Abstract
Background
While insulin has been central to the pathophysiology and treatment of patients with diabetes for the last 100 years, it has only been since 2007 that genetic variation in the INS gene has been recognised as a major cause of monogenic diabetes. Both dominant and recessive mutations in the INS gene are now recognised as important causes of neonatal diabetes and offer important insights into both the structure and function of insulin. It is also recognised that in rare cases, mutations in the INS gene can be found in patients with diabetes diagnosed outside the first year of life.
Scope of Review
This review examines the genetics and clinical features of monogenic diabetes resulting from INS gene mutations from the first description in 2007 and includes information from 389 patients from 292 families diagnosed in Exeter with INS gene mutations. We discuss the implications for diagnosing and treating this subtype of monogenic diabetes.
Major Conclusions
The dominant mutations in the INS gene typically affect the secondary structure of the insulin protein, usually by disrupting the 3 disulfide bonds in mature insulin. The resulting misfolded protein results in ER stress and beta-cell destruction. In contrast, recessive INS gene mutations typically result in no functional protein being produced due to reduced insulin biosynthesis or loss-of-function mutations in the insulin protein. There are clinical differences between the two genetic aetiologies, between the specific mutations, and within patients with identical mutations.
Keywords: Insulin gene, Insulin biosynthesis, Pathophysiology, Genetics, Neonatal diabetes, Monogenic diabetes
Abbreviations: ER, endoplasmic reticulum; IQR, interquartile range; MODY, maturity-onset diabetes of the young; NDM, neonatal diabetes mellitus; OHA, oral hypoglycaemic agent; PNDM, permanent neonatal diabetes; SRP, signal recognition particle; SU, sulfonylurea; TNDM, transient neonatal diabetes; UPR, unfolded protein response
Highlights
-
•
Dominant and recessive mutations in the insulin (INS) gene are important causes of neonatal diabetes.
-
•
Associated phenotypes are variable in terms of age at diabetes onset, birth weight and treatment requirements.
-
•
Dominant mutations affect the secondary structure of the insulin protein, resulting in beta-cell ER stress and destruction.
-
•
Recessive mutations result in reduced insulin biosynthesis or loss-of-function mutations of the insulin protein.
-
•
The studies of these forms of diabetes offer important insights into the structure, biosynthesis and secretion of insulin.
1. Introduction
The central role of insulin in the aetiology and treatment of diabetes has been established for 100 years. However, the role of the INS gene in the aetiology of diabetes has only recently been established. Though the INS gene was one of the earliest genes to be cloned (1980) [1,2], it did not have a significant role in the genetics of diabetes until the discovery of dominant heterozygous mutations causing neonatal diabetes in 2007/2008 [3,4]. This review examines the genetics and clinical features of monogenic diabetes resulting from INS gene mutations.
Insulin secreted from pancreatic beta-cells has a vital role in the regulation of energy homeostasis. Upon binding to its receptors in liver, muscle, and adipose tissue, insulin exerts its effects through complex signalling pathways that lead to energy deposition and preservation of energy stores. Insulin biosynthesis and secretion are tightly regulated to maintain blood glucose within a narrow physiological range, and failure of the beta-cells to produce adequate amounts of insulin leads to hyperglycaemia, as seen in patients with various subtypes of diabetes mellitus. Insulin is primarily expressed in the pancreatic beta-cells, and in the brain and thymus at low levels (reviewed in Hay and Docherty) [5]. These observations made the INS gene an ideal etiological candidate gene for isolated neonatal diabetes mellitus (NDM), which is diagnosed close to birth, typically in the first 6–9 months of life without the involvement of other organs.
Heterozygous INS mutations, both de novo and dominantly inherited, were the first to be shown to cause isolated neonatal diabetes [3,4]. Following this discovery, recessively inherited INS gene mutations were identified as a cause of NDM, particularly in patients born to consanguineous parents [6]. In recent years, the number and types of mutations identified have steadily increased. This review aims to provide an update on INS gene mutations and the associated phenotypes in monogenic diabetes based on published mutations and experience with 389 diabetes patients from 292 families with a mutation in the INS gene and who had their genetic testing done in the genetic laboratory in Exeter, UK (Table 1, Figure 1, Figure 2). The patients were referred from 62 countries from all continents.
Table 1.
Features of the Exeter cohort of patients with INS gene mutations. Median and IQR.
| n patients | n families | Birth weight (z-score) | Age at diagnosis (days) | |
|---|---|---|---|---|
| Dominant | 306 | 226 | −1.1 (−1.8 to 0.3), n = 205 | 119 (53–224), n = 274 |
| Recessive | 83 | 66 | −2.9 (−3.8 to −2.5), n = 59 | 7 (1–16), n = 72 |
Figure 1.
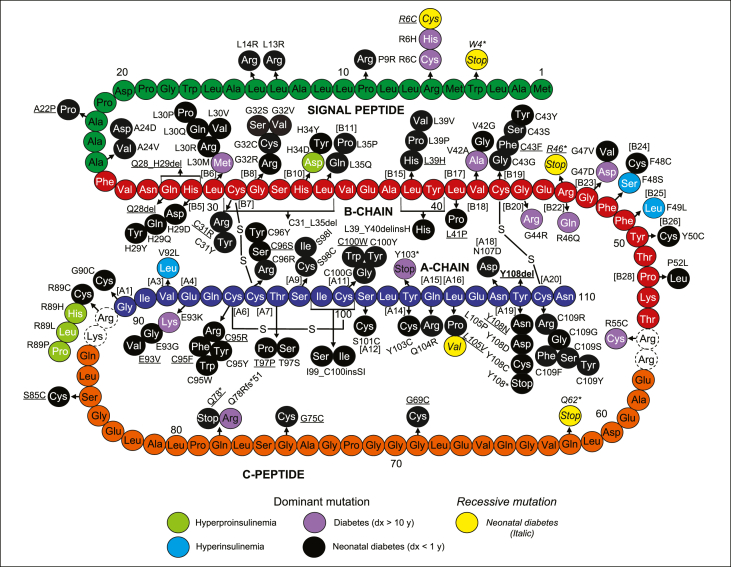
Diagram representing the amino acid sequence of human preproinsulin with signal peptide (green), B-chain (red), C-peptide (orange), A-chain (dark blue) with mutations identified in patients with NDM (black – dominant; yellow – recessive), diabetes diagnosed after 10 years of age (purple), hyperproinsulinemia (green), and hyperinsulinemia (blue). The dashed circles indicate the residues that are cleaved during the conversion of proinsulin to insulin. Novel mutations identified in the Exeter cohort are underlined.
Figure 2.
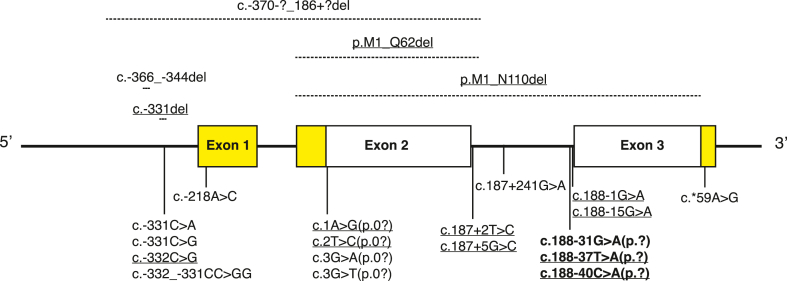
A schematic of the human INS gene showing the locations of homozygous and compound heterozygous mutations identified in patients with NDM. Mutations involving non-coding regions are displayed below the gene, whereas those affecting coding regions are shown above the gene. Three mutations displayed below the gene are dominant mutations affecting proinsulin folding (bold font). The protein-coding regions of the gene are shown in white, and the regions encoding the 5′- and 3′-untranslated regions of insulin mRNA are shown in yellow. Novel mutations identified in the Exeter cohort are underlined.
2. Molecular structure and biosynthesis of insulin
2.1. The human insulin gene
The human insulin gene (INS) spans approximately 1.5 Kb and is located on chromosome 11p15.5 [1,7]. The gene consists of three exons separated by two introns, of which only exon two and three are protein-coding [8]. Exon two encodes the signal peptide, the B-chain, and the N-terminal part of the connecting peptide of preproinsulin (amino acids 1–62). Exon three encodes the remainder of the connecting peptide and the A-chain of preproinsulin (amino acids 63–110), whereas exon one only has a regulatory role in the transcription of the INS gene, with the cyclic AMP response element, CRE3, embedded in this region [5] (Figure 2).
2.2. Insulin biosynthesis
The initial product of the translation of insulin mRNA is the insulin precursor preproinsulin, a single chain polypeptide, consisting of a signal peptide (amino acids 1–24), B-chain (amino acids 25–54), connecting peptide (amino acids 57–87), and A-chain (amino acids 90–110). Two dipeptides connect the N-terminal part of the connecting peptide to the B-chain (Arg-Arg) and the C-terminal part to the A-chain (Lys-Arg; Figure 1). The newly synthesised signal peptide binds the signal peptide recognition particle (SRP) in the cytosol, and the SRP-signal peptide complex interacts with the translocon on the membrane of the endoplasmic reticulum (ER) and directs the proinsulin into the lumen of the ER. During the transition of proinsulin through the ER, the molecule acquires its correct three-dimensional structure through the folding and formation of three stabilising disulfide bonds, a process catalysed by chemical chaperonins. The disulfide bond formation is essential for the characteristic structure of mature insulin, as these bonds connect the B-chain and the A-chain after the C-peptide is excised in the secretory granules. Two of these disulfide bonds connect the B-chain to the A-chain at B7-A7 and B19-A20, respectively, and the third disulfide bond is an A-chain intra-chain bond that connects A6 to A11. Only correctly folded proinsulin progresses from the ER to the Golgi apparatus. In the transGlogi network, the proinsulin is concentrated and sorted into immature secretory granules. In the secretory granules, the C-peptide is excised by cleaving the B-chain-C-peptide junction by proconvertase (PC) 1/3 and carboxypeptidase E (CPE) yielding des-31, 32-proinsulin and at the C-peptide-A-chain junction by PC2 and CPE yielding insulin, C-peptide, three molecules of Arginine, and one molecule of Lysine. Insulin is stored in the mature secretory granules and is secreted in equimolar amounts with C-peptide from the beta-cells in response to various metabolic signals [9].
3. INS gene mutations in diabetes
INS gene mutations are associated with a range of diabetes phenotypes. The first mutations to be described caused asymptomatic hyperproinsulinemia or hyperinsulinemia by affecting the excision of C-peptide from proinsulin or affecting parts of the protein involved in binding to the insulin receptor [10].
The important breakthrough in the role of INS gene mutations in diabetes came when the more common dominant and gain-of-function INS gene mutations were identified in patients with isolated permanent neonatal diabetes (PNDM) [3,4]. PNDM is distinct from the more common polygenic autoimmune type 1 diabetes, as the diabetes is usually diagnosed in the first 6 months of life and is caused by a single gene defect in one of >25 genes implicated to date [[11], [12], [13], [14]].
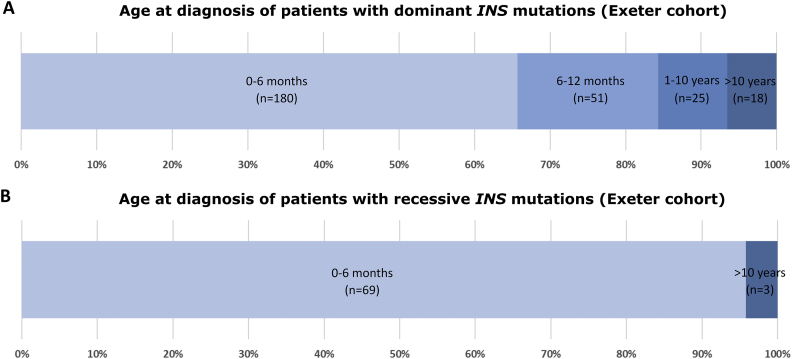
In the Exeter cohort, dominant INS gene mutations most commonly result in diabetes diagnosed in the first 6 months of life (66% of cases), with approximately 18% of patients diagnosed between 6 and 12 months of age (Figure 3A). In more than 60% of cases, the mutation appears to be de novo in origin (Figure 4), but INS gene mutations have also been identified in families with multiple affected family members across generations [3]. The diabetes is usually permanent and non-remitting. Dominant INS gene mutations are identified in 8% of patients diagnosed with diabetes before 6 months of age and they are the second most prevalent cause of PNDM, after KCNJ11 and ABCC8 mutations (two genes that encode the Kir6.2 and the SUR1 subunit of the ATP-sensitive potassium channel in the pancreatic beta cell that together account for 38% of neonatal diabetes cases [14]).
Figure 3.
A. Stacked bar chart representing age at diagnosis with diabetes in the Exeter cohort of patients with dominant INS gene mutations (age at diagnosis missing for 32 individuals). B. Stacked bar chart representing age at diagnosis with diabetes in the Exeter cohort of patients with recessive INS gene mutations (age at diagnosis missing for 11 individuals).
Figure 4.
Stacked bar chart representing inheritance of the dominant INS gene mutations in the Exeter cohort (inheritance could not be assessed for 106 individuals as DNA from both parents was not available).
Dominant INS gene mutations have been identified in children and adult patients diagnosed with non-autoimmune diabetes outside of infancy. The phenotype usually resembles type 1 diabetes with requirement for insulin treatment but some patients have a milder phenotype with preserved beta-cell function where OHA or even diet treatment is sufficient to achieve glycaemic targets [3,[15], [16], [17], [18], [19], [20], [21]]. The mutations in the latter patients are frequently inherited and the patients have a phenotype consistent with a diagnosis of maturity-onset diabetes of the young.
Recessive INS gene mutations were first described causing neonatal diabetes with most mutations being homozygous mutations in offspring of consanguineous parents [6]. The patients with NDM due to loss-of-function recessive mutations are effectively functional insulin-knockouts. Almost all diabetes caused by recessive INS gene mutations presents close to birth with only a minority causing diabetes outside the first 6 months (Figure 4) [6]. In consanguineous families, recessive INS gene mutations cause ~9% of NDM cases (in the Exeter cohort). While most recessive INS gene mutations cause PNDM, a proportion of the patients with homozygous variants affecting the INS promoter have transient diabetes (TNDM) that remits within the first 3 months of life [6]. In patients with TNDM due to abnormalities at 6q24 or mutations in KCNJ11 or ABCC8, diabetes relapse is seen in approximately 50% of patients close to puberty, but this is not as well described for recessive INS gene mutations [6,22]. The heterozygous parents with a single null allele usually have normal glucose tolerance but can present with gestational diabetes or diabetes later in life [23]. Further studies of these carriers could identify other genetic and non-genetic factors that contribute to the development of diabetes.
3.1. Characteristics of mutations
3.1.1. Dominant mutations
More than 90 different dominant mutations in the INS gene have been identified in probands with PNDM and diabetes diagnosed outside of infancy (Figure 1, Figure 2). In addition, three mutations have been found in patients with familial hyperinsulinemia and four mutations in familial hyperproinsulinemia. The dominantly acting mutations involve 48 of the 110 amino acids of the preproinsulin molecule and include missense (n = 77 and n = 7 in familial hyperinsulinemia and hyperproinsulinemia), in-frame deletions (n = 4), insertion (n = 1), nonsense (n = 2), indels (n = 3), frameshift (n = 1) mutations, and intronic pathogenic variants affecting the splicing of intron 2 (n = 3; Figure 1, Figure 2). The mutations affecting the coding sequence are located in all regions of the preproinsulin molecule. However, the majority are found in the A-chain and B-chain: Signal peptide (n = 8), B-chain (n = 38 and n = 2 in hyperinsulinemia and n = 1 in hyperproinsulinemia), A-chain (n = 35 and n = 1 in hyperinsulinemia), C-peptide (n = 5), B-chain/C-peptide junction (n = 1), and C-peptide/A-chain junction (n = 1 in PNDM and n = 3 in hyperproinsulinemia; Figure 1) with 23 residues reported to be affected by more than one different amino acid substitution. More than half of the identified mutations are located at amino acid residues directly involved in forming the three disulfide bonds of mature insulin or neighbouring amino acids. Mutations at five codons (A24, G32, F48, R89, and C96) together with the splice site mutation c.188-31G>A account for more than half of the identified probands, and thus, these positions seem to be mutation hotspots that affect proinsulin biosynthesis and function. More than 30 mutations reported so far are private, with only one proband identified with the specific mutation. Some mutations reported here are novel, previously unpublished mutations were identified in patients from the Exeter cohort: Signal-peptide (n = 1), B-chain (n = 6), C-peptide (n = 4), A-chain (n = 8), and two intronic mutations c.188-37T>A and c. 188-40C>A (Figure 1, Figure 2).
The majority of mutations are found in patients with a clinical phenotype of NDM, whereas a smaller group of mutations are only found in patients diagnosed with diabetes outside infancy and, in some cases, after 10 years of age [24]. Both groups of mutations are distributed across preproinsulin, but notably, mutations associated with NDM seem to more commonly affect amino acids directly involved in forming the three disulfide bonds of mature insulin. Interestingly, mutations at some of the hotspot codons (A24, G32, C43, R89, C96) are associated with some variation in diabetes severity, with phenotypes ranging from NDM to diabetes diagnosed outside infancy [24]. These differences have been observed both within families with several affected family members and between families with identical mutations, suggesting that other factors may play a role in the severity of the diabetes phenotype.
The diabetes-causing dominant INS mutations affect beta-cell viability and interfere with biosynthesis and secretion of insulin through distinct mechanisms. The mutations located in the signal peptide affect the processing of preproinsulin to proinsulin, whereas the mutations in the B-chain and A-chain negatively impact the structure of proinsulin, as they impede the formation of one of the three disulfide bonds of mature insulin. In many cases, the mutation leads to the insertion of an aberrant cysteine residue or substitution of a cysteine residue with another amino acid and directly compromises the correct formation or location of the cysteine–cysteine disulfide bonds between B7-A7, B19-A20, or A6-A11. The three dominant intronic mutations identified in NDM patients are all predicted to result in activation of a cryptic acceptor splicing site and a frameshift that causes the synthesis of a misfolded protein.
The mutant proinsulin accumulates within the beta-cell ER and leads to activation of the unfolded protein response (UPR), including suppression of synthesis of wild-type insulin and, ultimately, beta cell apoptosis [[25], [26], [27]]. Beta-cell death may be preceded by an early event of reduced insulin secretion due to complex formation between mutant and wild-type proinsulin with retention in the ER [28], and ER stress may develop as a consequence of both accumulation of wild-type and mutant proinsulin [26,[29], [30], [31], [32], [33]].
3.1.2. Recessive mutations
25 different homozygous or compound heterozygous mutations in the INS gene have been identified in probands with PNDM, TNDM, and in rare cases in patients diagnosed with diabetes outside infancy (Figure 2) [6,22,23,[34], [35], [36], [37], [38], [39]]. The pathophysiology of the recessive mutations is distinct from the dominantly acting mutations, as they lead to diabetes through reduced or no insulin biosynthesis. The specific disease mechanism is determined by the specific location of the mutation (see Figure 2). The mutations in the INS promoter (n = 7) reduce the transcription of the gene and may result in PNDM, TNDM, and diabetes diagnosed outside infancy. Other mutations result in loss of the translation initiation site (n = 4) and are predicted to alter mRNA translation or lead to no translation of that allele. Nonsense mutations (n = 3) lead to truncation of the protein. Three larger deletions involving variable stretches of the promoter, signal peptide, and B-chain have been reported (Figure 2) in patients with PNDM. Finally, a group of mutations (n = 6) in the terminal intron of the INS gene and 3′UTR affect mRNA stability.
Interestingly, mutations at three codons (R6, R46, and L105) have been identified in the heterozygous state in patients with a clinical diagnosis of NDM (p. L105P) or MODY (p. R46Q, p. R6C/H) and in the homozygous state in NDM (p. R6C, p. R46∗, p. L105V) [6,23,34].
4. Clinical characteristics and treatment of patients with INS gene mutations
4.1. Neonatal diabetes
In keeping with the expression of insulin in the developing pancreatic beta-cells, the phenotype of patients with NDM due to both dominant and recessive INS gene mutations is restricted to low birth weight due to intrauterine insulin deficiency and postnatal hyperglycaemia. The phenotype is more severe in patients with recessive INS mutations with diabetes onset within the first week of life and severe growth retardation compared with median diabetes onset after 4 months of age and less pronounced growth retardation in patients with dominant mutations (Table 1). This difference in phenotype is explained by the different genetic defects and associated pathophysiological events that lead to diabetes. In the developing fetus homozygous for null INS gene mutations, the total absence of insulin is likely present from the onset of endogenous insulin secretion during development, and therefore, all insulin-mediated growth of the fetus will be absent. Conversely, the negative impact of a dominant mutation on endogenous insulin secretion and beta cell viability is gradual and progressive, as it is an effect of the accumulation of mutant proinsulin in the beta cells ER, with a variable period of time before leading to a reduction in insulin secretion.
It is currently unclear which other factors may contribute to the variation of the diabetes phenotype. An early report of 29 parent-sibling pairs with dominant INS gene mutations suggested that diabetes was diagnosed at a younger age when there was paternal, rather than maternal, inheritance of the mutation [40]. However, this effect was not replicated in the large Exeter cohort, in which 72 subjects had inherited the mutations from an affected parent. The median age of diabetes onset was not significantly different in individuals who had inherited the mutation from an affected father (14 weeks, IQR 7–34) versus an affected mother (20 weeks, IQR 12–52, p-value = 0.406). Gender has been reported to influence the phenotype in the Akita mouse model (C96Y) of NDM due to a dominant INS gene mutation as the phenotype is markedly more severe in male mice than in females [32]. A gender dimorphism was also observed in the patients in the Exeter cohort, with lower birth weights in males than in females (median z score −1.26 (IQR −2 to −0.79) versus −0.88 (IQR −1.55 to −0.3), p = 0.001). However, the age of diagnosis was not significantly different between the two sexes (median age at onset of diabetes 13 weeks [IQR 7–23] in males versus 17 weeks [IQR 8–26] in females, p = 0.124).
There are few other clinical features associated with INS gene mutations other than the direct role of insulin on fetal growth and postnatal glucose. Premature development of cataract is a feature of the Munich pig (C95S) and this has also been reported in a minority of patients with dominant INS gene mutations [33,41].
Though the treatment of the large group of patients with NDM due to either a mutation in KCNJ11 or ABCC8 has changed fundamentally from mainly insulin to treatment with sulfonylurea, the genetic diagnosis in patients with NDM due to INS gene mutations has not led to a change in treatment. Patients are treated with insulin in replacement doses, with variable regimens starting with basal insulin alone, basal-bolus insulin using insulin pens, and treatment with insulin-pump with or without sensor augmentation [3,[42], [43], [44], [45], [46]].
Even though the genetic diagnosis is not currently associated with a change in treatment, identifying the causative mutation at an early stage is essential for genetic counselling and could also be essential for future treatments of this form of diabetes. It is speculated that early and aggressive insulin treatment could suppress endogenous insulin biosynthesis and minimise the toxic effect on the beta cells of mutant proinsulin. This could in turn support preservation of a pool of viable beta cells that would allow for, and be the fundament for future treatments with agents supporting the expression and biosynthesis of insulin from the healthy allele of the INS gene [47].
4.2. Diabetes diagnosed outside infancy
The phenotype of patients with diabetes onset outside infancy due to a dominant INS gene mutation is variable with associated clinical phenotypes that reflect minimal endogenous insulin secretion and relative early-onset diabetes in some patients, and in other patients, a milder phenotype with more modest treatment requirements and diabetes onset well beyond 10 years of age.
The group of patients diagnosed in early childhood has phenotypes resembling the features of patients with NDM due to an INS gene mutation, with the exception that beta-cell failures becomes manifest later in life. Consistent with this, some of the patients carry the same mutations as identified in patients with NDM [41,44,[48], [49], [50], [51], [52]]. The reason for the delay in diabetes onset is not well understood but it could be influenced by other genetic or non-genetic factors such as genetic mosaicism. Genetic testing of all patients with diabetes onset before 6–9 months of age is well established in most centres worldwide, enabling INS mutations to be easily detected in this age group, but this is not the case for those diagnosed later. These patients with dominant INS mutations often have no family history of early-onset diabetes, as in the majority of cases, they harbour de novo mutations and present with diabetes after 9 months. Therefore, the decision to perform genetic testing heavily relies on the lack of diabetes-associated autoantibodies, and the patients will frequently be misclassified as having early-onset autoimmune, polygenic type-1 diabetes [53].
A small group of mutations (Figure 1) are associated with diabetes diagnosed after 10 years of age. There is considerable phenotypic variation within this group of patients, both between mutations and among patients carrying the same mutation. The mutation p. R55C is associated with diabetic ketoacidosis at presentation, requiring insulin treatment. However, further studies of patients with this mutation could lead to new approaches for treating patients with this specific mutation. Other mutations, such as p. R46Q, are associated with preserved beta cell function and treatment requirements that range from diet intervention to insulin treatment [16,17,21]. There are insufficient data to document whether non-insulin regimens can sustain acceptable glycaemic control in the long term or if beta-cell function progressively declines, necessitating insulin treatment at some point. Treatments that aim to reduce the demand for endogenous insulin secretion could reduce the toxic effect of the mutant proinsulin on beta-cells and potentially postpone the development of manifest beta-cell failure. Interestingly, a novel case report suggested a marked response to metformin treatment in a sib-pair with the p. R46Q mutation [21]. This finding warrants replication in additional patients. In theory, treatment with selective sodium glucose co-transporter-2-inhibitors (SGLT2i) could be beneficial for these patients by reducing the glucose load to the beta cells. However, it remains to be tested in patients, and the risk of ketoacidosis in insulin-deficient patients may limit its usefulness. Conversely, treatment with medications that work through stimulation of endogenous insulin secretion, such as sulfonylurea, is unlikely to benefit this group of patients, as treatment may increase the secretion of the misfolded mutant insulin and accelerate the destruction of beta-cells. Treatment with GLP1-analogues has been suggested as a way of targeting beta cell stress following in vitro studies and studies in mice that suggest that this group of medications may inhibit beta cell apoptosis and alleviate beta cell stress [[54], [55], [56]]. However, GLP-1 analogues stimulate glucose-dependent insulin secretion, and the joint effect of these mechanisms on patients with dominant INS mutations has not been reported in patients.
INS gene mutations in diabetes diagnosed after 10 years of age are likely to be underdiagnosed as the phenotype is subtler than the phenotypes of patients with other subtypes of monogenic diabetes. Furthermore, the INS gene is not routinely tested in all centres testing for MODY, and the correct diagnosis could be missed on this basis [23,51].
5. Future perspectives for diagnosis and treatment of INS gene mutations
The detection of patients with INS mutations is likely to increase as molecular genetic testing becomes cheaper and part of routine clinical management. In patients diagnosed in the first 6 months of life, molecular genetic sequencing guiding management is already established, as routine care and most centres already sequence the INS gene routinely [57]. The major change will come in the increased detection of late-onset cases with dominant INS gene mutations, both de novo and inherited from an affected parent. This change will occur as it becomes routine to perform molecular genetic testing on all children, adolescents, and young adults diagnosed with diabetes when they are negative after being tested for 3 or 4 islet autoantibodies. This trend for testing all antibody-negative patients is supported by recent findings that 1 in 8 of these islet autoantibody negative patients will have monogenic diabetes [58].
The treatment of most patients with INS gene mutations, in which there is no or minimal insulin secretion, will include exogeneous insulin, and it will need to be accurately matched to glycaemia, food, and exercise. This treatment will increasingly occur, using novel technology to sense glucose and deliver insulin appropriately. An exciting future prospect includes the delivery of insulin by cell therapy potentially by beta-cells derived from stem cells. When these methods are available, patients who have absolute insulin deficiency due to recessive INS mutations will be ideal candidates for this therapy, as they are unlikely to suffer from an autoimmune attack of the beta-cell therapy, unlike patients with type 1 diabetes.
For the dominant INS mutations, it is likely that a specific molecular silencing of the mutant allele leaving a functioning normal allele would be sufficient to prevent diabetes developing. While appearing an attractive therapeutic option there are many barriers to this being implemented: there is not a safe way established to silence a mutant INS allele in animal models. Insulin injections offer a well-established alternative and risks for novel treatments will need to be known to be very low. Furthermore, this approach requires intact beta-cells, but currently most patients only present with diabetes when there is already considerable damage caused to the beta-cell.
Author contributions
JS conceived the review and drafted the first version of the manuscript. EDF conceived the review, analysed data, generated tables and figures, critically reviewed and edited the manuscript. HY made the figures, reviewed and edited the manuscript. SYP reviewed and edited the manuscript. GIB conceived the review, reviewed and edited the manuscript. ATH conceived the review, drafted the section on future perspectives, reviewed and edited the manuscript. All authors approved the final version of the manuscript.
Funding
The work of JS is funded by Steno Diabetes Center Aarhus through an unrestricted donation from the Novo Nordic Foundation. EDF is a Diabetes UK RD Lawrence fellow in diabetes research. The work of HY, SYP, and GIB is funded in part by NIH/NIDDK. The work of ATH funded by P30DK020595.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.molmet.2021.101280.
Contributor Information
Julie Støy, Email: julistoe@rm.dk.
Elisa De Franco, Email: E.De-Franco@exeter.ac.uk.
Honggang Ye, Email: hye@uchicago.edu.
Soo-Young Park, Email: sooyoung@uchicago.edu.
Graeme I. Bell, Email: gb11@uchicago.edu.
Andrew T. Hattersley, Email: A.T.Hattersley@exeter.ac.uk.
Conflict of interest
None.
Appendix A. Supplementary data
The following is the supplementary data to this article:
References
- 1.Bell G.I., Pictet R.L., Rutter W.J., Cordell B., Tischer E., Goodman H.M. Sequence of the human insulin gene. Nature. 1980;284(5751):26–32. doi: 10.1038/284026a0. [DOI] [PubMed] [Google Scholar]
- 2.Bell G.I., Swain W.F., Pictet R., Cordell B., Goodman H.M., Rutter W.J. Nucleotide sequence of a cDNA clone encoding human preproinsulin. Nature. 1979;282(5738):525–527. doi: 10.1038/282525a0. [DOI] [PubMed] [Google Scholar]
- 3.Støy J., Edghill E.L., Flanagan S.E., Ye H., Paz V.P., Pluzhnikov A. Insulin gene mutations as a cause of permanent neonatal diabetes. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(38):15040–15044. doi: 10.1073/pnas.0707291104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Colombo C., Porzio O., Liu M., Massa O., Vasta M., Salardi S. Seven mutations in the human insulin gene linked to permanent neonatal/infancy-onset diabetes mellitus. Journal of Clinical Investigation. 2008;118(6):2148–2156. doi: 10.1172/JCI33777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hay C.W., Docherty K. Comparative analysis of insulin gene promoters: implications for diabetes research. Diabetes. 2006;55(12):3201–3213. doi: 10.2337/db06-0788. [DOI] [PubMed] [Google Scholar]
- 6.Garin I., Edghill E.L., Akerman I., Rubio-Cabezas O., Rica I., Locke J.M. Recessive mutations in the INS gene result in neonatal diabetes through reduced insulin biosynthesis. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(7):3105–3110. doi: 10.1073/pnas.0910533107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Owerbach D., Bell G.I., Rutter W.J., Brown J.A., Shows T.B. The insulin gene is located on the short arm of chromosome 11 in humans. Diabetes. 1981;30(3):267–270. doi: 10.2337/diab.30.3.267. [DOI] [PubMed] [Google Scholar]
- 8.Steiner D.F., Chan S.J., Welsh J.M., Kwok S.C. Structure and evolution of the insulin gene. Annual Review of Genetics. 1985;19:463–484. doi: 10.1146/annurev.ge.19.120185.002335. [DOI] [PubMed] [Google Scholar]
- 9.Steiner D.F., Park S.Y., Støy J., Philipson L.H., Bell G.I. A brief perspective on insulin production. Diabetes, Obesity & Metabolism. 2009;11(Suppl 4):189–196. doi: 10.1111/j.1463-1326.2009.01106.x. [DOI] [PubMed] [Google Scholar]
- 10.Steiner D.F., Tager H.S., Nanjo K., Chan S.J., Rubenstein A.H. In: The metabolic basis of inherited disease. Scriver C., Beaudet A., Sly W., Valle D., editors. McGraw-Hill; New York: 1995. Familial syndromes of hyperproinsulinemia and hyperinsulinemia with mild diabetes; pp. 897–904. [Google Scholar]
- 11.Barbetti F., D'Annunzio G. Genetic causes and treatment of neonatal diabetes and early childhood diabetes. Best Practice & Research. Clinical Endocrinology & Metabolism. 2018;32(4):575–591. doi: 10.1016/j.beem.2018.06.008. [DOI] [PubMed] [Google Scholar]
- 12.Lemelman M.B., Letourneau L., Greeley S.A.W. Neonatal diabetes mellitus: an update on diagnosis and management. Clinics in Perinatology. 2018;45(1):41–59. doi: 10.1016/j.clp.2017.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sanyoura M., Philipson L.H., Naylor R. Monogenic diabetes in children and adolescents: recognition and treatment options. Current Diabetes Reports. 2018;18(8):58. doi: 10.1007/s11892-018-1024-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.De Franco E., Flanagan S.E., Houghton J.A., Lango Allen H., Mackay D.J., Temple I.K. The effect of early, comprehensive genomic testing on clinical care in neonatal diabetes: an international cohort study. Lancet. 2015;386(9997):957–963. doi: 10.1016/S0140-6736(15)60098-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Edghill E.L., Flanagan S.E., Patch A.M., Boustred C., Parrish A., Shields B. Insulin mutation screening in 1,044 patients with diabetes: mutations in the INS gene are a common cause of neonatal diabetes but a rare cause of diabetes diagnosed in childhood or adulthood. Diabetes. 2008;57(4):1034–1042. doi: 10.2337/db07-1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Molven A., Ringdal M., Nordbø A.M., Raeder H., Støy J., Lipkind G.M. Mutations in the insulin gene can cause MODY and autoantibody-negative type 1 diabetes. Diabetes. 2008;57(4):1131–1135. doi: 10.2337/db07-1467. [DOI] [PubMed] [Google Scholar]
- 17.Støy J., Olsen J., Park S.Y., Gregersen S., Hjørringgaard C.U., Bell G.I. In vivo measurement and biological characterisation of the diabetes-associated mutant insulin p.R46Q (GlnB22-insulin) Diabetologia. 2017;60(8):1423–1431. doi: 10.1007/s00125-017-4295-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boesgaard T.W., Pruhova S., Andersson E.A., Cinek O., Obermannova B., Lauenborg J. Further evidence that mutations in INS can be a rare cause of Maturity-Onset Diabetes of the Young (MODY) BMC Medical Genetics. 2010;11:42. doi: 10.1186/1471-2350-11-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meur G., Simon A., Harun N., Virally M., Dechaume A., Bonnefond A. Insulin gene mutations resulting in early-onset diabetes: marked differences in clinical presentation, metabolic status, and pathogenic effect through endoplasmic reticulum retention. Diabetes. 2010;59(3):653–661. doi: 10.2337/db09-1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Křížková K., Veverka V., Maletínská L., Hexnerová R., Brzozowski A.M., Jiráček J. Structural and functional study of the GlnB22-insulin mutant responsible for maturity-onset diabetes of the young. PLoS One. 2014;9(11) doi: 10.1371/journal.pone.0112883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Riar S.S., Toppings N.B., Donovan L.E. Glycemic impact of metformin in diabetes caused by heterozygous insulin gene mutation R46Q. Canadian Journal of Diabetes. 2020 doi: 10.1016/j.jcjd.2020.08.105. [DOI] [PubMed] [Google Scholar]
- 22.Demiral M., Demirbilek H., Çelik K., Okur N., Hussain K., Ozbek M.N. Neonatal diabetes due to homozygous INS gene promoter mutations: highly variable phenotype, remission and early relapse during the first 3 years of life. Pediatric Diabetes. 2020;21(7):1169–1175. doi: 10.1111/pedi.13079. [DOI] [PubMed] [Google Scholar]
- 23.Carmody D., Park S.Y., Ye H., Perrone M.E., Alkorta-Aranburu G., Highland H.M. Continued lessons from the INS gene: an intronic mutation causing diabetes through a novel mechanism. Journal of Medical Genetics. 2015;52(9):612–616. doi: 10.1136/jmedgenet-2015-103220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Støy J., Steiner D.F., Park S.Y., Ye H., Philipson L.H., Bell G.I. Clinical and molecular genetics of neonatal diabetes due to mutations in the insulin gene. Reviews in Endocrine & Metabolic Disorders. 2010;11(3):205–215. doi: 10.1007/s11154-010-9151-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hodish I., Liu M., Rajpal G., Larkin D., Holz R.W., Adams A. Misfolded proinsulin affects bystander proinsulin in neonatal diabetes. Journal of Biological Chemistry. 2010;285(1):685–694. doi: 10.1074/jbc.M109.038042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Park S.Y., Ye H., Steiner D.F., Bell G.I. Mutant proinsulin proteins associated with neonatal diabetes are retained in the endoplasmic reticulum and not efficiently secreted. Biochemical and Biophysical Research Communications. 2010;391(3):1449–1454. doi: 10.1016/j.bbrc.2009.12.090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rajan S., Eames S.C., Park S.Y., Labno C., Bell G.I., Prince V.E. In vitro processing and secretion of mutant insulin proteins that cause permanent neonatal diabetes. American Journal of Physiology. Endocrinology and Metabolism. 2010;298(3):E403–E410. doi: 10.1152/ajpendo.00592.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu M., Hodish I., Haataja L., Lara-Lemus R., Rajpal G., Wright J. Proinsulin misfolding and diabetes: mutant INS gene-induced diabetes of youth. Trends in Endocrinology and Metabolism. 2010;21(11):652–659. doi: 10.1016/j.tem.2010.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu M., Weiss M.A., Arunagiri A., Yong J., Rege N., Sun J. Biosynthesis, structure, and folding of the insulin precursor protein. Diabetes, Obesity & Metabolism. 2018;20(Suppl 2):28–50. doi: 10.1111/dom.13378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang H., Saint-Martin C., Xu J., Ding L., Wang R., Feng W. Biological behaviors of mutant proinsulin contribute to the phenotypic spectrum of diabetes associated with insulin gene mutations. Molecular and Cellular Endocrinology. 2020;518:111025. doi: 10.1016/j.mce.2020.111025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ron D. Proteotoxicity in the endoplasmic reticulum: lessons from the Akita diabetic mouse. Journal of Clinical Investigation. 2002;109(4):443–445. doi: 10.1172/JCI15020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang J., Takeuchi T., Tanaka S., Kubo S.K., Kayo T., Lu D. A mutation in the insulin 2 gene induces diabetes with severe pancreatic beta-cell dysfunction in the Mody mouse. Journal of Clinical Investigation. 1999;103(1):27–37. doi: 10.1172/JCI4431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Herbach N., Rathkolb B., Kemter E., Pichl L., Klaften M., de Angelis M.H. Dominant-negative effects of a novel mutated Ins2 allele causes early-onset diabetes and severe beta-cell loss in Munich Ins2C95S mutant mice. Diabetes. 2007;56(5):1268–1276. doi: 10.2337/db06-0658. [DOI] [PubMed] [Google Scholar]
- 34.Garin I., Perez de Nanclares G., Gastaldo E., Harries L.W., Rubio-Cabezas O., Castaño L. Permanent neonatal diabetes caused by creation of an ectopic splice site within the INS gene. PLoS One. 2012;7(1) doi: 10.1371/journal.pone.0029205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yildiz M., Akcay T., Aydin B., Akgun A., Dogan B.B., De Franco E. Emergence of insulin resistance following empirical glibenclamide therapy: a case report of neonatal diabetes with a recessive INS gene mutation. Journal of Pediatric Endocrinology & Metabolism. 2018;31(3):345–348. doi: 10.1515/jpem-2017-0325. [DOI] [PubMed] [Google Scholar]
- 36.Dusatkova L., Dusatkova P., Vosahlo J., Vesela K., Cinek O., Lebl J. Frameshift mutations in the insulin gene leading to prolonged molecule of insulin in two families with Maturity-Onset Diabetes of the Young. European Journal of Medical Genetics. 2015;58(4):230–234. doi: 10.1016/j.ejmg.2015.02.004. [DOI] [PubMed] [Google Scholar]
- 37.Demirbilek H., Arya V.B., Ozbek M.N., Houghton J.A., Baran R.T., Akar M. Clinical characteristics and molecular genetic analysis of 22 patients with neonatal diabetes from the South-Eastern region of Turkey: predominance of non-KATP channel mutations. European Journal of Endocrinology. 2015;172(6):697–705. doi: 10.1530/EJE-14-0852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ganesh R., Suresh N., Vasanthi T., Ravikumar K.G. Neonatal diabetes: a case series. Indian Pediatrics. 2017;54(1):33–36. doi: 10.1007/s13312-017-0993-6. [DOI] [PubMed] [Google Scholar]
- 39.Shaikh A.A., Shirah B., Alzelaye S. A homozygous mutation in the insulin gene (INS) causing autosomal recessive neonatal diabetes in Saudi families. Annals of Pediatric Endocrinology & Metabolism. 2020;25(1):42–45. doi: 10.6065/apem.2020.25.1.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fendler W., Borowiec M., Antosik K., Jaroszewska-Swiatek B., Szwalkiewicz-Warowicka E., Malecki M. Paternally inherited proinsulin mutations may result in earlier onset of monogenic diabetes mutation identity effect in monogenic diabetes. Diabetes Care. 2011;34(1):e9. doi: 10.2337/dc10-1142. [DOI] [PubMed] [Google Scholar]
- 41.Wasserman H., Hufnagel R.B., Miraldi Utz V., Zhang K., Valencia C.A., Leslie N.D. Bilateral cataracts in a 6-yr-old with new onset diabetes: a novel presentation of a known INS gene mutation. Pediatric Diabetes. 2016;17(7):535–539. doi: 10.1111/pedi.12335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Talaat I.M., Kamal N.M., Alghamdi H. Permanent neonatal DM in monozygotic twins with p.C109Y mutation in INS gene: first report from Saudi Arabia. Journal of Diabetes and Metabolism. 2014;5(2) [Google Scholar]
- 43.Ortolani F., Piccinno E., Grasso V., Papadia F., Panzeca R., Cortese C. Diabetes associated with dominant insulin gene mutations: outcome of 24-month, sensor-augmented insulin pump treatment. Acta Diabetologica. 2016;53(3):499–501. doi: 10.1007/s00592-015-0793-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dimova R., Tankova T., Gergelcheva I., Tournev I., Konstantinova M. A family with permanent neonatal diabetes due to a novel mutation in INS gene. Diabetes Research and Clinical Practice. 2015;108(2):e28–e30. doi: 10.1016/j.diabres.2015.02.021. [DOI] [PubMed] [Google Scholar]
- 45.Bee Y.M., Zhao Y., Ellard S., Hattersley A.T., Yap F. Permanent neonatal diabetes in siblings with novel C109Y INS mutation transmitted by an unaffected parent with somatic mosaicism. Pediatric Diabetes. 2014;15(4):324–328. doi: 10.1111/pedi.12104. [DOI] [PubMed] [Google Scholar]
- 46.Catli G., Abaci A., Flanagan S.E., Anik A., Ellard S., Bober E. Permanent neonatal diabetes caused by a novel mutation in the INS gene. Diabetes Research and Clinical Practice. 2013;99(1):e5–e8. doi: 10.1016/j.diabres.2012.10.009. [DOI] [PubMed] [Google Scholar]
- 47.Letourneau L.R., Carmody D., Philipson L.H., Greeley S.A.W. Early intensive insulin use may preserve β-cell function in neonatal diabetes due to mutations in the proinsulin gene. Journal of the Endocrine Society. 2018;2(1):1–8. doi: 10.1210/js.2017-00356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bonfanti R., Colombo C., Nocerino V., Massa O., Lampasona V., Iafusco D. Insulin gene mutations as cause of diabetes in children negative for five type 1 diabetes autoantibodies. Diabetes Care. 2009;32(1):123–125. doi: 10.2337/dc08-0783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lei S.Q., Wang J.Y., Li R.M., Chang J., Li Z., Ren L. MODY10 caused by c.309-314del CCAGCT insGCGC mutation of the insulin gene: a case report. American Journal of Translational Research. 2020;12(10):6599–6607. [PMC free article] [PubMed] [Google Scholar]
- 50.Johnson S.R., McGown I., Oppermann U., Conwell L.S., Harris M., Duncan E.L. A novel INS mutation in a family with maturity-onset diabetes of the young: variable insulin secretion and putative mechanisms. Pediatric Diabetes. 2018;19(5):905–909. doi: 10.1111/pedi.12679. [DOI] [PubMed] [Google Scholar]
- 51.Piccini B., Artuso R., Lenzi L., Guasti M., Braccesi G., Barni F. Clinical and molecular characterization of a novel INS mutation identified in patients with MODY phenotype. European Journal of Medical Genetics. 2016;59(11):590–595. doi: 10.1016/j.ejmg.2016.09.016. [DOI] [PubMed] [Google Scholar]
- 52.Moritani M., Yokota I., Tsubouchi K., Takaya R., Takemoto K., Minamitani K. Identification of INS and KCNJ11 gene mutations in type 1B diabetes in Japanese children with onset of diabetes before 5 years of age. Pediatric Diabetes. 2013;14(2):112–120. doi: 10.1111/j.1399-5448.2012.00917.x. [DOI] [PubMed] [Google Scholar]
- 53.Johansson B.B., Irgens H.U., Molnes J., Sztromwasser P., Aukrust I., Juliusson P.B. Targeted next-generation sequencing reveals MODY in up to 6.5% of antibody-negative diabetes cases listed in the Norwegian Childhood Diabetes Registry. Diabetologia. 2017;60(4):625–635. doi: 10.1007/s00125-016-4167-1. [DOI] [PubMed] [Google Scholar]
- 54.Buteau J., El-Assaad W., Rhodes C.J., Rosenberg L., Joly E., Prentki M. Glucagon-like peptide-1 prevents beta cell glucolipotoxicity. Diabetologia. 2004;47(5):806–815. doi: 10.1007/s00125-004-1379-6. [DOI] [PubMed] [Google Scholar]
- 55.Yusta B., Baggio L.L., Estall J.L., Koehler J.A., Holland D.P., Li H. GLP-1 receptor activation improves beta cell function and survival following induction of endoplasmic reticulum stress. Cell Metabolism. 2006;4(5):391–406. doi: 10.1016/j.cmet.2006.10.001. [DOI] [PubMed] [Google Scholar]
- 56.Farilla L., Bulotta A., Hirshberg B., Li Calzi S., Khoury N., Noushmehr H. Glucagon-like peptide 1 inhibits cell apoptosis and improves glucose responsiveness of freshly isolated human islets. Endocrinology. 2003;144(12):5149–5158. doi: 10.1210/en.2003-0323. [DOI] [PubMed] [Google Scholar]
- 57.Hattersley A.T., Greeley S.A.W., Polak M., Rubio-Cabezas O., Njølstad P.R., Mlynarski W. ISPAD Clinical Practice Consensus Guidelines 2018: the diagnosis and management of monogenic diabetes in children and adolescents. Pediatric Diabetes. 2018;19(Suppl 27):47–63. doi: 10.1111/pedi.12772. [DOI] [PubMed] [Google Scholar]
- 58.Carlsson A., Shepherd M., Ellard S., Weedon M., Lernmark Å., Forsander G. Absence of islet autoantibodies and modestly raised glucose values at diabetes diagnosis should lead to testing for MODY: lessons from a 5-year pediatric Swedish national cohort study. Diabetes Care. 2020;43(1):82–89. doi: 10.2337/dc19-0747. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.