Abstract
A fundamental question in developmental biology is how the progeny of stem cells become differentiated tissues. The Arabidopsis root is a tractable model to address this question due to its simple organization and defined cell lineages. In particular, the zone of dividing cells at the root tip—the root apical meristem—presents an opportunity to map the gene regulatory networks underlying stem cell niche maintenance, tissue patterning, and cell identity acquisition. To identify molecular regulators of these processes, studies over the last 20 years employed global profiling of gene expression patterns. However, these technologies are prone to information loss due to averaging gene expression signatures over multiple cell types and/or developmental stages. Recently developed high-throughput methods to profile gene expression at single-cell resolution have been successfully applied to plants. Here, we review insights from the first published single-cell mRNA sequencing and chromatin accessibility datasets generated from Arabidopsis roots. These studies successfully reconstruct developmental trajectories, phenotype cell identity mutants at unprecedented resolution, and reveal cell type-specific responses to environmental stimuli. The experimental insight gained from Arabidopsis paves the way to profile roots from additional species.
Keywords: Arabidopsis root, cell identity, chromatin accessibility, developmental trajectories, environmental response, single-cell RNA sequencing, transcriptomics
Single-cell mRNA sequencing and chromatin accessibility approaches are revolutionizing the study of plant development and environmental responses. Droplet-based technologies have been successfully applied to root and shoot tissues of model species.
Introduction
Sierra redwoods (Sequoia gigantea) are capable of reaching heights of 90 m after growing for over a thousand years. The formidable size and longevity of these trees are enabled by indeterminate organ growth mediated by stem cell niches in roots and shoots. The root apical meristem (RAM) is a zone of dividing cells that encompasses a stem cell niche at the root tip. These stem cells surround the quiescent center (QC), which comprises cells that divide infrequently and maintain the stem cell identity of adjacent cells (van den Berg et al., 1997; Drisch and Stahl, 2015). The discovery of the QC in the early 1950s by Lionel Clowes revolutionized our understanding of self-renewing cells in plants (Clowes, 1953; Dubrovsky and Barlow, 2015).
In contrast to redwoods, the small size and simplicity of the Arabidopsis thaliana root make it an ideal model for studying tissue patterning, cell differentiation, and organ development. Root cells are immobile and organized in concentric rings around a central vasculature. As the stem cells divide, new cells are added at the root tip. The root is therefore made up of longitudinal files of different cell types that represent a developmental timeline. This simple organization facilitated the early classification of Arabidopsis root cell type identities and developmental zones via morphological and histological characterization (Dolan et al., 1993).
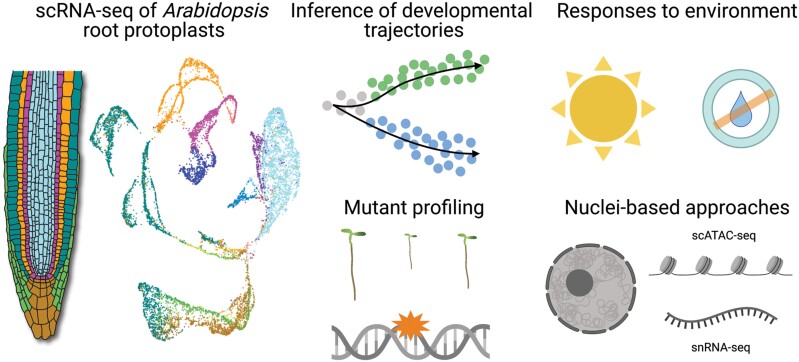
Beyond categorizing cells based on root anatomy, the rise of global transcriptomic sequencing opened up new opportunities to classify cell identity based on gene expression profiles (Birnbaum et al., 2003; Nawy et al., 2005; Brady et al., 2007; Li et al., 2016). However, bulk transcriptomic experiments conflate multiple cell types and/or developmental states (Birnbaum, 2018). Here, we review recent studies that profile Arabidopsis root transcriptomes at single-cell resolution and describe the potential of single-cell omics to revolutionize our understanding of cell identity and response to environmental stimuli in the RAM (Fig. 1).
Fig. 1.
Single-cell transcriptomics and chromatin accessibility experiments provide new insight into root development and environmental response. Single-cell mRNA sequencing (scRNA-seq) of Arabidopsis root protoplasts captures all major cell types and developmental stages. Since scRNA-seq requires cell destruction, developmental trajectories are inferred computationally. In addition to wild-type roots, scRNA-seq has been applied to mutants and roots which have been subjected to abiotic stress. In combination with scRNA-seq, single-cell chromatin accessibility (scATAC-seq) data generated from nuclei can be used to infer gene regulation underlying developmental processes and environmental responses. Beyond Arabidopsis, single-nuclei RNA-seq (snRNA-seq) is a promising approach to profile recalcitrant tissues from species such as maize or rice. The root illustration is modified from the Plant Illustrations repository (Sparks, 2017). All other panels were created with BioRender.com.
Bulk tissue approaches produce averaged gene expression information
The root consists of four major tissue types: the stele, ground tissue, epidermis, and root cap. Divisions of specific stem cells, also called initial cells or initials, give rise to the cell types that make up each of the tissues. For example, the cortex endodermis initial (CEI) divides twice to pattern the cortex and endodermis cell layers of the ground tissue. The first division produces the CEI daughter (CEID), which itself divides to produce one cortex cell and one endodermis cell (Drapek et al., 2017; Pierre-Jerome et al., 2018). The dividing CEI and CEID, as well as the orderly cell files that they produce, are easily observed with microscopy (Dolan et al., 1993). Thus, cell types and developmental zones can be determined by spatial location, and cell lineages are easily traced (Fig. 1). This is in contrast to development in animals, such as the zebrafish embryo, in which cells are mobile relative to each other (Farrell et al., 2018).
Despite the advantages of simple organization and immobile cells, global gene expression profiling experiments conducted on Arabidopsis roots over the last two decades required cell aggregation. Fluorescence activated cell sorting (FACS) enabled isolation of specific cell types for microarray analysis (Birnbaum et al., 2003; Brady et al., 2007) and RNA-seq experiments (Li et al., 2016), but required the generation of transgenic material, relied on the specificity of cell type markers, and necessitated the mixing of cells from different developmental stages. Careful hand dissection allowed profiling of cells from 12 longitudinal segments (Brady et al., 2007) or three morphological root developmental zones (Li et al., 2016), but this approach still required mixing cells of different lineages within each segment or zone. A major challenge remained: how can we increase the resolution at which gene expression changes are profiled? Observation of swift global gene expression changes or gene expression profiles that are specific to a small number of cells requires single-cell resolution.
Droplet-based technology revolutionizes single-cell transcriptomics
The first successful experiment to profile global gene expression at cellular resolution was published in 2009 on a single mouse blastomere (Tang et al., 2009). Several years later, Ken Birnbaum’s group pioneered single-cell transcriptomics in plants (Efroni et al., 2015, 2016), first by profiling 31 Arabidopsis root cells isolated by mouth pipette (Efroni et al., 2015). Beginning in 2015, microfluidics devices, first with homebrew set-ups such as Drop-seq (Macosko et al., 2015) and then with commercial platforms (e.g. 10X Genomics), allowed droplet-based profiling of thousands of cells in a single experiment. Experiments of this size are especially crucial to increase data depth given the technical challenges and noisy measurements associated with profiling the minuscule amount of mRNA present in a single cell.
The throughput realized by droplet-based technologies represents an opportunity to revolutionize the way we study developing organisms, particularly in how we classify different cell types versus different developmental states of the same cell type (Morris, 2019; Mayr et al., 2019; Rich-Griffin et al., 2020). For example, cell type and subtype identification previously based on morphology and markers developed to represent anatomical features can benefit from unbiased classifications based on global gene expression (Efroni and Birnbaum, 2016). Although droplet-based approaches were quickly and successfully applied to many types of human and animal cells (Stuart and Satija, 2019; Rozenblatt-Rosen et al., 2021), the suitability for plant cells, which require enzymatic digestion to remove cell walls, remained to be seen.
Large-scale single-cell transcriptomics pioneered with Arabidopsis roots
The first six studies to successfully apply droplet-based single-cell RNA-seq (scRNA-seq) to plants were published in 2019, all focused on the Arabidopsis primary root (Denyer et al., 2019; Jean-Baptiste et al., 2019; Ryu et al., 2019; Shulse et al., 2019; Turco et al., 2019; Zhang et al., 2019) (Fig. 1). Two additional Arabidopsis primary root studies (Shahan et al., 2020, Preprint; Wendrich et al., 2020) and one lateral root study (Gala et al., 2021) followed shortly thereafter. Of these nine publications, two generated data using Drop-seq (Shulse et al., 2019; Turco et al., 2019) while the rest used the commercial 10X Genomics platform. This body of work established several fundamental principles. First, overlapping cell distributions from biological replicates suggest a high degree of experimental reproducibility. Second, published, unsupervised clustering methodologies (Butler et al., 2018; Stuart et al., 2019) coupled with known, individual root cell type markers as well as known cell type gene expression profiles can successfully annotate datasets of several thousand cells. Importantly, comprehensive examination of transcriptional reporter expression of newly identified cell type markers established the utility of known cell type markers and gene expression profiles to accurately annotate root scRNA-seq data (Denyer et al., 2019; Wendrich et al., 2020). Third, all major cell types can be captured, most in proportions on a par with those reported from microscopy data (Cartwright et al., 2009; Wendrich et al., 2020).
Some of the most challenging cells to annotate in root scRNA-seq datasets have been the youngest cells of the RAM, which are more transcriptionally similar to each other than are older cells of each cell type (Denyer et al., 2019). However, single-cell datasets themselves represent an opportunity to identify and verify new cell type-specific markers that can in turn improve annotations (Denyer et al., 2019; Wendrich et al., 2020).
Developmental trajectories inferred at unprecedented resolution with scRNA-seq
Subsequent to assigning cell type labels, a major area of inquiry is the identification of genes with dynamic expression patterns across developmental time. These genes represent candidate regulators of tissue patterning and cell differentiation. For example, genes differentially expressed between cortex and endodermis, both of which are derived from the same stem cell, could identify new regulators of cell identity acquisition. Waves of gene expression provide a fine-grained view of transcriptomic changes undergone by a cell as it progresses along the pathway from stem cell to terminal differentiation. This resolution is an opportunity to identify regulators unique to each cell type as well as shared genes that control general developmental processes. Predicting regulatory connections between identified transcription factors is an exciting approach toward mapping gene regulatory networks (GRNs) underlying cell differentiation in each root tissue (Denyer et al., 2019).
Since current droplet-based scRNA-seq methods require tissue disruption and cell destruction, developmental trajectories must be computationally reconstructed for each root cell and tissue type. A common approach is to compute a pseudotime estimation for each cell in the dataset (Saelens et al., 2019). In this way, a developmental progression is inferred by ordering individual cells based on overlaps in their gene expression profiles (Fig. 1). Root data are especially amenable to this type of analysis since cell lineages are well defined.
A variety of pseudotime estimation tools are published and several have been used for Arabidopsis root data, including Monocle (Trapnell et al., 2014; Qiu et al., 2017), CytoTRACE (Gulati et al., 2020), Slingshot (Street et al., 2018), and scVelo (Bergen et al., 2020). These tools produce developmental progressions that reflect the dynamic expression of known genes in all four root tissue types (Denyer et al., 2019; Ryu et al., 2019; Jean-Baptiste et al., 2019; Shulse et al., 2019; Zhang et al., 2019; (Shahan et al., 2020, Preprint; Wendrich et al., 2020). The general reliability of pseudotime estimations was validated with transcriptional reporters for uncharacterized genes with dynamic expression patterns. For example, reporter expression is consistent with predicted expression in early or late developmental stages (Denyer et al., 2019; Shulse et al., 2019; Wendrich et al., 2020). Technical details regarding pseudotime estimations with plant data have recently been reviewed in detail elsewhere (Seyffurth et al., 2021; Shaw et al., 2021).
Profiling mutants at single-cell resolution enables unprecedented phenotypic insight
In addition to wild-type (WT) organs and organisms, scRNA-seq can be applied to mutant genotypes to characterize cell identity, tissue composition, and gene expression phenotypes. To test the ability of scRNA-seq data to reflect known phenotypes, Denyer et al. (2019) profiled shortroot-3 (shr-3), a previously characterized mutant with a strong phenotype in the ground tissue. In agreement with a known loss of endodermal identity, aggregation of the shr-3 data with those of the WT clearly shows a lack of shr-3 cells grouped with WT endodermis cells.
Box 1. Key developments in root single-cell sequencing.
• Successful application of scRNA-seq to Arabidopsis primary root tips
Eight papers established the applicability of droplet-based scRNA-seq to Arabidopsis primary roots (Denyer et al., 2019; Jean-Baptiste et al., 2019; Ryu et al., 2019; Shulse et al., 2019; Turco et al., 2019; Zhou et al., 2019; Shahan et al., 2020, Preprint; Wendrich et al., 2020). Transcriptional reporter expression of newly identified cell type markers established the reliability of scRNA-seq data themselves as well as the utility of known cell type markers and expression profiles for annotation (Denyer et al., 2019; Wendrich et al., 2020). In a different approach, Roszak et al. (2021) applied a plate-based scRNA-seq method, Switch Mechanism at the 5′ End of RNA Templates (Smart-seq), to profile 19 Arabidopsis root phloem cells. This work reconstructed the protophloem developmental trajectory to provide a detailed dissection of cell identity acquisition during tissue maturation.
• Developmental trajectories are successfully inferred from root scRNA-seq data
Since current scRNA-seq methods require tissue disruption, root developmental trajectories can be computationally reconstructed to identify candidate developmental regulators. Popular tools used by published root scRNA-seq papers include Monocle (Trapnell et al., 2014; Qiu et al., 2017), CytoTRACE (Gulati et al., 2020), Slingshot (Street et al., 2018), and scVelo (Bergen et al., 2020).
• scRNA-seq informs mutant phenotypes at unprecedented resolution
Strong cell identity and tissue composition phenotypes can be readily discerned from epidermis (Ryu et al., 2019) and ground tissue (Denyer et al., 2019; Shahan et al., 2020, Preprint) mutants. At single-cell resolution, new phenotypes can be characterized that were indiscernible with bulk tissue transcriptomics or morphological assessment.
• scRNA-seq informs transcriptional responses to environmental factors
Cell type- and developmental stage-specific responses to the environment can be assayed using single-cell omics approaches. So far, two studies have profiled Arabidopsis roots under different environmental conditions by altering sucrose levels in the growth media (Shulse et al., 2019) or performing a heat shock stress (Jean-Baptiste et al., 2019).
• Profiling root nuclei to generate transcriptome and open chromatin data
Isolation of nuclei allows for snRNA-seq and scATAC-seq to assay the transcriptome and open chromatin regions, respectively, while circumventing the need for cell dissociation. Several studies have demonstrated the applicability of these techniques to Arabidopsis roots (Dorrity et al., 2021, Preprint; Farmer et al., 2021) as well as more recalcitrant rice and maize roots (Marand et al., 2021; Zhang et al., 2021).
To examine epidermal phenotypes, Ryu et al. (2019) profiled the mutants root hair deficient6 (rhd6) and glabrous2 (gl2), the roots of which lack hair and non-hair cells, respectively. Both cell identity phenotypes were clearly reflected in the data. However, a deeper analysis of gene expression in the abnormal epidermal cells in both mutants indicated that hair cells in rhd6 were not entirely converted to non-hair cells and non-hair cells in gl2 were not entirely converted to hair cells. Together, these studies demonstrate that scRNA-seq captures expected cell identity alterations and can reveal subtle changes that are not easily discernible with morphological or bulk gene expression approaches.
Applications of scRNA-seq: environmental effects on cell identity
Another intriguing question in developmental biology is how cell identity and environmental responses affect one another. Bulk transcriptomic approaches have revealed that both cell identity and developmental stage influence stress-responsive gene expression (Dinneny et al., 2008; Mustroph et al., 2009; Iyer-Pascuzzi et al., 2011; Geng et al., 2013). For example, Dinneny et al. (2008) used FACS to examine the response to high salinity across radial cell layers. The majority of differentially expressed genes were affected in a single cell layer, with the highest number (48%) in the cortex. The same study also used dissection to examine the developmental stage-specific response to salinity along the longitudinal axis of the root. In this case, the elongation zone had the strongest transcriptional changes, which were associated with altered expression of cell wall components and radial swelling of the cortex (Dinneny et al., 2008). While this and other bulk studies have shown the promise of cell type-specific profiling of environmental changes, single-cell approaches are needed to determine if responses are specific to a cell type, a developmental stage, or both.
scRNA-seq has already been applied to several environmental perturbations. Shulse et al. (2019) performed scRNA-seq on 12 198 cells in the presence or absence of sucrose. Sucrose altered the composition of cell clusters in a manner consistent with phenotypic observations, provoking an increase in the number of epidermal hair cells. Additionally, differential expression analysis showed that nearly half of the genes affected were confined to a single cell type, whereas only 1% of genes were ubiquitously altered (Shulse et al., 2019). Another study profiled the response of 2085 cells to a 45 min heat shock stress as compared with a time-matched control (Jean-Baptiste et al., 2019). Heat shock caused dramatic changes in the transcriptome, which necessitated batch correction and integration in order to match cell identities between control and treated conditions. Subsequently, the authors identified 8526 genes with altered expression. These included known heat shock-responsive genes and identified a potential trade-off between induction of HEAT SHOCK PROTEIN 101 and cell identity markers such as COBL9 in the epidermis (Shulse et al., 2019). These studies highlight the potential for scRNA-seq to investigate environmental responses.
As throughput of scRNA-seq increases and costs decline, it will be possible to profile responses to environmental stimuli across multiple time points with biological replicates. A recent benchmark study identified pseudobulk approaches as top performing methods for differential expression in multi-condition experiments (Crowell et al., 2020), but this analysis requires biological replicates, which are costly to perform for scRNA-seq experiments. Increased cell numbers derived from biological replicates enabled coverage of the developmental progression across each cell lineage for untreated samples (Shahan et al., 2020, Preprint), therefore, as throughput increases, it should be possible to ask how stimuli-specific gene expression changes across the combination of cell type and developmental stage.
Bulk studies showed that only 15% of cell type-specific biological processes are maintained across different stress conditions (Dinneny et al., 2008; Iyer-Pascuzzi et al., 2011). Similar to the insight garnered from mutant analysis (Denyer et al., 2019; Ryu et al., 2019; Shahan et al., 2020, Preprint), scRNA-seq provides an opportunity to examine how the environment affects cell identity in more nuanced ways, which could reveal core aspects of cell identity and enable cell type-specific engineering of stress responses without comprising growth.
New applications: nuclei-based approaches
Although protoplast-based approaches have proven fruitful for the Arabidopsis RAM, protoplast isolation is typically performed using fresh tissues, and results in alterations of a subset of the transcriptome (Denyer et al., 2019). Nuclei-based approaches are emerging as an alternative and can be used to profile the transcriptome using single-nuclei RNA-seq (snRNA-seq) or to define open chromatin regions using single-cell sequencing of Assay for Transposase Accessible Chromatin (scATAC-seq) with either fresh or frozen tissues (Fig. 1). Several studies have applied these techniques to Arabidopsis roots as well as crops such as Zea mays (maize) and Oryza sativa (rice), which are more difficult to protoplast (Dorrity et al., 2021, Preprint; Farmer et al., 2021; Marand et al., 2021; Sunaga-Franze et al., 2021, Preprint; Zhang et al., 2021).
Farmer et al. (2021) profiled 10 548 Arabidopsis root nuclei using snRNA-seq and observed a relatively high concordance between protoplast and nuclei single-cell datasets, although fewer genes were detected per nucleus than per cell. Notably, several clusters of nuclei were present that were not easily discernible in a comparable protoplast sample. One nuclei-specific cluster corresponded to mature endodermal cells, which undergo suberization, probably making these cells recalcitrant to protoplasting (Andersen et al., 2015). Another nuclei-specific cluster was annotated as root cap, which undergoes programmed cell death upon terminal differentiation (Kumpf and Nowack, 2015).
Three studies have performed scATAC-seq using Arabidopsis roots (Dorrity et al., 2021, Preprint; Farmer et al., 2021; Marand et al., 2021). These datasets demonstrate that cell identity can be captured via accessible chromatin regions, albeit with largely distinct markers compared with those defined by scRNA-seq (Dorrity et al., 2021, Preprint). scATAC-seq can also be leveraged to identify cell- or cluster-specific transcription factor-binding sites to aid in GRN reconstruction (Dorrity et al., 2021, Preprint; Marand et al., 2021).
A major challenge with scATAC-seq is to determine the relationship between open chromatin regions and gene expression levels. Cell to cell correspondences appear to be at least partially recovered through the integration of independent scRNA-seq and scATAC-seq datasets (Farmer et al., 2021). However, dynamic changes in chromatin status and gene expression levels could be difficult to capture in this way. The emergence of paired RNA and ATAC assays, wherein both modalities can be measured from the same nucleus (Cao et al., 2018; Chen et al., 2019; Zhu et al., 2019), has great potential to overcome this limitation and unravel dynamic GRNs controlling cell identity and differentiation.
Conclusion and future perspectives
Breakthroughs in large-scale, single-cell omics technologies are revolutionizing the study of developmental biology. Profiling gene expression dynamics and chromatin accessibility at single-cell resolution in the RAM is an exciting opportunity to map GRNs underlying cell identity acquisition and fate stabilization, particularly in cell types that arise from asymmetric divisions of the same stem cell. Single-cell omics is also poised to uncover the interplay between cell identity and environmental responses.
The pioneering application of droplet-based scRNA-seq and scATAC-seq to Arabidopsis root tissue was aided by defined cell lineages and a suite of known cell type markers. The next challenge is to profile roots of species for which fewer molecular tools are available. Indeed, two recent studies pioneered droplet-based scRNA-seq in rice (Liu et al., 2021; Zhang et al., 2021). In a different approach to profile rare cell types, Omary et al. (2020) used a plate-based technology, molecular crowding single-cell RNA barcoding and sequencing (mcSCRB-seq), to identify new cell identities during stem-borne root initiation in tomato. Beyond roots, a major goal is to apply single-cell omics technologies to other plant organs. To date, droplet-based scRNA-seq has been successfully applied to shoot tissues of Arabidopsis, rice, maize, and tomato, the details of which are reviewed by Seyfferth et al. (2021). This body of work paves the way to apply single-cell approaches to non-model plant species and less-studied plant organs. However, challenges include the need to optimize protoplasting and/or nuclei isolation protocols and a paucity of established marker genes to assist cell type annotation.
With the rapid pace of single-cell data generation in plants, there is a growing need for a community effort to standardize experimental and analytical methods as well as data curation and accessibility. Such an effort is already underway in the form of the Plant Cell Atlas initiative (Rhee et al., 2019). Methods to integrate data generated across multiple labs provide the foundation to create comprehensive WT atlases for different organs and organisms. Integration of multi-omics data, such as transcriptomic and proteomic data, is also a major area of interest for atlas development. These community resources will be valuable to query genes of interest and inform new datasets generated from mutants and plants treated with hormones or subjected to stress.
Acknowledgements
We apologize to colleagues whose work was not cited due to space constraints. This work was funded by the US National Institutes of Health (NRSA postdoctoral fellowship 1F32GM136030-01 and MIRA 1R35GM131725) to RS and PNB, respectively; the US National Science Foundation (Postdoctoral Research Fellowships in Biology Program Grant no. IOS-2010686) to TMN; and the Howard Hughes Medical Institute to PNB as an Investigator. PNB is the co-founder and Chair of the Scientific Advisory Board of Hi Fidelity Genetics, Inc, a company that works on crop root growth.
References
- Andersen TG, Barberon M, Geldner N. 2015. Suberization—the second life of an endodermal cell. Current Opinion in Plant Biology 28, 9–15. [DOI] [PubMed] [Google Scholar]
- Bergen V, Lange M, Peidli S, Wolf FA, Theis FJ. 2020. Generalizing RNA velocity to transient cell states through dynamical modeling. Nature Biotechnology 38, 1408–1414. [DOI] [PubMed] [Google Scholar]
- Birnbaum KD. 2018. Power in numbers: single-cell RNA-seq strategies to dissect complex tissues. Annual Review of Genetics 52, 203–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnbaum K, Shasha DE, Wang JY, Jung JW, Lambert GM, Galbraith DW, Benfey PN. 2003. A gene expression map of the Arabidopsis root. Science 302, 1956–1960. [DOI] [PubMed] [Google Scholar]
- Brady SM, Orlando DA, Lee JY, Wang JY, Koch J, Dinneny JR, Mace D, Ohler U, Benfey PN. 2007. A high-resolution root spatiotemporal map reveals dominant expression patterns. Science 318, 801–806. [DOI] [PubMed] [Google Scholar]
- Butler A, Hoffman P, Smibert P, Papalexi E, Satija R. 2018. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nature Biotechnology 36, 411–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J, Cusanovich DA, Ramani V, et al. 2018. Joint profiling of chromatin accessibility and gene expression in thousands of single cells. Science 361, 1380–1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartwright DA, Brady SM, Orlando DA, Sturmfels B, Benfey PN. 2009. Reconstructing spatiotemporal gene expression data from partial observations. Bioinformatics 25, 2581–2587. [DOI] [PubMed] [Google Scholar]
- Chen S, Lake BB, Zhang K. 2019. High-throughput sequencing of the transcriptome and chromatin accessibility in the same cell. Nature Biotechnology 37, 1452–1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clowes FAL. 1953. The cytogenerative centre in roots with broad columellas. New Phytologist 52, 48–57. [Google Scholar]
- Crowell HL, Soneson C, Germain PL, Calini D, Collin L, Raposo C, Malhotra D, Robinson MD. 2020. muscat detects subpopulation-specific state transitions from multi-sample multi-condition single-cell transcriptomics data. Nature Communications 11, 6077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denyer T, Ma X, Klesen S, Scacchi E, Nieselt K, Timmermans MCP. 2019. Spatiotemporal developmental trajectories in the Arabidopsis root revealed using high-throughput single-cell RNA sequencing. Developmental Cell 48, 840–852.e5. [DOI] [PubMed] [Google Scholar]
- Dinneny JR, Long TA, Wang JY, Jung JW, Mace D, Pointer S, Barron C, Brady SM, Schiefelbein J, Benfey PN. 2008. Cell identity mediates the response of Arabidopsis roots to abiotic stress. Science 320, 942–945. [DOI] [PubMed] [Google Scholar]
- Dolan L, Janmaat K, Willemsen V, Linstead P, Poethig S, Roberts K, Scheres B. 1993. Cellular organisation of the Arabidopsis thaliana root. Development 119, 71–84. [DOI] [PubMed] [Google Scholar]
- Dorrity MW, Alexandre CM, Hamm M, Vigil A-L, Fields S, Queitsch C, Cuperus J. 2021. The regulatory landscape of Arabidopsis thaliana roots at single-cell resolution. bioRxiv. doi: 10.1101/2020.07.17.204792. [Preprint]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drapek C, Sparks EE, Benfey PN. 2017. Uncovering gene regulatory networks controlling plant cell differentiation. Trends in Genetics 33, 529–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drisch RC, Stahl Y. 2015. Function and regulation of transcription factors involved in root apical meristem and stem cell maintenance. Frontiers in Plant Science 6, 505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubrovsky JG, Barlow PW. 2015. The origins of the quiescent centre concept. New Phytologist 206, 493–496. [DOI] [PubMed] [Google Scholar]
- Efroni I, Birnbaum KD. 2016. The potential of single-cell profiling in plants. Genome Biology 17, 65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efroni I, Ip PL, Nawy T, Mello A, Birnbaum KD. 2015. Quantification of cell identity from single-cell gene expression profiles. Genome Biology 16, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efroni I, Mello A, Nawy T, Ip PL, Rahni R, DelRose N, Powers A, Satija R, Birnbaum KD. 2016. Root regeneration triggers an embryo-like sequence guided by hormonal interactions. Cell 165, 1721–1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farmer A, Thibivilliers S, Ryu KH, Schiefelbein J, Libault M. 2021. Single-nucleus RNA and ATAC sequencing reveals the impact of chromatin accessibility on gene expression in Arabidopsis roots at the single-cell level. Molecular Plant 14, 372–383. [DOI] [PubMed] [Google Scholar]
- Farrell JA, Wang Y, Riesenfeld SJ.Shekhar K, Regev A, Schier AF. 2018. Single-cell reconstruction of developmental trajectories during zebrafish embryogenesis. Science 360, eaar3131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gala HP, Lanctot A, Jean-Baptiste K, Guiziou S, Chu JC, Zemke JE, George W, Queitsch C, Cuperus JT, Nemhauser JL. 2021. A single cell view of the transcriptome during lateral root initiation in Arabidopsis thaliana. The Plant Cell doi: 10.1093/plcell/koab101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng Y, Wu R, Wee CW, Xie F, Wei X, Chan PM, Tham C, Duan L, Dinneny JR. 2013. A spatio-temporal understanding of growth regulation during the salt stress response in Arabidopsis. The Plant Cell 25, 2132–2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulati GS, Sikandar SS, Wesche DJ, et al. 2020. Single-cell transcriptional diversity is a hallmark of developmental potential. Science 367, 405–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer-Pascuzzi AS, Jackson T, Cui H, Petricka JJ, Busch W, Tsukagoshi H, Benfey PN. 2011. Cell identity regulators link development and stress responses in the Arabidopsis root. Developmental Cell 21, 770–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jean-Baptiste K, McFaline-Figueroa JL, Alexandre CM, Dorrity MW, Saunders L, Bubb KL, Trapnell C, Fields S, Queitsch C, Cuperus JT. 2019. Dynamics of gene expression in single root cells of Arabidopsis thaliana. The Plant Cell 31, 993–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumpf RP, Nowack MK. 2015. The root cap: a short story of life and death. Journal of Experimental Botany 66, 5651–5662. [DOI] [PubMed] [Google Scholar]
- Li S, Yamada M, Han X, Ohler U, Benfey PN. 2016. High-resolution expression map of the arabidopsis root reveals alternative splicing and lincRNA regulation. Developmental Cell 39, 508–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Liang Z, Feng D, Jiang S, Wang Y, Du Z, Li R, Hu G, Zhang P, Ma Y, Lohmann JU, Gu X. 2021. Transcriptional landscape of rice roots at the single-cell resolution. Molecular Plant 14, 384–394. [DOI] [PubMed] [Google Scholar]
- Macosko EZ, Basu A, Satija R, et al. 2015. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell 161, 1202–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marand AP, Chen Z, Gallavotti A, Schmitz RJ. 2021. A cis-regulatory atlas in maize at single-cell resolution. Cell 184, 1–15. [DOI] [PubMed] [Google Scholar]
- Mayr U, Serra D, Liberali P. 2019. Exploring single cells in space and time during tissue development, homeostasis and regeneration. Development 146, dev176727. [DOI] [PubMed] [Google Scholar]
- Morris SA. 2019. The evolving concept of cell identity in the single cell era. Development 146, dev169748. [DOI] [PubMed] [Google Scholar]
- Mustroph A, Zanetti ME, Jang CJ, Holtan HE, Repetti PP, Galbraith DW, Girke T, Bailey-Serres J. 2009. Profiling translatomes of discrete cell populations resolves altered cellular priorities during hypoxia in Arabidopsis. Proceedings of the National Academy of Sciences, USA 106, 18843–18848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawy T, Lee JY, Colinas J, Wang JY, Thongrod SC, Malamy JE, Birnbaum K, Benfey PN. 2005. Transcriptional profile of the Arabidopsis root quiescent center. The Plant Cell 17, 1908–1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omary M, Gil-Yarom N, Yahav C, Steiner E, Efroni I. 2020. A conserved superlocus regulates above- and belowground root initiation. bioRxiv doi: 10.1101/2020.11.11.377937. [Preprint]. [DOI] [PubMed] [Google Scholar]
- Pierre-Jerome E, Drapek C, Benfey PN. 2018. Regulation of division and differentiation of plant stem cells. Annual Review of Cell and Developmental Biology 34, 289–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu X, Mao Q, Tang Y, Wang L, Chawla R, Pliner HA, Trapnell C. 2017. Reversed graph embedding resolves complex single-cell trajectories. Nature Methods 14, 979–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee SY, Birnbaum KD, Ehrhardt DW. 2019. Towards building a plant cell atlas. Trends in Plant Science 24, 303–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rich-Griffin C, Stechemesser A, Finch J, Lucas E, Ott S, Schäfer P. 2020. Single-cell transcriptomics: a high-resolution avenue for plant functional genomics. Trends in Plant Science 25, 186–197. [DOI] [PubMed] [Google Scholar]
- Roszak P, Heo J, Blob B, et al. 2021. Analysis of phloem trajectory links tissue maturation to cell specialization. bioRxiv doi: 10.1101/2021.01.18.427084. [Preprint]. [DOI] [Google Scholar]
- Rozenblatt-Rosen O, Shin JW, Rood JE, Hupalowska A, Regev A, Heyn H; Human Cell Atlas Standards and Technology Working Group . 2021. Building a high-quality human cell atlas. Nature Biotechnology 39, 149–153. [DOI] [PubMed] [Google Scholar]
- Ryu KH, Huang L, Kang HM, Schiefelbein J. 2019. Single-cell RNA sequencing resolves molecular relationships among individual plant cells. Plant Physiology 179, 1444–1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saelens W, Cannoodt R, Todorov H, Saeys Y. 2019. A comparison of single-cell trajectory inference methods. Nature Biotechnology 37, 547–554. [DOI] [PubMed] [Google Scholar]
- Seyfferth C, Renema J, Wendrich JR, et al. 2021. Advances and opportunities of single-cell transcriptomics for plant research. Annual Review of Plant Biology 72, doi: 10.1146/annurev-arplant-081720-010120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahan R, Hsu C-W, Nolan TM, et al. 2020. A single cell Arabidopsis root atlas reveals developmental trajectories in wild type and cell identity mutants. bioRxiv doi: 10.1101/2020.06.29.178863. [Preprint]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw R, Tian X, Xu J. 2021. Single-cell transcriptome analysis in plants: advances and challenges. Molecular Plant 14, 115–126. [DOI] [PubMed] [Google Scholar]
- Shulse CN, Cole BJ, Ciobanu D, et al. 2019. High-throughput single-cell transcriptome profiling of plant cell types. Cell Reports 27, 2241–2247.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparks E. 2017. Arabidopsis root anatomy. 10.6084/m9.figshare.4688344.v1 [DOI]
- Street K, Risso D, Fletcher RB, Das D, Ngai J, Yosef N, Purdom E, Dudoit S. 2018. Slingshot: cell lineage and pseudotime inference for single-cell transcriptomics. BMC Genomics 19, 477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM 3rd, Hao Y, Stoeckius M, Smibert P, Satija R. 2019. Comprehensive integration of single-cell data. Cell 177, 1888–1902.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart T, Satija R. 2019. Integrative single-cell analysis. Nature Reviews. Genetics 20, 257–272. [DOI] [PubMed] [Google Scholar]
- Sunaga-Franze DY, Muino JM, Braeuning C, et al. 2021. Single-nuclei RNA-sequencing of plant tissues. bioRxiv doi: 10.1101/2020.11.14.382812. [Preprint]. [DOI] [PubMed] [Google Scholar]
- Tang F, Barbacioru C, Wang Y, et al. 2009. mRNA-Seq whole-transcriptome analysis of a single cell. Nature Methods 6, 377–382. [DOI] [PubMed] [Google Scholar]
- Trapnell C, Cacchiarelli D, Grimsby J, Pokharel P, Li S, Morse M, Lennon NJ, Livak KJ, Mikkelsen TS, Rinn JL. 2014. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nature Biotechnology 32, 381–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turco GM, Rodriguez-Medina J, Siebert S, et al. 2019. Molecular mechanisms driving switch behavior in xylem cell differentiation. Cell Reports 28, 342–351.e4. [DOI] [PubMed] [Google Scholar]
- van den Berg C, Willemsen V, Hendriks G, Weisbeek P, Scheres B. 1997. Short-range control of cell differentiation in the Arabidopsis root meristem. Nature 390, 287–289. [DOI] [PubMed] [Google Scholar]
- Wendrich JR, Yang B, Vandamme N, et al. 2020. Vascular transcription factors guide plant epidermal responses to limiting phosphate conditions. Science 370, eaay4970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang TQ, Chen Y, Liu Y, Lin WH, Wang JW. 2021. Single-cell transcriptome atlas and chromatin accessibility landscape reveal differentiation trajectories in the rice root. Nature Communications 12, 2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang TQ, Xu ZG, Shang GD, Wang JW. 2019. A single-cell RNA sequencing profiles the developmental landscape of arabidopsis root. Molecular Plant 12, 648–660. [DOI] [PubMed] [Google Scholar]
- Zhu C, Yu M, Huang H, Juric I, Abnousi A, Hu R, Lucero J, Behrens MM, Hu M, Ren B. 2019. An ultra high-throughput method for single-cell joint analysis of open chromatin and transcriptome. Nature Structural & Molecular Biology 26, 1063–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]