriptional integration of mitogenic and mechanical signals by Myc and YAP. Genes Dev. 2017;31:2017–2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morikawa Y, Heallen T, Leach J, Xiao Y and Martin JF. Dystrophin-glycoprotein complex sequesters Yap to inhibit cardiomyocyte proliferation. Nature. 2017;547:227–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morikawa Y, Zhang M, Heallen T, Leach J, Tao G, Xiao Y, Bai Y, Li W, Willerson JT and Martin JF. Actin cytoskeletal remodeling with protrusion formation is essential for heart regeneration in Hippo-deficient mice. Sci Signal. 2015;8:ra41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Banziger C, Soldini D, Schutt C, Zipperlen P, Hausmann G and Basler K. Wntless, a conserved membrane protein dedicated to the secretion of Wnt proteins from signaling cells. Cell. 2006;125:509–22. [DOI] [PubMed] [Google Scholar]
- 29.Bartscherer K, Pelte N, Ingelfinger D and Boutros M. Secretion of Wnt ligands requires Evi, a conserved transmembrane protein. Cell. 2006;125:523–33. [DOI] [PubMed] [Google Scholar]
- 30.Giudice J, Xia Z, Wang ET, Scavuzzo MA, Ward AJ, Kalsotra A, Wang W, Wehrens XH, Burge CB, Li W and Cooper TA. Alternative splicing regulates vesicular trafficking genes in cardiomyocytes during postnatal heart development. Nat Commun. 2014;5:3603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nord AS, Blow MJ, Attanasio C, Akiyama JA, Holt A, Hosseini R, Phouanenavong S, Plajzer-Frick I, Shoukry M, Afzal V, Rubenstein JL, Rubin EM, Pennacchio LA and Visel A. Rapid and pervasive changes in genome-wide enhancer usage during mammalian development. Cell. 2013;155:1521–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xiao Y, Hill MC, Zhang M, Martin TJ, Morikawa Y, Wang S, Moise AR, Wythe JD and Martin JF. Hippo Signaling Plays an Essential Role in Cell State Transitions during Cardiac Fibroblast Development. Dev Cell. 2018;45:153–169 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hu P, Liu J, Zhao J, Wilkins BJ, Lupino K, Wu H and Pei L. Single-nucleus transcriptomic survey of cell diversity and functional maturation in postnatal mammalian hearts. Genes Dev. 2018;32:1344–1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Siar CH, Nagatsuka H, Han PP, Buery RR, Tsujigiwa H, Nakano K, Ng KH and Kawakami T. Differential expression of canonical and non-canonical Wnt ligands in ameloblastoma. J Oral Pathol Med. 2012;41:332–9. [DOI] [PubMed] [Google Scholar]
- 35.Yuzugullu H, Benhaj K, Ozturk N, Senturk S, Celik E, Toylu A, Tasdemir N, Yilmaz M, Erdal E, Akcali KC, Atabey N and Ozturk M. Canonical Wnt signaling is antagonized by noncanonical Wnt5a in hepatocellular carcinoma cells. Mol Cancer. 2009;8:90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chae WJ and Bothwell ALM. Canonical and Non-Canonical Wnt Signaling in Immune Cells. Trends Immunol. 2018;39:830–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sklepkiewicz P, Shiomi T, Kaur R, Sun J, Kwon S, Mercer B, Bodine P, Schermuly RT, George I, Schulze PC and D’Armiento JM. Loss of secreted frizzled-related protein-1 leads to deterioration of cardiac function in mice and plays a role in human cardiomyopathy. Circ Heart Fail. 2015;8:362–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wei WY, Zhao Q, Zhang WZ, Wang MJ, Li Y, Wang SZ and Zhang N. Secreted frizzled-related protein 2 prevents pressure-overload-induced cardiac hypertrophy by targeting the Wnt/β-catenin pathway. Mol Cell Biochem. 2020;472:241–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vatner DE, Oydanich M, Zhang J, Babici D and Vatner SF. Secreted frizzled-related protein 2, a novel mechanism to induce myocardial ischemic protection through angiogenesis. Basic Res Cardiol. 2020;115:48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhai CG, Xu YY, Tie YY, Zhang Y, Chen WQ, Ji XP, Mao Y, Qiao L, Cheng J, Xu QB and Zhang C. DKK3 overexpression attenuates cardiac hypertrophy and fibrosis in an angiotensin-perfused animal model by regulating the ADAM17/ACE2 and GSK-3β/β-catenin pathways. J Mol Cell Cardiol. 2018;114:243–252. [DOI] [PubMed] [Google Scholar]
- 41.Barandon L, Dufourcq P, Costet P, Moreau C, Allieres C, Daret D, Dos Santos P, Daniel Lamaziere JM, Couffinhal T and Duplaa C. Involvement of FrzA/sFRP-1 and the Wnt/frizzled pathway in ischemic preconditioning. Circ Res. 2005;96:1299–306. [DOI] [PubMed] [Google Scholar]
- 42.Kobayashi K, Luo M, Zhang Y, Wilkes DC, Ge G, Grieskamp T, Yamada C, Liu TC, Huang G, Basson CT, Kispert A, Greenspan DS and Sato TN. Secreted Frizzled-related protein 2 is a procollagen C proteinase enhancer with a role in fibrosis associated with myocardial infarction. Nat Cell Biol. 2009;11:46–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Carroll KJ, Makarewich CA, McAnally J, Anderson DM, Zentilin L, Liu N, Giacca M, Bassel-Duby R and Olson EN. A mouse model for adult cardiac-specific gene deletion with CRISPR/Cas9. Proc Natl Acad Sci U S A. 2016;113:338–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kamizaki K, Doi R, Hayashi M, Saji T, Kanagawa M, Toda T, Fukada SI, Ho HH, Greenberg ME, Endo M and Minami Y. The Ror1 receptor tyrosine kinase plays a critical role in regulating satellite cell proliferation during regeneration of injured muscle. J Biol Chem. 2017;292:15939–15951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yoshikawa S, McKinnon RD, Kokel M and Thomas JB. Wnt-mediated axon guidance via the Drosophila Derailed receptor. Nature. 2003;422:583–8. [DOI] [PubMed] [Google Scholar]
- 46.Mao B, Wu W, Davidson G, Marhold J, Li M, Mechler BM, Delius H, Hoppe D, Stannek P, Walter C, Glinka A and Niehrs C. Kremen proteins are Dickkopf receptors that regulate Wnt/beta-catenin signalling. Nature. 2002;417:664–7. [DOI] [PubMed] [Google Scholar]
- 47.Bilic J, Huang YL, Davidson G, Zimmermann T, Cruciat CM, Bienz M and Niehrs C. Wnt induces LRP6 signalosomes and promotes dishevelled-dependent LRP6 phosphorylation. Science. 2007;316:1619–22. [DOI] [PubMed] [Google Scholar]
- 48.De A Wnt/Ca2+ signaling pathway: a brief overview. Acta Biochim Biophys Sin (Shanghai). 2011;43:745–56. [DOI] [PubMed] [Google Scholar]
- 49.Fu X, Khalil H, Kanisicak O, Boyer JG, Vagnozzi RJ, Maliken BD, Sargent MA, Prasad V, Valiente-Alandi I, Blaxall BC and Molkentin JD. Specialized fibroblast differentiated states underlie scar formation in the infarcted mouse heart. J Clin Invest. 2018;128:2127–2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Siwik DA, Pagano PJ and Colucci WS. Oxidative stress regulates collagen synthesis and matrix metalloproteinase activity in cardiac fibroblasts. Am J Physiol Cell Physiol. 2001;280:C53–60. [DOI] [PubMed] [Google Scholar]
- 51.Lafontant PJ, Burns AR, Donnachie E, Haudek SB, Smith CW and Entman ML. Oncostatin M differentially regulates CXC chemokines in mouse cardiac fibroblasts. Am J Physiol Cell Physiol. 2006;291:C18–26. [DOI] [PubMed] [Google Scholar]
- 52.Turner NA, Das A, Warburton P, O’Regan DJ, Ball SG and Porter KE. Interleukin-1alpha stimulates proinflammatory cytokine expression in human cardiac myofibroblasts. Am J Physiol Heart Circ Physiol. 2009;297:H1117–27. [DOI] [PubMed] [Google Scholar]
- 53.Shinde AV and Frangogiannis NG. Fibroblasts in myocardial infarction: a role in inflammation and repair. J Mol Cell Cardiol. 2014;70:74–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kanisicak O, Khalil H, Ivey MJ, Karch J, Maliken BD, Correll RN, Brody MJ, SC JL, Aronow BJ, Tallquist MD and Molkentin JD. Genetic lineage tracing defines myofibroblast origin and function in the injured heart. Nat Commun. 2016;7:12260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jiang X, Hao HX, Growney JD, Woolfenden S, Bottiglio C, Ng N, Lu B, Hsieh MH, Bagdasarian L, Meyer R, Smith TR, Avello M, Charlat O, Xie Y, Porter JA, Pan S, Liu J, McLaughlin ME and Cong F. Inactivating mutations of RNF43 confer Wnt dependency in pancreatic ductal adenocarcinoma. Proc Natl Acad Sci U S A. 2013;110:12649–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Barry ER, Morikawa T, Butler BL, Shrestha K, de la Rosa R, Yan KS, Fuchs CS, Magness ST, Smits R, Ogino S, Kuo CJ and Camargo FD. Restriction of intestinal stem cell expansion and the regenerative response by YAP. Nature. 2013;493:106–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Varelas X, Miller BW, Sopko R, Song S, Gregorieff A, Fellouse FA, Sakuma R, Pawson T, Hunziker W, McNeill H, Wrana JL and Attisano L. The Hippo pathway regulates Wnt/beta-catenin signaling. Dev Cell. 2010;18:579–91. [DOI] [PubMed] [Google Scholar]
- 58.Imajo M, Miyatake K, Iimura A, Miyamoto A and Nishida E. A molecular mechanism that links Hippo signalling to the inhibition of Wnt/beta-catenin signalling. EMBO J. 2012;31:1109–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Park HW, Kim YC, Yu B, Moroishi T, Mo JS, Plouffe SW, Meng Z, Lin KC, Yu FX, Alexander CM, Wang CY and Guan KL. Alternative Wnt Signaling Activates YAP/TAZ. Cell. 2015;162:780–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yu J, Chia J, Canning CA, Jones CM, Bard FA and Virshup DM. WLS retrograde transport to the endoplasmic reticulum during Wnt secretion. Dev Cell. 2014;29:277–91. [DOI] [PubMed] [Google Scholar]
- 61.Najdi R, Proffitt K, Sprowl S, Kaur S, Yu J, Covey TM, Virshup DM and Waterman ML. A uniform human Wnt expression library reveals a shared secretory pathway and unique signaling activities. Differentiation. 2012;84:203–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Herr P and Basler K. Porcupine-mediated lipidation is required for Wnt recognition by Wls. Dev Biol. 2012;361:392–402. [DOI] [PubMed] [Google Scholar]
- 63.Goodman RM, Thombre S, Firtina Z, Gray D, Betts D, Roebuck J, Spana EP and Selva EM. Sprinter: a novel transmembrane protein required for Wg secretion and signaling. Development. 2006;133:4901–11. [DOI] [PubMed] [Google Scholar]
- 64.Guder C, Philipp I, Lengfeld T, Watanabe H, Hobmayer B and Holstein TW. The Wnt code: cnidarians signal the way. Oncogene. 2006;25:7450–60. [DOI] [PubMed] [Google Scholar]
- 65.Morishita Y, Kobayashi K, Klyachko E, Jujo K, Maeda K, Losordo DW and Murohara T. Wnt11 Gene Therapy with Adeno-associated Virus 9 Improves Recovery from Myocardial Infarction by Modulating the Inflammatory Response. Sci Rep. 2016;6:21705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Moon J, Zhou H, Zhang LS, Tan W, Liu Y, Zhang S, Morlock LK, Bao X, Palecek SP, Feng JQ, Williams NS, Amatruda JF, Olson EN, Bassel-Duby R and Lum L. Blockade to pathological remodeling of infarcted heart tissue using a porcupine antagonist. Proc Natl Acad Sci U S A. 2017;114:1649–1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.He W, Zhang L, Ni A, Zhang Z, Mirotsou M, Mao L, Pratt RE and Dzau VJ. Exogenously administered secreted frizzled related protein 2 (Sfrp2) reduces fibrosis and improves cardiac function in a rat model of myocardial infarction. Proc Natl Acad Sci U S A. 2010;107:21110–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Barandon L, Couffinhal T, Ezan J, Dufourcq P, Costet P, Alzieu P, Leroux L, Moreau C, Dare D and Duplaa C. Reduction of infarct size and prevention of cardiac rupture in transgenic mice overexpressing FrzA. Circulation. 2003;108:2282–9. [DOI] [PubMed] [Google Scholar]
- 69.Zhang Z, Deb A, Zhang Z, Pachori A, He W, Guo J, Pratt R and Dzau VJ. Secreted frizzled related protein 2 protects cells from apoptosis by blocking the effect of canonical Wnt3a. J Mol Cell Cardiol. 2009;46:370–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Laeremans H, Hackeng TM, van Zandvoort MA, Thijssen VL, Janssen BJ, Ottenheijm HC, Smits JF and Blankesteijn WM. Blocking of frizzled signaling with a homologous peptide fragment of wnt3a/wnt5a reduces infarct expansion and prevents the development of heart failure after myocardial infarction. Circulation. 2011;124:1626–35. [DOI] [PubMed] [Google Scholar]
- 71.Saraswati S, Alfaro MP, Thorne CA, Atkinson J, Lee E and Young PP. Pyrvinium, a potent small molecule Wnt inhibitor, promotes wound repair and post-MI cardiac remodeling. PLoS One. 2010;5:e15521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Matsushima K, Suyama T, Takenaka C, Nishishita N, Ikeda K, Ikada Y, Sawa Y, Jakt LM, Mori H and Kawamata S. Secreted frizzled related protein 4 reduces fibrosis scar size and ameliorates cardiac function after ischemic injury. Tissue Eng Part A. 2010;16:3329–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mizutani M, Wu JC and Nusse R. Fibrosis of the Neonatal Mouse Heart After Cryoinjury Is Accompanied by Wnt Signaling Activation and Epicardial-to-Mesenchymal Transition. J Am Heart Assoc. 2016;5:e002457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Xiang FL, Fang M and Yutzey KE. Loss of beta-catenin in resident cardiac fibroblasts attenuates fibrosis induced by pressure overload in mice. Nat Commun. 2017;8:712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.van Amerongen R and Nusse R. Towards an integrated view of Wnt signaling in development. Development. 2009;136:3205–14. [DOI] [PubMed] [Google Scholar]
- 76.Niida A, Hiroko T, Kasai M, Furukawa Y, Nakamura Y, Suzuki Y, Sugano S and Akiyama T. DKK1, a negative regulator of Wnt signaling, is a target of the beta-catenin/TCF pathway. Oncogene. 2004;23:8520–6. [DOI] [PubMed] [Google Scholar]
- 77.Lescher B, Haenig B and Kispert A. sFRP-2 is a target of the Wnt-4 signaling pathway in the developing metanephric kidney. Dev Dyn. 1998;213:440–51. [DOI] [PubMed] [Google Scholar]
- 78.Wang Z, Cui M, Shah AM, Tan W, Liu N, Bassel-Duby R and Olson EN. Cell-Type-Specific Gene Regulatory Networks Underlying Murine Neonatal Heart Regeneration at Single-Cell Resolution. Cell Rep. 2020;33:108472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Dawson K, Aflaki M and Nattel S. Role of the Wnt-Frizzled system in cardiac pathophysiology: a rapidly developing, poorly understood area with enormous potential. J Physiol. 2013;591:1409–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.McQuate A, Latorre-Esteves E and Barria A. A Wnt/Calcium Signaling Cascade Regulates Neuronal Excitability and Trafficking of NMDARs. Cell Rep. 2017;21:60–69. [DOI] [PubMed] [Google Scholar]
- 81.Li G, Khandekar A, Yin T, Hicks SC, Guo Q, Takahashi K, Lipovsky CE, Brumback BD, Rao PK, Weinheimer CJ and Rentschler SL. Differential Wnt-mediated programming and arrhythmogenesis in right versus left ventricles. J Mol Cell Cardiol. 2018;123:92–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data and supporting materials have been provided with the published article, and all essential research materials are listed in the Major Resources Table in the Data Supplement. The sequencing data in this manuscript have been deposited in the Gene Expression Omnibus under accession number GSE179355. All other data are available from the corresponding author upon reasonable request.