Abstract
The presence of two meristematic cell populations in the root and shoot apex allows plants to grow indefinitely. Due to its simple and predictable tissue organization, the Arabidopsis root apical meristem remains an ideal model to study mechanisms such as stem cell specification, asymmetric cell division, and differentiation in plants. The root stem cell niche consists of a quiescent organizing centre surrounded by mitotically active stem cells, which originate all root tissues. The transcription factors PLETHORA, SCARECROW, and WOX5 form signalling hubs that integrate multiple inputs from an increasing number of proteins implicated in the regulation of stem cell niche function. Recently, locally produced auxin was added to the list of important mobile factors in the stem cell niche. In addition, protein–protein interaction data elegantly demonstrate how parallel pathways can meet in a common objective. Here we discuss how multiple networks converge to specify and maintain the root stem cell niche.
Keywords: Arabidopsis, development, JACKDAW, PLETHORA, quiescent centre, root, SCARECROW, SHORT-ROOT, stem cell niche, WOX5
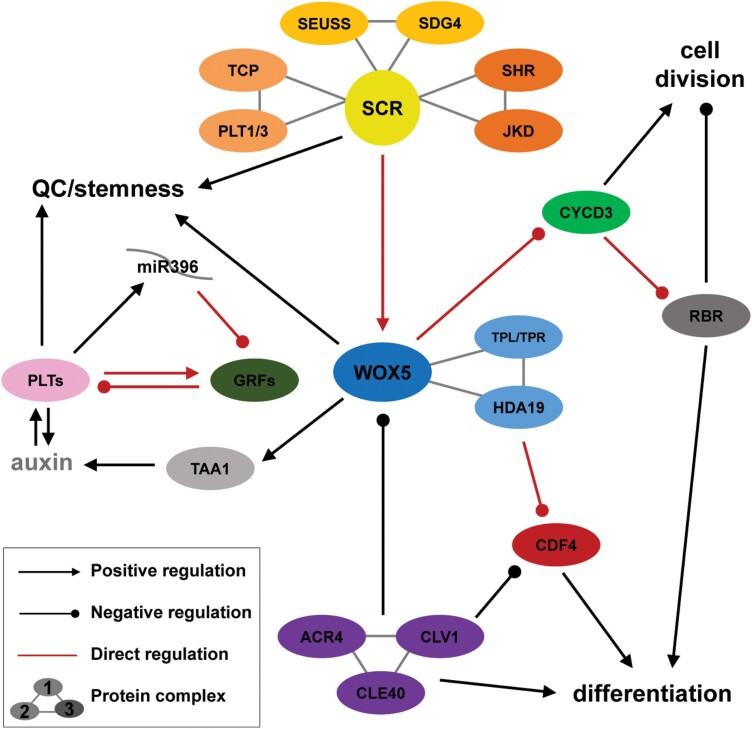
An updated overview of the main gene networks converging for the specification and maintenance of the Arabidopsis root stem cell niche.
Introduction
Unlike animals, plants display indefinite growth and produce organs throughout their life cycle. This is possible due the presence of two populations of stem cells located in the shoot and root apex. The Arabidopsis root apical meristem remains one of the best studied model systems in plants to address developmental questions like stem cell specification, differentiation, regeneration, cell–cell interaction, and cell cycle control. One aspect of the continued popularity of the Arabidopsis root meristem is the highly structured, simple, and predictable tissue organization. The root stem cell niche resides within the meristem and is composed of a group of infrequently dividing cells named the quiescent centre (QC), surrounded by mitotically active stem cells, also called initials (Dolan et al., 1993). Division of the initials produces daughter cells that remain in ordered cell files and together form all the tissues that compose the root in a stereotypic pattern. The initial cells together with the QC are considered a stem cell niche (SCN). Within the SCN, distal to the QC, the columella initials (or distal meristem) divide to form the mature differentiated columella cells. The latter are easily distinguished from the columella initials by the accumulation of starch-laden amyloplasts. Proximal to the QC are the vascular initials and the ground tissue initials. The vascular initials originate the stele after going through a round of formative division. The ground tissue initial daughter originates both cortex and endodermis. Finally, lateral to the QC are the epidermis/lateral root cap initials that, like the ground tissue initial, generate two tissues (Fig. 1) (Dolan et al., 1993).
Fig. 1.
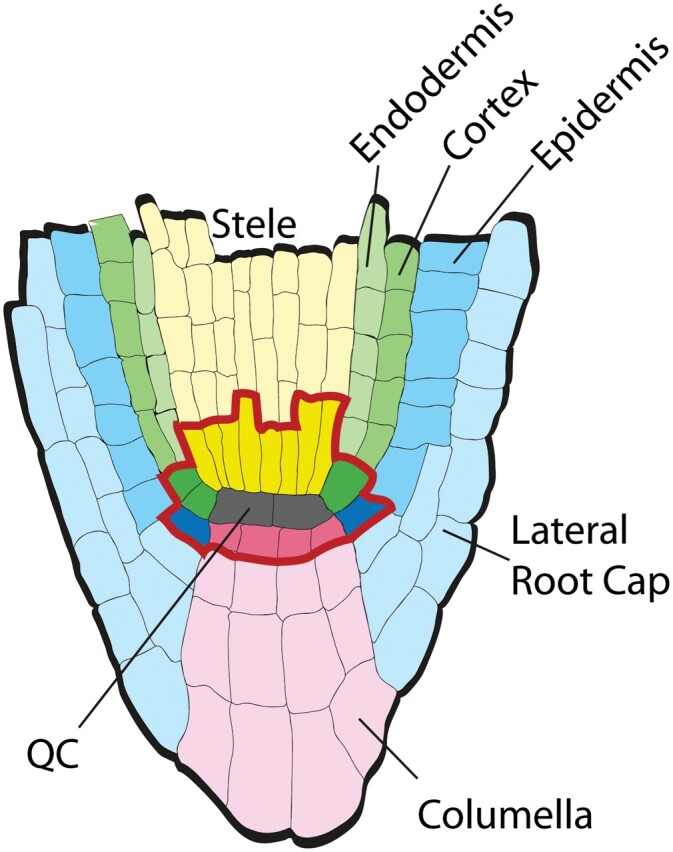
Schematic representation of the Arabidopsis root tip. The stem cell niche, enclosed by the red line, consists of a centrally localized quiescent centre (QC) surrounded by the tissue initials/stem cells. Vascular initials are above the QC, shown in dark yellow. Flanking the QC are two types of initials: the ground tissue initials, shown in dark green that originate the cortex and endodermis, and the epidermis/lateral root cap initials, shown in dark blue. Below the QC, shown in dark pink, are the columella initials.
Throughout this review, we dissect the complex puzzle underlying the specification and maintenance of the Arabidopsis root SCN into separate regulatory pieces. By exploring previous and recent data, we address the interaction between well-known factors such as PLETHORA, SCARECROW, and WOX5 in the establishment of a functional root organizing centre. Likewise, the relevance of intercellular communication within the SCN, the importance of quiescence, and the role of local auxin production are addressed. In addition, we explore the multiple complexes that have SCARECROW as a member, hypothesizing how these are formed and regulated within the cellular environment. Furthermore, we discuss in a comprehensive way how these networks connect and converge into a functional SCN (Table 1; Fig. 2).
Table 1.
Main genes involved in SCN regulation
| Gene | Description | Expression in post-embryonic root | SCN-related processes | References |
|---|---|---|---|---|
| WOX5 | Homeobox TF | Strongly expressed in QC and weakly expressed in the proximal meristem | QC specification and maintenance; maintenance of columella initials | Sarkar et al. (2007), Forzani et al. (2014), Pi et al. (2015), Shimotohno et al. (2018), Berckmans et al. (2020), Zhai et al. (2020) |
| CYCD3;3 | Cyclin | Lateral root cap, stele, distal columella, and epidermal initials | Columella formation and maintenance of columella initials | Forzani et al. (2014) |
| RBR1 | Retinoblastoma protein family | Whole root tip | QC quiescence; columella differentiation; ground tissue formative divisions | Wildwater et al. (2005), Wachsman et al. (2011), Cruz-Ramírez et al. (2012),Cruz-Ramírez et al. (2013) |
| SCR | GRAS family TF | U-domain | QC specification and maintenance; ground tissue formative divisions | Sabatini (2003), Cui et al. (2007), Cruz-Ramírez et al. (2012), Hirano et al. (2017), Long et al. (2017), Shimotohno et al. (2018), Zhai et al. (2020) |
| SHR | GRAS family TF | Expressed in the stele; protein moves to U-domain | QC specification and maintenance; ground tissue formative divisions | Helariutta et al. (2000), Nakajima et al. (2001), Cui et al. (2007), Welch et al. (2007), Long et al. (2015), Long et al. (2017) |
| BIB | BIRD C2H2-type zinc finger protein | QC, ground tissue initials, and mature cells | QC maintenance; ground tissue formative divisions |
Long et al. (2015) |
| JKD | BIRD C2H2-type zinc finger protein | QC, ground tissue initials and mature cells | QC specification and maintenance; maintenance of columella initials; ground tissue formative divisions | Welch et al. (2007), Long et al. (2015), Hirano et al. (2017), Long et al. (2017) |
| PLT’s | AP2/ERF TF | Diverse gradient patterns; all peaking at the SCN | QC/SCN specification and maintenance | Aida et al. (2004), Galinha et al. (2007), Mähönen et al. (2014), Santuari et al. (2016) |
| miR396 | MicroRNA | Combined miR396A and miR396B: QC, lateral root cap, columella initials, and mature cells | SCN maintenance | Rodriguez et al. (2015) |
| CDF4 | Dof zinc finger protein | Columella initials and mature cells | Columella differentiation | Pi et al. (2015) |
| ACR4 | Receptor kinase | Part of lateral root cap, columella initials, and mature cells | Columella differentiation | Stahl et al. (2009), Stahl et al. (2013), Berckmans et al. (2020) |
| CLV1 | Receptor kinase | Part of lateral root cap, columella initials, and mature cells | Columella differentiation | Stahl et al. (2013), Berckmans et al. (2020) |
| CLE40 | CLV3/ESR signal peptide family | Differentiating stele and columella | Columella differentiation | Stahl et al. (2009), Stahl et al. (2013), Berckmans et al. (2020) |
| HDA19 | Histone deacetylase | Whole root tip; reduced expression in the columella | Maintenance of columella initials | Pi et al. (2015) |
| SDG4 | Histone-lysine methyltransferase | SCN | QC specification and maintenance; maintenance of columella initials | Kumpf et al. (2014), Zhai et al. (2020) |
| SEU | LIM-domain-binding co-regulator | Whole root tip | QC specification and maintenance; maintenance of columella initials | Gong et al. (2016), Zhai et al. (2020) |
| TCP20/TCP21 | TCP family TF | Combined TCP20 and TCP21: whole root tip | QC specification and maintenance; maintenance of columella initials | Shimotohno et al. (2018) |
| TPL/TPR | TOPLESS co-repressor | TOPLESS is expressed in the whole root tip; reduced expression in the columella | Maintenance of columella initials | Pi et al. (2015) |
| TAA1 | Tryptophan aminotransferase | QC | Auxin biosynthesis; SCN maintenance | Stepanova et al. (2008), Brumos et al. (2018), Savina et al. (2020) |
TF, transcription factor.
Fig. 2.
Overview of the pathways regulating the stem cell niche. At the top are three protein complexes for which SCR is fundamental. These complexes are implicated in the direct positive transcriptional regulation of WOX5 in the QC. Note that data only support the assembly of SCR into individual complexes. Also, SCR can promote stemness and QC fate independently from WOX5. On the right is shown CYCD3, the expression of which is directly repressed by WOX5 in the QC and which participates in the phosphorylation of RBR, thereby inactivating it. Whereas CYCD3 promotes cell division, RBR—in its active form—represses cell division and promotes cell differentiation. On the left is shown TAA1, the expression of which is up-regulated by WOX5 and which in turn enhances auxin production within the QC. High levels of auxin increase PLT expression, which in turn promotes auxin production in a feed-forward loop. PLTs are required for QC specification and promote stemness of surrounding initials. In a feed-back loop, PLTs positively regulate expression of GRF and miR396, which directly post-transcriptionally repress GRFs in the SCN. Low levels of GRF can no longer directly repress PLTs, thereby assuring their high expression in the SCN. In the centre, WOX5 autonomously promotes QC fate and non-autonomously promotes stemness in the columella initials. WOX5 also forms a complex with TPL/TPR and HDA19 to repress the differentiation factor gene CDF4 in the QC. Shown at the bottom, the ACR4–CLV1–CLE40 signalling module represses WOX5 expression, whereas it promotes columella differentiation in a CDF4-independent manner. The red lines indicate direct transcriptional, post-transcriptional, or post-translational regulation based on experimental data.
WOX5 autonomous function
When we refer to the QC, the first gene that comes to mind is likely to be WUSCHEL RELATED HOMEOBOX 5 (WOX5). Expression of WOX5 begins at the globular stage of embryogenesis, in the hypophyseal cell. Upon division of the hypophyseal cell, WOX expression is restricted to the lens shaped cell that is the founder of the QC (Haecker et al., 2004; Sarkar et al., 2007). Post-embryonically, the promoter of this transcriptional factor is highly active in the QC and slightly active in the vascular initials. However, driving the expression of a green fluorescent protein (GFP)–WOX5 fusion protein under its endogenous promoter has shown that the WOX5 protein also localizes to the adjacent columella initial layer, indicating its mobile nature (Pi et al., 2015). The loss of function mutant wox5-1 displays accumulation of starch in the columella initials, indicating that these cells fail to maintain their undifferentiated state. On the other hand, plants ectopically expressing WOX5 develop multiple undifferentiated columella cell layers (Sarkar et al., 2007; Pi et al., 2015). In addition, wox5-1 also presents extra transverse divisions in the QC (Sarkar et al., 2007; Forzani et al., 2014). Importantly, there is only a clear effect on proximal stem cell differentiation when wox5-1 mutant is combined with mutations in SHORT-ROOT (SHR), SCARECROW (SCR), or PLETHORA1 (PLT1) and PLT2 genes (Sarkar et al., 2007). Therefore, these data indicate that the primary roles of WOX5 are the autonomous prevention of QC divisions and the non-autonomous maintenance of columella stem cell activity (Sarkar et al., 2007; Forzani et al., 2014; Pi et al., 2015).
RETINOBLASTOMA-RELATED (RBR) is a master regulator of the cell cycle. In its hypophosphorylated and active form, it prevents cell cycle progression. When phosphorylated by cyclin–cyclin-dependent kinase (CYC–CDKs) complexes, RBR is inactivated allowing cell cycle advancement (Boniotti and Gutierrez, 2001). Concordantly, CYCLIN B1;1 expression was observed frequently in QC cells of RBR RNA interference plants, indicating that QC cells divide more frequently than in the wild type (WT). These plants also developed additional columella stem cell layers. However, these did not originate by enhanced stem cell divisions, indicating a role for RBR in stem cell differentiation. This was corroborated by ectopic RBR expression that had the opposite effect and resulted in differentiation of columella stem cells (Wildwater et al., 2005). Using a clonal deletion system, it was shown that RBR confers its QC division and stem cell differentiation effects in a cell-autonomous manner (Wachsman et al., 2011). Tissue specific CYCLIN D genes are candidates to modulate local RBR activity. Indeed, it has been shown that the ectopic expression of CYCLIN D3;3 (CYCD3;3) under the WOX5 promoter induces divisions in the QC and even leads to reduction in the expression of the QC identity markers QC25 and QC46 (Sabatini, 2003; Forzani et al., 2014). Analysis of GFP reporter lines showed that CYCD3;3 is expressed in the QC of wox5-1 mutant but not in the QC of WT plants, indicating that the correct expression of WOX5 is sufficient to exclude CYCD3;3 expression from the QC. These results are further supported by chromatin immunoprecipitation (ChIP) experiments indicating that WOX5 binds to the CYCD3;3 promoter (Forzani et al., 2014). Together, these results demonstrate that at least part of the autonomous role of WOX5 in QC function is maintenance of quiescence through the regulation of CYCD3;3.
How important is quiescence?
The abolition of QC quiescence leading to partial loss of QC identity either in the wox5-1 mutant or upon ectopic CYCD3;3 expression in the QC, suggests that the QC and its function in stem cell maintenance strictly depend on quiescence (Sarkar et al., 2007; Forzani et al., 2014). However, other mutants and conditions that induce ectopic divisions in the QC indicate otherwise. For example, the mutant ethylene overproducer 1 (eto1) that overproduces ethylene, displays extra QC divisions but does not lose expression of the QC identity marker QC25 (Ortega-Martínez et al., 2007). Similarly, four mutants insensitive to abscisic acid (aba insensitive 1 (abi1), abi2, abi3, and abi5) also exhibit enhanced QC divisions, but unlike wox5-1 they do not lose the columella initials (Zhang et al., 2010). Furthermore, despite the QC divisions observed upon RBR depletion from the QC, these roots still express WOX5, and in normal conditions, neither root growth nor root SCN function is affected. Nevertheless, differences are seen when plants are treated with a DNA-damaging agent. Upon loss of quiescence, the RBR-depleted QC is more sensitive to DNA damage (displayed by higher lethality), and over time, root growth is more affected compared with WT plants (Cruz-Ramírez et al., 2013). These observations indicate that quiescence is not essential for maintenance of a functional QC or root apical meristem, since a dividing QC can still function as an organizing centre. Thus, it appears that the main function of WOX5 is the non-autonomous maintenance of stem cells. On the other hand, the QC quiescence that is mediated autonomously by WOX5, through the direct regulation of CYCD3 and indirect regulation of RBR, functions to protect the QC from stresses, exemplified by DNA damage induction experiments. In line with this, AP2/ERF subfamily X transcription factors ETHYLENE RESPONSE FACTOR 109 (ERF109) and ERF115 are induced by stress conditions, allowing the QC to divide in order to replenish affected stem cells (Heyman et al., 2013; Kong et al., 2018). ERF115 interacts with RBR thereby coupling at least part of its effect to the RBR–SCR module in regulating QC division (see below) (Zhou et al., 2019). These observations expand the idea that the QC acts as a reservoir, ready to replenish damaged stem cells when required, thereby securing a sustained root meristem.
QC communication is a two-way street
Investigation of the cell–cell communication in the stem cell niche using a modified version of the inducible callose deposition system (icals3m) (Vatén et al., 2011) strongly suggests that bilateral symplastic communication between the QC and the columella initials is essential for both QC and distal stem cell maintenance (Liu et al., 2017). Blocking QC symplastic traffic by WOX5 promoter-driven icals3m led to starch accumulation in QC and columella stem cells. In addition, QC cell division and misexpression of identity markers Q0608 (columella), J2341 (columella stem cells), and QC25 (QC) was observed. Interestingly, proWOX5::erGFP expression was not strongly reduced upon plasmodesmatal closure, suggesting that QC function is partially retained. These observations indicate that correct SCN function requires mutual and bilateral exchange of signals between QC and surrounding cells, rather than unilateral signalling from QC to stem cells. Nevertheless, the disorganized SCN that resulted from symplastic traffic disruption barely had an impact on meristem cell number or root growth, further supporting the idea that the SCN acts as a stem cell reservoir to support long-lasting root growth (Liu et al., 2017). These results provide an elegant molecular elaboration on early ablation experiments indicating the essential role of positional cues for fate determination (van den Berg et al., 1995, 1997).
WOX5 as a non-autonomous factor
To test the requirement of WOX5 mobility for columella stem cell maintenance, Pi and collaborators generated a non-mobile version of WOX5 fused to three copies of yellow fluorescent protein (YFP). Expressed from its own promoter, WOX5–3xYFP was unable to prevent the phenotype of starch accumulation in the columella initials when introduced in the wox5-1 mutant background. Despite this stem cell differentiation phenotype, expression of the QC identity marker QC184 was restored in the wox5-1 mutant indicating the WOX5–3xYFP fusion protein was functional. These results indicate that WOX5 mobility is required for columella stem cell maintenance but not for QC fate (Pi et al., 2015). To elucidate the function of WOX5 in stem cell maintenance, transcriptomics, ChIP, and protein–protein interaction experiments were performed. These revealed that WOX5 binds to the promoter region of CYCLING DOF FACTOR 4 (CDF4), where it forms a complex with TOPLESS/TOPLESS-RELATED (TPL/TPR) and HISTONE DEACETYLASE 19 (HDA19) proteins, leading to CDF4 transcriptional repression. CDF4 induces stem cell differentiation, as shown by transgenic lines that accumulate starch in columella stem cells upon ectopic expression of CDF4 either in the QC or in the columella initials. Interestingly, CDF4 exhibits a gradient expression pattern, peaking at the mature columella cells, displaying low levels in the columella initials and no expression in the QC. Based on these observations, an elegant model was proposed in which WOX5 and CDF4 form opposite gradients. In this model, CDF4 promotes the differentiation of columella cells whereas WOX5 autonomously represses CDF4 expression in the QC and non-autonomously in the columella initials, maintaining their stem cell status (Pi et al., 2015).
Recently, results that contradict the requirement of WOX5 mobility for columella stem cell maintenance were presented by Berckmans et al. (2020). In a similar wox5-1 complementation experiment with an immobilized WOX5, but now fused to two GFPs, the QC and columella stem cell defects were restored to WT levels (Berckmans et al., 2020). In their hands, also WOX5 fused to three GFPs was able to rescue the mutant phenotype, albeit partially. It was hypothesized that fusing WOX5 to the larger 3xGFP tag impaired its function, contradicting the previous observation of the fusion to 3xYFP that appeared to be generally functional in rescuing QC fate characteristics (Pi et al., 2015; Berckmans et al., 2020). Thus, it appears that a non-mobile, QC-localized WOX5 is sufficient for columella stem cell maintenance. In light of these data, stem cell maintenance warrants an independent factor from the QC to maintain stemness.
Columella differentiation is actively promoted by the CLAVATA3/ESR-RELATED40 (CLE40) peptide, which is expressed in the differentiating stele and columella cells, through its interaction with ARABIDOPSIS CRINKLY4 (ACR4) and CLAVATA1 (CLV1) receptor kinases. Single mutants cle40, acr4, and clv1 exhibit an extra layer of columella initials, and the acr4 mutant is insensitive to exogenous CLE40 treatment. Both receptors were shown to co-localize to the plasmodesmata, and form homo- and heterodimers (Stahl et al., 2009, 2013). Based on these observations, a ‘gating model’ was hypothesized: CLE40 secreted from differentiated columella cells binds and activates ACR4–CLV1 complexes located at the plasmodesmata, thereby blocking the traffic of ‘stemness’ factors between the QC/columella initials and mature columella cells (Stahl et al., 2013). However, recent experiments with exogenous CLE40 treatment did not appear to change the diffusion rate of fluorescent proteins between QC, columella initials, and mature cells. Also, WOX5–GFP mobility was not affected in cle40, acr4, or clv1 single mutants (Berckmans et al., 2020). These results indicate the CLE40–CLV1–ACR4 signalling module promotes columella differentiation independent of symplastic protein traffic control.
Upon CLE40 treatment, QC and columella initials accumulate starch; WOX5 and QC markers (QC184 and ALG42) are repressed in the QC and their expression relocated into vascular cells (Stahl et al., 2013; Berckmans et al., 2020). Interestingly, this shift of WOX5 expression towards the vasculature as a prelude to QC re-specification is not accompanied by an apical rearrangement of the CDF4 expression domain into the differentiating cells. This indicates that columella cells can also undergo differentiation in the absence of CDF4 (Pi et al., 2015; Berckmans et al., 2020).
The multiple undifferentiated columella cell layers that form upon induced overexpression of WOX5 fused to a glucocorticoid receptor (WOX5–GR) (Sarkar et al., 2007) could be caused by de-differentiation of mature columella cells, by ectopic cell division, or by both. Treating WOX5–GR-induced overexpression plants with differentiation-inducing CLE40 peptide indicated that only the first mature columella layer may undergo de-differentiation (Berckmans et al., 2020). However, reproduction of the WOX5–GR-inducible overexpression experiments with a focus on cell division dynamics suggested that the observed multiple stem cell phenotype is exclusively caused by proliferation of columella stem cells (Savina et al., 2020), which are subsequently maintained in their undifferentiated state.
In summary, contradictory data on the WOX5 mobility requirement for non-autonomous distal meristem maintenance highlighted the requirement of a downstream stemness factor from the QC.
Local auxin is key
For a long time it has been known that auxin is implicated in the control of patterning of the root SCN (Sabatini et al., 1999; Blilou et al., 2005). Exogenous auxin application (or ectopic auxin production) represses WOX5 and induces columella differentiation. This process depends on the transcriptional repressor AUXIN RESISTANT3 (IAA17/AXR3) and on two AUXIN RESPONSE FACTORS, ARF10 and ARF16, which were suggested to confine WOX5 expression to the QC (Ding and Friml, 2010). To address the importance of local auxin biosynthesis and transport for correct SCN function, a double mutant displaying impaired auxin biosynthesis in shoots and roots (wei8;tar2) (Stepanova et al., 2008) was used in grafting experiments (Brumos et al., 2018). By fusing WT shoots to wei8;tar2 mutant roots, it was demonstrated that the shoot-produced auxin that is transported to the root is unable to restore the root SCN function. The characteristic WT auxin distribution gradient, with an auxin maximum in the QC and lower auxin levels in mature columella cells, was not re-established in these chimeric plants, and consequently the root meristem was terminated. The same result was observed when full wei8;tar2 mutant plant shoots were locally treated with exogenous auxin. Even though the shoot treatment induced formation of adventitious and lateral roots, the root apical meristem was not maintained (Brumos et al., 2018). Conversely, the auxin gradient and SCN function of wei8;tar2 mutant roots was restored by local exogenous auxin application or by expression of auxin biosynthesis genes in the root. However, complementation occurred under the condition that the PIN-FORMED auxin efflux carriers were not disturbed. This shows that in roots locally supplied by an auxin source, the auxin transporters have the capacity to create the auxin maximum in the QC. When polar auxin transport was blocked, only QC-specific (WOX5 promoter driven) auxin biosynthesis was able to maintain the root meristem, confirming the importance of an auxin peak in the QC for SCN function (Brumos et al., 2018). These data agree with previous findings that upon ectopic callose-induced closure of QC plasmodesmata, the auxin gradient is lost and starch accumulation is observed in QC and columella initials. Apparently, disrupting QC symplastic traffic impairs local auxin biosynthesis, also evidenced by down-regulation of auxin biosynthetic genes. Again, exogenous auxin treatment or QC-specific promoter driven expression of a bacterial auxin biosynthesis gene rescued the QC and columella stem cell defects (Liu et al., 2017).
Upon induction of ectopic WOX5-GR expression, prior to any morphological change, the TRYPTOPHAN AMINOTRANSFERASE OF ARABIDOPSIS1 (TAA1) gene that is involved in auxin biosynthesis is up-regulated and has its expression domain expanded into the columella (Savina et al., 2020). A similar pattern was observed for the auxin polar transporters PIN1 and PIN4, suggesting that WOX5 ectopic expression impacts both auxin biosynthesis and transport. Mathematical modelling supported the hypothesis that WOX5-induced TAA1-mediated auxin biosynthesis is sufficient to explain the phenotypes observed in the columella. Absence of QC quiescence and the presence of one or two stem cells were predicted for a 50% reduction in TAA1-dependent synthesis in a WT background (Savina et al., 2020). Accordingly, wox5-1 might be an in vivo representation of this predicted phenotype, since it displays QC divisions, and indeed it was shown to have reduced TAA1 marker expression. Finally, the model predicted that a 50% reduction of TAA1-dependent auxin biosynthesis in a WOX5 overexpression line restores the columella phenotype back to WT. This was confirmed by partial restoration using a competitive inhibitor of TAA1-dependent auxin synthesis. Altogether these results reveal locally produced auxin to be a candidate for a non-autonomous signal originating in the QC, downstream of WOX5, to regulate distal meristem functioning (Savina et al., 2020).
SHORT-ROOT, SCARECROW, and JACKDAW: a ground tissue detour
SCR and SHR are two transcription factors from the GRAS family that are well known for their role in regulating the asymmetric division of the ground tissue stem cell daughter that generates cortex and endodermis. Single null mutants for either SCR or SHR (scr, shr) fail to perform the asymmetric cell division of the ground tissue initial daughter, ending up with a single layer of ground tissue, which has cortical characteristics in shr and mixed cortical–endodermal features in scr (Benfey et al., 1993; Scheres et al., 1995; Di Laurenzio et al., 1996; Helariutta et al., 2000). Previous work has shown that SHR is expressed in the stele, and its encoded protein moves one layer outward into the so-called U-domain (composed by QC, ground tissue initials, and endodermis), where it physically interacts with SCR. By forming a complex, SCR was reported to promote SHR nuclear localization, which leads to direct up-regulation of SCR and prevents SHR from moving to outer layers, thus restricting it to the U-domain (Helariutta et al., 2000; Nakajima et al., 2001; Cui et al., 2007). Several zinc-finger proteins of the C2H2 type, belonging to the so-called BIRD family, such as JACKDAW (JKD), NUTCRACKER (NUC), BALD-IBIS (BIB), and MAGPIE (MGP), which are expressed in the ground tissue, also play important roles in this process. For example, the jkd mutant displays ectopic cortical periclinal divisions, a phenotype intensified when JKD’s homolog BIB is knocked down in the jkd background (jkd;bib-i) (Welch et al., 2007; Long et al., 2015). In jkd;bib-i, SCR expression and SHR nuclear retention are compromised, which correlates to spreading of SHR to multiple layers outside its transcription domain. In line with that, it was shown that JKD and BIB can promote SHR nuclear localization, a process that is reinforced by SCR (Long et al., 2015).
The process regulating the formative division to generate cortex and endodermis is a puzzle that also involves RBR, CYCLIN D6;1 (CYCD6;1), CYCLIN-DEPENDENT KINASE B1 (CDKB1), and CDKA1, and additionally auxin (Sozzani et al., 2010; Cruz-Ramírez et al., 2012; Weimer et al., 2012). CYCD6;1 expression is directly and specifically up-regulated by the SHR–SCR complex in cells that undergo asymmetric cell division such as the ground tissue initial daughter. Consistently, null cycd6;1 mutants exhibit delayed cortex–endodermis formative divisions, while ectopic transactivation of CYCD6;1 in ground tissue produces an extra endodermal layer (Sozzani et al., 2010). RBR forms a ternary complex with SHR–SCR repressing some of their targets (i.e. MGP, NUC, and CYCD6;1). Consistently, scr complementation with SCRAxCxA (which has reduced binding to RBR) leads to expansion of the CYCD6;1 expression domain and the formation of an extra QC and ground tissue layer. In turn, CYCD6;1 interacts with CDKB1;1 and CDKB1;2 to phosphorylate and inactivate RBR, preventing RBR from co-repressing SCR targets, thereby forming a bistable circuit. SHR–SCR complex activity together with auxin-induced CYCD6;1 accumulation forms the basis of restricting the ground tissue formative division to the stem cell daughter (Cruz-Ramírez et al., 2012). Another study suggests that CYCD6;1 primarily acts through CDKA;1 to promote the formative divisions, in a dose-dependent manner. Accordingly, intermediate levels of CDKA activity were shown to be sufficient to promote symmetric cell division, whereas high levels of CDKA activity are required to completely inactivate RBR inducing formative divisions. This study also suggests that CDKBs would have a minor role in the formative division process, just backing up CDKA;1 function (Weimer et al., 2012). Dual luciferase reporter assays in Arabidopsis protoplasts have shown that the up-regulation of CYCD6;1 by SCR–SHR is counteracted by JKD and BIB, which matches the observations that the CYCD6;1 domain is expanded in jkd;bib-i roots. These results indicate that JKD and BIB restrict CYCD6;1 expression to ground tissue initials and daughters, preventing its expression in already differentiated endodermal and cortical cells (Long et al., 2015).
The concept of proteins being assembled into different complexes in order to promote specific processes is further supported and extended by Förster resonance energy transfer measured by fluorescence lifetime microscopy (FRET-FLIM) data. FRET-FLIM experiments show that SCR and SHR association is stronger in the ground tissue initial/daughter cells that undergo asymmetric cell division, compared with QC or endodermis. On the other hand, JKD–SCR association is stronger in endodermal cells compared with QC or ground tissue initial/daughter. In addition, a split-luciferase assay in HeLa cells demonstrated that addition of JKD competes with the SCR–SHR interaction, making it weaker, whereas SHR addition makes the SCR–JKD association stronger. Importantly, co-immunoprecipitation (CoIP) experiments in tobacco leaves show that SCR, SHR, and JKD can form a ternary complex, indicating that the FRET results may represent either the formation of distinct heterodimers or the assembly of a ternary complex with closer interaction between two of the partners (Long et al., 2017). Indeed, the crystal structure for a SHR–SCR–JKD complex suggests that, at least in the formation of this ternary complex, JKD only binds to SHR and not directly to SCR (Hirano et al., 2017). These and other results reinforce the idea that differential interactions between SHR, SCR, and JKD triggers different processes: whereas strong SHR–SCR interaction is associated with ground tissue asymmetric cell division, stronger SCR–JKD interaction is associated with endodermis specification.
Wasn’t this about SCN? Yes, QC function also requires SHR, SCR, and JKD
In a no less complex process, SHR, SCR, and JKD are also pivotal for QC specification and maintenance. Also, the heat shock transcription factor gene SCHIZORIZA (SCZ) was shown to genetically interact with SCR and SHR for SCN and ground tissue patterning, although the underlying mechanism remains unclear (Mylona et al., 2002; Pernas et al., 2010; ten Hove et al., 2010). The single mutants shr, scr, scz, and jkd display defective QC specification and maintenance, exemplified by a disorganized stem cell niche, accumulation of starch in the columella initials, and failure to correctly express QC identity markers such as QC25 and QC46. All four mutants have shorter meristems and roots, a phenotype that is less prominent in jkd, while shr and scr are unable to maintain root meristematic activity (Benfey et al., 1993; Scheres et al., 1995; Helariutta et al., 2000; Sabatini, 2003; Pernas et al., 2010; ten Hove et al., 2010; Welch et al., 2007). Whereas SCR expression is reduced in the shr mutant, transactivation of SCR in the QC of shr or scr only rescued the root stem cell niche defect of scr, indicating that both genes are required for correct QC specification and maintenance (Sabatini, 2003). It was also observed that from early heart stage onward, SCR is no longer expressed in jkd mutants, while JKD expression is only reduced in shr and scr mutants after embryogenesis. Taken together, SHR and JKD are required for SCR expression during and after embryogenesis, whereas SHR and SCR are only required for elevated JKD expression in post-embryonic roots. Furthermore, analysis of double mutants showed that jkd;scr displays a stronger phenotype compared with the scr single mutant, but jkd;shr has only slightly shorter roots than shr, suggesting that JKD exerts its function in root stem cell niche mostly through SHR (Welch et al., 2007).
The same FRET-FLIM experiments that showed stronger association of SHR–SCR and SCR–JKD in ground tissue initial/daughter and endodermal cells, respectively, have also shown that SHR and JKD associate more strongly in QC cells. Interestingly, whereas WOX5 expression is mostly absent in shr, scr, and jkd mutants, ectopic WOX5 promoter-driven JKD expression leads to expansion of the WOX5 expression domain towards vasculature. Given that the WOX5 promoter is by default weakly active in vascular initials, a feedforward loop was suggested to amplify WOX5 promoter activity in the vasculature. This effect was observed also in scr and shr backgrounds, indicating that ectopically expressed JKD alone can induce WOX5 expression. The enlargement of the WOX5 domain coincided with expansion of the QC identity markers QC25, QC46, and QC184, indicating that JKD is able to promote QC fate. Concordantly, dual-luciferase reporter experiments in tobacco leaves demonstrated that among all possible combinations, SHR–JKD was the one to more effectively enhance WOX5 promoter activity. Additionally, split-luciferase in HeLa cells indicated that SCR strengthens the association between SHR and JKD. Put together, these results suggest that, whether by forming a SHR–JKD heterodimer or by assembly of a ternary complex with SCR in a supporting role, an enhanced interaction between SHR and JKD is associated with QC specification and maintenance via WOX5 regulation (Long et al., 2017).
PLETHORA proteins maintain the QC in a redundant fashion
Another set of transcriptional factors necessary for QC specification and maintenance are the PLT proteins. Four PLTs (PLT1–4) have partially overlapping expression domains in the form of a gradient, peaking in the root stem cell niche. In addition, PLTs are also partially redundant in function, acting in a dose-dependent manner. Whereas single mutants do not display obvious phenotypes, plt1;plt2 double mutants display a strongly reduced root meristem that undergoes terminal differentiation, and plt1;plt2;plt3 triple mutants completely fail to develop a primary root (Aida et al., 2004; Galinha et al., 2007). Consistently, plt1;plt2 double mutants fail to correctly express the QC identity markers QC25, QC46, and QC184, and accumulate starch granules in the columella initials (Aida et al., 2004). Interestingly, despite the resemblance of plt1:plt2 mutants to scr and shr regarding the stem cell niche defects, SHR and SCR expression is not affected in plt1;plt2 mutants and vice versa. Concordantly, scr;plt1;plt2 and shr;plt1;plt2 triple mutants display earlier root meristem differentiation when compared with shr/scr single- or plt1;plt2 double-mutants (Aida et al., 2004). Collectively, these results suggest that PLTs and SHR/SCR act in parallel pathways converging to specify and maintain the QC and the root stem cell niche (Aida et al., 2004; Galinha et al., 2007; Santuari et al., 2016). PLTs were shown to be induced by auxin and to directly or indirectly regulate the expression of many genes involved in patterning, growth, and differentiation, including genes involved in auxin biosynthesis, response, and transport (Aida et al., 2004; Mähönen et al., 2014; Santuari et al., 2016). Besides the auxin–PLT feedforward cascade, additional PLT-regulated genes feed back on PLTs, thereby creating additional feedforward loops (Santuari et al., 2016). The GROWTH-REGULATING FACTOR–GRF-INTERACTING FACTOR (GRF–GIF) repressive module, consisting of interacting transcriptional repressors and co-regulators, is a candidate to control PLT levels through negative feedback (Rodriguez et al., 2015; Santuari et al., 2016; Ercoli et al., 2018). PLTs promote GRF and GIF expression, whereas GIF co-regulators are implicated in the direct repression of PLTs (Santuari et al., 2016; Ercoli et al., 2018). miR396 post-transcriptionally represses GRFs, whereas PLTs maintain high levels of miR396 within the SCN. Therefore miR396 excludes the GRFs from the SCN, restricting their activity to the transit-amplifying zone (Rodriguez et al., 2015; Ercoli et al., 2018). This interplay between PLT, GIF–GRF, and miR396 collaborates to establish the PLT gradients, consisting of reduced levels in the transit-amplifying zone and maxima at the SCN.
Parallel pathways converge in the QC
A recent study has described for the first time how PLT and SCR parallel pathways may converge into QC specification (Shimotohno et al., 2018). By means of yeast-two-hybrid (Y2H) and bimolecular fluorescence complementation (BiFC) in protoplasts, PLT1, PLT3, and SCR were shown to interact with two TEOSINTE-BRANCHED/CYCLOIDEA/PCNA proteins (TCP20 and TCP21). To map the interaction domains, Y2H and BiFC experiments were subsequently performed using truncated versions of TCP20 and TCP21, showing that PLTs and SCR bind to different regions of the TCPs. Additionally, CoIP in protoplasts transiently expressing tagged PLT3, TCP20, and SCR combinations showed that PLT3 and SCR only interact in the presence of TCP20. These results suggest that SCR, PLTs, and TCPs form a complex in planta. To understand the individual and combined dose effect of these genes on root development, several double, triple, and quadruple mutants were obtained, varying the allelic contribution of these four genes. For example, a double mutant homozygous for the scr allele and heterozygous for tcp20 (scr;tcp20+/−) displays an intermediary root length compared with a scr single mutant (longer root) and the double homozygous scr;tcp20 mutant (shorter root). A quadruple homozygous plt1;plt3;tcp20;scr3 mutant completely abolishes primary root growth (Shimotohno et al., 2018). The results are consistent with these genes acting in a dose-dependent manner for root length.
As expected, these mutants also present morphological defects in the root stem cell niche. The quadruple homozygous mutant completely loses the stereotypical stem cell niche pattern, whereas triple homozygous (plt1;tcp20;scr and plt3;tcp20;scr) mutants exhibit extra QC divisions and accumulate starch in columella initials earlier than observed in the scr single mutant. Morphological changes are already observed early during embryogenesis: quadruple homozygous mutants exhibit unusual divisions in the hypophyseal cell at the dermatogen stage, corresponding to the time and position of their expression. Concordantly, the WOX5 promoter is less active in a double tcp;scr compared with scr single mutant, and less expressed in quadruple heterozygous mutants compared with WT (Shimotohno et al., 2018). These results again reveal a gene dose output response, consistent with protein complex formation.
ChIP-seq data reported the occurrence of PLT2 binding at the WOX5 promoter (Santuari et al., 2016), suggesting that PLT1 and PLT3 may be mediating the output of the complex through direct regulation of WOX5 target expression. Supporting this hypothesis, it was demonstrated that PLT1 or PLT3 fused to GR, upon dexamethasone induction and in the presence of the protein synthesis inhibitor cycloheximide, were able to directly induce WOX5 expression. Mutating the PLT binding sites in the WOX5 promoter abolished its expression, indicating the relevance of the identified binding sites. Dual-luciferase assays in protoplasts showed that, although induced by PLT3 alone, WOX5 promoter activity was significantly enhanced when PLT3, TCP20, and SCR were combined. In addition, compared with WT promoter induction by PLT3–TCP20–SCR, the induction of the promoter harbouring mutated PLT-binding motifs was markedly reduced. Altogether, these data strongly support the idea that PLT proteins together with SCR form a TCP-containing complex in order to specify the QC and establish the root stem cell niche (Shimotohno et al., 2018).
Recently, Zhai and collaborators presented data supporting WOX5 regulation by SCR in a PLT1/2-independent manner, thereby elaborating on earlier data that indicated that WOX5 expression depends mainly on SCR/SHR, with PLT1/PLT2 playing only a minor role in confining WOX5 expression to the QC (Sarkar et al., 2007; Zhai et al., 2020). Previous data showed reduced fluorescence of SHR and SCR promoter and protein fusions in a seuss (seu) mutant background (Gong et al., 2016). The nuclear localized SEU protein is a transcriptional co-regulator that is broadly expressed in embryos and in post-embryonic root meristem. Indeed, the scr;seu and shr;seu double mutants do not display enhanced phenotypes compared with scr and shr single mutants. However, a seu;plt1;plt2 triple mutant presents shorter roots compared with plt1;plt2. These results suggest that SCR/SHR and SEU act in the same pathway in parallel to the PLT pathway (Gong et al., 2016).
WOX5 expression is both delayed and significantly reduced in a seu null mutant. Accordingly, seu displays phenotypes similar to the wox5 mutant, harbouring ectopic divisions in the QC, starch accumulation in columella initials, and strongly reduced expression of the QC184 identity marker. Also, seu;wox5 double mutants did not present enhanced defects compared with wox5, indicating that both genes act in the same pathway (Zhai et al., 2020). Interestingly, Y2H showed that SEU interacts with SCR but not SHR and the SCR–SEU interaction was confirmed by CoIP in Arabidopsis roots. ChIP–qPCR experiments showed enrichment of SCR and SEU binding at ~1100 bp upstream of the WOX5 transcription start site, with dramatically reduced binding for SEU in a scr mutant background. Also, dual-luciferase assays in tobacco leaves showed that WOX5 promoter activity is ~15% up-regulated by the SCR–SEU combination compared with SCR alone. These results indicate that SCR recruits and physically interacts with SEU to jointly bind to the WOX5 promoter (Zhai et al., 2020).
As a third player in this complex, the histone methyltransferase protein SET DOMAIN GROUP 4 (SDG4) was shown to interact with SEU by Y2H and split-luciferase in tobacco leaves. The null mutant sdg4 (originally ashr3-1) displays defects resembling those observed in seu (Kumpf et al., 2014). Concordantly, the SDG4–GFP fusion protein was observed during embryo development from dermatogen stage onwards in the nucleus of the hypophysis, whereas post-embryonically it was broadly expressed in the root meristem. SDG4 and SCR bind to different regions of SEU as shown by Y2H, while CoIP in tobacco leaves and Arabidopsis roots demonstrated that SCR and SDG4 can associate to SEU at the same time, and that SEU is required for SCR–SDG4 interaction. ChIP–qPCR experiments for SDG4–GFP showed enrichment of the same region in the WOX5 promoter as is observed to be bound by GFP–SCR and SEU–GFP. In addition, sdg4;seu;wox5 and sdg4;seu;scr triple mutants do not enhance QC defects observed in either single wox5 or scr mutant, respectively. Furthermore, WOX5–GFP overexpression is able to rescue the QC defects in a sdg4;seu;scr triple mutant. Finally, both sdg4 and seu mutants display reduced levels of H3K4me3 marks (associated with transcriptional activation) at the WOX5 promoter. Altogether, these results support a SCR–SEU–SDG4 complex that triggers H3K4 trimethylation of the WOX5 promoter, in order to promote its expression and consequently QC specification (Zhai et al., 2020).
Wrap up: the attractive SCARECROW
The research discussed above indicates the existence of at least three ternary complexes regulating QC specification and WOX5 expression. All of these include the SCR transcription factor interacting with two partners (PLT/TCP, SEU/SDG4 or SHR/JKD) (Long et al., 2017; Shimotohno et al., 2018; Zhai et al., 2020). The corresponding genes start to be expressed early during embryogenesis, around the time of or prior to WOX5 accumulation at the globular stage (Helariutta et al., 2000; Welch et al., 2007; Shimotohno et al., 2018; Zhai et al., 2020). Realizing that all these proteins display overlapping expression domains in the hypophysis/QC, it is intriguing to think how those complexes behave in this same cellular environment. Do all complexes co-exist or are there factors regulating the conditional assembly of one complex at the expense of the others? Considering that SCR is a key factor for all complexes, at least some competition could be expected. The reported protein–protein interaction experiments so far only addressed which SCR domain is used for the interaction with SHR, but not for the interaction with JKD, TCP20/21, and SEU (Hirano et al., 2017; Long et al., 2017; Shimotohno et al., 2018; Zhai et al., 2020). The evidence presented in literature only addresses and supports the assembly of individual complexes. However, we can speculate that if SCR uses different domains for its interactions, one unique SCR might associate to more than one complex at the same time. In line with this, it was previously demonstrated that SCR binds to RBR through a domain that is dispensable for its interaction with SHR (Cruz-Ramírez et al., 2012). Therefore, we can hypothesize on at least two possible scenarios. Considering that SCR appears to be crucial for recruitment and assembly of a SEU complex at ~1100 bp as well as for the TCP complex ~300 bp upstream of the WOX5 transcription start site (Shimotohno et al., 2018; Zhai et al., 2020), we can envision that SCR physically links both PLT–TCP and SDG4–SEU sub-complexes, and maybe even the SHR–JKD sub-complex, and mediates their interaction in the formation of a single multi-protein complex (Shimotohno et al., 2018; Zhai et al., 2020). In this way, the SDG4–SEU sub-complex would promote H3K4me3 methylation, making the WOX5 promoter region more accessible to other factors, including the PLT–TCP sub-complex, to effectively induce WOX5 expression. On the other hand, in the case of SCR only being assembled into separate complexes, those different complexes may perform specific roles during different subsequent phases of QC specification and WOX5 expression, thereby recycling SCR between the different complexes. For example, the SDG4–SEU–SCR complex could primarily establish chromatin activation marks at WOX5 promoter, and this would relax the chromatin allowing subsequent complexes such as JKD–SHR–SCR and PLT–TCP–SCR to access this region in order to promote and maintain WOX5 expression during and after embryogenesis.
The question is whether SCR itself can physically accommodate binding to all these proteins or whether there is a common additional scaffold protein(s) to accommodate all sub-complexes, or perhaps there is a hybrid version of these proposed mechanisms. In that respect it is also interesting to examine the role of the Mediator complex for which the subunit MED31 has been observed to compete with SHR for binding to SCR, in order to regulate downstream CYCD6;1 transcription in the ground tissue (Zhang et al., 2018). The large Mediator complex couples the enhancer-bound transcription factors to RNA polymerase II-dependent gene transcription, which includes epigenetic control of transcription (Buendía-Monreal and Gillmor, 2016). One way of testing these hypotheses is to determine whether SCR, while being part of one complex, can bind to proteins of the other complexes. A first candidate would be SHR, since the interaction between SCR and SHR was reported to be stronger than binding of SCR to JKD, RBR, or MED31 (Cruz-Ramírez et al., 2012; Long et al., 2017; Zhang et al., 2018). In addition, since SHR can enhance the association between JKD and SCR, SHR may act in stabilizing the interaction between SCR and other complexes (Long et al., 2017). Whereas these hypotheses are still purely speculative, future experiments will tell whether SCR can act as a core protein being assembled into multiple complexes simultaneously, or if SCR is recycled between complexes to exert its function in the QC and SCN regulation. Since the first ablation studies that showed the importance of organizer signalling for stem cell maintenance, detailed studies have provided insights into genes and networks acting in the SCN. Nevertheless, more is still to be discovered on the mechanisms acting in the short-range control of SCN function and stem cell differentiation.
Acknowledgements
We apologize to researchers whose work could not be cited here due to space limitations. We thank Anneke Horstman, Ikram Blilou, and Viola Willemsen for critical reading and comments. Research on this topic was supported by CNPq, National Council for Scientific and Technological Development—Brazil (203730/2017-8).
References
- Aida M, Beis D, Heidstra R, Willemsen V, Blilou I, Galinha C, Nussaume L, Noh YS, Amasino R, Scheres B. 2004. The PLETHORA genes mediate patterning of the Arabidopsis root stem cell niche. Cell 119, 109–120. [DOI] [PubMed] [Google Scholar]
- Benfey PN, Linstead PJ, Roberts K, Schiefelbein JW, Hauser MT, Aeschbacher RA. 1993. Root development in Arabidopsis: four mutants with dramatically altered root morphogenesis. Development 119, 57–70. [DOI] [PubMed] [Google Scholar]
- Berckmans B, Kirschner G, Gerlitz N, Stadler R, Simon R. 2020. CLE40 signaling regulates root stem cell fate. Plant Physiology 182, 1776–1792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blilou I, Xu J, Wildwater M, Willemsen V, Paponov I, Friml J, Heidstra R, Aida M, Palme K, Scheres B. 2005. The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature 433, 39–44. [DOI] [PubMed] [Google Scholar]
- Boniotti MB, Gutierrez C. 2001. A cell-cycle-regulated kinase activity phosphorylates plant retinoblastoma protein and contains, in Arabidopsis, a CDKA/cyclin D complex. The Plant Journal 28, 341–350. [DOI] [PubMed] [Google Scholar]
- Brumos J, Robles LM, Yun J, Vu TC, Jackson S, Alonso JM, Stepanova AN. 2018. Local auxin biosynthesis is a key regulator of plant development. Developmental Cell 47, 306–318.e5. [DOI] [PubMed] [Google Scholar]
- Buendía-Monreal M, Gillmor CS. 2016. Mediator: A key regulator of plant development. Developmental Biology 419, 7–18. [DOI] [PubMed] [Google Scholar]
- Cruz-Ramírez A, Díaz-Triviño S, Blilou I, et al. 2012. A bistable circuit involving SCARECROW-RETINOBLASTOMA integrates cues to inform asymmetric stem cell division. Cell 150, 1002–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz-Ramírez A, Díaz-Triviño S, Wachsman G, et al. 2013. A SCARECROW-RETINOBLASTOMA protein network controls protective quiescence in the Arabidopsis root stem cell organizer. PLoS Biology 11, e1001724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui H, Levesque MP, Vernoux T, Jung JW, Paquette AJ, Gallagher KL, Wang JY, Blilou I, Scheres B, Benfey PN. 2007. An evolutionarily conserved mechanism delimiting SHR movement defines a single layer of endodermis in plants. Science 316, 421–425. [DOI] [PubMed] [Google Scholar]
- Di Laurenzio L, Wysocka-Diller J, Malamy JE, Pysh L, Helariutta Y, Freshour G, Hahn MG, Feldmann KA, Benfey PN. 1996. The SCARECROW gene regulates an asymmetric cell division that is essential for generating the radial organization of the Arabidopsis root. Cell 86, 423–433. [DOI] [PubMed] [Google Scholar]
- Ding Z, Friml J. 2010. Auxin regulates distal stem cell differentiation in Arabidopsis roots. Proceedings of the National Academy of Sciences, USA 107, 12046–12051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolan L, Janmaat K, Willemsen V, Linstead P, Poethig S, Roberts K, Scheres B. 1993. Cellular organisation of the Arabidopsis thaliana root. Development 119, 71–84. [DOI] [PubMed] [Google Scholar]
- Ercoli MF, Ferela A, Debernardi JM, Perrone AP, Rodriguez RE, Palatnik JF. 2018. GIF transcriptional coregulators control root meristem homeostasis. The Plant Cell 30, 347–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forzani C, Aichinger E, Sornay E, Willemsen V, Laux T, Dewitte W, Murray JA. 2014. WOX5 suppresses CYCLIN D activity to establish quiescence at the center of the root stem cell niche. Current Biology 24, 1939–1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galinha C, Hofhuis H, Luijten M, Willemsen V, Blilou I, Heidstra R, Scheres B. 2007. PLETHORA proteins as dose-dependent master regulators of Arabidopsis root development. Nature 449, 1053–1057. [DOI] [PubMed] [Google Scholar]
- Gong X, Flores-Vergara MA, Hong JH, Chu H, Lim J, Franks RG, Liu Z, Xu J. 2016. SEUSS integrates gibberellin signaling with transcriptional inputs from the SHR-SCR-SCL3 module to regulate middle cortex formation in the Arabidopsis root. Plant Physiology 170, 1675–1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haecker A, Gross-Hardt R, Geiges B, Sarkar A, Breuninger H, Herrmann M, Laux T. 2004. Expression dynamics of WOX genes mark cell fate decisions during early embryonic patterning in Arabidopsis thaliana. Development 131, 657–668. [DOI] [PubMed] [Google Scholar]
- Helariutta Y, Fukaki H, Wysocka-Diller J, Nakajima K, Jung J, Sena G, Hauser MT, Benfey PN. 2000. The SHORT-ROOT gene controls radial patterning of the Arabidopsis root through radial signaling. Cell 101, 555–567. [DOI] [PubMed] [Google Scholar]
- Heyman J, Cools T, Vandenbussche F, et al. 2013. ERF115 controls root quiescent center cell division and stem cell replenishment. Science 342, 860–863. [DOI] [PubMed] [Google Scholar]
- Hirano Y, Nakagawa M, Suyama T, Murase K, Shirakawa M, Takayama S, Sun TP, Hakoshima T. 2017. Structure of the SHR-SCR heterodimer bound to the BIRD/IDD transcriptional factor JKD. Nature Plants 3, 17010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong X, Tian H, Yu Q, et al. 2018. PHB3 maintains root stem cell niche identity through ROS-responsive AP2/ERF transcription factors in Arabidopsis. Cell Reports 22, 1350–1363. [DOI] [PubMed] [Google Scholar]
- Kumpf R, Thorstensen T, Rahman MA, et al. 2014. The ASH1-RELATED3 SET-domain protein controls cell division competence of the meristem and the quiescent center of the Arabidopsis primary root. Plant Physiology 166, 632–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Xu M, Liang N, Zheng Y, Yu Q, Wu S. 2017. Symplastic communication spatially directs local auxin biosynthesis to maintain root stem cell niche in Arabidopsis. Proceedings of the National Academy of Sciences, USA 114, 4005–4010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long Y, Smet W, Cruz-Ramírez A, et al. 2015. Arabidopsis BIRD zinc finger proteins jointly stabilize tissue boundaries by confining the cell fate regulator SHORT-ROOT and contributing to fate specification. The Plant Cell 27, 1185–1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long Y, Stahl Y, Weidtkamp-Peters S, et al. 2017. In vivo FRET-FLIM reveals cell-type-specific protein interactions in Arabidopsis roots. Nature 548, 97–102. [DOI] [PubMed] [Google Scholar]
- Mähönen AP, Ten Tusscher K, Siligato R, Smetana O, Díaz-Triviño S, Salojärvi J, Wachsman G, Prasad K, Heidstra R, Scheres B. 2014. PLETHORA gradient formation mechanism separates auxin responses. Nature 515, 125–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mylona P, Linstead P, Martienssen R, Dolan L. 2002. SCHIZORIZA controls an asymmetric cell division and restricts epidermal identity in the Arabidopsis root. Development 129, 4327–4334. [DOI] [PubMed] [Google Scholar]
- Nakajima K, Sena G, Nawy T, Benfey PN. 2001. Intercellular movement of the putative transcription factor SHR in root patterning. Nature 413, 307–311. [DOI] [PubMed] [Google Scholar]
- Ortega-Martínez O, Pernas M, Carol RJ, Dolan L. 2007. Ethylene modulates stem cell division in the Arabidopsis thaliana root. Science 317, 507–510. [DOI] [PubMed] [Google Scholar]
- Pernas M, Ryan E, Dolan L. 2010. SCHIZORIZA controls tissue system complexity in plants. Current Biology 20, 818–823. [DOI] [PubMed] [Google Scholar]
- Pi L, Aichinger E, van der Graaff E, Llavata-Peris CI, Weijers D, Hennig L, Groot E, Laux T. 2015. Organizer-derived WOX5 signal maintains root columella stem cells through chromatin-mediated repression of CDF4 expression. Developmental Cell 33, 576–588. [DOI] [PubMed] [Google Scholar]
- Rodriguez RE, Ercoli MF, Debernardi JM, Breakfield NW, Mecchia MA, Sabatini M, Cools T, De Veylder L, Benfey PN, Palatnik JF. 2015. MicroRNA miR396 regulates the switch between stem cells and transit-amplifying cells in Arabidopsis roots. The Plant Cell 27, 3354–3366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabatini S, Beis D, Wolkenfelt H, et al. 1999. An auxin-dependent distal organizer of pattern and polarity in the Arabidopsis root. Cell 99, 463–472. [DOI] [PubMed] [Google Scholar]
- Sabatini S, Heidstra R, Wildwater M, Scheres B. 2003. SCARECROW is involved in positioning the stem cell niche in the Arabidopsis root meristem. Genes & Development 17, 354–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santuari L, Sanchez-Perez GF, Luijten M, et al. 2016. The PLETHORA gene regulatory network guides growth and cell differentiation in Arabidopsis roots. The Plant Cell 28, 2937–2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar AK, Luijten M, Miyashima S, Lenhard M, Hashimoto T, Nakajima K, Scheres B, Heidstra R, Laux T. 2007. Conserved factors regulate signalling in Arabidopsis thaliana shoot and root stem cell organizers. Nature 446, 811–814. [DOI] [PubMed] [Google Scholar]
- Savina MS, Pasternak T, Omelyanchuk NA, Novikova DD, Palme K, Mironova VV, Lavrekha VV. 2020. Cell dynamics in WOX5-overexpressing root tips: the impact of local auxin biosynthesis. Frontiers in Plant Science 11, 560169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres B, Laurenzio LD, Willemsen V, Hauser MT, Janmaat K, Weisbeek P, Benfey PN. 1995. Mutations affecting the radial organisation of the Arabidopsis root display specific defects throughout the embryonic axis. Development 121, 53–62. [Google Scholar]
- Shimotohno A, Heidstra R, Blilou I, Scheres B. 2018. Root stem cell niche organizer specification by molecular convergence of PLETHORA and SCARECROW transcription factor modules. Genes & Development 32, 1085–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sozzani R, Cui H, Moreno-Risueno MA, Busch W, Van Norman JM, Vernoux T, Brady SM, Dewitte W, Murray JA, Benfey PN. 2010. Spatiotemporal regulation of cell-cycle genes by SHORTROOT links patterning and growth. Nature 466, 128–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl Y, Grabowski S, Bleckmann A, et al. 2013. Moderation of Arabidopsis root stemness by CLAVATA1 and ARABIDOPSIS CRINKLY4 receptor kinase complexes. Current Biology 23, 362–371. [DOI] [PubMed] [Google Scholar]
- Stahl Y, Wink RH, Ingram GC, Simon R. 2009. A signaling module controlling the stem cell niche in Arabidopsis root meristems. Current Biology 19, 909–914. [DOI] [PubMed] [Google Scholar]
- Stepanova AN, Robertson-Hoyt J, Yun J, Benavente LM, Xie DY, Dolezal K, Schlereth A, Jürgens G, Alonso JM. 2008. TAA1-mediated auxin biosynthesis is essential for hormone crosstalk and plant development. Cell 133, 177–191. [DOI] [PubMed] [Google Scholar]
- ten Hove CA, Willemsen V, de Vries WJ, van Dijken A, Scheres B, Heidstra R. 2010. SCHIZORIZA encodes a nuclear factor regulating asymmetry of stem cell divisions in the Arabidopsis root. Current Biology 20, 452–457. [DOI] [PubMed] [Google Scholar]
- van den Berg C, Willemsen V, Hage W, Weisbeek P, Scheres B. 1995. Cell fate in the Arabidopsis root meristem determined by directional signalling. Nature 378, 62–65. [DOI] [PubMed] [Google Scholar]
- van den Berg C, Willemsen V, Hendriks G, Weisbeek P, Scheres B. 1997. Short-range control of cell differentiation in the Arabidopsis root meristem. Nature 390, 287–289. [DOI] [PubMed] [Google Scholar]
- Vatén A, Dettmer J, Wu S, et al. 2011. Callose biosynthesis regulates symplastic trafficking during root development. Developmental Cell 21, 1144–1155. [DOI] [PubMed] [Google Scholar]
- Wachsman G, Heidstra R, Scheres B. 2011. Distinct cell-autonomous functions of RETINOBLASTOMA-RELATED in Arabidopsis stem cells revealed by the Brother of Brainbow clonal analysis system. The Plant Cell 23, 2581–2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weimer AK, Nowack MK, Bouyer D, Zhao X, Harashima H, Naseer S, De Winter F, Dissmeyer N, Geldner N, Schnittger A. 2012. Retinoblastoma related1 regulates asymmetric cell divisions in Arabidopsis. The Plant Cell 24, 4083–4095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch D, Hassan H, Blilou I, Immink R, Heidstra R, Scheres B. 2007. Arabidopsis JACKDAW and MAGPIE zinc finger proteins delimit asymmetric cell division and stabilize tissue boundaries by restricting SHORT-ROOT action. Genes & Development 21, 2196–2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wildwater M, Campilho A, Perez-Perez JM, Heidstra R, Blilou I, Korthout H, Chatterjee J, Mariconti L, Gruissem W, Scheres B. 2005. The RETINOBLASTOMA-RELATED gene regulates stem cell maintenance in Arabidopsis roots. Cell 123, 1337–1349. [DOI] [PubMed] [Google Scholar]
- Zhai H, Zhang X, You Y, Lin L, Zhou W, Li C. 2020. SEUSS integrates transcriptional and epigenetic control of root stem cell organizer specification. The EMBO Journal 39, e105047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Han W, De Smet I, Talboys P, Loya R, Hassan A, Rong H, Jürgens G, Paul Knox J, Wang MH. 2010. ABA promotes quiescence of the quiescent centre and suppresses stem cell differentiation in the Arabidopsis primary root meristem. The Plant Journal 64, 764–774. [DOI] [PubMed] [Google Scholar]
- Zhang X, Zhou W, Chen Q, Fang M, Zheng S, Scheres B, Li C. 2018. Mediator subunit MED31 is required for radial patterning of Arabidopsis roots. Proceedings of the National Academy of Sciences, USA 115, E5624–E5633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W, Lozano-Torres JL, Blilou I, Zhang X, Zhai Q, Smant G, Li C, Scheres B. 2019. A jasmonate signaling network activates root stem cells and promotes regeneration. Cell 177, 942–956.e14. [DOI] [PubMed] [Google Scholar]