Abstract
The two yeast pheromone receptors, the a and α-factor receptors, share many functional similarities: both G protein-coupled receptors couple to the same downstream signal transduction pathway, and both receptors undergo feedback regulation involving increased phosphorylation on their C-terminal domains in response to ligand challenge. The present work, which focuses on the signaling mechanism controlling this feedback phosphorylation, indicates one striking difference. While the α-factor-induced phosphorylation of the α-factor receptor does not require activation of the downstream G protein-directed signaling pathway (B. Zanolari, S. Raths, B. Singer-Kruger, and H. Riezman, Cell 71:755–763, 1992), the a-factor-induced phosphorylation of the a-factor receptor (Ste3p) clearly does. Induced Ste3p phosphorylation was blocked in cells with disruptions of various components of the pheromone response pathway, indicating a requirement of pathway components extending from the G protein down through the mitogen-activated protein kinase (MAPK). Furthermore, Ste3p phosphorylation can be induced in the absence of the a-factor ligand when the signaling pathway is artificially activated, indicating that the liganded receptor is not required as a substrate for induced phosphorylation. While the activation of signaling is critical for the feedback phosphorylation of Ste3p, pheromone-induced gene transcription, one of the major outcomes of pheromone signaling, appears not to be required. This conclusion is indicated by three results. First, ste12Δ cells differ from cells with disruptions of the upstream signaling elements (e.g., ste4Δ, ste20Δ, ste5Δ, ste11Δ, ste7Δ, or fus3Δ kss1Δ cells) in that they clearly retain some capacity for inducing Ste3p phosphorylation. Second, while activated alleles of STE11 and STE12 induce a strong transcriptional response, they fail to induce a-factor receptor phosphorylation. Third, blocking of new pheromone-induced protein synthesis with cycloheximide fails to block phosphorylation. These findings are discussed within the context of a recently proposed model for pheromone signaling (P. M. Pryciak and F. A. Huntress, Genes Dev. 12:2684–2697, 1998): a key step of this model is the activation of the MAPK Fus3p through the Gβγ-dependent relocalization of the Ste5p-MAPK cascade to the plasma membrane. Ste3p phosphorylation may involve activated MAPK Fus3p feeding back upon plasma membrane targets.
Phosphorylation of the G protein-coupled receptors (GPCRs) family generally serves to negatively regulate receptor activity, i.e., functioning in desensitization. For the β2-adrenergic receptor (β2AR), a well-studied member of the GPCR family (30, 36), desensitization is initiated through Ser-Thr phosphorylation of the cytoplasmic, C-terminal regulatory tail domain (CTD) of the receptor by the dedicated kinase, the β2AR kinase (βARK). The binding of β-arrestin protein to the phosphorylated CTD then serves to attenuate receptor responsiveness both by uncoupling the liganded receptor from the G protein and by directing the binding of the liganded receptor into clathrin-coated pits, allowing it to be sequestered by endocytosis to the inside of the cell. This feedback control is regulated through the regulation of βARK. βARK is activated through interactions with the liganded receptor, through interactions with plasma membrane lipids, and through a binding interaction with the βγ subunit of the αβγ heterotrimeric G protein. Free βγ subunit, released from the heterotrimer through the actions of the liganded receptor, remains plasma membrane associated due to C-terminal prenylation of the γ subunit and thus serves as a plasma membrane binding site for cytosolic βARK. In addition to this βARK-mediated, so-called homologous desensitization, β2AR is also subject to heterologous desensitization, i.e., phosphorylation by protein kinase C and protein kinase A (PKA), which may be activated through the actions of a variety of different signaling pathways. PKA also mediates direct feedback control of β2AR; as adenylate cyclase is the primary downstream β2AR effector, agonistic stimulation of β2AR elevates cytoplasmic cyclic AMP levels and serves to activate PKA. β2AR provides a well-characterized paradigm for GPCR desensitization. The GPCR family, however, is huge and evolutionarily diverse, encompassing hundreds of different receptors in organisms throughout the eukaryotic kingdom. The few cases where the desensitization mechanism has been analyzed have revealed both a diversity of mechanisms and a reiteration of common themes (36). While ligand-stimulated CTD phosphorylation generally plays a central role, the molecular mechanisms that regulate this phosphorylation often stray from the β2AR paradigm. The work presented herein focuses on the mechanism underlying the ligand-induced phosphorylation of a yeast GPCR.
In the yeast Saccharomyces cerevisiae, two GPCRs function in the sexual conjugation of the two haploid mating types, a and α. Identifying and locating a potential mate involves tracking the signature pheromone of the mating partner to its source. α cells serve as the source for the secreted peptide pheromone α-factor, while a cells secrete the farnesylated peptide a-factor. The two GPCRs serve to detect these pheromones. The α-factor receptor (Ste2p), localized to the surface of a cells, detects α-factor and thereby its α cell mate, while the a-factor receptor (Ste3p), located on α cells, detects a-factor from nearby a cells. The ensuing cell-cell contact and cell fusion depend on a chemotropic response wherein new cell growth is polarized into a tapered mating projection directed toward the pheromone source. The pheromone receptors, through their detection of the external pheromone gradient, play a central role in this chemotropic response. Both receptors have relatively large CTDs which, although dispensable for the gross functioning of the receptor (ligand binding and G protein coupling), are required for receptor regulation and are required for the production of a normal mating projection (25) (A. F. Roth and N. G. Davis, unpublished results). The regulatory functions controlled by the pheromone receptor CTDs, namely, desensitization and endocytosis (2, 8, 25, 40, 42), likely help to shape this chemotropic response.
The elements which comprise the pheromone signaling pathway are now quite well understood (29, 47). Liganded receptors couple to a typical heterotrimeric G protein, which in turn activates a downstream mitogen-activated protein kinase (MAPK) cascade. Elements of the MAPK cascade, the MAPK kinase kinase Ste11p, the MAPK kinase Ste7p, and the MAPK Fus3p, are coordinated upon the scaffolding protein Ste5p. Propagation of the signal along the pathway involves a scheme of sequential activation wherein each kinase is activated through phosphorylation by the kinase immediately upstream. Two of the main outcomes of signaling, G1 cell cycle arrest and the transcriptional induction of a set of pheromone-responsive genes, are triggered by Fus3p-mediated activation of Far1p, the effector of cell cycle arrest, and Ste12p, the transcriptional activator for pheromone-responsive genes.
For the yeast G protein-coupled signaling pathway, the free βγ subunit of the G protein, not the Gα subunit, initiates downstream signaling. Reminiscent of βARK activation (described above), recent work with yeast indicates that a key signaling step may be the Gβγ-directed translocation of the Ste5p-MAPK complex to the plasma membrane (37). Once localized to the plasma membrane, the Ste5p-MAPK complex may be activated through an interaction with other plasma membrane-localized signaling components. Relevant in this regard is the PAK kinase homologue Ste20p, as well as both the Rho-like GTPase Cdc42p and its guanine nucleotide exchange factor Cdc24p.
Although the two pheromone receptors, Ste2p and Ste3p, have no obvious sequence homology, both activate the same downstream signal transduction pathway (1). Furthermore, regulation of the two receptors appears, superficially at least, to be similar. Both are subject to CTD phosphorylation: both show a constitutive level of CTD phosphorylation which increases upon exposure of the cells to the appropriate pheromone ligand (8, 40). Similarly, both receptors are also subject to two modes of ubiquitin-directed endocytosis (16, 43)—a constitutive, ligand-independent mode as well as a ligand-dependent uptake mode (8, 23, 45).
As with β2AR, multiple Ser-Thr residues within the pheromone receptor CTDs apparently serve as phosphoryl acceptors. For Ste2p, phosphorylation occurs at at least 10 distinct CTD sites (40, 56). As with β2AR, pheromone receptor CTD phosphorylation likely functions in desensitization: receptors with deletions of portions of the CTD or having mutated CTD serines manifest a hypersensitive response to pheromone challenge (2, 3, 25, 40). Details of this process, however, may stray from the β2AR paradigm, as clear homologues either for arrestin or for a βARK-like kinase are notably absent from the completed yeast genomic database.
For Ste2p, α-factor-induced phosphorylation does not require the instigation of G protein-mediated signal transduction from the liganded receptor: ligand-induced phosphorylation of Ste2p proceeds normally in cells lacking the heterotrimeric G protein (56). We find in the present work that quite a different story applies to the ligand-induced phosphorylation of the a-factor receptor. This phosphorylation does require the G protein heterotrimer. Indeed, not only is the G protein required but also required are many of the downstream signal transduction components. Based upon these results, we have proposed a model for the regulation of Ste3p phosphorylation wherein the key step is the localization of activated MAPK to the plasma membrane.
MATERIALS AND METHODS
Plasmids.
pND733 is a pRS316-based URA3/CEN/ARS plasmid (46) carrying a GAL1p-STE2-HA construct. Ste2p expressed from this construct has a single hemagglutinin (HA) epitope tag fused to its C terminus following Thr425 and replacing the Ste2p C-terminal six amino acid residues. Three Ycp50-based URA3/CEN/ARS plasmids carrying either wild-type STE12 (p650) (13) or the equivalent plasmid with one of two in-frame deletions within the STE12 coding sequence were used. The ste12Δ253-335 allele removes STE12 residues 253 through 335, and the ste12Δ255-354 allele removes residues 255 through 354 (24). pND882 carries the GAL1p-STE5-CTM construct from pGS5-CTM (37) on pRS316. The Ste5-CTM fusion has the single C-terminal transmembrane domain (CTM) of Snc2p fused to the Ste5p C terminus. pC1-H6 is a URA3/CEN/ARS GAL1p-STE12 isolate from a library of GAL1-driven yeast genomic fragments (P. Pryciak, personal communication). pRD-STE11-H3 is a URA3/CEN/ARS GAL1p-GST-STE11ΔN plasmid (32).
Strains.
Table 1 shows the strains used in this study. The GAL1p-STE3(M271I, M304I) allele was constructed by two sequential oligonucleotide site-directed mutageneses (26) of the GAL1p-STE3 URA3–integrating plasmid pSL1904. pSL1904 is identical to pSL1839 (44) except that the 760-bp GAL1,10 promoter fragment of pSL552 (1) substitutes for sequences 417 to 110 bp upstream of the STE3 open reading frame. To construct NDY867, the GAL1p-STE3(M271I, M304I) allele was integrated at the STE3 locus of NDY344 (44), displacing ste3Δ::LEU2 via the two-step gene replacement method (43).
TABLE 1.
Yeast strains
| Strain | Genotype | Reference or source | Classification of straina |
|---|---|---|---|
| SY1574 | MATα STE3 ura3 leu2 ade2-1oc ade1 his6 trp1am | 8 | |
| SY2638 | MATα ste3Δ::LEU2 | This work | A |
| NDY414 | MATα HIS3P::STE3 | This work | A |
| NDY647 | MATα HIS3P::STE3 ste20Δ::URA3 | This work | A |
| NDY675 | MATα HIS3P::STE3 ste4Δ | This work | A |
| NDY693 | MATα HIS3P::STE3 ste11Δ::URA3 | This work | A |
| NDY708 | MATα HIS3P::STE3 ste12Δ::LEU2 | This work | A |
| NDY847 | MATα HIS3P::STE3 fus3Δ::LEU2 | This work | A |
| NDY953 | MATα HIS3P::STE3 far1Δ | This work | A |
| NDY1022 | MATα HIS3P::STE3 fus3Δ::LEU2 kss1Δ::URA3 | This work | A |
| W303-1B | MATα STE3 ura3-1 leu2-3 his3-11,15 trp1-1 ade2-1 can1-100 | R. Rothstein laboratory | |
| NDY746 | MATα ste3Δ::LEU2 | This work | B |
| NDY753 | MATα HIS3P::STE3 | This work | B |
| T2-4D | MATα STE3 mfα1::LEU2 mfα2::LEU2 | 27 | B |
| NDY544 | MATα STE3 mfα1::LEU2 mfα2::LEU2 ste2Δ::TRP1 | This work | B |
| NDY598 | MATα STE3 mfα1::LEU2 mfα2::LEU2 HIS3p::STE2 | This work | B |
| RH268-1C | MATa end4-1 ura3 leu2 his4 bar1-1 | 39 | |
| NDY344 | MATα end4-1 ste3Δ::LEU2 | 44 | C |
| NDY867 | MATα end4-1 GAL1p-STE3(M271I, M304I) | This work | C |
| NDY795 | MATa end4-1 ste4Δ | This work | C |
| SY1793 | MATα STE3 mfa1Δ mfa2Δ FUS1P::HIS3 ura3-52 leu2-3,112 ade1 | 8 | |
| SY1937 | MATα STE3 ste2Δ mfa1Δ mfa2Δ::FUS1-LacZ | 2 | D |
| NDY787 | MATα STE3 ste2Δ mfa1Δ mfa2Δ::FUS1-LacZ rad16::GAL1p-STE4 | 2 | D |
| SY2011 | MATα ste3Δ ste2Δ mfa1Δ mfa2Δ::FUS1-LacZ | 2 | D |
| NDY691 | MATα ste3Δ::LEU2 mfa1Δ mfa2Δ rad16::GAL1p-STE4 | This work | D |
| NDY692 | MATα HIS3P::STE3 mfa1Δ mfa2Δ rad16::GAL1p-STE4 | This work | D |
| NDY711 | MATα HIS3P::STE3 mfa1Δ mfa2Δ | This work | D |
Strains designated A are isogenic to SY1574; strains designated B are isogenic to W303-1B; strains designated C are isogenic to RH268-1C; and strains designated D are isogenic to SY1793.
The HIS3P::STE2 and HIS3P::STE3 alleles have the mating type-specific, pheromone-inducible promoters of STE2 and STE3 replaced by the HIS3 promoter (HIS3P). For HIS3P::STE2, a 164-bp HIS3 fragment extending from −210 to −374 bp upstream of the HIS3 ATG initiator codon replaces STE2 sequences from −37 to −535 bp upstream of the STE2 initiator codon. For HIS3P::STE3, the same 164-bp HIS3P fragment replaces STE3 sequences −108 to −411 bp upstream of the STE3 initiator codon. The resulting HIS3P-controlled STE2 and STE3 alleles were chromosomally integrated via the two-step gene replacement strategy (43), replacing the ste2Δ::TRP1 and ste3Δ::LEU2 loci of NDY544 and SY2638, respectively.
A number of different STE genes were disrupted in the MATα HIS3P::STE3 strain NDY414. The unmarked ste4Δ allele deletes a 605-bp HindIII fragment from the STE4 coding sequence. For disruption of other STE functions, a panel of well-characterized knockout alleles was used: pEL45 for the ste20Δ::URA3 disruption (28), pSL1094 for the ste11Δ::URA3 disruption (48), and SUL16 for the ste12Δ::LEU2 disruption (13). Disruption of the genes for the two redundant MAPKs, Kss1p and Fus3p, was done in two steps. First, the fus3Δ::LEU2 allele from pYEE98 (10) was introduced into NDY414. Then, this fus3Δ::LEU2 strain was made kss1Δ::URA3 by use of pBC65 (8). Introduction of the unmarked far1Δ allele was done with p1054, which deletes a 1,025-bp HindIII-to-BamHI segment within the FAR1 coding sequence (C. Boone, personal communication). Plasmid pDH15 (19) was used for the generation of the rad16::GAL1p-STE4 strains.
Cell labeling, pheromone treatment, extract preparation, and immunoprecipitation.
Cells were pulse-labeled for 10 min with a mixture of [35S]methionine and [35S]cysteine and then chased for various times with excess cold amino acids as described previously (43). For a-factor treatment, a concentrated culture supernatant from yeast cells which overproduce a-factor was prepared (43). The a-factor activity of this preparation was estimated to be 400-fold more concentrated than that of an unconcentrated supernatant from a saturated culture of wild-type MATa cells. Pulse-labeled cells were treated for 15 min with 20 μl of one of three different a-factor dilutions per labeling reaction: concentrated a-factor (1×), a 1:10 dilution (0.1×), or a 1:100 dilution (0.01×) (YPD medium was used for dilutions). Mock-treated controls were treated with a concentrated supernatant prepared in parallel from isogenic mfa1Δ mfa2Δ cells (43). Following the a-factor treatment of the labeled cultures, cells were collected by centrifugation, extracts were prepared via glass bead disruption, and Ste3p was immunoprecipitated, all as described previously (43).
Pulse-labeling of cells expressing the GAL1-driven activated signaling alleles was done as described above except that 2% raffinose was substituted for glucose in the culture medium. Expression of the activating constructs was induced with 2% galactose added 100 min prior to the pulse-labeling. Cells were pulse-labeled for 10 min and then chased with cold amino acids for an additional 10 min as described above.
Phosphatase treatment of labeled protein extracts.
In some cases, 35S-labeled protein extracts were treated with phosphatase prior to immunoprecipitation. For this, the glass bead protein extraction method (43) was altered to reduce the sodium dodecyl sulfate (SDS) concentration within the final labeled protein extract. Culture aliquots corresponding to approximately 2 × 107 cells, pulse-labeled and chased as described above, were collected by centrifugation, resuspended in 15 μl of extraction buffer (40 mM Tris-Cl [pH 8.0], 8 M urea, 0.1 mM EDTA, 1% β-mercaptoethanol), transferred to a tube containing a 10-μl volume of acid-washed glass beads (212 to 300 μm in diameter; Sigma Chemical Co., St. Louis, Mo.), and vortexed for 15 s. An additional 15 μl of extraction buffer supplemented with 1.5% SDS was added, and cell lysis was completed with an additional 1 min of vortexing. Samples were immediately heated at 120°C for 5 min and then stored for several hours at 0°C prior to initiation of the phosphatase treatment. Just prior to the phosphatase treatment, samples were reheated at 100°C for 5 min, following which a 9-μl portion of the extract was diluted into 0.8 ml of phosphatase digestion buffer (20 mM sodium citrate [pH 6.0], 50 mM NaCl, 0.5 mM phenylmethylsulfonyl fluoride, 1 μg of leupeptin per ml, 1 μg of pepstatin per ml). Digestion was done with 0.5 U of potato acid phosphatase (Boehringer Mannheim Biochemicals, Indianapolis, Ind.) for 40 min at 25°C. Mock-digested control samples were diluted and incubated in parallel. Reactions were terminated, and samples were prepared for immunoprecipitation with the addition of 0.2 ml of concentrated immunoprecipitation buffer (0.5 M Tris-Cl [pH 8.0], 0.36% SDS, 10 mM EDTA, 0.5% Triton X-100). Immunoprecipitation was done as described previously (43).
Immunoblots.
Log-phase cultures of cells carrying the GAL1-regulated receptor genes, either plasmid STE2-HA (pND733) or chromosomally integrated GAL1p-STE3, were grown overnight in 2% raffinose medium as described previously (44). Receptor expression was induced with the addition of galactose to 2% for various times and subsequently repressed with the addition of glucose to 3%. At various times after glucose addition, cells were treated with pheromone. a-Factor treatment and mock pheromone treatment were as described previously (8), except that 0.5 volume of pheromone supernatant was added. For α-factor treatment, synthetic α-factor peptide (Sigma) was added to either 10−5 M or 10−7 M. Preparation of protein extracts for Western blotting, SDS-polyacrylamide gel electrophoresis (PAGE), and transfer to nitrocellulose were done as described previously (8). Ste2p was detected with monoclonal antibody (MAb) HA.11 (BAbCo, Berkeley, Calif.) specific for an HA epitope tag C-terminally fused to Ste2p (pND733). Ste3p was detected with affinity-purified rabbit polyclonal antibodies directed against the Ste3p CTD (43). Blots were developed with the ECL chemiluminescence system (Amersham Corp., Arlington Heights, Ill.). In some cases, prior to Western blotting, protein extracts were treated with potato acid phosphatase at a final concentration of 0.06 U/ml as described previously (43).
CNBr treatment.
Protein extracts were prepared for immunoblotting from MATα GALp-STE3(M271I, M304I) NDY867 cells essentially as described above except that 5 × 108 cells were used to make the extracts more concentrated. Extracts (20 μl) were digested in a 200-μl reaction mixture containing 70% formic acid and 2.5 M CNBr. Digestion was done at 25°C for 16 h in the dark. Protein was precipitated with trichloroacetic acid and prepared for SDS-PAGE and Western blot analysis as described previously (43). Some samples were treated with phosphatase as described above prior to SDS-PAGE.
β-Galactosidase assays.
MATα FUS1-LacZ cells harboring the GAL1-driven constitutively active STE gene constructs either as CEN plasmids or as a chromosomally integrated construct (rad16::GAL1p-STE4) were grown as overnight log-phase cultures in minimal synthetic medium supplemented with 0.2% yeast extract, as for pulse-labeling (43), with the exception that 2% raffinose was substituted for glucose. To induce the expression of the signaling component, 2% galactose was added for 2 h. Culture aliquots were then collected by centrifugation, cells were permeabilized, and β-galactosidase activity was determined as described previously (22).
RESULTS
Ligand-induced phosphorylation of the Ste3p CTD.
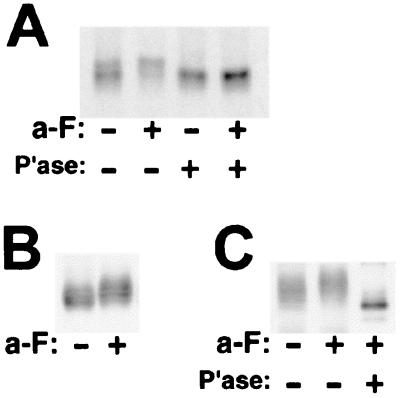
Ste3p is a short-lived protein: in cells growing at 30°C, Ste3p is degraded with a half-life of about 15 min (8). Rapid turnover occurs via rapid constitutive endocytosis, which removes the receptor from the cell surface and delivers it to the vacuole for degradation. This process is ligand independent, occurring whether or not the a-factor ligand is present. A consequence of such dynamic membrane trafficking is that, at any particular moment, much of the Ste3p in the cell is in transit—traveling through either the secretory pathway on its way to the cell surface or the endocytic pathway on its way to the vacuole. For studying ligand-induced phosphorylation, we routinely use a pulse-chase protocol to focus analysis on the subset of newly synthesized receptor proteins that is surface resident and therefore available for interaction with pheromone. In resting, unstimulated cells, Ste3p shows heterogeneous gel mobility (Fig. 1A). Treatment of these cells with the a-factor ligand results in a reduction in Ste3p mobility, with much of the receptor now running coherently as a single species (Fig. 1A). When protein extracts are subjected to phosphatase treatment prior to immunoprecipitation, the receptor from both a-factor-treated and untreated cells collapses to a single, faster-migrating species (Fig. 1A). Our conclusion is that in resting cells, Ste3p is subject to a constitutive level of phosphorylation that increases when cells are treated with a-factor.
FIG. 1.
The Ste3p CTD is subject to both constitutive and ligand-dependent phosphorylation. (A) a-factor treatment leads to increased Ste3p phosphorylation. MATα HIS3p-STE3 yeast cells (NDY414), which have the constitutive HIS3 promoter replacing the pheromone-regulated STE3 promoter, were pulse-labeled for 10 min with [35S]methionine, chased with excess cold methionine and cysteine for 1 min, and either mock treated (−) or treated with a 1× concentration (see Materials and Methods) of a-factor (a-F) (+) for an additional 15 min. Protein extracts prepared from the labeled cells were treated with potato acid phosphatase (P'ase) (+) or were mock treated (−) in parallel. Finally, Ste3p was immunoprecipitated with Ste3p-specific antibodies and subjected to SDS-PAGE and autoradiography. Quantitative phosphorimager analysis of this and parallel experiments indicates that the a-factor treatment results in no significant increase or decrease in the amount of radioactive Ste3p recovered, implying no major effect of the pheromone treatment on the rate of Ste3p turnover. (B) Ste3p retained at the cell surface in end4 mutant cells is subject to ligand-induced phosphorylation. Receptor synthesis was induced from a log-phase culture of MATα GAL1p-STE3(M271I, M304I) end4-1 yeast cells (NDY867) with a 45-min period of galactose addition. Glucose was then added to repress further synthesis. After 10 min, cultures were treated for 15 min with a-factor (+) or were mock treated (−) as described in Materials and Methods. Cells were grown at 30°C; while this temperature is permissive for end4-1 cell growth, endocytosis of both pheromone receptors remains strongly impaired (39, 43). Protein extracts prepared from these cells were subjected to SDS-PAGE and Western analysis with Ste3p-specific antibodies. (C) The Ste3p CTD provides a major locus for both constitutive and ligand-induced phosphorylation. The protein extracts prepared for panel B were treated with CNBr (see Materials and Methods) prior to SDS-PAGE and Western analysis. The M271I and M304I mutations remove methionyl sites of CNBr cleavage and augment the visualization of the CTD fragment: M304I removes a cleavage site internal to the CTD, and M271I eliminates a partial fragment from position 271 to the C terminus. Neither mutation perturbs Ste3p functioning in terms of mating, endocytosis, or phosphorylation (data not shown). To identify the electrophoretic position of a CTD that lacks phosphoryl modification, an additional protein extract was prepared in parallel to the two prepared for panel B. In this case, the requirement for increased concentrations of Ste3 antigen within the protein extract to be subjected to phosphatase digestion was accommodated with a 2-h galactose induction period prior to the 15 min a-factor treatment. (The intact Ste3p derived from this extract showed electrophoretic mobility identical to that of the Ste3p derived from the a-factor-treated culture shown in Fig. 1B) (data not shown). Following CNBr treatment and prior to SDS-PAGE and Western analysis, this sample was treated with phosphatase. For the SDS-PAGE analysis of the CNBr-treated protein extracts, 12% acrylamide gels were used (as opposed to the 8% gels used for the visualization of full-length Ste3p).
In addition to the radiolabeling approach of Fig. 1A, a-factor-dependent phosphorylation of unlabeled Ste3p may also be observed by Western blotting under conditions that block Ste3p constitutive endocytosis. By trapping Ste3p at the cell surface, the receptor remains available to the extracellular ligand. Blocking endocytosis also eliminates the potential loss of phosphorylated species to vacuolar turnover. When examined within the context of endocytosis-defective end4-1 cells (39), Ste3p shows a constitutive level of phosphorylation that increases with a-factor treatment (Fig. 1B). Identical treatment of wild-type (END4+) cells results in no clear effect of a-factor treatment on the receptor modification status (data not shown). As discussed above, this result likely is a consequence of the dynamic trafficking of Ste3p; receptor localized to intracellular compartments of the secretory and endocytic pathways is unavailable to the extracellular ligand.
GPCRs generally are subject to a desensitizing phosphorylation of Ser-Thr residues located within their regulatory CTDs. To test if the CTD provides a locus for Ste3p phosphorylation, we have examined the constitutive and ligand-induced phosphorylation present on a 183-residue C-terminal peptide fragment cleaved from the intact receptor by CNBr at Met288 (Met288 is located just C-terminal to the seventh predicted transmembrane domain). Protein extracts from a-factor-treated and from unstimulated cells (Fig. 1B) were treated with CNBr, and the released Ste3p CTD fragment was visualized by Western blotting (Fig. 1C; the Ste3p-specific antibody is directed against the Ste3p CTD). The Ste3p CTD isolated from a-factor-treated versus untreated cells shows changes in gel mobility (Fig. 1C) that mirror those seen for the intact receptor (Fig. 1B). The CTD isolated from unstimulated cells migrates as a heterogeneous cluster, consistent with heterogeneous constitutive phosphorylation. When isolated from the a-factor-stimulated cells, the heterogeneous CTD shows retarded gel mobility, consistent with the induced phosphorylation seen for the intact receptor. Both constitutive and ligand-induced CTD modifications are removed with phosphatase treatment of the peptide fragments (Fig. 1C). The CTD, we conclude, is a major locus for both constitutive and ligand-dependent phosphorylation of Ste3p.
Ligand-induced phosphorylation of Ste3p requires the Gβ subunit of heterotrimeric G protein.
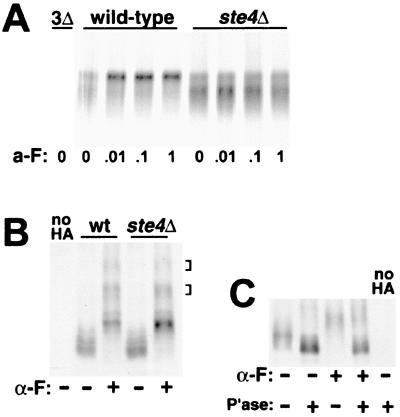
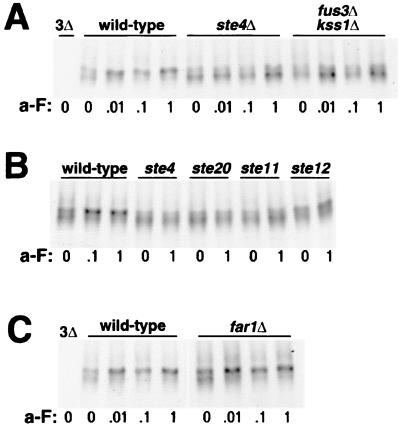
For the GPCRs, the first cytoplasmic signaling step initiated by the liganded receptor is the dissociation of the αβγ heterotrimeric G protein into the α and βγ subunits. The ligand-dependent phosphorylation of the α-factor receptor (Ste2p) does not require this step and proceeds unimpaired in cells deficient for the G protein components (56). To test if this is also true for Ste3p, we have used pulse-chase analysis to examine both constitutive and ligand-dependent Ste3p phosphorylation in ste4Δ cells. STE4 encodes the Gβ component of the G protein. As the βγ subunit is the primary transducer of the ligand signal in yeast, ste4Δ cells fail to mount a pheromone response. For this analysis, we have used strains that have the endogenous, pheromone-responsive STE3 promoter replaced by the constitutive HIS3 promoter. This strategy not only blocks the induced increase in transcription with a-factor treatment (15) but also, more importantly, eliminates the fivefold reduction in basal STE3 expression seen in resting cells with disruptions of elements of the pheromone signal transduction pathway; the resting activity of this pathway is required for the basal transcription of many pheromone-inducible genes (12). The HIS3 promoter is a relatively weak promoter (52), and HIS3-driven Ste3p expression is found to be two- to threefold higher than expression from the unstimulated STE3 promoter (A. Roth and N. Davis, unpublished results). STE4+ and ste4Δ HIS3P-STE3 MATα cells were mock treated or treated with one of three different a-factor concentrations for 10 min (Fig. 2A). In the STE4+ background, ligand-induced mobility shifts consistent with induced phosphorylation were equivalently seen over the entire 100-fold a-factor concentration range (these concentrations of the pheromone give a graded response when tested for induction of the pheromone-induced transcriptional response; see Fig. 8A) (data not shown). In ste4Δ cells, the a-factor-induced mobility shift was completely blocked at all concentrations (Fig. 2A). Thus, the ligand-induced phosphorylation of Ste3p quite clearly requires functional Gβ.
FIG. 2.
The ligand-dependent phosphorylation of the two pheromone receptors exhibits different requirements for the Gβ subunit. (A) The Gβ subunit is required for the a-factor-induced phosphorylation of Ste3p. MATα HIS3P-STE3 cells (wild type; NDY414) and isogenic ste4Δ cells (NDY675) were subjected to a pulse-chase protocol similar to that described in the legend to Fig. 1. Following a 10-min pulse with [35S]methionine and a 10-min chase with excess cold amino acids, cultures were treated with four different concentrations of a-factor (a-F) for 15 min: concentrated a-factor (1), a 10-fold dilution of concentrated a-factor (.1), a 100-fold dilution of concentrated a-factor (.01), and no a-factor (0). Ste3p was immunoprecipitated from the labeled protein extracts and then subjected to SDS-PAGE and autoradiography. As a control for the specificity of the anti-Ste3p antibodies used for immunoprecipitation, extracts from the isogenic ste3Δ strain SY2638 (3Δ) were processed in parallel. For this experiment, electrophoresis was done on a 20-cm gel format, as opposed to the 7.5-cm minigel format used in the other experiments. On this extended gel format, the presentation of Ste3p from pheromone-treated and untreated cells is much different from that on the minigel format (compare wild-type samples with those of Fig. 1A); with the extended format, heterogeneously modified receptor species are more widely spread. (B) The Gβ subunit is not required for α-factor-induced modifications of the α-factor receptor. The GAL1p-STE2-HA centromeric plasmid pND733 was introduced into the MATa end4-1 strain RH268-1C (wt) or the isogenic ste4Δ strain NDY795. Following 1 h of galactose-induced receptor expression, glucose was added, and cultures were further treated for 10 min with α-factor (α-F) added to 10−6 M (+) or were mock treated (−) in parallel. Cultures were maintained at 30°C throughout (see the legend to Fig. 1B). Protein extracts prepared from these cells were subjected to SDS-PAGE and then to Western analysis with an anti-HA.11 monoclonal antibody. As a control for cross-reaction of the antibody, extracts from RH268-1C cells transformed by the empty centromeric plasmid vector (no HA) were processed in parallel. The brackets at right indicate the positions of pheromone-dependent Ste2p modifications likely corresponding to ubiquitin-modified Ste2p (16). (C) The pheromone-induced modifications of Ste2p include phosphorylation. Protein extracts from the ste4Δ cells of panel B were treated with phosphatase (P'ase) (+) or mock treated in parallel with no added phosphatase (−) as described in Materials and Methods. Extracts were then subjected to SDS-PAGE and Western analysis as described for panel B.
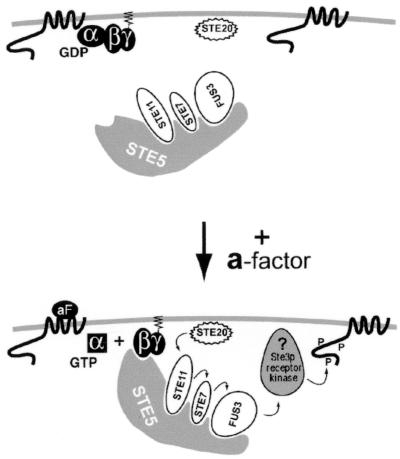
FIG. 8.
Model for the regulation of Ste3p phosphorylation by the pheromone signaling pathway. Results for Ste3p phosphorylation are explained here within the context of a model for Gβγ activation of the signaling pathway originally presented by Pryciak and Huntress (37). In resting MATα cells (top), the Ste5p-kinase complex localizes to both the cytoplasm and the nucleus. With a-factor (aF) stimulation (bottom), the liganded receptor catalyzes the exchange of nucleotide, GTP for GDP, on the Gα subunit, with the consequent dissociation of Gα from Gβγ. Free Gβγ now serves as a plasma membrane binding site for the Ste5p-kinase complex. Localized to the plasma membrane, the Ste5p-kinase complex is now subject to activating phosphorylation of Ste11p by the PAK kinase Ste20p. This process leads to the sequential activation of the Ste5p-bound kinases: first Ste7p and then the MAPK Fus3p. Finally, activated Fus3p is expected to phosphorylate and thereby activate a plasma membrane-localized target that stimulates the phosphorylation of Ste3p. In this depiction, an unidentified receptor kinase is shown as the Fus3p target. Alternatively, as discussed in the text, Fus3p may act directly to phosphorylate the receptor protein.
In light of the effect of the ste4Δ mutation on the ligand-dependent phosphorylation of Ste3p, we have sought to reconfirm that the ligand-dependent phosphorylation of Ste2p is indeed G protein independent (56). We have monitored the α-factor-induced phosphorylation of Ste2p in both STE4+ and ste4Δ cells (Fig. 2B and C). For this purpose, an HA epitope-tagged version of Ste2p has been examined by Western blotting of extracts from α-factor-treated or mock-treated cells. In addition, this construct has the pheromone-inducible STE2 promoter replaced by the GAL1 promoter, eliminating pheromone pathway effects on receptor expression levels. Extracted from unstimulated MATa cells, Ste2p shows heterogeneous electrophoretic mobility, a result presumably of both heterogeneous constitutive phosphorylation (40, 56) and heterogeneous glycosylation (25). Treatment with α-factor induces dramatic changes in Ste2p modification (Fig. 2B), consistent with known effects of the pheromone on receptor phosphorylation and ubiquitination levels (16). Phosphatase treatment collapses the modified receptor to species of more rapid gel mobility, indicating that phosphorylation is a major component of the α-factor-induced mobility changes (Fig. 2C). As previously reported (56), α-factor-induced phosphorylation proceeds unimpaired in ste4Δ cells (Fig. 2B), indicating that Ste2p differs from Ste3p in that G protein-mediated signaling clearly is not required for the induced phosphorylation of Ste2p.
Ste3p phosphorylation is induced when the signaling pathway is artificially activated.
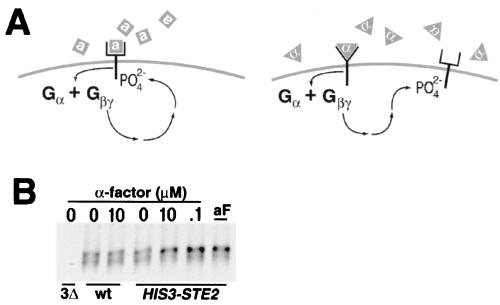
To assess if the STE4 dependence reflected a need for induction of the downstream signaling pathway, we have tested if Ste3p phosphorylation could be induced in trans via α-factor induction of the Ste2p signaling. While the two receptors normally function in different cell types, both couple to the same downstream signaling components (1). We reasoned that if the activation of the downstream signaling cascade was all that was required for the induced phosphorylation of Ste3p, then its activation through α-factor stimulation of Ste2p should also lead to Ste3p phosphorylation. For this experiment, we have constructed the MATα strain NDY598, which artificially expresses both pheromone receptors: Ste3p expressed from its natural, α-specific promoter and Ste2p artificially expressed from the HIS3 promoter. In addition, NDY598 also has disruptions of the two α-factor-encoding loci, MFα1 and MFα2, obviating any autocrine Ste2p stimulation (Fig. 3A). Again, we have monitored Ste3p phosphorylation via immunoprecipitation from in vivo pulse-labeled cells. Added α-factor is without effect on Ste3p phosphorylation in control MATα strains lacking the α-factor receptor (Fig. 3B). In cells that express Ste2p, however, a clear induction of Ste3 phosphorylation is seen (Fig. 3B). Indeed, the induced changes in Ste3p gel mobility induced at two concentrations of the α-factor ligand are identical to those seen when the a-factor ligand is added (Fig. 3B). In addition to indicating a role for the activated signaling pathway in the induced phosphorylation of Ste3p, this experiment also demonstrates that liganded a-factor receptor is not the obligate substrate for phosphorylation: phosphorylation proceeds normally even when the normal receptor ligand is not present.
FIG. 3.
Ste3p phosphorylation may be induced in trans via α-factor stimulation of an ectopically expressed α-factor receptor. (A) Experimental design. At left is shown an approximate mechanism for the induced phosphorylation of Ste3p that occurs in wild-type MATα cells treated with a-factor. The binding of a-factor pheromone (gray squares) to the a-factor receptor elicits a Gβγ-dependent mechanism which leads to receptor phosphorylation. At right is outlined the logic of an experiment that tests if G protein-dependent signaling activated by the binding of α-factor (gray triangles) to Ste2p may substitute for the a-factor requirement in inducing Ste3p phosphorylation. This experiment was done with the MATα HIS3P-STE2 mfα1Δ mfα2Δ strain NDY598. Thus, these cells constitutively express both yeast pheromone receptors but neither of the two pheromones. (B) trans-Activation of G protein signaling bypasses the a-factor requirement for induced phosphorylation of Ste3p. NDY598 cells were labeled within a pulse-chase regimen identical to that used in the experiment shown in Fig. 2A, i.e., 10 min of pulse-labeling followed by 10 min of chase, followed by 15 min of pheromone treatment. Pheromone concentrations used were as follows: concentrated a-factor (aF), 0.1 × 10−6 M α-factor (.1), 10 × 10−6 M α-factor (10), or no pheromone (0). As a control, NDY544 (wt), a strain isogenic to NDY598 but lacking the HIS3P-STE2 construct, was pulse-labeled and treated with 10 × 10−6 M α-factor (10) or mock treated (0). Ste3p was immunoprecipitated from the labeled protein extracts and then subjected to SDS-PAGE and autoradiography. As a control for antibody specificity, an extract from the isogenic ste3Δ strain NDY746 (3Δ) was processed in parallel.
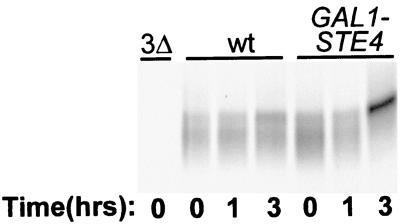
We next asked if it might be possible to dispense with the ligand altogether for the induced phosphorylation of Ste3p. Most of the pheromone responses, i.e., G1 arrest, induction of pheromone-responsive genes, and even mating, can be induced through Ste4p overproduction from a GAL1p-STE4 construct (6, 33, 54). A 3-h period of galactose induction for GAL1p-STE4 cells resulted in clear effects on Ste3p phosphorylation, with the receptor showing mobility changes typical of a-factor-induced phosphorylation (Fig. 4). Receptors from similarly treated isogenic cells lacking GAL1p-STE4 failed to show the same changes (Fig. 4). Thus, pathway activation, either through the action of α-factor on ectopically expressed Ste2p (Fig. 3B) or through Ste4p overproduction, suffices to induce Ste3p phosphorylation, bypassing the requirement for the a-factor ligand.
FIG. 4.
Ligand-independent activation of the signaling pathway via Gβ overproduction leads to induced phosphorylation of Ste3p. Two isogenic MATα HIS3P-STE3 strains were used: NDY711 (wt) and NDY692, a version having an integrated GAL1p-STE4 construct. Galactose (2%) was added to cells growing in raffinose medium to induce Ste4p overproduction. At 20 min prior to the indicated times, culture aliquots were removed and subjected to 10 min of pulse-labeling with [35S]methionine, followed by 10 min of chase with excess cold methionine and cysteine. Cultures labeled for the 0-h time point were never exposed to galactose. Radiolabeled Ste3p was immunoprecipitated from protein extracts derived from these cells and then subjected to SDS-PAGE and autoradiography. As a control for antibody specificity, an extract from the isogenic ste3Δ strain NDY691 (3Δ) was processed in parallel. For this experiment, SDS-PAGE was done with the extended gel format (see the legend to Fig. 2A).
Requirement for the distal components of the signaling pathway.
A key element within the pheromone signaling pathway is the MAPK Fus3p. Through phosphorylation of the transcriptional activator Ste12p and the Dig1p-Dig2p corepressor system, Fus3p activates pheromone-dependent transcription (7, 20, 49). Through phosphorylation of the Cdk inhibitor Far1p, Fus3p serves to institute pheromone-dependent cell cycle arrest in G1 (34). Fus3p is activated as an end point of a cascade of sequential kinase activation steps. Elements within this cascade include the PAK kinase Ste20p, the MAPK kinase kinase Ste11p, and the MAPK kinase Ste7p. Fus3p, together with two kinases immediately upstream, Ste11p and Ste7p, is coordinately bound to the kinase scaffolding protein Ste5p. Fus3p is considered to be the dedicated MAPK for the pheromone signaling pathway (31). However, in fus3Δ cells, Fus3p function can be partly replaced by Kss1p, a homologous MAPK apparently dedicated to the filamentation response. Due to the redundant actions of Kss1p, testing of the MAPK requirement for various pheromone responses generally is done with strains having both MAPK genes deleted, i.e., fus3Δ kss1Δ cells.
In Fig. 5A, fus3Δ kss1Δ cells are compared to both wild-type and ste4Δ cells with regard to pheromone-induced Ste3p phosphorylation. As in previous experiments (Fig. 1A and 2C), phosphorylation was assessed in terms of the gel mobility shifts seen for Ste3p immunoprecipitated from extracts of pulse-labeled cells treated with various a-factor dosages. The usual a-factor-induced upward mobility shift is seen for wild-type cells treated with the three concentrations of a-factor (Fig. 5A). The receptor from treated fus3Δ kss1Δ cells was indistinguishable from that from ste4Δ cells: no induction of phosphorylation was apparent at any of the pheromone concentrations tested (Fig. 5A). Like the ste4Δ mutation, the fus3Δ kss1Δ double deletion wholly blocks the receptor phosphorylation response, indicating that, like the Gβ subunit, the MAPK is also required. In parallel experiments, we have also tested cells with fus3Δ and kss1Δ single deletions. Both were indistinguishable from wild-type cells; i.e., both failed to block induced Ste3p phosphorylation (data not shown). Thus, as with other pheromone responses, FUS3 and KSS1 can contribute redundantly to the induction of Ste3p phosphorylation.
FIG. 5.
Participation of the pheromone signal transduction chain components in the ligand-induced phosphorylation of Ste3p. The signaling-competent wild-type MATα HIS3P-STE3 strain NDY414 and isogenic strains with disruptions of various elements of the pheromone signaling pathway were pulse-labeled and treated with the concentrations of a-factor described in the legend to Fig. 2A. Following pheromone treatment, labeled Ste3p was immunoprecipitated from protein extracts and subjected to SDS-PAGE and autoradiography. As in previous experiments, the ste3Δ strain SY2638 (3Δ) was processed in parallel as a control for the specificity of the Ste3p antibodies. (A) The redundant pair of MAPKs, Fus3p and Kss1p, is required for the ligand-induced phosphorylation of Ste3p. The gel mobilities of Ste3p isolated from a-factor-treated wild-type cells, ste4Δ cells (NDY675), or fus3Δ kss1Δ cells (NDY1022) were compared. Four different a-factor (a-F) concentrations were used: concentrated a-factor (1), 10-fold-diluted a-factor (.1), 100-fold-diluted a-factor (.01), or no a-factor (0). (B) Involvement of the PAK kinase homologue Ste20p, the MAPK kinase kinase Ste11p, and the pheromone-dependent transcriptional activator Ste12p in Ste3p ligand-induced phosphorylation. The gel mobilities of Ste3p isolated from a-factor-treated or mock-treated wild-type cells, ste4Δ cells (NDY675), ste20Δ cells (NDY647), ste11Δ cells (NDY693), or ste12Δ cells (NDY708) were compared. Pulse-labeled cells for each were treated with concentrated a-factor (1) or were mock treated (0). Wild-type cells were additionally treated with a 10-fold dilution of concentrated a-factor (.1). (C) Far1p, a protein required both for pheromone-induced G1 arrest and for morphogenesis, is not required for the ligand-induced phosphorylation of Ste3p. The gel mobilities of Ste3p isolated from a-factor-treated or mock-treated wild-type and far1Δ cells (NDY953) were compared as described for panel A.
We have also tested other elements of the signaling pathway for their involvement in the induced phosphorylation of Ste3p. In Fig. 5B, the gel mobility of Ste3p from ste20Δ, ste11Δ, and ste12Δ cells is compared to that of the receptor from isogenic wild-type and ste4Δ cells. For this experiment, cultures were treated only with the highest a-factor concentration (1×) (Fig. 5A). Both ste20Δ and ste11Δ cells behaved identically to ste4Δ cells, showing no discernible effect of the pheromone treatment on Ste3p gel mobility (Fig. 5B). ste12Δ cells showed an intermediate phenotype: while Ste3p was shifted less and presumably was less extensively phosphorylated than in wild-type cells, there was nonetheless a clear effect of the a-factor treatment on the receptor modification state. (This intermediate phenotype of ste12Δ cells is explored more fully below; see Fig. 6). In addition to these components of the signaling pathway, two other components, Ste5p and Ste7p, have also been tested. Like ste4Δ, ste20Δ, ste11Δ, and fus3Δ kss1Δ cells, ste5Δ and ste7Δ cells also failed to mount a-factor-induced phosphorylation of Ste3p (data not shown).
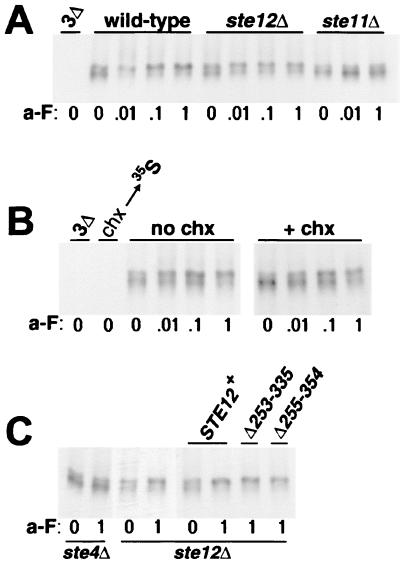
FIG. 6.
Pheromone-induced gene expression is not required for induced phosphorylation of Ste3p. (A) Ste12p is only partially required. Cells from the MATα HIS3P-STE3 strain NDY414 as well as from the isogenic ste12Δ (NDY708) and ste11Δ (NDY693) strains were pulse-labeled, treated with a-factor (a-F), and processed for immunoprecipitation as described in the legend to Fig. 2A. As a control for antibody specificity, a labeled extract from the ste3Δ strain SY2638 (3Δ) was processed in parallel. (B) Ligand-induced Ste3p phosphorylation does not require new protein synthesis. Following 10 min of pulse-labeling of MATα HIS3P-STE3 cells (NDY753) with [35S]methionine, the culture was split: half was treated with 50 μg of cycloheximide per ml (+ chx) added together with cold chase amino acids for 10 min prior to the 15-min a-factor treatment, and half received no cycloheximide (no chx) but was otherwise treated identically. A labeled extract from the ste3Δ strain NDY746 was processed in parallel (3Δ). As a test of the efficacy of the cycloheximide treatment, an aliquot of the NDY753 culture was pretreated with 50 μg of cycloheximide per ml 10 min prior to [35S]methionine pulse-labeling (chx→35S). Strain NDY753 (isogenic to W303-1B) was used for this experiment instead of strain NDY414, due to the relative insensitivity of NDY414 cells to cycloheximide treatment. NDY753 cells differ somewhat from NDY414 cells in terms of the dose response for a-factor-induced Ste3p phosphorylation: NDY753 cells require at least 10-fold-higher a-factor levels to achieve Ste3p mobility shifts equivalent to those seen in the NDY414 background (compare to Fig. 2A or 5). Panels labeled “no chx” and “+ chx” were derived from the same gel exposed for different periods of time. For the former panel, a threefold-longer autoradiographic exposure was required to attain the level of band intensity equivalent to that in the latter panel. This result likely reflects an effect of cycloheximide treatment on the normally rapid Ste3p turnover rate. Previous work with the α-factor receptor (Ste2p) indicated that α-factor-induced vacuolar turnover was dramatically slowed in cycloheximide-treated cells, with the receptor accumulating within a prevacuolar endosomal compartment (17). (C) ste12 mutants, which maintain the basal rate of pheromone transcription, restore wild-type Ste3p phosphorylation to ste12Δ cells. MATα HIS3P-STE3 cells that were either ste4Δ (NDY675) or ste12Δ (NDY708) were subjected to the a-factor treatment and pulse-chase described in the legend to Fig. 2A. In addition, some of the ste12Δ cells were transformed with one of three plasmids: the centromeric plasmid p650, which carries wild-type STE12, or the equivalent plasmid carrying one of two in-frame ste12 deletion mutants (Δ253-335 or Δ255-354).
We have also tested if FAR1 is required for the induced phosphorylation of Ste3p. Far1p participates in two aspects of the pheromone response: G1 arrest and the morphogenetic chemotropism of the arrested cell toward the pheromone source (34, 35, 50). far1Δ cells, tested as described above, behave the same as wild-type cells in terms of induced Ste3p phosphorylation (Fig. 5C), indicating that the pheromone effects on phosphorylation are independent of the pheromone effects on either the cell cycle or the morphogenetic response.
Lack of a requirement for pheromone-induced gene expression.
A major outcome of pheromone signaling is the induction of new gene transcription mediated by the transcriptional activator Ste12p. Most of the known pheromone responses, e.g., G1 arrest, the development of a mating projection, and cell-cell agglutination, require this induced transcription. For these responses, ste12 null mutations show the same absolute block as that seen with null mutations in the other pathway genes (47). In Fig. 5B, however, the effect of the ste12Δ mutation appeared to differ from that of ste4Δ, ste20Δ, or ste11Δ in terms of the impact upon the induced phosphorylation of Ste3p: ste12Δ cells showed only a partial block. In the experiment shown in Fig. 6A, we have examined the ste12 defect more extensively, comparing Ste3p phosphorylation in ste12Δ mutant cells to that in both isogenic wild-type cells and ste11Δ mutant cells. For ste11Δ cells, as with the other signaling mutants tested in Fig. 5, induced Ste3p phosphorylation was completely blocked at all pheromone concentrations (Fig. 6A). ste12Δ cells clearly differed: ligand-induced changes in Ste3p phosphorylation were apparent even at the lowest concentration of a-factor used (Fig. 6A). Thus, in terms of Ste3p phosphorylation, ste12Δ cells are capable of mounting a significant response to the pheromone. Nonetheless, this response is somewhat impaired relative to that of wild-type cells: in ste12Δ cells, a portion of the newly synthesized receptor resisted shifting to the hyperphosphorylated form (slow electrophoretic species) over the entire 100-fold a-factor concentration range (Fig. 6A).
The differential requirement for STE12 versus the other STE functions suggests that pheromone-induced transcription likely is not an essential part of the signaling mechanism that links ligand binding to induced phosphorylation. As a further check of this possibility, we have examined the effect of cycloheximide on ligand-induced Ste3p phosphorylation. The addition of cycloheximide prior to the pheromone challenge should block the protein synthesis of all pheromone-responsive gene products. As a control, cycloheximide added 10 min prior to [35S]methionine labeling fully blocked the incorporation of the label into new protein (Fig. 6B). However, when added prior to a-factor treatment, cycloheximide failed to block the pheromone-induced phosphorylation of Ste3p; Ste3p phosphorylation looked quite the same in the presence of cycloheximide as in its absence (Fig. 6B). We conclude that induced Ste3p phosphorylation does not require pheromone-induced protein synthesis. This result, along with the incomplete block seen in ste12Δ cells (Fig. 6A), serves to eliminate models wherein induced phosphorylation results primarily from the induced synthesis of either the responsible kinase or a positive regulator of the kinase.
If pheromone-induced transcription is not an essential part of the signaling mechanism leading to the induced phosphorylation of Ste3p, what explains the partial defect seen in ste12Δ cells? Ste12p is required not only for pheromone-induced transcription but also for the maintenance of the basal rate of transcription of a number of mating-related genes, including GPA1, SST2, FUS1, FUS2, and FUS3. In ste12Δ cells unstimulated by pheromone, the basal rate of expression of these genes is reduced 5- to 10-fold relative to that in wild-type cells (47). One explanation for the partial defect in Ste3p phosphorylation seen in ste12Δ cells is that some participant in the signaling mechanism leading to Ste3p phosphorylation depends on Ste12p for its basal expression. With the basal expression of this protein depressed, the activation of phosphorylation may be inefficient. To test this possibility, we have used two ste12 mutant alleles that, while specifically defective for pheromone-induced transcription, remain competent for the maintenance of the basal rate of transcription of pheromone-inducible genes (24). The two ste12 deletion mutants, Δ253-335 and Δ255-354, fail to complement a ste12Δ allele for mating (24), presumably reflecting their inability to induce pheromone-dependent transcription. However, given their ability to sustain the basal transcription rate, we have examined if they might restore full induction of Ste3p phosphorylation to a ste12Δ strain. In Fig. 6C, a-factor-induced Ste3p phosphorylation is compared in ste12Δ cells and ste12Δ cells transformed with plasmids carrying either wild-type STE12 or one of the two ste12 deletion mutants (Δ253-335 or Δ255-354). Both deletion mutants behave like wild-type STE12 in terms of this response—both fully restore the induced phosphorylation of Ste3p. Thus, the partial defect manifested by ste12Δ cells likely does result from the failure to maintain basal transcription levels.
Effects of different dominantly activated STE gene constructs.
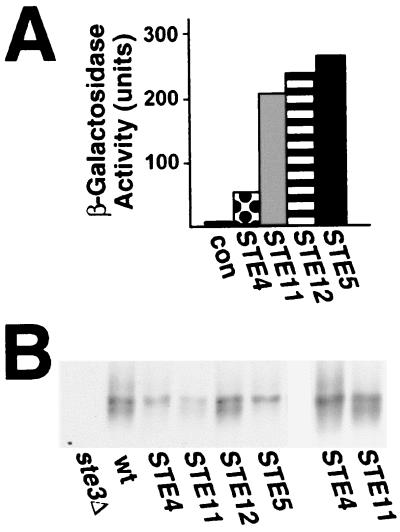
We have shown (Fig. 4) that activation of the signaling pathway by Ste4p overproduction induces Ste3p phosphorylation. Next, we tested the effects on Ste3p phosphorylation of pathway activation at other steps. Three activating constructs, in addition to GAL1p-STE4, were obtained: a GAL1p-STE5-CTM construct, a GAL1p-STE11ΔN construct, and a GAL1p-STE12 construct. Like that of Ste4p, Ste12p overproduction also leads to pheromone-independent activation of the pheromone response (9). The GAL1p-STE11ΔN and GAL1p-STE5-CTM constructs overproduce dominantly activated mutant alleles of STE11 and STE5. STE11ΔN lacks the N-terminal Ste11p regulatory domain (32, 38), and STE5-CTM adds a plasma membrane-targeting transmembrane domain to the Ste5p C terminus (37). All four activating constructs are grossly similar in their effects: all lead to pheromone-independent G1 arrest, transcriptional activation of FUS1, and partial suppression of the sterility associated with deletion of the pheromone receptor gene.
Following a 2-h period of galactose-induced synthesis, all four constructs induced the expression of the pheromone-responsive FUS1-LacZ reporter to levels that exceeded those produced through treatment of the cells with a-factor at concentrations which fully induce Ste3p phosphorylation (Fig. 7A). The effects of the activating constructs on Ste3p phosphorylation were assessed following this same 2-h galactose induction period. As shown previously (Fig. 4), GAL1p-STE4 induces an Ste3p mobility shift identical to that induced by the pheromone (Fig. 7B). A similar shift is seen for the receptor from cells expressing the STE5-CTM allele. Like GAL1p-STE4, GAL1p-STE5-CTM apparently bypassed the a-factor requirement in the induction of Ste3p phosphorylation. The expression of the other two activating constructs, GAL1p-STE11ΔN and GAL1p-STE12, failed to produce a change in Ste3p gel mobility.
FIG. 7.
Effect of dominantly activated STE gene constructs on both pheromone-dependent transcription and induced Ste3p phosphorylation. Four different GAL1-driven constructs were used: the GAL1p-STE4 construct was chromosomally integrated, while the other three were introduced into an isogenic strain as CEN/ARS plasmids: GAL1p-STE5-CTM (pND882), GAL1p-STE11ΔN (pRD-STE11-H3), and GAL1p-STE12(pC1-H6). The wild-type control strain, which lacked an activated signaling allele, carried the CEN/ARS vector pRS316 (46). (A) Each of the activating constructs strongly induces the expression of the pheromone-responsive gene FUS1. The MATα FUS1-LacZ strain SY1937 transformed by the four plasmids mentioned above or the isogenic rad16::GAL1p-STE4 strain NDY787 was treated with 2% galactose for 2 h, following which the amount of β-galactosidase expressed was measured (see Materials and Methods). β-Galactosidase activity measured from cells expressing the four different dominantly activated STE gene constructs was compared to the baseline level derived from SY1937 cells transformed with pRS316 (con). Reported values represent the average of two separate determinations (for each data point, the two activity measurements differed from one another by less than 20%). A 1-h treatment of SY1937 cells with a 0.1× concentration of a-factor (see Fig. 2A) resulted in the expression of 35 U of β-galactosidase activity (data not shown). (B) Differential effect of the activated alleles on the induced phosphorylation of Ste3p. Cells of the MATα HIS3P-STE3 strain NDY711 transformed by the four plasmids mentioned above or of the isogenic HIS3P-STE3 rad16::GAL1p-STE4 strain NDY692 growing in raffinose medium were induced by the addition of 2% galactose for 2 h. The final 20 min of the galactose induction period consisted of 10 min of pulse-labeling and then 10 min of chase with excess cold amino acids. Ste3p was immunoprecipitated from protein extracts prepared from these cells and then subjected to SDS-PAGE and autoradiography. Cells expressing the four different dominantly activated STE gene constructs were compared to NDY711 cells transformed by the empty vector pRS316 (wt). As a control, isogenic ste3Δ cells (SY1817) were labeled and processed in parallel. The panel on the right is a longer autoradiographic exposure of the two lanes expressing the activated alleles of STE4 and STE11 (poor Ste3p labeling is routinely seen in cells expressing the STE11ΔN construct).
The different abilities of the four dominantly activated STE gene constructs to induce Ste3p phosphorylation do not correlate with their potency in terms of activation of the signaling pathway (Fig. 7A). Instead, the key difference appears to be the position within the pathway at which signaling is instigated. GAL1p-STE4 and GAL1p-STE5-CTM activate the pathway at earlier steps than does GAL1p-STE11ΔN or GAL1p-STE12. As Ste11p is required for the induction of Ste3p phosphorylation (Fig. 5B), it seems a bit surprising that the dominantly activated GAL1p-STE11ΔN construct fails to induce this response. As discussed below, however, this paradox is explained within the context of a model wherein the key event for induced Ste3p phosphorylation is the translocation of the Ste5p-kinase complex to the plasma membrane.
DISCUSSION
Different kinase systems operate on the two pheromone receptors.
While the two yeast pheromone receptors share many functional similarities, the present work indicates a clear difference between the two in terms of the mechanisms used for inducing ligand-dependent phosphorylation. For Ste2p, the α-factor-induced increase in receptor phosphorylation does not depend upon the activation of the downstream signaling pathway; pheromone-induced phosphorylation is unimpeded in cells with disruptions of the individual G protein subunits or when receptor expression is artificially forced in a/α diploid cells which lack the expression of key signaling components: Gpa1p (Gα), Ste4p (Gβ), Ste18p (Gγ), and Ste5p (56). For Ste3p, a-factor-induced phosphorylation depends not only on the G protein β subunit (Ste4p) (Fig. 2A) but also on most of the downstream signaling components, including Ste20p (the PAK kinase homologue), Ste5p (the kinase cascade scaffolding protein), and the individual components of the MAPK cascade (i.e., Ste11p, Ste7p, and Fus3p/Kss1p). Gene disruptions of each of these pathway elements share the same phenotype in terms of Ste3p phosphorylation—induced phosphorylation is wholly blocked. For the induced phosphorylation of Ste3p, we conclude that the pheromone signal must be transduced along the signaling pathway from the G protein through the MAPK.
Furthermore, not only is the downstream signaling pathway required, but also its activation is sufficient for inducing Ste3p phosphorylation: phosphorylation of the unliganded a-factor receptor could be induced both through α-factor stimulation of cells artificially expressing both receptors (Fig. 3B) or through the ligand-independent activation of the signaling pathway by GAL1p-STE4 or GAL1p-STE5-CTM (Fig. 4 and 7B). Examination of Ste2p in contexts where the signaling pathway is activated artificially has failed to show evidence of induced Ste2p phosphorylation (data not shown). For Ste2p, the liganded receptor appears to participate more directly in fostering its own feedback phosphorylation. Liganded Ste2p may serve to directly activate the receptor kinase; alternatively, the liganded receptor may be the obligate substrate for the receptor kinase.
The difference in the regulatory mechanisms underlying the ligand-induced phosphorylation of the two receptors suggests that the kinases responsible for the phosphorylation may also differ. A recent report provides strong evidence that the two redundant, plasma membrane-localized type I casein kinases, Yck1p and Yck2p (51, 53), may be the kinases responsible for Ste2p phosphorylation (18). In a yck1Δ yck2-2ts strain at the restrictive temperature (yck1Δ yck2Δ cells are inviable) (41), both constitutive phosphorylation and α-factor-induced phosphorylation of Ste2p are strongly impaired (18). We have tested the yck1Δ yck2-2ts mutant for effects on Ste3p phosphorylation. Although Yck1p and Yck2p do participate in the phosphorylation of the Ste3p CTD, unlike Ste2p, the Ycks clearly do not constitute the major kinase system acting upon Ste3p. Indeed, constitutive Ste3p phosphorylation and ligand-induced Ste3p phosphorylation monitored as in the present work appear to be only minimally affected in the yck mutant background at a restrictive temperature (Y. Feng and N. Davis, unpublished data). Thus, the differential regulation which we have described for the two receptors in the present work likely does reflect the actions of two different kinase systems.
Regulation of ligand-induced phosphorylation.
While Ste3p phosphorylation clearly responds to the action of the pheromone signaling pathway, this response stands out in terms of its lack of a requirement for pheromone-induced gene expression. The transcriptional response is a major outcome of pheromone signaling, and most of the other pheromone responses (e.g., G1 arrest, polarized morphogenesis, and cell-cell agglutination and fusion) depend on it. Three results presented here indicate that Ste3p phosphorylation differs. First, while gene disruption of most of the components of the pheromone signaling pathway abolishes induced phosphorylation, disruption of STE12 does not: ste12Δ cells clearly respond to a-factor treatment with increased Ste3p phosphorylation (Fig. 6A). Indeed, the partial impairment seen in ste12Δ cells apparently does not result from failed transcriptional induction (Fig. 6C). Second, induced Ste3p phosphorylation is not blocked by cycloheximide, indicating that this response does not depend on pheromone-induced new protein synthesis (Fig. 6B). Third, while GAL1p-STE11-ΔN and GAL1p-STE12 constructs strongly and rapidly induce pheromone-responsive transcription (Fig. 7A), neither functions to induce Ste3p phosphorylation (Fig. 7B).
The signaling component thought to operate just prior to Ste12p in the pathway is the MAPK. As the induced phosphorylation of Ste3p is fully blocked in fus3Δ kss1Δ cells, the breakpoint between elements that are absolutely required and those that are not appears to occur between Fus3p/Kss1p and Ste12p. In other words, the pathway from the G protein through the MAPK appears to be absolutely required for the ligand-induced phosphorylation of Ste3p, while downstream elements controlling either induced transcription (Ste12p) or G1 arrest (Far1p) are not.
What accounts for the partial impairment of induced Ste3p phosphorylation seen in ste12Δ cells? The ability of the ste12Δ253-335 and ste12Δ255-354 alleles to restore the capacity of ste12Δ cells to fully induce Ste3p phosphorylation (Fig. 6C) indicates that the relevant ste12Δ defect likely is the lost capacity for maintaining the basal level of expression of some required component(s). Possible candidates for the affected components are the Fus3p and Kss1p MAPKs. FUS3 expression strongly depends on Ste12p both for pheromone induction and for its basal level of expression (47). Furthermore, recent whole-genome expression analyses indicate a role for Ste12p in the expression of KSS1 as well (C. Roberts, B. Nelson, and C. Boone, personal communication). As Fus3p and Kss1p are redundantly required for induced Ste3p phosphorylation, mutants with reduced expression of these MAPKs would be expected to show impaired Ste3p phosphorylation.
Relevant to the present discussion are recent findings suggesting how the pheromone signal may be transduced from the G protein to the Ste5p-kinase complex (11, 21, 37, 55). The RING-H2 domain of Ste5p has been shown to be a site for potential interaction with Gβγ (11, 21). Recent results suggest that this Gβγ-Ste5p interaction directs a pheromone-induced relocalization of the Ste5p-kinase complex to the plasma membrane from its resting position in the cytoplasm and the nucleus (Fig. 8) (37). Pryciak and Huntress (37) have proposed that this Gβγ-dependent relocalization of the Ste5p-kinase complex is the key step in the transmission of the signal from G protein to downstream elements. Relocalized to the plasma membrane, the Ste5p-kinase becomes available for activation by plasma membrane-resident PAK kinase Ste20p. Ste20p phosphorylates and activates Ste11p (the MAPK kinase kinase), which in turn phosphorylates and activates Ste7p (the MAPK kinase), which then phosphorylates and activates Fus3p (the MAPK).
Given the efficacy of the STE5-CTM allele in inducing Ste3p phosphorylation, translocation of the Ste5p complex to the plasma membrane likely is an important step in this response. As all three elements of the MAPK cascade are also required for the induced phosphorylation of Ste3p, we surmise that activated Fus3p may also play a central role. Why, then, does the activated STE11 allele fail to induce Ste3p phosphorylation? Like GAL1p-STE4 or GAL1p-STE5-CTM, GAL1p-STE11ΔN also is expected to lead to Fus3p activation. However, in this case, activated Fus3p is not expected to be plasma membrane localized. Indeed, the GAL1p-STE11ΔN construct does not induce the plasma membrane relocalization of the Ste5p-kinase complex (37). Thus, for Ste3p phosphorylation, the activation of Fus3p may not be sufficient: activated Fus3p also may need to be localized to the plasma membrane. The need for plasma membrane localization suggests that the key MAPK target substrate for induced phosphorylation also may be plasma membrane localized.
A model for how the pheromone-induced phosphorylation of Ste3p might be regulated is presented in Fig. 8. The liganded receptor catalyzes the exchange of guanyl nucleotides on the Gα subunit of the G heterotrimer, with the consequent dissociation of Gβγ from Gα. Gβγ attracts the Ste5p-kinase complex to the plasma membrane, where the kinase cascade can be activated by Ste20p. Fus3p is activated via the sequential activation of Ste5p-bound kinases of the MAPK cascade. Active, plasma membrane-localized Fus3p now may phosphorylate some plasma membrane target, leading to Ste3p phosphorylation. In Fig. 8, we suggest that this target could be the receptor kinase. In such a model, Fus3p-dependent phosphorylation would be expected to activate the receptor kinase, increasing its activity toward the Ste3p CTD substrate. Another attractive possibility would have Fus3p directly acting upon the a-factor receptor. While the mammalian MAPK consensus site, (L/P)-X-(S/T)-P (5), is not found in the Ste3p sequence, three Ste3p CTD sites conform to a less stringent (S/T)-P consensus site (5, 14). Perhaps the phosphorylation of one or several of these sites by Fus3p activates the Ste3p CTD as a substrate for some second kinase responsible for the bulk of the ligand-induced phosphorylation. Experiments for testing the involvement of these potential Fus3p (S/T)-P sites in the induced phosphorylation of Ste3p are planned.
A signaling pathway wherein activated Fus3p feeds back upon plasma membrane substrates provides a mechanism potentially useful in other aspects of the pheromone response. Indeed, a recent report suggests that a similar feedback phosphorylation mechanism may control the pheromone-dependent phosphorylation of the clathrin light chain (4). Like induced Ste3p phosphorylation, clathrin light-chain phosphorylation depends on MAPK activity and can be induced through Gβ overproduction. Furthermore, the authors argue against a role for a pheromone-dependent transcriptional response in this induction, given the rapid kinetics of clathrin light-chain phosphorylation upon pheromone challenge (4). More generally, such a feedback mechanism could participate in directing many of the localized, cell surface changes that occur in mating cells in anticipation of cell-cell fusion (29). The central role suggested for free Gβγ in directing the plasma membrane localization of the Ste5p-kinase complex allows for local activation of MAPK in physical proximity to the liganded receptor. With such a mechanism, areas of the cell surface receiving the strongest pheromone input can be marked through the phosphorylation of Fus3p plasma membrane targets—an attractive way of establishing the directional cues that properly orient the mating projection toward the source of the pheromone.
ACKNOWLEDGMENTS
We thank Charlie Boone for the collection of STE knockout plasmids, Peter Pryciak for the collection of dominantly activated signaling constructs, and Stan Fields for the ste12 mutants. We also thank Charlie Boone, Linyi Chen, and Amy Roth for helpful comments on the manuscript.
This work was supported by a grant from the National Science Foundation (MCB 95-06839).
REFERENCES
- 1.Bender A, Sprague G F., Jr Yeast peptide pheromones, a-factor and α-factor, activate a common response mechanism in their target cells. Cell. 1986;47:929–937. doi: 10.1016/0092-8674(86)90808-1. [DOI] [PubMed] [Google Scholar]
- 2.Boone C, Davis N G, Sprague G F., Jr Mutations that alter the third cytoplasmic loop of the a-factor receptor lead to a constitutive and hypersensitive phenotype. Proc Natl Acad Sci USA. 1993;90:9921–9925. doi: 10.1073/pnas.90.21.9921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen Q, Konopka J B. Regulation of the G-protein-coupled α-factor pheromone receptor by phosphorylation. Mol Cell Biol. 1996;16:247–257. doi: 10.1128/mcb.16.1.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chu D S, Pishvaee B, Payne G S. A modulatory role for clathrin light chain phosphorylation in Golgi membrane protein localization during vegetative growth and during the mating response of Saccharomyces cerevisiae. Mol Biol Cell. 1999;10:713–726. doi: 10.1091/mbc.10.3.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clark-Lewis I, Sanghera J S, Pelech S L. Definition of a consensus sequence for peptide substrate recognition by p44mpk, the meiosis-activated myelin basic protein kinase. J Biol Chem. 1991;266:15180–15184. [PubMed] [Google Scholar]
- 6.Cole G M, Stone D E, Reed S I. Stoichiometry of G protein subunits affects the Saccharomyces cerevisiae mating pheromone signal transduction pathway. Mol Cell Biol. 1990;10:510–517. doi: 10.1128/mcb.10.2.510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cook J G, Bardwell L, Kron S J, Thorner J. Two novel targets of the MAP kinase Kss1 are negative regulators of invasive growth in the yeast Saccharomyces cerevisiae. Genes Dev. 1996;10:2831–2848. doi: 10.1101/gad.10.22.2831. [DOI] [PubMed] [Google Scholar]
- 8.Davis N G, Horecka J L, Sprague G F., Jr Cis- and trans-acting functions required for endocytosis of the yeast pheromone receptors. J Cell Biol. 1993;122:53–65. doi: 10.1083/jcb.122.1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dolan J W, Fields S. Overproduction of the yeast STE12 protein leads to constitutive transcriptional induction. Genes Dev. 1990;4:492–502. doi: 10.1101/gad.4.4.492. [DOI] [PubMed] [Google Scholar]
- 10.Elion E A, Grisafi P L, Fink G R. FUS3 encodes a cdc2+/CDC28-related kinase required for the transition from mitosis into conjugation. Cell. 1990;60:649–664. doi: 10.1016/0092-8674(90)90668-5. [DOI] [PubMed] [Google Scholar]
- 11.Feng Y, Song L Y, Kincaid E, Mahanty S K, Elion E A. Functional binding between Gβ and the LIM domain of Ste5 is required to activate the MEKK Ste11. Curr Biol. 1998;8:267–278. doi: 10.1016/s0960-9822(98)70108-3. [DOI] [PubMed] [Google Scholar]
- 12.Fields S, Chaleff D T, Sprague G F., Jr Yeast STE7, STE11, and STE12 genes are required for expression of cell-type-specific genes. Mol Cell Biol. 1988;8:551–556. doi: 10.1128/mcb.8.2.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fields S, Herskowitz I. Regulation by the yeast mating-type locus of STE12, a gene required for cell-type-specific expression. Mol Cell Biol. 1987;7:3818–3821. doi: 10.1128/mcb.7.10.3818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gartner A, Jovanovic A, Jeoung D I, Bourlat S, Cross F R, Ammerer G. Pheromone-dependent G1 cell cycle arrest requires Far1 phosphorylation, but may not involve inhibition of Cdc28-Cln2 kinase, in vivo. Mol Cell Biol. 1998;18:3681–3691. doi: 10.1128/mcb.18.7.3681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hagen D C, McCaffrey G, Sprague G F., Jr Evidence the yeast STE3 gene encodes a receptor for the peptide pheromone a factor: gene sequence and implications for the structure of the presumed receptor. Proc Natl Acad Sci USA. 1986;83:1418–1422. doi: 10.1073/pnas.83.5.1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hicke L, Riezman H. Ubiquitination of a yeast plasma membrane receptor signals its ligand-stimulated endocytosis. Cell. 1996;84:277–287. doi: 10.1016/s0092-8674(00)80982-4. [DOI] [PubMed] [Google Scholar]
- 17.Hicke L, Zanolari B, Pypaert M, Rohrer J, Riezman H. Transport through the yeast endocytic pathway occurs through morphologically distinct compartments and requires an active secretory pathway and Sec18p/N-ethylmaleimide-sensitive fusion protein. Mol Biol Cell. 1997;8:13–31. doi: 10.1091/mbc.8.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hicke L, Zanolari B, Riezman H. Cytoplasmic tail phosphorylation of the α-factor receptor is required for its ubiquitination and internalization. J Cell Biol. 1998;141:349–358. doi: 10.1083/jcb.141.2.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Horecka J, Sprague G F., Jr Identification and characterization of FAR3, a gene required for pheromone-mediated G1 arrest in Saccharomyces cerevisiae. Genetics. 1996;144:905–921. doi: 10.1093/genetics/144.3.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hung W, Olson K A, Breitkreutz A, Sadowski I. Characterization of the basal and pheromone-stimulated phosphorylation states of Ste12p. Eur J Biochem. 1997;245:241–251. doi: 10.1111/j.1432-1033.1997.00241.x. [DOI] [PubMed] [Google Scholar]
- 21.Inouye C, Dhillon N, Thorner J. Ste5 RING-H2 domain: role in Ste4-promoted oligomerization for yeast pheromone signaling. Science. 1997;278:103–106. doi: 10.1126/science.278.5335.103. [DOI] [PubMed] [Google Scholar]
- 22.Jarvis E E, Hagen D C, Sprague G F., Jr Identification of a DNA segment that is necessary and sufficient for α-specific gene control in Saccharomyces cerevisiae: implications for regulation of α-specific and a-specific genes. Mol Cell Biol. 1988;8:309–320. doi: 10.1128/mcb.8.1.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jenness D D, Spatrick P. Down regulation of the α-factor pheromone receptor in S. cerevisiae. Cell. 1986;46:345–353. doi: 10.1016/0092-8674(86)90655-0. [DOI] [PubMed] [Google Scholar]
- 24.Kirkman-Correia C, Stroke I L, Fields S. Functional domains of the yeast STE12 protein, a pheromone-responsive transcriptional activator. Mol Cell Biol. 1993;13:3765–3772. doi: 10.1128/mcb.13.6.3765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Konopka J B, Jenness D D, Hartwell L H. The C-terminus of the S. cerevisiae α-pheromone receptor mediates an adaptive response to pheromone. Cell. 1988;54:609–620. doi: 10.1016/s0092-8674(88)80005-9. [DOI] [PubMed] [Google Scholar]
- 26.Kunkel T A, Roberts J D, Zakour R A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- 27.Kurjan J. α-Factor structural gene mutations in Saccharomyces cerevisiae: effects on α-factor production and mating. Mol Cell Biol. 1985;5:787–796. doi: 10.1128/mcb.5.4.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leberer E, Dignard D, Harcus D, Thomas D Y, Whiteway M. The protein kinase homologue Ste20p is required to link the yeast pheromone response G-protein beta gamma subunits to downstream signalling components. EMBO J. 1992;11:4815–4824. doi: 10.1002/j.1460-2075.1992.tb05587.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leberer E, Thomas D Y, Whiteway M. Pheromone signalling and polarized morphogenesis in yeast. Curr Opin Genet Dev. 1997;7:59–66. doi: 10.1016/s0959-437x(97)80110-4. [DOI] [PubMed] [Google Scholar]
- 30.Lefkowitz R J. G protein-coupled receptors. III. New roles for receptor kinases and β-arrestins in receptor signaling and desensitization. J Biol Chem. 1998;273:18677–18680. doi: 10.1074/jbc.273.30.18677. [DOI] [PubMed] [Google Scholar]
- 31.Madhani H D, Styles C A, Fink G R. MAP kinases with distinct inhibitory functions impart signaling specificity during yeast differentiation. Cell. 1997;91:673–684. doi: 10.1016/s0092-8674(00)80454-7. [DOI] [PubMed] [Google Scholar]
- 32.Neiman A M, Herskowitz I. Reconstitution of a yeast protein kinase cascade in vitro: activation of the yeast MEK homologue STE7 by STE11. Proc Natl Acad Sci USA. 1994;91:3398–3402. doi: 10.1073/pnas.91.8.3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nomoto S, Nakayama N, Arai K, Matsumoto K. Regulation of the yeast pheromone response pathway by G protein subunits. EMBO J. 1990;9:691–696. doi: 10.1002/j.1460-2075.1990.tb08161.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Peter M, Gartner A, Horecka J, Ammerer G, Herskowitz I. FAR1 links the signal transduction pathway to the cell cycle machinery in yeast. Cell. 1993;73:747–760. doi: 10.1016/0092-8674(93)90254-n. [DOI] [PubMed] [Google Scholar]
- 35.Peter M, Herskowitz I. Direct inhibition of the yeast cyclin-dependent kinase Cdc28-Cln by Far1. Science. 1994;265:1228–1231. doi: 10.1126/science.8066461. [DOI] [PubMed] [Google Scholar]
- 36.Pitcher J A, Freedman N J, Lefkowitz R J. G protein-coupled receptor kinases. Annu Rev Biochem. 1998;67:653–692. doi: 10.1146/annurev.biochem.67.1.653. [DOI] [PubMed] [Google Scholar]
- 37.Pryciak P M, Huntress F A. Membrane recruitment of the kinase cascade scaffold protein Ste5 by the Gβγ complex underlies activation of the yeast pheromone response pathway. Genes Dev. 1998;12:2684–2697. doi: 10.1101/gad.12.17.2684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ramer S W, Elledge S J, Davis R W. Dominant genetics using a yeast genomic library under the control of a strong inducible promoter. Proc Natl Acad Sci USA. 1992;89:11589–11593. doi: 10.1073/pnas.89.23.11589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Raths S, Rohrer J, Crausaz F, Riezman H. end3 and end4: two mutants defective in receptor-mediated and fluid-phase endocytosis in Saccharomyces cerevisiae. J Cell Biol. 1993;120:55–65. doi: 10.1083/jcb.120.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reneke J E, Blumer K J, Courchesne W E, Thorner J. The carboxy-terminal segment of the yeast α-factor receptor is a regulatory domain. Cell. 1988;55:221–234. doi: 10.1016/0092-8674(88)90045-1. [DOI] [PubMed] [Google Scholar]
- 41.Robinson L C, Hubbard E J, Graves P R, DePaoli-Roach A A, Roach P J, Kung C, Haas D W, Hagedorn C H, Goebl M, Culbertson M R, et al. Yeast casein kinase I homologues: an essential gene pair. Proc Natl Acad Sci USA. 1992;89:28–32. doi: 10.1073/pnas.89.1.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rohrer J, Benedetti H, Zanolari B, Riezman H. Identification of a novel sequence mediating regulated endocytosis of the G protein-coupled α-pheromone receptor in yeast. Mol Biol Cell. 1993;4:511–521. doi: 10.1091/mbc.4.5.511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Roth A F, Davis N G. Ubiquitination of the yeast a-factor receptor. J Cell Biol. 1996;134:661–674. doi: 10.1083/jcb.134.3.661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Roth A F, Sullivan D M, Davis N G. A large PEST-like sequence directs the ubiquitination, endocytosis, and vacuolar degradation of the yeast a-factor receptor. J Cell Biol. 1998;142:949–961. doi: 10.1083/jcb.142.4.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schandel K A, Jenness D D. Direct evidence for ligand-induced internalization of the yeast α-factor pheromone receptor. Mol Cell Biol. 1994;14:7245–7255. doi: 10.1128/mcb.14.11.7245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sikorski R S, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sprague G F, Jr, Thorner J W. Pheromone response and signal transduction during the mating process of Saccharomyces cerevisiae. In: Jones E W, Pringle J R, Broach J R, editors. The molecular and cellular biology of the yeast Saccharomyces cerevisiae: gene expression. Vol. 2. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1992. pp. 657–744. [Google Scholar]
- 48.Stevenson B J, Rhodes N, Errede B, Sprague G F., Jr Constitutive mutants of the protein kinase STE11 activate the yeast pheromone response pathway in the absence of the G protein. Genes Dev. 1992;6:1293–1304. doi: 10.1101/gad.6.7.1293. [DOI] [PubMed] [Google Scholar]
- 49.Tedford K, Kim S, Sa D, Stevens K, Tyers M. Regulation of the mating pheromone and invasive growth responses in yeast by two MAP kinase substrates. Curr Biol. 1997;7:228–238. doi: 10.1016/s0960-9822(06)00118-7. [DOI] [PubMed] [Google Scholar]
- 50.Valtz N, Peter M, Herskowitz I. FAR1 is required for oriented polarization of yeast cells in response to mating pheromones. J Cell Biol. 1995;131:863–873. doi: 10.1083/jcb.131.4.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vancura A, Sessler A, Leichus B, Kuret J. A prenylation motif is required for plasma membrane localization and biochemical function of casein kinase I in budding yeast. J Biol Chem. 1994;269:19271–19278. [PubMed] [Google Scholar]
- 52.Velculescu V E, Zhang L, Zhou W, Vogelstein J, Basrai M A, Bassett D E, Jr, Hieter P, Vogelstein B, Kinzler K W. Characterization of the yeast transcriptome. Cell. 1997;88:243–251. doi: 10.1016/s0092-8674(00)81845-0. [DOI] [PubMed] [Google Scholar]
- 53.Wang P C, Vancura A, Mitcheson T G, Kuret J. Two genes in Saccharomyces cerevisiae encode a membrane-bound form of casein kinase-1. Mol Biol Cell. 1992;3:275–286. doi: 10.1091/mbc.3.3.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Whiteway M, Hougan L, Thomas D Y. Overexpression of the STE4 gene leads to mating response in haploid Saccharomyces cerevisiae. Mol Cell Biol. 1990;10:217–222. doi: 10.1128/mcb.10.1.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Whiteway M S, Wu C, Leeuw T, Clark K, Fourest-Lieuvin A, Thomas D Y, Leberer E. Association of the yeast pheromone response G protein beta gamma subunits with the MAP kinase scaffold Ste5p. Science. 1995;269:1572–1575. doi: 10.1126/science.7667635. [DOI] [PubMed] [Google Scholar]
- 56.Zanolari B, Raths S, Singer-Kruger B, Riezman H. Yeast pheromone receptor endocytosis and hyperphosphorylation are independent of G protein-mediated signal transduction. Cell. 1992;71:755–763. doi: 10.1016/0092-8674(92)90552-n. [DOI] [PubMed] [Google Scholar]